Publisher Summary
This chapter outlines the progress made to meet delivery challenges and the clinical applications of antisense technology and highlights the limitations of antisense agents in therapeutics. Antisense technology presents an opportunity to manipulate gene expression within cells to treat an endless number of diseases and is a powerful tool for studying gene function. The antisense approach utilizes antisense agents to fight various diseases by regulating the expression of a specific factor, the presence of which actually causes the particular disease. Liposomes, polyconjugates, and other biodegradable polymeric carriers have emerged as the leading platform for the systemic delivery of RNA interference (RNAi) therapeutics and offer considerable promise for diseases of the liver, solid cancers, as well as potentially enhanced vaccines, infectious diseases, and immune cell-related disorders. This chapter surveys the agents employed in antisense technologies and discusses the various mechanisms of gene silencing. It focuses on those techniques that employ oligonucleotides composed of both modified and unmodified DNA or RNA nucleotides, and on RNAi.
Antisense technology presents an opportunity to manipulate gene expression within the cells to treat various diseases, and acts as a powerful tool for studying gene function utilizing antisense agents to manage the diseases by regulating the expression of the specific factor that actually causes the particular disease. Highly specific and effective gene silencing of any disease can be achieved by an accurate knowledge of the target mRNA sequence and rational design of its complementary antisense agents for the downregulation of its protein message. This technology has been successfully used for the treatment of cancer, HIV, and other mutating viral diseases. The technology uses agents like antisense oligonucleotides, ribozymes, short interfering RNA (siRNA), microRNA (miRNA), apatamers, and others. The purpose of this chapter is to survey different antisense technologies and the various mechanisms of gene silencing. The chapter also highlights therapeutic limitations, benefits, clinical applications, and various advances to meet delivery challenges of these agents, and gives due consideration to the regulatory and patent aspects.
7.1. Introduction
Antisense technology presents an opportunity to manipulate gene expression within cells to treat an endless number of diseases and is a powerful tool for studying gene function. The antisense approach utilizes antisense agents to fight various diseases by regulating the expression of a specific factor, the presence of which actually causes that particular disease. Some antisense approaches have evolved over the last few decades, explicitly the introduction of antisense oligonucleotides (AS ODNs) by Stephenson and Zamecnik in the late 1970s [1], the description of ribozymes by Cech and colleagues in the 1980s [2], and the demonstration of short interfering RNA (siRNA) by Fire and Mello in the 1990s [3]. Recently, microRNA (miRNA) replacement therapy has emerged as a new approach to treat human diseases like cancer and various neurodegenerative diseases. Replacement therapy involves the reintroduction of a synthetic version of a natural miRNA that gets depleted in the diseased tissue [4].
Highly specific and effective gene silencing of any disease can be achieved by an accurate knowledge of the target mRNA sequence and rational design of its complementary antisense agents for the downregulation of its protein message. Thus, these are being extensively explored for personalized therapy of cancer, HIV, and other mutating viral diseases [5], [6], [7], [8]. Gene silencing also has a great potential as a chemosensitizing agent to overcome the difficulties of drug resistance and dose-limiting toxicities of chemotherapeutic agents [9]. This technique differs from that used with conventional drugs in that it precisely checks the formation of disease-causing protein by downregulating its expression, rather than relieving the symptoms of the disease after its manifestation. Moreover, this technique can be differentiated from the genetic approach by its action on the mRNA, expressing the disease-causing protein rather than acting on a particular faulty gene. The success of this approach relies on understanding the correct sequence of RNA that carries the protein message responsible for the disease of interest. Fortunately, the completion of the human genome project endows us with a rich source of information on target genes, for the rational design of antisense drugs within hours, for research and clinical trials.
This chapter anticipates the accomplishment of the therapeutic use of oligonucleotides and siRNA. One AS ODN, Fomivirsen, is marketed under the trade name Vitravene® by ISIS pharmaceuticals as a local injection to treat retinitis [10]. Another similar approach to inhibit proteins is via specific three-dimensional complex-structured molecules called aptamers. Pigatinib (MacugenTM) is an FDA-approved aptamer for the treatment of wet macular degeneration [10]. Currently, many antisense agents are under clinical trials and many others at preclinical stage are in a queue to enter clinics for various applications such as cancer, HIV, age-related macular degeneration (AMD), and respiratory syncytial virus, as well as rare diseases like pachyonychia congenita. Conversely, some antisense agents, like Bevasiranib of Acuity Pharmaceuticals and Sirna-027 of Sirna Therapeutics, have recently been terminated at phase III and phase II, respectively, of clinical trials (Table 7.1, Table 7.2, Table 7.3 ).
Table 7.1.
Clinical Status of siRNA Formulations
| S. No. | Company and Strategic Alliances | Product Details | Clinical Status as on Dec. 2009 | Drug Target/Tissue | Indication | Route of Delivery | Delivery System |
|---|---|---|---|---|---|---|---|
| 1. | Acuity/later licensed by Opko | siRNA Cand5/Bevasiranib | Terminated at Phase III | VEGF/Eye | AMD | Intravitreal injections | Naked siRNA |
| VEGF/Eye | Diabetic retinopathy | Intravitreal injections | Naked siRNA | ||||
| 2. | Sirna Therapeutics/Later acquired by Allergan | Sirna-027/Now AGN-745 | Terminated at Phase II | VEGF/Eye | AMD | Intravitreal injections | Naked siRNA |
| 3. | Silence Therapeutics/Quark/Pfizer | RTP-801i | Phase II | VEGF/Eye | AMD | Intravitreal injections | Naked siRNA |
| Atu027 | Phase I | Targets PKN3 molecule in cancer cells | Cancer | Intravenous | siRNA incorporated in AtuPLEX delivery platform | ||
| AKIi-5 | Phase I/II | P53 gene/kidney | Acute kidney injury in kidney transplantation | Intravenous | Chemically modified siRNA with AtuRNAi technology | ||
| DGFi | Phase I/II | Delayed graft function in kidney transplantation | Intravenous | ||||
| 4. | Alnylam Pharmaceuticals | ALN-RSV01 | Phase III | RSV nucleocapsid /lungs | Respiratory syncytial virus (RSV) infection | Intranasal | Naked siRNA |
| ALN-VSP | Phase I | Kinesin spindle protein (KSP) and VEGF/liver | Liver cancer | Intravenous | Two siRNA molecules formulated in lipid nanoparticles | ||
| 5. | Nucleonics | NUC B1000 | Phase I | 4 HBV genes/liver | Hepatitis B antiviral agent | Intravenous | Plasmid DNA formulated in cationic lipid delivery system |
| 6. | TransDerm (Santa Cruz, CA) | TD101 | Phase Ib | Targets the N171K mutant form of the gene/skin | Pachyonychia congenita | Topical | Two delivery methods:
|
| 7. | Calando Pharmaceuticals | CALAA-01 | Phase I | M2 subunit of ribonucleotide reductase/solid tumors | Anticancer | Intravenous | RONDEL (RNAi/oligonucleotide nanoparticle delivery) |
| 8. | MDRNA Inc. | MDR-03030 | Preclinical phase | Targets conserved in region of the influenza viral genome | Acts on influenza viral genome; has the ability to mutate around the compound | Intranasal | Combined UsiRNAs with DiLA2 delivery platform |
| 9. | Benitec | rHIV7-shl-TAR-CCR5RZ | Phase I | HIV tat/rev gene, TAT-responsive elements, CCR5 receptors/targets stem cells | AIDS-related lymphoma | Systemic | DNA-based plasmid expressing anti-HIV RNA |
Reference: http://clinicaltrials.gov
Table 7.2.
Clinical Status of AS ODN Formulations
| S. No. | Developer/Partner | Reference | Product Details | Clinical Status as on Dec. 2009 | Drug Target | Indication | Route of Delivery | Delivery System |
|---|---|---|---|---|---|---|---|---|
| 1. | AVI Biopharma, Inc. | http://www.avibio.com/ | AVI-4658 | Phase Ib/II | Exon 51 | Duchenne muscular dystrophy | Intramuscular | Morpholino-oligomer |
| 2. | Neopharm | http://www.neopharm.com/ | LErafAON-ETU | Phase I | Raf-1 protein | Neoplasms | Intravascular infusion | Liposomes |
| 3. | ISIS/Novartis | http://www.isispharm.com/ | Vitravene (Fomivirsen) | Marketed | CMV IE | CMV retinitis | Ocular | PS |
| ISIS/OncoGeneX | OGX-427 OGX-011 |
Phase I Phase II |
Hsp 27 Clusterin |
Bladder cancer Cancer |
Intravesical instillation | MBO MBO |
||
| Isis/Genzyme | Mipomersen | Phase III | apoB-100 | Cardiovascular | Subcutaneous injection | AS ODN drug | ||
| Isis/Bristol Myers Squibb | BMS-PCSK9Rx | Preclinical | PCSK9 | Cardiovascular | Not available | AS ODN drug | ||
| Isis/Alsa, MDA | ISIS-SOD1Rx | Preclinical | superoxide dismutase, or SOD1 | Amyotrophic lateral sclerosis | Not available | AS ODN drug | ||
| Isis/antisense | ACHN-490 | Preclinical | growth hormone receptor, or GHr | Acromegaly | Not available | AS ODN drug | ||
| Isis/Excaliard | EXC001 | Phase I | Antifibrotic | Not available | AS ODN drug | |||
| ISIS/Teva | ATL/TV1102 | Phase II | CD49d | Multiple sclerosis | Not available | MOE | ||
| ISIS/iCo Therapeutics Inc/ISIS | iCo-007 | Phase I | c-Raf | Diabetic retinopathy | Not available | Phosphothiorate AS ODN | ||
| Isis Pharmaceuticals | ISIS 104838 | Phase II | TNF-alpha messenger RNA | Rheumatoid arthritis | Subcutaneous injection | Phosphothiorate AS ODN | ||
| ISIS 113715 | Phase I | Protein tyrosine phosphatase 1B | Type 2 diabetes mellitus | Subcutaneous injection | Phosphothiorate AS ODN | |||
| ISIS 3521 | Phase II | Pkc-Alpha | Metastatic breast cancer | Intravenous infusion | Phosphothiorate AS ODN | |||
| ISIS 5132 | Phase II | C-Raf kinase | Metastatic breast cancer | Intravenous infusion | Phosphothiorate AS ODN | |||
| Alicaforsen (ISIS 2302) | Phase III | ICAM-1 | Crohn's disease | Not available | Phosphothiorate AS ODN | |||
| ISIS-CRPRx | Phase I | CRP | Cardiovascular | Not available | Phosphothiorate AS ODN | |||
| ISIS-SGLT2Rx | Phase I | SGLT2 | Type 2 diabetes | Not available | Phosphothiorate AS ODN | |||
| 4. | Enzon Pharmaceuticals, Inc | http://www.enzon.com | EZN-2968 | Phase I | anti-HIF-1α LNA | Carcinoma, lymphoma | Intravenous | LNA AS ODN |
| 5. | Genta Incorporated | http://www.genta.com | Genasense (G3139, Oblimerson sodium) | Phase III | Bcl2 | Solid tumors | Subcutaneous/intravenous infusion | Phosphothiorate AS ODN |
| Inex/Genta | G4460/LR 3001 | Phase II | C-myb | Cancer | Intravenous infusion | Phosphothiorate AS ODN | ||
| 6. | Lorus Therapeutics | http://www.lorusthera.com | GTI-2040 | Phase I/II | R2 component of ribonucleotide reductase (RNR) | Renal cell carcinoma | Intravenous | Phosphothiorate AS ODN |
| GTI-2501 | Phase I/II | R1 component of ribonucleotide reductase (RNR) | Cancer | Intravenous | Phosphothiorate AS ODN | |||
| 7. | Lilly | http://clinicaltrials.gov | LY2181308 | Phase I/II | Survivin | Cancer | Intravenous infusion | LOE gapmers |
| http://clinicaltrials.gov | LY2275796 | Phase I | eIF-4E | Cancer | Intravenous infusion | LOE | ||
| 8. | Topigen Pharmaceuticals | http://www.topigen.com | ASM8 | Phase I/II | CCR3 and common beta chain of IL-3/5 and GM-CSF receptors | Asthma | Inhalation | Proprietary oligonucleotide technology |
| 9. | Aegera Therapeutics | http://www.aegera.com | AEG35156 | Phase IIB | XIAP mRNA | Chemosensitization of cancer cells | Intravenous infusion | MBO |
| 10. | Santaris Pharma | http://clinicaltrials.gov | SPC3647 | Phase I | miRNA 122 | Hepatitis C | Not available | LNA AS ODN |
| 11. | VasGene Therapeutics | http://www.vasgene.com | Veglin | Phase I/II | VEGF | Cancer | Intravenous infusion | Phosphothiorate AS ODN |
| 12. | Antisense Pharma | http://www.antisense-pharma.com | AP 12009 | Phase III | TGF-β2 | Cancer | Intratumorally | Phosphothiorate AS ODN |
| 13. | MethylGene/MGI Pharma/British Biotech | http://clinicaltrials.gov | MG 98 | Phase II (Not in trials) | DNA methyltransferase | Cancer | Not available | MBO |
| 14. | Eleos, Inc. | Aezea® (Cenersen) | Phase II | p53 | Acute myelogenous leukemia | Intravenous infusion | Phosphothiorate AS ODN | |
| 15. | Epigenesis/Genta | EPI-2010 (RASON) | Phase II (Not in trials) | Adenosine A1 receptor | Asthma | Not available | Phosphothiorate AS ODN | |
| 16. | Idera Pharmaceuticals/Merck | IMO-2055 | Phase Ib | TLR9 agonist | Non-small cell lung cancer and colorectal cancer | Subcutaneous | Immunomodulatory oligonucleotide |
Table 7.3.
Clinical Status of Ribozymes and Aptamers Formulations
| S. No. | Developer/Partner | Product Details | Clinical Status as on Dec. 2009 | Drug Target | Indication | Route of Delivery |
|---|---|---|---|---|---|---|
| A. Aptamers | ||||||
| 1. | Ophthotech Corporation | ARC1905 | Phase I | Anti-C5 | AMD | Intravenous |
| E10030 | Phase I | PDGF | AMD | Intravitreal injections | ||
| 2. | Eyetech Pharmaceuticals/Pfizer | Pegaptanib sodium (Macugen) | Marketed (FDA approved) | VEGF | AMD | Intravitreal injections |
| Eyetech Pharmaceuticals | EYE001 | Phase II/Phase III, completed in 2002 | VEGF | Macular degeneration, choroidal neovascularization | Intravitreal injections | |
| 3. | National Heart, Lung, and Blood Institute (NHLBI) | REG1 | Phase I | Dual effect of antifactor IX and antidrug | Anticoagulation | Intravenous |
| 4. | Noxxon Pharma AG | NOX-E36 | Phase I | Monocyte chemotactic protein-1 (MCP-1) | Chronic inflammatory diseases, Type 2 diabetes mellitus, systemic lupus erythematosus | Intravenous and subcutaneous injection |
| Noxxon Pharma AG; German Federal Ministry of Education and Research | NOX-A12 | Phase I | Inhibitor of stromal cell-derived factor-1 (SDF-1) | Autologous stem cell transplantation | Intravitreal injections | |
| Archemix Corp. | ARC1779 | Phase II | vWF (Von Willebrand factor) | Platelet disorders | Intravenous infusion | |
| B. Ribozymes | ||||||
| 1. | Johnson & Johnson Pharmaceutical Research & Development, L.L.C./Tibotec Pharmaceutical Limited | OZ1 | Phase II | Anti-HIV-1 gene | HIV infections | Not available |
| 2. | Jonsson Comprehensive Cancer Center National Cancer Institute (NCI) | RPI.4610 | Phase II | Anti-FLT-1 | Kidney cancer | Subcutaneous injection |
Reference: http://clinicaltrials.gov
The major obstacle in navigating these molecules for regulatory approval is efficient delivery to the desired site. However, this challenge can be met by better understanding of the various formidable barriers encountered, from the site of delivery to the site of action of antisense drugs. Two major limitations, insufficient delivery to target cells and off-target side effects, can be addressed by designing a suitable delivery system, by chemical modifications such as using structural modifications or nanocarriers, or by conjugation with receptor-specific targeting ligands, or combinations thereof [11], [12]. Thus, the purpose of this chapter is to highlight the limitations of antisense agents in therapeutics, the progress made to meet delivery challenges, and the clinical applications of antisense technology. This chapter surveys the agents employed in antisense technologies and discusses the various mechanisms of gene silencing. The emphasis will be on those techniques that employ oligonucleotides composed of both modified and unmodified DNA and/or RNA nucleotides, and another major antisense technology called RNA interference, or RNAi.
7.2. The Evolution of Antisense Drug Technology
Since its discovery, antisense technology has continued to progress rapidly. In this section, we review our knowledge of antisense drug delivery, from discovery to application.
7.2.1. History
It was discovered in the late 1970s that the expression of a specific gene product could be inhibited using a short complementary DNA sequence [1]. This led to intensive research on the antisense approach. In 1978, the concept of antisense technique came into view after the discovery of single-stranded DNA molecules, known as antisense oligodeoxynucleotide (AS ODNs), by Zamecnik and Stephenson [1]. Since then, new applications of antisense technology have continued to develop rapidly. In early developmental stages, blockage of target protein expression was achieved by administering whole DNA or RNA locally as therapeutics [13]. Then, single-stranded DNA molecules (AS ODNs) were first locally administered for the treatment of tumors [1]. However, the mechanism of their antitumor activity was never elucidated, and their activity also varied with their size and structure. Compared to studies done on DNA, more extensive research has been reported on using RNA and polyribonucleotides as medicinal agent. In early stages of development, double-stranded polyriboinosine–polyribonocytidine was the most extensively studied polynucleotide [14]. Miller et al. were the first to try modifying the phosphate backbone of oligonucleotides to improve their properties, and they synthesized the first chemically modified oligonucleotide belonging to the class of methyl phosphotriester oligonucleotides [15], [16]. Thereafter, ribozymes, another class of catalytic oligonucleotides, appeared as a new tool to investigate gene expression. Depending on the structure, ribozymes can degrade or modify the target mRNA to produce correct sequence [17], [18]. AS ODNs and ribozymes were already in clinical practice as genetic therapeutics long before small interfering RNAs (siRNAs) were developed as potential medicinal agents. In 1992, Fire et al. were the first to describe the RNAi as a mechanism of action of AS ODNs for the destruction of target mRNA [19]. They are a natural cellular defense mechanism, by virtue of which the presence of double-stranded viral DNA triggers the mRNA degradation. Introduction of 20- to 23- nucleotide-long siRNA could exhibit antiviral activity by blocking the expression of viral proteins, which led to the progress of siRNA in therapeutics to block the production of disease-related proteins.
7.2.2. Present Scenario
Presently, three decades after the emergence of the antisense concept, the basics of this technology and the key steps to challenges in therapeutics are well comprehensible. The main attention of researchers and pharmaceutical industries today is to make this technology available for its therapeutic applications. Thus, today's focus is not only to design an antisense molecule with good affinity and specificity or to predict its in vivo effect, but also to practically approach its formulation, taking care of the ultimate pharmacological and toxicological aspects at the initial stage of development to avoid rejection at the final clinical phases. Incredible progress has been made in the rational design and appropriate selection of antisense agents, its formulations, delivery carriers, doses, and dosage regimens, and most importantly in the design of preclinical and clinical trials [20], [21], [22]. Since the discovery of antisense technology, there have been numerous advances both in the structure and properties of oligonucleotides used as therapeutic agents. Compared to that of whole DNA or RNA, today RNAi is achieved using siRNA, dsRNA (double-stranded RNA), and shRNA (short hairpin RNA). Several modifications such as novel bases, sugars, conjugates, and chimeric technology have been tried to improve the pharmacokinetic and pharmacodynamic properties of these oligonucleotides [11], [12], [20], [21], [22]. Because the biological activity of an AS ODN at the site of action is dependent on factors such as its concentration, concentration of mRNA, rate of synthesis and degradation of mRNA, and the type of terminating mechanism, various strategies have been employed to improve the properties of systemically administered oligonucleotides. Further, a more potent gene therapy, ribozymes, has gained more attention than AS ODNs [23], [24], [25]. Ribozymes are enzymes that cleave single-stranded regions of RNA by transesterification or hydrolysis of a phosphodiester bond. To achieve RNAi, either ribozyme-encoding sequences are incorporated into plasmids, or chemically modified minimum ribozyme structure is administered [26]. Chemical modifications have also been tried to synthesize nuclease-resistant ribozyme drugs [27]. Moreover, earlier used AS ODNs exhibit gene silencing after entering the nucleus, but some newer ones and siRNA need not enter the nucleus; this leads to posttranscriptional gene silencing by degrading target mRNA in the cytoplasm itself, making the delivery challenge a little simpler than it was before with AS ODNs [18]. The evolution of different antisense technologies has not closed the path of oglionucelotides; rather the progress and improvements in the oligonucleotides have hastened the pace of newer antisense agents to reach the therapeutic platform. Currently, there is swift progress in systematic research and development, while obviating the downside faced previously by AS ODNs at the earlier growth and clinical steps.
7.3. Strategies of Transcriptional Arrest
Downregulation of mRNA expression is made possible either via transcriptional arrest of the RNA complementary to the disease-related protein or through the posttranscriptional gene silencing (PTGS) phenomenon. Transcriptional arrest is an alternative to inhibiting mRNA expression by AS ODNs posttranscription. The transcriptional arrest of double-stranded DNA can be achieved by two distinct strategies, namely, strand invasion and triple-strand formation [28]. Until recently, triple-strand formation has been the most commonly used strategy to induce transcriptional arrest. Triple strands are formed by involving Watson–Crick hydrogen bonds between the third strand and the complementary strand of DNA duplex [28]. Homopyrimidine oligonucleotides are capable of inhibiting transcription via triple-strand formation [29], [30]. Several modifications in oligonucleotides have been tried to improve binding to duplex DNA via triplex formation with high affinity and specificity [31], [32], [33]. Strand invasion, though not a widely studied strategy to induce transcriptional arrest, is being used by certain oligonucleotides, such as peptide nucleic acids (PNAs), to inhibit transcription [34]. PTGS is the phenomenon in which antisense agents act through degrading the transcribed target mRNA to prevent translation into the complementary protein (Fig. 7.1 ).
Figure 7.1.
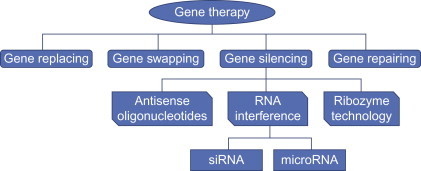
Various strategies of gene therapy.
Antisense molecules lead to the manipulation and/or modification of DNA or RNA through a number of different mechanisms to partially or completely eliminate the normal cellular processing of the genetic message of a gene. Accomplishment of this knockdown or knockout is the major challenge presented by the antisense technique. To achieve clinically approved status, it is essential to have a better understanding of the various pharmaceutical and pharmacological considerations, availability of the active moiety to act at the desired site, therapeutically effective concentration, its formulation into an appropriate dosage form, and different barriers to reaching the target of interest [21], [22].
7.4. Barriers to Oligonucleotide and siRNA Delivery
Though antisense technology holds great therapeutic potential, several barriers (Fig. 7.2 ) often impede delivery of AS ODNs and siRNA to their site of action. The barriers encountered by naked oligonucleotides and siRNA are different from those encountered by the ones associated with various nanocarriers. To elicit a pharmacological response, an antisense agent must reach the tissue of action from the general circulation, invade the diseased cells there, and interact with the complementary mRNA, following endosomal release, thereby inhibiting the expression of the desired protein. However, the large size and ionic nature of the oligonucleotides and siRNA impede them from efficiently traversing the various biological membranes [22]. Here we discuss the various barriers encountered by the administered AS ODNs, which have also been studied extensively for siRNA, and the possible ways to overcome these barriers for efficient delivery. The ultimate target and the action of AS ODNs and siRNA are the same, and thus the barriers, challenges, and remedies discussed for one applies more or less for the other too.
Figure 7.2.
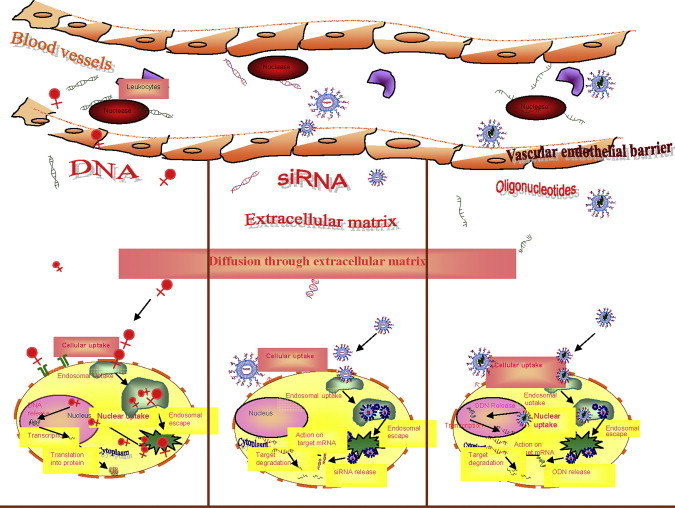
Potential physiological barriers to antisense drug delivery.
7.4.1. Physiological Barriers
While traveling from the site of administration to the site of action, antisense agents cross various physiological enzymes and compartments that affect the extent and efficiency of their delivery to the target.
7.4.1.1. Degradation by Nucleases
Following administration, the first biological barrier an oligonucleotide faces is presented by the nuclease activity in blood and tissues. Within 1 min, nearly 70% of the administered antisense molecule degrades, resulting in low gene silencing [35]. Chemical modifications and use of nonviral vectors can drastically improve the stability of the oligonucleotide toward nucleases in the biological system [18], [36], [37], [38], [39]. Remarkable modifications can be done at the 2′-OH position of pentose sugars and the 3′ half of the siRNA structure. The substitution of sulfur for oxygen to form phosphorothioate oligonucleotides is the most common chemical modification to improve stability toward the nucleases [36]. Further, 2′-OH modifications, locked nucleic acids (LNAs), PNAs, morpholino compounds, and hexitol nucleic acids (HNAs) can also improve mRNA stability toward nucleases. In addition, prolonged pharmacological action has been observed after the inclusion of a six-carbon sugar instead of ribose, 2′-F and 2′-OMe modifications, and use of gapmers [18], [36]. Similarly, cationic lipids and polymers readily complex with the anionic antisense molecules by electrostatic interaction, thereby protecting the oligonucleotides from degradation by nucleases [40], [41].
7.4.1.2. Glomerular Filtration, Hepatic Metabolism, and RES Uptake
Following administration into general circulation, oligonucleotides—or, more specifically, oligonucleotides associated with nanocarriers of size greater than 200 nm—are subjected to phagocytosis by mature macrophages residing in the tissues of the reticuloendothelial system (RES), such as the liver, spleen, and lungs [42]. Nevertheless, particles smaller than 100 nm leak out from the intercellular junction of capillary endothelium to the interstitial space of hepatic sinusoid because of hepatic uptake, and get trapped by the hepatic Kupffer cells there. Colloidal complexes of AS ODN and siRNA with polymers or lipids of high-charge density get destabilized as aggregates due to the presence of negatively charged serum proteins. Both size and charge of these complexes determine their clearance from the circulation [43], [44], [45], [46]. A coating of polyethylene glycol (PEG) helps in making these nanocarrier complexes long circulating by neutralizing the surface charge and imparting a protective hydrophilic sheath around it [47], [48]. Thus, hepatic clearance and the RES uptake of nanocarrier-associated antisense agent can be avoided by carefully monitoring the size and charge of the final complex, which should be around 100 nm and near to neutral respectively, to avoid opsonization. The large uptake of antisense agents by tissues with fenestrated vasculature, liver, and spleen can be beneficial while targeting such molecules to these tissues. Also, oligonucleotides smaller than 5 nm (70 kD in molecular weight) undergo rapid clearance from the body through glomerular filtration. This glomerular filtration can be avoided by manipulating the size of the antisense molecules by incorporating them into a suitable nanocarrier system and attaching with targeting ligands [18].
7.4.1.3. Endothelial Barrier
The endothelial cells that line the vascular lumen present a barrier to the AS ODN–based therapy, as the oligonucleotides need to cross the endothelium before being delivered to the tissue parenchymal cells. The endothelial cells tightly adhere to the underlying extracellular matrix via integrins and to each other via several adhesion molecules, forming tight intercellular junctions with very small intercellular spaces. Small oligonucleotides travel across the endothelium via a paracellular route involving imperfections in these intercellular junctions [49]. However, in certain tissues such as those of the liver and spleen, these endothelial intercellular spaces are relatively larger than in any other body tissue, allowing access of even large oligonucleotides. AS ODNs also traverse across these endothelial cells transcellularly via claveolin-based transcytosis [50]. This transcellular transport is size independent, allowing passage of both small and large oligonucleotide molecules. Cell-penetrating peptides, targeting ligands, or molecular conjugates, can be used to facilitate passage of AS ODNs across endothelial lining [18].
7.4.2. Cellular Barrier
To exert its action, an antisense agent needs to enter the cell and then reach the actual target. To do so, it faces some of the following challenges.
7.4.2.1. Cell Entry
Nonviral vector–AS ODN complexes, by being highly cationic, bind nonspecifically to negatively charged cell membranes and are easily taken up by cells of the RES by endocytosis or membrane fusion [51]. This nonspecific cellular uptake by nontarget tissues results in severe toxic manifestations due to unwanted protein expression. This nonspecific cellular uptake can be reduced by coating the nonviral vector–AS ODN complexes with PEG and by attaching cell-specific targeting ligands such as transferrin [52], folate [53], surface receptor–specific antibodies [54], and so on. Coupling with membrane-permeable peptides like transportan and penetratin also enhances the cellular internalization [55]. Coating with PEG not only reduces uptake by the RES but also reduces uptake by target cells. Hence, it is more rational to use cell-specific targeting ligands along with PEG coating. However, use of receptor-specific antibodies can evoke immunogenic manifestations. Therefore, these antibodies must be suitably tailored before being used as targeting ligands [56].
7.4.2.2. Endosomal Release
Once an AS ODN reaches the target cells, escape from pericellular vesicles (endosomes) or lysosomes is required for transfection [57]. Hence, the transfection efficiency of nonviral vectors depends on cellular internalization as well as the endosomal escape of the active moiety to reach the actual target [57], [58]. Two strategies are widely used to enhance the endosomal escape. The first uses fusogenic lipids or peptides to rupture lysosomal membranes, by forming pores in membranes [59], [60]. A pH-sensitive liposome system, such as Lipofectin, composed of cationic lipids along with a fusogenic helper lipid such as DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), has been reported, which readily releases the entrapped oligonucleotides at low-pH environments [61]. Interaction of the cationic lipids of these liposomes with the anionic lipids of the cell membranes (endosomal membrane) results in phase separation, thereby creating DOPE-rich regions that form pores in the membranes, causing membrane destabilization [62], [63]. The second strategy involves using delivery systems that possess high buffering capacity. This prevents acidification of the endosomes, resulting in disruption of the endosomal membrane [64], [65]. Polyethylenimine is used as a buffering agent in such polymer–AS ODN complexes. Also endo-osmolytic agents like chloroquine and a higher concentration of other osmotic agents like glycerol, sucrose, or polyvinyl pyrrolidone (PVP) have been shown to aid osmolysis of endosomes.
7.4.2.3. Nuclear Localization
Following endosomal escape, release from the cationic complex of siRNA in cytoplasm and nuclear localization in AS ODNs is required for interaction with target mRNA to inhibit related protein expression. Addition of an anionic lipid can displace negatively charged siRNA or AS ODN from the cationic lipid–polymer-oligonucleotide complex [66], [67]. This release is attributed to the multivalent nature of the anionic lipid and the electrostatic and hydrophilic–hydrophobic interactions of the lipids. This lipid mixing results in charge neutralization, allowing the diffusion of the cationic lipids away from the oligonucleotide. Thus, the anionic lipid competes with the anionic oligonucleotide for the cationic lipid, displacing the oligonucleotide. In the cationic polymer–AS ODN complex, cationic polymers like polyethyleneimine (PEI) or poly-l-lysine (PLL) accelerates nuclear localization by preventing cytosolic degradation of the complex. The extent and duration of the oligonucleotides' response depends upon the step limiting the rate of cellular uptake, intracellular trafficking, endosomal release, and nuclear entry. This step can be controlled by the conjugation of cell-penetrating signal peptides, endosomal release signal peptides, or nuclear localization signal peptides to the oligonucleotides. However, siRNA polyplexes with reducible polymers, such as polyethyleneglycol-poly-l-lysine (PEG-PLL) block copolymers with disulfide crosslinking, are preferred for cytosol-specific degradation, to release siRNA in cytoplasm to act on target mRNA in cytosol [68].
7.4.2.4. Inhibition of Protein Expression
After nuclear localization, the AS ODN must bind to the complementary mRNA, thereby downregulating the related protein expression. The transfection efficiency of these cationic lipid–polymer-oligonucleotide complexes depends on the lipid/polymer-to-oligonucleotide ratio [57]. Hence, the lipid/polymer-to-oligonucleotide ratio must be optimized to achieve maximum inhibition of mRNA expression. Inclusion of tissue-specific promoters can be tried to inhibit mRNA expression within the therapeutic window [69], [70]. Formulation with the appropriate concentration decided after a sound understanding of the pharmacokinetics and biodistribution of a particular antisense molecule and the appropriate modifications to overcome undesired barriers, can provide further value to antisense therapeutics.
7.4.3. Immunological Barrier
The body has an inbuilt mechanism to fight against invading foreign bodies such as pathogens. This resistance is conferred by two distinct mechanisms, namely, innate immunity and adaptive immunity, and presents a major barrier to intracellular AS ODN delivery. An individual is born with innate immunity, which is mediated by receptors that bind to conserved structures called pathogen-associated molecular patterns (PAMPs), common to many pathogens [71]. Thus, innate immunity is of major concern in case of viral vector–based antisense drug delivery. On the other hand, adaptive immunity an individual acquires after birth, on exposure to disease-causing pathogens, and it is mediated by T and B lymphocytes. When a pathogen invades the body, it interacts with specific surface receptors on T and B lymphocytes, causing their activation and production of effector T cells and antibodies that neutralize the pathogen. Many researchers have reported dangerous immune responses with AS ODNs [72]. When phosphorothioates were administered to monkeys as a large, one-time injection, they triggered a systemic and lethal inflammation by activating complement. They also stimulated a dramatic increase in immunoglobulin secretion within 24 h and increased the expression of activation markers such as MHC class II. CpG (cytosine and guanine) separated by phosphate-containing phosphorothioates augment natural killer (NK) cell activity, modulate T cell function, and may stimulate the release of several members of the interleukin family.
7.5. Molecular Mechanisms of AS ODN Interactions
AS ODNs bind to specific mRNA, thereby downregulating its expression and that of the encoded protein. However, the mechanisms by which these oligonucleotides interact with the complementary mRNA and induce a biological effect are complex and difficult to elucidate completely. The ultimate goal of an antisense agent is to suppress or completely block the production of the related gene product. This means that, in the process of transition from DNA sequence to amino acid sequence, the normal transcription and translation apparatus must be affected. The formation of a protein product involves three distinct steps (Fig. 7.3 ).
Figure 7.3.
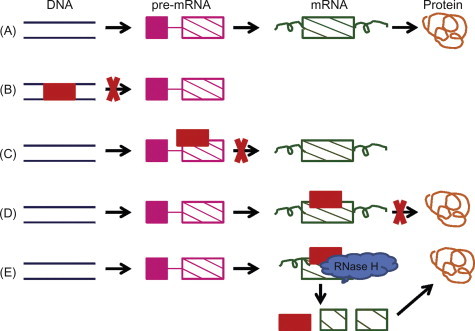
Various strategies available for antisense knockdown. (A) The normal process of protein synthesis involving transcription and translation. (B) Transcriptional arrest by DNA-targeted agents. (C) Prevention of mature mRNA formation by pre-mRNA targeting. (D) Translational arrest by interruption of the translation apparatus. (E) Prevention of translation by RNase H enzyme.
In the first step, the sense strand of the DNA is transcribed into a pre-mRNA. In the second, the pre-mRNA is converted into a mature mRNA via the simultaneous action of three separate processes viz. 5′ capping, intron excision, and polyadenylation. Finally, in the third step the mRNA is transported to the ribosomes for translation into the appropriate polypeptide. Thus, antisense drugs can act by inhibiting any of these steps involved in normal protein production. Although activation of RNase H enzyme activity is thought to be the mechanism of action for the majority of the AS ODNs, many still exert their biological effect via other reported mechanisms, and next we discuss these mechanisms in detail.
7.5.1. Induction of RNase H
RNase H is an endogenous enzyme that cleaves the RNA strand of an RNA–DNA duplex [73]. This is the most widely used and validated mechanism for the knockdown of mRNA, resulting in more than 80% reduction in mRNA and protein expression. However, the precise mechanism by which the RNase H enzyme recognizes a duplex has not been elucidated completely. The RNase H cleavage sites are found near the translational initiation codon and the 3′ and 5′ untranslated regions. RNase H is found in both the nucleus and the cytoplasm of all cells [74]. Its regular function is to remove RNA primers from Okazaki fragments during DNA replication. Hence, oligonucleotides that act via RNase H activation must be designed carefully. Although the requirement for an AS ODN to inhibit a protein expression is not known precisely, modifications in sugar moiety such as sugar type and its orientation are thought to influence RNase H activation [75], [76], [77], [78]. Modifications that result in DNA-like oligonucleotides support RNase H activity, while changes resulting in RNA-like oligonucleotides do not support RNase H activity. Modifications in oligonucleotide backbone also alter RNase H activity [79], [80], [81]. However, the favored design is to use chimeric molecules, such as a single ribonucleotide, which can be bound to its complementary oligonucleotide backbone, which then serves as a substrate for RNase H [36]. These RNase H-dependent oligonucleotides can inhibit protein expression by binding to any region of the complementary mRNA.
7.5.2. Inhibition of 5′ Capping
5′ capping is an essential step in the protein synthesis cascade, stabilizing and translating an mRNA into a mature polypeptide sequence [82]. Although this is an effective mechanism of inhibiting mRNA translation, it is not an established mechanism of action of AS ODNs due to the inaccessibility of the 5′ end of mRNA to capping. The lone exceptions so far validated are the morpholino-oligonucleotides [83]. When placed near the 5′ end of the mRNA, morpholino-oligonucleotides have been shown to specifically reduce translation.
7.5.3. Inhibition of Splicing
Splicing is an important and specific step in the translation of an mRNA to a protein and requires spliceosomes. The AS ODNs act by binding to specific splicing sequence of mRNA, thereby inhibiting its translation and the production of related protein [84], [85]. 2′-O-methyl phosphorothioate oligonucleotides have been reported to inhibit protein expression by inhibiting splicing [86], [87].
7.5.4. Translational Arrest
Many AS ODNs bind to the translational initiation codon, thereby inducing translational arrest. PNAs and morpholino-oligonucleotides have been reported to act via translational arrest by binding to the translational initiation codon or 5′-UTR [88], [89]. Translational arrest has been reported to be a mechanism of action to inhibit replication of viruses like HIV and vesicular stomatitis virus [90], [91].
7.5.5. Inhibition of Polyadenylation
Polyadenylation is an intermediate step in the protein synthesis requiring addition of long tracts of polyadenylate to the pre-mRNA molecules, thereby stabilizing it. Capping of the 3′-terminal of pre-mRNA could inhibit polyadenylation and destabilize it. However, to date no study reports polyadenylation as a mechanism of action for antisense drugs [92].
7.5.6. Steric Block
This mechanism involves physical blockage of the RNA, thereby preventing protein expression by an RNA–DNA duplex formation. This can be achieved by binding to the 5′ end or the translational initiation codon of mRNA [93]. Other RNA-processing events such as nuclear splicing and polyadenylation are also inhibited by steric blockade of mRNA. PNAs and morpholino-oligonucleotides have been reported to inhibit mRNA translation by steric blockade [93], [94].
7.5.7. Activation of Double-Strand RNase
Some AS ODNs inhibit mRNA translation by activating a double-strand RNase enzyme called Dicer, thereby cleaving a dsRNA [95]. RNAi is an antisense mechanism of action that utilizes the enzyme Dicer to promote hydrolysis of the target RNA. siRNA oligonucleotide duplexes have been reported to inhibit protein expression through RNAi pathway by activation of double-strand RNase [96]. The potency, maximal effectiveness, duration of action, and sequence specificity of siRNA oligonucleotide duplexes have been found to be comparable to those of RNase H-dependent oligonucleotides [97]. Also, the activity of siRNA oligonucleotides has been found to be affected by the secondary structure of the target mRNA. The RNAi pathway (Fig. 7.4 ) involves several complicated steps, which we discuss here in brief [96].
-
1.
The process of RNAi is activated by exposure to dsRNA precursors. In the initiation step, in the presence of ATP and RNase-III-type endonucleases, the dsRNA precursors are processed into smaller nucleotides called siRNAs having length from 21 to 23 nucleotides.
-
2.
The resulting siRNAs are subsequently incorporated into a multiprotein complex known as the RNA-induced silencing complex (RISC).
-
3.
In the successive step, in the presence of ATP the siRNA molecules undergo unwinding with the aid of helicases, while getting processed into RISC. This activates the complex, leaving only the antisense strand of siRNA associated with the RISC, while the sense strand gets degraded by the exoribonucleases in cytoplasm.
-
4.
The activated RISC then directs the siRNA toward its complementary mRNA sequence.
-
5.
After binding of siRNA to complementary mRNA sequence, the RISC complex cleaves the target mRNA with the help of Argonaute enzymes associated with the RISC.
-
6.
The cleaved mRNA is then degraded by nucleases in the cytoplasm, thereby inhibiting the expression of the related protein, while the freed RISC complex recycles to cleave more of the target mRNA sequences.
Figure 7.4.
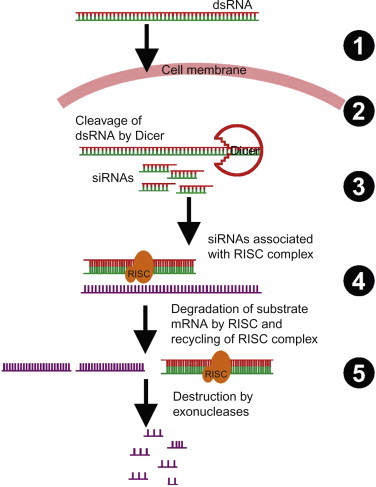
Basic steps involved in the mechanism of RNAi.
7.6. Types of Antisense Agents
To date, a broad array of disease targets have been explored utilizing various antisense agents. AS ODNs, ribozymes, aptamers, and siRNA are the available techniques to achieve suppression or elimination of a genetic message related to a particular disease. Oligonucleotide-based antisense techniques represent the first clinically successful approach to target an ocular disease. None of the antisense agent has become available for systemic applications. AS ODNs and siRNA, being large, ionic, and structurally similar to natural nucleic acids, cannot be used per se as genetic medicines. Hence, to serve as effective drugs, these must possess some desirable pharmacokinetic and pharmacodynamic properties, which vary with their oligonucleotide length, sequence, and chemical class [98], [99]. Various modifications in the basic structure of AS ODNs have been tried to improve their properties and reduce toxicities while maintaining their target specificity and imparting resistance to nucleases. To elicit a biological response, an AS ODN must be absorbed from the site of administration and distributed to various tissues with maximum uptake by the target cells while being resistant to any chemical or enzymatic degradation.
Amendments that can be protective to nucleases while maintaining the desired characteristics of antisense effect can be introduced in DNA as well as RNA nucleotides (Fig. 7.5 ) by alteration in the base and modifications in the phosphate backbone [100]. Further, the 2′-OH group can also be tailored in RNA nucleotides. Synthetically modified AS ODNs can be grouped into three broad categories viz. first-, second-, and third-generation AS ODNs.
Figure 7.5.
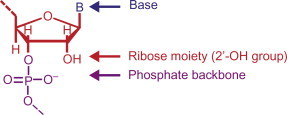
Possible sites for chemical modification of AS ODNs to improve their properties. Note that the 2′-OH site is only available in RNA.
7.6.1. First-Generation AS ODNs
These include phosphorothioate oligonucleotides (Fig. 7.6A ) synthesized by replacing one of the nonbridging oxygen atoms in the phosphate backbone with a sulfur atom [101] and methylphosphonates prepared by replacing a nonbridging oxygen atom with a methyl group at each phosphorus in the oligonucleotide chain.
Figure 7.6.
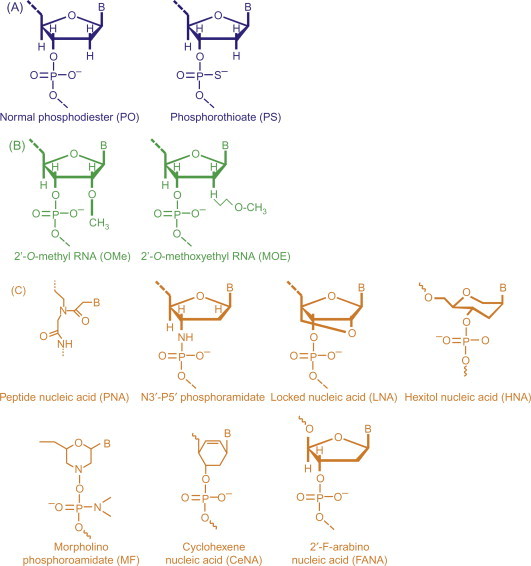
Representation of three generations of chemically modified AS ODNs for use in therapeutics. (A) First-generation AS ODNs. (B) Second-generation antisense ribonucleotides modified at the 2′ hydroxyl by adding a methyl (OMe) or a methoxyethyl (MOE) group. (C) Third-generation modifications involving a variety of sites including the entire backbone as in the peptide nucleic acid (PNA), a backbone substitution as in the N3′-P5′ phosphoroamidate (PA), the conformational lock in the locked nucleic acid (LNA), or the substituted ring in the hexitol nucleic acid (HNA) or morpholino phosphoroamidate (MF) or cyclohexene nucleic acid (CeNA) or 2′-F-arabino nucleic acid (FANA).
Methylphosphonates are neutral with excellent stability in biological milieu [101], but cannot activate RNase H activity [102]. Their cellular uptake occurs by adsorptive endocytosis [103] and not by membrane diffusion [104]. Phosphorothioate oligonucleotides are the most widely studied oligonucleotides and were the first to be synthesized chemically [105]. This modification was primarily tried to improve the stability of AS ODNs toward nucleases, but phosphorothioate oligonucleotides were found to be cytotoxic at high concentrations by binding nonspecifically to certain proteins [106]. These induce an antisense effect by an RNase H-dependent mechanism [81], [107]. Phosphorothioate oligonucleotides had a half-life of up to 10 h in human serum compared to only 1 h for an unmodified oligonucleotide of the same sequence [108], and are taken up by receptor-mediated endocytosis into the cells [109]. However, due to the negatives associated with first-generation AS ODNs, such as large size and chemical and enzymatic instability, second- and third-generation AS ODNs were developed.
7.6.2. Second-Generation AS ODNs
These include RNA oligonucleotides with alkyl modifications at the 2′ position of the ribose sugar such as 2′-O-methyl RNA (OMe-RNA) and 2′-O-methoxyethyl RNA (MOE-RNA) and were synthesized by replacing the 2′-OH group with a methyl or a methoxyethyl group, respectively (Fig. 7.6B). These oligonucleotides were designed to address issues like nonspecific protein binding and cytotoxicity associated with phosphorothioate AS ODNs, and they are more resistant to nucleases than phosphorothioates. However, the major drawbacks associated with these oligonucleotides are their poor elimination properties and RNase H-independent antisense mechanism of action [110]. These agents are only effective through the steric blockade mechanism.
7.6.3. Third-Generation AS ODNs
These include gapmer AS ODNs like PNAs, LNAs, N3′-P5′ phosphoroamidate (PA), HNAs, 2′-F-arabino nucleic acids, cyclohexene nucleic acid, caged nucleic acids, and others, as shown in Fig. 7.6C. A gapmer contains a central block of deoxynucleotides sufficient to induce RNase H cleavage flanked by blocks of 2′-O-methyl-modified ribonucleotides that protect the internal block from nuclease degradation [111]. These AS ODNs have increased thermal stability in hybridization and enhanced target recognition but do not support RNase H activity. These are also comparatively less toxic than first- or second-generation oligonucleotides as they show low interaction with plasma proteins.
One of the earliest and most studied third-generation constructs for antisense are PNAs . PNAs are AS ODNs in which the sugar phosphate backbone is replaced completely by polyamide linkages comprising repeating N-(2-aminoethyl)glycine units attached to nucleobases via methylene carbonyl linkers [111], [112]. These possess increased stability and favorable hybridization [113] due to absence of negative charges on the PNA oligomers, but do not support the RNase H antisense mechanism. These exert antisense effect through steric blockade and can bind to both RNA and transcription factors [114], [115]. N3′-P5′ PA morpholino-oligonucleotides are synthesized by substituting the deoxyribose moiety with a morpholino ring, and the charged phosphodiester linkage with a neutral PA linkage [116]. These are biologically stable [117] and possess efficient antisense activity that is RNase H independent. These are comparatively less toxic than first- or second-generation oligonucleotides and show low interaction with plasma proteins. LNA is a new and promising third-generation modification composed of nucleotides that are “locked” into a single conformation via a 2′-O, 4′-C methylene linkage in 1,2:5,6-di-O-isopropylene-α-d-allofuranose [117]. These possess remarkably increased thermodynamic stability and enhanced nucleic acid recognition.
7.6.4. Ribozymes
A decade after the appearance of AS ODNs, another enzymatic molecule called ribozyme was described in tetrahymena thermophilia as an antisense agent [2]. Ribozymes are RNA enzymes having the potential to process RNA and thus can act to knock down the gene expression. Hammerhead ribozyme has been explored extensively for its catalytic efficiency due to the catalytic core in its structure and for its sequence-specific binding capacity to RNA due to two flanking sequences [24], [25]. The structure of hammerhead ribozyme has two regions: a catalytic core for cleavage and two flanking sequences confirming binding and specificity. Shorter flanking sequences of 6–10 nucleotides present a more rapid turnover rate. Ribozymes can lead to the activation of RNase degradation through dsRNA recognition [118]. Many ribozyme formulations are under different phases of clinical trials (Table 7.3).
7.6.5. Aptamers
Aptamers can be considered chemical antibodies having the properties of nucleotide-based therapies. These can be used to knock down the expression of target extracellular as well as cytoplasmic proteins. Aptamers are short stretches of RNA or DNA with a specific three-dimensional structure that can form complexes with the target protein to inhibit its expression by blocking its activity [119]. It is not essential for the aptamers to be complementary to the target mRNA; instead its three-dimensional tertiary and quaternary structures determine the specificity and binding capabilities. Aptamers can be well utilized as nonimmunogenic alternatives of antibodies even at 1000 times higher doses. Aptamers are acquiescent to the amendments that apply to the other nucleotide-based antisense agents. Thus, aptamers can be tailored according to various required modifications while safeguarding their structure, specifically, the binding region [119], [120].
7.6.6. siRNA and miRNA
siRNA is a 20- to 25-nucleotide long dsRNA that triggers cellular RNAi for degradation of the complementary mRNA in the cytoplasm and required sequence-specific translational arrest. siRNA and oligonucleotides both are being extensively explored for therapeutic targeting and have their own pros and cons. Although similar in the sense of acting as antisense, they differ in many aspects from each other (Table 7.4 ).
Table 7.4.
AS ODNs Versus RNAi
| S. No. | AS ODNs | RNAi |
|---|---|---|
| 1. | Single-stranded 12–22 mer DNA oligonucleotides complementary to the target mRNA sequence silence the expression of the target gene. | 19- to 23-nucleotide long double-stranded siRNAs target gene silencing. |
| 2. | AS ODNs exert a gene-silencing effect mostly by steric inhibition of translation by the ribosomal complex or by activating RNase H to cleave the target RNA molecule. | The mechanism involves sequential cleavage of long dsRNA by the enzyme Dicer RNase III into siRNAs, which are then incorporated into a complex termed RISC to target the degradation of mRNA transcript. |
| 3. | These can either act on DNA to interfere in mRNA transcription or may interact with mRNA. | siRNA specifically interferes at the posttranscriptional phase to perform the gene-silencing action. |
| 4. | Target sequence identification and oligonucleotide design is difficult due to unknown secondary RNA structure. | Target sequence identification and oligo design is easier than for AS ODNs. |
| 5. | They require higher concentration to exert their action. | Gene silencing occurs at much lower concentration. |
| 6. | Gene silencing induced by oligonucleotides is short lived. | Stable incorporation of siRNA into RISC leads to prolonged gene silencing. |
| 7. | AS ODNs result in a less potent gene-silencing effect than siRNA. | siRNA results in significantly greater gene-silencing effect at such a lower concentration than AS ODNs that it becomes difficult even to detect them. |
| 8. | Being highly target specific, AS ODNs induce many fewer “off-target” effects. | Though highly target specific, siRNAs may induce significant “off-target” effects, depending on the length and siRNA design. |
| 9. | AS ODNs can cross the cell membrane comparatively faster as compared to siRNAs. | Because of their large molecular mass (twice that of single-stranded AS ODNs) and high negative charge, siRNAs do not readily cross the cell membrane. |
| 10. | Mostly AS ODNs need to enter the nucleus for effective gene silencing. | siRNA does not require nuclear access and exhibits its action by target mRNA degradation in the cytoplasm. |
doi:10.1093/ndt/gfn095 and doi:10.1016/j.drudis.2008.03.014
RNAi is more potent than antisense in general and makes selection of a candidate sequence much easier [17], [18], [19], [20]. Mammalian cells have single-stranded RNA, and the introduction of viral long dsRNA (>30 nt) can trigger RNAi and initiate a potent antiviral response by inhibition of protein synthesis through mRNA degradation. This natural defense mechanism has been utilized as an antisense technology to fight a tremendous number of diseases by the introduction or expression of siRNAs [20]. Presence of long dsRNA can trigger the RNAi pathway, which leads to the activation of Dicer enzyme to cut long dsRNA into short RNA fragments called siRNA [118]. These siRNA fragments degrade the complementary mRNA to prevent its expression into undesired protein. siRNAs can be produced chemically as well as enzymatically and introduced directly into the cell with or without minimal interferon response as compared to the long dsRNA. Since 1998, many studies have been executed to evaluate the therapeutic potential of siRNA, and pharmaceutical industries have invested billions on this technology through licensing delivery platforms and strategic alliances in the development of RNAi-based therapeutic products. Initially, the studies were limited to local administration of siRNA for the treatment of topical, specifically ocular, diseases, but presently vigorous efforts are in progress to cover the treatment of almost all the incurable diseases like cancer, hepatitis, AIDS, various other mutating viral diseases, and respiratory disorders, through various routes of administration, including topical, inhalation, and systemic [10], [121], [122], [123], [124], [125], [126], [127]. Increasing figure of products in clinical and preclinical stages is a sign of foreseen breakthrough in pharmaceutical and biotechnology market. Besides siRNA, a few other short RNAs like miRNA and Piwi-associated RNA (piRNA) have also been identified [128]. The folding of long single-stranded RNA sequences (encoded by specific genes and function in repressing mRNA translation or degradation) into intramolecular hairpins containing imperfectly base-paired segments led to the formation of miRNAs. piRNAs are also from the long single-stranded precursors and its function is associated with Piwi subfamily of Argonaute proteins. A large number of tiny noncoding RNAs have been discovered since 1990, and this continues. [126]. Of these, siRNA was first identified because of their potential to regulate gene expression. Recently, miRNAs have been shown to regulate critical biological processes from growth and development, to oncogenesis and host–pathogen interaction in higher eukaryotes. miRNA is a natural molecule, also consisting of dsRNA with short single-stranded ends. Primary miRNA is transcribed from DNA and folds into a hairpin. The Drosha enzyme cuts the hairpin from the rest of the transcript, forming pre-miRNA. The Dicer enzyme cuts away the loop, forming the mature double-stranded miRNA. The double strand loads onto a ribonucleoprotein complex (miRNP), which includes the Argonaute protein, and Argonaute cleaves one strand of the dsRNA, incorporating the uncleaved single strand into the mature complex. This complex inhibits translation of partially complementary mRNA [128], [129], [130].
The behavior of the two classes, siRNA and miRNA, is the same. miRNAs are encoded in the genome and are naturally used by cells to regulate gene expression. siRNAs, on the other hand, are the affector molecules of the RNAi pathway and are generated from the cleavage of dsRNA. Each can cleave perfectly complementary mRNA targets and decrease the expression of partially complementary targets. However, the major difference between endogenous siRNA and miRNA (Table 7.5 ) seems to be that the precursor of endogenous siRNA is a long dsRNA, whereas the precursor of a miRNA is hairpin-shaped RNA.
Table 7.5.
siRNA Versus miRNA
| S. No. | siRNA | miRNA |
|---|---|---|
| 1. | siRNA is synthesized from double-stranded segments of matched mRNA via RNA-dependent RNA polymerase. | miRNA is synthesized from an unmatched segment of RNA precursor featuring a hairpin foldback structure. |
| 2. | The precursor of endogenous siRNA is a long dsRNA. | The precursor of a miRNA is hairpin-shaped RNA. |
| 3. | Each dsRNA precursor gives rise to numerous different siRNAs. | Each hairpin is processed to ultimately accumulate a single miRNA molecule from one arm of each hairpin precursor molecule. |
| 4. | siRNAs may be endogenous or exogenously derived from viruses. | miRNAs are entirely endogenous. |
| 5. | siRNAs generally exhibit less sequence conservation. | Sequences of the mature miRNAs and their hairpin precursors are usually evolutionarily conserved. |
| 6. | siRNAs are synthesized from RNA-dependent RNA polymerase and processed from Dicer enzymes. | miRNAs are synthesized from RNA polymerase-II and processed from Drosha and Dicer enzymes. |
| 7. | The main function of siRNAs is cleavage of mRNA. | Mainly, miRNAs inhibit protein synthesis by blocking mRNA translation; however, cleavage of mRNA may also be there. |
| 8. | siRNAs are affecter molecules of RNAi pathway. | miRNAs are encoded in the genome to regulate natural gene expression. |
| 9. | siRNAs are involved in natural cellular defense mechanism. | miRNAs regulate critical biological processes from growth and development, to oncogenesis and host–pathogen interaction. |
| 10. | siRNAs often perfectly correspond to the sequences of known or predicted mRNAs, transposons, or regions of heterochromatic DNA. | miRNAs rarely correspond perfectly to the sequences of mRNAs targets and are derived from loci distinct from those of their mRNA targets. |
Endogenous silencing small RNAs are termed miRNAs when they are genetically encoded. They have the potential to arise from foldback structures characteristic of miRNA precursor hairpins. siRNAs are similar small RNAs that do not appear to correspond to protein-coding regions and do not have the potential to arise from hairpins characteristic of miRNA precursors, and yet they are expressed at sufficiently high endogenous levels to be detected on RNA blots; there is a theory that they might be processed from long dsRNA. As progress is made in understanding the role of miRNA in biological milieu, these therapeutic molecules are promising for targeting various diseases, including various neurodegenerative diseases that do not yet have any effective therapies and conventional druggable targets. However, traditional antisense and novel siRNA oligonucleotides or miRNA all typically need chemical modifications and formulation into clinically suitable, safe, and effective drug delivery vehicles for stability and tissue targeting. To achieve this, it is essential to understand the in vivo effect of these molecules.
7.7. Pharmacokinetics and Pharmacodynamics
The development of various sensitive analytical methods to selectively quantify oligonucleotides in biological systems has made it possible to study the metabolism of these compounds easily [131], [132], [133]. However, because the pharmacokinetics of oligonucleotides has been reported to be sequence independent, data from one sequence can be used to understand pharmacokinetics of the entire class [134], [135]. Under pharmacokinetics, we discuss the kinetics of antisense drugs, clearance of drugs from the site of action, and its ultimate pharmacological activity. Because phosphorothioate oligonucleotides are the most widely studied, we discuss the AS ODN kinetics with respect to the pharmacokinetics of phosphorothioate oligonucleotides [135], [136].
7.7.1. Pharmacokinetics
AS ODNs are designed specifically and selectively to inhibit translation of target mRNA and expression of related protein [137], [138]. To elucidate the safety of AS ODNs, their biological activity as a function of dose, rate of distribution, and mechanism of clearance from the body must be established. Because phosphorothioate oligonucleotides were the first synthetically prepared antisense nucleotides, their pharmacokinetics, and hence their efficacy and safety, has been widely studied and reported [134], [135], [136]. Their pharmacokinetics has been found to be independent of their physical and chemical properties. Following injection, AS ODNs bind to various proteins, distribute to various tissues, and finally get cleared from the body. The kidney, liver, and other organs of the RES are the major organs of distribution for AS ODNs and siRNA [135], [136], [137], and have benefitted from their ability to target to these sites. Certain siRNA formulations also accumulate in subcutaneous tumors through enhanced permeability and retention due to their leaky vasculature [135]. The reported data demonstrate that the pattern of absorption, distribution, and clearance of phosphorothioates in various species such as mouse, rat, dog, and monkey is similar and independent of oligonucleotide sequence and route of administration [135], [136], [137], [138], [139]. With intravenous administration, the concentration of oligonucleotides in plasma decreases rapidly, with distribution half lives of 30–80 min, whereas with intravenous infusion, oligonucleotide concentration [134], [136] increases linearly as the dose increases [140]. Intravenous administration of oligonucleotides also bypasses the absorption barriers to antisense drug delivery, usually encountered with other routes of administrations. However, rapid intravenous injection results in hemolytic effects due to high local concentrations of oligonucleotides. Hence, slow intravenous administration is advantageous over rapid injection. When considering tissue distribution of phosphothiorate (PS) oligonucleotides, highest concentrations are found in the liver and kidney, followed by the spleen and lymph nodes [140], [141], [142]. Tissue uptake of these oligonucleotides can be readily increased using long-term continuous infusion, which extends the exposure of PS oligonucleotides to the target tissues. However, their clearance from the organs of distribution is relatively slow, requiring a 3-times-a-week treatment regime, thereby ensuring an enhanced biological effect.
7.7.2. Elimination
This includes both metabolism and excretion of AS ODNs. Plasma and tissue exonucleases account majorly for the degradation of oligonucleotides in blood and organs of distribution, respectively [133], [143]. These exonucleases cleave oligonucleotides either at the 3′ or 5′ end, liberating smaller nucleotides, each shortened by a single nucleotide. The metabolism of many AS ODNs follows the same pathway as the endogenous nucleic acids. Immediately following intravenous administration, 35% of the AS ODN degrades within 10 min [140]. In tissue metabolism, the metabolites, being smaller, excrete more slowly than the parent oligonucleotide [144]. These low-molecular-weight metabolites are ultimately excreted from the body through urinary and fecal excretion. However, excretion via bile has also been suggested for AS ODNs.
7.7.3. Protein Binding
PS oligonucleotides have been reported to bind readily with plasma proteins such as albumin and α2-macroglobuline, inhibiting their rapid excretion from the body [145]. This protein binding is a nonspecific electrostatic interaction dependent on salt type and pH of the surrounding biological milieu. The PS oligonucleotides have been reported to bind to α2-macroglobuline with higher affinity than albumin. These oligonucleotides have also been reported to bind to other plasma proteins as well, but with lower affinity, and binding of the PS oligonucleotides to the thrombin-binding sites of these plasma proteins results in severe hematological risks [146]. However, this protein binding of PS oligonucleotides has been found to be reversible and to occur both in plasma and tissues. Tissue distribution of these oligonucleotides has been attributed to the protein binding within the organ and on the cell surface, constituting the organ resulting in higher tissue uptake of oligonucleotides. Thus, a decrease in protein binding of oligonucleotides results in a decreased tissue disposition and an increased urinary excretion. Certain drugs such as aspirin have been reported to displace PS oligonucleotides from plasma proteins, increasing their rate of excretion and decreasing the magnitude of their biological effect [147].
7.7.4. Effect of Route of Administration
AS ODNs can be administered locally or systemically to elicit a biological effect. Local delivery is advantageous over systemic administration as it avoids distribution into nontarget tissues, resulting in reduced side effects associated with unwanted tissue distribution. The antisense agents can be administered through oral, nasal, rectal vaginal, pulmonary, topical, intravenous, subcutaneous, intradermal, or intrathecal routes [135], [136], [142], [148], [149], [150]. Bioavailability of PS oligonucleotides has been found to be very poor following oral administration because of their large size, and hydrophilic and ionic nature [136], [149]. PS oligonucleotides are rapidly absorbed from the site of injection following intradermal or subcutaneous administration, but not following intrathecal or pulmonary administrations [142], [148], [149].
7.7.5. Pharmacodynamics
The cellular distribution of AS ODNs accounts for their pharmacodynamic effect. This requires suborgan distribution of the oligonucleotides to the target cells and subcellular distribution to the binding site in target mRNA. The onset and duration of antisense activity decides the dose of AS ODN and its frequency of administration. The dosing of AS ODNs is dependent on their structural chemistry, with first-generation AS ODNs being administered three times a week and second-generation AS ODNs being administered once a week. The therapeutic effect of an AS ODN in an organ or tissue weakens in parallel to its elimination from that organ or tissue [151], [152].
7.8. Formulation Considerations for Antisense Drug Delivery
Naked AS ODNs and siRNA, being large and ionic, cannot diffuse freely across the cell membrane, and hence, to facilitate their entry to intracellular targets, a suitable delivery system is required. The degree of biological effect of an AS ODN is highly dependent on the delivery vector used; these are generally categorized into viral and nonviral vectors [12]. Viral vectors usually have better transfection efficiency; however, nonviral vectors have been found to be superior to viral vectors in terms of toxicity, immunogenicity, and insertional mutagenesis [153]. Here we discuss these vectors in detail. Generally, a delivery vector includes a cationic group for efficient loading of oligonucleotide, a nonionic group for steric hindrance, an endosomolytic group for endosomal escape, and a targeting ligand for site-specific delivery [20]. The delivery system should be sufficiently large, in other words, greater than 5 nm, to avoid clearance by glomerular filtration. Simultaneously, it should be greater than 100 nm, to avoid leakage to interstitial spaces of hepatic sinusoid and entrapment by hepatic Kuffer cells [20]; but smaller than 200 nm, to avoid uptake by organs of the RES, such as the liver and spleen. Thus, the size requirement for systemic delivery of these delivery systems is about 100 nm.
7.8.1. Viral Vectors
Intracellular delivery of AS ODNs using a viral vector is called transduction. A viral vector usually consists of a viral genome with deletions in some or all essential genes, into which a transgene is inserted. Viral vectors provide high tissue specificity and result in highly efficient oligonucleotide expression. However, they pose severe safety risks owing to their oncogenic potential, and immunogenic effects, and are still being used widely for AS ODN delivery. In this section, we discuss some of the most commonly employed viral vectors, such as retrovirus, lentivirus, herpesvirus, adenovirus, and adeno-associated virus. However, some viral vectors like herpesvirus and poxvirus have also been used to carry AS ODNs in a few applications.
7.8.1.1. Retrovirus
Retroviruses are the most widely used RNA viruses for delivering AS ODNs and were the first vectors to be developed for intracellular gene delivery. These infect the host cells via the help of the enzyme transcriptase and require dividing cells to achieve high transduction. Hence, replication defective vectors are used for transducing the host cells. These vectors require integration into the host genome, resulting in a sustained expression of vector. However, this integration is highly nonspecific, and by integrating into the host genome at random positions these vectors possess high potential for mutagenic consequences. They possess high transduction efficiency and can carry oligonucleotides up to 8 kB without expression of viral proteins [154], [155].
7.8.1.2. Lentivirus
Lentiviruses are a subclass of retroviruses and hence are also RNA viruses. These have recently been developed as viral vectors and have the potential to infect both dividing and nondividing cells. This feature is unique to lentiviruses that use integrase enzyme to transduce host cells. These vectors also require integration into the host genome for the expression of the vector. Oligonucleotides of size up to 8 kB can be packed into a lentiviral vector. They possess high transduction efficiency and mutagenic potential [154], [155]. HIV-based lentiviral vectors have been successfully tried against AIDS [155].
7.8.1.3. Adenovirus
Adenovirus is a DNA virus commonly used as an AS ODN vector. Replication deficient adenoviruses with a deleted E1A region are used as viral vectors. The E1A region is essential for the replication of these viruses. Such vectors can infect a cell only once and can infect both dividing and nondividing cells. They possess high transduction efficiency and can carry oligonucleotides of size up to 8 kB [154], [155]. As with retrovirus and lentivirus, adenoviruses do not integrate into the host genome and hence are not replicated during cell division. Thus, these vectors possess low mutagenic potential but may pose severe immunological risks due to the expression of viral proteins in the host cells following vector administration.
7.8.1.4. Adeno-Associated Viruses
Adeno-associated virus is a small virus that infects humans and other species and requires coinfection with either adenovirus or herpesvirus for replication. These can infect both dividing and resting cells, with site-specific integration into the host genome, and so they possess low mutagenic potential. These vectors cannot incorporate oligonucleotides larger than 5 kB but are capable of infecting multiple types of cells [154], [156].
7.8.2. Nonviral Delivery Techniques
Transfection is the term used to describe intracellular delivery of antisense agents using nonviral vectors. The oncogenic consequences and immunological risks associated with the viral vectors have led to the development of novel nonviral vectors for antisense drug delivery. Although nonviral vectors are safer than viral vectors, they impart low and transient transfection efficiency. Along with the chemical modifications, the conjugation and/or incorporation of targeting ligands like peptides, monoclonal antibodies, and so on, are essential to achieve the desired therapeutic effect. Cationic lipids/polymers and cell-penetrating peptides are commonly used to design these delivery vectors (Table 7.6 ).
Table 7.6.
Nonviral Delivery Systems for Antisense Drug Delivery
| S. No. | Delivery System | Composition | Route of Administration | Characteristics | References |
|---|---|---|---|---|---|
| 1. | Cationic lipid-based vectors: liposomes/lipoplexes | Lipids such as lipofectin, RNAifect, oligofectamine, lipofectamine, cardiolipin, and transIT TKO are commonly used transfection reagents composed of cationic lipids and colipids like ceramide carbomoyl spermine (CCS) and dioleoyl phosphatidylethanolamine (DOPE), dioleoyl phosphatidylcholine (DOPC), N-(1-(2,3-dioleoyloxy)propyl)-N,N,N-trimethylammonium chloride (DOTAP), 3ss[N-(N′,N′ dimethylaminoethane)carbamoyl]-cholesterol (DC-CHOL), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), N-[1-(2,3-dioleyloyx) propyl]-N-N-N-trimethyl ammonium chloride (DOTMA), dioctadecyldimethylammonium bromide (DODAB)cholesterol, etc. | Intravenous, intracerebroventricular, intravaginal | Have improved pharmacokinetic properties with reduced systemic toxicity, but may precipitate acute immune responses Protect oligonucleotides from degradation and provide controlled drug delivery |
[12], [155], [156], [157], [158], [159], [160], [161] |
| 2. | Polymeric micelles | Polymers like poly(aspartate hydrazone adriamycin), poly(ethylene oxide)-block-poly(aspartic acid), pluronics, poly(N-isopropylacrylamide), poly(ethylene glycol)–block-poly(ethylenimine), N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers, etc. | Intravenous, intraperitoneal | Protect oligonucleotides from degradation and provide controlled drug delivery | [160], [162], [163], [164], [165] |
| 3. | Polymeric nanoparticles | Polymers like gelatin, chitosan, albumin, sodium alginate, poly(lactide-co-glycolide), polyanhydrides, hyaluronic acid, cyclodextrin, gold nanoparticles, silica nanoparticles, etc. | Intravenous | Have improved pharmacokinetic properties with reduced systemic toxicity but may cause acute immune responses, protect oligonucleotides from degradation, and provide controlled drug delivery | [12], [155], [160], [166], [167], [168], [169], [170] |
| 4. | Lipid-polymer hybrid systems | Consists of hybrid systems like liposome-entrapped polylysine-condensed DNA, lipid-coated precondensed polylysine-DNA, poly(propylacrylic acid)-coated cationic lipid–DNA conjugate, etc. | Intravenous | Provide better protection against nucleases and more efficient transfection than uncoated lipoplexes or polyplexes | [160], [171], [172], [173], [174] |
| 5. | Peptide-based delivery systems | Peptides like tetra-amine spermine, PLL, protamine, histone, oligoarginine, streptavidin, aptamers, receptor-specific monoclonal antibodies | Intrathecal, intravenous | Provide site-specific delivery of AS ODNs | [12], [155], [160], [175], [176], [177] |
| 6. | Hydrogels | Polymers like polyacrylic acid, pluronic, PEI, PEG, hyaluronic acid, polyvinyl alcohol, chitosan, polyhydroxyethyl methacrylate, polyvinyl pyrrolidone silk-elastin, etc. | Intravenous | Provide controlled drug delivery in response to pH, temperature, ionic strength, electric field, or specific analyte concentration differences | [160], [166], [178], [179], [180], [181] |
| 7. | Hydrodynamic injection | High pressure across the cell membrane, resulting in the permeation of AS ODNs | Intravenous, intravascular | Injects AS ODNs directly into the diseased cells. Minimized side effects associated with unwanted tissue distribution, but is an invasive method | [12], [155], [160], [182], [183] |
| 8. | Electroporation | Uses externally applied electrical field to facilitate penetration of AS ODNs across the cell membrane | Transdermal, intratissue | Provides site-specific delivery of AS ODNs, but causes high cell mortality | [155], [160], [184], [185] |
| 9. | Ultrasound-mediated antisense drug delivery | Makes use of ultrasound waves of optimum strength and for an optimal time to facilitate permeation of AS ODNs into the target cells | Intratumoral, intravenous, transdermal | Provides noninvasive site-specific delivery of AS ODNs, but results in low transfection | [160], [186], [187] |
| 10. | Light-mediated antisense drug delivery | Makes use of photolabile compounds that block the bioactivity of AS ODNs until exposed to near-ultraviolet light | Intravenous | Protects oligonucleotides from degradation and provides noninvasive site-specific delivery | [188], [189], [190], [191] |
The positive charge of these delivery systems facilitates complexation with negatively charged oligonucleotides or siRNA and ionic interaction with cell membranes.
7.8.2.1. Cationic Lipid-Based Vectors
These include lipidic delivery systems to enhance transfection efficiency like liposomes and lipoplexes with or without any surface modifications. Liposomes are spherical vesicles composed of a central aqueous compartment enclosed within a phospholipid bilayer. Cationic lipids in combination with neutral lipids are used to formulate liposomes to deliver AS ODNs and siRNA intracellularly [155]. Neutral lipids are used to facilitate fusion with cell membranes. The cationic lipids widely used in formulating cationic liposomes include DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) and DOTMA (1,2-dioleoyl-3-trimethylammonium-propane) [12], [157], [159]. However, lipoplexes, along with cationic lipids, contain anionic amphiphilic molecules, facilitating the release of negatively charged oligonucleotides from the lipoplex, thus assisting in nuclear localization of oligonucleotides [12], [160], [161], [162]. Overall neutral charge of the formulation reduces the biological toxic effects of cationic lipids Lipofectin RNAifect, Oligofectamine, Lipofectamine, TransIT TKO, synthetically cationized cholesterol, and natural analogues of cardiolipin are the most widely explored lipids for improving the delivery, safety, and efficacy of antisense drugs [12], [160]. Thiocholesterol-based cationic lipids have been used as components in self-assembled cationic micelles and nanolipoparticles, which can efficiently entrap anionic oligonucleotides and deliver them intracellularly [12], [157], [163]. However, because this complex is unstable, it should be prepared immediately before use.
7.8.2.2. Cationic Polymer-Based Vectors
Complexes of polymers with DNA are called polyplexes. Amphiphilic block copolymers spontaneously self-assemble to form core shell–type micelles in aqueous media called polymeric micelles. Their solid core shell, small size, and modifiable surface make them a suitable carrier for AS ODNs [162]. These cationic hydrophilic copolymers efficiently entrap anionic oligonucleotides. Some examples of such copolymers are PEG-block-polylysine copolymer, PEG-block-polyethylenimine copolymer, and poly(lactic-co-glycolicacid)/PLGA-block-polyarginine copolymer [162], [164], [165], [166], [167]. Polymeric nanoaparticulate systems have also been reported for intracellular delivery of AS ODNs. These proteinaceous biopolymer-based nanoparticles are biocompatible, and their surfaces can be modified for improved transfection efficiencies. Examples of such biopolymers include atelocollagen, gelatine, sodium alginate, and hyaluronic acid. Many authors have also reported chitosan-based nanoparticles for antisense drug delivery [12], [157], [162], [168]. Cyclodextrin-based nanoparticles have also been developed by Mark Davis for efficient intracellular siRNA delivery to tumors, and this has recently been approved by the FDA [169]. This is a three-component system comprising a cyclodextrin-containing polymer (CDP), PEG as a steric stabilization agent, and human transferrin (Tf) as a targeting ligand. This system has been reported to improve the diseased state in tumor-bearing mice. Nanoparticles of inorganic material, like silica and gold with or without surface modifications, to deliver antisense drugs intracellularly have also been reported by several authors [162], [170], [171], [172].
7.8.2.3. Lipid–Polymer Hybrid Systems
Lipid–polymer hybrid systems have also been reported to deliver ODNs intracellularly. These include ODNs precondensed with polycations and then coated with either cationic or anionic lipids, or amphiphilic polymers with or without helper lipids. Polypeptides, such as PLL, protamine, histone, and so on, have been used to precondense ODNs to form polyplexes that are then coated with a lipid layer to form a lipid–polymer hybrid system [162], [173], [174], [175], [176]. Such systems are also addressed as LPD (lipid-polycation-DNA) systems. AS ODNs are better protected in these lipid-coated polyplexes. These systems appear to be more efficient in transfection than lipoplexes ex vivo and are equally active in vivo [162], [173], [174].
7.8.2.4. Peptide-Based Delivery Systems
One approach to deliver antisense agents intracellularly is to use synthetic or natural peptides. This system makes use of small proteins such as enzymes, receptors, or antibodies to complex with antisense drugs and may provide site-specific delivery of these agents [12], [157], [162], [177]. An example of such a peptide is tetra-amine spermine [178], [179]. Schiffelers et al. have reported direct siRNA uptake to tumor neovasculature by coupling of integrin-binding RGD peptide to PEGylated PEI [12]. MPG (N-methylpurine DNA glycosylase), a short amphipathic peptide, has been utilized to form stable nanoparticles for efficient cellular delivery of siRNA [157].
7.8.2.5. Hydrogels
Hydrogels are hydrophilic polymeric networks capable of imbibing large amounts of water or biological fluids. When placed in aqueous media, hydrogels swell to form insoluble three-dimensional networks via chemical crosslinks (tie-points, junctions) or physical crosslinks, such as entanglements or crystallites. These networks are composed of either homopolymers or copolymers and are used to control drug release in reservoir-based or controlled release systems or as carriers in swelling-controlled release devices. Hydrogels can be used to modulate antisense drug release in response to pH, temperature, ionic strength, electric field, or specific analyte concentration differences, thus making them enviro-intelligent and stimuli-sensitive delivery systems [162]. Antisense activity of oligonucleotides immobilized in a cationic crosslinked poly(ethylene oxide) (PEO) and PEI nanogel has been evaluated in a cell model [180]. A poly[1-vinyl-2-pyrrolidinone-co-(2-hydroxyethyl methacrylate)] hydrogel has been reported as a potential carrier for AS ODNs [181]. Also, cationic agarose-based hydrogel has been developed to deliver AS ODNs targeting the mRNA of TNF-α for the prevention of arthritis in animal models [182]. In hydrogels, release can be designed to occur within specific areas of the body (e.g., within a certain pH of the digestive tract or within cancerous cells) and also to specific sites (using adhesive or cell-receptor specific gels with modified hydrogel surface). Hydrogels can be prepared from polymers like polyacrylic acid, pluronic, PEI, PEG, agarose, etc. [162], [168], [180], [181], [182], [183].
7.8.2.6. Hydrodynamic Injection
Injecting AS ODNs in a physiological buffer locally to the diseased site effectively concentrates them at the site of injection. The method is simple, reproducible, and highly efficient, with transfection efficiency dependent on the anatomy of the target organ, the injection volume, and the injection rate [162], [184]. Being nonspecific, hydrodynamic delivery can be used for intracellular delivery of any water-soluble compounds, small colloidal particles, or even viruses. This technique allows direct transfer of substances into cytoplasm without endocytosis. Hydrodynamic renal vein injection of an AS ODN against connective tissue growth factor has been reported to treat renal fibrosis in rats [185]. However, due to the large injection volumes and invasiveness of the technique, it is not widely used clinically. Recently, an improvement in the precision and reproducibility of this technique has been reported, using a computer-controlled catheter-guided injection device. Successful gene delivery to the liver, kidney, and muscles of rodents has been observed with this device [12], [156].
7.8.2.7. Electroporation
Electroporation is a technique for local delivery of AS ODNs, making use of an externally applied electrical field to increase the electrical conductivity and permeability of the cellular plasma membrane by creating localized pores in the membrane. Treatment of tissue with hyaluronidase prior to injection of AS ODNs may significantly enhance transfection due to improved distribution of ODNs within the tissue [157], [162]. The technique has been reported to successfully deliver phosphorothioate-modified ODNs against c-myc proto-oncogene of U937 cells [186] and fluorescein-labeled AS ODNs to the promoters of the proto-oncogene c-myb (24-mer) transdermally [187]. However, the technique has limited use because of high cell mortality and suffers from drawbacks such as difficulty in transfecting cells in a large area of tissues due to a limited effective range of approximately 1 cm between the electrodes. Furthermore, surgery is required to place the electrodes into deep-seated organs. Process parameters like voltage, length, and number of electric pulses can be optimized to get maximum transfection with minimum cell mortality.
7.8.2.8. Ultrasound-Mediated Antisense Drug Delivery
Ultrasound-mediated antisense drug delivery is a noninvasive physical method of transfection that makes use of ultrasound waves of optimum strength for optimal time to facilitate permeation of AS ODNs into the target cells. The ultrasound creates pores in the cellular membrane, facilitating passive diffusion of ODNs across the membrane. The transfection efficiency is dependent on the size and local concentration of ODNs, and better transfection is obtained when complexes of ONDs with cationic lipids are used [162], [188]. The technique can be readily used for site-specific delivery of AS ODNs to soft internal tissues. The method has been reported to deliver digoxigenin-labelled AS ODN to treat prostate cancer in nude mice [189]. The only drawback associated with ultrasound-mediated antisense drug delivery is low transfection efficiency.
7.8.2.9. Light-Mediated Antisense Drug Delivery
Caged oligonucleotides are one of the most recently reported chemically modified AS ODNs for targeted oligonucleotide delivery. Monroe et al. and Coll et al. described the photocaging spatiotemporal strategy, which makes use of a photolytic chromophore for rapid release of a biologically active substrate on exposure to light [190], [191]. This caging involves covalent attachment of photolabile compounds to effector nucleic acid species that block the bioactivity until triggered by near-ultraviolet light. Photocaging also protects the effector nucleic acid from nuclease and blocks its native bioactivity until exposed to near-ultraviolet light. This phenomenon was first used by Kaplan et al. to release an inducer in the biological environment [192]. The various types of cage compounds commonly used with an effector nucleic acid are summarized in Fig. 7.7 .
Figure 7.7.
Common cage compounds (A) NB (nitrobenzyl), (B) DMNB (dimethoxy-nitrobenzyl), (C) NPP (nitrophenylpropyl), (D) NPE (nitrophenylethyl), (E) DMNPE (dimethoxy-nitrophenylethyl), (F) NPOM (6-nitropiperonyloxymethyl), (G) pHP (p-hydroxyphenacyl), (H) benzoin, and (I) coumarinyl.
Recently, Deiters's Group has made NPOM (nitropiperonyloxymethyl)-caged dT phosphoramidite for light-mediated delivery of phosphoramidite oligonucleotides commercially available [193].
7.8.2.10. Other Delivery Techniques
In addition to the delivery technologies discussed earlier, some other practices have also been reported, including the use of nonprotein alternatives to antibiotics, that is, selected neucleotide binding aptamers for the treatment of prostate cancer. In addition to that, repeated branched dendritic structures, specifically polycationic dendrimers, can bind strongly with oligonucleotides/siRNA for better nuclease resistance and efficient gene silencing. Also, human mesenchymal stem cells are inevitable as a viable nonimmunogenic cellular delivery system [194]. Moreover, many pharma companies are licensing their FDA-approved delivery platforms to the specialized players of antisense technology in the market: ALZET pumps [195], Minicircle DNA technology, DiLA2 platform, PRINT nanoparticle technologies, to name a few (Table 7.1).
7.8.3. Route of Administration
The first step in developing vectors for antisense drug delivery is to choose an effective route of administration, depending on the intended use of the antisense agent, that is, whether local or systemic. The potential of AS ODNs and siRNA has been investigated for applications through different routes, for example, the skin, lungs, and mucosal membranes like intravaginal, ocular, and nasal, and various systemic delivery routes [10], [126], [141]. Furthermore, there are some recent papers reporting the study of siRNA delivery through oral route for targeting macrophages [121].
7.8.3.1. Local Applications
Local delivery of antisense drugs, such as by electroporation or hydrodynamic intravenous injection, can reduce general problems associated with systemic administration, for example, clearance from the body, toxicity due to unwanted tissue distribution, and reduced transfection due to low cellular uptake; but they suffer from limitations like cytotoxicity at the application site and the requirement of large injection volume [157], [162]. PEI-associated siRNA has been delivered via electroporation in rodents for the treatment of collagen-induced arthritis [196]. Further, intrathecal injection of antisense agents have been explored for CNS delivery for a number of applications like knockdown of serotonin transporters in mice brain, treatment of various diseases like chronic neuropathic pain, and formalin-induced nociception [197], [198]. Some ocular diseases can be treated by intravitreal injection of antisense agents to treat eye diseases like AMD, inhibition of ocular neurovascularization, and angiogenesis [199], [200], [201], [202]. An AS ODN formulation, Vitravene, is available in the market for the treatment of retinitis. siRNA is also being evaluated as an inhalation therapy for lung disorders and systemic applications [203]. Moreover, intranasal administration of siRNA has been investigated in rodents for targeting heme oxygenase-1 to enhance ischemia-reperfusion-induced lung apoptosis and for the treatment of respiratory virus infection [8]. However, many diseases require systemic administration, and hence a special consideration should be given to the rational design of an antisense agent and development of its delivery vector.
7.8.3.2. Systemic Applications
As discussed earlier, systemic administration of siRNA and AS ODNs is a big challenge, because they have to cross many obstacles on the way to the target. Many efforts have been made in previous and ongoing research to judicially modify antisense agents and to develop an effective formulation for the desired application. Therapeutic silencing of various endogenous genes by systemic administration by the delivery of modified siRNAs was studied. To study antitumor activity, protamine-antibody–mediated siRNA was injected intravenously and intratumorally in rodents [204]. siRNA-targeting Fas protein protects mice against renal ischemia-reperfusion injury and fulminant hepatitis [205], [206]. Caspase 8 siRNA was targeted to liver via systemic administration to prevent acute liver failure in mice [207].
7.9. Applications of Antisense Drugs
Antisense technology can potentially be applied to treat various diseases, especially viral infections and cancers. However, with rapid progress in antisense technology, the technique has found newer applications to treat several other infections, which we discuss here in detail.
7.9.1. Genetic Research
AS ODNs are now being widely used in basic research to elucidate the general splicing patterns of various genes, which in turn may help in treating genetic disorders [99], [208]. Oligonucleotides splice out specific introns from a pre-mRNA by interfering with the assembly of a spliceosome. A spliceosome is complex comprising various proteins and small nuclear RNA (snRNA) and is formed for each splicing event. Thus, AS ODNs can be used to elucidate the various pathways involved in the synthesis of a protein and to determine the various possible mutagenic splicing sites alterations that result in inappropriate protein production. In genetic disorders, point mutations result in new splice sites within the introns, directing the splicing machinery to rupture the splicing pathways, resulting in the formation of a defective mRNA and an inappropriate protein product [209]. Blocking of these splice sites using AS ODNs thus prevents inappropriate protein expression. However, oligonucleotides must not activate RNase H enzyme when used to alter these splicing patterns. This is usually achieved by inducing certain chemical modifications in the AS ODNs such that the oligonucleotide RNA duplex is not recognized by the RNase H [210]. Examples of such enzymes include methylphosphonate derivatives, O-alkyl oligonucleotides, and PNAs [211]. Using antisense technology, O-alkyl oligonucleotides or morpholino-oligonucleotides have been reported to treat genetic blood disorder β-thelassemia completely [212], [213].
7.9.2. CNS Protein Function
The application of antisense technology to investigate protein function in the living brain has recently been reported. This technology has been successfully used to study central nervous system (CNS) proteins such as transmembrane receptors, ion channels, transporters, G proteins, and growth factors [214]. Many of these belong to large families with closely related subtypes and isoforms are generally nondistinguishable using common assay techniques. By using AS ODNs that can bind to one specific subtype, however, it is possible to distinguish them, as a sufficient degree of diversity exists in the mRNA sequences of these closely related proteins. Phosphodiester oligonucleotides exhibit considerably more stability in cerebrospinal fluid (CSF) than in plasma but require frequent administrations in large quantity to obtain significant antisense inhibitory effects [215]. Phosphorothioate oligonucleotides, though having good stability in CSF and producing the desired antisense effect, are quite toxic [216], [217]. AS ODNs exert their inhibitory effect through RNase H mechanisms in the brain [218]. PNAs and LNAs have also been tried to inhibit inappropriate protein expression [219], [220]. As AS ODNs cannot readily cross the blood–brain barrier, invasive methods such as intraventricular, intraparenchymal, and intrathecal administrations have also been tried to deliver AS ODNs to the brain, but these result in tissue injury and hemorrhage [142], [221]. Antisense technology has been reported to treat brain disorders such as Alzheimer's disease [222], [223], pain [224], [225], and affective disorders [226] successfully.
7.9.3. Inhibition of Specific Enzymes or Receptors
Being selective and specific, AS ODNs can be readily used to inhibit expression of a particular enzyme or a receptor belonging to a large family with closely related subtypes. Inhibition of acetyl cholinesterase enzyme is the molecular target for the treatment of diseases like Alzheimer's disease [227] and myasthenia gravis [228]. Advances in research concerning Alzheimer's disease and myasthenia gravis have demonstrated alternative splicing variants of acetyl cholinesterase enzyme being involved in the etiology of these diseases. Thus, only AS ODN therapy could be useful in the management of these diseases. AS ODNs have also been used successfully to characterize the various members of D2 dopamine receptors [229]. This technology has also been employed to differentiate between different opioid receptors viz. mu, delta, and kappa [230]. Similarly, AS ODNs have been successfully utilized to study the various phosphatases involved in cellular growth–control pathways and thus help in the screening of potential anticancerous agents [231], [232], [233]. These enzymes regulate diverse cellular processes such as metabolism, ion-channel activity, and membrane transport as well as learning and memory. Thus, inhibition of many protein kinases using AS ODNs have been used to treat related disorders in humans.
7.9.4. Inflammatory Diseases
Inflammation is the body's protective response to physical or microbial threat, characterized by redness, swelling, heat, and pain; it results in increased blood flow, increased capillary permeability, plasma protein leakage, and migration of leukocytes to the site of injury. When inflammation is deregulated, disease or even death can happen. In acute inflammatory processes cells like eosinophils, neutrophils, monocytes, and macrophages migrate to the site of injury in response to chemotactic factors such as platelet-activating factor, leukotrienes, and cytokines. In inflammation, antisense technology can be used to demonstrate the relative importance of various signaling components at the molecular level in a controlled manner. Hence, this technology is useful both in defining the role of a particular mediator in the process of inflammation and in the screening of potential anti-inflammatory drugs [138], [234], [235]. The technology has been reported to downregulate successfully the molecular targets involved in various inflammatory diseases such as Crohn's disease [236], inflammatory bowel disease [237], renal allograft rejection [238], and psoriasis [239].
7.9.5. Respiratory Diseases
For the treatment of various diseases of the respiratory tract, novel respirable antisense drugs called RASONS are used [240], [241], [242]. These RASONS can deliver antisense medicines effectively and safely, even to deep lungs, by interacting with the unique pulmonary surfactant of alveolar epithelial cells, resulting in enhanced cellular uptake of the oligonucleotides. These are effective even at very low doses and can be delivered using any of the delivery devices, whether nebulizer or dry powder inhaler or metered dose inhalers. RASONS have been successfully used to treat respiratory diseases such as asthma [240], [242], influenza [243], bronchitis [242], pulmonary fibrosis [242], pneumonia [242], and lung cancer [244].
7.9.6. Cancer Chemotherapy
Numerous antisense drugs are currently being investigated to treat various cancers in humans [245]. Here AS ODNs have the additional advantage of being less toxic than conventional anticancerous medicines. A combination therapy of antisense drugs with conventional medications may result in a reduction of dose and dose-related side effects for both therapies. Phosphorothioates were the first AS ODNs to be used in combination with cisplatin for the treatment of bladder cancer [246], [247]. Since then, several antisense drugs have been investigated for various cancers like colon cancer [248], [249], lymphatic leukemia [250], lung cancer [244], [251], testicular cancer [252], and lymphoma [250].
7.9.7. Renal and Cardiovascular Diseases
AS ODNs can be used to target specific components in the blood vessel wall that influence the pathophysiological mechanisms in renal and cardiovascular disorders. Several reports document the role of the tissue kallikrein-kinin system in the pathogenesis of hypertension. Reduced urinary kallikrein has been observed in hypertensive subjects. Hence, systemic delivery of the human tissue kallikrein gene results in a decrease in blood pressure and may assuage even glomerular sclerosis [253], [254]. However, antisense technology has been successfully used to elucidate the role of tissue kallikrein-kinin system in blood pressure regulation. Intracerebroventricular injection of AS ODNs against kininogen mRNA or bradykinin B2-receptor mRNA results in an increase in blood pressure. However, injection of antisense against the B1-receptor causes a decrease in blood pressure. Thus, antisense technology is useful in elucidating the role of bradykinin B1 and B2 receptors in the central regulation of blood pressure [253].
7.9.8. Viral Infections
It has always been difficult to design and develop antiviral drugs with low toxicity and improved specificity. A rational strategy to fight against viral diseases is to inhibit the genes implicated in various viral infections with the help of AS ODNs. Antisense technology can be easily applied to viruses as they encode unique proteins, very different from the ones encoded by normal cells. Vitravene® (fomivirsen) was the first antiviral AS ODN used intravitreally to treat cytomegalovirus retinitis [10]. The various viruses being investigated as potential targets of antisense drugs include human papillomavirus [255], human immunodeficiency virus-2 (HIV-2) [256], hepatitis-B virus [257], influenza A virus [258], and herpes simplex virus (HSV) [259]. Traditional antiviral drugs are competitive antagonists binding to the disease-causing proteins and subsequently blocking the actions of natural agonists, resulting in toxicological manifestations. Viral proteins essential for the replication of viruses are the prime targets of these antisense drugs. AS ODNs, being selective and specific, readily bind to the target mRNA sequence, thereby downregulating the expression of related proteins and causing the death of the virus.
7.10. Benefits of Antisense Drugs in Therapeutics
Antisense molecules that mediate RNAi can be synthesized chemically in the laboratory and then introduced into cells to achieve targeted gene silencing. This opens up enormous possibilities for using these as potential drug candidates. Following are the key features highlighting the advantages of antisense technology, specifically siRNA:
-
•
siRNA is a potent and highly specific therapeutic moiety [5].
-
•
The traditional drugs have limited targets, whereas due to the completion of the human genome project, antisense agents like siRNA can be designed for unlimited disease targets [5], [10], [18], [130].
-
•
Most of the drugs act for the symptomatic relief of the disease by inhibiting the disease-causing factor. However, siRNA therapy does not allow the formation of disease-causing elements and hence acts to remove the root cause of the disease [5], [10], [18], [130].
-
•
siRNA can be explored as a prophylactic agent for various known epidemic diseases.
-
•
Antisense technology can be utilized for the prevention and treatment of deadly diseases like SAARS and swine flu, by sequence-specific design complementary to the disease-causing genes of these viruses.
-
•
Some diseases are caused by mutation in a single allele of the gene, and siRNA can be designed to act on that particular allele without affecting the normal allele [5].
-
•
The difficulty in treating some viral diseases like HIV is the changing viral mutation. Thus, a drug effective at a particular time will not be effective for the next viral mutation. siRNA can be designed according to the particular gene mutation; moreover, a pool of more than one mutant-specific siRNA can be incorporated in a single delivery agent for delivering at one time [18].
-
•
Once a carrier is approved for siRNA delivery, different mutant-specific siRNAs can be formulated without changing the carrier system [5].
-
•
Cancer-like diseases are highly patient specific. Thus, siRNA can be explored as personalized therapy to benefit an individual patient [5].
-
•
Antisense drugs have the potential to act as a chemosensitizing agent and can prevent multidrug resistance by blocking the resistance-causing component [9].
7.11. Regulatory Aspects and Guidelines for Targeted siRNA Delivery
After three decades of antisense research, to date only two antisense agents have been approved by regulatory bodies, that too for local application only. However, a large number of diseases require systemic administration for the desired application. Intense research is in progress to make these agents available in the market for systemic use. Leaders of antisense technology can learn from the reasons for unsuccessful entry into clinics and the drawbacks of products like AEG35156, Bevasiranib, and AGN-749. Despite these ups and downs, more than 13 siRNA products, 47 AS ODNs, 3 ribozymes, and 9 aptamers have gained entry into clinical studies, and some others are still at the preclinical stage. Although FDA has not yet issued any guidelines for the targeted delivery of siRNA, several pharmaceutical and biotechnology-based companies involved in developing siRNA delivery techniques have laid down some internal specifications for maximizing the reliable guidelines for validated design, process, and evaluation of siRNA formulations [127], [260]. Presented here are some rules and conventions based on the practical understanding of siRNA design and delivery performance. They should be followed while developing antisense formulations in order to achieve regulatory approval and gain entry to the antisense therapeutic market.
1. Determine a Rational Design of siRNA to Get the Right Strand into RISC
At present, many computer software programs with optimum selection rules are available for specific siRNA designs. siRNA should be designed with excellent specificity to target mRNA with required chemical stability and pharmacologic effectiveness. Rational siRNA design schemes are being developed that are based on an understanding of RNAi biochemistry and on naturally occurring miRNA function. The difference in thermodynamic stability of two strands of siRNA can determine which strand will be directed towards RISC. This is taken into consideration while designing siRNA.
2. A Pool of Alleles Is Better Than a Single One
Several siRNAs or shRNAs should give the same phenotypic outcome, but similar off-target effects can be achieved with a pool of different triggers. It is critical to correlate this phenotypic outcome with the effectiveness of suppression. Examining target protein levels will help determine the effective siRNAs out of the pool of several siRNAs or shRNAs. Moreover, siRNAs or shRNAs that do not affect the target protein act as negative controls. Arguably, one could use a “scrambled” siRNA or shRNA for this purpose. It is better to use such scrambled siRNAs, which are known to enter the RNAi pathway effectively without any biological activity. Some examples of such target-validated RNAs are RNA-targeting luciferase, green fluorescent protein, and other marker genes.
3. Work at the Lowest Possible Concentrations
siRNA effectiveness depends on its specificity toward the RISC enzyme complex. This specificity is affected by the high enzyme-to-substrate ratio. Thus, siRNA concentration should be titrated to have a correlation between degree of suppression and phenotype outcome. In fact, 50% of the suppression of the expression has been observed in some siRNAs, for example, in the Hela cell at a very low concentration of 500 pM.
4. Physical and Chemical Properties
Besides the design of siRNA and its cocktails, siRNA formulations modified as conjugates, complexes, or with delivery carriers must meet some criteria: ensuring reproducibility of reassembly of functional complexes, incorporation efficiency, zeta potential, polydispersity, and no aggregation in 50% mouse/human serum, with chemical stability data of the assembly for >30 days, can provide confidence in the formulation.
5. Activity in Cell-Based Assays
Final effectiveness of formulation can be predicted on the basis of specifications designed for a particular formulation. To augment the triumph of final formulation, many companies like Sirna Therapeutics have revealed some specifications. Explicitly these state that there should be greater than 50% reduction in target mRNA levels by target siRNA at concentrations <10 nM in media containing 10% serum. But control siRNA at a concentration of <10 nM in media containing 10% serum should reduce target mRNA levels less than 10%. There should be more than a fivefold window between IC50 of target gene silencing and IC50 for reduction in viability. Activity of the delivery system should be reported in at least three cell lines relevant to the delivery system under evaluation. Targeting moiety in a delivery platform should be validated for targeting by (1) using cell lines with differential expression of the targeted receptor and (2) using assemblies with “active” and “inactive or mutant” targeting moieties.
6. In Vivo Performance in Mouse Models
In vivo performance of the siRNA-containing delivery assemblies should be evaluated in suitable animal models. A comparison of a single intravenous dose at 1, 3, and 9 mg/kg for target siRNA-containing delivery assemblies with control should be provided. Delivery platform with “targeting moeity” as well as assemblies incorporating “inactive/mutant” targeting moieties should be evaluated to provide an evidence of effective targeting. There should be more than 50% reduction in target mRNA levels in target tissue at 1 mg/kg dose by target siRNA by 24–48 h and less than 10% reduction in target mRNA levels in target tissue at 1 mg/kg dose by control siRNA by 24–48 h. Demonstration of RNAi-induced off-target effects by less than 10-fold cytokine induction in 2 and 24 h at 3 mg/kg and less than 10-fold increases in ALT and AST at 3 mg/kg with no effect on body weight, blood clinical chemistry, and hematology data in 48 h is also essential.
7. Targeted Delivery
Prior understanding of the nature of the targeting mechanism (passive or active) helps to prove the active targeting and cellular internalization in vitro by blocking with the appropriate soluble ligand or receptor. Incorporated targeted moeity should be able to compete with the ligands like transferrin, folate, and galactosamine for binding to the target cell. Controls for in vivo active targeting experiments should include a nonfunctional targeting moiety of similar class, molecular weight, and pI, like irrelevant IgG. Materials without a control targeting ligand are not good controls. In addition, unrelated cell lines that do not express the receptor are not good controls. There should be at least fivefold greater in vivo gene silencing in target cells using actively targeted materials than that observed with the negative targeting controls.
7.12. Patent Trends
A mark of the astonishing potential of antisense technology is the surge in the number of patents applied for in the past few years, which must take into account the 90 granted US patents and the 745 US applications on siRNA only in the year 2009 itself [261]. The trend in the patents is toward gene silencing, preparation techniques for judicious design, and modifications to improve specificity, nuclease resistance, transfection efficiency, and targeting of the molecules. At present, the focus of antisense research is toward delivering an adequate amount in the right cells at the right time through an appropriate siRNA design and labeling it with suitable delivery system. Commercial benefits can be fully exploited with sensible and executed patent strategies based on an understanding of the existing patent coverage. Many antisense technology–based formulations are now being developed with a view toward patentability in the USA, and there are key differences between US and international patent systems that may be relevant to international patenting of RNAi technology. Patents on most recent antisense technology based on siRNA can be grasped by analyzing the strengths and weaknesses of patents of established technologies, such as monoclonal antibodies, gene therapy, and AS ODNs. Successful patent position is also being addressed in therapeutics characteristics and platform technologies of antisense technology for the treatment of various disorders like allergy, ocular angiogenesis, cancer, cardiovascular problems, CNS disorders, and AIDS. As well, patents for targeting and delivery technologies like nanoparticles, both polymer and lipid based; electroporation-based delivery, carrier mediated–delivery, biodegradable cationic polymer–based delivery, and others, are being explored. A glance at the US patent scenario for a few major companies involved in siRNA technology is presented in Table 7.7 [261].
Table 7.7.
Patent Scenario in the USA for Major Companies Involved in siRNA Therapeutics
| S. No. | Company | Trend of Patents/Applications | Approximate Number of Granted Patents | Approximate Number of Applications |
|---|---|---|---|---|
| 1. | Sirna Therapeutics |
|
25 | 202 |
| 2. | Dharmacon |
|
35 | 141 |
| 3. | Alnylam |
|
17 | 26 |
| 4. | Ambion |
|
1 | 8 |
| 5. | Silence Therapeutics |
|
1 | 4 |
| 6. | Calando Therapeutics |
|
1 | 2 |
| 7. | Nucleonics |
|
0 | 5 |
| 8. | Roche |
|
10 | 9 |
| 9. | Alcon |
|
3 | 40 |
| 10. | MDRNA |
|
0 | 3 |
7.13. Future Directions
Antisense therapeutics, though being explored for a tremendous number of diseases and disorders, is more biased toward anticancer therapeutics. Due to the differences between normal and tumor vasculature, the highest concentration is achieved around the tumor vasculature, and thus targeting genes within the endothelial and supporting cells has become an attractive antitumor strategy. Moreover, the applications for liposome technology have also been explored more in the fields of cancer and vaccines. A coherent utilization of the liposomes for antisense agents has thus become a hot area for many leading companies involved in RNAi therapeutics. In fact, liposomes, polyconjugates, and other biodegradable polymeric carriers have emerged as the leading platform for the systemic delivery of RNAi therapeutics and offer considerable promise for diseases of the liver, solid cancers, as well as potentially enhanced vaccines, infectious diseases, and immune cell-related disorders. Because the drugs based on liposomal and biodegradable polymeric carriers systems are already on the market, a big leap forward should occur toward the development of polymer- and liposome-based RNAi formulations to make a swift and safer dosage form; from a regulatory perspective. An attention toward the development of an attractive and feasible field of RNAi-based vaccines is also the need of the hour. Many flu vaccines, like that for severe acute respiratory syndrome (SARS), swine flu, and its further mutating varieties, can be developed and put into one pool to cover the closer probable mutations of the existing mutant variety in a single formulation. Because these antisense formulations can have potential side effects and can lose their viability in the biological milieu, extensive but satisfactory pharmacological and toxicological studies should be performed at the initial development and preclinical stages so as to avoid failures at the later clinical phases. Many experiments and evaluations have already been performed on the eldest antisense technology of oligonucleotides. Thus, all the shortcomings and profits from such studies should be taken into account while developing newer antisense-based formulations. Highly specific agents like siRNA can boom as agents for individualized therapies. Principal companies that are actively involved in RNAi-based therapeutics should focus on investing in developing these as personalized therapeutic agents for patients suffering from either major disorders or rare ones that have occurred because of an individual-specific gene defect or mutation. As a potent agent effective at very low doses, siRNA is a strikingly suitable agent to be delivered via inhalation route. Also many studies done on inhalation of various genes can support the development of stable and effective aerosolizable or nebulizable siRNA formulations. Simplicity of siRNA design, its specificity, potency, availability of human genome information, feasibility of fabrication into required sequence, and applications for an endless number of disease-related expressions offers the broadest application in almost every area of therapeutics. Thus, these features should be sincerely evaluated through specialized distribution of antisense-oriented research to various respected companies, industries, and institutes. We request their required collaboration, cooperation, and support of one another in order to present this long-awaited technology as a practically available therapy to humankind.
7.14. Acknowledgment
The authors acknowledge financial assistance from All India Council for Technical Education, and TIFAC CORE in NDDS, Government of India, New Delhi, for providing research facilities to the team.
References
- 1.Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov Today. 2009;14:1123–1129. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Dowdy S. The future of personalized cancer treatment: an entirely new direction for RNAi delivery. http://www.medicalnewstoday.com/articles/150367.php (accessed December 2009).
- 6.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 7.Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronso RT, Knipe DM. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 8.Bitko V, Musiyenko A, Shulyayev O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 9.MacDiarmid JA, Amaro-Mugridge NB, Madrid-Weiss J, Sedliarou I, Wetzel S, Kochar K. Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat Biotechnol. 2009;27:643–651. doi: 10.1038/nbt.1547. [DOI] [PubMed] [Google Scholar]
- 10.Fattal E, Bochot A. Antisense oligonucleotides, aptamers and siRNA: promises for the treatment of ocular diseases. Arch Soc Esp Oftalmol. 2006;81:3–6. [PubMed] [Google Scholar]
- 11.Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 12.Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharm. 2008;6:651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crooke ST. Preliminary studies on genetic engineering: the uptake of oligonucleotides and RNA by Novikoff Hepatoma Ascites Cells. Baylor College of Medicine, School of Graduate Studies; Houston, TX: 1972. [Google Scholar]
- 14.Colby C, Chamberlin MJ, Duesberg PH, Simon MI. Specificity of interferon induction. In: Beers Jun RF, Braun W, editors. Biological effects of polynucleotides. Springer-Verlag; New York, NY: 1971. pp. 79–87. [Google Scholar]
- 15.Ts'o PO, Miller PS, Greene JJ. Nucleic acid analogues with targeted delivery at chemotherapeutic agents. In: Cheng YC, Goz B, Minkoff M, editors. Development of target-oriented anticancer drugs. Raven Press; New York, NY: 1983. pp. 189–206. [Google Scholar]
- 16.Barrett JC, Miller PS, Ts'o PO. Inhibitory effect of complex formation with oligodeoxyribonucleotide ethyl phosphodiesters on transfer ribonucleic acid aminoacylation. Biochemistry. 1974;13:4897–4906. doi: 10.1021/bi00721a004. [DOI] [PubMed] [Google Scholar]
- 17.Scanlon KJ. Anti-genes: siRNA, ribozymes and antisense. Curr Pharm Biotechnol. 2004;5:415–420. doi: 10.2174/1389201043376689. [DOI] [PubMed] [Google Scholar]
- 18.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajeri PB, Singh SK. siRNAs: their potential as therapeutic agents—Part I. Designing of siRNAs. Drug Discov Today. 2009;14:851–858. doi: 10.1016/j.drudis.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Urakami T, Oku N. Current status of siRNA delivery technology and siRNA drug development. Open Drug Delivery J. 2007;1:20–27. [Google Scholar]
- 21.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juliano R, Bauman J, Kang H, Ming X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol Pharm. 2009;6:686–695. doi: 10.1021/mp900093r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couture LA, Stinchcomb DT. Anti-gene therapy: the use of ribozymes to inhibit gene function. Cell. 1996;12:510–515. doi: 10.1016/s0168-9525(97)81398-4. [DOI] [PubMed] [Google Scholar]
- 24.Haseloff J, Gerlach WL. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 25.Uhlenbeck OC. Using ribozymes to cleave RNAs. In: Crooke S, Lebleu B, editors. Antisense research and applications. CRC Press; Boca Raton, FL: 1993. pp. 83–89. [Google Scholar]
- 26.Parthasarathy R, Cote GJ, Gogel RF. Hammerhead ribozyme mediated inactivation of mutant RET in medullary thyroid carcinoma. Cancer Res. 1999;59:3911–3914. [PubMed] [Google Scholar]
- 27.Heidenreich O, Benseler F, Fahrenholz A, Eckstein F. High activity and stability of hammerhead ribozymes containing 2′-modified pyrimidine nucleotides and phosphorothioates. J Biol Chem. 1994;269:2131–2138. [PubMed] [Google Scholar]
- 28.Hélène C. Control of gene expression by triple-helix-forming oligonucleotides—the antigene strategy. In: Crooke S, Lebleu B, editors. Antisense research and applications. CRC Press; Boca Raton, FL: 1993. pp. 375–385. [Google Scholar]
- 29.Moser HE, Dervan PB. Sequence specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 30.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;24:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 31.Faruqi AF, Egholm M, Glazer PM. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cell. Proc Natl Acad Sci USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blommers MJ, Natt F, Jahnke W, Cuenoud B. Dual recognition of double stranded DNA by 2′-aminoethoxy-modified oligonucleotides—the solution structure of an intramolecular triplex obtained by NMR spectroscopy. Biochemistry. 1998;37:17714–17725. doi: 10.1021/bi9816352. [DOI] [PubMed] [Google Scholar]
- 33.Faruqi AF, Krawezyk SH, Matteucci MD, Glazer PM. Potassium-resistant triple helix formation and improved intracellular gene targeting by oligodeoxyribonucleotides containing 7-deazaxanthine. Nucleic Acids Res. 1997;25:633–640. doi: 10.1093/nar/25.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen PE, Egholm M, Berg RH, Buchardt O. Peptide nucleic acids (PNA). DNA analogues with a polyamide backbone. In: Crooke S, Lebleu B, editors. Antisense research and applications. CRC Press; Boca Raton, FL: 1993. pp. 375–385. [Google Scholar]
- 35.Mahato RI, Kawabata K, Takakura Y, Hashida M. In vivo disposition characteristics of plasmid DNA complexed with cationic liposomes. J Drug Targeting. 1995;3:149–157. doi: 10.3109/10611869509059214. [DOI] [PubMed] [Google Scholar]
- 36.Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 37.Juliano RL, Dixit VR, Kang H, Kim TY, Miyamoto Y, Xu D. Epigenetic manipulation of gene expression: a toolkit for cell biologists. J Cell Biol. 2005;169:847–857. doi: 10.1083/jcb.200501053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borchard G. Chitosans for gene delivery. Adv Drug Deliv Rev. 2001;52:145–150. doi: 10.1016/s0169-409x(01)00198-3. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Zhang PC, Lu HF, Ma N, Wang S, Mao HQ. New polyphosphoramidate with a spermidine side chain as a gene carrier. J Control Release. 2002;83:157–168. doi: 10.1016/s0168-3659(02)00180-3. [DOI] [PubMed] [Google Scholar]
- 40.Yi SW, Yune TY, Kim TW, Chung H, Choi YW, Kwon IC. A cationic lipid emulsion/DNA complex as a physically stable and serum-resistant gene delivery system. Pharm Res. 2000;17:314–320. doi: 10.1023/a:1007553106681. [DOI] [PubMed] [Google Scholar]
- 41.Sternberg B, Sorgi FL, Huang L. New structures in complex formation between DNA and cationic liposomes visualized by freeze-fracture electron microscopy. FEBS Lett. 1994;356:361–366. doi: 10.1016/0014-5793(94)01315-2. [DOI] [PubMed] [Google Scholar]
- 42.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasic D, Martin F, editors. Stealth liposomes. CRC Press; Boca Raton, FL: 1995. [Google Scholar]
- 44.Litzinger DC, Brown JM, Wala I, Kaufman SA, Han GY, Farrell CL. Fate of cationic liposomes and their complex with oligonucleotive in vivo. Biochim Biophys Acta. 1996;1281:139–149. doi: 10.1016/0005-2736(95)00268-5. [DOI] [PubMed] [Google Scholar]
- 45.Mahato RI, Kawabata K, Nomura T, Takakur Y, Hashida M. Physicochemical and pharmacokinetic characteristics of plasmid DNA/cationic liposomes complexes. J Pharm Sci. 1995;84:1267–1271. doi: 10.1002/jps.2600841102. [DOI] [PubMed] [Google Scholar]
- 46.Thierry AR, Lunardi-Iskandar Y, Bryant JL, Rabinovich P, Gallo RC, Mahan LC. Systemic gene therapy: biodistribution and long-term expression of a transgene in mice. Proc Natl Acad Sci USA. 1995;92:9742–9746. doi: 10.1073/pnas.92.21.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodle MC, Martin FJ, Yan-Young A, Redemann CT. Liposomes with enhanced circulation time. 2000;US5013556.
- 48.Bailon P, Won CY. PEG-modified biopharmaceuticals. Expert Opin Drug Deliv. 2009;6:1–16. doi: 10.1517/17425240802650568. [DOI] [PubMed] [Google Scholar]
- 49.Rippe B, Rosengren BI, Carlsson O, Venturoli D. Transendothelial transport: the vesicle controversy. J Vasc Res. 2002;39:375–390. doi: 10.1159/000064521. [DOI] [PubMed] [Google Scholar]
- 50.Schubert W, Frank PG, Razani B, Park DS, Chow CW, Lisanti MP. Caveolae-deficient endothelial cells show defects in the uptake and transport of albumin in vivo. J Biol Chem. 2001;276:48619–48622. doi: 10.1074/jbc.C100613200. [DOI] [PubMed] [Google Scholar]
- 51.Uyechi LS, Gagne L, Thurston G, Szoka FC., Jr. Mechanism of lipoplex gene delivery in mouse lung: binding and internalization of fluorescent lipid and DNA components. Gene Ther. 2001;8:828–836. doi: 10.1038/sj.gt.3301461. [DOI] [PubMed] [Google Scholar]
- 52.Kircheis R, Wightman L, Schreiber A, Robitza B, Rossler V, Kursa M. Polyethylenimine/DNA complexes shielded by transferrin target gene expression to tumors after systemic application. Gene Ther. 2001;8:28–40. doi: 10.1038/sj.gt.3301351. [DOI] [PubMed] [Google Scholar]
- 53.Reddy JA, Abburi C, Hofland H, Howard SJ, Vlahov I, Wils P. Folate-targeted, cationic liposome-mediated gene transfer into disseminated peritoneal tumors. Gene Ther. 2002;9:1542–1550. doi: 10.1038/sj.gt.3301833. [DOI] [PubMed] [Google Scholar]
- 54.Balyasnikova IV, Yeomans DC, McDonald TB, Danilov SM. Antibody-mediated lung endothelium targeting: in vivo model on primates. Gene Ther. 2002;9:282–290. doi: 10.1038/sj.gt.3301657. [DOI] [PubMed] [Google Scholar]
- 55.Abes R, Arzumanov AA, Moulton HM, Abes S, Ivanova GD, Iversen PL. Cell-penetrating-peptide-based delivery of oligonucleotides: an overview. Biochem Soc Trans. 2007;35:775–779. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- 56.Harding JA, Engbers CM, Newman MS, Goldstein NI, Zalipsky S. Immunogenicity and pharmacokinetic attributes of poly(ethylene glycol)-grafted immunoliposomes. Biochim Biophys Acta. 1997;1327:181–192. doi: 10.1016/s0005-2736(97)00056-4. [DOI] [PubMed] [Google Scholar]
- 57.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 58.Lechardeur D, Lukacs GL. Intracellular barriers to non-viral gene transfer. Curr Gene Ther. 2002;2:183–194. doi: 10.2174/1566523024605609. [DOI] [PubMed] [Google Scholar]
- 59.Lima MC, Simões S, Pires P, Gaspar R, Slepushkin V, Duzgunes N. Gene delivery mediated by cationic liposomes: from biophysical aspects to enhancement of transfection. Mol Membr Biol. 1999;16:103–109. doi: 10.1080/096876899294823. [DOI] [PubMed] [Google Scholar]
- 60.Pereira FB, Goni FM, Nieva J. Liposome destabilization induced by the HIV-1 fusion peptide Effect of a single amino acid substitution. FEBS Lett. 1995;362:243–246. doi: 10.1016/0014-5793(95)00257-a. [DOI] [PubMed] [Google Scholar]
- 61.Kaneda Y, Uchida T, Kim J, Ishiura M, Okada Y. The improved efficient method for introducing macromolecules into cells using HVJ (Sendai virus) liposomes with gangliosides. Exp Cell Res. 1987;173:56–69. doi: 10.1016/0014-4827(87)90331-4. [DOI] [PubMed] [Google Scholar]
- 62.Farhood H, Serbina N, Huang L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim Biophys Acta. 1995;1235:289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
- 63.Litzinger DC, Huang L. Phosphatidylethanolamine liposomes: drug delivery, gene transfer and immunodiagnostic applications. Biochim Biophys Acta. 1992;1113:201–227. doi: 10.1016/0304-4157(92)90039-d. [DOI] [PubMed] [Google Scholar]
- 64.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Remy JS, Sirlin C, Vierling P, Behr JP. Gene transfer with a series of lipophilic DNA-binding molecules. Bioconjug Chem. 1994;5:647–654. doi: 10.1021/bc00030a021. [DOI] [PubMed] [Google Scholar]
- 66.Xu Y, Szoka FC., Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 67.Johnson-Saliba M, Jans DA. Gene therapy: optimising DNA delivery to the nucleus. Curr Drug Targets. 2001;2:371–399. doi: 10.2174/1389450013348245. [DOI] [PubMed] [Google Scholar]
- 68.Jere D, Arote R, Jiang HL, Kim YK, Cho MH, Cho CS. Bioreducible polymers for efficient gene and siRNA delivery. Biomed Mater. 2009;20:1–4. doi: 10.1088/1748-6041/4/2/025020. [DOI] [PubMed] [Google Scholar]
- 69.Wang CY, Huang L. Highly efficient DNA delivery mediated by pH-sensitive immunoliposomes. Biochemistry. 1989;28:9508–9514. doi: 10.1021/bi00450a039. [DOI] [PubMed] [Google Scholar]
- 70.Shockett PE, Schatz DG. Diverse strategies for tetracycline-regulated inducible gene expression. Proc Natl Acad Sci USA. 1996;93:5173–5176. doi: 10.1073/pnas.93.11.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Medzhitov R, Janeway C., Jr. Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 72.Varga LV, Toth S, Novak I, Falus A. Antisense strategies: functions and applications in immunology. Immunol Lett. 1999;69:217–224. doi: 10.1016/s0165-2478(99)00082-6. [DOI] [PubMed] [Google Scholar]
- 73.Walder RY, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci USA. 1988;85:5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crum C, Johnson JD, Nelson A, Roth D. Complementary oligodeoxynucleotide mediated inhibition of tobacco mosaic virus RNA translation in vitro. Nucleic Acids Res. 1988;16:4569–4581. doi: 10.1093/nar/16.10.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL. Uniformly modified 2′-deoxy-2′-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 76.Sproat BS, Lamond AI, Beijer B, Neuner P, Ryder U. Highly efficient chemical synthesis of 2′-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989;17:3373–3386. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morvan F, Rayner B, Imbach JL. Alpha-oligonucleotides: a unique class of modified chimeric nucleic acids. Anticancer Drug Des. 1991;6:521–529. [PubMed] [Google Scholar]
- 78.Gagnor C, Rayner B, Leonetti JP, Imbach JL, Lebleu B. Alpha-DNA.IX: parallel annealing of alpha-anomeric oligodeoxyribonucleotides to natural mRNA is required for interference in RNase H mediated hydrolysis and reverse transcription. Nucleic Acids Res. 1989;17:5107–5114. doi: 10.1093/nar/17.13.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maher LJ, III, Wold B, Dervan PB. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989;245:725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- 80.Stein CA, Cheng YC. Antisense oligonucleotides as therapeutic agents-is the bullet really magical? Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 81.Mirabelli CK, Bennett CF, Anderson K, Crooke ST. In vitro and in vivo pharmacologic activities of antisense oligonucleotides. Anticancer Drug Des. 1991;6:647–661. [PubMed] [Google Scholar]
- 82.Baker BF, Monia BP. Novel mechanisms for antisense-mediated regulation of gene expression. Biochim Biophys Acta. 1999;1489:3–18. doi: 10.1016/s0167-4781(99)00146-3. [DOI] [PubMed] [Google Scholar]
- 83.Summerton J. Morpholino antisense oligomers: the case for an RNase H independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 84.Kulka M, Smith CC, Aurelian L, Fishelevich R, Meade K, Miller P. Site specificity of the inhibitory effects of oligo(nucleoside methylphosphonate)s complementary to the acceptor splice junction of herpes simplex virus type 1 immediate early mRNA 4. Proc Natl Acad Sci USA. 1989;86:6868–6872. doi: 10.1073/pnas.86.18.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McManaway ME, Neckers LM, Loke SL, al-Nasser AA, Redner RL, Shiramizu BT. Tumor-specific inhibition of lymphoma growth by an antisense oligonucleotide. Lancet. 1990;335:808811. doi: 10.1016/0140-6736(90)90934-w. [DOI] [PubMed] [Google Scholar]
- 86.Sierakowska H, Sambade MJ, Agrawal S, Kole R. Repair of thalassemic human b-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knudsen H, Nielsen PE. Antisense properties of duplex- and triplex-forming PNAs. Nucleic Acids Res. 1996;24:494–500. doi: 10.1093/nar/24.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gambacorti-Passerini C, Mologni L, Bertazzoli C, Coutre P, Marchesi E, Grignani F. In vitro transcription and translation inhibition by anti-promyelocytic leukemia (PML)/retinoic acid receptor alpha and anti-PML peptide nucleic acid. Blood. 1996;88:1411–1417. [PubMed] [Google Scholar]
- 90.Agrawal S, Goodchild J, Civeira MP, Thornton AH, Sarin PS, Zamecnik PC. Oligodeoxynucleoside phosphoramidates and phosphorothioates as inhibitors of human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:7079–7083. doi: 10.1073/pnas.85.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lemaitre M, Bayard B, Lebleu B. Specific antiviral activity of a poly(l-lysine)-conjugated oligodeoxyribonucleotide sequence complementary to vesicular stomatitis virus N protein mRNA initiation site. Proc Natl Acad Sci USA. 1987;84:648–652. doi: 10.1073/pnas.84.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chiang MY, Chan H, Zounes MA, Freier SM, Lima WF, Bennett CF. Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J Biol Chem. 1991;266:18162–18171. [PubMed] [Google Scholar]
- 93.Lebleu B, Moulton HM, Abes R, Ivanova GD, Abes S, Stein DA. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv Drug Deliv Rev. 2008;60:517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xanthos JB, Kofron M, Wylie C, Heasman J. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development. 2001;128:167–180. doi: 10.1242/dev.128.2.167. [DOI] [PubMed] [Google Scholar]
- 95.Lima WF, Crooke ST. Cleavage of single strand RNA adjacent to RNA–DNA duplex regions by Escherichia coli RNase H1. J Biol Chem. 1997;272:27513–27516. doi: 10.1074/jbc.272.44.27513. [DOI] [PubMed] [Google Scholar]
- 96.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 97.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 98.Crooke ST. Progress in evaluation of the potential of antisense technology. Antisense Res Dev. 1994;4:145–146. doi: 10.1089/ard.1994.4.145. [DOI] [PubMed] [Google Scholar]
- 99.Crooke ST. Oligonucleotide therapeutics. In: Wolff ME, editor. vol. 1. John Wiley; New York, NY: 1995. pp. 863–900. (Burger's medicinal chemistry and drug discovery). [Google Scholar]
- 100.Kurreck J. Antisense technologies: improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 101.Miller PS, Yano J, Yano E, Carroll C, Jayaraman K, Ts'o PO. Nonionic nucleic acid analogues. Synthesis and characterization of dideoxyribonucleoside methylphosphonates. Biochemistry. 1979;18:5134–5143. doi: 10.1021/bi00590a017. [DOI] [PubMed] [Google Scholar]
- 102.Miller PS, Bhan P, Cushman CD, Kean JM, Levis JT. Antisense oligonucleoside methylphosphonates and their derivatives. Nucleosides Nucleotides Nucleic Acids. 1991;10:37–46. [Google Scholar]
- 103.Tonkinson JL, Stein CA. Patterns of intracellular compartmentalization, trafficking and acidification of 5′-fluorescein labeled phosphodiester and phosphorothioate oligodeoxynucleotides in HL60 cells. Nucleic Acids Res. 1994;22:4268–4275. doi: 10.1093/nar/22.20.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shoji Y, Akhtar S, Periasamy A, Herman B, Juliano RL. Mechanism of cellular uptake of modified oligodeoxynucleotides containing methylphosphonate linkages. Nucleic Acids Res. 1991;19:5543–5550. doi: 10.1093/nar/19.20.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DeClerq E, Eckstein F, Merigan TC. Interferon induction increased through chemical modification of a synthetic polyribonucleotide. Science. 1969;165:1137–1139. doi: 10.1126/science.165.3898.1137. [DOI] [PubMed] [Google Scholar]
- 106.Matsukura M, Shinozuka K, Zon G, Mitsuya H, Reitz M, Cohen JS. Phosphorothioate analogs of oligodeoxynucleotides: inhibitors of replication and cytopathic effects of human immunodeficiency virus. Proc Natl Acad Sci USA. 1987;84:7706–7719. doi: 10.1073/pnas.84.21.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stein CA, Cohen JS. Phosphorothioate oligodeoxynucleotide analogues. In: Cohen JS, editor. Oligodeoxynucleotides: antisense inhibitors of gene expression. CRC Press; Boca Raton, FL: 1989. pp. 97–117. [Google Scholar]
- 108.Campbell JM, Bacon TA, Wickstrom E. Oligodeoxynucleoside phosphorothioate stability in subcellular extracts, culture media, sera, and cerebrospinal fluid. J Biochem Biophys Methods. 1990;20:259–267. doi: 10.1016/0165-022x(90)90084-p. [DOI] [PubMed] [Google Scholar]
- 109.Loke SL, Stein CA, Zhang XH, Mori K, Nakanishi M, Subasinghe C. Characterization of oligonucleotide transport into living cells. Proc Natl Acad Sci USA. 1989;86:3474–3478. doi: 10.1073/pnas.86.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J Biol Chem. 1997;272:11994–20000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 111.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 112.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 113.Jensen KK, Orum H, Nielsen PE, Norden B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry. 1997;36:5072–5077. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]
- 114.Vickers TA, Griffith MC, Ramasamy K, Risen LM, Freier SM. Inhibition of NF-kappa B specific transcriptional activation by PNA strand invasion. Nucleic Acids Res. 1995;23:3003–3008. doi: 10.1093/nar/23.15.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mologni L, Marchesi E, Nielsen PE, Gambacorti-Passerini C. Inhibition of promyelocytic leukemia (PML)/retinoic acid receptor-α and PML expression in acute promyelocytic leukemia cells by anti-PML peptide nucleic acid. Cancer Res. 2001;61:5468–5473. [PubMed] [Google Scholar]
- 116.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 117.Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB, Weller DD. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 1996;6:267–272. doi: 10.1089/oli.1.1996.6.267. [DOI] [PubMed] [Google Scholar]
- 118.Rayburn ER, Zhang R. Antisense, RNAi, and gene silencing strategies for therapy: mission possible or impossible? Drug Discov Today. 2008;13:513–521. doi: 10.1016/j.drudis.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hicke BJ, Stephens AW. Escort aptamers: a delivery service for diagnosis and therapy. J Clin Invest. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an l-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lv W, Zhang C, Hao J. RNAi technology: a revolutionary tool for the colorectal cancer therapeutics. World J Gastroenterol. 2006;12:4636–4639. doi: 10.3748/wjg.v12.i29.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release. 2008;129:107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 125.Lomas-Neira JL, Chung CS, Wesche DE, Perl M, Ayala A. In vivo gene silencing (with siRNA) of pulmonary expression of MIP-2 versus KC results in divergent effects on hemorrhage-induced, neutrophilmediated septic acute lung injury. J Leukoc Biol. 2005;77:846–853. doi: 10.1189/jlb.1004617. [DOI] [PubMed] [Google Scholar]
- 126.Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Großhans H, Filipowicz W. The expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 129.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hannon GJ, Ross JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 131.Leeds JM, Graham MJ, Troung L, Cummins LL. Quantitation of phosphorothioate oligonucleotides in human plasma. Anal Biochem. 1996;235:36–43. doi: 10.1006/abio.1996.0088. [DOI] [PubMed] [Google Scholar]
- 132.Griffey RH, Greig MJ, Gaus HJ, Liu K, Monteith D, Winniman M. Characterization of oligonucleotide metabolism in vivo via liquid chromatography/electrospray tandem mass spectrometry with a quadrupole ion trap mass spectrometer. J Mass Spectrom. 1997;32:305–313. doi: 10.1002/(SICI)1096-9888(199703)32:3<305::AID-JMS482>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 133.Cohen AS, Bourque AJ, Wang BH, Smisek DL, Belenky A. A nonradioisotope approach to study the in vivo metabolism of phosphorothioate oligonucleotides. Antisense Nucleic Acid Drug Dev. 1997;7:13–22. doi: 10.1089/oli.1.1997.7.13. [DOI] [PubMed] [Google Scholar]
- 134.Leeds JM, Geary RS, Henry SP, Glover J, Shanahan W, Fitchett J. Pharmacokinetic properties of phosphorothioate oligonucleotides. Nucleosides Nucleotides. 1997;16:1689–1693. [Google Scholar]
- 135.Geary RS, Leeds JM, Henry SP, Monteith DK, Levin AA. Antisense oligonucleotide inhibitors for the treatment of cancer: 1. Pharmacokinetic properties of phosphorothioate oligodeoxynucleotides. Anticancer Drug Des. 1997;12:383–393. [PubMed] [Google Scholar]
- 136.Nicklin PL, Craig SJ, Phillips JA. Pharmacokinetic properties of phosphorothioates in animals-absorption, distribution, metabolism and elimination. In: Crooke ST, editor. vol. 131. Springer-Verlag; Berlin: 1998. pp. 141–168. (Antisense research and application: handbook of experimental pharmacology). [Google Scholar]
- 137.Crooke ST. Basic principles of antisense therapeutics. In: Crooke ST, editor. vol. 131. Springer-Verlag; Berlin: 1998. pp. 1–50. (Antisense research and application: handbook of experimental pharmacology). [Google Scholar]
- 138.Bennett CF, Condon TP. Use of antisense oligonucleotides to modify inflammatory processes. In: Crooke ST, editor. vol. 131. Springer-Verlag; Berlin: 1998. pp. 371–394. (Antisense research and application: handbook of experimental pharmacology). [Google Scholar]
- 139.Geary RS, Leeds JM. Pharmacokinetic properties of phosphorothioate oligonucleotides in humans. In: Crooke ST, editor. vol. 131. Springer-Verlag; Berlin: 1998. pp. 217–232. (Antisense research and application: handbook of experimental pharmacology). [Google Scholar]
- 140.Geary RS, Leeds JM, Fitchett J, Burckin T, Truong L, Spainhour C. Pharmacokinetics and metabolism in mice of a phosphorothioate oligonucleotide antisense inhibitor of C-raf-1 kinase expression. Drug Metab Dispos. 1997;25:1272–1282. [PubMed] [Google Scholar]
- 141.Grindel JM, Musick TJ, Jiang Z, Roskey A, Agrawal S. Pharmacokinetics and metabolism of an oligodeoxynucleotide phosphorothioate (GEM91®) in cynomolgus monkeys following intravenous infusion. Antisense Nucleic Acid Drug Dev. 1998;8:43–52. doi: 10.1089/oli.1.1998.8.43. [DOI] [PubMed] [Google Scholar]
- 142.Phillips JA, Craig SJ, Bayley D, Christian RA, Geary R, Nicklin PL. Pharmacokinetics, metabolism, and elimination of a 20-mer phosphorothioate oligodeoxynucleotide (cgp 69846a) after intravenous and subcutaneous administration. Biochem Pharmacol. 1997;54:657–668. doi: 10.1016/s0006-2952(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 143.Temsamani J, Roskey A, Chaix C, Agrawal S. In vivo metabolic profile of a phosphorothioate oligodeoxyribonucleotide. Antisense Nucleic Acid Drug Dev. 1997;7:159–165. doi: 10.1089/oli.1.1997.7.159. [DOI] [PubMed] [Google Scholar]
- 144.Cummins LL, Winniman M, Gaus HJ. Phosphorothioate oligonucleotide metabolism: characterization of the “N+”-mer by CE and HPLC-ES/MS. Bioorg Med Chem Lett. 1997;7:1225–1230. [Google Scholar]
- 145.Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- 146.Henry SP, Giclas PC, Leeds J, Pangburn M, Auletta C, Levin AA. Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J Pharmacol Exp Ther. 1997;281:810–816. [PubMed] [Google Scholar]
- 147.Agrawal S, Zhang X, Cai Q, Kandimalla ER, Manning A, Jiang Z. Effect of aspirin on protein binding and tissue disposition of oligonucleotide phosphorothioate in rats. J Drug Targeting. 1998;5:303–312. doi: 10.3109/10611869808995883. [DOI] [PubMed] [Google Scholar]
- 148.Cossum PA, Truong L, Owens SR, Markham PM, Shea JP, Crooke ST. Pharmacokinetics of a 14C-labeled phosphorothioate oligonucleotide, ISIS 2105, after intradermal administration to rats. J Pharmacol Exp Ther. 1994;269:89–94. [PubMed] [Google Scholar]
- 149.Nicklin PL, Bayley D, Giddings J, Craig SJ, Cummins LL, Hastewell JG. Pulmonary bioavailability of a phosphorothioate oligonucleotide (CGP 64128A): comparison with other delivery routes. Pharm Res. 1998;15:583–591. doi: 10.1023/a:1011934011690. [DOI] [PubMed] [Google Scholar]
- 150.Beck GF, Irwin WJ, Nicklin PL, Akhtar S. Interactions of phosphodiester and phosphorothioate oligonucleotides with intestinal epithelial Caco-2 cells. Pharm Res. 1996;13:1028–1037. doi: 10.1023/a:1016002606705. [DOI] [PubMed] [Google Scholar]
- 151.Graham MJ, Crooke ST, Monteith DK, Cooper SR, Lemonidis KM, Stecker KK. In vivo distribution and metabolism of a phosphorothioate oligonucleotide within rat liver after intravenous administration. J Pharmacol Exp Ther. 1998;286:447–458. [PubMed] [Google Scholar]
- 152.Zhang H, Cook J, Nickel J, Yu R, Stecker K, Myers K. Reduction of liver Fas expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nat Biotechnol. 2000;18:862–867. doi: 10.1038/78475. [DOI] [PubMed] [Google Scholar]
- 153.Li SD, Huang L. Non-viral is superior to viral gene delivery. J Control Release. 2007;123:181–183. doi: 10.1016/j.jconrel.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 154.Verma IM, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 155.Raizada MK, Katovich MJ, Wang H, Berecek KH, Gelband CH. Is antisense gene therapy a step in the right direction in the control of hypertension? Am J Physiol Heart Circ Physiol. 1999;277:H423–H432. doi: 10.1152/ajpheart.1999.277.2.H423. [DOI] [PubMed] [Google Scholar]
- 156.Flotte TR, Carter BJ. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 157.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McKeon J, Khaledi MG. Evaluation of liposomal delivery of antisense oligonucleotide by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr A. 2003;1004:39–46. doi: 10.1016/s0021-9673(03)00721-0. [DOI] [PubMed] [Google Scholar]
- 159.Meng J, Wang H, Hou Z, Chen T, Fu J, Ma X. Novel anion liposome-encapsulated antisense oligonucleotide restores susceptibility of methicillin-resistant Staphylococcus aureus and rescues mice from lethal sepsis by targeting mecA. Antimicrob Agents Chemother. 2009;53:2871–2878. doi: 10.1128/AAC.01542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Meidan VM, Glezer J, Salomon S, Sidi Y, Barenholz Y, Cohen JS. Specific lipoplex-mediated antisense against Bcl-2 in breast cancer cells: a comparison between different formulations. J Liposome Res. 2006;16:27–43. doi: 10.1080/08982100500528685. [DOI] [PubMed] [Google Scholar]
- 161.Resina S, Abes S, Turner JJ, Prevot P, Travo A, Clair P. Lipoplex and peptide-based strategies for the delivery of steric-block oligonucleotides. Int J Pharm. 2007;344:96–102. doi: 10.1016/j.ijpharm.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 162.Gao X, Kim KS, Liu D. Nonviral gene delivery: what we know and what is next. AAPS J. 2007;9:E92–E104. doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huang Z, Jr., Li W, Jr., MacKay Jr JA, Jr., Szoka Jr F., Jr. Thiocholesterol-based lipids for ordered assembly of bioresponsive gene carriers. Mol Ther. 2005;11:409–417. doi: 10.1016/j.ymthe.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 164.Bae Y, Jang WD, Nishiyama N, Fukushima S, Kataoka K. Multifunctional polymeric micelles with folate-mediated cancer cell targeting and pH-triggered drug releasing properties for active intracellular drug delivery. Mol Biosyst. 2005;1:242–250. doi: 10.1039/b500266d. [DOI] [PubMed] [Google Scholar]
- 165.Kwon GS, Okano T. Soluble self-assembled block copolymers for drug delivery. Pharm Res. 1999;16:597–600. doi: 10.1023/a:1011991617857. [DOI] [PubMed] [Google Scholar]
- 166.Jeong JH, Kim SH, Kim SW, Park TG. Polyelectrolyte complex micelles composed of c-raf antisense oligodeoxynucleotide−poly(ethylene glycol) conjugate and poly(ethylenimine): effect of systemic administration on tumor growth. Bioconjug Chem. 2005;16:1034–1037. doi: 10.1021/bc0497315. [DOI] [PubMed] [Google Scholar]
- 167.Vinogradov SV, Batrakova EV, Li S, Kabanov AV. Mixed polymer micelles of amphiphilic and cationic copolymers for delivery of antisense oligonucleotides. J Drug Targeting. 2004;12:517–526. doi: 10.1080/10611860400011927. [DOI] [PubMed] [Google Scholar]
- 168.Narayani R. Polymeric delivery systems in biotechnology: a mini review. Trends Biomater Artif Organs. 2007;21:14–19. [Google Scholar]
- 169.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 170.Han G, Ghosh P, De M, Rotello VM. Drug and gene delivery using gold nanoparticles. Nanobiotechnology. 2007;3:40–45. [Google Scholar]
- 171.Bharali DJ, Klejbor I, Stachowiak EK, Dutta P, Roy I, Kaur N. Organically modified silica nanoparticles: a nonviral vector for in vivo gene delivery and expression in the brain. Proc Natl Acad Sci USA. 2005;102:11539–11544. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Peng J, He X, Wang K, Tan W, Li H, Xing X. An antisense oligonucleotide carrier based on amino silica nanoparticles for antisense inhibition of cancer cells. Nanomedicine. 2006;2:113–120. doi: 10.1016/j.nano.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 173.Gao X, Huang L. Potentiation of cationic liposome-mediated gene delivery by polycations. Biochemistry. 1996;35:1027–1036. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]
- 174.Sorgi FL, Bhattacharya S, Huang L. Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther. 1997;4:961–968. doi: 10.1038/sj.gt.3300484. [DOI] [PubMed] [Google Scholar]
- 175.Lee RJ, Huang L. Folate-targeted, anionic liposome-entrapped polylysine-condensed DNA for tumor cell-specific gene transfer. J Biol Chem. 1996;271:8481–8487. doi: 10.1074/jbc.271.14.8481. [DOI] [PubMed] [Google Scholar]
- 176.Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4:891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- 177.Penichet ML, Kang YS, Pardridge WM, Morrison SL, Shin SU. An antibody-avidin fusion protein specific for the transferrin receptor serves as a delivery vehicle for effective brain targeting: initial applications in anti-HIV antisense drug delivery to the brain. J Immunol. 1999;163:4421–4426. [PubMed] [Google Scholar]
- 178.Ahmed OAA, Adjimatera N, Pourzand C, Blagbrough IS. N4,N9-dioleoyl spermine is a novel nonviral lipopolyamine vector for plasmid DNA formulation. Pharm Res. 2005;22:972–980. doi: 10.1007/s11095-005-4592-1. [DOI] [PubMed] [Google Scholar]
- 179.Hosseinkhani H, Azzam T, Tabata Y, Domb AJ. Dextran-spermine polycation: an efficient nonviral vector for in vitro and in vivo gene transfection. Gene Ther. 2004;11:194–203. doi: 10.1038/sj.gt.3302159. [DOI] [PubMed] [Google Scholar]
- 180.Bronich TK, Cherry T, Vinogradov SV, Eisenberg A, Kabanov VA, Kabanov AV. Self-assembly in mixtures of poly(ethylene oxide)-graft-poly(ethyleneimine) and alkyl sulphates. Langmuir. 1998;14:6101–6106. [Google Scholar]
- 181.Lou X, Garrett KL, Rakoczy PE, Chirila TV. Synthetic hydrogels as carriers in antisense therapy: preliminary evaluation of an oligodeoxynucleotide covalent conjugate with a copolymer of 1-vinyl-2-pyrrolidinone and 2-hydroxyethyl methacrylate. J Biomater Appl. 2001;15:307–320. doi: 10.1106/LVPH-0P1F-V947-RWD1. [DOI] [PubMed] [Google Scholar]
- 182.Dong L, Xia S, Chen H, Chen J, Zhang J. Spleen-specific suppression of TNF-α by cationic hydrogel-delivered antisense nucleotides for the prevention of arthritis in animal models. Biomaterials. 2009;30:4416–4426. doi: 10.1016/j.biomaterials.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 183.Vinogradov SV, Bronich TK, Kabanov AV. Nanosized cationic hydrogels for drug delivery: preparation, properties and interactions with cells. Adv Drug Deliv Rev. 2002;54:135–147. doi: 10.1016/s0169-409x(01)00245-9. [DOI] [PubMed] [Google Scholar]
- 184.Sud T, Li D. Hydrodynamic gene delivery: its principles and applications. Mol Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- 185.Yokoi H, Mukoyama M, Nagae T, Mori K, Suganami T, Sawai K. Reduction in connective tissue growth factor by antisense treatment ameliorates renal tubulointerstitial fibrosis. J Am Soc Nephrol. 2004;15:1430–1440. doi: 10.1097/01.asn.0000130565.69170.85. [DOI] [PubMed] [Google Scholar]
- 186.Bergan R, Connell Y, Fahmy B, Neckers L. Electroporation enhances c-myc antisense oligodeoxynucleotide efficacy. Nucleic Acids Res. 1993;21:3567–3573. doi: 10.1093/nar/21.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zewert TE, Pliquett UF, Langer R, Weaver JC. Transdermal transport of DNA antisense oligonucleotides by electroporation. Biochem Biophys Res Commun. 1995;212:286–2892. doi: 10.1006/bbrc.1995.1968. [DOI] [PubMed] [Google Scholar]
- 188.Gravas S, Mamoulakis C, Rioja J, Tzortzis V, Reijke T, Wijkstra H. Advances in ultrasound technology in oncologic urology. Urol Clin North Am. 2009;36:133–145. doi: 10.1016/j.ucl.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 189.Haag P, Frauscher F, Gradl J, Seitz A, Schafer G, Lindner JR. Microbubble-enhanced ultrasound to deliver an antisense oligodeoxynucleotide targeting the human androgen receptor into prostate tumours. J Steroid Biochem Mol Biol. 2006;102:103–113. doi: 10.1016/j.jsbmb.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 190.Casey JP, Blidner RA, Monroe WT. Caged siRNAs for spatiotemporal control of gene silencing. Mol Pharm. 2009;6:669–685. doi: 10.1021/mp900082q. [DOI] [PubMed] [Google Scholar]
- 191.Zavaglia D, Normand N, Brewis N, O'Hare P, Favrot MC, Coll J. VP22-mediated and light-activated delivery of an anti-c-raf1 antisense oligonucleotide improves its activity after intratumoral injection in nude mice. Mol Ther. 2003;8:840–845. doi: 10.1016/j.ymthe.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 192.Kaplan JH, Forbush B, Hoffman JF. Rapid photolytic release of adenosine 5′-triphosphate from a protected analog—utilization by na-k pump of human red blood-cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 193.Young DD, Lusic H, Lively MO, Yoder JA, Deiters A. Gene silencing in mammalian cells with light-activated antisense agents. Chem Bio Chem. 2008;9:2937–2940. doi: 10.1002/cbic.200800627. [DOI] [PubMed] [Google Scholar]
- 194.Brink PR. Stem cells as a non-immunogenic vehicle for delivery of siRNA. Third Annual RNAi for Developing Targeted Therapeutics, November 2–3, 2009, Boston Massachusetts.
- 195.Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schiffelers RM, Xu J, Storm G, Woodle MC, Scaria PV. Effects of treatment with small interfering RNA on joint inflammation in mice with collagen-induced arthritis. Arthritis Rheum. 2005;52:1314–1318. doi: 10.1002/art.20975. [DOI] [PubMed] [Google Scholar]
- 197.Barclay J, Patel S, Dorn G, Wotherspoon G, Moffatt S, Eunson L. Functional downregulation of P2X3 receptor subunit in rat sensory neurons reveals a significant role in chronic neuropathic and inflammatory pain. J Neurosci. 2004;22:8139–8147. doi: 10.1523/JNEUROSCI.22-18-08139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Thakker DR, Natt F, Husken D, van der Putten H, Maier R, Hoyer D. siRNA-mediated knockdown of the serotonin transporter in the adult mouse brain. Mol Psychiatry. 2005;10:782–789. doi: 10.1038/sj.mp.4001687. [DOI] [PubMed] [Google Scholar]
- 199.Bochot A, Couvreur P, Fattal E. Intavitreal administration of antisense oligonucleotides: potential of liposomal delivery. Prog Retin Eye Res. 2000;19:131–147. doi: 10.1016/s1350-9462(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 200.Reich S, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;30:210–216. [PubMed] [Google Scholar]
- 201.Kim B, Tang Q, Biswas PS, Xu J, Schiffelers RM, Xie FY. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes. Am J Pathol. 2004;165:2177–2185. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tolentino MJ, Brucker AJ, Fosnot J, Ying GS, Wu IH, Malik G. Intravitreal injection of vascular endothelial growth factor small interfering RNA inhibits growth and leakage in a nonhuman primate, laser induced model of choroidal neovascularization. Retina. 2004;24:132–138. doi: 10.1097/00006982-200402000-00018. [DOI] [PubMed] [Google Scholar]
- 203.Durcan N, Murphy C, Cryan SA. Inhalable siRNA: potential as a therapeutic agent in the lungs. Mol Pharm. 2008;5:559–566. doi: 10.1021/mp070048k. [DOI] [PubMed] [Google Scholar]
- 204.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 205.Hamar P, Song E, Kokeny G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 207.Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci USA. 2003;100:7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Moore MJ, Query CC, Sharp PA. Splicing precursors to mRNAs by the spliceosomes. In: Gesteland RF, Atkins JF, editors. RNA world. Cold Spring Harbor Lab; Cold Spring Harbor, NY: 1993. pp. 303–358. [Google Scholar]
- 209.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 210.Wu H, Lima WF, Crooke ST. Properties of clones and expressed human RNase H1. J Biol Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 211.Miller PS. Oligonucleoside methylphosphonates—synthesis and properties. In: Stein CA, Krieg A, editors. Applied antisense oligonucleotide technology. Wiley-Liss; New York, NY: 1998. pp. 3–22. [Google Scholar]
- 212.Sazani P, Kole R. Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J Clin Invest. 2003;112:481–486. doi: 10.1172/JCI19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Svasti S, Suwanmanee T, Fucharoen S, Moulton HM, Nelson MH, Maeda N. RNA repair restores hemoglobin expression in IVS2–654 thalassemic mice. Proc Natl Acad Sci USA. 2009;106:1205–1210. doi: 10.1073/pnas.0812436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ho SP, Hartig PR. Antisense oligonucleotides for target validation in the CNS. Curr Opin Mol Ther. 1999;1:336–343. [PubMed] [Google Scholar]
- 215.Whitesell L, Geselowitz D, Chavany C, Fahmy B, Walbridge S, Alger JR. Stability, clearance, & disposition of intraventricularly administered oligodeoxynucleotides: implications for therapeutic application within the central nervous system. Proc Natl Acad Sci USA. 1993;90:4665–4669. doi: 10.1073/pnas.90.10.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Peng HS, Livanov V, Zhang W, Li J, Lesher T. Modification of phosphorothioate oligonucleotides yields potent analogs with minimal toxicity for antisense experiments in the CNS. Brain Res Mol Brain Res. 1998;62:1–11. doi: 10.1016/s0169-328x(98)00185-5. [DOI] [PubMed] [Google Scholar]
- 217.Skutella T, Stohr T, Probst JC, Ramalho-Ortigao FJ, Holsboer F, Jirikowski GF. Antisense oligonucleotides for in vivo targeting of corticotropin-releasing hormone mRNA: comparison of phosphorothiotate and 3′-inverted probe performance (1994) Horm Metab Res. 1994;26:460–464. doi: 10.1055/s-2007-1001733. [DOI] [PubMed] [Google Scholar]
- 218.Ho SP, Bao Y, Lesher T, Conklin D, Sharp D. Regulation of the angiotensin type-1 receptor by antisense oligonucleotides occurs through an RNase H-type mechanism. Brain Res Mol Brain Res. 1999;65:23–33. doi: 10.1016/s0169-328x(98)00326-x. [DOI] [PubMed] [Google Scholar]
- 219.Aldrian-Herrada G, Desarmenien MG, Orcel H, Boissin-Agasse L, Mery J, Brugidou J. A peptide nucleic acid (PNA) is more rapidly internalized in cultured neurons when coupled to a retro-inverso delivery peptide. The antisense activity depresses the target mRNA and protein in magnocellular oxytocin neurons. Nucleic Acids Res. 1998;26:4910–4916. doi: 10.1093/nar/26.21.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Salmi P, Kela J, Good F, Wengel J, Wahlestedt C. Locked nucleic acids (LNA): a novel and efficacious class of oligonucleotides for antisense knockdown in vivo. Soc Neurosci. 1999;25:2003–2011. [Google Scholar]
- 221.Yaksh TL, Rudy TA. Chronic catheterization of the spinal subarachnoid space. Physiol Behav. 1976;17:1031–1036. doi: 10.1016/0031-9384(76)90029-9. [DOI] [PubMed] [Google Scholar]
- 222.Troy CM, Rabacchi SA, Friedman WJ, Frappier TF, Brown K, Shelanski ML. Caspase-2 mediates neuronal cell death induced by β-amyloid. J Neurosci. 2000;20:1386–1392. doi: 10.1523/JNEUROSCI.20-04-01386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Senechal Y, Larmet Y, Dev KK. Unraveling in vivo functions of amyloid precursor protein: insights from knockout and knockdown studies. Neurodegenerative Dis. 2006;3:134–147. doi: 10.1159/000094772. [DOI] [PubMed] [Google Scholar]
- 224.Khasar SG, Gold MS, Levine JD. A tetrodotoxin-resistant sodium current mediates inflammatory pain in the rat. Neurosci Lett. 1998;256:17–20. doi: 10.1016/s0304-3940(98)00738-1. [DOI] [PubMed] [Google Scholar]
- 225.Stone LS, Vulchanova L. The pain of antisense: in vivo application of antisense oligonucleotides for functional genomics in pain and analgesia. Adv Drug Deliv Rev. 2003;55:1081–1112. doi: 10.1016/s0169-409x(03)00105-4. [DOI] [PubMed] [Google Scholar]
- 226.Sibille E, Sarnyai Z, Benjamin D, Ga J, Baker H, Toth M. Antisense inhibition of 5-hydroxytryptamine2a receptor induces an antidepressant-like effect in mice. Mol Pharmacol. 1997;52:1056–1063. doi: 10.1124/mol.52.6.1056. [DOI] [PubMed] [Google Scholar]
- 227.Cohen O, Erb C, Ginzberg D, Pollak Y, Seidman S, Shoham S. Neuronal overexpression of “readthrough” acetylcholinesterase is associated with antisense-suppressible behavioral impairments. Mol Psychiatry. 2002;7:874–885. doi: 10.1038/sj.mp.4001103. [DOI] [PubMed] [Google Scholar]
- 228.Argov Z, McKee D, Agus S, Brawer S, Shlomowitz N, Yoseph OB. Treatment of human myasthenia gravis with oral antisense suppression of acetylcholinesterase. Neurology. 2007;69:699–700. doi: 10.1212/01.wnl.0000267884.39468.7a. [DOI] [PubMed] [Google Scholar]
- 229.Surmeier DJ, Eberwine J, Wilson CJ, Cao Y, Stefani A, Kitai ST. Dopamine receptor subtypes colocalize in rat striatonigral neurons. Proc Natl Acad Sci USA. 1992;89:10178–10182. doi: 10.1073/pnas.89.21.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Standifer KM, Rossi GC, Pasternak GW. Differential blockade of opioid analgesia by antisense oligodeoxynucleotides directed against various G protein alpha subunits. Mol Pharmacol. 1996;50:293–298. [PubMed] [Google Scholar]
- 231.Ramalingam A, Hirai A, Thompson EA. Glucocorticoid inhibition of fibroblast proliferation and regulation of the cyclin kinase inhibitor p21Cip1. Mol Endocrinol. 1997;11:577–586. doi: 10.1210/mend.11.5.9923. [DOI] [PubMed] [Google Scholar]
- 232.Rogatsky I, Trowbridge JM, Garabedian MJ. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol. 1997;17:3181–3193. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cha HH, Cram EJ, Wang EC, Huang AJ, Kasler HG, Firestone GL. Glucocorticoids stimulate p21 gene expression by targeting multiple transcriptional elements within a steroid responsive region of the p21waf1/cip1 promoter in rat hepatoma cells. J Biol Chem. 1998;273:1998–2007. doi: 10.1074/jbc.273.4.1998. [DOI] [PubMed] [Google Scholar]
- 234.Bennett CF. Regulation of the immune response with antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 1999;9:423–427. [Google Scholar]
- 235.Stepkowski S. Application of antisense oligodeoxynucleotides for organ transplantation. Transplant Proc. 1998;30:2142–2145. doi: 10.1016/s0041-1345(98)00567-3. [DOI] [PubMed] [Google Scholar]
- 236.Yacyshyn BR, Bowen-Yacyshyn MB, Jewell L, Tami JA, Bennett CF, Kisner DL. A placebo-controlled trial of ICAM-1 antisense oligonucleotide in the treatment of Crohn's disease. Gastroenterology. 1998;114:1133–1142. doi: 10.1016/s0016-5085(98)70418-4. [DOI] [PubMed] [Google Scholar]
- 237.Bruce R, Yacyshyn BR. Antisense oligonucleotide treatment of inflammatory bowel diseases. In: Phillips IN, editor. 2nd ed. vol. 106. Humana Press; Totowa, NJ: 2004. pp. 295–305. (Methods in molecular medicine). [DOI] [PubMed] [Google Scholar]
- 238.Haller H, Dragun D, Miethke A, Park JK, Weis A, Lippoldt A. Antisense oligonucleotides for ICAM-1 attenuate reperfusion injury and renal failure in the rat. Kidney Int. 1996;50:473–480. doi: 10.1038/ki.1996.338. [DOI] [PubMed] [Google Scholar]
- 239.White PJ, Atley LM, Wraight CJ. Antisense oligonucleotide treatments for psoriasis. Expert Opin Biol Ther. 2004;4:75–81. doi: 10.1517/14712598.4.1.75. [DOI] [PubMed] [Google Scholar]
- 240.Nyce JW, Metzger WJ. DNA antisense therapy for asthma in an animal model. Nature. 1997;385:721–725. doi: 10.1038/385721a0. [DOI] [PubMed] [Google Scholar]
- 241.Tanaka M, Nyce JW. Respirable antisense oligonucleotides: a new drug class for respiratory disease. Respir Res. 2001;2:5–9. doi: 10.1186/rr32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Nyce JW. Respirable antisense oligonucleotides as novel therapeutic agents for asthma and other pulmonary diseases. Expert Opin Investig Drugs. 1997;6:1149–1156. doi: 10.1517/13543784.6.9.1149. [DOI] [PubMed] [Google Scholar]
- 243.Dreyfus DH, Tompkins SM, Fuleihan R, Ghoda LY. Gene silencing in the therapy of influenza and other respiratory diseases: targeting to RNase P by use of External Guide Sequences (EGS) Biologics. 2007;1:425–432. [PMC free article] [PubMed] [Google Scholar]
- 244.Nishio K, Fukuoka K, Fukumoto H, Sunami T, Iwamoto Y, Suzuki T. Mitogen-activated protein kinase antisense oligonucleotide inhibits the growth of human lung cancer cells. Int J Oncol. 1999;14:461–469. doi: 10.3892/ijo.14.3.461. [DOI] [PubMed] [Google Scholar]
- 245.Crooke ST. Basic principles. In: Crooke S, editor. Antisense technology: principles, strategies and applications. Marcel Dekker; New York, NY: 2001. [Google Scholar]
- 246.Mizutani Y, Fukumoto M, Bonavida B, Yoshida O. Enhancement of sensitivity of urinary bladder tumor cells to cisplatin by c-myc antisense oligonucleotide. Cancer. 1994;74:2546–2554. doi: 10.1002/1097-0142(19941101)74:9<2546::aid-cncr2820740924>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 247.Sklar MD, Prochownik EV. Modulation of cis-platinum resistance in Friend erythroleukemia cells by c-myc. Cancer Res. 1991;51:2118–2123. [PubMed] [Google Scholar]
- 248.Hasegawa S, Koshikawa N, Momiyama N, Moriyama K, Ichikawa Y, Ishikawa T. Matrilysin-specific antisense oligonucleotide inhibits liver metastasis of human colon cancer cells in a nude mouse model. Int J Cancer. 1998;76:812–816. doi: 10.1002/(sici)1097-0215(19980610)76:6<812::aid-ijc8>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 249.Ishiguro T, Nagawa H, Naito M, Tsuruo T. Inhibitory effect of ATF3 antisense oligonucleotide on ectopic growth of ht29 human colon cancer cells. Jpn J Cancer Res. 2000;91:833–836. doi: 10.1111/j.1349-7006.2000.tb01021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Szegedi I, Katona K, Horvath A, Molnar A, Aradi J, Kiss C. Bcl-2 antisense oligonucleotide inhibits the proliferation of childhood leukemia/lymphoma cells of the B-cell lineage. Pathol Oncol Res. 2008;14:275–279. doi: 10.1007/s12253-008-9076-2. [DOI] [PubMed] [Google Scholar]
- 251.Zangemeister-Wittke U, Schenker T, Luedke GH, Stahel RA. Synergistic cytotoxicity of bcl-2 antisense oligodeoxynucleotides and etoposide, doxorubicin and cisplatin on small-cell lung cancer cell lines. Br J Cancer. 1998;78:1035–1042. doi: 10.1038/bjc.1998.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Arriola EL, Rodriguez LA, Hickman JA, Chresta C. M. Bcl-2 overexpression results in reciprocal down-regulation of Bcl-X (L) and sensitizes human testicular germ cell tumours to chemotherapy-induced apoptosis. Oncogene. 1999;18:1457–1464. doi: 10.1038/sj.onc.1202420. [DOI] [PubMed] [Google Scholar]
- 253.Chao J, Chao L. Kallikrein gene therapy in hypertension, cardiovascular and renal diseases. Gene Ther Mol Biol. 1998;1:301–308. [Google Scholar]
- 254.Crooke S, Lebleu B. Antisense research and applications. CRC Press; Boca Raton, FL: 1993. [Google Scholar]
- 255.Wong YF, Chung TKH, Cheung TH, Lam SK, Chang AMZ. Effects of human papillomavirus type-specific antisense oligonucleotides on cervical cancer cells containing papillomavirus type 16. Med Oncol. 1994;11:149–151. doi: 10.1007/BF02999867. [DOI] [PubMed] [Google Scholar]
- 256.Pokrovsky AG, Plyasunova OA, Blinov VM, Abramova TV, Svinarchuk FP, Vlassov VV. Anti-HIV action of antisense and sense oligodeoxynucleotides and its derivatives. Int Conf AIDS. 1992;8:32. [Google Scholar]
- 257.Yao ZQ, Zhou YX, Feng XM, Chen ZX. Specific inhibition of hepatitis b virus gene expression by an antisense oligonucleotide in vitro. Acta Virol. 1995;39:227–230. [PubMed] [Google Scholar]
- 258.Mizuta T, Fujiwara M, Abe T, Miyano-Kurosaki N, Yokota T, Shigeta S. Inhibitory effects of an antisense oligonucleotide in an experimentally infected mouse model of influenza a virus. Biochem Biophys Res Commun. 2000;279:158–161. doi: 10.1006/bbrc.2000.3924. [DOI] [PubMed] [Google Scholar]
- 259.Vinogradov SV, Suzdaltseva Y, Alakhov VY, Kabanov AV. Inhibition of herpes simplex virus 1 reproduction with hydrophobized antisense oligonucleotides. Biochem Biophys Res Commun. 1994;203:959–966. doi: 10.1006/bbrc.1994.2275. [DOI] [PubMed] [Google Scholar]
- 260.http://www.sirna.com
- 261.http://patft.uspto.gov/netahtml/PTO/search-adv.htm20. Accessed December 2009.


