Abstract
Hepatocellular carcinoma (HCC) is the most commonly diagnosed cancer and the leading cause of cancer mortality. Several lines of evidence have demonstrated the aberrant expression of long noncoding RNAs (lncRNAs) in carcinogenesis and their universal regulatory properties. A thorough understanding of lncRNA regulatory roles in HCC pathology would contribute to HCC prevention and treatment. In this study, we identified a novel human lncRNA, LNC-HC, with significantly reduced levels in hepatic tumors from patients with HCC. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide) assays as well as colony formation and wound healing experiments showed that LNC-HC significantly inhibited the proliferation of the HCC cell line Huh7. Xenograft transplantation of LNC-HC-overexpressing Huh7 cells in nude mice resulted in the production of smaller tumors. Mechanistically, LNC-HC inhibited the proliferation of HCC cells by directly interacting with hsa-miR-183-5p. LNC-HC rescued the expression of five tumor suppressors, including AKAP12, DYRK2, FOXN3, FOXO1, and LATS2, that were verified as target genes of hsa-miR-183-5p. Overall, human LNC-HC was identified as a novel tumor suppressor that could inhibit HCC cell proliferation in vitro and suppress tumor growth in vivo by competitively binding hsa-miR-183-5p as a competing endogenous RNA (ceRNA). These findings suggest that LNC-HC could be a biomarker of HCC and provide a novel therapeutic target for HCC treatment.
Keywords: LNC-HC, hsa-miR-183-5p, hepatocellular carcinoma, molecular mechanism
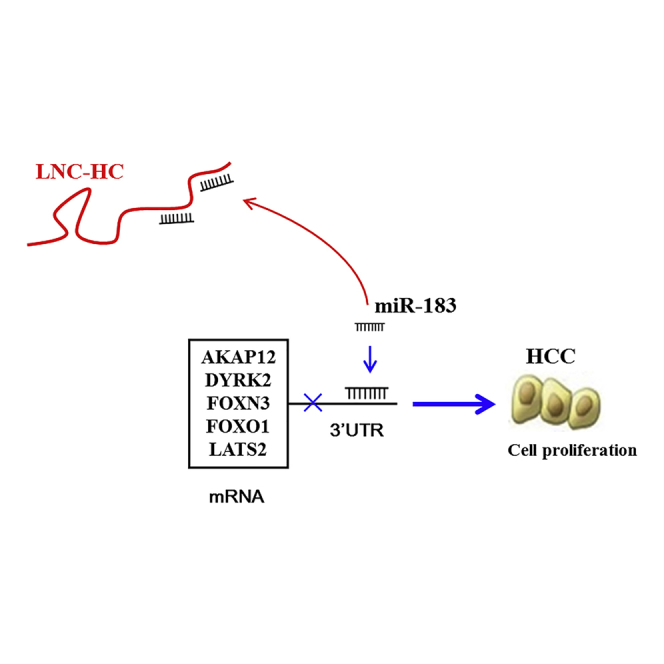
Graphical Abstract
Introduction
Hepatocellular carcinoma (HCC) is a commonly diagnosed malignancy and the leading cause of cancer death worldwide and it has shown an increasing incidence compared to that of other cancers.1, 2, 3 Although improvements have been made in surgical and therapeutic techniques, the 5-year survival rate of HCC patients is still unsatisfactory. The molecular mechanisms underlying liver cancer pathology need to be further explored.
Long noncoding RNAs (lncRNAs) are a type of RNAs that are longer than 200 nt and traditionally defined as not having translation potential; however, recent studies have shown that some lncRNAs could produce short functional peptides.4,5 An increasing number of studies have verified that lncRNAs play a large role in biological processes. Some lncRNAs function as coregulators of proteins that directly combine with coregulator proteins to regulate downstream tumor-related gene expression.6, 7, 8, 9, 10, 11, 12 Furthermore, another class of lncRNAs functions as competing endogenous RNAs (ceRNAs).13, 14, 15 This class of lncRNAs sequesters microRNAs by acting as a sponge and controls the expression of microRNA-targeted oncogenes or tumor repressors. A well-studied lncRNA, lncRNA-ATB, inducesepithelial-mesenchymal transition (EMT) and the invasion of cancer cells through upregulating ZEB1 and ZEB2 expression by acting as a ceRNA for miR-200.13 In addition, the tumorigenic lncRNA HULC also functions as a ceRNA through sequestering miR-372, resulting in the upregulation of an oncogene in liver cancer.14 These findings revealed that the aberrant expression of lncRNAs plays a role in cell proliferation, angiogenesis, and tumor development. The identification of novel lncRNAs and the exploration of their functions and regulatory molecular mechanisms could provide promising therapeutic targets for liver cancer.
In our previous study, a novel rat lncRNA, lnc-HC, was identified in rat hepatocytes as a regulator of hepatocyte cholesterol catabolism.16,17 Mechanistically, lnc-HC and its coregulator hnRNPA2B1 form the lncRNA-protein complex and decrease the stability of mRNAs encoding Cyp7a1 and Abca1, which are critical enzymes that contribute to cholesterol catabolism. Furthermore, we decided to identify the human homolog of lnc-HC and explore its role in biological processes of the human body. In the current study, a novel human lncRNA was identified, named LNC-HC. Interestingly, human LNC-HC is remarkably decreased in HCC tumor tissues. LNC-HC inhibits HCC cell proliferation both in vitro and in vivo. Mechanistically, LNC-HC upregulates the expression of tumor repressors by functioning as a ceRNA to sequester hsa-miR-183-5p. Our findings suggest that LNC-HC could be regarded as a novel biomarker of HCC and could provide a novel therapeutic target for HCC treatment.
Results
Identification of the Novel Human lncRNA LNC-HC
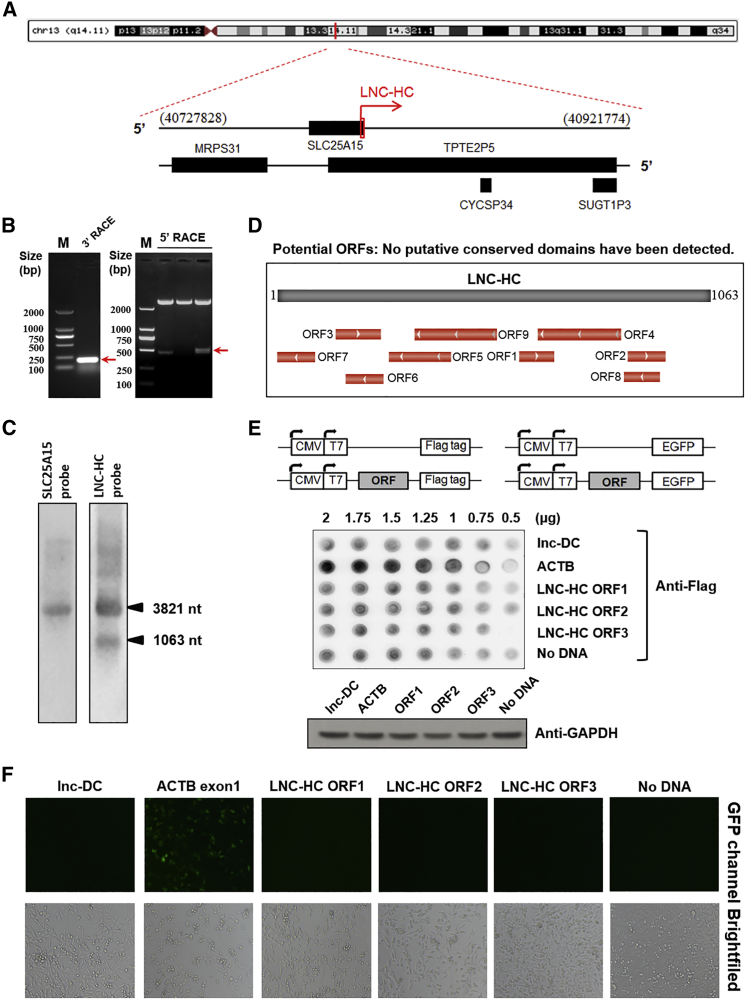
Previously, we identified a lncRNA in rat liver, lnc-HC, that plays a role in fine-tuning hepatic cholesterol and fatty acid metabolism.16,17 Since then, we further identified the human homolog of lnc-HC, LNC-HC (GenBank: MN026163). A 1,226-nt fragment of LNC-HC was first identified by a BLAST (Basic Local Alignment Search Tool) search in the UCSC (University of California Santa Cruz) database with the homologous rat lnc-HC gene sequence, and LNC-HC was found to be located on chromosome 13 (Figure 1A). The full-length sequence of human LNC-HC was confirmed to be 1,063 nt in length by SMART RACE technology (Figure 1B; Table S1). LNC-HC overlaps with the 3′ UTR of protein-encoding gene SLC25A15. In accordance, northern blotting detected two clear transcripts (1,063 and 3,821 nt) with LNC-HC-probe and one transcript (3,821 nt) with SLC25A15-probe (Figure 1C). These results confirmed that LNC-HC is a real transcript and is overlapping with the 3′ UTR of SLC25A15.
Figure 1.
Identification of LNC-HC as a Novel Human lncRNA
(A) Location of LNC-HC in the human genome. (B) The PCR products obtained from 3′-RACE (left panel) and 5′-RACE (right panel). Lane M, DL2000 DNA marker. (C) Northern blotting identification of LNC-HC as a novel transcript and expressed differentially from overlapping gene SLC25A15. (D) Predicted ORFs of LNC-HC by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). (E) The predicted ORFs of LNC-HC, the full length of lnc-DC, and the first exon of ACTB were cloned into pcDNA3.1+ with C-terminal FLAG/EGFP (upper panel). Immunoblotting of FLAG-fusion protein in LO2 cells transfected with the recombinant plasmids, taking GAPDH as the loading control (lower panel). (F) Fluorescence microscopic images of the EGFP-fusion protein expression.
To determine the protein coding potential of LNC-HC, we performed informatics analysis and protein expression experiments. First, the predicted open reading frames (ORFs) of LNC-HC are short and did not show any conserved protein domains among various species (Figure 1D; Table S2). Second, there was no protein coding potential as determined by the coding potential calculator (http://cpc2.cbi.pku.edu.cn/). Experimentally, we cloned the three ORFs of LNC-HC predicted with sense strand into the pcDNA3.1+ vector with a C-terminal FLAG or EGFP tag (Figures 1E and S1). ACTB is a control of the protein-coding gene and lnc-DC is a control of lncRNA. Immunoblotting results showed that FLAG-tag protein was hardly detected in the LNC-HC group (Figure 1E, lower panel). EGFP-fusion protein showed the similar results (Figure 1F). Based on bioinformatics analysis and protein expression experiments, we confirmed that LNC-HC is a lncRNA and is the human homolog of the previously identified rat lnc-HC.
LNC-HC Is Decreased in Hepatic Tumors in HCC
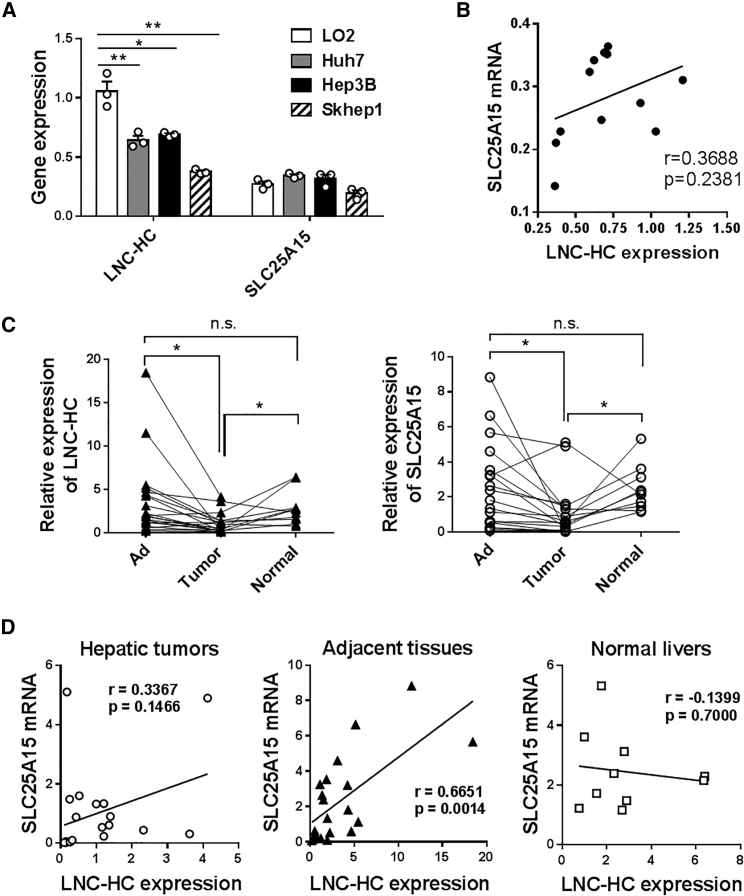
Quantitative real-time PCR results showed that LNC-HC was significantly decreased in three types of HCC cell lines, including Huh7, Hep3B and Skhep1, compared with the human hepatocyte cell line LO2; SLC25A15 showed no differences among these groups of cells (Figure 2A). In addition, there was no significant correlation between LNC-HC and SLC25A15 expression in these four cell lines (Figure 2B). Among the 20 hepatic tumor tissues, the paired 20 adjacent non-tumor tissues and 10 normal liver tissues from 20 patients with HCC, LNC-HC was significantly decreased in the tumor tissues compared with the adjacent and normal liver tissues; SLC25A15 showed a similar expression pattern (Figure 2C). To check the correlation of the expression between LNC-HC and SLC25A15 in clinical samples, a detailed analysis was performed. The two genes expressions were significantly correlated in adjacent tissues; however, no correlation was observed in tumor tissues and normal ones (Figure 2D). According to the median ratio of LNC-HC expression, HCC patients were divided into LNC-HClow and LNC-HChigh groups. Clinicopathological analysis suggested that LNC-HC expression was more likely correlated with tumor size (p = 0.1409) and tumor-node-metastasis (TNM) stage (p = 0.075) as compared with other parameters, including age, sex, metastasis, carcinoembryonic antigen (CEA), and alpha-fetoprotein (AFP) levels (Table 1). These findings suggest that LNC-HC might be a potential tumor suppressor in HCC.
Figure 2.
LNC-HC Expression Was Significantly Reduced in Both HCC Cell Lines and Clinical Hepatic Tumor Tissues
(A) Quantitative real-time PCR detection of LNC-HC and SLC25A15 mRNA levels in the human hepatocyte cell line LO2 and the HCC cell lines Huh7, Hep3B, and Skhep1. (B) Correlation analysis of LNC-HC and SLC25A15 expression in four types of cell lines at the mRNA level. (C) Quantitative real-time PCR detection of LNC-HC and SLC25A15 expression in 20 hepatic tumor tissues, 20 paired adjacent non-tumor tissues, and 10 paired normal tissues from HCC patients. (D) Correlation analysis of LNC-HC and SLC25A15 expression in clinical liver samples at the mRNA level. Ad, adjacent non-tumor tissues. ACTB was used for normalization in the quantitative real-time PCR assay. ∗p < 0.05, ∗∗p < 0.01. n.s., not significant. Values are expressed as the means ± SEM.
Table 1.
The Association between LNC-HC Expression and Clinicopathological Characteristics of the 20 HCC Patients
| Variable | All Patients (n) |
LNC-HC Expression |
p Value | |
|---|---|---|---|---|
| Low | High | |||
| All cases | 20 | 10 | 10 | |
| Age (years) | ||||
| 15 | 8 | 7 | >0.9999 | |
| ≥60 | 5 | 2 | 3 | |
| Sex | ||||
| Male | 16 | 8 | 8 | >0.9999 |
| Female | 4 | 2 | 2 | |
| Tumor size (cm) | ||||
| 6 | 1 | 5 | 0.1409 | |
| ≥5 | 14 | 9 | 5 | |
| TNM stage | ||||
| I | 2 | 1 | 1 | 0.075 |
| II | 4 | 0 | 4 | |
| III | 12 | 7 | 5 | |
| IV | 2 | 2 | 0 | |
| Lymphatic metastasis | ||||
| Yes | 2 | 1 | 1 | >0.9999 |
| No | 18 | 9 | 9 | |
| Distant metastasis | ||||
| Yes | 2 | 1 | 1 | >0.9999 |
| No | 18 | 9 | 9 | |
| CEA (ng/mL) | ||||
| 13 | 5 | 8 | 0.3498 | |
| ≥5 | 7 | 5 | 2 | |
| AFP (ng/mL) | ||||
| 10 | 4 | 6 | 0.6563 | |
| ≥400 | 10 | 6 | 4 | |
TNM, tumor-node-metastasis; CEA, carcinoembryonic antigen; AFP, alpha-fetoprotein.
LNC-HC Inhibits Cell Proliferation In Vitro
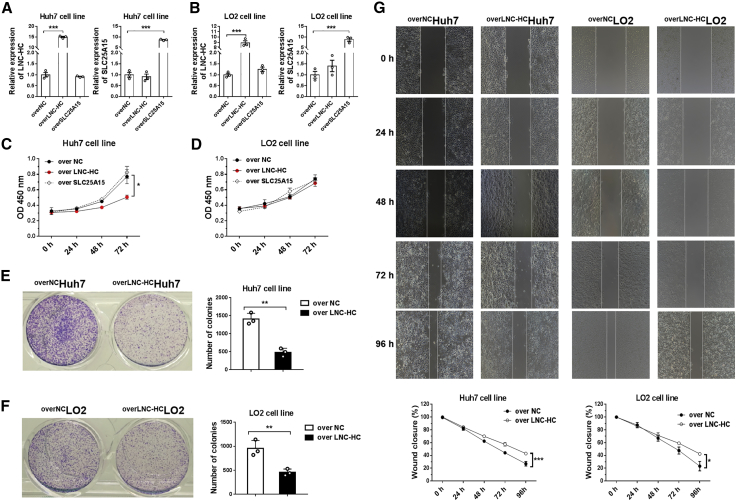
To test the biological effects of LNC-HC on cell growth, LNC-HC and SLC25A15 were overexpressed in Huh7 and LO2 cells to generate overLNC-HCHuh7/LO2 and overSLC25A15Huh7/LO2 cells, respectively (Figures 3A and 3B). An MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide) assay showed that LNC-HC overexpression significantly inhibited the proliferation of Huh7 cells in vitro compared with overexpressed negative control (NC) cells; SLC25A15 overexpression had no effect (Figure 3C). Moreover, the overexpression of LNC-HC and SLC25A15 did not affect the growth of LO2 cells within 72 h (Figure 3D). Consistently, LNC-HC overexpression suppressed Huh7 and LO2 cell colony formation (Figures 3E and 3F). A wound healing study showed that LNC-HC inhibited the migration of Huh7 and LO2 cells (Figure 3G). These results indicated that LNC-HC suppresses Huh7 cell growth in vitro.
Figure 3.
LNC-HC Suppressed Cell Proliferation and Migration
(A) Quantitative real-time PCR detection of LNC-HC and SLC25A15 expression in LNC-HC or SLC25A15 stably overexpressing Huh7 cells. (B) Quantitative real-time PCR detection of LNC-HC and SLC25A15 in the indicated stably overexpressing LO2 cells. (C) MTT assay measurement of the proliferation of the indicated Huh7 cells at different time points (0, 24, 48, and 72 h). (D) MTT measurement of the proliferation of LO2 cells at different time points (0, 24, 48, and 72 h). (E) Colony formation of overNCHuh7 and overLNC-HCHuh7 cells (left) and quantification histogram (right). (F) Colony formation of overNCLO2 and overLNC-HCLO2 cells (left) and quantification histogram (right). (G) Detection of the migration of overNCHuh7/LO2 and overLNC-HCHuh7/LO2 cells by using a wound healing assay (up) and quantification histogram (low). ACTB was utilized for normalization in the quantitative real-time PCR. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Values are expressed as the means ± SEM.
LNC-HC Inhibits Tumor Growth in a Mouse Xenograft Model
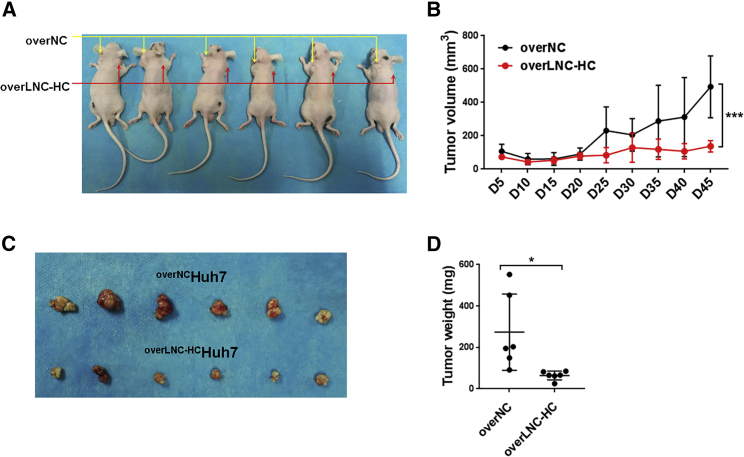
To check the effect of LNC-HC on tumor growth in vivo, we generated a xenograft tumor model by subcutaneously injecting overNCHuh7 and overLNC-HCHuh7 cells into BALB/c nude mice. Compared with overNCHuh7 cells, overLNC-HCHuh7 cells grew more slowly, as indicated by their smaller tumor volumes (Figures 4A–4C) and tumor weights (Figure 4D). These results showed that LNC-HC significantly -suppresses tumor growth in vivo.
Figure 4.
LNC-HC Inhibited Tumor Growth in a Xenograft Mouse Model
(A) BALB/c nude mice (aged 4 weeks, male, n = 6) were subcutaneously injected with 5 × 106 overNCHuh7 (left side) or overLNC-HCHuh7 cells (right side) per mouse. (B) Tumor volumes were measured every 5 days from day 5 to day 45. On day 45, the mice were sacrificed. (C and D) The tumors from model mice were collected (C) and weighed (D). ∗p < 0.05, ∗∗∗p < 0.001. Values are expressed as the means ± SEM.
LNC-HC Interacts with hsa-miR-183-5p
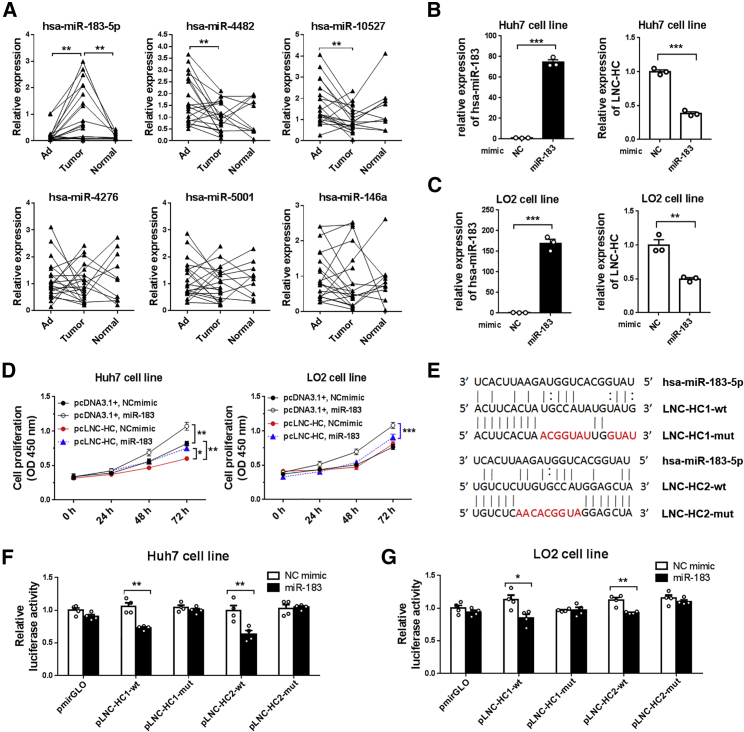
To determine the regulatory mechanism involved in the effects of LNC-HC on the antitumor process, we first examined the possibility of a ceRNA-like pattern. In the prediction of the interaction between microRNAs and LNC-HC by the miRDB software (http://mirdb.org/), six microRNA candidates with higher scores were obtained, including hsa-miR-183-5p, -4482, -10527, -4276, -5001, and -146a. Quantitative real-time PCR results suggested that hsa-miR-183-5p (hereafter referred to as miR-183) was significantly increased in tumor tissues compared with adjacent and normal tissues, while miR-4482 and miR-10527 expression were decreased in tumors compared with adjacent tissues (Figure 5A). Among the six candidates, miR-183 expression in tumor tissues indicated that it might interact with LNC-HC to form a ceRNA regulatory system. In vitro experiments showed that the miR-183 mimic significantly decreased LNC-HC expression both in Huh7 and LO2 cells (Figures 5B and 5C), whereas LNC-HC had no effect on miR-183 expression (Figures S2A and S2B). In addition, the miR-183 mimic facilitated Huh7 and LO2 cell proliferation; miR-183 inhibited the suppression by LNC-HC of the proliferation of Huh7 cells (Figure 5D). To verify the direct interaction between miR-183 and LNC-HC, we constructed dual-luciferase reporter gene vectors that contained wild-type and mutant miR-183-targeted LNC-HC sequences (1 and 2) using pmirGLO as a basic plasmid, which are referred to as pLNC-HC1-wt/mut and pLNC-HC2-wt/mut, respectively (Figure 5E). After the transfection of pLNC-HC1/2-wt/mut and the miR-183 mimic, we found that miR-183 decreased the luciferase activity of pLNC-HC1-wt and pLNC-HC2-wt but had no effect on the mutant plasmids in both Huh7 and LO2 cells (Figures 5F and 5G). These findings suggested that miR-183 directly targets LNC-HC through binding regions 1 and 2; the two genes, a lncRNA and a microRNA, may form a ceRNA system to coregulate downstream gene expression.
Figure 5.
miR-183 Directly Interacted with LNC-HC
(A) Quantitative real-time PCR detection of the expression of the indicated microRNAs in hepatic tumor samples. (B) Quantitative real-time PCR detection of miR-183 and LNC-HC expression in Huh7 cells transfected with the negative control mimic (NC, 50 nM) or miR-183 mimic (miR-183, 50 nM) for 24 h. (C) Quantitative real-time PCR detection of miR-183 and LNC-HC expression in LO2 cells transfected with the NC mimic (50 nM) or miR-183 mimic (50 nM) for 24 h. (D) MTT assay of the cell proliferation under LNC-HC or miR-183 overexpression. (E) Schematic diagram of wild-type and site-directed mutant of the miR-183 binding sites 1 and 2 within the LNC-HC sequence. (F) Dual-luciferase reporter gene assay of the Huh7 cells cotransfected with pLNC-HC1/2-WT/mut and NC/miR-183 mimic. (G) Luciferase activity of LO2 cells cotransfected with pLNC-HC1/2-WT/mut and NC/miR-183 mimic. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Values are expressed as the means ± SEM.
LNC-HC/miR-183 Coregulate the Expression of Tumor Repressors as ceRNA
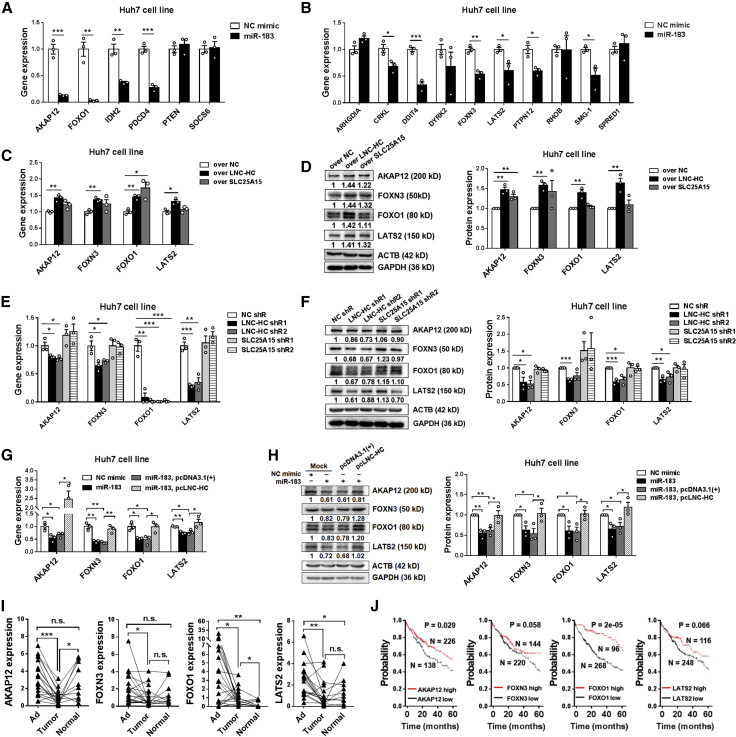
To verify the ceRNA function of LNC-HC and miR-183, we predicted 16 tumor repressors that were directly targeted by miR-183 using miRDB software. Among these candidates, 6 genes (AKAP12,18, 19, 20 FOXO1,21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 IDH2,35, 36, 37 PDCD4,38, 39, 40, 41, 42, 43, 44, 45, 46 PTEN,47,48 and SOCS635,49) have previously been reported as miR-183 targets; the other 10 genes (ARHGDIA, CRKL, DDIT4, DYRK2, FOXN3, LATS2, PTPN12, RHOB, SMG-1, and SPRED1) were the predicted candidates with the highest scores without experimental verification. In Huh7 cells, four of the verified genes (including AKAP12, FOXO1, IDH2, and PDCD4) and six of the predicted genes (including CRKL, DDIT4, FOXN3, LATS2, PTPN12, and SMG-1) were significantly suppressed by miR-183 (Figures 6A and 6B). In LO2 cells, five of the verified genes (including AKAP12, FOXO1, IDH2, PDCD4, and SOCS6) and four of the predicted genes (including DDIT4, DYRK2, LATS2, and SPRED1) were suppressed by miR-183 (Figures S3A and S3B). Next, we examined the effect of LNC-HC on the expression of these candidate genes. In Huh7 cells, LNC-HC overexpression increased the expression of four genes both at the mRNA and protein levels, including AKAP12, FOXN3, FOXO1, and LATS2 (Figures 6C and 6D). In contrast, LNC-HC knockdown decreased the expression of these genes (Figures 6E and 6F). In LO2 cells, LNC-HC increased four genes, including AKAP12, DYRK2, FOXO1, and LATS2 (Figures S3C and S3D), while LNC-HC knockdown decreased these four genes (Figures S3E and S3F). To further elucidate the ceRNA regulation, we overexpressed miR-183 and LNC-HC expression in cells. The results showed that LNC-HC could completely rescue the suppressive effect of miR-183 on expression of the four genes Huh7 cells, including AKAP12, FOXN3, FOXO1, and LATS2 (Figures 6G and 6H). Meanwhile, the expressions of four genes, including AKAP12, DYRK2, FOXO1, and LATS2, were rescued by LNC-HC overexpression in LO2 cells (Figures S3G and S3H).
Figure 6.
LNC-HC and miR-183 Coregulated Downstream Tumor Repressor Expression as a ceRNA Pattern
(A and B) Quantitative real-time PCR analysis of indicated expression of genes in NC/miR-183 mimic-transfected Huh7 cells. (A) Genes already verified as miR-183 targets. (B) Genes predicted to be targets of miR-183. (C) Quantitative real-time PCR detection of gene expression in overLNC-HCHuh7 and overSLC25A15Huh7 cells. (D) Western blotting analysis of the indicated gene expression in overLNC-HCHuh7 and overSLC25A15Huh7 cells (left) and quantification histograms (right). (E) Quantitative real-time PCR detection of miR-183-targeted gene expression in LNC-HC or SLC25A15 knockdown Huh7 cells. (F) Western blotting analysis of indicated gene expression in LNC-HC or SLC25A15 knockdown Huh7 cells (left) and quantification histogram (right). (G) Quantitative real-time PCR detection of indicated genes in Huh7 cells transfected with NC/miR-183 mimic and pcDNA3.1+/pcLNC-HC for 24 h. (H) Western blotting analysis of indicated gene expression in Huh7 cells transfected with NC/miR-183 mimic and pcDNA3.1+/pcLNC-HC (left) and quantification histogram (right). (I) Quantitative real-time PCR detection of LNC-HC-targeted genes in hepatic tumor samples. (J) Kaplan-Meier plots of median overall survival data for patients with liver cancer (n = 364) by using Kaplan-Meier Plotter. Western blotting shows one representative result and quantitative data from three independent experiments. ACTB was the loading control in the western blotting assay. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Values are expressed as the means ± SEM.
To further check the effect of LNC-HC and miR-183 target genes on Huh7 cell proliferation, an MTT assay and colony formation and wound healing experiments were performed under the condition of LNC-HC overexpression and knockdown of indicated genes, including AKAP12, FOXN3, FOXO1, and LATS2. Each gene could be significantly knocked down by small interfering RNAs (siRNAs) (Figure S4A). MTT results showed that knockdown of FOXN3 and LATS2 could significantly counteract the suppression effect of LNC-HC on the proliferation of Huh7 cells; knockdown of AKAP12 and FOXO1 has no obvious effect on that within 96 h (Figure S4B). Similarly, knockdown of FOXN3 and LATS2 increased the number of colony formations as compared with the LNC-HC overexpression group (Figures S5A and S5B). A wound healing assay suggested that only FOXN3 knockdown strongly suppressed the metastasis of Huh7 cells under LNC-HC overexpression, while the other three genes showed no obvious effect within 96 h (Figures S6A and S6B). These results doubly confirmed that LNC-HC suppresses HCC cell proliferation; in addition, they suggested the critical roles of LNC-HC/miR-183/FOXN3 and LATS2 pathways on cell proliferation.
To verify the association between LNC-HC and its target genes, we further checked their expressions in hepatic tissues from HCC patients. These genes were all significantly decreased in tumors compared to those in adjacent tissues (Figures 6I and S7A). The Kaplan-Meier analysis showed that high mRNA level of AKAP12, FOXN3, FOXO1, and LATS2 associated with better patients median overall survival in patients with liver cancer (Figure 6J); however, a high level of DYRK2 mRNA indicated poor patient survival (Figure S7B). With regard to the correlation between the expression of LNC-HC and its target genes, three genes (AKAP12, FOXN3, and LATS2) were significantly correlated with LNC-HC in hepatic tumors; only LATS2 was significantly correlated with LNC-HC both in tumor and in adjacent tissues. AKAP12 and LATS2 showed a significant correlation with LNC-HC in all samples containing tumor, adjacent tissues, and normal tissues (Table 2). These findings verified that LNC-HC and miR-183 regulate downstream tumor repressors through a ceRNA pattern both in vitro and in vivo.
Table 2.
Correlation between LNC-HC and Target Genes in Hepatic Tumors from Patients with HCC
| Gene | Correlation Analysis between LNC-HC and Target Genes Expression |
|||||||
|---|---|---|---|---|---|---|---|---|
| Tumor |
Adjacent |
Normal |
All Samples |
|||||
| r Value | p Value | r Value | p Value | r Value | p Value | r Value | p Value | |
| AKAP12 | 0.5889 | 0.0063∗∗ | 0.2208 | 0.3495 | 0.1634 | 0.6520 | 0.385 | 0.0058∗∗ |
| DYRK2 | 0.3774 | 0.1009 | −0.0338 | 0.8875 | 0.5823 | 0.0774 | 0.2357 | 0.0995 |
| FOXN3 | 0.5045 | 0.0233∗ | −0.0361 | 0.8798 | 0.4152 | 0.2328 | 0.1822 | 0.2054 |
| FOXO1 | 0.2604 | 0.2675 | 0.2017 | 0.3939 | −0.5475 | 0.1014 | 0.276 | 0.0524 |
| LATS2 | 0.5376 | 0.0145∗ | 0.5278 | 0.0168∗ | 0.0013 | 0.9972 | 0.5121 | 0.0001∗∗∗ |
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
In the current study, we identified a novel human lncRNA, LNC-HC, that is the human homolog of rat lnc-HC. We verified its function as an inhibitor of HCC cell growth in vitro and its role as a tumor suppressor in vivo. Mechanistically, LNC-HC upregulates the expression of a cluster of tumor repressors, including AKAP12, DYRK2, FOXN3, FOXO1, and LATS2, by sequestering miR-183 as a ceRNA.
LNC-HC is the human homolog of rat lnc-HC and possesses a high degree of synteny, indicating it has a good conservation. SMART RACE and northern blotting confirmed that LNC-HC is a novel transcript of 1,063 nt. Both bioinformatics analysis and FLAG/EGFP-tag fusion protein expression indicated that LNC-HC has no protein coding potential. These results confirmed that LNC-HC functions as a lncRNA.
We further explored the function of human LNC-HC in HCC development. LNC-HC is significantly decreased in HCC hepatic tumors as well as in three types of HCC cell lines, including Huh7, Hep3B, and Skhep1. The mRNA expression of SLC25A15, a protein-encoding gene that overlaps with LNC-HC, was likewise significantly decreased in tumor tissues. However, expression of the two genes demonstrated a lack of correlation in HCC cells and HCC tumors, suggesting their independent expression patterns under these two conditions and different pathways in modulating cancer pathology; in addition, the two genes showed strong correlation in adjacent tissues, suggesting their potential cooperation in regulating tumorigenesis and indicating the aberrant gene profile presented in adjacent tissues. Although there is no significance regarding the correlation between LNC-HC expression and clinicopathological characteristics (p > 0.05), a low level of LNC-HC was clearly correlated with TNM stage and tumor size to a certain extent. The correlation tends to be significant with a bigger sample size. We further focused on LNC-HC by excluding the effects of SLC25A15. We constructed LNC-HC-overexpressing stable cells and utilized SLC25A15 overexpression as a control. The MTT assay showed that overLNC-HCHuh7 cells showed a slower proliferation rate over 72 h compared with overNCHuh7 and overSLC25A15Huh7 cells; however, no detectable change in proliferation was identified in LO2 cells. This might be because LO2 cells are a normal hepatocyte cell line with an increased baseline of LNC-HC level and thus have a normal proliferation rate. Nonetheless, during a longer cultivation, the growth difference between overNCLO2 and overLNC-HCLO2 cells was much more obvious, as indicated by the colony formation assay. In addition to the in vitro evidence, an in vivo mouse model further established the antitumor effect of LNC-HC.
The ceRNA hypothesis was proposed by Salmena et al.50 in 2011. Several lines of evidence in this field have shown the role of ceRNA in the fine-tuning of regulatory pathways; nonetheless, only a few studies have reported the role of ceRNAs in liver cancer pathology.51, 52, 53, 54, 55, 56 A full understanding of the ceRNAs involved in HCC, including those formed by lncRNAs and microRNAs, could provide new insights and new therapeutic targets for HCC. In our study, miR-183 was identified as the coregulator of LNC-HC. In agreement with previous studies, miR-183 expression was dramatically increased in HCC tumor tissues and is considered an oncomiRNA.29,46,49,57, 58, 59, 60 Interestingly, we also identified two microRNAs, miR-4482 and miR-10527, that were predicted to target LNC-HC and were significantly reduced in HCC tumors. Previous studies have not reported these two microRNAs in relationship to cancer development. In Huh7 cells, miR-183 significantly promoted cell growth; LNC-HC overexpression strongly eliminated this effect. In LO2 cells, LNC-HC overexpression resulted in no changes in cell proliferation compared with the control group during 72 h; however, it significantly suppressed cell proliferation compared with the miR-183-transfected overNCLO2 group. This difference between Huh7 and LO2 cells may due to the fact that the LO2 cells had a slower proliferation rate and transformed into a carcinoma cell type when miR-183 was overexpressed; under this condition, the tumor suppressor function of LNC-HC was obvious.
Furthermore, we also explored the LNC-HC/miR-183 ceRNA pathway in regulating downstream tumor repressors. As a novel gene, it is difficult to analyze the correlation between LNC-HC expression and patient survival with liver cancer. We thereupon checked its target genes, including AKAP12, DYRK2, FOXN3, FOXO1, and LATS2, and the HCC patient survival rate by using the public RNA sequencing (RNA-seq) database Kaplan-Meier Plotter. Except for DYRK2, the high mRNA levels of AKAP12, FOXN3, FOXO1, and LATS2 predicted the longer patient survival. This is in accordance with the results shown in Figure 6, LNC-HC/miR-183 coregulate the four genes in Huh7 cells; DYRK2 could be controlled only in LO2 cells. The discrepancy of DYRK2 suggests a potential variation during cancer pathogenesis. Supposing that DYRK2 overexpression is a compensatory effect induced by a high degree of malignancy, it could be explained that decreased DYRK2 predicts longer patient survival. However, one existing study reported that depletion of DYRK2 promoted HCC cell proliferation.61 This group further showed that a low expression of DYRK2 predicted a poor prognosis for HCC patients (n = 86). Due to the above contradiction, more studies need to be conducted to detail the function and expression pattern of DYRK2 during HCC pathogenesis. AKAP12 and FOXN3 have been reported to be decreased in HCC tissues compared to adjacent hepatic tissues and to function as tumor suppressors.20,62, 63, 64 In our study, AKAP12 and FOXN3 were verified to be correlated with LNC-HC expression in HCC tumors but not in paired-adjacent and normal samples. These observations suggested that the function of LNC-HC was different under different conditions, such as normal and carcinogenic treatments. LATS2 is an influential molecule with strong antitumor effects. It play roles in mitochondrial function and the Wnt/β-catenin pathway,65 AMPK-Mfn2 pathway,66 EMT,67,68 Hippo pathway69,70 and the YAP/TAZ system.71, 72, 73 Here, LATS2 was strongly correlated with LNC-HC not only in tumors and adjacent liver tissues, but also in all samples containing tumor, adjacent tissue, and normal liver tissue. When combined, the results of correlation analysis with the MTT, colony formation, and wound healing assays under LNC-HC overexpression and target gene knockdown, it could be concluded that the LNC-HC/ miR-183/FOXN3 and LATS2 pathways play a vital role in HCC cell proliferation and HCC pathology.
Overall, we identified LNC-HC as a novel human lncRNA and described its antitumor function in HCC. The identification of LNC-HC not only reveals new regulatory pathways important for understanding HCC pathology, but it also provides new insights into therapeutic possibilities for the prevention and treatment of HCC.
Materials and Methods
SMART RACE
The human homologous nucleotide fragment of rat lnc-HC was obtained through a BLAST search of the UCSC database. Then, 5′- and 3′-RACE of LNC-HC were conducted using the BD SMART RACE cDNA amplification kit (Clontech Laboratories, Mountain View, CA, USA) according to the manufacturer’s instructions. The primers used for 5′-RACE of LNC-HC were 5′-CTCCCAACCTAAGTTTCCGTTCCTCA-3′ (5-GSP) and 5′-CGTTCCTCATTTCCACTTGCCTCAGA-3′ (5-NGSP). The primers used for 3′-RACE of LNC-HC were 5′-AATCACAGAGCCAGAAGCCAGAGGG-3′ (3-GSP) and 5′-GAGGCAAGTGGAAATGAGGAACGG-3′ (3-NGSP).
Bioinformatics Analysis
The ORF prediction of LNC-HC was conducted using the ORF Finder online software (https://www.ncbi.nlm.nih.gov/orffinder/). The predicted LNC-HC ORFs are depicted in Table S2. The conserved protein domains were verified by comparison to the predicted LNC-HC ORFs obtained from a protein BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of the nonredundant protein sequences (nr) and reference proteins (refseq_protein) databases. The translation potential of LNC-HC was determined with a coding potential calculator (http://cpc2.cbi.pku.edu.cn/).
Northern Blotting
Northern blotting analysis was performed by using total RNA (30 μg) from the LO2 cell line, with digoxigenin (DIG)-labeling SLC25A15-probe or LNC-HC-probe. Detection was conducted with the DIG-High Prime DNA labeling and detection starter kit II (Roche, Basel, Switzerland), according to the manufacturer’s instructions. The primers used for the SLC25A15 DIG-labeling probe are 5′-TCCAATCCTGCTATCCAGGC-3′ and 5′-CCGGCTCAGTTCATAGCCAC-3′, and for LNC-HC the DIG-labeling probe are 5′-AGTGTTCCTAAGCAGCCTGT-3′ and 5′-TTCCTCATTTCCACTTGCC-3′.
Construction of EGFP/FLAG Fusion Protein Expression Vectors and Protein Expression Assay
The C-terminal EGFP and FLAG tag were inserted into the pcDNA3.1+ vector between the EcoRI and XhoI restriction sites (Figure S1). Then, the predicted ORF1–3 of LNC-HC (from the transcription start site to the end of ORFs without a stop codon) was inserted into the pcDNA3.1-EGFP/FLAG vector between the BamHI and EcoRI sites. The first exon sequence of protein-coding gene ACTB (171 bp, 48 bp before the ATG start codon and without a stop codon) was the control of the protein coding gene. Lnc-DC (621 bp, full-length sequence, without a stop codon) was the control of lncRNAs. The primers for amplifying target sequence are shown in Table S3. These vectors were transfected into LO2 cells separately with Lipofectamine 2000 (Invitrogen, Waltham, MA, USA), according to the manufacturer’s instructions. After 48 h, FLAG-tag plasmid-transfected cells were collected for immunoblotting, whereas EGFP-tag plasmid-transfected cells were visualized by fluorescence microscopy (Olympus, Tokyo, Japan). To detect the FLAG-tag fusion protein expression, cell lysates were loaded in nitrocellulose (NC) membrane at the indicated level (2, 1.75, 1.5, 1.25, 1, 0.75, and 0.5 μg). GAPDH (10 μg of protein from each group of cells) was the loading control. Mouse anti-FLAG antibody (CMCTAG, Shanghai, China) and mouse anti-GAPDH antibody (CMCTAG, Shanghai, China) were the primary antibodies.
Clinical Samples and Cell Lines
Twenty hepatic tumor tissues, 20 paired adjacent non-tumor tissues, and 10 paired normal tissues were obtained from patients with HCC who underwent surgical treatment at The First Affiliated Hospital of Xi’an Jiaotong University (Xi’an, China). Adjacent non-tumor tissues were defined as tissues >1 cm and <5 cm from the tumor boundary. Normal tissues were defined as tissues >5 cm from the tumor boundary. Signed informed consents were collected and the study was approved by the Ethics Committee of The First Affiliated Hospital of Xi’an Jiaotong University. LO2 cells were maintained in RPMI 1640 medium (HyClone, South Logan, UT, USA) containing 10% fetal bovine serum (FBS) (ExCell Bio, Shanghai, China). Huh7 cells were maintained in DMEM high-glucose medium (HyClone, South Logan, UT, USA) supplemented with 10% FBS (ExCell Bio, Shanghai, China).
RNA Isolation and Quantitative Real-Time PCR
Total RNA was isolated from stored HCC liver samples, LO2 cells, and Huh7 cells. RNA was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA). The extraction was performed with the chloroform-isopropanol method. RNA quality and concentration were measured using a NanoDrop instrument. cDNAs were generated by the reverse transcription of mRNAs with 5 μg of total RNA per sample using the RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). The microRNA cDNAs were reverse transcribed with 2 μg of total RNA per sample using a Mir-X miRNA qRT-PCR SYBR kit (Clontech Laboratories, Mountain View, CA, USA). Quantitative real-time PCR was performed using SYBR Green qPCR master mix (Roche, Basel, Switzerland) in an Agilent Mx3005P instrument. Primer information for the mRNA quantitative real-time PCR is shown in Table S4. Primer information for the microRNA quantitative real-time PCR is shown in Table S5.
Western Blotting
Protein was isolated from cells by using cell lysis buffer (Beyotime Biotechnology, Shanghai, China). Protein from different cells were separated by 8% SDS-PAGE gel and transferred onto a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA) according to the standard process. Protein expression was analyzed by using rabbit anti-ACTB antibody (Santa Cruz Biotechnology, Shanghai, China, 1:500), mouse anti-GAPDH antibody (CMCTAG, Shanghai, China), rabbit anti-AKAP12 antibody (Proteintech, Wuhan, Hubei, China, 1:1,000), mouse anti-DYRK2 antibody (R&D Systems, Minneapolis, MN, USA, 1:1,000), rabbit anti-FOXN3 antibody (Abcam, Cambridge, MA, USA, 1:1,000), rabbit anti-FOXO1 antibody (Proteintech, Wuhan, Hubei, China, 1:1,000), and rabbit anti-LATS2 antibody (Proteintech, Wuhan, Hubei, China, 1:1,000).
Gene Overexpression and Knockdown
The expression vectors were constructed first. Full-length LNC-HC was obtained by PCR with PrimeSTAR GXL DNA polymerase (Takara Bio, Beijing, China) and inserted into the pcDNA3.1+ vector between the EcoRI and XhoI sites to obtain pcLNC-HC; the SLC25A15 coding region was inserted into the pcDNA3.1+ vector between HindIII and BamHI sites to obtain pcSLC25A15. The primers are shown in Table S6. For gene knockdown, short hairpin RNAs (shRNAs) for LNC-HC and SLC25A15 knockdown were ordered from GenePharma (Shanghai, China). The target sequences of the LNC-HC shRNAs and the SLC25A15 shRNAs are shown in Table S6. After the transfection of pcDNA3.1+, pcLNC-HC, pcSLC25A15, NC shRNAs, LNC-HC shRNAs, and SLC25A15 shRNAs using Lipofectamine 2000 (Invitrogen, Waltham, CA, USA) for 24 h, LO2 and Huh7 cells were treated with G418 (600 μg/mL in LO2 and 800 μg/mL in Huh7) for a period of 30 days to select the stable cell lines overNCLO2/Huh7, overLNC-HCLO2/Huh7, overSLC25A15LO2/Huh7, NCshRLO2/Huh7, LNC-HCshRLO2/Huh7, and SLC25A15shRLO2/Huh7, respectively.
Cell Proliferation Assay
An MTT assay was used to measure cell proliferation. In detail, 1 × 104 Huh7 or LO2 cells were seeded per well in a 96-well plate. The cells were transfected with the NC mimic (50 nM) or miR-183 mimic (50 nM) by using Lipofectamine RNA iMAX reagent (Invitrogen, Waltham, CA, USA) and the pcDNA3.1+ or pcLNC-HC vector by using Lipofectamine 2000 reagent (Invitrogen, Waltham, CA, USA) according to the manufacturer’s instructions. To check the cooperative effect of LNC-HC overexpression and knockdown of its target genes, Huh7 cells were transfected with pcDNA3.1+ or pcLNC-HC vector and NC siRNA (50 nM) or the indicated siRNA (50 nM). At different time points (0, 24, 48, and 72 h) the cells were incubated with 0.5 mg/mL MTT solution at 37°C for 4 h. Then, the culture medium was changed to DMSO (150 μL per well). The optical absorbance was measured at a wavelength of 450 nm. The sequence information for the miR-183 mimic, NC mimic, siRNAs targeting AKAP12, FOXN3, FOXO1, and LATS2 and NC siRNA are shown in Table S7. The siRNAs were ordered from Genepharma (Shanghai, China).
Mouse Xenograft Tumor Model
Animal experiments were approved by the Institutional Animal Ethics Committee of the Xi’an Jiaotong University School of Medicine (permission ID XJ2006Y039; Xi’an Jiaotong University, Xi’an, China) and were conducted in accordance with the European Communities Council Directive 2010/63/EU for the protection of animals used for scientific purposes. Male BALB/c nude mice (n = 6, aged 4 weeks) were ordered from the Medical Experimental Animal Center, Xi’an Jiaotong University. For the xenograft tumor model, 5 × 106 overNCHuh7 or overLNC-HCHuh7 cells were subcutaneously injected into the left or right flanks of nude mice on day 1. After 5 days, the tumor volume and body weight were detected every 5 days. On day 45, the mice were sacrificed, and the tumors were collected and measured.
Dual-Luciferase Reporter Gene Assay
The LNC-HC DNA fragments targeted by hsa-miR-183-5p (wild-type and mutant) were ordered from Sangon Biotech (Shanghai, China). They were inserted into the pmirGLO vector between the NheI and XhoI sites to generate pLNC-HC1-WT, pLNC-HC1-mut, pLNC-HC2-WT, and pLNC-HC2-mut. LO2 and Huh7 cells were seeded into 48-well plates and cotransfected with different pLNC-HC-target plasmids (100 ng) and NC/miR-183 mimic (50 nM) for 24 h. The relative luciferase activity of each cell group was measured using the Dual-luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Survival Analysis
The effects of AKAP12, DYRK2, FOXN3, FOXO1, and LATS2 on the overall survival of patients with liver cancer (n = 364) were analyzed by using the online RNA-seq database Kaplan-Meier Plotter (http://kmplot.com/analysis/).74
Statistical Analysis
Quantitative data were expressed as the means ± SEM. The statistical analysis of the differences between groups was conducted by using an unpaired Student’s t test. The difference in xenograft tumor growth between the overNCHuh7 and overLNC-HCHuh7 injection groups was analyzed by two-way ANOVA. The Fisher’s exact test was performed to compare the association between LNC-HC expression and clinical parameters. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 were considered statistically significant.
Author Contributions
Study design: X.L. and S.L. Analysis and interpretation of the data: X.L. Drafting of the manuscript: X.L. and S.L. Critical revision of the manuscript: X.L., S.L. and E.K.O. Obtained funding: X.L. and L.L. Performance of the experiments: X.L., N.W., L.W., and X.D. Collection of the clinical samples: K.Q and H.L. Preparation and assistant work: J.R., B.W., L.L., and Q.N. Necessary technical guides: D.G., S.C., J.M., and D.L.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (81600679 to X.L. and 31801083 to L.L.); the China Postdoctoral Science Foundation (2016M592778 to X.L.); the Fundamental Research Funds for the Central Universities (xjj2017051 to X.L.); and the Shaanxi Postdoctoral Science Foundation (2016BSHEDZZ98 to X.L.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.03.008.
Contributor Information
Xi Lan, Email: lanxi.2016@xjtu.edu.cn.
Shemin Lu, Email: lushemin@xjtu.edu.cn.
Supplemental Information
References
- 1.Fu J., Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Cronin K.A., Lake A.J., Scott S., Sherman R.L., Noone A.M., Howlader N., Henley S.J., Anderson R.N., Firth A.U., Ma J. Annual Report to the Nation on the Status of Cancer, part I: national cancer statistics. Cancer. 2018;124:2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J.Z., Chen M., Chen D., Gao X.C., Zhu S., Huang H., Hu M., Zhu H., Yan G.R. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell. 2017;68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z.Z., Huang L., Wu Y.H., Zhai W.J., Zhu P.P., Gao Y.F. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat. Commun. 2016;7:12598. doi: 10.1038/ncomms12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan S.X., Wang J., Yang F., Tao Q.F., Zhang J., Wang L.L., Yang Y., Liu H., Wang Z.G., Xu Q.G. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 9.Cao C., Sun J., Zhang D., Guo X., Xie L., Li X., Wu D., Liu L. The long intergenic noncoding RNA UFC1, a target of microRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-catenin in HCC cells. Gastroenterology. 2015;148:415–426.e18. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Quagliata L., Matter M.S., Piscuoglio S., Arabi L., Ruiz C., Procino A., Kovac M., Moretti F., Makowska Z., Boldanova T. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F., Yuan J.H., Wang S.B., Yang F., Yuan S.X., Ye C., Yang N., Zhou W.P., Li W.L., Li W., Sun S.H. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 12.Zhu P., Wang Y., Huang G., Ye B., Liu B., Wu J., Du Y., He L., Fan Z. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 13.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T., Xie J., Shen C., Cheng D., Shi Y., Wu Z., Deng X., Chen H., Shen B., Peng C. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 16.Lan X., Yan J., Ren J., Zhong B., Li J., Li Y., Liu L., Yi J., Sun Q., Yang X. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64:58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 17.Lan X., Wu L., Wu N., Chen Q., Li Y., Du X., Wei C., Feng L., Li Y., Osoro E.K. Long noncoding RNA lnc-HC regulates PPARγ-mediated hepatic lipid metabolism through miR-130b-3p. Mol. Ther. Nucleic Acids. 2019;18:954–965. doi: 10.1016/j.omtn.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Piao H.Y., Guo S., Wang Y., Zhang T., Zheng Z.C., Zhao Y. LINC00163 inhibits the invasion and metastasis of gastric cancer cells as a ceRNA by sponging miR-183 to regulate the expression of AKAP12. Int. J. Clin. Oncol. 2020 doi: 10.1007/s10147-019-01604-w. Published online January 1, 2020. [DOI] [PubMed] [Google Scholar]
- 19.Kang J., Kim W., Lee S., Kwon D., Chun J., Son B., Kim E., Lee J.M., Youn H., Youn B. TFAP2C promotes lung tumorigenesis and aggressiveness through miR-183- and miR-33a-mediated cell cycle regulation. Oncogene. 2017;36:1585–1596. doi: 10.1038/onc.2016.328. [DOI] [PubMed] [Google Scholar]
- 20.Goeppert B., Schmezer P., Dutruel C., Oakes C., Renner M., Breinig M., Warth A., Vogel M.N., Mittelbronn M., Mehrabi A. Down-regulation of tumor suppressor A kinase anchor protein 12 in human hepatocarcinogenesis by epigenetic mechanisms. Hepatology. 2010;52:2023–2033. doi: 10.1002/hep.23939. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L., Su X., Li B., Chu C., Sun H., Zhang N., Han B., Li C., Zou B., Niu Y., Zhang R. PM2.5 exposure impairs sperm quality through testicular damage dependent on NALP3 inflammasome and miR-183/96/182 cluster targeting FOXO1 in mouse. Ecotoxicol. Environ. Saf. 2019;169:551–563. doi: 10.1016/j.ecoenv.2018.10.108. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki R., Amatya V.J., Kushitani K., Kai Y., Kambara T., Takeshima Y. miR-182 and miR-183 promote cell proliferation and invasion by targeting FOXO1 in mesothelioma. Front. Oncol. 2018;8:446. doi: 10.3389/fonc.2018.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Q., Sang L., Du S. Long noncoding RNA LINC00261 regulates endometrial carcinoma progression by modulating miRNA/FOXO1 expression. Cell Biochem. Funct. 2018;36:323–330. doi: 10.1002/cbf.3352. [DOI] [PubMed] [Google Scholar]
- 24.Hou Y., Fu L., Li J., Li J., Zhao Y., Luan Y., Liu A., Liu H., Li X., Zhao S., Li C. Transcriptome analysis of potential miRNA involved in adipogenic differentiation of C2C12 myoblasts. Lipids. 2018;53:375–386. doi: 10.1002/lipd.12032. [DOI] [PubMed] [Google Scholar]
- 25.Ichiyama K., Gonzalez-Martin A., Kim B.S., Jin H.Y., Jin W., Xu W., Sabouri-Ghomi M., Xu S., Zheng P., Xiao C., Dong C. The microRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity. 2016;44:1284–1298. doi: 10.1016/j.immuni.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebremedhn S., Salilew-Wondim D., Hoelker M., Rings F., Neuhoff C., Tholen E., Schellander K., Tesfaye D. MicroRNA-183-96-182 cluster regulates bovine granulosa cell proliferation and cell cycle transition by coordinately targeting FOXO1. Biol. Reprod. 2016;94:127. doi: 10.1095/biolreprod.115.137539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebremedhn S., Salilew-Wondim D., Ahmad I., Sahadevan S., Hossain M.M., Hoelker M., Rings F., Neuhoff C., Tholen E., Looft C. MicroRNA expression profile in bovine granulosa cells of preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. PLoS ONE. 2015;10:e0125912. doi: 10.1371/journal.pone.0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Quan H., Wang S., Li X., Che X. miR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol. 2015;36:8121–8126. doi: 10.1007/s13277-015-3550-8. [DOI] [PubMed] [Google Scholar]
- 29.Leung W.K., He M., Chan A.W., Law P.T., Wong N. Wnt/β-catenin activates miR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015;362:97–105. doi: 10.1016/j.canlet.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 30.Nozaki T., Ohura K. Regulation of miRNA during direct reprogramming of dental pulp cells to insulin-producing cells. Biochem. Biophys. Res. Commun. 2014;444:195–198. doi: 10.1016/j.bbrc.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 31.McLoughlin H.S., Wan J., Spengler R.M., Xing Y., Davidson B.L. Human-specific microRNA regulation of FOXO1: implications for microRNA recognition element evolution. Hum. Mol. Genet. 2014;23:2593–2603. doi: 10.1093/hmg/ddt655. [DOI] [PubMed] [Google Scholar]
- 32.Tang H., Bian Y., Tu C., Wang Z., Yu Z., Liu Q., Xu G., Wu M., Li G. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr. Cancer Drug Targets. 2013;13:221–231. doi: 10.2174/1568009611313020010. [DOI] [PubMed] [Google Scholar]
- 33.Xie L., Ushmorov A., Leithäuser F., Guan H., Steidl C., Färbinger J., Pelzer C., Vogel M.J., Maier H.J., Gascoyne R.D. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119:3503–3511. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 34.Myatt S.S., Wang J., Monteiro L.J., Christian M., Ho K.K., Fusi L., Dina R.E., Brosens J.J., Ghaem-Maghami S., Lam E.W. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X.J., Zhang D.L., Fu C., Wei B.Z., Li G.J. miR-183 modulates multi-drug resistance in hepatocellular cancer (HCC) cells via miR-183-IDH2/SOCS6-HIF-1α feedback loop. Eur. Rev. Med. Pharmacol. Sci. 2016;20:2020–2027. [PubMed] [Google Scholar]
- 36.Tanaka H., Sasayama T., Tanaka K., Nakamizo S., Nishihara M., Mizukawa K., Kohta M., Koyama J., Miyake S., Taniguchi M. MicroRNA-183 upregulates HIF-1α by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J. Neurooncol. 2013;111:273–283. doi: 10.1007/s11060-012-1027-9. [DOI] [PubMed] [Google Scholar]
- 37.Vohwinkel C.U., Lecuona E., Sun H., Sommer N., Vadász I., Chandel N.S., Sznajder J.I. Elevated CO2 levels cause mitochondrial dysfunction and impair cell proliferation. J. Biol. Chem. 2011;286:37067–37076. doi: 10.1074/jbc.M111.290056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y.Y., Zeng Y., Zheng J.Y., Huang C.L., Wang X.P. [miR-183 regulates proliferation of SW1990 pancreatic cancer cell line by targeting PDCD4] Sichuan Da Xue Xue Bao Yi Xue Ban. 2016;47:691–696. [PubMed] [Google Scholar]
- 39.Cheng Y., Xiang G., Meng Y., Dong R. miRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod. Biol. 2016;16:225–233. doi: 10.1016/j.repbio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Li C., Deng L., Zhi Q., Meng Q., Qian A., Sang H., Li X., Xia J. MicroRNA-183 functions as an oncogene by regulating PDCD4 in gastric cancer. Anticancer. Agents Med. Chem. 2016;16:447–455. doi: 10.2174/1871520615666150914114237. [DOI] [PubMed] [Google Scholar]
- 41.Wei C., Song H., Sun X., Li D., Song J., Hua K., Fang L. miR-183 regulates biological behavior in papillary thyroid carcinoma by targeting the programmed cell death 4. Oncol. Rep. 2015;34:211–220. doi: 10.3892/or.2015.3971. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y.Y., Zheng J.Y., Liu J., Huang C.L., Zhang W., Zeng Y. miR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed. Pharmacother. 2015;70:151–157. doi: 10.1016/j.biopha.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Yang M., Liu R., Li X., Liao J., Pu Y., Pan E., Yin L., Wang Y. miRNA-183 suppresses apoptosis and promotes proliferation in esophageal cancer by targeting PDCD4. Mol. Cells. 2014;37:873–880. doi: 10.14348/molcells.2014.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu W., Gao T., Shen J., Sun Y., Zheng X., Wang J., Ma J., Hu X.Y., Li J., Hu M.J. MicroRNA-183 inhibits apoptosis and promotes proliferation and invasion of gastric cancer cells by targeting PDCD4. Int. J. Clin. Exp. Med. 2014;7:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 45.Ren L.H., Chen W.X., Li S., He X.Y., Zhang Z.M., Li M., Cao R.S., Hao B., Zhang H.J., Qiu H.Q., Shi R.H. MicroRNA-183 promotes proliferation and invasion in oesophageal squamous cell carcinoma by targeting programmed cell death 4. Br. J. Cancer. 2014;111:2003–2013. doi: 10.1038/bjc.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Fu H., Xu C., Tie Y., Xing R., Zhu J., Qin Y., Sun Z., Zheng X. miR-183 inhibits TGF-β1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010;10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Ma Z., Liu X., Zhang C., Hu Y., Ding L., Qi P., Wang J., Lu S., Li Y. miR-183-5p is required for non-small cell lung cancer progression by repressing PTEN. Biomed. Pharmacother. 2019;111:1103–1111. doi: 10.1016/j.biopha.2018.12.115. [DOI] [PubMed] [Google Scholar]
- 48.Sarver A.L., Li L., Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 49.Li Z.B., Li Z.Z., Li L., Chu H.T., Jia M. miR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3208–3217. [PubMed] [Google Scholar]
- 50.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M., Guan H., Liu Y., Gan X. lncRNA ZEB1-AS1 reduces liver cancer cell proliferation by targeting miR-365a-3p. Exp. Ther. Med. 2019;17:3539–3547. doi: 10.3892/etm.2019.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu X., Li Y., Kong D., Hu L., Liu D., Wu J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J. Cell. Physiol. 2019;234:10800–10808. doi: 10.1002/jcp.27903. [DOI] [PubMed] [Google Scholar]
- 53.Wu L.L., Cai W.P., Lei X., Shi K.Q., Lin X.Y., Shi L. NRAL mediates cisplatin resistance in hepatocellular carcinoma via miR-340-5p/Nrf2 axis. J. Cell Commun. Signal. 2019;13:99–112. doi: 10.1007/s12079-018-0479-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H., Huo X., Yang X.R., He J., Cheng L., Wang N., Deng X., Jin H., Wang N., Wang C. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S.P., Xu H.X., Yu Y., He J.D., Wang Z., Xu Y.J., Wang C.Y., Zhang H.M., Zhang R.X., Zhang J.J. lncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang L., Du W.W., Yang X., Chen K., Ghanekar A., Levy G., Yang W., Yee A.J., Lu W.Y., Xuan J.W. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013;27:907–919. doi: 10.1096/fj.12-220905. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Q.H., Sun H.M., Zheng R.Z., Li Y.C., Zhang Q., Cheng P., Tang Z.H., Huang F. Meta-analysis of microRNA-183 family expression in human cancer studies comparing cancer tissues with noncancerous tissues. Gene. 2013;527:26–32. doi: 10.1016/j.gene.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Liang Z., Gao Y., Shi W., Zhai D., Li S., Jing L., Guo H., Liu T., Wang Y., Du Z. Expression and significance of microRNA-183 in hepatocellular carcinoma. ScientificWorldJournal. 2013;2013:381874. doi: 10.1155/2013/381874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashmawy A.M., Elgeshy K.M., Abdel Salam E.T., Ghareeb M., Kobaisi M.H., Amin H.A.A., Sharawy S.K., Abdel Wahab A.H.A. Crosstalk between liver-related microRNAs and Wnt/β-catenin pathway in hepatocellular carcinoma patients. Arab J. Gastroenterol. 2017;18:144–150. doi: 10.1016/j.ajg.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Bharali D., Jebur H.B., Baishya D., Kumar S., Sarma M.P., Masroor M., Akhter J., Husain S.A., Kar P. Expression analysis of serum microRNA-34a and microRNA-183 in hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2018;19:2561–2568. doi: 10.22034/APJCP.2018.19.9.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X., Xu P., Ni W., Fan H., Xu J., Chen Y., Huang W., Lu S., Liang L., Liu J. Downregulated DYRK2 expression is associated with poor prognosis and oxaliplatin resistance in hepatocellular carcinoma. Pathol. Res. Pract. 2016;212:162–170. doi: 10.1016/j.prp.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Hayashi M., Nomoto S., Kanda M., Okamura Y., Nishikawa Y., Yamada S., Fujii T., Sugimoto H., Takeda S., Kodera Y. Identification of the A kinase anchor protein 12 (AKAP12) gene as a candidate tumor suppressor of hepatocellular carcinoma. J. Surg. Oncol. 2012;105:381–386. doi: 10.1002/jso.22135. [DOI] [PubMed] [Google Scholar]
- 63.Xia W., Ni J., Zhuang J., Qian L., Wang P., Wang J. miR-103 regulates hepatocellular carcinoma growth by targeting AKAP12. Int. J. Biochem. Cell Biol. 2016;71:1–11. doi: 10.1016/j.biocel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 64.Sun J., Li H., Huo Q., Cui M., Ge C., Zhao F., Tian H., Chen T., Yao M., Li J. The transcription factor FOXN3 inhibits cell proliferation by downregulating E2F5 expression in hepatocellular carcinoma cells. Oncotarget. 2016;7:43534–43545. doi: 10.18632/oncotarget.9780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L., Li S., Wang R., Chen C., Ma W., Cai H. Anti-tumor effect of LATS2 on liver cancer death: role of DRP1-mediated mitochondrial division and the Wnt/β-catenin pathway. Biomed. Pharmacother. 2019;114:108825. doi: 10.1016/j.biopha.2019.108825. [DOI] [PubMed] [Google Scholar]
- 66.Song J., Zhao W., Lu C., Shao X. Retracted. Cancer Cell Int. 2019;19:359. doi: 10.1186/s12935-019-1084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han L.L., Yin X.R., Zhang S.Q. miR-650 promotes the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by directly inhibiting LATS2 expression. Cell. Physiol. Biochem. 2018;51:1179–1192. doi: 10.1159/000495495. [DOI] [PubMed] [Google Scholar]
- 68.Han L.L., Yin X.R., Zhang S.Q. miR-103 promotes the metastasis and EMT of hepatocellular carcinoma by directly inhibiting LATS2. Int. J. Oncol. 2018;53:2433–2444. doi: 10.3892/ijo.2018.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bao L., Yuan L., Li P., Bu Q., Guo A., Zhang H., Cui N., Liu B. A FUS-LATS1/2 axis inhibits hepatocellular carcinoma progression via activating Hippo pathway. Cell. Physiol. Biochem. 2018;50:437–451. doi: 10.1159/000494155. [DOI] [PubMed] [Google Scholar]
- 70.Zhang S., Wang J., Wang H., Fan L., Fan B., Zeng B., Tao J., Li X., Che L., Cigliano A. Hippo cascade controls lineage commitment of liver tumors in mice and humans. Am. J. Pathol. 2018;188:995–1006. doi: 10.1016/j.ajpath.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yi J., Lu L., Yanger K., Wang W., Sohn B.H., Stanger B.Z., Zhang M., Martin J.F., Ajani J.A., Chen J. Large tumor suppressor homologs 1 and 2 regulate mouse liver progenitor cell proliferation and maturation through antagonism of the coactivators YAP and TAZ. Hepatology. 2016;64:1757–1772. doi: 10.1002/hep.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee D.H., Park J.O., Kim T.S., Kim S.K., Kim T.H., Kim M.C., Park G.S., Kim J.H., Kuninaka S., Olson E.N. LATS-YAP/TAZ controls lineage specification by regulating TGFβ signaling and Hnf4α expression during liver development. Nat. Commun. 2016;7:11961. doi: 10.1038/ncomms11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo C., Wang X., Liang L. LATS2-mediated YAP1 phosphorylation is involved in HCC tumorigenesis. Int. J. Clin. Exp. Pathol. 2015;8:1690–1697. [PMC free article] [PubMed] [Google Scholar]
- 74.Saito Y., Li L., Coyaud E., Luna A., Sander C., Raught B., Asara J.M., Brown M., Muthuswamy S.K. LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature. 2019;569:275–279. doi: 10.1038/s41586-019-1126-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.