Abstract
Foodborne illness is a serious public health concern. There are over 200 known microbial, chemical, and physical agents that are known to cause foodborne illness. Efforts are made for improved detection, control and prevention of foodborne pathogen in food, and pathogen associated diseases in the host. Several commonly used approaches to control foodborne pathogens include antibiotics, natural antimicrobials, bacteriophages, bacteriocins, ionizing radiations, and heat. In addition, probiotics offer a potential intervention strategy for the prevention and control of foodborne infections. This review focuses on the use of probiotics and bioengineered probiotics to control foodborne pathogens, their antimicrobial actions, and their delivery strategies. Although probiotics have been demonstrated to be effective in antagonizing foodborne pathogens, challenges exist in the characterization and elucidation of underlying molecular mechanisms of action and in the development of potential delivery strategies that could maintain the viability and functionality of the probiotic in the target organ.
Keywords: Probiotics, Bioengineered probiotics, Foodborne pathogens, Prevention, Gut health, Antibiotic resistance, Probiotic action, Immune response, Delivery of probiotics, Safety of probiotics
1. Introduction
Foodborne illness is a serious public health concern. The global burden of foodborne illness is currently unknown. However, the World Health Organization (WHO) reported that in 2005, 1.8 million people died from diarrheal diseases, largely due to contaminated food and water (Greig and Ravel, 2009, Newell et al., 2010). In the United States, the Centers for Diseases Control and Prevention (CDC) estimates that each year there are about 48 million cases of foodborne infections with 128,000 hospitalizations and 3000 deaths (Scallan et al., 2011). There are over 200 known microbial, chemical, or physical agents that can result in illness when consumed (Newell et al., 2010). Of these, microbial source comprising of bacterial, viral, and fungal is of major concern. CDC estimates that of all the foodborne infections, 44% of the hospitalizations and deaths are attributed to 31 known pathogens (Scallan et al., 2011). In light of this serious public health crisis, efforts have been directed toward the detection, control, and prevention of well-recognized foodborne pathogens and diseases in the food chain. It is estimated that a reduction in foodborne illness by 10% would keep about 5 million Americans from getting sick each year (Scallan et al., 2011). With increasing trend in consumer preference for safe and wholesome food, probiotics offer an effective and alternative intervention strategy to control foodborne illnesses.
1.1. Overview of foodborne pathogens and diseases
The etiologic agents for foodborne infections comprise bacterial, viral, parasitic, and fungal (Table 5.1 ). These pathogens have the potential to cause significant morbidity or mortality, and have low infective dose, high virulence potential, ubiquitous in nature, and are stable in food products.
Table 5.1.
List of major foodborne microbial pathogens
| Bacterial | Viral | Parasitic | Fungal |
|---|---|---|---|
| Aeromonas hydrophila | Aichivirus | Cryptosporidium parvum | Alexandrium tamarense |
| Arcobacter butzleri | Astrovirus | Cyclospora cayatenesis | Aspergillus spp. |
| Bacillus cereus/subtilis/licheniformis | Calcivirus (Norovirus) | Entamoeba histolytica | Fusarium spp. |
| Brucella abortus/melitensis/suis | Hepatitis A virus | Giardia intestinalis/lamblia | Microcystis aeruginosa |
| Campylobacter jejuni/coli | Hepatitis E virus | Isospora belli | Penicillium spp. |
| Clostridium botulinum | Rotavirus | Taenia saginata/solium | |
| Clostridium perfringens | Toxoplasma gondii | ||
| Cronobacter sakazakii/malonaticus/turicensis | Trichinella spiralis | ||
| Escherichia coli (pathogenic) | |||
| Listeria monocytogenes | |||
| Mycobacterium paratuberculosis | |||
| Plesiomonas spp. | |||
| Salmonella enterica | |||
| Shigella spp. | |||
| Staphylococcus aureus | |||
| Vibrio cholera/parahaemolyticus/vulnificus/fluvialis | |||
| Yersinia enterocolitica |
Of all the pathogens listed in Table 5.1, CDC estimates that the majority of the illnesses, hospitalizations, and deaths are caused by five known pathogens, which include Norovirus, nontyphoidal Salmonella, Clostridium perfringens, Campylobacter spp., and Staphylococcus aureus.
These pathogens enter into the food system through contaminated raw materials, water, humans, meat animals, wild life, and insect vectors. There are several factors that affect the trends in the occurrence of foodborne illness: large-scale production and wide distribution of food, globalization of food supply, eating outside of the home, microbial genomic diversification yielding the emergence of new pathogens, and growing population of at-risk consumers. The diseases caused by these pathogens have different consequences and sequel (Table 5.2 ).
Table 5.2.
List of common foodborne pathogens, incubation period, symptoms, and possible food sources
| Foodborne pathogens | Possible food source | Incubation period | Symptoms |
|---|---|---|---|
| Bacillus cereus | Meats, milk, rice, potatoes, pasta, vegetables, and cheese | 30 min to 15 h | Diarrhea, abdominal cramps, nausea, and vomiting |
| Campylobacter jejuni | Raw milk, eggs, poultry, raw beef, water, cake icing | 1–7 days | Nausea, abdominal cramps, diarrhea, headache |
| Clostridium botulinum | Low-acid canned foods, meats, sausage, fish | 12–36 h | Nausea, vomiting, dry mouth, diarrhea, fatigue, headache, double vision, slurred speech, respiratory distress, flaccid paralysis |
| Clostridium perfringens | Undercooked meats, roast beef, and gravies | 8–24 h | Abdominal cramps, diarrhea, dehydration |
| Cryptosporidium parvum | Contaminated water or milk, person-to-person transmission, raw or undercooked food | 2–10 days | Watery diarrhea accompanied by mild stomach cramping, nausea, loss of appetite |
| Escherichia coli O157:H7 and Shiga toxin producing E. coli (STEC) | Ground beef, raw milk, undercooked beef, apple, green leafy vegetables | 2–4 days | Hemorrhagic colitis, hemolytic uremic syndrome |
| Giardia lamblia | Contaminated soil, water, food, or surfaces | 1–2 weeks | Diarrhea, loose or watery stool, stomach cramps, and lactose intolerance |
| Hepatitis A | Water, fruits, vegetables, iced drinks, shellfish, and salads | 4–6 weeks | Fever, malaise, nausea, abdominal discomfort, hepatitis, jaundice |
| Listeria monocytogenes | Contaminated vegetables, milk, cheese, meat, sea food, smoked fish, ready-to-eat foods | 2 days to 3 weeks | Meningitis, septicemia, miscarriage, stillbirth, neonatal listeriosis |
| Norwalk, Norwalk-like, or Norovirus | Raw oysters, shellfish, water and ice, salads, frosting, person-to-person contact | 12–60 h | Nausea, vomiting, diarrhea, abdominal cramps |
| Nontyphoidal Salmonella serovars | Meat, poultry, eggs, milk products | 12–24 h | Nausea, diarrhea, abdominal pain, fever, headache, chills, prostration |
| Staphylococcus aureus | Custard or cream-filled baked goods, ham, poultry dressing, gravy, eggs, potato salad, cream sauces, sandwich fillings | 1–6 h | Severe vomiting, diarrhea, abdominal cramping |
| Shigella spp. | Salads, raw vegetables, dairy products, poultry | 12–50 h | Abdominal pain, cramps, fever, vomiting |
| Toxoplasma gondii | Domestic cat, bird or rodent feces, raw or undercooked food | 5–23 days | Swollen lymph glands, fever, headache, muscle aches, abortion in pregnant women. Severe infection in immunocompromised people and unborn babies |
| Vibrio parahaemolyticus/vulnificus | Fish, shellfish, oysters | 4 h to 4 days | Diarrhea, abdominal cramps, nausea, vomiting, headache, fever, and chills |
| Yersinia spp. | Raw milk, chocolate milk, water, pork, raw meats | 1–3 days | Enterocolitis, may mimic appendicitis |
1.2. Various strategies to control pathogens
Foodborne pathogens sicken more than 48 million Americans annually, that is, 1 in 7 in the U.S. population (Scallan et al., 2011). Reducing foodborne illnesses by 10% would keep about 5 million Americans from getting sick every year. Food safety concerns are also elevated because of consumers demand for high quality, low preservatives, and minimally processed convenient ready-to-eat meals. Such foods are highly vulnerable to contamination and heighten public health safety concerns. This has led to the development of several control strategies both by the government and the industry. Strategies to control foodborne infections can be classified as preharvest and postharvest interventions. Traditionally, much of the research effort was aimed at improving the safety of meat products as postslaughter sanitation and product formulations. However, the continual incidence of outbreaks and increase in knowledge about the pathogens have led to the development of preharvest intervention strategies (Doyle & Erickson, 2012). Preharvest intervention step is a logical food safety approach that allows reduced levels of pathogen loads in the incoming raw materials (Callaway, Anderson, et al., 2003, Soon et al., 2011).
Antibiotics are known to alter the microbiological ecology of the intestinal tract (Callaway, Edrington, et al., 2003). This led to the prophylactic use of antibiotics in animal agriculture to control disease and improve animal growth rate and efficiency. With emergence of antibiotic resistance among pathogens, research has focused on the use of naturally occurring antimicrobials as an alternative to control foodborne pathogens in live animals and foods. Antimicrobials comprise organic acids, essential oils, plant extracts, bacteriocins, probiotics, and bacteriophages (Baugher and Klaenhammer, 2011, Callaway, Anderson, et al., 2003, Hassan et al., 2012, Negi, 2012). Chemical rinses using organic acids that are generally recognized as safe such as acetic, lactic, and citric acids are commonly used in the meat industry to rinse animal carcasses and produce (fruits and vegetables) (Sirsat, Muthaiyan, & Ricke, 2009). These acids reduce the pH of the food and hence control the growth of microorganisms. Lactic acid is most effective when applied at higher temperatures and at a concentration of 2–4% (Sirsat et al., 2009). These organic acids are sometimes used in combination with oxidizing agents such as hydrogen peroxide to enhance their antimicrobial efficacies. Essential oils extracted from clove, cinnamon, thyme, and oregano, and their components have been used in the control of foodborne pathogens such as nontyphoidal Salmonella (Johny, Hoagland, & Venkitanarayanan, 2010), Escherichia coli O157:H7 (Amalaradjou et al., 2010), and Listeria monocytogenes in microbiological growth media, on live animals or food systems (Hyldgaard, Mygind, & Meyer, 2012). Unlike antibiotics, essential oils have multifaceted antimicrobial effects thereby making it difficult for the bacteria to develop resistance.
Microbial contamination can also be controlled by the use of microbicidal treatments, such as ionizing radiations, and heating. Application of nonthermal methods such as high hydrostatic pressure, high-intensity pulsed electric fields, oscillating magnetic fields, intense light pulse, photosensitization, or a combination of above (hurdle approach) has also been shown to be effective (Luksienė and Zukauskas, 2009, Morris et al., 2007). One of the most common physical methods of decontamination is irradiation (Radomyski et al., 1994, Smith and Pillai, 2004). Food irradiation destroys the indigenous flora and prolongs shelf life of products during storage. Food is exposed to doses of ionizing radiation sufficient enough to create positive and negative charges to kill bacteria in the food. The type of physical method used and the dosage of the treatment depend on the type of food matrix to be decontaminated.
Biological methods of control include the use of bacteriophages, bacteriocins, and probiotics. In food matrix, these components can be present naturally or added extrinsically. These biological agents can be used at the pre- and postharvest phase to prevent bacterial contamination (Hagens & Loessner, 2007). Bacteriophages are viruses that can infect and kill bacteria and are considered alternatives to antimicrobials for use in the food industry (García, Martínez, Obeso, & Rodríguez, 2008) and for therapeutic application to treat diseases (Fischetti, 2008, Fischetti, 2010, Hanlon, 2007). However, bacteriophages have narrow target spectra, and some can be used only against a particular strain. This high degree of specificity allows phages to be used against targeted microorganisms in a mixed population without affecting the microbial ecosystem (Callaway, Anderson, et al., 2003). Bacteriophages have been used to control foodborne pathogens in farm animals against specific pathogens (Hagens and Loessner, 2010, LeJeune and Wetzel, 2007, Wall et al., 2010). In addition, several phages or phage cocktails have been approved by the FDA for use as food additive in ready-to-eat meat or for application in cattle/poultry prior to slaughter (Hagens & Loessner, 2010).
The use of probiotics, prebiotics, and synbiotics (combination of prebiotics and probiotics) has also gained increased attention in recent years. The use of microflora to reduce pathogen load in the gut is termed as a probiotic strategy (Callaway, Anderson, et al., 2003). Probiotic techniques involve the introduction of a normal microbial population into the gut to provide a nutrient (prebiotic) that is limiting and allows the growth of a specific subset of the gut microflora. The goal of this approach is to fill all the niches available in the gut so as to exclude the establishment of pathogenic microbes (Doyle and Erickson, 2006, Gaggia et al., 2010, Patterson and Burkholder, 2003). Due to the increased concern about the emergence in antibiotic resistance, use of probiotics provides an effective alternative to combat foodborne illnesses (Baugher and Klaenhammer, 2011, Dobson et al., 2012, Hassan et al., 2012).
In addition, vaccine and antibody therapy also offer a viable option to reduce the burden of foodborne illness. Vaccine therapy involves the stimulation of the animal's immune system to limit pathogen colonization. Two types of vaccines can be employed to immunize food animals: the use of killed or inactivated bacterial cells or live attenuated cells. This approach has been used in poultry to reduce the colonization of Salmonella using Salmonella-specific antibodies (De Buck et al., 2005, Tellez et al., 2001). Similar approach in cattle and swine has shown promising results by enhancing immunoglobulin, IgA, IgG, and IgM in serum and by reducing pathogen carriage (House, Bishop, Parry, Dougan and Wain, 2001, House, Wain, et al., 2001, Mastroeni et al., 2001). Along with these intervention strategies, good animal management or good agriculture practices are equally crucial to the production of healthy animals and agricultural products to ensure food safety.
1.3. Antibiotics and bacterial resistance
Antibiotics have been widely used in animal agriculture to control disease and to increase animal growth rate or efficiency. Although antibiotics are used to target specific bacteria, the specificity can sometimes be too narrow to be highly effective. Therefore, in several occasions, broad-spectrum antibiotics are often included in animal rations. Such treatments can disrupt the intestinal microbial ecosystem and can lead to the establishment of opportunistic pathogens. This could also impose deleterious effects on animal health, performance, and food safety. In addition, the use of antibiotics in human and veterinary medicine to treat infectious diseases has led to the rise and spread of antibiotic resistance (Callaway, Anderson, et al., 2003, Callaway, Edrington, et al., 2003). Increased antibiotic-resistant strains can pose a significant public health hazard yielding increased frequency of treatment failures, severity of infection, prolonged duration of sickness, increase in systemic infections, and increased hospitalizations and mortality (Newell et al., 2010). Antibiotic use in plants, animals, and humans for health-promoting purposes can lead to emergence and dissemination of resistant bacteria and resistance genes. Since antibiotic resistance can spread horizontally, the use of antibiotics in one ecological compartment can have a consequence on the resistance status in another (Kruse and Sorum, 1994, Newell et al., 2010).
Food of plant or animal origin can be a source of both antibiotic-resistant bacteria and resistance genes. The presence of antibiotic-resistant bacteria in food presents a direct hazard to food handlers and consumers equally. Additionally, resistant traits can be transferred from bacteria of food origin to human pathogens directly or via a commensal resulting in an indirect hazard (Newell et al., 2010). In addition, antimicrobial resistance can also arise due to continued exposure of bacteria to antimicrobial residues in food. Furthermore, the different routes through which bacteria acquire resistance are complex. There is an increasing evidence of incidence of antibiotic-resistant bacteria in food, highlighting the importance of antibiotic-resistant foodborne pathogens and their infections. Several antibiotic-resistant strains of Salmonella (Threlfall, 2000), Campylobacter (FAO/WHO/OIE, 2003), Shigella, Vibrio, and S. aureus (de Boer et al., 2009), E. coli (Walsh et al., 2008), and Enterococci (FAO/WHO/OIE, 2003) have been reported for foods of plant and animal origin (Newell et al., 2010). Several of these strains have also been reported to be multidrug resistant. Infections caused by such strains are an important health problem. An important step in curbing antimicrobial resistance is the enforcement of prudent use of antimicrobial agents in all sectors of animal and food production (FAO/WHO/OIE, 2007) and to employ alternative strategies to control food pathogens.
1.4. Role of probiotics and their potential contribution to gut health and disease prevention
At present, the interrelationship between diet and health is well established. The traditional role of diet is to provide the nutrients essential for metabolism. However, over the last few decades, this idea has evolved. It is now established that besides meeting metabolic needs, the diet also helps promote health and the state of well-being of the individual. This change in concept of the dietary role of foods has led to the development of a new class of dietary products called functional foods. A food can be considered functional if it beneficially affects target function in the body besides its nutritional value (Figueroa-Gonzalez, Quijano, Ramirez, & Cruz-Guerrero, 2011). Probiotics fall into the category of functional foods (Nagpal et al., 2012). These are nonpathogenic microorganisms that confer a health benefit on the host and prevent some diseases when administered in adequate amounts (Fric, 2007). The widely used probiotics include lactobacilli, bifidobacter, bacilli, yeast, and some nonpathogenic species within the genera of Escherichia (i.e., E. coli Nissle 1917), Enterococcus, and Bacillus. However, the most common probiotics belong to the genera Lactobacillus and Bifidobacterium. Probiotics in general exert beneficial effects in three ways: (i) Provide end products of anaerobic fermentation of carbohydrates such as organic acids that can be utilized by the host. These end products once absorbed into the blood stream are able to influence human mood, energy level, and even cognitive abilities. (ii) Effectively compete with pathogen colonization to exclude them from causing disease, and (iii) stimulate host immune system by producing specific polysaccharides.
These health benefits are generally strain specific (not species- or genera specific); thus in most cases, a cocktail of probiotics is used to gain the most benefits. Probiotics cocktail can also ensure health benefits, in the event, if one strain fails. To control foodborne pathogens, probiotics are either used in competitive exclusion or as defined cultures. Competitive exclusion involves the extrinsic administration of probiotics to food animals for intestinal colonization. Probiotic bacteria also affect the composition and function of intestinal microbial population (O'Toole & Cooney, 2008). The predominant presence of the probiotic in the gut prevents the pathogens access to the ecological niche, interfering with the attachment of pathogens to the gut and subverting the eventual infection process (Gaggia et al., 2010). Besides physical displacement of pathogens, several probiotics also produce bacteriocins, which are antimicrobial peptides that inactivate pathogens. Additionally, probiotics stimulate the immune system and help in mounting protective response against pathogen interaction with host cells (Gill & Prasad, 2008). Probiotic-induced enhanced butyrate production can also increase resistance against diseases through activation of antimicrobial host defense peptide (Floch, 2010, Sunkara et al., 2011).
Although the use of probiotics is promising, there are several challenges for the industrial production of probiotic-based functional foods: (i) improvement in production techniques for large-scale manufacturing; (ii) cost-effective production strategy; (iii) enhancement of probiotic viability during storage, manufacturing, and transit in the gastrointestinal tract; and (iv) development of efficient delivery system. Strategies to overcome these challenges must be multidisciplinary and will help make the process cost-effective and beneficial to both the producers and consumers.
1.5. Safety assessments of probiotics for therapy
Bacteria are deliberately added to food either to enhance product flavor (starter cultures) or to enhance health benefits (functional additives). Probiotic bacteria are used for the prevention or treatment of various diseases; thus, the probiotic organisms must be nonpathogenic to the host prior to its consideration for use. Commonly used probiotic bacteria include members of the genera Lactococcus, Lactobacillus, and Bifidobacterium. Lactic acid bacteria are generally considered safe; however, some species of Lactobacillus and Bacillus have been associated with opportunistic infections in patients with underlying conditions resulting in endocarditis, bacteremia, and liver abscess (Boyle, Robins-Browne, & Tang, 2006). Presumably, the pathogenic property is associated with a specific strain rather than with the species in general. Thus, the strain-specific characterization is essential to prove the absence of pathogenicity.
Prior to the approval of a probiotic for human use, it is essential that the bacteria be screened for potential pathogenicity and virulence traits (Sanders et al., 2010). Providing evidence for the absence of virulence properties is relatively straightforward in elucidating the pathogenic potential. Besides phenotypic characterization, it is also essential to genetically screen potential candidates for use as probiotics. Another critical consideration is the scope for antimicrobial resistance. In addition to being sensitive to antibiotics, it is also essential that the probiotic bacteria do not carry any transferrable antibiotic resistance genes, which can serve as genetic reservoirs for other potentially pathogenic bacteria. Besides acquisition of antibiotic-resistant genes, there is also the risk for uptake of virulence genes from pathogens that coinhabit the intestinal tract at the same time. However, there is no evidence in the literature of such event taking place in the gut. This could partly be due to the transient colonization of the gut by probiotics. Scientific Committee on Animal Nutrition (SCAN) has established a guideline (a decision tree, Fig. 5.1 ) for the approval of a probiotic strain based on antibiotic resistance (SCAN, 2002). Similar screening strategies are also employed for the approval of a probiotic strain for use as a feed additive. Considering all the factors that are essential in the assessment of safety of probiotics, it is paramount that the general conclusion that “probiotics are safe” cannot be broadly made. Prior to the use of a probiotic or probiotic cocktail in foods or dietary supplement, they need to be determined to be safe for the general population. When intended for use as drugs, the safety assessment must balance risk with benefit (Sanders et al., 2010). The FAO has outlined a guideline for approval of a probiotic strain for use in food (Fig. 5.2 ).
Figure 5.1.
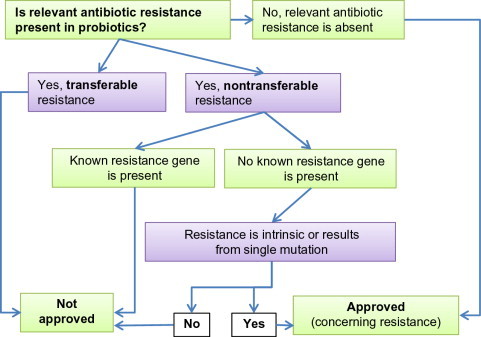
Decision network for approval of a probiotic additive based on resistance to antibiotics (SCAN, 2002).
Redrawn from SCAN (2002).
Figure 5.2.
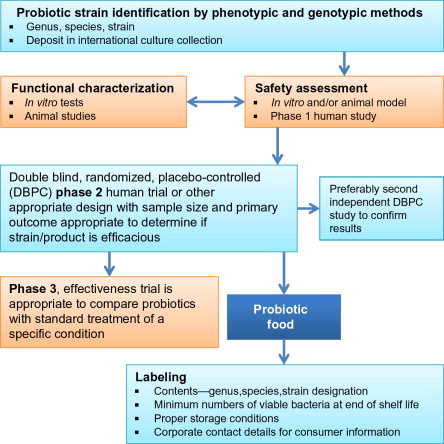
Food and Agriculture Organization of the United Nations (FAO) and World Health Organization (WHO) guidelines for evaluation of probiotics for food use. (ftp://ftp.fao.org/es/esn/food/wgreport2.pdf).
2. Probiotics
The term probiotic means “for life” and is associated with bacteria that exert beneficial effects on humans and animals. This was first observed by Eli Metchnikoff in 1907 who suggested that the dependence of intestinal microbes on food makes it possible for them to develop measures to modify the gut flora and to replace the harmful microbes with useful microbes. The initial works of E. Metchnikoff and H. Tissier in the early twentieth century set the stage for the elucidation of the beneficial effects of probiotics and their multitude of applications in human health as summarized in a recent review article (Bron, van Baarlen, & Kleerebezem, 2012). The increasing body of scientific evidence that demonstrates the beneficial effects of probiotics on health and disease prevention and treatment has made probiotics increasingly important as part of human nutrition and has led to a surge in the demand for probiotics in clinical applications (Deshpande et al., 2011, Ng et al., 2009, Vanderpool et al., 2008) and as functional foods (Ly et al., 2011, Nagpal et al., 2012).
2.1. Definition and classifications
Probiotics have been defined based on their intent of use. Fuller (1989) defined probiotics as “a live microbial feed supplement which beneficially affects the host animal by improving its intestinal balance.” This definition highlighted the microbial nature of probiotics. Similarly, Huis in't Veld, Havenaar, and Marteau (1994) defined probiotics as “a viable mono or mixed culture of bacteria which, when applied to animal or man, beneficially affects the host by improving the properties of the indigenous flora.” A more recent definition accepted by the FAO/WHO (2002) defines probiotics as “live microorganisms which, when administered in adequate amounts confer a health benefit on the host.” These definitions tend to reiterate the basic definition that probiotics are live microorganisms that in adequate dose can be beneficial to humans.
Probiotics can be classified based on their ability to colonize the intestine as resident or transient. Resident strains are those that are common inhabitants of the human digestive tract, and probiotic supplements containing these strains are able to re-establish in the intestinal tract. Resident strains may have least antagonistic effect on other beneficial resident strains in the intestinal tract. Transient strains pass through the system and do not re-establish themselves. Certain transient strains may not be effective when used as monocultures and hence in most cases are combined with other resident strains to enhance their efficacy. Taxonomically, probiotics must be identified by their genus, species, and strain as is done with other bacteria. The commonly used probiotics include the members of the genus Lactobacillus, Bifidobacteria, Streptococcus, Enterococcus, Leuconostoc, and yeast (Saccharomyces). Among these, the resident strains include Lactobacillus acidophilus, Lactobacillus salivarius, Bifidobacterium bifidum, Bifidobacterium infantis, Bifidobacterium longum, Bifidobacterium animalis, Streptococcus faecalis, and Streptococcus faecium. Transient strains include Lactobacillus casei, Lactobacillus rhamnosus GG, Lactobacillus bulgaricus, Lactobacillus yoghurti, Lactobacillus brevis, Lactobacillus kefir, Lactobacillus delbrueckii, Lactobacillus plantarum, Streptococcus lactis, and Streptococcus thermophilus.
2.2. General health benefits of probiotics
Bacteria should possess certain characteristics to be identified as a probiotic. Table 5.3 lists the criteria that are essential for a bacterium to be classified as a probiotic. To be able to produce desired beneficial effects, it has been established that a dose of 5 billion colony forming unit/day has been recommended for at least 5 days (Gronlund et al., 1999, Williams, 2010). Probiotics either as mono or mixed cultures mainly consisting of Lactobacilli have been used for human consumption in a variety of foods such as fermented milks (yogurt), chesses, fruit juices, chocolates, wine, and sausages. Mixed cultures are highly desirable because they may have synergistic effects, and moreover, if one fails, others still can exert beneficial effects.
Table 5.3.
Criteria of an ideal probiotic
|
It is well established that probiotics have several beneficial attributes (Dicks and Botes, 2010, Nagpal et al., 2012, Williams, 2010). Those include lactose metabolism and food digestion, production of antimicrobial peptides and control of enteric infections, antimycotic effects, anticarcinogenic properties, immunologic enhancement, enhancement of short-chain fatty acid (SCFA) production, antiatherogenic and cholesterol-lowering attributes, regulatory role in allergy (Thomas et al., 2011), protection against vaginal or urinary tract infections, increased nutritional value, maintenance of epithelial integrity and barrier, stimulation of repair mechanism in cells, and maintenance and reestablishment of a well-balanced indigenous intestinal and respiratory microbial communities.
2.3. Mechanism of probiotic action
Several mechanisms have been proposed regarding action of probiotics. Some of the major attributes are discussed below (Table 5.4 ).
Table 5.4.
Health benefits of probiotic bacteria and their proposed mechanisms
| Health benefits | Proposed mechanism |
|---|---|
| Resistance to enteric pathogens |
|
| Aid in lactose metabolism |
|
| Small bowel bacterial overgrowth |
|
| Immune system modulation |
|
| Anticolon cancer effect |
|
| Decreased detoxification/excretion of toxic microbial metabolites |
|
| Antiallergic activity (eczema or atopic dermatitis, asthma) |
|
| Blood lipids, heart disease |
|
| Urogenital infections |
|
| Necrotizing enterocolitis |
|
| Rotavirus gastroenteritis |
|
| Inflammatory bowel disease |
|
| Crohn's disease |
|
Adapted from Nagpal et al. (2012)
2.3.1. Enhancing barrier function
The intestinal barrier function is an important defensive mechanism of the intestinal epithelium to maintain its protective effects to protect against invading pathogens and other harmful agents (Ohland & MacNaughton, 2010). The barrier function is maintained by several mechanisms that include mucus secretion, chloride and water secretion, and maintenance of cell–cell tight junctions (Thomas & Ockhuizen, 2012). Disruption in the barrier function can lead to various conditions such as inflammatory bowel disease (IBD), coeliac disease, enteric infections, and other autoimmune diseases (Ng et al., 2009). Several probiotics have been shown to protect the epithelial barrier and prevent mucosal damage triggered by food antigens, enteric pathogens, drugs, and proinflammatory cytokines (O'Hara & Shanahan, 2007). These protective effects are mediated through several mechanisms either directly or indirectly through alteration of gut microflora populations.
The first barrier that the intestinal bacteria and pathogens meet is the mucus. The entire length of the intestinal tract is lined by goblet cells. The percent of goblet cells increase from duodenum (4%) to descending colon (∼ 16%) relative to the epithelial cells (Goto & Kiyono, 2012). Intestinal microflora also regulates the goblet cell populations in the gut. Goblet cells secrete mucin that are resistant to proteolysis and form a protective gel layer over the epithelial surface. Intestinal bacteria and pathogens have to penetrate the mucus layer to reach the epithelial cells during infection. Several microorganisms have developed diverse methods to degrade mucus either to aid in invasion or for uptake of mucus-derived nutrients (Aristoteli and Willcox, 2003, Ohland and MacNaughton, 2010). Additionally, several studies have also reported that the mucus layer is significantly thinner in areas of inflammation thus compromising the barrier and allowing for increased bacterial adherence and infiltration (Swidsinski et al., 2007). Probiotics enhance the barrier by promoting mucus secretion. Several Lactobacillus species have increased mucin expression in in vitro cell culture models and have blocked pathogenic E. coli adherence and invasion (Mack, Ahrne, Hyde, Wei, & Hollingsworth, 2003). Similarly, in vivo experiments in rats fed with probiotic cocktail VSL#3 for 7 days demonstrated a significant increase in mucin secretion (Caballero-Franco, Keller, De Simone, & Chadee, 2007). Thus, enhanced mucus production by probiotics in vitro and in vivo could be used as a protective strategy to augment the intestinal barrier function.
Once the bacteria come across the mucus layer, they find binding sites on the epithelium for colonization/attachment. Pathogenic bacteria, however, proceed to penetrate or damage the epithelium to cause disease. The enterocytes express pattern recognition receptors such as Toll-like receptors (TLRs) that sense the presence of conserved bacterial motifs and initiate cascade of proinflammatory signals (Franchi, Wamer, Viani, & Nunez, 2009). These receptors are present intracellularly and basolaterally on enterocytes. Therefore, only after a pathogen breaches the barrier, they can come in contact with the receptors, which can differentiate between commensal and pathogenic bacteria in the gut (Franchi et al., 2012, Franklin and Latz, 2012). Enterocyte cell–cell adhesion is an essential component of the intestinal barrier. Several components make up the cell–cell junctional complexes such as tight junctions, adherens junctions, gap junctions, and desmosomes (Turner, 2009). These intercellular junctional complexes help maintain the epithelial barrier permeability and its integrity (Groschwitz and Hogan, 2009, Marchiando et al., 2010, Ohland and MacNaughton, 2010). Regulation of tight junctions and the associated epithelial permeability is essential to maintain the epithelial barrier function. However, pathogens have evolved different mechanisms to cross the epithelial barrier demonstrating the critical role the barrier plays in maintenance of homeostasis and prevention of inflammation (Goto & Kiyono, 2012). Chronic inflammation has been shown to be associated with altered tight junction barrier function that can promote pathogen access to the basolateral side of the epithelial barrier.
Several studies have demonstrated that pretreatment with probiotic bacteria can inhibit the loss of permeability associated with tight junction alteration caused by stress, infection, or proinflammatory cytokines (Ait-Belgnaoui et al., 2006, Dahan et al., 2003, Ewaschuk et al., 2008, Sherman et al., 2005). It has been shown that probiotics can directly alter epithelial barrier function by altering the structure and function of tight junctions. Resta-Lenert and Barrett, 2003, Resta-Lenert and Barrett, 2006 found that pretreatment with S. thermophilus and L. acidophilus independently decreased the permeability of cell monolayers formed by intestinal cells of HT-29 and Caco-2. This study also demonstrated that probiotics altered the expression of several proteins that are structural components of tight junctions thereby decreasing permeability. Another study by Yan et al. (2007) demonstrated that certain proteins produced by probiotic bacteria can interact with mammalian cell signaling proteins and lead to alteration in tight junction function and permeability. Two such proteins produced by L. rhamnosus (p40 and p75) inhibited apoptosis in enterocytes and promoted survival. In addition, these proteins also altered structural components of the cell junctional complexes and enhanced barrier function (Table 5.5 ).
Table 5.5.
Summary of probiotics effects on epithelial barrier function in vitro and in vivo
| Barrier function and probiotic | Effect | Model | Reference |
|---|---|---|---|
| Mucous layer | |||
| Lactobacillus | ↑ MUC2 and/or 3 expression | Caco-2, HT29 | Kim et al. (2008), Mattar et al. (2002) |
| VSL#3 | ↑ MUC2, 3, and 5AC expression (no effect on MUC1) | HT29 | Otte and Podolsky (2004) |
| VSL#3 | ↑ MUC1, 2, and 3 expression and secretion | Rat | Caballero-Franco et al. (2007) |
| Tight junctions | |||
| S. thermophilus, L. acidophilus | ↑ TER, ↓ permeability; activation of occludins, ZO-1, ERK 1/2 | HT29, Caco-2 | Resta-Lenert and Barrett (2003) |
| B. infantis | ↑ TER, ↓ permeability; ↑ ZO-1, occludin, ↓ claudin-2 expression; prevent IFN-γ and TNF-α effects | T84 | Ewaschuk et al. (2008) |
| Escherichia coli Nissle | ↑ ZO-1 expression; prevent DSS-induced decrease in permeability and illness | DSS-treated mouse | Ukena et al. (2007) |
| Saccharomyces cerevisiae (Boulardii) | Prevent EHEC-induced apoptosis | T84 | Dalmasso et al. (2006) |
| L. rhamnosus p40, p75 | Inhibit cytokine-induced apoptosis | YAMC, HT29, mouse colon explant | Yan et al. (2007), Yan and Polk (2011) |
| L. rhamnosus p40, p75 | Inhibit H2O2-induced ↓ TER and ↑ permeability | Caco-2, HT29, T84 | Seth, Yan, Polk, and Rao (2008) |
↑, increased; ↓, decreased; EcN, Escherichia coli Nissle; EHEC, enterhemorrhagic E. coli; TER, transepithelial resistance; ZO, zonula occludens; TJ, tight junction; DSS, dextran sodium sulfate. VSL#3 contains four Lactobacillus spp. (L. acidophilus, L. casei, L. plantatarum, L. delbrueckii), three Bifidobacterium spp. (B. infantis, B. longum, B. breve), and one Streptococcus salivarius subsp. thermophilus.
Adapted from Ohland and MacNaughton (2010).
2.3.2. Immunomodulation
The intestine is the first site for foreign antigen encounter. Therefore, the intestine has developed a tightly regulated mechanism to protect against pathogen invasion. The intestinal immune system is made up of several lymphoid organs collectively called as the gut-associated lymphoid tissue. This is the largest collection of lymphoid tissues in the body and consists of the mesenteric lymph nodes, Peyer's patches, isolated lymphoid follicles, lymphocytes, and dendritic cells (Forchielli and Walker, 2005, Hakansson and Molin, 2011, Newberry and Lorenz, 2005). Microbial colonization of the gut affects the composition of immune cell populations. Several studies have demonstrated that bacterial colonization of the gut led to an increase in the number of intraepithelial lymphocytes, germinal centers with antibody-producing cells, and serum antibody concentrations (Hakansson & Molin, 2011). This demonstrates the complex relationship that exists between the intestinal immune system and the gut microbiota (Bron et al., 2012).
Several in vivo and in vitro studies have demonstrated the immunostimulatory effects that probiotics have on the intestinal immune system (Gill & Prasad, 2008). Probiotics and probiotics-derived products are detected by the specialized membranous cells (M cells). Antigens taken up by the M cells are processed by the antigen-processing cells (APCs) and presented to naïve T cells. The type of cytokine secreted, phenotype, and activation of APCs determine the lineage of the T cell that is produced, namely T helper 1 (Th1), T helper 2 (Th2), or the T regulatory (Treg) cells. Activation of Th1 cells leads to production of IFN-γ, TNF-α, and IL-2 which leads to the development of cell-mediated and cytotoxic immunity. Th2 cells activated by APCs mainly secrete IL-4, IL-5, and IL-13 which promote antibody production. Treg cells secrete IL-10 and TGF-β, which downregulate activities of both Th1 and Th2 cells and help maintain homeostasis in the intestine (Gill & Prasad, 2008).
Probiotics have been demonstrated to modulate the innate and acquired immune responses. The innate immune system forms the first line of defense against pathogens. The major components of the innate immune system include epithelial cells, phagocytic cells (monocytes, macrophages, neutrophils), and natural killer cells. Several clinical trials have demonstrated that probiotics enhance the phagocytic activity of peripheral blood leukocytes (Gill, 2003). Healthy subjects administered with L . johnsonii, L. rhamnosus, or B. lactis demonstrated an enhanced phagocytic capacity of peripheral blood leukocytes. The increase in phagocytic ability was also found to be dose dependent and lasted for several weeks after cessation of probiotic intake (Gill & Rutherfurd, 2001b). Probiotics also can activate neutrophils through increased expression of phagocytosis receptors (Pelto, Isolauri, Lilius, Nuutila, & Salminen, 1998) and an increased oxidative burst or microbicidal capacity of leukocytes (Parra, de Morentin, Cobo, Mateos, & Martinez, 2004). Similarly, consumption of probiotics by healthy subjects led to an increase in the number and activity of NK cells and increased phagocytic action in animals fed with probiotic supplements (Cross, 2002). Probiotic-mediated gut health is attributed to the stimulation of epithelial innate immunity. Administration of probiotic mixture VSL#3 exhibited anti-inflammatory effect by stimulation of epithelial-derived TNF-α production and activation of NF-κB (Pagnini et al., 2010).
Besides modulating the innate immune response, probiotics have also been shown to augment the acquired immune responses through induction of cell-mediated immunity. Intake of probiotics has led to an increase in antibody responses to natural infections and immunizations. A randomized trial in children with rotavirus administered with L. rhamnosus GG demonstrated an increase in specific mucosal and serum antibody responses (Kaila et al., 1992). Similarly, administration of probiotics following immunization with Salmonella vaccine in subjects led to a significantly higher specific serum IgA and IgA-mediated cell responses (Linkamster, Rochat, Saudan, Mignot, & Aeschlimann, 1994). These observations indicate that probiotic strains exhibit adjuvant properties increasing the efficacy of antibody production and immune responses to immunizations. Probiotics mediate these effects through increased transport of antigenic materials across the gut mucosa and upregulation of antigen-presenting molecules and costimulatory molecules in immune cells (Gill and Prasad, 2008, Hakansson and Molin, 2011).
An important component of the immune response mediators are cytokines. They are the largest and the most pleiotropic group of mediators. They are responsible for initiation, maintenance, and resolution of innate and acquired immune responses. Several studies have demonstrated that ability of specific probiotic strains to enhance cytokine production and influence both innate and acquired immune responses. Probiotic administration has been reported to enhance levels of IFN-γ, IFN-α, and IL-12 in healthy subjects (Arunachalam, Gill, & Chandra, 2000). Long-term consumption of probiotic containing yogurt has been shown to increase production of IL-1β, IL-6, IL-10, IFN-γ, IL-1, TNF-α, IL-10, IL-12, IL-18, and TGF-β by mononuclear cells and dendritic cells (Cross, 2002, Gill and Guarner, 2004, Niers et al., 2005). Several studies have provided direct evidence that the administration of specific probiotic strains can help in the prevention and treatment of gastrointestinal infections. Studies conducted in animals have also demonstrated the ability of probiotics to enhance serum and mucosal antibodies, phagocytic cell function and NK cell activity, and resistance to infection with pathogens (Gill and Prasad, 2008, Hakansson and Molin, 2011) (Table 5.6 ).
Table 5.6.
Immunomodulatory effects of probiotics
| Immune function and probiotic | Effect | Model | Reference |
|---|---|---|---|
| L. casei | ↑ Levels of IgA + and IL-6-producing cells in the lamina propria | Mouse | Galdeano and Perdigon (2006) |
| L. casei | No change | Monoassociated mouse | Martins et al. (2009) |
| B. animalis, E. coli EMO | ↑ Total sIgA | Monoassociated mouse | Martins et al. (2009) |
| B. bifidum and B. infantis | ↑ Levels of rotavirus-specific sIgA | Mouse | Qiao et al. (2002) |
| B. lactis | ↑ Levels of EHEC-specific sIgA | Mouse | Shu and Gill (2001) |
| L. casei | ↑ Levels of EHEC- or Shiga toxin-specific sIgA leading to increased survival | Rabbit | Ogawa et al. (2001) |
| L. helveticus | ↑ Lamina propria IgA + B cells and sIgA | Rat | LeBlanc, Fliss, and Matar (2004) |
| Saccharomyces cerevisiae (Boulardii) | ↑ Total sIgA | Conventional and monoassociated mouse | Martins et al. (2009) |
↑, increased; ↓, decreased; EHEC, enterhemorrhagic E. coli; sIgA, secretogy IgA.
Adapted from Ohland and MacNaughton (2010).
2.3.3. Antimicrobial action and cytoprotective effects
Probiotics exert their antimicrobial and cytoprotective effects on host intestinal epithelium directly through the production of antimicrobial factors and indirectly through the increase in expression of host cell antimicrobial peptides, enhancement of barrier function, and immunomodulation. This section specifically discusses the antimicrobials produced by probiotics and their antagonistic effect on pathogens. The intestinal cells produce two main classes of antimicrobial peptides, namely, defensins and cathelicidins. Cathelicidins are constitutively produced by intestinal epithelial cells to aid in host defense against pathogens (Kelsall, 2008). The only stimulus that appears to induce cathelicidin production is butyrate that is produced by intestinal flora (Schauber et al., 2003) and also by probiotics (Floch, 2010, Sunkara et al., 2011). Shigella infection (dysentery) in rabbits was significantly reduced by feeding butyrate to induce cathelicidin production (Raqib et al., 2006).
Defensins produced by the intestinal cells can be classified as α-defensins and β-defensins. α-Defensins are produced by small bowel Paneth cells (HD-5 and HD-6), and β-defensins are expressed by epithelial cells throughout the intestine (kBD-1 through 4). These defensins exhibit antimicrobial activity against a wide variety of bacteria, fungi, and viruses. Defensins are constitutively produced in the intestines to keep pathogens from reaching the epithelium (Wehkamp et al., 2004). In vitro studies with probiotics have demonstrated that Caco-2 cells upon stimulation by E. coli Nissle, E. coli strain DSM 17252, and several Lactobacilli in the cocktail VSL#3 led to an increased expression and secretion of human β defensin-2 (hBD-2) (Schlee et al., 2008, Wehkamp et al., 2004). The underlying mechanism through which probiotics stimulated hBD-2 production was elucidated using specific inhibitors. It was observed that hBD-2 secretion was enhanced through activation of MAP kinases (Schlee et al., 2008). Besides activation of MAP kinases, it was also observed that E. coli Nissle flagellin also stimulated hBD-2 production (Schlee et al., 2007).
In addition to stimulating the production of host antimicrobial peptides, probiotics by themselves produce several antimicrobial compounds such as bactericidal peptides and SCFAs that can directly inactivate pathogens (Dobson et al., 2012, Hassan et al., 2012). These secreted factors can be considered an integral part of the intestinal barrier. SCFAs include acetic and lactic acid which reduce the luminal pH resulting in the growth inhibition of some pathogens including enterohemorrhagic E. coli (EHEC) and Salmonella enterica serovar Typhimurium in vitro. Additionally, SCFAs produced by probiotics have also been shown to decrease Shiga toxin gene expression by E. coli O157:H7 (Carey et al., 2008, Fayol-Messaoudi et al., 2005). Furthermore, SCFAs can also disrupt the outer membrane of Gram-negative pathogens such as EHEC, Pseudomonas aeruginosa, and S. Typhimurium thereby inhibiting pathogen growth. Increased permeabilization of the outer membrane of pathogens also potentiates the activity of other antimicrobial molecules by aiding in their penetration of the cell wall (Alakomi et al., 2000). Besides lactic acid, the predominant antimicrobial activity in Lactobacilli is through the production of antimicrobial peptides, bacteriocins, and microcins (Fayol-Messaoudi et al., 2005). Bacteriocins and microcins are peptides with bactericidal or bacteriostatic activity produced in a strain-specific manner by probiotics (Lievin-Le Moal & Servin, 2006). Bacteriocins are peptides produced by Gram-positive bacteria while microcins are produced by Gram-negative bacteria. Bacteriocins permeabilize the cytoplasmic membrane of bacteria leading to disruption of cell wall synthesis and formation of pores eventually leading to cell death. Microcins, on the other hand, target the enzymes that are involved in DNA or RNA structure and synthesis or protein synthesis enzymes (Duquesne, Petit, Peduzzi, & Rebuffat, 2007). Collectively, these antimicrobial compounds protect the intestinal barrier by rapidly eliminating pathogens from the gut.
Bacteriocin ABP-118 produced by L. salivarius has been shown to inhibit the growth of Bacillus, Listeria, Staphylococcus, and Enterococcus species. However, this bacteriocin did not affect the growth of most Lactobacillus species thereby providing a selective advantage for intestinal colonization of probiotic and commensal bacteria (Flynn et al., 2002). Similarly, Lacticin 3147 produced by L. lactis has been shown to Clostridium difficile as a potential therapy; however, this bacteriocin inhibited other resident bacteria including Lactobacillus and Bifidobacteria (Rea et al., 2007). In a separate study, Banerjee, Merkel, and Bhunia (2009) showed that the soluble factors produced by probiotic Lactobacillus delbrueckii subsp. bulgaricus were able to neutralize C. difficile toxin, thereby preventing cytotoxicity in a cell culture model. L. delbrueckii has been demonstrated to produce hydrogen peroxide that can inactivate pathogens by oxidation. L. delbrueckii also produces lactic acid, heat-sensitive, and heat-resistant bacteriocins. The heat-resistant bacteriocin has been observed to inhibit the growth of S. thermophilus (van de Guchte, Ehrlich, & Maguin, 2001). Similarly, certain Bifidobacterium strains produce lipophilic molecules that have been shown to inhibit the viability of E. coli, Klebsiella pneumonia, Yersinia pseudotuberculosis, S. aureus, and S. Typhimurium. Additionally, this antimicrobial compound has also been demonstrated to prevent invasion of Caco-2 cells by S. Typhimurium and can also kill intracellular S. Typhimurium in a therapeutic model (Lievin et al., 2000).
Probiotics also exert their antimicrobial and cytoprotective effect by inhibiting pathogen adherence to the intestinal epithelium (Collado et al., 2009, Sherman et al., 2009). Probiotics inhibit pathogen adherence by competing for the binding sites on the epithelial cells. Pretreatment of HEp-2 and T84 cell lines with L. rhamnosus and L. acidophilus significantly reduced the binding of enteropathogenic E. coli (EPEC) and EHEC to the monolayers. In addition, probiotic pretreatment also reduced EHEC-induced increase in permeability and helped maintain monolayer integrity (Johnson-Henry et al., 2008, Sherman et al., 2005). Probiotic bacteria or its cell surface components also inhibited E. coli O157:H7 adhesion to Caco-2 cells (Johnson-Henry et al., 2007, Medellin-Pena and Griffiths, 2009) or virulence-associated gene expression (Medellin-Pena, Wang, Johnson, Anand, & Griffiths, 2007). Lactobacillus strains have also been shown to compete directly with pathogens such as Salmonella species, for binding sites on human mucins or Caco-2 cell surfaces (Gueimonde, Jalonen, He, Hiramatsu, & Salminen, 2006). Besides physical displacement of pathogens, E. coli Nissle has been shown to secrete a nonbacteriocin component that may act either on the pathogen or on the host cell to inhibit adherence of several pathogens (Altenhoefer et al., 2004). Similar studies performed in a rat model suffering from chronic psychological stress that were administered with L. rhamnosus and L. helveticus demonstrated reduced commensal bacterial adherence and translocation (Zareie et al., 2006). Thus, inhibition of pathogen adherence is another mechanism through which probiotic bacteria prevent intestinal infection. Altogether, probiotic exerts its cytoprotective effect in the intestinal tract through their ability to enhance intestinal barrier function, immune modulation, toxin binding and neutralization, and inactivation and prevention of pathogen attachment (Table 5.7 ).
Table 5.7.
In vivo effect of bacteriocins against enteric pathogens
| Bacteriocin | Producer strain | Animal model | Activity | Reference |
|---|---|---|---|---|
| Bacteriocin B602 | Paenibacillus polymyxa NRRL B-30509 | Chicken | Inhibitory to Campylobacter jejuni | Stern et al. (2005) |
| Mutacin B-Ny266 | Streptococcus mutans Ny266 | Mice | Reduced mortality due to S. aureus | Mota-Meira, Morency, and Lavoie (2005) |
| Enterocin A | Enterococcus faecium EK13 | Japanese quails | Reduced Salmonella concentration and associated intestinal damage | Cigankova, Laukova, Guba, and Nemcova (2004) |
| E-760 | Enterococcus sp. NRRL B-30745 | Broiler chicks | Reduced colonization by C. jejuni | Line et al. (2008) |
| Bacteriocin E 50-52 | Enterococcus faecium NRRL B-30746 | Broilers | Reduced colonization by C. jejuni and Salmonella enteritidis | Svetoch et al. (2008) |
| Bacteriocin OR7 | Lactobacillus salivarius NRRL B-30514 | Turkeys | Reduced Campylobacter concentrations | Cole et al. (2006) |
| Bacteriocin PPB CCM 7420 | Enterococcus faecium CCM 7420 | Rabbits | Reduced coagulase positive Staphylococcus in the cecum | Simonova et al. (2009) |
Adapted from Bogovic-Matijasic and Rogelj (2011).
3. Interaction of Gut Microbiota and Probiotics
When a probiotic is administered orally, it first encounters various harsh environments in the gut. Therefore, it is essential that probiotics survive in the intestinal tract in significant numbers to produce beneficial effects. Survival of the probiotic is dependent on multiple factors such as stress response, metabolism, pH homeogenesis, cell wall maintenance, and fatty acid synthesis (Breton et al., 2002, Sanders, 2011). Gram-positive probiotic bacteria use several mechanisms to help them survive in the gut. These include proton pumps, amino acid decarboxylation, and electrogenic transport systems that aid in acid resistance, changes in the structure of their cell envelop, chaperones involved in repair of damaged proteins, and incremental expression of regulators promoting global gene responses (Cotter & Hill, 2003). In addition, several probiotics have also been shown to modulate their gene expression in vitro under simulated gastrointestinal tract environment or in vivo in mouse model (Bron et al., 2004, Bron et al., 2006).
Once the probiotics survive the transit through stomach and intestine, it interacts with the major component of the gastrointestinal tract that is the resident commensal flora consisting of 500–1000 of different microbial species (Collado et al., 2009). One of the earliest investigations into the interaction between probiotics and the resident microbiota by Tannock et al. (2000) revealed that administration of L. rhamnosus DR20 resulted in modest fluctuations in resident Lactobacillus and Bifidobacterium numbers. Most subjects ceased shedding the probiotic strain once the administration was stopped. However, one subject continued to shed the probiotic strain for over 2 months after the test period, indicating that there are interhost variables such as bacterium–host interactions. Probiotics are commonly applied in companion and farm animals as growth enhancers (Patterson and Burkholder, 2003, Swanson and Fahey, 2006). It has been demonstrated that administration of a cocktail of Lactobacilli, Bifidobacteria, Enterococci, and Pediococci improved weight gain in broiler chickens associated with an increase in Bifidobacterium, Lactobacilli, and Gram-positive cocci populations (Mountzouris et al., 2007). A similar study in piglets administered with Enterococcus faecium strain reduced Enterococcus faecalis population in the intestine of weaning pigs, but the total numbers of E. faecium remained unchanged, suggesting that the E. faecium strain introduced had displaced part of the same species (Vahjen, Taras, & Simon, 2007). Another study performed in mice demonstrated that administration of L. casei and L. plantarum affected the overall diversity of the murine intestinal Lactobacilli but not the overall bacterial community structure (Fuentes et al., 2008). Furthermore, an increase in the population of Lactobacilli related to the acidophilus complex was observed.
Use of animal models provides us with an insight into the interaction between probiotics and the resident flora; however, studies in humans are essential if that species is the desired host. Few studies have been done on humans. Alterations in gut microflora have been reported in humans with irritable bowel syndrome (IBS) (Kassinen et al., 2007, Satokari et al., 2003), and administration of multispecies probiotic supplement was able to alleviate IBS with a stabilization of the gut microbiota over time (Kajander et al., 2007). Besides treating inflammatory conditions, probiotics have also been efficacious in the treatment of infectious diarrhea (Benchimol and Mack, 2004, Guandalini, 2008a, Guandalini, 2008b). There are several ways in which probiotics can alter the microbiota, which include competition for nutrients, changes in microenvironment, production of growth substrates, direct antagonism, competitive exclusion, barrier function, immune stimulation, and reduction of inflammation (O'Toole & Cooney, 2008). Studies using transcriptional microarrays have shown that introducing a probiotic into the mouse gut changed the metabolic pathway of the endogenous microbiota (Sonnenburg, Chen, & Gordon, 2006). Gnotobiotic mice colonized by Bacteroides thetaiotaomicron were challenged with B. animalis or L. casei, resulting in shifts in gene expression pattern of B. thetaiotaomicron. Many of the altered genes were found to be involved in carbohydrate metabolism (Sonnenburg et al., 2006). This suggests that probiotics alter resident microbiota population or gene expression through competition for substrate availability and by altering the dynamics of carbohydrate utilization (Keeney & Finlay, 2011).
Probiotics can also alter the microenvironment of microbiota through a diverse range of metabolic pathway outcomes. Colonization of germ-free mice by microbiota from human baby exposed to Lactobacilli strains resulted in microbiome modification measured by selected culture regimes (Claus et al., 2008). This was also associated with changes in fecal levels of choline, acetate, ethanol, unconjugated bile acids, and cecal concentrations of SCFAs. Besides changing the microenvironment, several probiotic strains are also known to produce vitamins and growth factors. The enhanced availability of such growth factors can also modulate the diversity of intestinal microbiota. Probiotic bacteria also impact the resident microbiota by direct antagonism. Natural competition between probiotics and opportunistic pathogens could also be mediated through the production of bacteriocins that are exploited to modulate microbiota. The indirect mechanisms through which probiotics modulate inherent microbiota include enhancement of intestinal barrier function that can alter release rates of host-derived micronutrients and suppression of proinflammatory cytokines resulting in a reduction of gut inflammation (Zyrek et al., 2007). Reduction in inflammation can alter the gut environment sufficiently to impact on the microbiota. Additionally, administration of probiotics can also bolster the innate and acquired immune responses leading to subtle changes in the overall composition of the gut microbiota (Gill and Rutherfurd, 2001a, Gill and Rutherfurd, 2001c). There is adequate information in literature to indicate that administration of probiotics in high dosage impacts the diversity of gut microflora (Preidis et al., 2012). However, detailed studies are required to understand the interplay of diet, microbiota, and host factors in determining the outcomes to allow its manipulation.
4. Wild-Type and Bioengineered Probiotics to Control Foodborne Enteric Pathogens
Foodborne pathogens are a major concern worldwide due to increased mortality and morbidity (Flint et al., 2005). Thus, various affordable intervention strategies including improved food-processing methods along with probiotic-based natural and functional food systems must be developed to protect people against foodborne infections. Probiotics are live nonpathogenic microorganisms that are administered to maintain and to improve intestinal microbial balance and also to protect the consumers from untoward infection from pathogens. Among the various etiologic agents, bacterial, viral, and mycotoxins are of major concerns. Several studies have demonstrated the efficacy of either wild-type or recombinant probiotics against foodborne pathogens thereby help improving animal health and preventing foodborne infections (Bhunia, 2012, Dobson et al., 2012, Paton et al., 2006, Salminen et al., 2010). This section discusses the various probiotic-based intervention strategies in controlling foodborne pathogens. Mechanism of probiotic-mediated antimicrobial action in gastrointestinal tract is illustrated in Fig. 5.3 .
Figure 5.3.
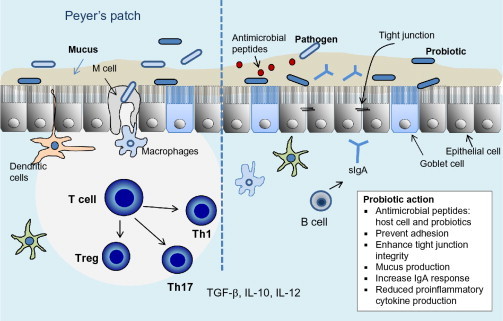
Mechanism of probiotic action against foodborne pathogens in the gastrointestinal tract depicting immunological and cellular responses.
4.1. Probiotics to control bacterial pathogens
4.1.1. E. coli O157:H7
EHEC is commonly implicated in foodborne illness. Control of E. coli O157:H7, a predominant EHEC, is particularly important because of its low infectious dose, acid tolerance, and is harbored in healthy cattle (Ferens and Hovde, 2011, Soon et al., 2011). Among the many intervention strategies investigated, probiotics are found to be effective. Lema, Williams, and Rao (2001) demonstrated the efficacy of several probiotics including L. acidophilus, L. casei, L. fermentum, L. plantarum, and E. faecium in reducing E. coli O157:H7 shedding by sheep. The microbial supplements were fed with freeze-dried fermentation products of the probiotics for a period of 7 weeks. The animal group fed with the probiotic supplement showed a significantly lower numbers of E. coli O157:H7 shedding in the feces and an increase in average daily weight gain and feed conversion. In addition to reducing fecal shedding, administration of Bifidobacteria in mice showed a decrease in Shiga toxin production. Mice that were fed with Bifidobacterium breve had a significant reduction in body weight loss and mortality compared to the control group demonstrating that B. breve is capable of protecting mice from E. coli O157:H7 infection (Asahara et al., 2004). A study conducted by Stephens, Loneragan, Karunasena, and Brashears (2007) also demonstrated that direct-fed probiotics consisting of L. acidophilus at different doses led to a dose-dependent decrease in fecal shedding and presence on the hide. In addition to the use of Lactobacillus, studies using nonpathogenic E. coli such as E. coli 1307 and Nissle strains also inhibited bacterial growth and Shiga toxin production by STEC (Reissbrodt et al., 2009).
Studies using in vitro cell culture models have also been used to elucidate the efficacy of probiotics in controlling E. coli O157:H7 (Sherman et al., 2005). Intestinal cell monolayers (HEp-2 and T84) were exposed to L. acidophilus R0052 and L. rhamnosus R0011 followed by infection with EHEC or EPEC (E. coli O157:H7 or E. coli O127:H6), respectively. Following infection, the adherence and cytotoxicity induced by EHEC and EPEC were examined. Exposure to the probiotic strains significantly reduced pathogen attachment to the monolayers. In addition, the probiotics also protected the monolayers from pathogen-induced loss of transepithelial resistance and tight junction integrity (Sherman et al., 2009). Besides reducing the adherence and cell injury, L. acidophilus cell-free spent media resulted in downregulation of several virulence genes involved in the colonization of EHEC (Medellin-Pena et al., 2007). Similar studies conducted using E. coli O157:H7-infected BALB/c mice demonstrated that preexposure to L. paracasei resulted in an upregulation of dendritic cells and helper T cell, antibody production, and downregulation of proinflammatory cytokines yielding enhanced protection of the intestinal integrity (Tsai, Cheng, & Pan, 2010).
In addition to using wild-type probiotics, Paton et al. (2005) generated a recombinant nonpathogenic E. coli carrying chimeric lipopolysaccharide capable of binding enterotoxin produced by enterotoxigenic E. coli (ETEC). The recombinant probiotic was able to neutralize 93% of enterotoxin in culture lysates of diverse ETEC strains (Paton et al., 2006). Similarly, a recombinant L. casei strain carrying the K88 fimbriae from ETEC was able to reduce the attachment of ETEC to porcine intestinal brush border in a dose-dependent manner and to reduce infection in a mice model (Wen et al., 2012). These studies demonstrate that bioengineered probiotics can be used in the targeted control of specific enteropathogens.
4.1.2. Salmonella
Salmonella infection is the leading cause of foodborne gastroenteritis in humans worldwide and is commonly associated with raw or uncooked poultry and eggs, and fruits and vegetables (Foley et al., 2008, Weill et al., 2006). In the United States, nontyphoidal Salmonella is responsible for 11% of total foodborne illness and 35% of hospitalizations (Scallan et al., 2011). Salmonella utilizes numerous virulence factors to initiate infection and colonizes effectively on the epithelial cells in the gut (Ahmer & Gunn, 2011). Therefore, control strategies have to be applied along the food production chain to prevent the entry into human food supply (Vandeplas, Dubois Dauphin, Beckers, Thonart, & Théwis, 2010). Several studies have demonstrated that inoculation of cultures of one or several probiotic strains into broiler chickens may inhibit Salmonella contamination (Audisio et al., 2000, Higgins et al., 2007, Higgins et al., 2008, Van Coillie et al., 2007). Neonatal broiler chicks were challenged with Salmonella Enteritidis and then challenged with Lactobacillus probiotic culture at different doses orally. Lactobacillus significantly reduced Salmonella incidence in chicks by 85% (Higgins et al., 2008). Similar study conducted in grower pigs challenged with S. enterica serotype Typhimurium and administered with L. plantarum resulted in a reduction in fecal shedding of the pathogen. In addition, probiotic feeding also improved the performance of the pigs (Gebru et al., 2010). L. rhamnosus was also able to reduce epithelial cells stress induced by heat or cytotoxicity induced by S. Typhimurium infection in a cultured epithelial cell model (Burkholder & Bhunia, 2009).
Commercially available probiotic cocktails were evaluated for their ability to inhibit Salmonella colonization in neonatal broiler chickens and turkey poults. Administration of probiotic cultures (FloraMax, IVS-Wynco LLC, Springdale, AR) significantly reduced Salmonella counts in the tonsils and ceca of chickens and poults (Menconi et al., 2011). Furthermore, administration of Lactobacillus reuteri strain that produced reuterin (bacteriocin) significantly reduced Salmonella populations and increased the survival rate in chicks (Zhang, Li, & Li, 2012). Probiotic Bacillus subtilis DSM 17299 was also able to significantly reduce cecal loads of Salmonella (Knap et al., 2011). An in vitro gut fermentation cellular model was used to evaluate the protective effect of probiotics against Salmonella. Addition of Bacillus thermophilus RBL67 to Salmonella in the reactors of the colonic fermentation model revealed a protective effect on epithelial integrity and increased the transepithelial resistance by 58% (Zihler, Gagnon, Chassard, & Lacroix, 2011). Probiotic has also been effective against antibiotic-resistant S. Typhimurium DT104 (Asahara et al., 2011). Administration of L. casei Shirota in mice challenged with S. Typhimurium significantly reduced the pathogen growth and subsequent extraintestinal dissemination. The increase in concentration of organic acids and lowering of pH in the intestine were thought to reduce probiotic colonization, which correlated with the antimicrobial activity (Asahara et al., 2011).
The mechanism underlying the antibacterial effect of Lactobacillus is multifactorial and involves lowering of the pH, production of lactic acid, and production of bacteriocins, nonbacteriocins, and nonlactic compounds (Dobson et al., 2012). It was observed that L. johnsonii La1, L. rhamnosus GG, L. casei Shirota YIT9029, L. casei DN-114001, and L. rhamnosus GR1 dramatically reduced the viability of S. enterica serovar Typhimurium through the production of nonlactic acid molecules, while the complete inhibition of Salmonella growth was observed to be due to a pH-lowering effect (Fayol-Messaoudi et al., 2005). In another study, soluble factors produced by B. bifidum also suppressed gene expressions in S. Typhimurium that are required for adhesion and invasion for systemic spread (Bayoumi & Griffiths, 2012). In vivo study using a mouse model demonstrated that continued administration of L. casei CRL diminished Salmonella counts in the intestine as well as its spread outside this organ. The probiotic-associated immunomodulatory effect involved both the innate and adaptive immune responses. Probiotic administration reduced neutrophil infiltration, activated phagocytic activity, increased IgA + cells, and released sIgA specific to the pathogen in the intestinal fluid (de LeBlanc, Castillo, & Perdigon, 2010). Besides antimicrobial actions, probiotics also increased feed conversion and the performance in chickens and turkey poults.
4.1.3. Campylobacter
In poultry, Campylobacter is another predominant bacterial pathogen that is responsible for numerous outbreaks. Several probiotic strains have been evaluated for their efficacy in controlling Campylobacter. Human colon T84 and embryonic Int-407 epithelial cells were pretreated with Lactobacillus strains and then infected with C. jejuni. It was observed that L. helveticus R0052 reduced C. jejuni invasion into T84 cells and Int-407 cells by 35–41% and 55%, respectively. In addition, L. helveticus R0052 adhered efficiently to the epithelial cells suggesting that the inhibition of pathogen invasion could be due to competitive exclusion (Wine, Gareau, Johnson-Henry, & Sherman, 2009). In in vivo experiments conducted using a defined human microbiota-associated BALB/c mice were orally infected with either C. jejuni or Salmonella and then subsequently challenged with probiotic Lactobacilli and Bifidobacteria. Probiotics were able to enhance colonization resistance by successfully excluding both pathogens from mice and also increased proliferation of lymphocytes against Salmonella antigens. This study indicates that the probiotic administration reversed the immunosuppressive activity of Salmonella in BALB/c mice (Wagner, Johnson, & Rubin, 2009). Baffoni et al. (2012) evaluated the use of synbiotics to control C. jejuni in poultry. Prebiotic galacto-oligosaccharide was used with probiotic B. longum subsp. longum PCB133, and this synbiotic significantly reduced C. jejuni population in poultry feces thereby highlighting the positive effect of employing the synbiotic approach to reduce pathogen loads.
4.1.4. L. monocytogenes
Among the various pathogens causing foodborne illness, hospitalizations, and deaths, Listeria is responsible for 19% of the associated deaths (Scallan et al., 2011). Upon arrival in the gastrointestinal tract, L. monocytogenes invades the intestinal epithelium and disseminates from the mesenteric lymph nodes to the spleen and liver (Vazquez-Boland et al., 2001). During bacteremia, the organism reaches to liver, spleen, gall bladder, brain, and placenta. In the placenta, it can cause villous necrosis and microabscesses resulting in preterm abortion and infection of the fetus leading to stillbirth or neonatal listeriosis (Bakardjiev et al., 2006, Jiao et al., 2011). Several probiotics have been evaluated for their ability to control L. monocytogenes infection. dos Santos et al. (2011) evaluated the monoassociation of L. delbrueckii in gnotobiotic mice and their effect on Listeria colonization. Administration of L. delbrueckii was capable of protecting the mice against death caused by L. monocytogenes and also led to a faster clearance of the bacteria from various organs such as the liver, spleen, peritoneal cavity, and the gut. Additionally, probiotic-fed mice also showed an increase in IFN-γ and IL-10 signifying the role for effector molecules in probiotic-induced cytoprotection. Similar results were obtained in a study where rats were fed with L. casei Shirota strain. In addition to reducing Listeria populations in the gut, spleen, liver, and feces, the probiotic strain also increased cellular immunity as determined by the delayed-type hypersensitivity response against heat-killed L. monocytogenes (de Waard, Garssen, Bokken, & Vos, 2002). Puertollano et al. (2008) evaluated the immunomodulatory effects of L. plantarum against L. monocytogenes infections in mice. Administration of L. plantarum in mice infected with L. monocytogenes resulted in a reduction in the production of proinflammatory cytokines which circumvented Listeria-mediated cytotoxicity. Several species of Lactobacillus and Bifidobacterium were also able to inhibit L. monocytogenes infection in a cell culture model (Corr, Gahan, & Hill, 2007). Later, the anti-infective property of probiotic L. salivarius was shown to be due to the production of a bacteriocin that protected mice from listeriosis when challenged with L. monocytogenes (Corr, Li, et al., 2007). In addition to the use of wild-type probiotics, Koo, Amalaradjou, and Bhunia (2012) generated a recombinant L. paracasei strain to control L. monocytogenes infection in a cell culture model. The recombinant probiotic was designed to express Listeria adhesion protein, an essential virulence factor (Burkholder and Bhunia, 2010, Jagadeesan et al., 2010) aiding Listeria in transepithelial translocation during intestinal phase of infection. Preexposure of intestinal monolayers to the recombinant probiotic followed by Listeria infection led to a reduction in adhesion and paracellular translocation by 44% and 46%, respectively. The recombinant probiotic also protected the monolayers from Listeria-mediated cytotoxicity and tight junction compromise. The use of such recombinant probiotics can help in the targeted elimination of enteric pathogens.
4.1.5. Miscellaneous bacterial pathogens
Probiotics have been evaluated for the control of C. perfringens in turkey poults (Rahimi, Kathariou, Grimes, & Siletzky, 2011). Administration of Primalac, a commercial probiotic cocktail to turkey poults, significantly reduced cecal C. perfringens counts compared to the control. In another study, a recombinant Pichia pastoris containing C. perfringens alpha toxin gene significantly increased weight gain, feed efficiency, sero conversion, and an absence of adverse reactions in histopathological evaluation of broiler chicks (Gil de los Santos, Storch, Fernandes, & Gil-Turnes, 2012). Probiotic bacteria have also been used to control Vibrio parahaemolyticus attachment in cell culture model. L. plantarum AS1 was shown to attach efficiently to HT-29 cells and reduce V. parahaemolyticus attachment by competitive exclusion and displacement (Satish Kumar et al., 2011). A recombinant probiotic expressing a chimeric lipopolysaccharide was able to bind to cholera toxin and protected infant mice challenged with virulent V. cholera (Focareta, Paton, Morona, Cook, & Paton, 2006). Probiotics have also been shown to be effective against other pathogens such as Shigella sonnei, S. aureus, E. faecalis, Proteus mirabilis, and P. aeruginosa (Varma, Dinesh, Menon, & Biswas, 2010).
4.2. Probiotics to control viral pathogens
Probiotics have also been used to control viral infections. Gnotobiotic pigs fed with L. acidophilus and L. reuteri enhanced IFN-γ and IL-4 responses in serum and decreased human Rotavirus infection (Wen et al., 2009). Protection against rotavirus-induced diarrhea was also induced by administration of L. paracasei expressing variable domain of llama heavy-chain antibody fragments (Pant et al., 2006). Another major enteric virus responsible for 58% of foodborne illness is Norovirus (Marshall and Bruggink, 2011, Mattison, 2011). Probiotic-fermented milk containing L. casei Shirota strain has been evaluated for its efficacy in controlling norovirus gastroenteritis in a health service facility. A total of 77 people were enrolled in the study. Intake of probiotic-fermented milk by the treatment group resulted in a reduction in the mean duration of fever after the onset of gastroenteritis (Nagata et al., 2011). Wang, Yu, Gao, and Yang (2012) generated a recombinant Lactobacillus strain expressing the hemagglutinin of the avian influenza virus H5N1. Oral administration of this recombinant probiotic BALB/c mice triggered both mucosal and systemic immune responses. There was an increase in anti-HA IgA and anti-HA IgG levels with an associated increase in IL-4 production. Recombinant L. casei expressing human Lactoferrin was shown to exhibit antibacterial and antiviral activities in in vitro and in vivo models (Chen et al., 2010).
Receptor mimetics has been used as a strategy to control foodborne pathogens. Many enteric pathogens recognize oligosaccharides expressed on host cells as receptors for toxins and adhesion factors. This interaction between the ligand and the receptor is essential for the initiation of infection process. This critical step in the infection process can be adapted to develop intervention strategies. This involves the expression of molecular mimics of host oligosaccharides in probiotic bacteria to control enteric pathogens (Paton, Morona, & Paton, 2010). Such designer probiotics can bind bacterial toxins to competitively inhibit pathogen binding to its receptor in the gut. Such receptor mimic probiotics have been developed against STEC and ETEC (Paton et al., 2005, Paton et al., 2006), Cholera (Focareta et al., 2006), and C. jejuni (Yuki et al., 2004). In addition to the enteric pathogens, probiotics have also been developed against many other pathogens and infectious agents summarized in Table 5.8 .
Table 5.8.
Lactic acid bacteria-based intervention strategies (vaccines)
| Vaccine target | Vehicle | Antigen | Model | Effect | Reference |
|---|---|---|---|---|---|
| Helicobacter pylori | L. plantarum | Urease B | Mouse | Reduction in colonization | Corthesy, Boris, Isler, Grangette, and Mercenier (2005) |
| Tetanus | Lactococcus lactis | Tetanus toxin fragment C | Mouse | Survival after tetanus toxin challenge | Robinson et al. (2004) |
| Streptococcus pneumoniae | L. plantarum and L. helveticus | PsaA | Mouse | Reduction in nasal colonization | Oliveira et al. (2006) |
| ETEC | L. acidophilus | K99 | Pig | Inhibition of adhesion | Tarahomjoo (2012) |
| SARS-associated coronavirus | L. casei | Spike antigen segments | Mouse | Viral neutralizing antibody elicited | Lee et al. (2006) |
| Rotavirus | L. lactis | VP7 | Mouse | Neutralizing antibody against VP7 | Perez, Eichwald, Burrone, and de Mendoza (2005) |
| Group B Streptococcus | L. lactis | Pilus | Mouse | Survival of offspring from vaccinated mothers after infectious challenge | Buccato et al. (2006) |
Adapted from Wells and Mercenier (2008).
4.3. Probiotics to neutralize toxins in food
Fungal pathogens could be controlled by inactivation of their toxins (mycotoxins) that cause foodborne infections. Mycotoxins are of clinical importance due to their ability to induce acute and chronic toxicity in the host (Kabak, Dobson, & Var, 2006). These mycotoxins can be mutagenic, carcinogenic, teratogenic, and immunosuppressive in nature (Salminen et al., 2010). Probiotics are known to bind and neutralize toxins. This attribute of the probiotic has been exploited in the reduction of dietary exposure to fungal toxins. Specific strains of probiotics have proved to be highly effective in removing mycotoxins in model systems. Armando et al. (2012) have demonstrated that Saccharomyces cerevisiae strains can adsorb ochratoxin and zearalenone in a mycotoxin binding assay under simulated gastrointestinal conditions. It was also observed that toxin binding was a function of cell wall thickness and binding occurs through physical adsorption. Similar studies conducted using a food system (grape juice) also demonstrated the ability of Saccharomyces strains to adsorb ochratoxin (Bejaoui, Mathieu, Taillandier, & Lebrihi, 2004).
Rats fed with aflatoxin-containing diet showed that administration of Lactobacilli casei and L. reuteri significantly reduced the oxidative stress induced by aflatoxins in comparison to the control group that did not receive any probiotics (Hathout et al., 2011). Similar experiment conducted in rats fed with different doses of aflatoxin (AFB-1) followed by administration with L. reuteri demonstrated significantly lower levels of AFB-1 in the intestine than in the control group. This study demonstrated that probiotic strains can bind to toxin in the intestine and act as a barrier to toxin-induced cell cytotoxicity (Hernandez-Mendoza, González-Córdova, Vallejo-Cordoba, & Garcia, 2011). Besides aflatoxin, ochratoxin, and zearalenone, other fungal toxins such as patulin and fumonisin can also be neutralized by probiotics. Several probiotic strains such as Propionibacterium, L. rhamnosus GG, L. plantarum, L. casei Bacillus species, and E. faecium were demonstrated to bind and neutralize mycotoxins (Danicke and Doll, 2010, Hernandez-Mendoza et al., 2009, Hernandez-Mendoza et al., 2011, Khanafari et al., 2007, Niderkorn et al., 2009, Topcu et al., 2010).
5. Delivery System for Probiotics to the Gut
When using probiotics for control of infections, it is essential that they survive the acidic gastric pH, the intestinal enzymes, bile, and alkaline pH to be able to exert their protective effect. This is critical when using recombinant probiotic to ensure production of the foreign protein at the target site. In general, probiotic supplements have been delivered as capsules, tablets, or powders. The choice of delivery matrix is also important to ensure probiotics activity and viability (Hajela, Nair, Abraham, & Ganguly, 2012).
Probiotic encapsulation technology (PET) has emerged as the principal method for delivery of probiotics. A wide range of microbes have been immobilized using semipermeable and biocompatible materials that modulate their delivery and release. Probiotics are either entrapped or encapsulated. Entrapment refers to the trapping of material within or throughout a matrix. Encapsulation involves the formation of a continuous coating around the inner matrix that is present within the capsule wall as a core of the encapsulated material. Encapsulation helps to stabilize cells, enhancing their viability and stability during production, storage, and handling. PET helps in the controlled and continuous delivery of probiotic cells in the gut. The biomaterials used in the encapsulation of probiotics include alginate, carrageenan, gelatin, chitosan, whey proteins, cellulose acetate phthalate, locust bean gum, and starches (Anal and Singh, 2007, Chandramouli et al., 2004, Krasaekoopt et al., 2003).
Alginate gels are insoluble in acidic media and hence protect probiotics from gastric acidity (Gbassi, Vandamme, Ennahar, & Marchioni, 2009). Carrageenan is used for its ability to form gels that can entrap the probiotics bacteria (Doleyres et al., 2004, Doleyres et al., 2002). Cellulose acetate phthalate is insoluble at pH below 5 but soluble when pH is greater than 6. This is especially advantageous for the stability of encapsulated probiotic in the stomach and for controlled release in the intestine (Anal & Singh, 2007). Microencapsulation of probiotics is performed by freeze-drying, extrusion, or emulsification. Freeze-drying results in the formation of concentrated dry powder that has longer shelf life but do not protect probiotics from gastric fluid or bile (Rokka & Rantamaki, 2010). It is essential to provide probiotics with physical barriers to protect them from the adverse conditions encountered in the gut. For this purpose, extrusion and emulsification techniques intended to develop gel beads or capsules were applied (Lacroix, Paquin, & Arnaud, 1990). Extrusion technique involves the mixture of probiotics with the hydrocolloid finally resulting in the formation of gelled droplets called beads (Gouin, 2004). Emulsification involves the mixture of probiotics with a continuous phase resulting in a water-in-oil emulsion. This technique has been used to develop encapsulated probiotics (Shima, Morita, Yamashita, & Adachi, 2006). Emulsification results in the formation of oily or aqueous droplets called capsules (Gbassi & Vandamme, 2012).
Alginate microcapsules coated with chitosan were evaluated for their ability to increase gastrointestinal viability of B. breve. Chitosan stabilizes the alginate microcapsules at pH above 3. Coating with chitosan also increased the survival of B. breve in simulated gastric fluid and prolonged its release upon exposure to intestinal pH (Cook, Tzortzis, Charalampopoulos, & Khutoryanskiy, 2011). Calinescu and Mateescu (2008) developed a tablet dosage system based on carboxylated methyl high amylose starch (CM-HAS) and chitosan excipient for probiotic colon delivery. CM-HAS ensures protection in the gastric pH while chitosan helps with slow release of the probiotic in the intestine. This encapsulation of L. rhamnosus resulted in an improved percentage of delivered bacteria in simulated intestinal conditions. To enhance the slow and sustained release of probiotics in the gut, a coacervate extended-release microparticulate delivery system was developed (Alli, 2011). Core mucoadhesive L. rhamnosus was prepared using hypermellose and subsequent enteric coating with hypermellose phthalate. Hypermellose has good mucoadhesive and release rate controlling properties, which are preferred in mucoadhesive formulations. Use of such microparticles in simulated intestinal conditions exhibited appreciable mucoadhesion compared to the freeze-dried cultures.
In addition to the use of the above-mentioned matrices, DNA-based gels have also been evaluated as a vehicle for oral delivery of probiotics. DNA-based gels were initially prepared to protect lactic acid bacteria from extreme conditions in commercial yogurts (Jonganurakkun et al., 2003, Jonganurakkun et al., 2006). The DNA forms complexes with gelling agents such as gelatin and κ-carrageenan at low pH. This resulted in the formation of two kinds of gel: hydrogels and complex gels. In a hydrogel, the bacteria are fixed in the gel when the yogurt containing bacteria is added to the precooled mixture of DNA and gelling agent. In a complex gel, a mixture of yogurt and cacao oil is added to the gel solution containing the DNA and the gelling agent to form the encapsulated probiotic. It was seen that complex gel was able to protect the probiotic from adverse simulated gastric conditions. Hydrogels in addition to protecting from acidic conditions were also more stable during 4 °C storage which corresponds to the storage temperature at which the product would be maintained for prolonged storage (Jonganurakkun et al., 2006). Thus, DNA-based hydrogels offer a potential delivery system for oral administration of probiotics.
6. Conclusion and Future Perspectives
Probiotics have been extensively studied in in vitro and in vivo models. Ample evidence is documented to support the potential application of probiotics for the prevention and treatment of enteric infections. Health benefits of probiotics include the prevention of diarrhea, atopic eczema, antibiotic-associated diarrhea, traveler's diarrhea, prevention of dental carries, colorectal cancers, and treatment of IBD. However, as with any potential intervention strategies, probiotics also have a safety concern. Therefore, it is essential that toxic and metabolic effects of probiotics on humans are assessed for patient safety, especially for critically ill or immunocompromised patients. Significant challenges still exist in the effective application of probiotics in pathogen control. Future studies are essential to define optimal doses and their correct combinations of various probiotic species based on their molecular mode of antimicrobial action. There is also a need for improvement of production techniques to understand and develop better approaches to probiotic delivery and bioavailability in the gut as we move from theoretical benefits to clinical application. Now with the possibility to express different molecules in probiotic bacteria, such as enzymes, cytokines, receptor mimics, adhesion molecules, antibodies, and host targeting molecules, future research can help optimize applications and develop biologically contained strains to support clinical trials. Thus, probiotic bacteria can be used to control and prevent pathogen colonization in the food animals to improve food safety and a realistic therapeutic option in humans to control enteric pathogens.
References
- Ahmer B.M.M., Gunn J.S. Interaction of Salmonella spp. with the intestinal microbiota. Frontiers in Microbiology. 2011;2:101. doi: 10.3389/fmicb.2011.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Belgnaoui A., Han W., Lamine F., Eutamene H., Fioramonti J., Bueno L. Lactobacillus farciminis treatment suppresses stress induced visceral hypersensitivity: A possible action through interaction with epithelial cell cytoskeleton contraction. Gut. 2006;55:1090–1094. doi: 10.1136/gut.2005.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakomi H.L., Skytta E., Saarela M., Mattila-Sandholm T., Latva-Kala K., Helander I.M. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Applied and Environmental Microbiology. 2000;66:2001–2005. doi: 10.1128/aem.66.5.2001-2005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alli S.M.A. Preparation and characterization of a coacervate extended-release microparticulate delivery system for Lactobacillus rhamnosus. International Journal of Nanomedicine. 2011;6:1699–1707. doi: 10.2147/IJN.S19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoefer A., Oswald S., Sonnenborn U., Enders C., Schulze J., Hacker J. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunology and Medical Microbiology. 2004;40:223–229. doi: 10.1016/S0928-8244(03)00368-7. [DOI] [PubMed] [Google Scholar]
- Amalaradjou M.A.R., Baskaran S.A., Ramanathan R., Johny A.K., Charles A.S., Valipe S.R. Enhancing the thermal destruction of Escherichia coli O157:H7 in ground beef patties by trans-cinnamaldehyde. Food Microbiology. 2010;27:841–844. doi: 10.1016/j.fm.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Anal A.K., Singh H. Recent advances in microencapsulation of probiotics for industrial applications and targeted delivery. Trends in Food Science and Technology. 2007;18:240–251. [Google Scholar]
- Aristoteli L.P., Willcox M.D.P. Mucin degradation mechanisms by distinct Pseudomonas aeruginosa isolates in vitro. Infection and Immunity. 2003;71:5565–5575. doi: 10.1128/IAI.71.10.5565-5575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armando M.R., Pizzolitto R.P., Dogi C.A., Cristofolini A., Merkis C., Poloni V.L. Adsorption of ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness. Journal of Applied Microbiology. 2012;113:256–264. doi: 10.1111/j.1365-2672.2012.05331.x. [DOI] [PubMed] [Google Scholar]
- Arunachalam K., Gill H.S., Chandra R.K. Enhancement of natural immune function by dietary consumption of Bifidobacterium lactis (HN019) European Journal of Clinical Nutrition. 2000;54:263–267. doi: 10.1038/sj.ejcn.1600938. [DOI] [PubMed] [Google Scholar]
- Asahara T., Shimizu K., Nomoto K., Hamabata T., Ozawa A., Takeda Y. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157: H7. Infection and Immunity. 2004;72:2240–2247. doi: 10.1128/IAI.72.4.2240-2247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara T., Shimizu K., Takada T., Kado S., Yuki N., Morotomi M. Protective effect of Lactobacillus casei strain Shirota against lethal infection with multi-drug resistant Salmonella enterica serovar Typhimurium DT104 in mice. Journal of Applied Microbiology. 2011;110:163–173. doi: 10.1111/j.1365-2672.2010.04884.x. [DOI] [PubMed] [Google Scholar]
- Audisio M.C., Oliver G., Apella M.C. Protective effect of Enterococcus faecium J96, a potential probiotic strain, on chicks infected with Salmonella pullorum. Journal of Food Protection. 2000;63:1333–1337. doi: 10.4315/0362-028x-63.10.1333. [DOI] [PubMed] [Google Scholar]
- Baffoni L., Gaggìa F., Di Gioia D., Santini C., Mogna L., Biavati B. A Bifidobacterium-based synbiotic product to reduce the transmission of C. jejuni along the poultry food chain. International Journal of Food Microbiology. 2012;157:156–161. doi: 10.1016/j.ijfoodmicro.2012.04.024. [DOI] [PubMed] [Google Scholar]
- Bakardjiev A.I., Theriot J.A., Portnoy D.A. Listeria monocytogenes traffics from maternal organs to the placenta and back. PLoS Pathogens. 2006;2:623–631. doi: 10.1371/journal.ppat.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P., Merkel G.J., Bhunia A.K. Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells. Gut Pathogens. 2009;1:8. doi: 10.1186/1757-4749-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugher J.L., Klaenhammer T.R. Invited review: Application of omics tools to understanding probiotic functionality. Journal of Dairy Science. 2011;94:4753–4765. doi: 10.3168/jds.2011-4384. [DOI] [PubMed] [Google Scholar]
- Bayoumi M.A., Griffiths M.W. In vitro inhibition of expression of virulence genes responsible for colonization and systemic spread of enteric pathogens using Bifidobacterium bifidum secreted molecules. International Journal of Food Microbiology. 2012;156:255–263. doi: 10.1016/j.ijfoodmicro.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Bejaoui H., Mathieu F., Taillandier P., Lebrihi A. Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains. Journal of Applied Microbiology. 2004;97:1038–1044. doi: 10.1111/j.1365-2672.2004.02385.x. [DOI] [PubMed] [Google Scholar]
- Benchimol E.I., Mack D.R. Probiotics in relapsing and chronic diarrhea. Journal of Pediatric Hematology/Oncology. 2004;26:515–517. doi: 10.1097/01.mph.0000133291.11443.a1. [DOI] [PubMed] [Google Scholar]
- Bhunia A.K. Bioengineered probiotics—A solution to broaden probiotics efficacy. Journal of Nutrition and Food Sciences. 2012;2:e105. [Google Scholar]
- Bogovic-Matijasic B., Rogelj I. Springer; New York: 2011. Bacteriocins of probiotics and enteric cytoprotection. [Google Scholar]
- Boyle R.J., Robins-Browne R.M., Tang M.L. Probiotic use in clinical practice: What are the risks? The American Journal of Clinical Nutrition. 2006;83:1256–1264. doi: 10.1093/ajcn/83.6.1256. [DOI] [PubMed] [Google Scholar]
- Breton Y.L., Maze A., Hartke A., Lemarinier S., Auffray Y., Rince A. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Current Microbiology. 2002;45:434–439. doi: 10.1007/s00284-002-3714-9. [DOI] [PubMed] [Google Scholar]
- Bron P.A., Grangette C., Mercenier A., de Vos W.M., Kleerebezem M. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. Journal of Bacteriology. 2004;186:5721–5729. doi: 10.1128/JB.186.17.5721-5729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron P.A., Molenaar D., Vos W.M., Kleerebezem M. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. Journal of Applied Microbiology. 2006;100:728–738. doi: 10.1111/j.1365-2672.2006.02891.x. [DOI] [PubMed] [Google Scholar]
- Bron P.A., van Baarlen P., Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nature Reviews. Microbiology. 2012;10:66–78. doi: 10.1038/nrmicro2690. U90. [DOI] [PubMed] [Google Scholar]
- Buccato S., Maione D., Rinaudo C.D., Volpini G., Taddei A.R., Rosini R. Use of Lactococcus lactis expressing pili from group B Streptococcus as a broad-coverage vaccine against streptococcal disease. The Journal of Infectious Diseases. 2006;194:331–340. doi: 10.1086/505433. [DOI] [PubMed] [Google Scholar]
- Burkholder K., Bhunia A. Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathogens. 2009;1:14. doi: 10.1186/1757-4749-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder K.M., Bhunia A.K. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation, and induces expression of LAP receptor Hsp60. Infection and Immunity. 2010;78:5062–5073. doi: 10.1128/IAI.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Franco C., Keller K., De Simone C., Chadee K. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2007;292:G315–G322. doi: 10.1152/ajpgi.00265.2006. [DOI] [PubMed] [Google Scholar]
- Calinescu C., Mateescu M.A. Carboxymethyl high amylose starch: Chitosan self-stabilized matrix for probiotic colon delivery. European Journal of Pharmaceutics and Biopharmaceutics. 2008;70:582–589. doi: 10.1016/j.ejpb.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Callaway T.R., Anderson R.C., Edrington T.S., Elder R.O., Genovese K.J., Bischoff K.M. Preslaughter intervention strategies to reduce food-borne pathogens in food animals. Journal of Animal Science. 2003;81:E17–E23. doi: 10.2527/2003.812553x. [DOI] [PubMed] [Google Scholar]
- Callaway T.R., Edrington T.S., Rychlik J.L., Genovese K.J., Poole T.L., Jung Y.S. Ionophores: Their use as ruminant growth promotants and impact on food safety. Current Issues in Intestinal Microbiology. 2003;4:43–51. [PubMed] [Google Scholar]
- Carey C.M., Kostrzynska M., Ojha S., Thompson S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157: H7. Journal of Microbiological Methods. 2008;73:125–132. doi: 10.1016/j.mimet.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Chandramouli V., Kailasapathy K., Peiris P., Jones M. An improved method of microencapsulation and its evaluation to protect Lactobacillus spp. in simulated gastric conditions. Journal of Microbiological Methods. 2004;56:27–35. doi: 10.1016/j.mimet.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Chen H.L., Lai Y.W., Chen C.S., Chu T.W., Lin W., Yen C.C. Probiotic Lactobacillus casei expressing human lactoferrin elevates antibacterial activity in the gastrointestinal tract. BioMetals. 2010;23:543–554. doi: 10.1007/s10534-010-9298-0. [DOI] [PubMed] [Google Scholar]
- Cigankova V., Laukova A., Guba P., Nemcova R. Effect of enterocin A on the intestinal epithelium of Japanese quails infected by Salmonella duesseldorf. Bulletin of the Veterinary Institute in Pulawy. 2004;48:25–27. [Google Scholar]
- Claus S.P., Tsang T.M., Wang Y.L., Cloarec O., Skordi E., Martin F.P. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Molecular Systems Biology. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole K., Farnell M.B., Donoghue A.M., Stern N.J., Svetoch E.A., Eruslanov B.N. Bacteriocins reduce Campylobacter colonization and alter gut morphology in turkey poults. Poultry Science. 2006;85:1570–1575. doi: 10.1093/ps/85.9.1570. [DOI] [PubMed] [Google Scholar]
- Collado M.C., Isolauri E., Salminen S., Sanz Y. The impact of probiotic on gut health. Current Drug Metabolism. 2009;10:68–78. doi: 10.2174/138920009787048437. [DOI] [PubMed] [Google Scholar]
- Cook M.T., Tzortzis G., Charalampopoulos D., Khutoryanskiy V.V. Production and evaluation of dry alginate-chitosan microcapsules as an enteric delivery vehicle for probiotic bacteria. Biomacromolecules. 2011;12:2834–2840. doi: 10.1021/bm200576h. [DOI] [PubMed] [Google Scholar]
- Corr S.C., Gahan C.G., Hill C. Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunology and Medical Microbiology. 2007;50:380–388. doi: 10.1111/j.1574-695X.2007.00264.x. [DOI] [PubMed] [Google Scholar]
- Corr S., Li Y., Riedel C.U., O'Toole P.W., Hill C., Gahan C.G.M. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy B., Boris S., Isler P., Grangette C., Mercenier A. Oral immunization of mice with lactic acid bacteria producing Helicobacter pylori urease B subunit partially protects against challenge with Helicobacter felis. The Journal of Infectious Diseases. 2005;192:1441–1449. doi: 10.1086/444425. [DOI] [PubMed] [Google Scholar]
- Cotter P.D., Hill C. Surviving the acid test: Responses of gram-positive bacteria to low pH. Microbiology and Molecular Biology Reviews. 2003;67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M.L. Microbes versus microbes: Immune signals generated by probiotic lactobacilli and their role in protection against microbial pathogens. FEMS Immunology and Medical Microbiology. 2002;34:245–253. doi: 10.1111/j.1574-695X.2002.tb00632.x. [DOI] [PubMed] [Google Scholar]
- Dahan S., Dalmasso G., Imbert V., Peyron J.F., Rampal P., Czerucka D. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infection and Immunity. 2003;71:766–773. doi: 10.1128/IAI.71.2.766-773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso G., Loubat A., Dahan S., Calle G., Rampal P., Czerucka D. Saccharomyces boulardii prevents TNF-alpha-induced apoptosis in EHEC-infected T84 cells. Research in Microbiology. 2006;157:456–465. doi: 10.1016/j.resmic.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Danicke S., Doll S. A probiotic feed additive containing spores of Bacillus subtilis and B. licheniformis does not prevent absorption and toxic effects of the Fusarium toxin deoxynivalenol in piglets. Food and Chemical Toxicology. 2010;48:152–158. doi: 10.1016/j.fct.2009.09.032. [DOI] [PubMed] [Google Scholar]
- de Boer E., Zwartkruis-Nahuis J.T.M., Wit B., Huijsdens X.W., de Neeling A.J., Bosch T. Prevalence of methicillin-resistant Staphylococcus aureus in meat. International Journal of Food Microbiology. 2009;134:52–56. doi: 10.1016/j.ijfoodmicro.2008.12.007. [DOI] [PubMed] [Google Scholar]
- De Buck J., Van Immerseel F., Haesebrouck F., Ducatelle R. Protection of laying hens against Salmonella Enteritidis by immunization with type 1 fimbriae. Veterinary Microbiology. 2005;105:93–101. doi: 10.1016/j.vetmic.2004.10.008. [DOI] [PubMed] [Google Scholar]
- de LeBlanc A.D., Castillo N.A., Perdigon G. Anti-infective mechanisms induced by a probiotic Lactobacillus strain against Salmonella enterica serovar Typhimurium infection. International Journal of Food Microbiology. 2010;138:223–231. doi: 10.1016/j.ijfoodmicro.2010.01.020. [DOI] [PubMed] [Google Scholar]
- de Waard R., Garssen J., Bokken G., Vos J.G. Antagonistic activity of Lactobacillus casei strain Shirota against gastrointestinal Listeria monocytogenes infection in rats. International Journal of Food Microbiology. 2002;73:93–100. doi: 10.1016/s0168-1605(01)00699-7. [DOI] [PubMed] [Google Scholar]
- Deshpande G., Rao S., Patole S. Progress in the field of probiotics: Year 2011. Current Opinion in Gastroenterology. 2011;27:13–18. doi: 10.1097/MOG.0b013e328341373e. [DOI] [PubMed] [Google Scholar]
- Dicks L.M.T., Botes M. Probiotic lactic acid bacteria in the gastro-intestinal tract: Health benefits, safety and mode of action. Beneficial Microbes. 2010;1:11–29. doi: 10.3920/BM2009.0012. [DOI] [PubMed] [Google Scholar]
- Dobson A., Cotter P.D., Ross R.P., Hill C. Bacteriocin production: A probiotic trait? Applied and Environmental Microbiology. 2012;78:1–6. doi: 10.1128/AEM.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doleyres Y., Fliss I., Lacroix C. Continuous production of mixed lactic starters containing probiotics using immobilized cell technology. Biotechnology Progress. 2004;20:145–150. doi: 10.1021/bp020096w. [DOI] [PubMed] [Google Scholar]
- Doleyres Y., Paquin C., LeRoy M., Lacroix C. Bifidobacterium longum ATCC 15707 cell production during free- and immobilized-cell cultures in MRS-whey permeate medium. Applied Microbiology and Biotechnology. 2002;60:168–173. doi: 10.1007/s00253-002-1103-8. [DOI] [PubMed] [Google Scholar]
- dos Santos L., Santos M., de Souza Silva H., Arantes R., Nicoli J., Vieira L. Monoassociation with probiotic Lactobacillus delbrueckii UFV-H2b20 stimulates the immune system and protects germfree mice against Listeria monocytogenes infection. Medical Microbiology and Immunology. 2011;200:29–38. doi: 10.1007/s00430-010-0170-1. [DOI] [PubMed] [Google Scholar]
- Doyle M., Erickson M. Reducing the carriage of foodborne pathogens in livestock and poultry. Poultry Science. 2006;85:960–973. doi: 10.1093/ps/85.6.960. [DOI] [PubMed] [Google Scholar]
- Doyle M.P., Erickson M.C. Opportunities for mitigating pathogen contamination during on-farm food production. International Journal of Food Microbiology. 2012;152:54–74. doi: 10.1016/j.ijfoodmicro.2011.02.037. [DOI] [PubMed] [Google Scholar]
- Duquesne S., Petit V., Peduzzi J., Rebuffat S. Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. Journal of Molecular Microbiology and Biotechnology. 2007;13:200–209. doi: 10.1159/000104748. [DOI] [PubMed] [Google Scholar]
- Ewaschuk J.B., Diaz H., Meddings L., Diederichs B., Dmytrash A., Backer J. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2008;295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- FAO/WHO. (2002). Guidelines for the evaluation of probiotics in food. In Food and agriculture organization of united nations and world health organization. Working Group Report. London, Ontario.
- FAO/WHO/OIE. (2003). Joint FAO/WHO/OIE expert workshop on non-human antimicrobial usage and antimicrobial resistance: scientific assessment. Geneva, Switzerland, December 1–5, 2003. http://whqlibdoc.who.int/hq/2004/WHO_CDS_CPE_ZFK_2004.7.pdf.
- FAO/WHO/OIE. (2007). Joint FAO/WHO/OIE expert meeting on critically important antimicrobials report of the FAO/WHO/OIE expert meeting. FAO Headquarters, Rome, Italy, November 26–30, 2007. ftp://ftp.fao.org/docrep/fao/010/i0204e/i0204e00.pdf.
- Fayol-Messaoudi D., Berger U.N., Coconnier-Polter M.H., Lievin-Le Moal V., Servin A.L. pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against Salmonella enterica serovar Typhimurium. Applied and Environmental Microbiology. 2005;71:6008–6013. doi: 10.1128/AEM.71.10.6008-6013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferens W.A., Hovde C.J. Escherichia coli O157:H7: Animal reservoir and sources of human infection. Foodborne Pathogens and Disease. 2011;8:465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Gonzalez I., Quijano G., Ramirez G., Cruz-Guerrero A. Probiotics and prebiotics—Perspectives and challenges. Journal of the Science of Food and Agriculture. 2011;91:1341–1348. doi: 10.1002/jsfa.4367. [DOI] [PubMed] [Google Scholar]
- Fischetti V.A. Bacteriophage lysins as effective antibacterials. Current Opinion in Microbiology. 2008;11:393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti V.A. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. International Journal of Medical Microbiology. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J.A., Van Duynhoven Y.T., Angulo F.J., DeLong S.M., Braun P., Kirk M. Estimating the burden of acute gastroenteritis, foodborne disease, and pathogens commonly transmitted by food: An international review. Clinical Infectious Diseases. 2005;41:698–704. doi: 10.1086/432064. [DOI] [PubMed] [Google Scholar]
- Floch M.H. The effect of probiotics on host metabolism: The microbiota and fermentation. Journal of Clinical Gastroenterology. 2010;44:S19–S21. doi: 10.1097/MCG.0b013e3181dd4fb7. [DOI] [PubMed] [Google Scholar]
- Flynn S., van Sinderen D., Thornton G.M., Holo H., Nes I.F., Collins J.K. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp salivarius UCC118. Microbiology (UK) 2002;148:973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- Focareta A., Paton J.C., Morona R., Cook J., Paton A.W. A recombinant probiotic for treatment and prevention of cholera. Gastroenterology. 2006;130:1688. doi: 10.1053/j.gastro.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Foley S.L., Lynne A.M., Nayak R. Salmonella challenges: Prevalence in swine and poultry and potential pathogenicity of such isolates. Journal of Animal Science. 2008;86:E149–E162. doi: 10.2527/jas.2007-0464. [DOI] [PubMed] [Google Scholar]
- Forchielli M.L., Walker W.A. The role of gut-associated lymphoid tissues and mucosal defence. The British Journal of Nutrition. 2005;93:S41–S48. doi: 10.1079/bjn20041356. [DOI] [PubMed] [Google Scholar]
- Franchi L., Kamada N., Nakamura Y., Burberry A., Kuffa P., Suzuki S. NLRC4-driven production of IL-1 beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nature Immunology. 2012;13:449–456. doi: 10.1038/ni.2263. U445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Wamer N., Viani K., Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunological Reviews. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin B.S., Latz E. For gut's sake: NLRC4 inflammasomes distinguish friend from foe. Nature Immunology. 2012;13:429–431. doi: 10.1038/ni.2289. [DOI] [PubMed] [Google Scholar]
- Fric P. Probiotics and prebiotics—Renaissance of a therapeutic principle. Central European Journal of Medicine. 2007;2:237–270. [Google Scholar]
- Fuentes S., Egert M., Jimenez-Valera M., Ramos-Cormenzana A., Ruiz-Bravo A., Smidt H. Administration of Lactobacillus casei and Lactobacillus plantarum affects the diversity of murine intestinal lactobacilli, but not the overall bacterial community structure. Research in Microbiology. 2008;159:237–243. doi: 10.1016/j.resmic.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Fuller R. Probiotics in man and animals. The Journal of Applied Bacteriology. 1989;66:365–378. [PubMed] [Google Scholar]
- Gaggia F., Mattarelli P., Biavati B. Probiotics and prebiotics in animal feeding for safe food production. International Journal of Food Microbiology. 2010;141:S15–S28. doi: 10.1016/j.ijfoodmicro.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Galdeano C.M., Perdigon G. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clinical and Vaccine Immunology. 2006;13:219–226. doi: 10.1128/CVI.13.2.219-226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García P., Martínez B., Obeso J.M., Rodríguez A. Bacteriophages and their application in food safety. Letters in Applied Microbiology. 2008;47:479–485. doi: 10.1111/j.1472-765X.2008.02458.x. [DOI] [PubMed] [Google Scholar]
- Gbassi G.K., Vandamme T. Probiotic encapsulation technology: From microencapsulation to release into the gut. Pharmaceutics. 2012;4:149–163. doi: 10.3390/pharmaceutics4010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gbassi G.K., Vandamme T., Ennahar S., Marchioni E. Microencapsulation of Lactobacillus plantarum spp in an alginate matrix coated with whey proteins. International Journal of Food Microbiology. 2009;129:103–105. doi: 10.1016/j.ijfoodmicro.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Gebru E., Lee J.S., Son J.C., Yang S.Y., Shin S.A., Kim B. Effect of probiotic-, bacteriophage-, or organic acid-supplemented feeds or fermented soybean meal on the growth performance, acute-phase response, and bacterial shedding of grower pigs challenged with Salmonella enterica serotype Typhimurium. Journal of Animal Science. 2010;88:3880–3886. doi: 10.2527/jas.2010-2939. [DOI] [PubMed] [Google Scholar]
- Gil de los Santos J.R., Storch O.B., Fernandes C.G., Gil-Turnes C. Evaluation in broilers of the probiotic properties of Pichia pastoris and a recombinant P. pastoris containing the Clostridium perfringens alpha toxin gene. Veterinary Microbiology. 2012;156:448–451. doi: 10.1016/j.vetmic.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Gill H.S. Probiotics to enhance anti-infective defences in the gastrointestinal tract. Best Practice and Research. Clinical Gastroenterology. 2003;17:755–773. doi: 10.1016/s1521-6918(03)00074-x. [DOI] [PubMed] [Google Scholar]
- Gill H.S., Guarner F. Probiotics and human health: A clinical perspective. Postgraduate Medical Journal. 2004;80:516–526. doi: 10.1136/pgmj.2003.008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill H., Prasad J. Probiotics, immunomodulation, and health benefits. In: Bosze Z., editor. Vol. 606. Springer-Verlag; Berlin: 2008. pp. 423–454. (Bioactive components of milk). [DOI] [PubMed] [Google Scholar]
- Gill H.S., Rutherfurd K.J. Viability and dose-response studies on the effects of the immunoenhancing lactic acid bacterium Lactobacillus rhamnosus in mice. The British Journal of Nutrition. 2001;86:285–289. doi: 10.1079/bjn2001402. [DOI] [PubMed] [Google Scholar]
- Gill H.S., Rutherfurd K.J. Immune enhancement conferred by oral delivery of Lactobacillus rhamnosus HN001 in different milk-based substrates. The Journal of Dairy Research. 2001;68:611–616. doi: 10.1017/s0022029901005155. [DOI] [PubMed] [Google Scholar]
- Gill H.S., Rutherfurd K.J. Probiotic supplementation to enhance natural immunity in the elderly: Effects of a newly characterized immunostimulatory strain Lactobacillus rhamnosus HN001 (DR20 (TM)) on leucocyte phagocytosis. Nutrition Research. 2001;21:183–189. [Google Scholar]
- Goto Y., Kiyono H. Epithelial barrier: An interface for the cross-communication between gut flora and immune system. Immunological Reviews. 2012;245:147–163. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- Gouin S. Microencapsulation: Industrial appraisal of existing technologies and trends. Trends in Food Science and Technology. 2004;15:330–347. [Google Scholar]
- Greig J.D., Ravel A. Analysis of foodborne outbreak data reported internationally for source attribution. International Journal of Food Microbiology. 2009;130:77–87. doi: 10.1016/j.ijfoodmicro.2008.12.031. [DOI] [PubMed] [Google Scholar]
- Gronlund M.M., Lehtonen O.P., Eerola E., Kero P. Fecal microflora in healthy infants born by different methods of delivery: Permanent changes in intestinal flora after cesarean delivery. Journal of Pediatrics Gastroenterology and Nutrition. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- Groschwitz K.R., Hogan S.P. Intestinal barrier function: Molecular regulation and disease pathogenesis. The Journal of Allergy and Clinical Immunology. 2009;124:3–20. doi: 10.1016/j.jaci.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guandalini S. Probiotics for children with diarrhea—An update. Journal of Clinical Gastroenterology. 2008;42:S53–S57. doi: 10.1097/MCG.0b013e3181674087. [DOI] [PubMed] [Google Scholar]
- Guandalini S. Acute diarrhea in children in Europe: Do we know how to treat it? Journal of Pediatrics Gastroenterology and Nutrition. 2008;46:S77–S80. doi: 10.1097/MPG.0b013e31816f7ad2. [DOI] [PubMed] [Google Scholar]
- Gueimonde M., Jalonen L., He F., Hiramatsu M., Salminen S. Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Research International. 2006;39:467–471. [Google Scholar]
- Hagens S., Loessner M. Application of bacteriophages for detection and control of foodborne pathogens. Applied Microbiology and Biotechnology. 2007;76:513–519. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- Hagens S., Loessner M.J. Bacteriophage for biocontrol of foodborne pathogens: Calculations and considerations. Current Pharmaceutical Biotechnology. 2010;11:58–68. doi: 10.2174/138920110790725429. [DOI] [PubMed] [Google Scholar]
- Hajela N., Nair G., Abraham P., Ganguly N. Health impact of probiotics—Vision and opportunities. Gut Pathogens. 2012;4:1–6. doi: 10.1186/1757-4749-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson A., Molin G. Gut microbiota and inflammation. Nutrients. 2011;3:637–682. doi: 10.3390/nu3060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon G.W. Bacteriophages: An appraisal of their role in the treatment of bacterial infections. International Journal of Antimicrobial Agents. 2007;30:118–128. doi: 10.1016/j.ijantimicag.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Hassan M., Kjos M., Nes I.F., Diep D.B., Lotfipour F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. Journal of Applied Microbiology. 2012 doi: 10.1111/j.1365-2672.2012.05338.x. [DOI] [PubMed] [Google Scholar]
- Hathout A.S., Mohamed S.R., El-Nekeety A.A., Hassan N.S., Aly S.E., Abdel-Wahhab M.A. Ability of Lactobacillus casei and Lactobacillus reuteri to protect against oxidative stress in rats fed aflatoxins-contaminated diet. Toxicon. 2011;58:179–186. doi: 10.1016/j.toxicon.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hernandez-Mendoza A., Garcia H.S., Steele J.L. Screening of Lactobacillus casei strains for their ability to bind aflatoxin B-1. Food and Chemical Toxicology. 2009;47:1064–1068. doi: 10.1016/j.fct.2009.01.042. [DOI] [PubMed] [Google Scholar]
- Hernandez-Mendoza A., González-Córdova A.F., Vallejo-Cordoba B., Garcia H.S. Effect of oral supplementation of Lactobacillus reuteri in reduction of intestinal absorption of aflatoxin B1 in rats. Journal of Basic Microbiology. 2011;51:263–268. doi: 10.1002/jobm.201000119. [DOI] [PubMed] [Google Scholar]
- Higgins J.P., Higgins S.E., Vicente J.L., Wolfenden A.D., Tellez G., Hargis B.M. Temporal effects of lactic acid bacteria probiotic culture on Salmonella in neonatal broilers. Poultry Science. 2007;86:1662–1666. doi: 10.1093/ps/86.8.1662. [DOI] [PubMed] [Google Scholar]
- Higgins S.E., Higgins J.P., Wolfenden A.D., Henderson S.N., Torres-Rodriguez A., Tellez G. Evaluation of a Lactobacillus-based probiotic culture for the reduction of Salmonella enteritidis in neonatal broiler chicks. Poultry Science. 2008;87:27–31. doi: 10.3382/ps.2007-00210. [DOI] [PubMed] [Google Scholar]
- House D., Bishop A., Parry C., Dougan G., Wain J. Typhoid fever: Pathogenesis and disease. Current Opinion in Infectious Diseases. 2001;14:573–578. doi: 10.1097/00001432-200110000-00011. [DOI] [PubMed] [Google Scholar]
- House D., Wain J., Ho V.A., Diep T.S., Chinh N.T., Bay P.V. Serology of typhoid fever in an area of endemicity and its relevance to diagnosis. Journal of Clinical Microbiology. 2001;39:1002–1007. doi: 10.1128/JCM.39.3.1002-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in't Veld J.H.J., Havenaar R., Marteau P. Establishing a scientific basis for probiotic R&D. Trends in Biotechnology. 1994;12:6–8. doi: 10.1016/0167-7799(94)90004-3. [DOI] [PubMed] [Google Scholar]
- Hyldgaard M., Mygind T., Meyer R.L. Essential oils in food preservation: Mode of action, synergies, and interactions with food matrix components. Frontiers in Microbiology. 2012;3:12. doi: 10.3389/fmicb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesan B., Koo O.K., Kim K.P., Burkholder K.M., Mishra K.K., Aroonnual A. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology (UK) 2010;156:2782–2795. doi: 10.1099/mic.0.036509-0. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Zhang W., Ma J., Wen C., Wang P., Wang Y. Early onset of neonatal listeriosis. Pediatrics International. 2011;53:1034–1037. doi: 10.1111/j.1442-200X.2011.03442.x. [DOI] [PubMed] [Google Scholar]
- Johnson-Henry K.C., Donato K.A., Shen-Tu G., Gordanpour A., Sherman P.A. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157: H7-induced changes in epithelial barrier function. Infection and Immunity. 2008;76:1340–1348. doi: 10.1128/IAI.00778-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Henry K.C., Hagen K.E., Gordonpour M., Tompkins T.A., Sherman P.M. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157: H7 adhesion to epithelial cells. Cellular Microbiology. 2007;9:356–367. doi: 10.1111/j.1462-5822.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- Johny A.K., Hoagland T., Venkitanarayanan K. Effect of subinhibitory concentrations of plant-derived molecules in increasing the sensitivity of multidrug-resistant Salmonella enterica serovar Typhimurium DT104 to antibiotics. Foodborne Pathogens and Disease. 2010;7:1165–1170. doi: 10.1089/fpd.2009.0527. [DOI] [PubMed] [Google Scholar]
- Jonganurakkun B., Liu X.D., Nodasaka Y., Nomizu M., Nishi N. Survival of lactic acid bacteria in simulated gastrointestinal juice protected by a DNA-based complex gel. Journal of Biomaterials Science, Polymer Edition. 2003;14:1269–1281. doi: 10.1163/156856203322553482. [DOI] [PubMed] [Google Scholar]
- Jonganurakkun B., Nodasaka Y., Sakairi N., Nishi N. DNA-based gels for oral delivery of probiotic bacteria. Macromolecular Bioscience. 2006;6:99–103. doi: 10.1002/mabi.200500199. [DOI] [PubMed] [Google Scholar]
- Kabak B., Dobson A.D.W., Var I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Critical Reviews in Food Science and Nutrition. 2006;46:593–619. doi: 10.1080/10408390500436185. [DOI] [PubMed] [Google Scholar]
- Kaila M., Isolauri E., Soppi E., Virtanen E., Laine S., Arvilommi H. Enhancement of the circulating antibody secreting cell response in human diarrhea by a human Lactobacillus strain. Pediatric Research. 1992;32:141–144. doi: 10.1203/00006450-199208000-00002. [DOI] [PubMed] [Google Scholar]
- Kajander K., Krogius-Kurikka L., Rinttila T., Karjalainen H., Palva A., Korpela R. Effects of multispecies probiotic supplementation on intestinal microbiota in irritable bowel syndrome. Alimentary Pharmacology and Therapeutics. 2007;26:463–473. doi: 10.1111/j.1365-2036.2007.03391.x. [DOI] [PubMed] [Google Scholar]
- Kassinen A., Krogius-Kurikka L., Makivuokko H., Rinttila T., Paulin L., Corander J. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Keeney K.M., Finlay B.B. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Current Opinion in Microbiology. 2011;14:92–98. doi: 10.1016/j.mib.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall B.L. Innate and adaptive mechanisms to control pathological intestinal inflammation. Journal of Pathology. 2008;214:242–259. doi: 10.1002/path.2286. [DOI] [PubMed] [Google Scholar]
- Khanafari A., Soudi H., Miraboulfathi M., Osboo R.K. An in vitro investigation of aflatoxin B1 biological control by Lactobacillus plantarum. Pakistan Journal of Biological Sciences. 2007;10:2553–2556. doi: 10.3923/pjbs.2007.2553.2556. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Kim J.S., Kim Y.J., Oh Y.K., Kim I.Y., Chee Y.J. Conjugated linoleic acids produced by Lactobacillus dissociates IKK-gamma and Hsp90 complex in Helicobacter pylori-infected gastric epithelial cells. Laboratory Investigation. 2008;88:541–552. doi: 10.1038/labinvest.2008.16. [DOI] [PubMed] [Google Scholar]
- Knap I., Kehlet A.B., Bennedsen M., Mathis G.F., Hofacre C.L., Lumpkins B.S. Bacillus subtilis (DSM17299) significantly reduces Salmonella in broilers. Poultry Science. 2011;90:1690–1694. doi: 10.3382/ps.2010-01056. [DOI] [PubMed] [Google Scholar]
- Koo O.K., Amalaradjou M.A.R., Bhunia A.K. Recombinant probiotic expressing Listeria adhesion protein attenuates Listeria monocytogenes virulence in vitro. PLoS One. 2012;7:e29277. doi: 10.1371/journal.pone.0029277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasaekoopt W., Bhandari B., Deeth H. Evaluation of encapsulation techniques of probiotics for yoghurt. International Dairy Journal. 2003;13:3–13. [Google Scholar]
- Kruse H., Sorum H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Applied and Environmental Microbiology. 1994;60:4015–4021. doi: 10.1128/aem.60.11.4015-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix C., Paquin C., Arnaud J.P. Batch fermentation with entrapped growing cells of Lactobacillus casei optimization of the rheological properties of the entrapment gel matrix. Applied Microbiology and Biotechnology. 1990;32:403–408. [Google Scholar]
- LeBlanc J., Fliss I., Matar C. Induction of a humoral immune response following an Escherichia coli O157: H7 infection with an immunomodulatory peptidic fraction derived from Lactobacillus helveticus-fermented milk. Clinical and Diagnostic Laboratory Immunology. 2004;11:1171–1181. doi: 10.1128/CDLI.11.6.1171-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Poo H., Han D.P., Hong S.P., Kim K., Cho M.W. Mucosal immunization with surface-displayed severe acute respiratory syndrome coronavirus spike protein on Lactobacillus casei induces neutralizing antibodies in mice. Journal of Virology. 2006;80:4079–4087. doi: 10.1128/JVI.80.8.4079-4087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeJeune J.T., Wetzel A.N. Preharvest control of Escherichia coli O157 in cattle. Journal of Animal Science. 2007;85:E73–E80. doi: 10.2527/jas.2006-612. [DOI] [PubMed] [Google Scholar]
- Lema M., Williams L., Rao D.R. Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157: H7 in lambs by feeding microbial feed supplement. Small Ruminant Research. 2001;39:31–39. doi: 10.1016/s0921-4488(00)00168-1. [DOI] [PubMed] [Google Scholar]
- Lievin V., Peiffer I., Hudault S., Rochat F., Brassart D., Neeser J.R. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievin-Le Moal V., Servin A.L. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: Mucins, antimicrobial peptides, and microbiota. Clinical Microbiology Reviews. 2006;19:315–337. doi: 10.1128/CMR.19.2.315-337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line J.E., Svetoch E.A., EruslanoV B.V., Perelygin V.V., Mitsevich E.V., Mitsevich I.P. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrobial Agents and Chemotherapy. 2008;52:1094–1100. doi: 10.1128/AAC.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkamster H., Rochat F., Saudan K.Y., Mignot O., Aeschlimann J.M. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunology and Medical Microbiology. 1994;10:55–63. doi: 10.1111/j.1574-695X.1994.tb00011.x. [DOI] [PubMed] [Google Scholar]
- Luksienė Z., Zukauskas A. Prospects of photosensitization in control of pathogenic and harmful micro-organisms. Journal of Applied Microbiology. 2009;107:1415–1424. doi: 10.1111/j.1365-2672.2009.04341.x. [DOI] [PubMed] [Google Scholar]
- Ly N.P., Litonjua A., Gold D.R., Celedon J.C. Gut microbiota, probiotics, and vitamin D: Interrelated exposures influencing allergy, asthma, and obesity? The Journal of Allergy and Clinical Immunology. 2011;127:1087–1094. doi: 10.1016/j.jaci.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D.R., Ahrne S., Hyde L., Wei S., Hollingsworth M.A. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52:827–833. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiando A.M., Graham W.V., Turner J.R. Epithelial barriers in homeostasis and disease. Annual Review of Pathology: Mechanisms of Disease. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- Marshall J.A., Bruggink L.D. The dynamics of norovirus outbreak epidemics: Recent insights. International Journal of Environmental Research and Public Health. 2011;8:1141–1149. doi: 10.3390/ijerph8041141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F.S., Silva A.A., Vieira A.T., Barbosa F.H.F., Arantes R.M.E., Teixeira M.M. Comparative study of Bifidobacterium animalis, Escherichia coli, Lactobacillus casei and Saccharomyces boulardii probiotic properties. Archives of Microbiology. 2009;191:623–630. doi: 10.1007/s00203-009-0491-x. [DOI] [PubMed] [Google Scholar]
- Mastroeni P., Chabalgoity J.A., Dunstan S.J., Maskell D.J., Dougan G. Salmonella: Immune responses and vaccines. The Veterinary Journal. 2001;161:132–164. doi: 10.1053/tvjl.2000.0502. [DOI] [PubMed] [Google Scholar]
- Mattar A.F., Teitelbaum D.H., Drongowski R.A., Yongyi F., Harmon C.M., Coran A.G. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatric Surgery International. 2002;18:586–590. doi: 10.1007/s00383-002-0855-7. [DOI] [PubMed] [Google Scholar]
- Mattison K. Norovirus as a foodborne disease hazard. Advances in Food and Nutrition Research. 2011;62:1–39. doi: 10.1016/B978-0-12-385989-1.00001-6. [DOI] [PubMed] [Google Scholar]
- Medellin-Pena M.J., Griffiths M.W. Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Applied and Environmental Microbiology. 2009;75:1165–1172. doi: 10.1128/AEM.01651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medellin-Pena M.J., Wang H.F., Johnson R., Anand S., Griffiths M.W. Probiotics affect virulence-related gene expression in Escherichia coli O157: H7. Applied and Environmental Microbiology. 2007;73:4259–4267. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi A., Wolfenden A.D., Shivaramaiah S., Terraes J.C., Urbano T., Kuttel J. Effect of lactic acid bacteria probiotic culture for the treatment of Salmonella enterica serovar Heidelberg in neonatal broiler chickens and turkey poults. Poultry Science. 2011;90:561–565. doi: 10.3382/ps.2010-01220. [DOI] [PubMed] [Google Scholar]
- Morris C., Brody A.L., Wicker L. Non-thermal food processing/preservation technologies: A review with packaging implications. Packaging Technology and Science. 2007;20:275–286. [Google Scholar]
- Mota-Meira M., Morency H., Lavoie M.C. In vivo activity of mutacin B-Ny266. The Journal of Antimicrobial Chemotherapy. 2005;56:869–871. doi: 10.1093/jac/dki295. [DOI] [PubMed] [Google Scholar]
- Mountzouris K.C., Tsirtsikos P., Kalamara E., Nitsch S., Schatzmayr G., Fegeros K. Evaluation of the efficacy of a Probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora composition and metabolic activities. Poultry Science. 2007;86:309–317. doi: 10.1093/ps/86.2.309. [DOI] [PubMed] [Google Scholar]
- Nagata S., Asahara T., Ohta T., Yamada T., Kondo S., Bian L. Effect of the continuous intake of probiotic-fermented milk containing Lactobacillus casei strain Shirota on fever in a mass outbreak of norovirus gastroenteritis and the faecal microflora in a health service facility for the aged. The British Journal of Nutrition. 2011;106:549–556. doi: 10.1017/S000711451100064X. [DOI] [PubMed] [Google Scholar]
- Nagpal R., Kumar A., Kumar M., Behare P.V., Jain S., Yadav H. Probiotics, their health benefits and applications for developing healthier foods: A review. FEMS Microbiology Letters. 2012 doi: 10.1111/j.1574-6968.2012.02593.x. in press. [DOI] [PubMed] [Google Scholar]
- Negi P.S. Plant extracts for the control of bacterial growth: Efficacy, stability and safety issues for food application. International Journal of Food Microbiology. 2012;156:7–17. doi: 10.1016/j.ijfoodmicro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Newberry R.D., Lorenz R.G. Organizing a mucosal defense. Immunological Reviews. 2005;206:6–21. doi: 10.1111/j.0105-2896.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- Newell D.G., Koopmans M., Verhoef L., Duizer E., Aidara-Kane A., Sprong H. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. International Journal of Food Microbiology. 2010;139(Suppl.):S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.C., Hart A.L., Kamm M.A., Stagg A.J., Knight S.C. Mechanisms of action of probiotics: Recent advances. Inflammatory Bowel Diseases. 2009;15:300–310. doi: 10.1002/ibd.20602. [DOI] [PubMed] [Google Scholar]
- Niderkorn V., Morgavi D.P., Aboab B., Lemaire M., Boudra H. Cell wall component and mycotoxin moieties involved in the binding of fumonisin B(1) and B(2) by lactic acid bacteria. Journal of Applied Microbiology. 2009;106:977–985. doi: 10.1111/j.1365-2672.2008.04065.x. [DOI] [PubMed] [Google Scholar]
- Niers L.E.M., Timmerman H.M., Rijkers G.T., van Bleek G.M., van Uden N.O.P., Knol E.F. Identification of strong interleukin-10 inducing lactic acid bacteria which down-regulate T helper type 2 cytokines. Clinical and Experimental Allergy. 2005;35:1481–1489. doi: 10.1111/j.1365-2222.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Shimizu K., Nomoto K., Takahashi M., Watanuki M., Tanaka R. Protective effect of Lactobacillus casei strain Shirota on Shiga toxin-producing Escherichia coli O157: H7 infection in infant rabbits. Infection and Immunity. 2001;69:1101–1108. doi: 10.1128/IAI.69.2.1101-1108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara A.M., Shanahan F. Gut microbiota: Mining for therapeutic potential. Clinical Gastroenterology and Hepatology. 2007;5:274–284. doi: 10.1016/j.cgh.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Ohland C.L., MacNaughton W.K. Probiotic bacteria and intestinal epithelial barrier function. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2010;298:G807–G819. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- Oliveira M.L.S., Arêas A.P.M., Campos I.B., Monedero V., Perez-Martínez G., Miyaji E.N. Induction of systemic and mucosal immune response and decrease in Streptococcus pneumoniae colonization by nasal inoculation of mice with recombinant lactic acid bacteria expressing pneumococcal surface antigen A. Microbes and Infection. 2006;8:1016–1024. doi: 10.1016/j.micinf.2005.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole P.W., Cooney J.C. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdisciplinary Perspectives on Infectious Diseases. 2008;2008:175285. doi: 10.1155/2008/175285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte J.M., Podolsky D.K. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2004;286:G613–G626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- Pagnini C., Saeed R., Bamias G., Arseneau K.O., Pizarro T.T., Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:454–459. doi: 10.1073/pnas.0910307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant N., Hultberg A., Zhao Y.F., Svensson L., Pan-Hammarstrom Q., Johansen K. Lactobacilli expressing variable domain of llama heavy-chain antibody fragments (lactobodies) confer protection against rotavirus-induced diarrhea. The Journal of Infectious Diseases. 2006;194:1580–1588. doi: 10.1086/508747. [DOI] [PubMed] [Google Scholar]
- Parra M.D., de Morentin B.E.M., Cobo J.M., Mateos A., Martinez J.A. Daily ingestion of fermented milk containing Lactobacillus casei DN114001 improves innate-defense capacity in healthy middle-aged people. Journal of Physiology and Biochemistry. 2004;60:85–91. doi: 10.1007/BF03168444. [DOI] [PubMed] [Google Scholar]
- Paton A.W., Jennings M.P., Morona R., Wang H., Focareta A., Roddam L.F. Recombinant probiotics for treatment and prevention of enterotoxigenic Escherichia coli diarrhea. Gastroenterology. 2005;128:1219–1228. doi: 10.1053/j.gastro.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Paton A.W., Morona R., Paton J.C. Designer probiotics for prevention of enteric infections. Nature Reviews. Microbiology. 2006;4:193–200. doi: 10.1038/nrmicro1349. [DOI] [PubMed] [Google Scholar]
- Paton A.W., Morona R., Paton J.C. Bioengineered bugs expressing oligosaccharide receptor mimics: Toxin-binding probiotics for treatment and prevention of enteric infections. Bioengineered Bugs. 2010;1:172–177. doi: 10.4161/bbug.1.3.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J., Burkholder K. Application of prebiotics and probiotics in poultry production. Poultry Science. 2003;82:627–631. doi: 10.1093/ps/82.4.627. [DOI] [PubMed] [Google Scholar]
- Pelto L., Isolauri E., Lilius E.M., Nuutila J., Salminen S. Probiotic bacteria down-regulate the milk-induced inflammatory response in milk-hypersensitive subjects but have an immunostimulatory effect in healthy subjects. Clinical and Experimental Allergy. 1998;28:1474–1479. doi: 10.1046/j.1365-2222.1998.00449.x. [DOI] [PubMed] [Google Scholar]
- Perez C.A., Eichwald C., Burrone O., de Mendoza D. Rotavirus vp7 antigen produced by Lactococcus lactis induces neutralizing antibodies in mice. Journal of Applied Microbiology. 2005;99:1158–1164. doi: 10.1111/j.1365-2672.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- Preidis G.A., Saulnier D.M., Blutt S.E., Mistretta T.-A., Riehle K.P., Major A.M. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. The FASEB Journal. 2012;26:1960–1969. doi: 10.1096/fj.10-177980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano E., Puertollano M.A., Cruz-Chamorro L., de Cienfuegos G.A., Ruiz-Bravo A., de Pablo M.A. Orally administered Lactobacillus plantarum reduces pro-inflammatory interleukin secretion in sera from Listeria monocytogenes infected mice. The British Journal of Nutrition. 2008;99:819–825. doi: 10.1017/S0007114507832533. [DOI] [PubMed] [Google Scholar]
- Qiao H.P., Duffy L.C., Griffiths E., Dryja D., Leavens A., Rossman J. Immune responses in rhesus rotavirus-challenged balb/c mice treated with bifidobacteria and prebiotic supplements. Pediatric Research. 2002;51:750–755. doi: 10.1203/00006450-200206000-00015. [DOI] [PubMed] [Google Scholar]
- Radomyski T., Murano E.A., Olson D.G., Murano P.S. Elimination of pathogens of significance in food by low-dose irradiation: A review. Journal of Food Protection. 1994;57:73–86. doi: 10.4315/0362-028X-57.1.73. [DOI] [PubMed] [Google Scholar]
- Rahimi S., Kathariou S., Grimes J.L., Siletzky R.M. Effect of direct-fed microbials on performance and Clostridium perfringens colonization of turkey poults. Poultry Science. 2011;90:2656–2662. doi: 10.3382/ps.2011-01342. [DOI] [PubMed] [Google Scholar]
- Raqib R., Sarker P., Bergman P., Ara G., Lindh M., Sack D.A. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea M.C., Clayton E., O'Connor P.M., Shanahan F., Kiely B., Ross R.P. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. Journal of Medical Microbiology. 2007;56:940–946. doi: 10.1099/jmm.0.47085-0. [DOI] [PubMed] [Google Scholar]
- Reissbrodt R., Hammes W.P., dal Bello F., Prager R., Fruth A., Hantke K. Inhibition of growth of Shiga toxin-producing Escherichia coli by nonpathogenic Escherichia coli. FEMS Microbiology Letters. 2009;290:62–69. doi: 10.1111/j.1574-6968.2008.01405.x. [DOI] [PubMed] [Google Scholar]
- Resta-Lenert S., Barrett K.E. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resta-Lenert S., Barrett K.E. Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130:731–746. doi: 10.1053/j.gastro.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Robinson K., Chamberlain L.M., Lopez M.C., Rush C.M., Marcotte H., Le Page R.W.F. Mucosal and cellular immune responses elicited by recombinant Lactococcus lactis strains expressing tetanus toxin fragment C. Infection and Immunity. 2004;72:2753–2761. doi: 10.1128/IAI.72.5.2753-2761.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka S., Rantamaki P. Protecting probiotic bacteria by microencapsulation: Challenges for industrial applications. European Food Research and Technology. 2010;231:1–12. [Google Scholar]
- Salminen S., Nybom S., Meriluoto J., Collado M.C., Vesterlund S., El-Nezami H. Interaction of probiotics and pathogens—Benefits to human health? Current Opinion in Biotechnology. 2010;21:157–167. doi: 10.1016/j.copbio.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Sanders M.E. Impact of probiotics on colonizing microbiota of the gut. Journal of Clinical Gastroenterology. 2011;45:S115–S119. doi: 10.1097/MCG.0b013e318227414a. [DOI] [PubMed] [Google Scholar]
- Sanders M.E., Akkermans L.M.A., Haller D., Hammerman C., Heimbach J., Hormannsperger G. Safety assessment of probiotics for human use. Gut Microbes. 2010;1:164–185. doi: 10.4161/gmic.1.3.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satish Kumar R., Kanmani P., Yuvaraj N., Paari K.A., Pattukumar V., Arul V. Lactobacillus plantarum AS1 binds to cultured human intestinal cell line HT-29 and inhibits cell attachment by enterovirulent bacterium Vibrio parahaemolyticus. Letters in Applied Microbiology. 2011;53:481–487. doi: 10.1111/j.1472-765X.2011.03136.x. [DOI] [PubMed] [Google Scholar]
- Satokari R.M., Vaughan E.E., Smidt H., Saarela M., Matto J., de Vos W.M. Molecular approaches for the detection and identification of bifidobacteria and lactobacilli in the human gastrointestinal tract. Systematic and Applied Microbiology. 2003;26:572–584. doi: 10.1078/072320203770865882. [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L. Foodborne illness acquired in the United States—Major pathogens. Emerging Infectious Diseases. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scientific Committee on Animal Nutrition. 2002. Opinion of the Scientific Committee on Animal Nutrition on the criteria for assessing the safety of micro-organisms resistant to antibiotics of human clinical and veterinary importance. July 2001, updated April 2002. European Commission, Brussels.
- Schauber J., Svanholm C., Termen S., Iffland K., Menzel T., Scheppach W. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: Relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Harder J., Koten B., Stange E.F., Wehkamp J., Fellermann K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clinical and Experimental Immunology. 2008;151:528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Wehkamp J., Altenhoefer A., Oelschlaeger T.A., Stange E.F., Fellermann K. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infection and Immunity. 2007;75:2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Yan F., Polk D.B., Rao R.K. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2008;294:G1060–G1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P.M., Johnson-Henry K.C., Yeung H.P., Ngo P.S.C., Goulet J., Tompkins T.A. Probiotics reduce enterohemorrhagic Escherichia coli O157: H7-and enteropathogenic E.coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infection and Immunity. 2005;73:5183–5188. doi: 10.1128/IAI.73.8.5183-5188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P.M., Ossa J.C., Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutrition in Clinical Practice. 2009;24:10–14. doi: 10.1177/0884533608329231. [DOI] [PubMed] [Google Scholar]
- Shima M., Morita Y., Yamashita M., Adachi S. Protection of Lactobacillus acidophilus from the low pH of a model gastric juice by incorporation in a W/O/W emulsion. Food Hydrocolloids. 2006;20:1164–1169. [Google Scholar]
- Shu Q., Gill H.S. A dietary probiotic (Bifidobacterium lactis HN019) reduces the severity of Escherichia coli O157: H7 infection in mice. Medical Microbiology and Immunology. 2001;189:147–152. doi: 10.1007/s430-001-8021-9. [DOI] [PubMed] [Google Scholar]
- Simonova M.P., Laukova A., Chrastinova L., Strompfova V., Faix S., Vasilkova Z. Enterococcus faecium CCM7420, bacteriocin PPB CCM7420 and their effect in the digestive tract of rabbits. Czech Journal of Animal Science. 2009;54:376–386. [Google Scholar]
- Sirsat S.A., Muthaiyan A., Ricke S.C. Antimicrobials for foodborne pathogen reduction in organic and natural poultry production. Journal of Applied Poultry Research. 2009;18:379–388. [Google Scholar]
- Smith J.S., Pillai S. Irradiation and food safety. Food Technology. 2004;58:48–55. [Google Scholar]
- Sonnenburg J.L., Chen C.T.L., Gordon J.I. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biology. 2006;4:2213–2226. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon J.M., Chadd S.A., Baines R.N. Escherichia coli O157:H7 in beef cattle: On farm contamination and pre-slaughter control methods. Animal Health Research Reviews. 2011;12:197–211. doi: 10.1017/S1466252311000132. [DOI] [PubMed] [Google Scholar]
- Stephens T.P., Loneragan G.H., Karunasena E., Brashears M.M. Reduction of Escherichia coli O157 and Salmonella in feces and on hides of feedlot cattle using various doses of a direct-fed microbial. Journal of Food Protection. 2007;70:2386–2391. doi: 10.4315/0362-028x-70.10.2386. [DOI] [PubMed] [Google Scholar]
- Stern N.J., Svetoch E.A., Eruslanov B.V., Kovalev Y.N., Volodina L.I., Perelygin V.V. Paenibacillus polymyxa purified bacteriocin to control Campylobacter jejuni in chickens. Journal of Food Protection. 2005;68:1450–1453. doi: 10.4315/0362-028x-68.7.1450. [DOI] [PubMed] [Google Scholar]
- Sunkara L.T., Achanta M., Schreiber N.B., Bommineni Y.R., Dai G., Jiang W. Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression. PLoS One. 2011;6:e27225. doi: 10.1371/journal.pone.0027225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetoch E.A., Levchuk V.P., Pokhilenko V.D., Eruslanov B.V., Mitsevich E.V., Mitsevich I.P. Inactivating methicillin-resistant Staphylococcus aureus and other pathogens by use of bacteriocins OR-7 and E 50-52. Journal of Clinical Microbiology. 2008;46:3863–3865. doi: 10.1128/JCM.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.S., Fahey G.J. Prebiotic impacts on companion animals. In: Gibson G.R., Rastall R.A., editors. Prebiotics: Development and applications. John Wiley & Sons; Chichester, UK: 2006. pp. 213–236. [Google Scholar]
- Swidsinski A., Loening-Baucke V., Theissig F., Engelhardt H., Bengmark S., Koch S. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G.W., Munro K., Harmsen H.J.M., Welling G.W., Smart J., Gopal P.K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Applied and Environmental Microbiology. 2000;66:2578–2588. doi: 10.1128/aem.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarahomjoo S. Development of vaccine delivery vehicles based on lactic acid bacteria. Molecular Biotechnology. 2012;51:183–199. doi: 10.1007/s12033-011-9450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez G., Petrone V.M., Escorcia M., Morishita T.Y., Cobb C.W., Villaseñor L. Evaluation of avian-specific probiotic and Salmonella enteritidis-, Salmonella typhimurium-, and Salmonella heidelberg-specific antibodies on cecal colonization and organ invasion of Salmonella enteritidis in broilers. Journal of Food Protection. 2001;64:287–291. doi: 10.4315/0362-028x-64.3.287. [DOI] [PubMed] [Google Scholar]
- Thomas D.J., Husmann R.J., Villamar M., Winship T.R., Buck R.H., Zuckermann F.A. Lactobacillus rhamnosus HN001 attenuates allergy development in a pig model. PLoS One. 2011;6:e16577. doi: 10.1371/journal.pone.0016577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.V., Ockhuizen T. New insights into the impact of the intestinal microbiota on health and disease: A symposium report. The British Journal of Nutrition. 2012;107:S1–S13. doi: 10.1017/S0007114511006970. [DOI] [PubMed] [Google Scholar]
- Threlfall E.J. Epidemic Salmonella typhimurium DT 104—A truly international multiresistant clone. The Journal of Antimicrobial Chemotherapy. 2000;46:7–10. doi: 10.1093/jac/46.1.7. [DOI] [PubMed] [Google Scholar]
- Topcu A., Bulat T., Wishah R., Boyaci I.H. Detoxification of aflatoxin B-1 and patulin by Enterococcus faecium strains. International Journal of Food Microbiology. 2010;139:202–205. doi: 10.1016/j.ijfoodmicro.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Tsai Y.T., Cheng P.C., Pan T.M. Immunomodulating activity of Lactobacillus paracasei subsp. paracasei NTU 101 in enterohemorrhagic Escherichia coli O157H7-infected mice. Journal of Agricultural and Food Chemistry. 2010;58:11265–11272. doi: 10.1021/jf103011z. [DOI] [PubMed] [Google Scholar]
- Turner J.R. Intestinal mucosal barrier function in health and disease. Nature Reviews. Immunology. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Ukena S.N., Singh A., Dringenberg U., Engelhardt R., Seidler U., Hansen W. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. 2007;2:1308. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahjen W., Taras D., Simon O. Effect of the probiotic Enterococcus faecium NCIMB10415 on cell numbers of total Enterococcus spp., E. faecium and E. faecalis in the intestine of piglets. Current Issues in Intestinal Microbiology. 2007;8:1–7. [PubMed] [Google Scholar]
- Van Coillie E., Goris J., Cleenwerck I., Grijspeerdt K., Botteldoorn N., Van Immerseel F. Identification of lactobacilli isolated from the cloaca and vagina of laying hens and characterization for potential use as probiotics to control Salmonella Enteritidis. Journal of Applied Microbiology. 2007;102:1095–1106. doi: 10.1111/j.1365-2672.2006.03164.x. [DOI] [PubMed] [Google Scholar]
- van de Guchte M., Ehrlich S.D., Maguin E. Production of growth-inhibiting factors by Lactobacillus delbrueckii. Journal of Applied Microbiology. 2001;91:147–153. doi: 10.1046/j.1365-2672.2001.01369.x. [DOI] [PubMed] [Google Scholar]
- Vandeplas S., Dubois Dauphin R., Beckers Y., Thonart P., Théwis A. Salmonella in chicken: Current and developing strategies to reduce contamination at farm level. Journal of Food Protection. 2010;73:774. doi: 10.4315/0362-028x-73.4.774. [DOI] [PubMed] [Google Scholar]
- Vanderpool C., Yan F., Polk D.B. Mechanisms of probiotic action: Implications for therapeutic applications in inflammatory bowel diseases. Inflammatory Bowel Diseases. 2008;14:1585–1596. doi: 10.1002/ibd.20525. [DOI] [PubMed] [Google Scholar]
- Varma P., Dinesh K.R., Menon K.K., Biswas R. Lactobacillus fermentum isolated from human colonic mucosal biopsy inhibits the growth and adhesion of enteric and foodborne pathogens. Journal of Food Science. 2010;75:M546–M551. doi: 10.1111/j.1750-3841.2010.01818.x. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland J.A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W. Listeria pathogenesis and molecular virulence determinants. Clinical Microbiology Reviews. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R.D., Johnson S.J., Rubin D.K. Probiotic bacteria are antagonistic to Salmonella enterica and Campylobacter jejuni and influence host lymphocyte responses in human microbiota-associated immunodeficient and immunocompetent mice. Molecular Nutrition and Food Research. 2009;53:377–388. doi: 10.1002/mnfr.200800101. [DOI] [PubMed] [Google Scholar]
- Wall S.K., Zhang J., Rostagno M.H., Ebner P.D. Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Applied and Environmental Microbiology. 2010;76:48–53. doi: 10.1128/AEM.00785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C., Duffy G., Nally P., O'Mahony R., McDowell D.A., Fanning S. Transfer of ampicillin resistance from Salmonella Typhimurium DT104 to Escherichia coli K12 in food. Letters in Applied Microbiology. 2008;46:210–215. doi: 10.1111/j.1472-765X.2007.02288.x. [DOI] [PubMed] [Google Scholar]
- Wang Z., Yu Q., Gao J., Yang Q. Mucosal and systemic immune responses induced by recombinant Lactobacillus spp. expressing the hemagglutinin of the avian influenza virus H5N1. Clinical and Vaccine Immunology. 2012;19:174–179. doi: 10.1128/CVI.05618-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J., Harder J., Wehkamp K., Wehkamp-von Meissner B., Schlee M., Enders C. NF-kappa B- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: A novel effect of a probiotic bacterium. Infection and Immunity. 2004;72:5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill F.-X., Guesnier F., Guibert V., Timinouni M., Demartin M., Polomack L. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003) Journal of Clinical Microbiology. 2006;44:700–708. doi: 10.1128/JCM.44.3.700-708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.M., Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nature Reviews. Microbiology. 2008;6:349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K., Azevedo M.S.P., Gonzalez A., Zhang W., Saif L.J., Li G.H. Toll-like receptor and innate cytokine responses induced by lactobacilli colonization and human rotavirus infection in gnotobiotic pigs. Veterinary Immunology and Immunopathology. 2009;127:304–315. doi: 10.1016/j.vetimm.2008.10.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L.-J., Hou X.-L., Wang G.-H., Yu L.-Y., Wei X.-M., Liu J.-K. Immunization with recombinant Lactobacillus casei strains producing K99, K88 fimbrial protein protects mice against enterotoxigenic Escherichia coli. Vaccine. 2012;30:3339–3349. doi: 10.1016/j.vaccine.2011.08.036. [DOI] [PubMed] [Google Scholar]
- Williams N.T. Probiotics. American Journal of Health-System Pharmacy. 2010;67:449–458. doi: 10.2146/ajhp090168. [DOI] [PubMed] [Google Scholar]
- Wine E., Gareau M.G., Johnson-Henry K., Sherman P.M. Strain-specific probiotic (Lactobacillus helveticus) inhibition of Campylobacter jejuni invasion of human intestinal epithelial cells. FEMS Microbiology Letters. 2009;300:146–152. doi: 10.1111/j.1574-6968.2009.01781.x. [DOI] [PubMed] [Google Scholar]
- Yan F., Cao H.W., Cover T.L., Whitehead R., Washington M.K., Polk D.B. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Polk D.B. Probiotics and immune health. Current Opinion in Gastroenterology. 2011;27:496–501. doi: 10.1097/MOG.0b013e32834baa4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki N., Susuki K., Koga M., Nishimoto Y., Odaka M., Hirata K. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain–Barré syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11404–11409. doi: 10.1073/pnas.0402391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareie M., Johnson-Henry K., Jury J., Yang P.C., Ngan B.Y., McKay D.M. Probiotics prevent bacterial translocation and improve intestinal barrier function in rats following chronic psychological stress. Gut. 2006;55:1553–1560. doi: 10.1136/gut.2005.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li R., Li J. Lactobacillus reuteri ATCC 55730 and L22 display probiotic potential in vitro and protect against Salmonella-induced pullorum disease in a chick model of infection. Research in Veterinary Science. 2012;93:366–373. doi: 10.1016/j.rvsc.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Zihler A., Gagnon M., Chassard C., Lacroix C. Protective effect of probiotics on Salmonella infectivity assessed with combined in vitro gut fermentation-cellular models. BMC Microbiology. 2011;11:264. doi: 10.1186/1471-2180-11-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyrek A.A., Cichon C., Helms S., Enders C., Sonnenborn U., Schmidt M.A. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKC zeta redistribution resulting in tight junction and epithelial barrier repair. Cellular Microbiology. 2007;9:804–816. doi: 10.1111/j.1462-5822.2006.00836.x. [DOI] [PubMed] [Google Scholar]


