Publisher Summary
Viral taxonomists have developed a system of classification and nomenclature that embraces all viruses. From the operational point of view, viruses are divided into those that affect vertebrate animals, insects, plants, and bacteria. The chapter discusses the viruses of vertebrate animals, although some of these viruses, the arthropod-borne viruses (arboviruses), also replicate in insects or other arthropods. Several hundred species of viruses have been recovered from humans, who are the best-studied vertebrate hosts and new ones are still occasionally discovered. Somewhat fewer have been recovered from each of the common species of farm and companion animals and from the commonly used laboratory animals. To simplify the study of this vast number of infectious agents, these are sorted into groups that share certain common properties. The most important criteria for classification are the physical and chemical characteristics of the virion and the mode of replication of the virus. The chapter explains a formal viral taxonomy, which is based on these criteria.
It is reasonable to assume that all organisms are infected with viruses: vertebrate and invertebrate animals—including mammals, birds, reptiles, amphibia, helminths, and arthropods—as well as plants, algae, fungi, protozoa, bacteria, and mycoplasmas. Since every species that has been intensively searched has yielded several different viruses belonging to several viral families (including some that are host species-specific), it is not unreasonable to believe that the number of distinct species of viruses on earth may be greater than the number of species of all other living things. There are even some “satellite” viruses the replication of which depends on other viruses.
All viruses, whatever their cellular hosts, share the features summarized in Table 1-1; hence, viral taxonomists have developed a system of classification and nomenclature that embraces all viruses. From the operational point of view, however, we can divide viruses into those that affect vertebrate animals, insects, plants, and bacteria, respectively. We are concerned with the viruses of vertebrate animals, although some of these viruses, the arthropod-borne viruses (arboviruses), also replicate in insects or other arthropods.
Several hundred species of viruses have been recovered from humans, who are the best-studied vertebrate hosts, and new ones are still occasionally discovered. Somewhat fewer have been recovered from each of the common species of farm and companion animals, and from the commonly used laboratory animals. In order to simplify the study of this vast number of infectious agents we need to sort them into groups that share certain common properties. The most important criteria for classification are the physical and chemical characteristics of the virion and the mode of replication of the virus. Before discussing formal viral taxonomy, which is based on these criteria, a simpler, clinically useful classification based on modes of transmission from one host to another will be described.
CLASSIFICATION BASED ON EPIDEMIOLOGICAL CRITERIA
Viruses of animals can be transmitted by ingestion, inhalation, injection (including arthropod bites), contact (including coitus), or congenially. In discussing the pathogenesis and epidemiology of viral infections we will frequently make use of the following terminology as a useful way of classifying viruses.
Enteric viruses are usually acquired by ingestion and replicate primarily in the intestinal tract. The term is restricted to viruses that tend to remain localized in the intestinal tract, rather than becoming generalized. Enteric viruses important in veterinary practice include the rotaviruses, coronaviruses, enteroviruses, and some adenoviruses.
Respiratory viruses are usually acquired by inhalation and replicate in the respiratory tract. The term is usually restricted to those viruses that remain localized in the respiratory tract, and includes the orthomyxoviruses and rhinoviruses, and some of the paramyxoviruses, coronaviruses, and adenoviruses.
Arboviruses (arthropod-borne viruses) infect arthropods that ingest vertebrate blood; they replicate in the arthropod's tissues and can then be transmitted by bite to susceptible vertebrates. Viruses that belong to six families are included: all orbiviruses (a genus of the reoviruses), most bunyaviruses, flaviviruses, togaviruses and rhabdoviruses, and African swine fever virus.
CLASSIFICATION BASED ON PHYSICOCHEMICAL CRITERIA
Although classification on epidemiological criteria is convenient for certain purposes, each group contains viruses with very disparate physicochemical and biological properties. As knowledge of the properties of viruses increased it became clear that the physicochemical properties of the virion and the strategy of replication provided a more fundamental basis for viral classification. Classification into major groups called families, and the subdivision of families into genera, has now reached a position of substantial international agreement.
The primary criteria for delineation of families are (1) the kind of nucleic acid and the strategy of viral replication, and (2) the morphology of the virion, including its size, shape (see Fig. 2-1 ), symmetry of the nucleocapsid, and the presence or absence of an envelope, all of which are readily determined by electron microscopy.
FIG. 2-1.
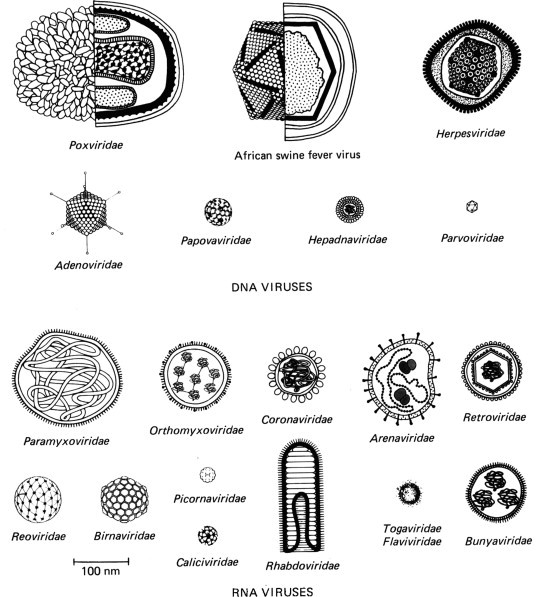
Diagram illustrating the shapes and sizes of animal viruses. The virions are drawn to scale, but artistic license has been used in representing their structure. In some, the cross-sectional structure of capsid and envelope are shown, with a representation of the genome; with the very small virions, only their size and symmetry are depicted.
Subdivision of families into genera depends on criteria that vary for different families. Genera contain from one to over a hundred species, usually defined by antigenic differences. The rationale for subdivision of genera into species is controversial. Most virologists agree that to be designated as distinct species, two viruses should differ substantially in nucleic acid sequence. However, there is as yet no consensus on how such differences should be quantified, and perhaps weighted. Antigenic differences are widely agreed to be of considerable importance. There is less unanimity about whether viruses distinguishable only by neutralization tests should be regarded as separate species, or merely as serotypes within a species. The effects of this ongoing debate on the designation of viral species should not be a major concern for the student, since we will use the common vernacular terms to distinguish viruses that cause different diseases.
Monoclonal antibodies are proving of great value in the differentiation of viruses at the species level and below: types, subtypes, strains, and variants—terms that have no generally accepted taxonomic status. At the research rather than the routine diagnostic level, the composition of the nucleic acid of the viral genome—as revealed by molecular hybridization, oligonucleotide fingerprinting or restriction endonuclease mapping, electrophoresis in gels (especially useful for RNA viruses with segmented genomes), and nucleotide sequence analysis—is used to identify species and to distinguish minor differences between viral strains and mutants.
NOMENCLATURE
Since 1966 the classification and nomenclature of viruses at the higher taxonomic levels (families and genera) has been systematically organized by the International Committee on Taxonomy of Viruses. The highest taxonomic group among viruses is the family; families are named with a suffix -viridae. Subfamilies have the suffix -virinae; genera the suffix -virus. The prefix may be another latin word or a sigla, i.e., an abbreviation derived from some initial letters. Latinized family, subfamily, and generic names are written in italics; vernacular terms derived from them are written in roman letters. For example, the term poxviruses is used to designate members of the family Poxviridae. It is still customary to use vernacular terms rather than latinized binomials for viral species, e.g., foot-and-mouth disease virus.
THE FAMILIES OF DNA VIRUSES
A brief description of each family of viruses of vertebrates is given below and summarized in Tables 2-1 and 2-2 , in order to orient the reader before embarking upon Part I of this book. Most families are allocated separate chapters in Part II, which deals with the role of the viruses in diseases of veterinary importance. We begin with the DNA viruses.
TABLE 2-1.
Properties of the Virions of the Families of DNA Viruses
| Virion | |||||||
|---|---|---|---|---|---|---|---|
| Nucleocapsid | Genomea | ||||||
| Family | Diameter (nm) | Envelope | Symmetry | Capsomers | Transcriptase | Natureb | Size (kb or kbp) |
| Papovaviridae | 45, 55c | – | Icosahedral | 72 | – | ds, circular | 5, 8c |
| Adenoviridae | 70 | – | Icosahedral | 252 | – | ds, linear | 30–37 |
| Herpesviridae | 150 | + | Icosahedral | 162 | – | ds, linear | 120–220 |
| Poxviridae | 300–450 × 170–260 | +d | Complex | — | + | ds, linear | 130–280 |
| African swine | 220 | +d | Icosahedral | 1892 | + | ds, linear | 150 |
| fever virus | |||||||
| Parvoviridae | 20 | – | Icosahedral | 32 | – | ss, (–), linear | 5 |
| Hepadnaviridae | 45 | – | Icosahedral | ? | +e | ds, circularf | 3.2 |
Genome of DNA viruses is invariably a single molecule.
ds, Double-stranded; ss, single-stranded; sense of single-stranded nucleic acid (+) or (–).
Lower figures, Polyomavirus; higher figures, Papillomavirus.
Not essential for infectivity.
Reverse transcriptase.
Circular molecule is double-stranded for most of its length but contains a single-stranded region.
TABLE 2-2.
Properties of the Virions of the Families of RNA Viruses
| Virion | |||||||
|---|---|---|---|---|---|---|---|
| Nucleocapsid | Genome | ||||||
| Family | Diameter (nm)a | Envelope | Symmetry | Capsomers | Transcriptase | Natureb | Size (kb or kbp) |
| Picornaviridae | 25 | – | Icosahedral | 60 | – | ss, 1, (+) | 7.5–8.5 |
| Caliciviridae | 35 | – | Icosahedral | 32 | – | ss, 1, (+) | 7.9 |
| Togaviridae | 65 | +c | Icosahedral | ? | – | ss, 1, (+) | 12 |
| Flaviviridae | 45 | +c | Icosahedral | ? | – | ss, 1, (+) | 12 |
| Orthomyxoviridae | 100 | + | Helical | — | + | ss, 8, (–) | 13.6 |
| Paramyxoviridae | 180 | + | Helical | — | + | ss, 1, (–) | 18–20 |
| Coronaviridae | 100 | +c | Helical | — | – | ss, 1, (+) | 17–24 |
| Arenaviridae | 120 | +c | Helical | — | + | ss, 2, (–) | 10–14 |
| Bunyaviridae | 110 | +c | Helical | — | + | ss, 3, (–) | 13.5–21 |
| Retroviridae | 100 | +c | Icosahedral | ? | +d | ss, 1, (+) | 2 × (3.5–9)e |
| Rhabdoviridae | 180 × 75 | + | Helical | — | + | ss, 1, (–) | 13–16 |
| Filoviridae | 790–970 × 80 | + | Helical | — | + | ss, 1, (–) | 12.7 |
| Reoviridae | 60–80 | – | Icosahedral | 32, 92f | + | ds, 10 to 12g | 17–22 |
| Birnaviridae | 60 | – | Icosahedral | 92 | + | ds, 2 | 7 |
Some enveloped viruses are very pleomorphic and sometimes filamentous.
All molecules linear; ss, single-stranded; ds, double-stranded; 1 to 12, number of molecules in genome; (+) or (–), sense of single-stranded nucleic acid.
No matrix protein.
Reverse transcriptase.
Genome is diploid, two identical molecules being held together by hydrogen bonds at their 5’ ends.
Inner capsid of Orbivirus, Rotavirus, and Reovirus, 32; outer capsid of Reovirus, 92.
Reovirus and Orbivirus, 10; Rotavirus, 11; Colorado tick fever virus, 12.
Papovaviridae
The papovaviruses (sigla: pa = papilloma; po = polyoma; va = vacuolating agent) are small nonenveloped icosahedral viruses which replicate in the nucleus and may transform infected cells. In the virion their nucleic acid occurs as a cyclic double-stranded molecule, which is infectious. There are two genera: Papillomavirus (wart viruses) (55 nm diameter) have a larger genome (8 kbp) which may persist in an episomal form in transformed cells; Polyomavirus (45 nm in diameter) has a smaller genome (5 kbp) which may persist in cells integrated into the host cell DNA.
Papillomaviruses occur in many species of animals, but each is usually of narrow host specificity. Bovine papillomaviruses, which cause cutaneous papillomas of cattle and sarcoids of horses, and the virus of canine oral papillomatosis, are among the most important. The genus Polyomavirus includes SV40 (from rhesus monkeys) and mouse polyoma virus, both of which have been very useful models for the study of viral tumorigenesis.
Adenoviridae
The adenoviruses (adeno = gland) are nonenveloped icosahedral viruses 70 nm in diameter, whose genome consists of a single linear molecule of dsDNA, 30–37 kbp. They replicate in the nucleus. Adenoviruses are usually associated with infection of the respiratory tract, the intestinal tract, and occasionally the eye. Many are characterized by prolonged subclinical infection.
There are two genera, Mastadenovirus and Aviadenovirus. Mastadenoviruses that are animal pathogens include equine adenovirus, bovine adenoviruses, infectious canine hepatitis virus, and canine adenovirus type 2. Pathogenic aviadenoviruses include those which cause egg drop syndrome and marble spleen disease. Some adenoviruses of other species of mammals and birds are also pathogenic. Some of the adenoviruses of humans, cattle, and chickens cause malignant tumors when inoculated into newborn hamsters and have been used in experimental studies on tumorigenesis, but none causes tumors in nature.
Herpesviridae
The herpesviruses (herpes = creeping) have enveloped virions 120–200 nm in diameter, with icosahedral nucleocapsids about 100 nm in diameter. Their genome is a single linear molecule of dsDNA, 120–220 kbp. They replicate in the nucleus and mature by budding through the nuclear membrane, thus acquiring an envelope. This large family includes many important veterinary and human pathogens, and has been subdivided into three subfamilies. Alphaherpesvirinae includes infectious bovine rhinotracheitis virus, bovine mammillitis virus, B-virus, pseudorabies virus, equine rhinopneumonitis and coital exanthema viruses, herpesviruses of cats, dogs, and chickens, and herpes simplex types 1 and 2 and varicella virus of humans. Betaherpesvirinae comprises the cytomegaloviruses, which are highly host-specific viruses of humans, cattle, pigs, horses, mice, guinea pigs, and other animals. Gammaherpesvirinae are often associated with tumors, and include baboon and chimpanzee herpesviruses, Marek's disease virus of chickens, turkey herpesvirus, Herpesvirus ateles, Herpesvirus saimiri of monkeys, and Epstein–Barr (EB) virus in humans.
A feature of all herpesvirus infections is lifelong persistence of the virus in the body, usually in latent form. Excretion may occur continuously or intermittently without disease, or episodes of recurrent clinical disease and recurrent excretion may occur years after the initial infection.
Iridoviridae and African Swine Fever Virus
Originally a number of large icosahedral DNA viruses that affect insects, nematodes, fish, and amphibians were grouped together with African swine fever virus into the family Iridoviridae. In 1984 African swine fever virus was removed from the family Iridoviridae, but it has not yet been allocated to any other family. The family Iridoviridae (irido = iridescent) was defined on the characteristics of certain insect viruses, but it includes two genera that affect vertebrates. They are the largest icosahedral viruses (capsid, 125 nm in diameter), and those that affect vertebrates have enveloped virions 200–250 nm in diameter. The genome is a single linear molecule of dsDNA, 150 kbp. Replication occurs in the cytoplasm, using a virion-associated transcriptase, but nuclear involvement is required for viral DNA synthesis.
The genus Ranavirus includes several members that affect amphibians; and a member of the genus Lymphocystivirus produces lymphocystis disease of fish. African swine fever virus is an important pathogen of swine, which can be transmitted by contact and also by ticks.
Poxviridae
The poxviruses (pock = pustule) are the largest and most complex viruses of vertebrates. The virions are brick shaped, measuring 300–450 × 170–260 nm in all genera except Pampoxvirus, in which virions are ovoid, 220–300 × 140–170 nm. All have an inner core which contains a single linear molecule of dsDNA, 130–280 kbp. Unlike all other DNA viruses of vertebrates except the iridoviruses and African swine fever virus, poxviruses replicate in the cytoplasm; mRNA is transcribed by a virion-associated transcriptase.
The family is divided into two subfamilies, one of which, Chordopoxvirinae, comprises the poxviruses of vertebrates. This subfamily is divided into six genera, all of which include animal pathogens. The genus Orthopoxvirus includes cowpox, camelpox, ectromelia (mousepox), rabbitpox (a variant of vaccinia virus), and monkeypox viruses. Variola virus, which caused human smallpox, and vaccinia virus, used to control that disease, also belong to this genus. Avipoxvirus includes many specific bird poxviruses; Capripoxvirus includes the viruses of sheeppox, goatpox, and lumpyskin disease of cattle. Leporipoxvirus includes myxoma and rabbit fibroma viruses, while the genus Suipoxvirus has only one member, swinepox virus. Parapoxvirus includes contagious pustular dermatitis, pseudocowpox (milker's nodules), and bovine papular stomatitis viruses.
Parvoviridae
Parvoviruses (parvus = small) are only about 20 nm in diameter, have icosahedral symmetry and a genome of ssDNA, 5 kb. The virions are relatively heat stable, and most species have a narrow host range. Members of two genera affect vertebrates. Animal pathogens of the genus Parvovirus include feline panleukopenia, mink enteritis, and Aleutian mink disease viruses, and canine, bovine, goose, porcine, and murine parvoviruses. Members of the genus Dependovirus are defective viruses, which depend on adenovirus for replication. They occur in birds, cattle, horses, and dogs, as well as humans, but are not known to cause disease.
Hepadnaviridae
Human hepatitis B virus and related viruses of other animals form a family for which the name Hepadnaviridae (hepa = liver; dna) has been proposed. The virion is a spherical particle 42 nm in diameter, consisting of a 27-nm icosahedral core within a closely adherent outer capsid that contains cellular lipids, glycoproteins, and a virus-specific surface antigen (HBsAg). The genome is a small, circular, partially double-stranded DNA molecule, which consists of a long (3.2 kb) and a short (1.7–2.8 kb) strand.
The hepadnaviruses replicate in the nucleus of hepatocytes and cause hepatitis, which may progress to cirrhosis and primary hepatocellular carcinoma. The most important species is human hepatitis B virus, but hepadnaviruses also occur in Pekin ducks, woodchucks, and squirrels.
THE FAMILIES OF RNA VIRUSES
Picornaviridae
The Picornaviridae (name originally derived from poliovirus, insensitivity to ether, coxsackievirus, orphan virus, rhinovirus, and ribonucleic acid (omitting one “r”), but conveniently also consistent with the sigla, pico = small; rna = ribonucleic acid) comprise small nonenveloped icosahedral viruses 25–30 nm in diameter, which contain a single molecule of (+) sense ssRNA (7.5–8.5 kb), and replicate in the cytoplasm.
There are four genera. Two genera, Enterovirus and Rhinovirus, include large numbers of species that affect domestic animals, often without causing significant disease. An enterovirus causes swine vesicular disease, which may be confused with foot-and-mouth disease. Porcine enterovirus 1 causes polioencephalomyelitis. These two genera also include several important human pathogens. The only species in the genus Aphthovirus is foot-and-mouth disease virus, which occurs as seven types and many antigenically distinct subtypes. It is among the most important of all viruses in veterinary medicine. The genus Cardiovirus comprises the viruses of encephalomyelocarditis of swine and rodents.
Caliciviridae
The caliciviruses (calix = cup) were first classified as a genus of the family Picornaviridae, but were later found to differ enough in morphology and mode of replication to warrant classification as a separate family. They are slightly larger than picornaviruses (diameter 35–40 nm) but have a genome of similar size. Most species have a narrow host range. Several are of veterinary importance: vesicular exanthema viruses of swine, San Miguel sea lion virus, and feline calicivirus. Caliciviruses have also been isolated from vesicular lesions of dogs, mink, dolphins, calves, and chimpanzees. Virions with caliciviruslike morphology have been demonstrated in the feces of humans, calves, pigs, and sheep with diarrhea.
Togaviridae
The togaviruses (toga = cloak) are small spherical enveloped viruses 60–70 nm in diameter, containing (+) sense ssRNA (12 kb) enclosed within an icosahedral core. They replicate in the cytoplasm and mature by budding from cell membranes. Three genera, Alphavirus, Pestivirus, and Arterivirus, contain species of veterinary importance. Alphavirus contains many species, all of which are arthropod-transmitted. Important alphaviruses that infect both livestock and humans include eastern, western, and Venezuelan equine encephalitis viruses. In nature the alphaviruses usually produce inapparent viremic infections of birds, mammals, or reptiles, but in domestic animals and humans generalized disease or encephalitis can sometimes result.
Non-arthropod-borne togaviruses include several important animal pathogens. The genus Pestivirus includes bovine virus diarrhea virus, hog cholera virus, and border disease virus of sheep. The genus Arterivirus includes only equine arteritis virus. The genus Rubivirus contains only one species, rubella virus of humans. The ungrouped non-arthropod-borne togaviruses include lactic dehydrogenase virus and simian hemorrhagic fever virus.
Flaviviridae
Formerly classified with the togaviruses, this group (flavi = yellow) has now been given family status, principally because of a distinctly different in strategy of replication. In most other respects, including their transmission by arthropods, the flaviviruses resemble the Alphavirus genus of the family Togaviridae, but they are somewhat smaller (40–50 nm in diameter). There is only one genus, Flavivirus.
Flavivirus diseases of veterinary importance include louping ill, turkey meningoencephalitis, and Wesselsbron disease. Central European tick-borne encephalitis virus produces an inapparent infection in ruminants but may be excreted in the milk and cause disease in humans who drink contaminated milk. Other important human pathogens include the viruses of yellow fever, dengue, and several kinds of arthropod-borne encephalitis.
Orthomyxoviridae
The orthomyxoviruses (myxo = mucus) are spherical or filamentous RNA viruses 80–120 nm in diameter, with a helical nucleocapsid enclosed within an envelope acquired by budding from the plasma membrane. Their genome consists of seven or eight segments of (–) sense ssRNA (total size, 13.6 kb), and is associated with a viral transcriptase. The envelope is studded with peplomers, which are of two kinds: a hemagglutinin and a neuraminidase.
The family includes two important species: influenzavirus A, which infects birds, horses, and swine, as well as humans; and influenzavirus B, which is a human pathogen only. Influenza C virus is a specifically human pathogen, which has seven segments of RNA in its genome, but rarely causes serious disease. Influenza A viruses of animals (swine, birds) and humans undergo genetic reassortment, to generate novel subtypes (“antigenic shift”) which cause major pandemics of influenza in humans.
Paramyxoviridae
The paramyxoviruses have a large, roughly spherical enveloped virion 150–300 nm in diameter, with a helical nucleocapsid. Their genome consists of a single linear molecule of (–) sense ssRNA (18–20 kb), and the virion contains a transcriptase. The envelope contains two glycoproteins: a hemagglutinin (in some species with neuraminidase activity also) and fusion protein.
The family Paramyxoviridae includes the etiological agents of some of the most important veterinary diseases: canine distemper, rinderpest of cattle, and Newcastle disease of chickens. It is subdivided into three genera: Paramyxovirus, Morbillivirus (morbilli = measles), and Pneumovirus (pneumo = lung). Viruses of the genera Paramyxovirus and Pneumovirus cause respiratory infections and occasionally generalized infections [Paramyxovirus: Newcastle disease virus, parainfluenza 1 virus (Sendai virus of mice), parainfluenza 3 virus of cattle, and other parainfluenza viruses of cattle and birds; Pneumovirus: bovine and ovine respiratory syncytial viruses, pneumonia virus of mice]. Viruses of the genus Morbillivirus usually cause generalized infections: rinderpest of cattle, peste-des-petits-ruminants of sheep and goats, and canine distemper, as well as measles.
Coronaviridae
The coronaviruses (corona = crown) are somewhat pleomorphic viruses 75–160 nm in diameter, with widely spaced, pear-shaped peplomers in their lipoprotein envelope. This lacks a matrix protein, and encloses a core of undetermined symmetry with a single linear molecule of (+) sense ssRNA, 17–24 kb. Animal pathogens include calf coronavirus (neonatal diarrhea), transmissible gastroenteritis virus of swine, hemagglutinating encephalomyelitis virus of swine, feline infectious peritonitis virus, infectious bronchitis virus of fowl, and turkey bluecomb virus.
Arenaviridae
Areanviruses (arena = sand) acquired their name because of the presence of ribosomes (resembling grains of sand in thin sections examined with the electron microscope) incorporated within pleomorphic enveloped virions 50–300 nm in diameter, which contain no matrix protein. Their genome consists of two pieces of (–) sense or ambisense ssRNA (total size, 10–14 kb), each held in a circular configuration by “sticky ends,” with an associated transcriptase. All cause natural inapparent infections of rodents.
None of them are of veterinary importance, but humans may occasionally contract a serious generalized disease, e.g., Lassa fever or lymphocytic choriomeningitis. Lassa, Machupo, and Junin viruses are “biosafety level 4” pathogens, i.e., they may be worked with in the laboratory only under maximum biocontainment conditions.
Bunyaviridae
Over 100 bunyaviruses (Bunyamwera is a locality in Africa) comprise the largest single group of arboviruses. The enveloped virions are 90–120 nm in diameter, with a nucleocapsid of helical symmetry. Their genome consists of three pieces of (–) sense or ambisense ssRNA (total size, 13.5–21 kb), each held in a circular configuration, and there is an associated transcriptase. They replicate in the cytoplasm and bud from Golgi membranes. Because of their segmented genome, bunyaviruses readily undergo genetic reassortment, which may produce “antigenic shift.” Some strains have been shown to undergo antigenic drift as well.
All have wild-animal reservoir hosts and some are transovarially transmitted in mosquitoes, with a high frequency. The genus Phlebovirus includes the important pathogen of sheep and humans, Rift Valley fever virus, and the genus Nairovirus includes Nairobi sheep disease virus. Recently it has been shown that the important human disease, hemorrhagic fever with renal syndrome, is caused by a bunyavirus which does not appear to be arthropod-transmitted but is enzootic in rodents.
Retroviridae
This name (sigla: re = reverse; tr = transcriptase) is used for a large family of enveloped viruses 80–100 nm in diameter, with a complex structure and an unusual enzyme, reverse transcriptase. Uniquely among viruses, the genome is diploid, consisting of an inverted dimer of (+) sense ssRNA (2 × 3.5–9 kb). The dsDNA copy of the viral genome transcribed by the viral reverse transcriptase is integrated into the cellular DNA as an essential part of the replication cycle. Proviral DNA is found in the DNA of all normal cells of many species of animals and may under certain circumstances be induced to produce virus. These are known as endogenous retroviruses. Others, called exogenous retroviruses, are transmitted horizontally.
The family Retroviridae is subdivided into three families, two of which contain pathogens of veterinary importance. The subfamily Oncovirinae (onkos = tumor) includes the oncogenic retroviruses: bovine leukemia virus, feline leukemia and sarcoma viruses, baboon, gibbon, and woolly monkey leukemia-sarcoma viruses, as well as avian reticuloendotheliosis, avian leukemia and sarcoma viruses, and the mouse mammary tumor virus. The subfamily Lentivirinae (lenti = slow) includes viruses responsible for several slowly developing, often fatal diseases: maedi–visna and progressive pneumonia of sheep, caprine arthritis-encephalitis, equine infectious anemia, and human acquired immune deficiency syndrome (AIDS). Spumavirinae (spuma = foam) includes the “foamy agents,” which are a problem when they contaminate cultured cells, but are not recognized as pathogens.
Rhabdoviridae
The rhabdoviruses (rhabdos = rod) are bullet-shaped viruses, about 180 × 75 nm, containing a single molecule of (–) sense ssRNA (13–16 kb), which is associated with a transcriptase. Their helical capsid is enclosed within a shell to which is closely applied an envelope with peplomers. The virion matures by budding through the plasma membrane. Animal pathogens in the genus Vesiculovirus include vesicular stomatitis, Chandipura, Piry, and Isfahan viruses. The genus Lyssavirus includes rabies virus and several serologically related agents from Africa. Ungrouped rhabdoviruses that cause diseases in animals include bovine ephemeral fever virus and several rhabdoviruses of fish.
Reoviridae
The family name is a sigla, respiratory enteric orphan virus, reflecting the fact that members of the first discovered genus, Reovirus, were found in both the respiratory and intestinal tract of humans and most animals, but were not associated with any disease. The distinctive feature of the family is that the virions contain dsRNA, in 10, 11, or 12 segments (total size, 17–22 kbp). The virions are nonenveloped icosahedrons 60–80 nm in diameter. There are two other genera, each of which causes infections of veterinary importance: Orbivirus (orbi = ring) and Rotavirus (rota = wheel). Orbiviruses are arboviruses that include bluetongue viruses of sheep, epizootic hemorrhagic disease virus of deer, and African horse-sickness virus. The genus Rotavirus includes viruses that are important causes of diarrhea in domestic animals and humans.
Birnaviridae
This recently named family (sigla: bi = two; rna) contains viruses with nonenveloped virions of icosahedral symmetry, 60 nm in diameter, which replicate in the cytoplasm. The genome consists of two segments of linear dsRNA (total size, 7 kbp). Animal pathogens include infectious bursal disease virus of chickens and infectious pancreatic necrosis virus of fish.
OTHER VIRUSES
There are a number of other viruses that belong to groups that are as yet unclassified. They are the proposed family Filoviridae, the toroviruses, the astroviruses, the unclassified virus of Borna disease, and the still mysterious agents that cause the subacute spongiform encephalopathies, which include scrapie.
Filoviridae
The “biosafety level 4” pathogens, Ebola and Marburg viruses, that cause African hemorrhagic fever of humans, have been tentatively allocated to a new family, Filoviridae. In many respects they resemble rhabdoviruses, but the virions are pleomorphic and sometimes very long (filum = thread), maximun infectivity being associated with a particle 790–970 nm long and 80 nm wide. Their genome is a single molecule of (–) sense ssRNA, 12.7 kb. They are known only by the sporadic infections and occasional nosocomial epidemics produced in humans in Africa.
Toroviruses
The name Toroviridae (torus = object shaped like a donut) has been suggested for hitherto undescribed viruses that are associated with diarrhea in horses (Berne virus) and calves (Breda virus). The virions are enveloped and disk-shaped (35 × 170 nm) and contain a nucleocapsid of presumed helical symmetry. The genome is a single molecule of (+) sense ssRNA, 20 kb.
Astroviruses
Astrovirus (astro = star) is a name accorded unofficially to viruses with small spherical virions with a characteristic star-shaped surface pattern, which have been visualized by electron microscopy in the feces of calves, lambs, and humans. Their genome consists of one molecule of ssRNA about the same size as that of the picornaviruses, but unlike picornaviruses, there are only two capsid proteins.
Borna Disease Virus
An encephalomyelitis of horses and sheep, called Borna disease (Borna is a town in Germany where the disease was first observed) is caused by an unclassified enveloped RNA virus.
Subacute Spongiform Encephalopathies
The causative agents of the subacute spongiform encephalopathies, which include the sheep disease scrapie and the human diseases kuru and Creutzfeldt–Jakob syndrome, remain enigmatic. Their infectivity is highly resistant to inactivation by physical and chemical agents, and they appear to be nonimmunogenic. They may be small and unusual viruses, or they may not be viruses at all, as the term is currently used. All produce slow infections with incubation periods measured in months or even years, followed by progressive disease, which leads inexorably to death from a degenerative condition of the brain characterized by a spongiform appearance.
PROVING A CAUSAL RELATIONSHIP BETWEEN VIRUS AND DISEASE
Before we embark upon the analysis of viral diseases of domestic animals, it will be useful to discuss briefly the problem of establishing causal relationships between a virus and a disease. One of the great landmarks in the scientific study of infectious diseases was the development of what have come to be called the Henle–Koch postulates of causation. They were originally drawn up for bacteria and protozoa, not viruses, and were revised in 1937 by Rivers, who developed a modified set of criteria to cover viruses. With the advent of cell culture for viral diagnosis in the early 1950s, a large number of new viruses were discovered. Many were not associated with disease and were referred to as “orphan” viruses or “viruses in search of disease.” Later the problem arose of determining whether viruses were causally involved in various chronic diseases including cancer, a problem that is still of major interest. Since the relevant disease may not be reproducible by experimental inoculation of animals, virologists have had to evaluate the probability of “guilt by association,” a difficult procedure that relies to a significant degree on epidemiological and serological methods.
Serological criteria for proving a causal relationship between virus and disease were first formulated by Evans (Table 2-3 ) in an assessment of the relationship of EB virus to infectious mononucleosis, a human disease, at a time when there was no method of isolation of the virus.
TABLE 2-3.
Criteria for Causation: A Unified Concept Appropriate for Viruses as Causative Agents, Based on the Henle–Koch Postulatesa
|
From A. S. Evans, Yale J. Biol. Med.49, 175 (1976).
Evans’ serological criteria are difficult to apply. With ubiquitous viruses that only occasionally cause diseases such as tumors or chronic neurological disorders, the presence of antibodies in a population is impossible to interpret; an antibody response might precede the development of the disease by so long a period that the relationship is blurred. Antibody may even play a role in the pathogenesis of the disease, or it might appear to develop after the onset of disease, e.g., in bovine leukemia (Chapter 31). Nevertheless, the criteria set out in Table 2-3 provide a useful guide to investigators, which if followed carefully can minimize the number of false attributions of causation.
FURTHER READING
- Andrewes C.H., Periera H.G., Wildy P. 4th edition. Baillière; London: 1978. “An Atlas of Mammalian. [Google Scholar]
- Brown F. Structural analysis of animal virus genomes. The classification and nomenclature of viruses: Summary of results of meetings of the International Committee on Taxonomy of Viruses in Sendai, September 1984. 1986;25:41. [Google Scholar]
- Fraenkel-Conrat H., Wagner R.R., editors. Plenum Press; New York: 1982–. (“The Viruses”). (a continuing series) [Google Scholar]
- Matthews R.E.F., Wagner R.R., editors. CRC Press; Boca Raton, Florida: 1983. (“A Critical Appraisal of Viral Taxonomy”). [Google Scholar]
- Matsudaira R.E.F. Classification and nomenclature of viruses: Fourth report of the International Committee on Taxonomy of Viruses. Intervirology. 1983;12:1. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- “Portraits of Viruses”—a series of historical reviews edited by F. Fenner and A. J. Gibbs and published in Intervirology, 1979 (11, 137; 201), 1980 (13, 133; 257), 1981 (15, 57; 121; 177), 1982 (18, 1), 1983 (20, 61), 1984 (22, 1; 121), 1986 (25, 117).


