Abstract
This chapter discusses the fundamental concepts, terminology, and practice of pathology as the discipline dedicated to the understanding of causes, mechanisms, and effects of diseases. It describes some key terms, definitions, and concepts, presents historical human approaches to diseases, and provides an overview of current diagnostic practice and a vision for new interface with applied molecular biology. Pathology refers to the specialty of medical science concerned with the cause, development, structural/functional changes, and natural history associated with diseases. Disease refers to a definable deviation from a normal phenotype (observable characteristics due to genome and environment), evident via patient complaints (symptoms), and/or the measurements of a careful observer (signs). The cause of the disease is referred to as its etiology. One disease entity can have more than one etiology, and one etiology can lead to more than one disease. Each disease entity develops through a series of mechanistic chemical and cellular steps. This stepwise process of disease development is referred to as its pathogenesis. Pathogenesis can refer to the changes in the structure or function of an organism at the gross/clinical level and the stepwise molecular abnormalities leading to changes in cellular and tissue function. The presentation of a disease to a clinician is in the form of a human patient with variably specific complaints (symptoms), to which the examining physicians can add diagnostic sensitivity and specificity by making observations (screening for signs of diseases).
“…Future discoveries will not likely be made by morphologists ignorant of molecular biologic findings, or by biologists unaware or scornful of morphologic data, but by those willing and capable of integrating them through a team approach….”
Rosai [1]
INTRODUCTION
This chapter will discuss the fundamental concepts, terminology, and practice of pathology as the discipline dedicated to the understanding of causes, mechanisms, and effects of diseases. A section on key terms, definitions, and concepts is followed by sections on historical human approaches to diseases, an overview of current diagnostic practice, and a vision for new interface with applied molecular biology.
TERMS, DEFINITIONS, AND CONCEPTS
Pathology (from the Greek word pathología, meaning the study of suffering) refers to the specialty of medical science concerned with the cause, development, structural/functional changes, and natural history associated with diseases. Disease refers to a definable deviation from a normal phenotype (observable characteristics due to genome and environment), evident via patient complaints (symptoms), and/or the measurements of a careful observer (signs). The cause of the disease is referred to as its etiology (from the Greek word meaning the study of cause). One disease entity can have more than one etiology, and one etiology can lead to more than one disease. Each disease entity develops through a series of mechanistic chemical and cellular steps. This stepwise process of disease development is referred to as its pathogenesis (from the Greek word meaning generation of suffering). Pathogenesis can refer to the changes in the structure or function of an organism at the gross/clinical level, and it can refer to the stepwise molecular abnormalities leading to changes in cellular and tissue function.
The presentation of a disease to a clinician is in the form of a human patient with variably specific complaints (symptoms), to which the examining physicians can add diagnostic sensitivity and specificity by making observations (screening for signs of diseases). These phenotypic (measurable characteristic) abnormalities reflect the interaction of the genotype (cytogenetic and nucleic acid sequence/expression) of the patient and his/her environment. Patient workup uses present illness history with reference to past medical history, review of other organ systems for other abnormalities, review of family history, physical examination, radiographic studies, clinical laboratory studies (for example, peripheral blood or CSF specimens), and anatomic pathology laboratory studies (for example, tissue biopsy or pleural fluid cytology specimens). As you will see from other chapters in this book, the ability to rapidly and inexpensively screen for chromosomal translocations, copy number variation, genetic variation, and abundance of mRNA and miRNA is adding substantial molecular correlative information to the workup of diseases.
The differential diagnosis represents the set of possible diagnoses that could account for symptoms and signs associated with the condition of the patient. The conclusion of the workup generally results in a specific diagnosis which meets a set of diagnostic criteria, and which explains the patient's symptoms and phenotypic abnormalities. Obviously, arrival at the correct diagnosis is a function of the examining physician and pathologist (fund of knowledge, experience, alertness), the prevalence of the disease in question in the particular patient (age, race, sex, site), and the sensitivity/specificity of the screening tests used (physical exam, vital signs, blood solutes, tissue stains, genetic assays). The pathologic diagnosis represents the best estimate currently possible of the disease entity affecting the patient, and is the basis for downstream follow-up and treatment decisions. The diagnosis implies a natural history (course of disease, including chronicity, functional impairment, survival) that most patients with this disease are expected to follow. Be aware that not all patients with a given disease will naturally follow the same disease course, so differences in patient outcome do not necessarily correspond to incorrect diagnosis. Variables that independently correlate with clinical outcome differences are called independent prognostic variables, and are routinely assessed in an effort to predict the natural history of the disease in the patient. It is also important to note that medical therapies for specific diseases do not always work. Variables that independently correlate with (predict) responses to therapy are called independent predictive variables.
Diagnosis of a disease and development of an effective therapy for that disease do not require knowledge of the underlying etiology or pathogenesis. For example, Wegener's granulomatosis was understood by morphology and outcomes to be a lethal disease without treatment, yet responsive to cyclophosphamide and corticosteroids, before it was found to be an autoimmune disease targeting neutrophil cytoplasmic protein PR3 (Figure 11.1 ). However, understanding the molecular and cellular pathogenesis of a disease allows development of screening methods to determine risk for clinically unaffected individuals, as well as mechanistic approaches to specific therapy.
Figure 11.1.
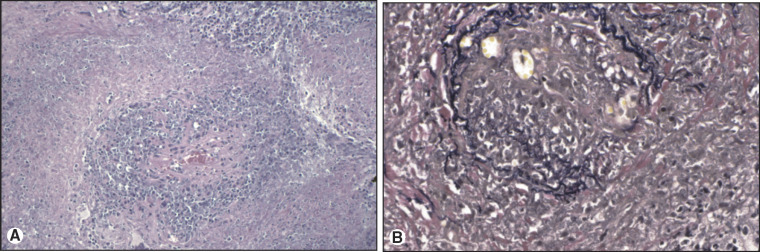
Wegener's granulomatosis of the lung. (A) H&E of Wegener's granulomatosis of lung. Necrosis, granulomatous inflammation, and vasculitis are identified. (B) Elastin stain of Wegener's granulomatosis of lung. Elastica disruption of the arterial wall supports a diagnosis of vasculitis.
The pathologist is that physician or clinical scientist who specializes in the art and science of medical risk estimation and disease diagnosis, using observations at the clinical, gross, body fluid, light microscopic, immunophenotypic, ultrastructural, cytogenetic, and molecular levels. Clearly, the pathologist has a duty to master any new concepts, factual knowledge, and technology that can aid in the estimation of risk for unaffected individuals, the statement of accurate and timely diagnosis, accurate prognosis, and accurate prediction of response to therapy for affected individuals.
A BRIEF HISTORY OF APPROACHES TO DISEASE
The ability of H. sapiens to adapt and thrive has been due in part to the ability of humans to remember the past, respect tradition, recognize the value of new observations, develop tools/symbols, manipulate the environment, anticipate the future, and role-specialize in a social structure. The history of human understanding of diseases has progressed at variable rates, depending on the good and bad aspects of these human characteristics.
Concepts and Practices Before the Scientific Revolution
Our understanding of ancient attitudes toward diseases is limited by the historical written record. Thus, the start point for written medical history corresponds to around 1700 BC for Mesopotamian rules in the code of Hammurabi, and around 1550 BC for the analogous Egyptian rules in the Ebers papyrus. By definition, these philosophers, theologians, and physicians had access and assets to allow a written record, and materials and storage sufficient for the written records to survive. The Mesopotamian records indicate a deity-driven and demon-driven theory and empirical practice by recognized professional physicians. In this context, the prevailing thought was that “Disease was caused by spirit invasion, sorcery, malice, or the breaking of taboos; sickness was both judgment and punishment” [2].
The Greek medical community evolved a theory of disease related to natural causes and effects, with less emphasis on deity-driven theory. The Hippocratic Corpus includes “On the Sacred Disease” (circa 400 BC), which rejected a divine origin for diseases, and postulated a natural rather than supernatural basis for disease etiology (“…nowise more divine nor more sacred than other diseases, but has a natural cause … like other affections.”). Aristotle (384–322 BC) wrote broadly on topics including logic, biology, physics, metaphysics, and psychology. To Aristotle, observations led to a description of causes, or first principles, which in turn could be used logically in syllogisms to predict future observations. We would agree with these basic notions of induction and deduction. However, there was a different background philosophical construct regarding the nature of matter and causality (four elements, four humors, and four causes, including a final or teleologic purpose). We would recognize Aristotle's “efficient” cause of a disease as its etiology. Alexander the Great's conquest of Egypt in the 4th century BC led to Greek (Ptolemaic) leadership of Egypt from 305 BC to 30 BC, with development of the Alexandrian library and University. Faculty such as Euclid developed geometric models of vision (Optica), and Herophilus described human anatomy by direct dissection and observation (Greek medicine apparently allowed dissection in Alexandria, including vivisection of the condemned). During the Roman imperial era, Galen (129–207 AD) used dissection and observation of other animals such as the macaque (human dissection was illegal) to extrapolate to human anatomy and physiology. Like Aristotle's approach, Galen's approach to patients, diseases, and treatments was guided by philosophical constructs of four humors (blood, phlegm, yellow bile, and black bile) and the resulting temperaments (buoyant, sluggish, quick-tempered, and melancholic) due to humoral imbalances. It is thought that many of Galen's texts were destroyed with the Alexandrian library before the 7th century AD, but a subset was preserved and translated by Middle Eastern scholars. These ancient classic texts were then retranslated into Latin and Greek when printing houses developed in the 15th century (for instance, Hippocrates' De Natura Hominis, circa 1480 AD, and Galen's Therapeutica, circa 1500 AD).
The historical picture of the Greco-Roman understanding of disease is one of empirical approaches to diseases based on inaccurate understanding of anatomy, physiology, and organ/cellular pathology. Greek medicine became less superstitious and more natural cause-and-effect oriented, yet philosophy still trumped direct observation, such that evidence was constrained to fit the classic philosophical constructs. Some of the concepts sound familiar; for example, normal represents equilibrium and disease represents disequilibrium. However, we would differ on what variables are in disequilibrium (the historical humors, numbers, and opposites versus contemporary chemical and kinetic equilibria).
Following the collapse of the Western Roman Empire in 476 AD, the classic texts of Aristotle, Hippocrates, and Galen were protected, translated, and built upon in the Byzantine and Arab societies of the near East, and in Spain during the Muslim/Moorish period through the 11th century. During these middle ages for Western Europe outside Spain, there was apparently a retreat to pre-Hellenistic beliefs in supernatural forces that intervened in human affairs, with protecting saints and relics for disease prevention and therapy. Centers of medical learning following the Spanish Muslim model developed in Montpellier, France, and in Salerno, Italy, beginning in the 11th century.
The Scientific Revolution
Aristotle's concept of induction from particulars to general first principles, then use of syllogistic logic to predict particulars, evolved into the scientific method during the Renaissance. Ibn Alhazen's (al-Haitham's) work on the physics of optics in the 11th century challenged Euclid's concepts of vision from the Alexandrian era. (Euclid thought that the eye generated the image, rather than light reflected from the object being received by the eye). In the 13th century, Roger Bacon reinforced this use of observation, hypothesis, and experimentation. The printing press (Gutenberg, 1440 AD) allowed document standardization and reproduction, such that multiple parallel university and city libraries could afford to have similar collections of critical texts, facilitating scholarly publications in journals. Access to publications in libraries and universities led to a system of review, demonstration, discussion, and consensus regarding new scientific findings.
The concept of the body as an elegant machine was captured not only by 15th and 16th centuries Renaissance artists like da Vinci and Michelangelo, but also by anatomists and pathologists interested in the structure/function of health and disease. The ancient models of Aristotle and Galen had become sacrosanct, and newer, evidence-based models were considered heretical to some degree. So it was somewhat revolutionary when Vesalius dissected corpses, compared them with Galenic descriptions, and published De humani corporis fabrica libri septem (1543 AD; “Seven Books on the Structure of the Human Body”; the Fabrica), challenging and correcting 16th century understanding of normal human anatomy. Vesalius's successors (Colombo, Fallopius, and Eustachius) further improved the accuracy of human anatomic detail. Thus, correction of Galen's anatomical inaccuracies (including the rete mirabilis at the base of the brain, the five lobed liver, and curved humerus) required at least 13 centuries for challenge, scientific disproof, and eventual medical community acceptance.
The Scientific Revolution describes the progressive change in attitude of scientists and physicians toward understanding of the natural world, health, and disease. This revolution began circa 1543 AD, when Copernicus published arguments for a heliocentric universe and Vesalius published the Fabrica series on human anatomy. By the 17th century, Galileo, Kepler, Newton, Harvey, and others had used this observation-based, matter-based, and mathematical law-based perspective to develop a scientific approach similar to our own modern approach of testing hypotheses with experimental data and statistics. In human biology, the investigation of structure led to studies of function, initially of human cardiovascular physiology, for example, Harvey's Anatomical Exercise Concerning the Motion of the Heart and Blood in Animals (1628). Whereas Galen conceived of parallel but unconnected arteries and veins, with continuous blood production in the liver and continuous blood consumption in the periphery, Harvey demonstrated that blood was pumped by the heart through arteries, through tissue capillaries, to veins, and then back to the heart in a circle (circulation). Correction of these and other Galenic physiological inaccuracies (such as nasal secretions representing the filtrate of cerebral ventricle fluid) thus required at least 14 centuries before challenge, scientific disproof, and eventual medical community acceptance.
The scientific method facilitates empirical, rational, and skeptical approaches to observational data, and minimizes human dependence on non-evidence-based traditional models. In spite of the scientific method, physicians are still human, and the medical community still shows an inertial reluctance to adapt to new information when it disrupts traditional paradigms. Recent examples would include reluctance to accept an etiologic role for the H. pylori bacterium in peptic ulcer disease [3], and reluctance to offer less than radical mastectomy for primary breast carcinoma.
Discovery of the Microscopic World
Before the use of lenses to magnify objects, it was physically impossible to make observations on objects smaller than the resolving limit of the human eye (about 0.1–0.25 mm). Thus, prokaryotic and eukaryotic cells, tissue architecture, and comparisons of normal and disease microanatomy were philosophical speculation until description of the mathematics of optics and lens design. In a real sense, optical technology was rate-limiting for the development of the fields of tissue anatomy, cellular biology, and microbiology. Concepts of optics were written as early as 300 BC in Alexandria (Optica, Euclid). Clear glass (crystallo) was developed in Venice in the 15th century. A compound microscope was invented by Janssen in 1590 AD. Microanatomy and structural terminology was begun by Malpighi (1661 AD), who examined capillaries in frog lung, trachea tubes for airflow in silkworms, and stomata in plant leaves. Robert Hooke used a compound microscope to describe common objects in Micrographia (1665 AD). Antony Van Leeuwenhoek used self-made simple magnifying lenses to count the threads in cloth in a Dutch dry-goods store, then later published descriptions of bacteria (termed animalcules), yeast, and algae, beginning in 1673 AD. Yet the relevance of these observations in microanatomy and microbiology to human diseases required changes in conceptual understanding of the etiology and pathogenesis of diseases. For example, it was 200 years after Van Leeuwenhoek that the common bacterium Streptococcus was recognized as the etiologic agent of puerperal fever in post-partum women.
Critical Developments During the 19th Century
Cellular Pathology, Germ Theory, and Infectious Etiologic Agents
The relevance of microanatomy and microbiology to human disease required expansion of conceptual understanding to include morphologic changes in diseased cells and tissues, as well as recognition of an etiologic role for microorganisms. Rokitansky's gross correlates with clinical disease (A Manual of Pathological Anatomy, 1846, discussed in [4]), Paget's surgical perspective on gross pathology [5], and Virchow's morphologic correlates with clinical disease [6] were critical to the development of clinico-pathologic correlation, and served to create a role for pathologists to specialize in autopsy and tissue diagnosis. Virchow's description of necrotizing granulomatous inflammation, the morphologic correlate of infections caused by mycobacteria such as TB and leprosy, preceded the discovery of the etiologic agents years later by Hansen (M. leprae, 1873 [7]) and Koch (M. tuberculosis, 1882 [8]), reviewed in [9].
The causal relationship between microorganisms and clinical disease required scientific demonstration and logical proof before medical community acceptance. For example, it took two centuries for the common bacterium Streptococcus pyogenes to be recognized as the etiologic agent of puerperal fever in post-partum women. Identification of this cause-and-effect relationship required an initial recognition of unusual clinical outcomes (clusters of post-partum deaths), then correlation of puerperal fever clusters with obstetrician habits [10,11], then Semmelweiss's experimental demonstration in 1847 that hand-washing reduced the incidence of puerperal fever [12], then the demonstration that particular bacteria (Streptococci) are regularly associated with the clinical disease (Koch, circa 1870), and finally by culture of the organism from the blood of patients with the disease (Pasteur, 1879).
During the 19th century, critical causal associations between microorganisms and infectious diseases were proven and accepted. Technical improvements in microscopes (Abbe condenser, apochromatic lenses, oil immersion lenses), the development of culture media, and the development of histochemical stains no doubt made it possible for Koch to identify M. tuberculosis in 1882 [8]. To be emphasized is the process of recognition, first of all of the variables associated with the clinical disease, then the scientific demonstration of a causal relationship between one or more of these variables with the clinical disease. This latter step was enunciated as Koch's postulates (1890): (i) the bacteria must be present in every case of the disease, (ii) the bacteria must be isolated from a diseased individual and grown in pure culture, (iii) the specific disease must be reproduced when a pure culture is inoculated into a healthy susceptible host, and (iv) the same bacteria must be recoverable from the experimentally infected host.
Organic Chemistry
Prior to 1828, organic (carbon-containing) compounds were thought to derive from living organisms, and it was thought that they could never be synthesized from nonliving (inorganic) material. This concept (termed vitalism) was disproven by the in vitro synthesis of urea by F. Wohler in 1828 [13]. Work from this era initiated the field of organic chemistry. Predictable rules for in vitro and in vivo organic reactions, structural theory, modeling, separation technologies, and accurate measurement subsequently allowed the chemical description of natural products, and the chemical synthesis of both natural products and synthetic compounds. In addition to setting the stage for a systematic understanding of cellular biochemistry and physiology, organic chemistry set the stage for laboratory synthesis of natural products, such as dyes, vitamins, hormones, proteins, and nucleic acids. In that era, the textile industry was the main consumer of dyes. Access to imported natural product dyes from plants was predictable for seafaring nations, but not for landlocked nations. In parallel with natural product extraction and purification was the recognition that aniline from coal (Perkin, 1856) could be modified to generate a spectrum of colors, catalyzing the development of the German dye industry in the last half of the 19th century. Some of these synthetic dyes were found useful for histochemical staining.
Histotechnology
Morphologic diagnosis requires thin (3–5 microns), contrast-rich (requiring dyes) sections of chemically fixed (cross-linked or precipitated) tissue. Thin sections allow the passage of light through the tissue, but reduce overlapping of cells in the light path. Thus, technologies had to develop for cutting and staining of thin fixed tissue sections (reviewed in [14]). Work leading to our current technique for tissue solidification in paraffin wax was first described by Klebs in 1869. Prototypes of our current mechanical microtome for making thin (~5 micron thick) tissue sections was developed by Minot in 1885. Precursor work leading to our current technique for tissue fixation with diluted formalin was first described by Blum in 1893.
Histochemical stains developed in parallel with dye technology for the textile industry. Botanists used carmine as a stain in 1849, and subsequently Gerlack applied carmine to stain brain tissue in 1858. The current hematoxylin dye used in tissue histochemistry was originally extracted from logwood trees from Central America for the dye industry (to compete with indigo). Metallic ion mordants made oxidized hematoxylin (hematein) colorfast in textiles, and a protocol for tissue staining was published by Boehmer in 1865. Similar to the hematoxylin story, semisynthetic analine dye technology was adapted by histochemists from 1850–1900. Many of these dyes are still routinely used for recognition of tissue structure, peripheral blood cells, and microorganisms, including hematoxylin, eosin, methylene blue, Ziehl-Neelsen, Gram, van Gieson, Mallory trichrome, and Congo red [14].
The most commonly used stains for general tissue diagnosis are the hematoxylin and eosin (H&E) stains, which provide a wealth of nuclear and cytoplasmic detail not visible in an unstained section. Supplemental histochemical stains demonstrate specific structures and organisms: collagen and muscle (trichome), elastin (Verhoff-von Giessen), glycogen/mucin (periodic acid-Schiff, PAS), mucin (PAS diastase, mucicarmine, alcian blue), fungi (Gemori methenamine silver, GMS), mycobacteria (Ziehl-Neelsen, Fite), and bacteria (Gram, Warthin-Starry). Each of these stains is inexpensive ($10–$50), fast (minutes to hours), and automatable, making them extremely valuable for service diagnostic pathology use.
Light microscopy lens technology matured during the last half of the 19th century. Critical were Abbe's introductions of the apochromatic lens system to eliminate chromatic aberration (different focal lengths for different wavelengths of visible light) in 1868, a novel condenser for compound microscopes (to provide better illumination at high magnification) in 1870, and an oil immersion lens (for 10 × objective magnification with visible light) in 1878.
By 1900, maturation of tissue fixation chemistry, histochemical stain protocols, and light microscope technology had evolved into the workhorse technique for evaluation of morphologic abnormalities in tissue examination in anatomic pathology labs, and for the evaluation of morphologic features of microorganisms in microbiology labs. The scope of this chapter is limited to pathology, but it should be clear to the reader that the momentous discoveries of deep general anesthetics (Long, 1841; Morton, 1846), commercial electricity (Edison, 1882), and radiography (Roentgen, 1895) also contributed to the development of the modern medical specialties of diagnostic pathology and laboratory medicine.
Developments During the 20th Century
Humoral and Cellular Immunology
The development of antisera in the 20th century for therapeutic purposes (for instance, treatment of diptheria) led to progressive understanding of the antibody, the efferent arm of the humoral immune response. Similarly, tissue transplantation experiments led to the recognition of cellular rejection [15] due to thymus-derived T-cells. Antibodies and T-cells cooperate to react to foreign (non-self) molecules, common examples being allergic responses, viral infections, and organ transplants. Antibodies were found to be made by B-cells and plasma cells, and were found to be exquisitely specific for binding to their particular antigens (ligands) either in solid phase or in solution. The analogous T-cell receptor (TCR) recognizes a ligand made up of a 15–20mer peptide presented by self MHC (HLA in human) molecules on the surface of antigen-presenting cells (macrophages, dendritic cells, activated B-cells). B- and T-cells activate and proliferate when exposed to non-self proteins, but not to self proteins, attesting to tolerance to self. When B- and T-cell self-tolerance breaks down, autoimmune diseases can result (including myasthenia gravis, Grave's disease, and lupus erythematosus). Antibodies (immunoglobulins) were found to be heterodimers of 50 kD heavy chains and 25 kD light chains, folded together so that a highly variable portion defines the antigen-binding site, and a constant portion defines the isotype (IgM, IgD, IgG, IgE, or IgA). Similarly, T-cell receptors were found to be heterodimers of immunoglobulin-like molecules with a highly variable portion for ligand binding, and a constant portion, but without different isotypes. The range of antigen-binding specificities in a normal mammal is extensive, perhaps infinite, subtracting out only those self proteins to which the animal is tolerant. The genes encoding immunoglobulins and T-cell receptors were sequenced and, surprisingly, the extensive variation of specificities was due to a unique system of gene rearrangements of polymorphic V, D, and J gene segments with random nucleotide addition at the junctions [16,17]. This system allows generation of literally billions of different Ig and TCR binding specificities.
Polyclonal antibodies raised in other species (goat, mouse, rabbit) against an antigen can be used to detect the antigen in diffusion gels (Ochterlony, western blot), in solution (ELISA), and in tissue sections. Use of fluorescent-tagged antibodies for frozen section immunohistochemistry was developed first [18], and immunofluorescence (IF) is still routinely used in renal pathology and dermatopathology. Peroxidase-tagged secondary antibodies and DAB chemistry were developed to generate a stable chromogen in the tissue, and this is now the primary method for detecting antigens in formalin-fixed tissue sections. Improved antibody binding specificity and industrial production required monoclonal antibodies, which required the development of mouse plasmacytomas/myelomas and cell fusion protocols [19]. The net result is that commercial antibodies are now available for antigens of both clinical and research interest, including antibodies specific enough to distinguish minimally modified variant antigens (for instance, phosphorylated versus dephosphorylated proteins).
Natural Product Chemistry and the Rise of Clinical Laboratories
Diseases due to dietary deficiencies (like scurvy) and hormonal imbalances (like diabetes mellitus) were described clinically long before they were understood pathologically. Dietary deficiency diseases prompted searches for the critical metabolic cofactors, so-called vital amines or vitamins. Xerophthalmia was linked to retinol (vitamin A) deficiency in 1917 (McCollum). Rickets was linked to calcitriol (vitamin D) deficiency in 1926. Beri-beri was linked to a deficiency of thiamine (vitamin B1) in 1926. Scurvy was linked to ascorbic acid (vitamin C) deficiency in 1927. Pellagra in the United States was linked to niacin deficiency in 1937. Pernicious anemia was linked to cobalamin (vitamin B12) deficiency in 1948. It is currently unusual to see morphologic features of these diseases in this country. Diseases due to nondietary physiologic imbalances prompted isolation of hormones. For example, hyperthyroidism and hypothyroidism were linked to thyroxine imbalances in 1915, and table salt was iodized starting in 1917. Diabetes mellitus was linked to insulin deficiency in 1921, nonhuman insulin was industrially purified and marketed soon thereafter, and recombinant human insulin was marketed in 1982.
These examples highlight the ability of 20th century chemists to fractionate, purify, synthesize, measure bioactivity, and manufacture these compounds for safe use by humans. Study of diseases due to deficiencies and excesses of single molecule function led to a mechanistic understanding of biochemistry and physiology, with resultant interconnected reaction pathways of byzantine complexity (now referred to as systems biology). Clinical demand for body fluid levels of ions (such as sodium, potassium chloride, and bicarbonate), glucose, creatinine, hormones (such as thyroxine and parathyroid hormone), albumin, enzymes (related to liver and cardiac function), and antibodies (reactive to ASO, Rh, ABO, and HLA antigens) led to the development of clinical laboratories in chemistry, endocrinology, immunopathology, and blood banking. Functional assays for coagulation cascade status were developed, as were methods for estimating blood cell concentration and differential, leading to coagulation and hematology laboratories. Serologic and cell activation assays to define HLA haplotype led to HLA laboratories screening donors and recipients in anticipation of bone marrow and solid organ transplants. Culture medium-based screening for infectious agents led to dedicated clinical microbiology laboratories, which are beginning to incorporate nucleic acid screening technologies for speciation and prediction of treatment response. The clinical laboratories now play a critical, specialized role in inpatient and outpatient management, and their high test volumes (a 700-bed hospital may perform 5 million tests per year) have catalyzed computer databases for central record-keeping of results.
Natural Product Chemistry: Nucleic Acids
The previous vignettes indicate a scientific approach to natural products of the steroid and protein types, but do not indicate how proteins are encoded, what accounts for variation in the same protein in the population, or how inherited diseases are inherited. It turns out that the instruction set for protein sequence is defined by DNA sequence. The role of nucleoproteins as a genetic substance was alluded to by Miescher in 1871, and was shown by Avery to be the pneumococcal transforming principle in 1944. The discovery of X-ray crystallography in 1912 made it possible for Franklin, Wilkins, and Gosling to study DNA crystal structure [20,21], and led to the description of the antiparallel double helix of DNA by Watson and Crick in 1953 [22]. This seminal event in history facilitated dissection of the instruction set for an organism, with recognition that 3-base codons specified amino acids in 1961 [23], and description of the particular codons encoding each amino acid in 1966 [24]. Demonstration of in situ hybridization in 1969 [25,26] made it possible to localize specific DNA or RNA sequences within the cells of interest. Recognition and purification of restriction endonucleases and DNA ligases, and the development of cloning vectors, made it possible to clone individual sequences, leading to methods for synthesis of natural products (such as recombinant human insulin in 1978 [27]). Chemical methods were developed for sequencing DNA [28,29], initially with radioisotope-tagged nucleotide detection in plate gels, then with fluorescent nucleotide detection in 1986 [30]. Subsequent conversion to capillary electrophoresis and computer scoring of sequence output allowed high throughput protocols which generated the human genome sequence by 2001 [31,32]. The development of polymerase chain reaction (PCR) [33] made it possible to quickly screen for length polymorphisms (identity testing, donor:recipient ratio after bone marrow transplant, and microsatellite instability) and sequence abnormalities (translocations, rearrangements, insertions, deletions, substitutions) in a targeted fashion. Quantitative PCR methods using fluorescent detection of amplicons made it possible to study DNA and RNA copy number, and to mimic Northern blots/oligo microarrays in estimating RNA transcript abundance for cluster analysis.
CURRENT PRACTICE OF PATHOLOGY
Diseases can be distinguished from each other based on differences at the molecular, cellular, tissue, fluid chemistry, and/or individual organism level. One hundred and fifty years of attention to the morphologic and clinical correlates of diseases has led to sets of diagnostic criteria for the recognized diseases, as well as a reproducible nomenclature for rapid description of the changes associated with newly discovered diseases. Sets of genotypic and phenotypic abnormalities in the patient are used to determine a diagnosis, which then implies a predictable natural history and can be used to optimize therapy by comparison of outcomes among similarly afflicted individuals. The disease diagnosis becomes the management variable in clinical medicine, and management of the clinical manifestations of diseases is the basis for day-to-day activities in clinics and hospitals nationwide. The pathologist is responsible for integration of the data obtained at the clinical, gross, morphologic, and molecular levels, and for issuing a clear and logical statement of diagnosis.
Clinically, diseases present to front-line physicians as patients with sets of signs and symptoms. Symptoms are the patient's complaints of perceived abnormalities. Signs are detected by examination of the patient. The clinical team, including the pathologist, will work up the patient based on the possible causes of the signs and symptoms (the differential diagnosis). Depending on the differential diagnosis, the workup typically involves history-taking, physical examination, radiographic examination, fluid tests (blood, urine, sputum, stool), and possibly tissue biopsy.
Radiographically, abnormalities in abundance, density or chemical microenvironment of tissues allows distinction from surrounding normal tissues. Traditionally, the absorption of electromagnetic waves by tissues led to summation differences in exposure of silver salt photographic film. Tomographic approaches such as CT (1972) and NMR (1973) complemented summation radiology, allowing finely detailed visualization of internal anatomy in any plane of section. In the same era, ultrasound allowed visualization of tissue with density differences, such as a developing fetus or gallbladder stones. More recently, physiology of neoplasms can be screened with positron emission tomography (PET, 1977) for decay of short half-life isotopes such as fluorodeoxyglucose. Neoplasms with high metabolism can be distinguished physiologically from adjacent low-metabolism tissues, and can be localized with respect to normal tissues by pairing PET with standard CT. The result is an astonishingly useful means of identifying and localizing new space-occupying masses, assigning a risk for malignant behavior and, if malignant, screening for metastases in distant sites. This technique is revolutionizing the preoperative decision making of clinical teams, and improves the likelihood that patients undergo resections of new mass lesions only when at risk for morbidity from malignant behavior or interference with normal function.
Pathologically, disease is diagnosed by determining whether the morphologic features match the set of diagnostic criteria previously described for each disease. Multivolume texts are devoted to the gross and microscopic diagnostic criteria used for diagnosis, prognosis, and prediction of response to therapy [1,34,35]. Pathologists diagnose disease by generating a differential diagnosis, then finding the best fit for the clinical presentation, the radiographic appearance, and the pathologic (both clinical lab and morphologic) findings. Logically, the Venn diagram of the clinical, radiologic, and pathologic differential diagnoses should overlap. Unexpected features expand the differential diagnosis and may raise the possibility of previously undescribed diseases. For example, Legionnaire's disease, human immunodeficiency virus (HIV), Hantavirus pneumonia, and severe acute respiratory syndrome (SARS) are examples of newly described diagnoses during the last 30 years. The mental construct of etiology (cause), pathogenesis (progression), natural history (clinical outcome), and response to therapy is the standard approach for pathologists thinking about a disease. A disease may have one or more etiologies (initial causes, including agents, toxins, mutagens, drugs, allergens, trauma, or genetic mutations). A disease is expected to follow a particular series of events in its development (pathogenesis), and to follow a particular clinical course (natural history). Disease can result in a temporary or lasting change in normal function, including patient death. Multiple diseases of different etiologies can affect a single organ, for example, infectious and neoplastic diseases involving the lung. Different diseases can derive from a single etiology, for example, emphysema, chronic bronchitis, and small cell lung carcinoma in long-term smokers. The same disease (for instance, emphysema of the lung) can derive from different etiologies (emphysema from α-1-antitrypsin deficiency or cigarette smoke).
Modern surgical pathology practice hinges on morphologic diagnosis, supplemented by special stains, immunohistochemical stains, cytogenetics, and clinical laboratory findings, as well as the clinical and radiographic findings. Sections that meet all of these criteria are diagnostic for the disease. If some, but not all, of the criteria are present to make a definitive diagnosis, the pathologist must either equivocate or make an alternate diagnosis. Thus, a firm grasp of the diagnostic criteria and the instincts to rapidly create and sort through the differential diagnosis must be possessed by the service pathologist.
The tissue diagnosis has to make sense, not only from the morphologic perspective, but from the clinical and radiographic vantage points as well. It is both legally risky and professionally erosive to make a clinically and pathologically impossible diagnosis. In the recent past, limited computer networking meant numerous phone calls to gather the relevant clinical and radiographic information to make an informed morphologic diagnosis. For example, certain diseases such as squamous and small cell carcinomas of the lung are extremely rare in nonsmokers. Thus, a small cell carcinoma in the lung of a nonsmoker merits screening for a nonpulmonary primary site. Fortunately for pathologists, computing and networking technologies now allow us access to preoperative clinical workups, radiographs/reports, clinical laboratory data, and prior pathology reports. All of these data protect pathologists by providing them with the relevant clinical and radiographic information, and protect patients by improving diagnostic accuracy. Just as research scientists “…ignore the literature at their peril…”, diagnostic pathologists “… ignore the presentation, past history, workup, prior biopsies, and radiographs at their peril. …”
There are limitations to morphologic diagnosis by H&E stains. First, lineage of certain classes of neoplasms (including small round blue cell tumors, clear cell neoplasms, spindle cell neoplasms, and undifferentiated malignant neoplasms) is usually clarified by immunohistochemistry, frequently by cytogenetics (when performed), and sometimes by electron microscopy. Second, there are limitations inherent in a snapshot biopsy or resection. Thus, the etiology and pathogenesis can be obscure or indeterminate, and rates of growth, invasion, or timing of metastasis cannot be inferred. Third, the morphologic changes may not be specific for the underlying molecular abnormalities, particularly the rate-limiting (therapeutic target) step in the pathogenesis of a neoplasm. For example, Ret gain of function mutations in a medullary thyroid carcinoma will require DNA level screening to determine germline involvement, familial risk, and presence or absence of a therapeutic target. Fourth, the same morphologic appearance may be identical for two different diseases, each of which would be treated differently. For example, there is no morphologic evidence by H&E stain alone to distinguish host lymphoid response to Hepatitis C viral (HCV) antigens from host lymphoid response to allo-HLA antigens in a liver allograft. This is obviously a major diagnostic challenge when the transplant was done for HCV-related cirrhosis, and the probability of recurrent HCV infection in the liver allograft is high.
Paraffin section immunohistochemistry has proven invaluable in neoplasm diagnosis for clarifying lineage, improving diagnostic accuracy, and guiding customized therapy. If neoplasms are poorly differentiated or undifferentiated, the lineage of the neoplasm may not be clear. For example, sheets of undifferentiated malignant neoplasm with prominent nucleoli could represent carcinoma, lymphoma, or melanoma. To clarify lineage, a panel of immunostains is performed for proteins that are expressed in some of the neoplasms, but not in others. Relative probabilities are then used to lend support (rule in) or exclude (rule out) particular diagnoses in the differential diagnosis of these several morphologically similar undifferentiated neoplasms. The second role is to make critical distinctions in diagnosis that cannot be accurately made by H&E alone. Examples of this would include demonstration of myoepithelial cell loss in invasive breast carcinoma but not in its mimic, sclerosing adenosis (Figure 11.2 ), or loss of basal cells in invasive prostate carcinoma (Figure 11.3 ). The third role of immunohistochemistry is to identify particular proteins, such as nuclear estrogen receptor (ER) (Figure 11.4 ) or the plasma membrane HER2 proteins (Figure 11.5 ), both of which can be targeted with inhibitors rather than generalized systemic chemotherapy. Morphology remains the gold standard in this diagnostic process, such that immunohistochemical data support or fail to support the H&E findings, not vice versa.
Figure 11.2.
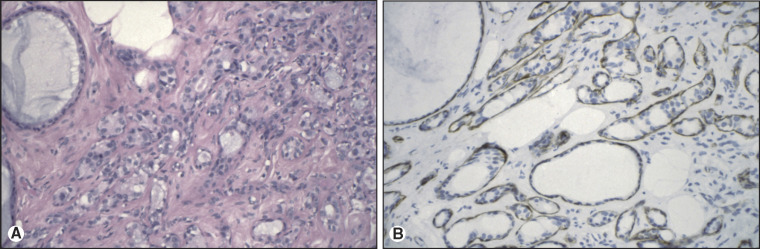
Sclerosing adenosis of breast. (A) H&E of sclerosing adenosis of breast. By H&E alone, the differential diagnosis includes infiltrating ductal carcinoma and sclerosing adenosis. (B) Actin immunostain of sclerosing adenosis of breast. Actin immunoreactivity around the tubules of interest supports a diagnosis of sclerosing adenosis and serves to exclude infiltrating carcinoma.
Figure 11.3.
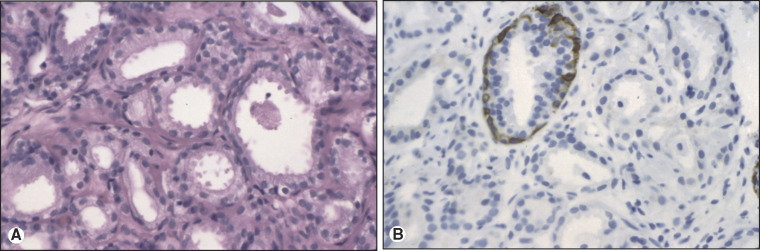
Invasive adenocarcinoma of prostate. (A) H&E of invasive adenocarcinoma of prostate. By H&E alone, the differential diagnosis includes invasive adenocarcinoma and adenosis. (B) High molecular weight cytokeratin immunostain of invasive adenocarcinoma of prostate. Loss of high molecular weight cytokeratin (34βE12) immunoreactivity around the glands of interest supports a diagnosis of invasive adenocarcinoma.
Figure 11.4.
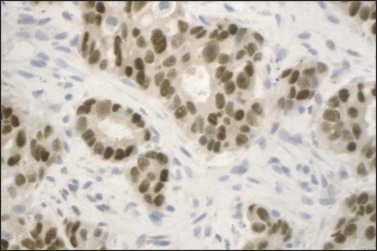
Estrogen receptor immunostain of breast carcinoma. Strong nuclear immunoreactivity for ER is noted, guiding use of ER inhibitor therapy.
Figure 11.5.
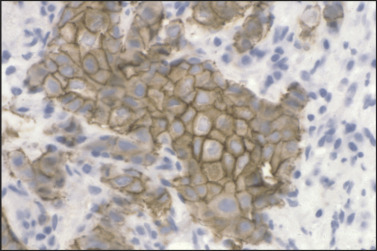
HER2/c-erbB2 immunostain of breast carcinoma. Strong plasma membrane immunoreactivity for c-erbB2/HER2 is noted, guiding use of either anti-HER2 antibody or HER2 kinase inhibitor therapy.
Probability and statistics are regular considerations in immunohistochemical interpretation, since very few antigens are tissue-specific or lineage-specific. Cytokeratin is positive in carcinomas, but also in synovial and epithelioid sarcomas. This example may imply aspects of the lineage of these two sarcomas that may be helpful in our categorization of these neoplasms. Another example would be the diagnosis of small cell carcinoma in the lung of a nonsmoker. Because lung primary small cell carcinoma is extremely uncommon, in non-smokers, this diagnosis would prompt the pathologist to inquire about screening results for other, nonpulmonary, sites. Likewise, immunohistochemistry results are always put into the context of the morphologic, clinical, and radiographic findings. For example, an undifferentiated CD30(+) neoplasm of the testis supports embryonal carcinoma primary in the testis, whereas a lymph node effaced by sclerotic bands with admixed CD30(+) Reed-Sternberg cells supports nodular sclerosing Hodgkin's disease.
Demand for both diagnostic accuracy and report promptness has increased as hospitals come under increasing financial pressure to minimize length of patient stay. Hospitals now manage all but the sickest patients as outpatients. Minimally invasive approaches for the acquisition of tissue samples for diagnosis use flexible endoscopic biotomes or hollow needles that sample 1–2 mm diameter tissue specimens. Multidisciplinary conferences function almost real-time with respect to the initial biopsies. Together, these changes have forced modern pathologists to make critical diagnoses on progressively smaller biopsy specimens, sometimes bordering on the amounts seen in cytopathology aspirates, and to do this in a timely fashion. This requires a clear understanding of the limitations to development of an accurate diagnosis and a willingness on the part of the pathologist to request repeat biopsy for additional tissue when it is necessary for accurate diagnosis.
Diagnostic criteria involving electron microscopic ultrastructure found relevance for the evaluation of neoplasms described as small round blue cell tumors, spindle cell tumors, melanocytic tumors, and neuroendocrine/neuroblastic tumors, as well as delineation of ciliary ultrastructural abnormalities in primary ciliary dyskinesia. Current approaches to these neoplasms are now generally approached using paraffin section immunohistochemistry. Electron microscopy is currently used mainly for nephropathology, for evaluation of ciliary axonemes, for rare cases where immunohistochemistry is not diagnostic, and where demonstration of premelanosomes, neuroendocrine granules, or amyloid is diagnostic.
Adequate sampling of a lesion is critical to making an accurate diagnosis. Undercall diagnostic discrepancies are frequently due to sampling of a small portion of a large lesion that is unrepresentative of the most abnormal portion of the lesion. Insufficient sampling can result in an equivocal diagnosis or, worse, an inaccurate diagnosis. Empirical rules have been adopted over the decades to ensure statistically adequate sampling of masses and organs such as transurethral resections of prostate, soft tissue sarcomas, and heart allograft biopsies.
In spite of the limitations and statistical uncertainties relating to morphologic diagnosis, a wealth of information is conveyed to a service pathologist in a tried-and-true H&E section [36]. Analogous to the fact that a plain chest X-ray is the sum total of all densities in the beam path, the morphologic changes in diseased cells and tissues are the morphologic sum total of all of the disequilibria in the abnormal cells. For most neoplastic diseases, morphologic criteria are sufficient to predict the risk of invasion and metastasis (the malignant potential), the pattern of metastases, and the likely clinical outcomes. For example, the etiology and pathogenesis in small cell lung carcinoma can be inferred (cigarette smoking, with carcinogen-induced genetic mutations) and the outcome predicted (early metastasis to regional nodes and distant organs, with high probability of death within 5 years of diagnosis). New molecular data for both neoplastic and non-neoplastic diseases will most likely benefit unaffected individuals by estimating disease risk, and will most likely benefit patients by defining the molecular subset for morphologically defined diagnostic entities, thus guiding individualized therapy.
THE FUTURE OF DIAGNOSTIC PATHOLOGY
Diagnostic pathology will continue to use morphology and complementary data from protein (immunohistochemical) and nucleic acid (cytogenetics, in situ hybridization, DNA sequence, and RNA abundance) screening assays. New data will be integrated into the diagnostic process by reducing the cost and turnaround time of current technologies, and by development of new technologies, some of which are described.
Individual Identity
For transplant candidates, major histocompatability complex (MHC, HLA in human) screening is evolving from cellular assays and serology toward sequencing of the alleles of the class I and II HLA loci. Rapid sequencing of these alleles in newborn cord blood would allow databasing of the population's haplotypes, facilitating perfect matches for required bone marrow or solid organ transplants.
Rapid Cytogenetics
Current uses of in situ hybridization to screen for viruses (such as EBV), light chain restriction (in B lymphomas), and copy number variation (for instance, HER2 gene amplification) demonstrate the benefit of in situ nucleic acid hybridization assays. It is possible that interphase FISH/CISH will become rapid enough to be used in the initial diagnostic workup of certain patients, including for sarcoma-specific translocations, ploidy analysis in hydatidiform moles, and gene amplification of receptor tyrosine kinase genes.
Rapid Nucleic Acid Sequence and RNA Abundance Screening
Current uses of nucleic acid screening for bcr-abl translocation, donor:recipient ratios after bone marrow transplant, microsatellite instability, quantitative viral load (for EBV, BK, CMV, and others), and single gene mutations (for CFTR, Factor 2, α-1-antitrypsin) demonstrate the benefit of nucleic acid screening in diagnosis and management. It is possible that each new neoplasm will be promptly defined as to ploidy, translocations, gene copy number differences, DNA mutations, and RNA expression cluster subset, allowing residual disease screening as well as individualized therapy.
Computer-Based Prognosis and Prediction
Current uses of morphology, immunohistochemistry, and molecular pathology demonstrate their benefit through improved diagnostic accuracy. However, diagnosis, extent of disease, and molecular subsets are currently imperfect estimators of prognosis and response to therapy. Relational databases which correlate an individual's demographic data, family history, concurrent diseases, morphologic features, immunophenotype, and molecular subset, and which integrate disease prevalence by age, sex, and ethnicity using Bayesian probabilities, should improve accuracy of prognosis and prediction of response to therapy. As risk correlates are developed, it is possible that healthy individuals will be screened and given risk estimates for development of different diseases.
Normal Ranges and Disease Risks by Ethnic Group
Current uses of normal ranges for serum chemistry assumes a similar bell-curve distribution across ages, sexes, and races. This may be true for most but not all analytes. Computer reference databases will likely generate normal ranges specific for the particular age/sex/ethnicity of individual patients. Similarly, familial risk for an inherited disease may vary by ethnic group, and this variation should be used in Bayesian calculations to define risk for unaffected at-risk family members.
Individual Metabolic Differences Relevant to Drug Metabolism
Current uses of liver and renal impairment to guide drug dosage demonstrate the benefit of using patient physiology to customize therapy. It is likely that individual differences in enzymatic metabolism of particular drugs (for instance, warfarin or tamoxifen) will be defined at the enzyme sequence level, and that gene haplotype data will be determined for new patients prior to receipt of these drugs.
Serum Biomarkers
Current uses of prostate specific antigen (PSA) to screen for prostate carcinoma and its recurrence demonstrates the benefit of serum biomarkers in common neoplasms. It is likely that high-sensitivity screening of single and clustered serum analytes will lead to improved methods for early detection and persistence of neoplasms, autoimmune diseases, and infections.
CONCLUSION
Pathologists consider each disease to have a natural, mechanical, physicochemical basis. Each disease has an etiology (initial cause), a pathogenesis (stepwise progression), and a natural history with effects on normal function (clinical outcome). Pathologists collect the data needed to answer patients' and clinicians' questions, simply phrased as “what is it?” (diagnosis), “how it going to behave?” (prognosis), and “how do I treat it?” (prediction of response to therapy). Instincts and diagnostic criteria, as well as the optical, mechanical, chemical, and computing technologies described previously, are the basis for modern service pathology. As the human genome is deciphered, and as the complex interactions of cellular biochemistry are refined, risk of disease in unaffected individuals will be calculable, disease diagnosis will be increasingly accurate and prognostic, and molecular subsets of morphologically defined disease entities will be used to guide customized therapy for individual patients. It is a great time in history to be a pathologist.
REFERENCES
- 1.Rosai J. Rosai and Ackerman's Surgical Pathology. Mosby; 2004. [Google Scholar]
- 2.Porter R. The Greatest Benefit to Mankind: A Medical History of Humanity. 1st ed. W. W. Norton & Co.; New York, NY: 1998. [Google Scholar]
- 3.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 4.Jay V. The legacy of Karl Rokitansky. Arch Pathol Lab Med. 2000;124:345–346. doi: 10.5858/2000-124-0345-TLOKR. [DOI] [PubMed] [Google Scholar]
- 5.Paget J. Lectures on Surgical Pathology. Brown, Green, and Longmans; London: 1853. [Google Scholar]
- 6.Virchow R. Cellular Pathology. August Hirschwald; Berlin: 1858. [Google Scholar]
- 7.Hansen G. Investigations concerning the etiology of leprosy. Norsk Mag. Laegervidenskaben. 1874;4:1–88. [Google Scholar]
- 8.Koch R. The etiology of tuberculosis. Berliner Klinische Wochenschrift. 1882;15:221–230. [Google Scholar]
- 9.Turk JL. Rudolf Virchow—Father of cellular pathology. J R Soc Med. 1993;86:688–689. doi: 10.1177/014107689308601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes O. Contagiousness of puerperal fever. New England Quarterly Journal of Medicine. 1843;1:503–530. [PubMed] [Google Scholar]
- 11.Dunn PM. Oliver Wendell Holmes (1809–1894) and his essay on puerperal fever. Arch Dis Child Fetal Neonatal Ed. 2007;92:F325–F327. doi: 10.1136/adc.2005.077578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raju TN. Ignac Semmelweis and the etiology of fetal and neonatal sepsis. J Perinatol. 1999;19:307–310. doi: 10.1038/sj.jp.7200155. [DOI] [PubMed] [Google Scholar]
- 13.Wohler F. Annalen der Physik und Chemie. 1828:88. [Google Scholar]
- 14.Gal AA. In search of the origins of modern surgical pathology. Adv Anat Pathol. 2001;8:1–13. doi: 10.1097/00125480-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Medawar PB. The immunology of transplantation. Harvey Lect. 1956:144–176. [PubMed] [Google Scholar]
- 16.Early P, Huang H, Davis M. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 17.Leder P, Max EE, Seidman JG. Recombination events that activate, diversify, and delete immunoglobulin genes. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):859–865. doi: 10.1101/sqb.1981.045.01.103. [DOI] [PubMed] [Google Scholar]
- 18.Coons AH, Kaplan MH. Localization of antigen in tissue cells; improvements in a method for the detection of antigen by means of fluorescent antibody. J Exp Med. 1950;91:1–13. doi: 10.1084/jem.91.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 20.Wilkins MH, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953;171:738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 21.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 22.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 23.Crick FH, Barnett L, Brenner S. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 24.Nirenberg M, Caskey T, Marshall R. The RNA code and protein synthesis. Cold Spring Harb Symp Quant Biol. 1966;31:11–24. doi: 10.1101/sqb.1966.031.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Gall JG, Pardue ML. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci USA. 1969;63:378–383. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci USA. 1969;64:600–604. doi: 10.1073/pnas.64.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goeddel DV, Kleid DG, Bolivar F. Expression in Escherichia coli of chemically synthesized genes for human insulin. Proc Natl Acad Sci USA. 1979;76:106–110. doi: 10.1073/pnas.76.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith LM, Sanders JZ, Kaiser RJ. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 31.Lander ES, Linton LM, Birren B. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 32.Venter JC, Adams MD, Myers EW. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 33.Mullis K, Faloona F, Scharf S. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 34.Mills S. Sternberg's Diagnostic Surgical Pathology. 2004. Lippincott, Philadelphia. [Google Scholar]
- 35.Fletcher C. Diagnostic Histopathology of Tumors. Churchill Livingstone; 2007. [Google Scholar]
- 36.Rosai J. The continuing role of morphology in the molecular age. Mod Pathol. 2001;14:258–260. doi: 10.1038/modpathol.3880295. [DOI] [PubMed] [Google Scholar]


