Abstract
In this work we provide preclinical data to support initiation of a first-in-human trial for sickle cell disease (SCD) using an approach that relies on reversal of the developmental fetal-to-adult hemoglobin switch. Erythroid-specific knockdown of BCL11A via a lentiviral-encoded microRNA-adapted short hairpin RNA (shRNAmiR) leads to reactivation of the gamma-globin gene while simultaneously reducing expression of the pathogenic adult sickle β-globin. We generated a refined lentiviral vector (LVV) BCH-BB694 that was developed to overcome poor vector titers observed in the manufacturing scale-up of the original research-grade LVV. Healthy or sickle cell donor CD34+ cells transduced with Good Manufacturing Practices (GMP)-grade BCH-BB694 LVV achieved high vector copy numbers (VCNs) >5 and gene marking of >80%, resulting in a 3- to 5-fold induction of fetal hemoglobin (HbF) compared with mock-transduced cells without affecting growth, differentiation, and engraftment of gene-modified cells in vitro or in vivo. In vitro immortalization assays, which are designed to measure vector-mediated genotoxicity, showed no increased immortalization compared with mock-transduced cells. Together these data demonstrate that BCH-BB694 LVV is non-toxic and efficacious in preclinical studies, and can be generated at a clinically relevant scale in a GMP setting at high titer to support clinical testing for the treatment of SCD.
Keywords: Sickle cell disease, HbS, hemoglobinopathies, gene therapy, lentiviral vector, BCL11A, hemoglobin switch, fetal hemoglobin, shRNAmiR, RNA interference
Introduction
Sickle cell disease (SCD) is the most common monogenic disease in the world.1 Around 2.1% of adults globally are carriers,2 and an estimated 100,000 individuals are affected in the United States alone.3 SCD is an inherited disorder caused by the E6V missense mutation [rs334] in the β-globin gene, leading to the production of mutant sickle hemoglobin S (HbS). Homozygosity of this mutation, coinheritance with β-globin thalassemic variants (β0 or β+ mutations), and the hemoglobin C (HbC) mutation (E6K) in the second β-globin allele all lead to SCD.4, 5, 6, 7 HbS has an increased propensity to polymerize under low oxygen conditions, which causes formation of sickled, inflexible red blood cells (RBCs). Sickled and sickle-polymer-containing RBCs are associated with hemolytic anemia, vasculopathy, and vaso-occlusive events (VOEs) that are characteristic for SCD and subsequently result in serious acute and chronic complications.
Treatments to reduce SCD complications include fluid administration, acute analgesia for VOEs, and daily administration of hydroxyurea, which has been used for more than 20 years to increase fetal hemoglobin (HbF) expression.8 In some cases, erythrocyte transfusions are administered to improve oxygen delivery in the presence of sickle RBCs. These treatments are largely symptomatic and transient, and the only available curative therapy for SCD is hematopoietic stem cell transplantation (HSCT).9 Around 2,000 SCD patients in the world have received HSCT from an allogeneic donor, and survival rates can exceed 90% in Europe and the United States, with the best outcomes in SCD attained using histocompatibility leukocyte antigen (HLA)-matched related donors.10,11 Only a minority of SCD patients can benefit from this therapy because of the unavailability of HLA-matched unaffected related donors.12, 13, 14 Genetically engineering autologous cells offers two major benefits over allogeneic HSCT: it eliminates the need to find a suitable hematopoietic stem cell (HSC) donor, and it eliminates the risks of graft-versus-host-disease (GVHD) and immune-mediated graft rejection, both serious complications of allogeneic HSCT. Gene therapy has been used to successfully treat multiple rare genetic conditions, including adenosine deaminase deficiency,15 X-linked severe combined immunodeficiency (SCID),16, 17, 18 X-linked chronic granulomatous disease,19 Wiskott-Aldrich syndrome,20,21 childhood cerebral adrenoleukodystrophy,22,23 and metachromatic leukodystrophy.24 More recently, ex vivo lentiviral vector (LVV)-based gene therapy has shown promise as a treatment for severe SCD and β-thalassemia.25, 26, 27, 28, 29 In previous and ongoing trials, this approach relies on the regulated expression of β-globin, a modified anti-sickling β-globin (HbAT87Q), or transgenic expression of γ-globin in erythroid cells. Several clinical trials that rely on a similar strategy but use different beta-like globin variants are ongoing.
An alternative approach to gene therapy for SCD aims at reversing the fetal-to-adult hemoglobin switch by interfering with the transcriptional repressor BCL11A. BCL11A was first identified as a potent regulator of the hemoglobin switch in genome-wide association studies (GWASs) in healthy individuals with higher levels of HbF.30, 31, 32 In erythroid cells, BCL11A functions as a developmental stage-specific repressor of HbF expression,33 but it is also essential for B lymphocyte development, and more recently was identified by us and others as critical for HSC function.34, 35, 36 A proof-of-concept study showing phenotypic correction of SCD has been reported in a transgenic mouse model of SCD with genetic deletion of Bcl11a, which displayed pancellular induction of HbF.37 Interfering with BCL11A takes advantage of this developmentally regulated physiological switch and has the potential added benefit over gene addition strategies of not only providing therapeutically relevant levels of functional γ-globin, but simultaneously reducing expression of the adult β-globin gene, thereby concurrently and coordinately reducing the concentration of HbS in erythrocytes.38 The increase in HbF, which itself has potent anti-sickling properties,39,40 and concomitant reduction in HbS provide an extremely powerful approach to attenuating the sickling tendency of RBCs.
We have previously reported on an LVV that mediates potent erythroid-specific knockdown of BCL11A via RNA interference (RNAi) using a microRNA (miRNA)-adapted short hairpin RNA (shRNAmiR).35,41 In order to circumvent poor titers seen during large-scale production of the original research construct, we transferred the hairpin cassette into an LVV backbone that has consistently demonstrated enhanced titer characteristics.26,29,42 We present data on the safety and efficacy of this LVV and show large-scale validation experiments using clinical-grade LVV to genetically modify SCD patient cells. The data presented here form the basis for an ongoing NIH-funded pilot and feasibility clinical trial (ClinicalTrials.org: NCT03282656).
Results
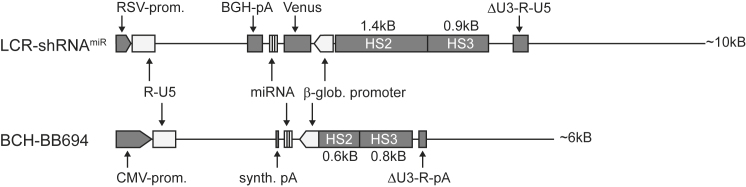
Design and Production of the BCH-BB694 Anti-BCL11A LVV for HbF Induction
Inducing HbF is a promising approach to correcting the phenotype of β-thalassemic or SCD erythrocytes.43 We have previously designed an LVV (LCR-shRNAmiR) for erythroid-specific BCL11A knockdown, which resulted in robust induction of γ-globin and increased production of HbF.35 With the aim of conducting Investigational New Drug (IND)-enabling studies and to initiate a clinical trial, we performed pilot large-scale production of purified LVV. HEK293T cells were expanded and transfected with helper constructs and the plasmid pLCR-shRNAmiR. After clarification, nuclease digestion, and filtration, vector particles were purified by ion exchange chromatography and concentrated by tangential flow filtration before final filtration and vialing. The titer of this vector in pilot production runs was lower than desired, which prompted us to test the therapeutic shRNAmiR cassette in an alternative LVV backbone, which was previously used in clinical trials.26,29 Similar to the research vector, this optimized self-inactivating (SIN) third generation LVV BCH-BB694 expresses the therapeutic shRNAmiR targeting BCL11A under transcriptional control of regulatory elements derived from the β-globin locus (Figure 1). The regulatory elements are comprised of portions of DNase hypersensitive sites 2 and 3 (HS2 and HS3) of the β-globin locus control region (LCR) fused to the minimal proximal promoter element from the β-globin gene. The Venus fluorescent reporter present in LCR-shRNAmiR is not present in this clinical BCH-BB694 LVV. The optimized backbone is substantially smaller in size (6 versus 10 kB) and was associated with a nearly 5-fold increase in titers as assessed in three independent production runs (Figure S1), with the titer of the final Good Manufacturing Practices (GMP) grade vector batch being 6.47E+8 transducing units/ml (TU/ml). In a side-by-side comparison between the original LCR-shRNAmiR and the refined BCH-BB694 LVVs, we confirmed comparable levels of HbF induction per vector insertion. CD34+ cells from two different healthy donors (HDs) were left unmodified (mock) or transduced with an LVV containing a non-targeting control hairpin (NT), the original LCR-shRNAmiR research LVV, or BCH-BB694 LVV (Figure S2). After erythroid differentiation in liquid culture in vitro, the levels of HbF induction were determined by high-performance liquid chromatography (HPLC) on the total cell population, which owing to the lack of a selectable marker included untransduced cells. At similar vector copy numbers (VCNs; ∼1 copy/diploid genome [c/dg]), the fractions of HbF were comparable for LCR-shRNAmiR and BCH-BB694 LVVs, ranging between 23% and 33% in these mixed cell populations. For comparison with previous experiments, cells transduced with LCR-shRNAmiR were additionally sorted by flow cytometry for Venus expression (data not shown), and induction levels of HbF were between 44% and 69% of total hemoglobin, consistent with previous findings.35
Figure 1.
Comparison of the LCR-shRNAmiR and the New BCH-BB694 Lentiviral Vector
The representation shows the plasmid form of the vectors to scale. Both vectors contain the same shRNAmiR sequence targeting BCL11A embedded in a microRNA scaffold,35,41 which is expressed under control of the β-globin promoter and regulatory elements derived from hypersensitive sites 2 and 3 (HS2 and HS3) of the human β-globin locus control region (LCR). Elements that are identical are light gray; elements that differ between vectors are indicated in dark gray. The total length of the plasmid is indicated on the right. BGH-pA, bovine growth hormone poly-adenylation signal; CMV-prom, cytomegalovirus promoter; HS2 and HS3, DNase hypersensitive sites 2 and 3 derived from the β-globin LCR; miRNA, short hairpin RNA embedded in miRNA-derived flanking sequences; RSV-prom, Rous sarcoma virus promoter; synth-pA, synthetic poly-A-signal;79 ΔU3, self-inactivating 3′ long terminal repeat (LTR).
Assessment of Transduction Efficacy and HbF Induction Using HD and SCD CD34+ Cells In Vitro
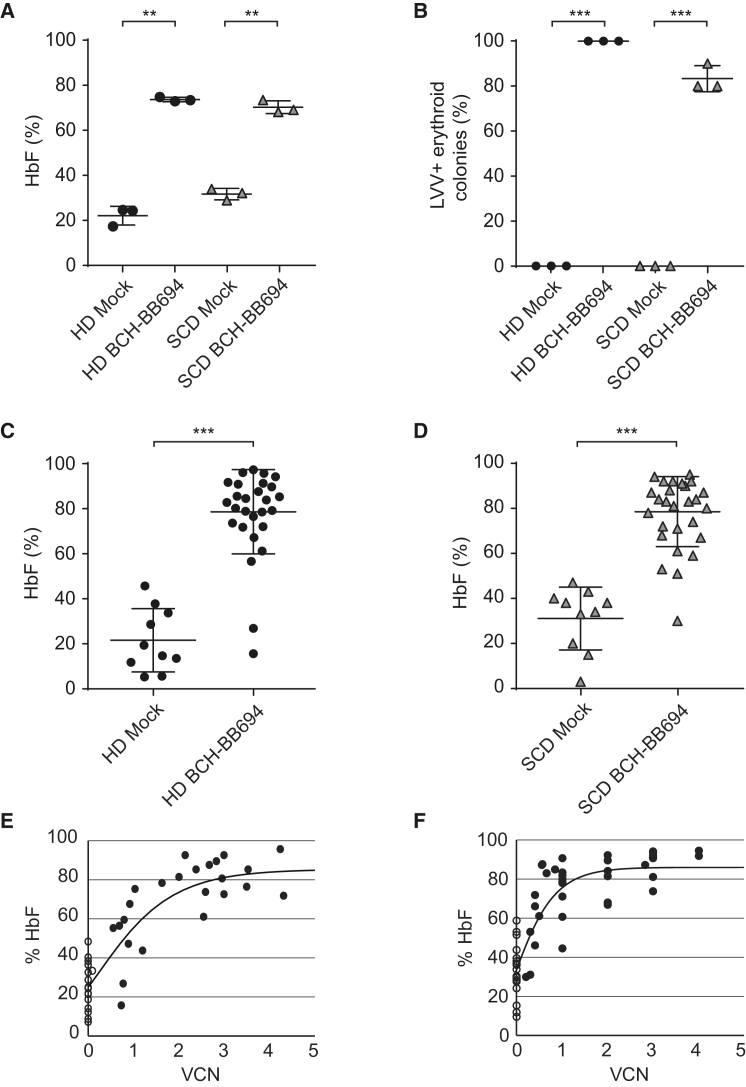
To further characterize the performance of the BCH-BB694 LVV, we transduced human mobilized peripheral blood (mPB) CD34+ cells from HD and SCD donors at a multiplicity of infection (MOI) of 25. After transduction the cells were kept in culture for 2 weeks under conditions supporting erythroid differentiation.44,45 Ion exchange-HPLC (IE-HPLC) analysis was performed to monitor expression of adult type (HbA or HbS) hemoglobin or HbF (Figures 2A and 2B), and the VCN was assessed by qPCR. The levels of HbF in mock-treated control groups were 22% and 32%, on average, for HD and SCD groups, respectively. BCH-BB694 LVV-transduced samples showed significant induction of HbF averaging 74% and 70% for HD and SCD groups at VCNs of 2.78 ± 0.08 and 1.16 ± 0.03 c/dg, respectively. To further assess the rate of transduction, we utilized progenitor colony-forming assays as a surrogate readout for HSC transduction and to assess HbF induction at the clonal level. Transduced CD34+ cells were plated in semisolid media under conditions supporting myeloid and erythroid colony formation. The total number of colonies was similar between mock and transduced groups (Figure S3). Individual erythroid colonies were picked and analyzed by HPLC (Figures 2C and 2D). Similar to the results from erythroid differentiation cultures, the mean HbF in mock-treated and BCH-BB694 LVV-transduced HD cells was 22% and 79%, respectively, at a mean VCN of 3.5 c/dg in the transduced colonies. Transduced CD34+ cells from SCD patients showed 31% HbF in mock and 79% HbF on average in BCH-BB694 LVV-transduced groups at a VCN of 1.95 c/dg. Nearly all the analyzed colonies from the transduced groups contained integrated vector and showed high HbF levels, demonstrating highly efficient transduction of HSPC. We additionally investigated the correlation between the levels of HbF and the VCNs in individual erythroid colonies derived from HD or SCD CD34+ cells (Figures 2E and 2F). The observed distribution demonstrated a non-linear correlation between VCN and HbF induction in addition to some degree of variegation between transduced clones. High HbF induction was seen in colonies with only a single vector integrant (∼30% increase in HbF over mock control per VCN), which is indicative of the potency of the BCH-BB694 LVV vector, and colonies containing higher VCNs showed additional moderate but non-linear increase in HbF.
Figure 2.
In Vitro Assays with Human CD34+ Cells
Human CD34+ cells from a healthy donor (HD) and a sickle cell disease (SCD) donor were transduced with BCH-BB694. (A) The proportion of HbF (A) of total hemoglobin was assessed by ion exchange HPLC in bulk erythroid liquid cultures. The percentage of HbF was calculated based on peak areas. The average VCN was measured by qPCR: VCN HD, 2.78 ± 0.08 copies per diploid genome (c/dg); SCD, 1.16 ± 0.03 c/dg. (B) Colony assays were performed, and the fraction of transduced colonies containing the vector was assessed in individual replicates. (C and D) HbF induction was analyzed in individual erythroid colonies by ion exchange HPLC for HD (C) or SCD (D) samples. (E and F) The percentage of HbF (%HbF) in each colony plotted as a function of the vector copy number (VCN) per diploid genome in individual erythroid colonies for HD (E) or SCD (F) samples. Open circles indicate mock groups; closed circles indicate BCH-BB694-transduced groups. R2 = 0.83 and 0.79, respectively. The average VCN on pooled colonies was HD: 3.54 ± 0.82 and SCD: 1.95 ± 0.11 c/dg. Two-sided unpaired t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005
Quantification of Vector-Mediated Genotoxicity Using the In Vitro Immortalization Assay
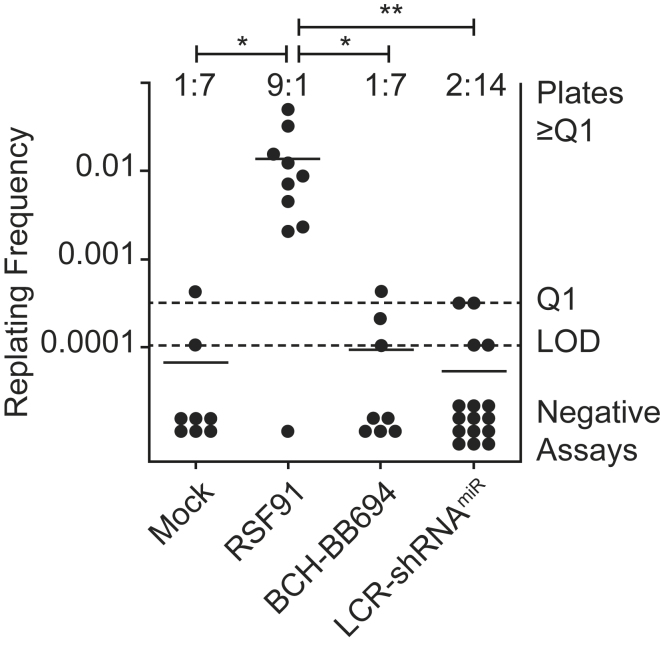
We next assessed the potential of the LCR-shRNAmiR and BCH-BB694 LVVs for insertional mutagenesis using an in vitro immortalization assay (IVIM).46 This quantitative assay detects the rate of immortalization of primary lineage-negative mouse bone marrow (BM) cells caused by insertional mutagenesis. The assay includes a positive control vector RSF91 previously demonstrated to induce immortalization.47 At MOIs of up to 500 and VCNs of ∼4–11 (mean 7.5 c/dg), there was no difference in the frequency of immortalization of BCH-BB694 LVV-transduced cells in comparison with mock-transduced cells, whereas the positive control showed the expected high rate of immortalization in this assay (Figure 3). In addition, BCH-BB694 LVV-transduced cells showed no differences in proliferation or viability compared with mock-transduced cells, indicating the absence of detectable signs of cellular toxicity (Figure S4). These results indicate low genotoxicity of the BCH-BB694 LVV.
Figure 3.
IVIM Assay
The in vitro immortalization (IVIM) assay was performed to assess the genotoxic potential of the BCH-BB694 and LCR-shRNAmiR gene therapy vectors. Untransduced (mock) and RSF91-transduced cells served as negative and positive controls, respectively. Each dot represents an independent assay replicate. The frequency of assays with a detectable immortalization event is shown above the plot, and negative assays showed no cell growth in replating assays. The y axis indicates the frequency of immortalization events per integrated vector copy. LOD, limit of detection; Q1, threshold for quantification. Statistical test: Fisher’s exact test, ∗p ≤ 0.05, ∗∗p ≤ 0.01.
Competitive Transplantation Assay for the Detection of Phenotoxicity Due to BCL11A Knockdown
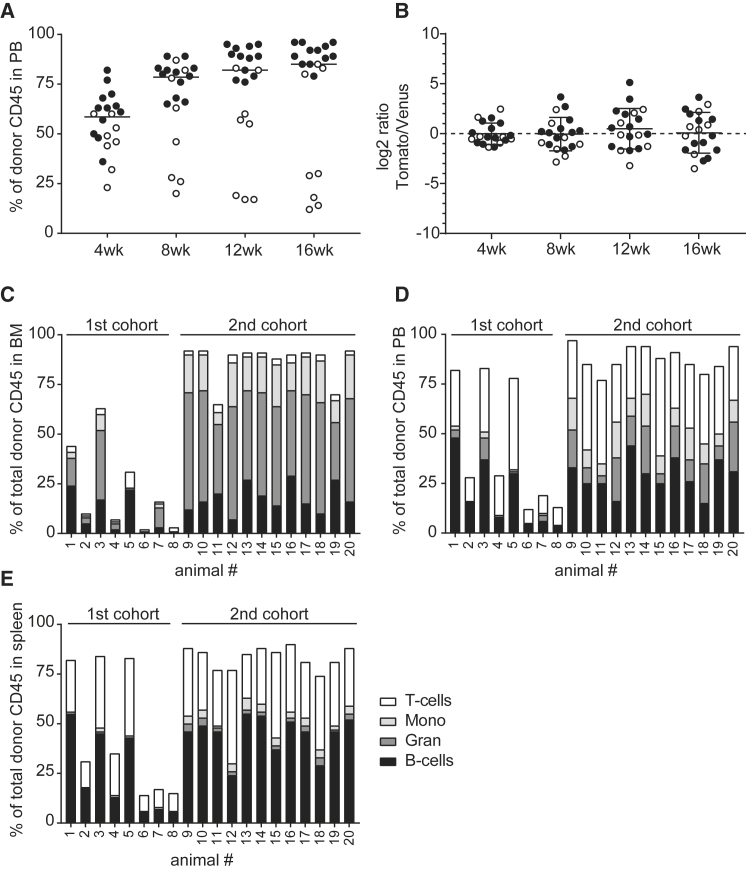
We and others have previously shown that the transcription factor BCL11A is essential for the engraftment of HSCs.35,36 Knockdown of BCL11A in all hematopoietic cells is associated with a rapid and near-complete loss of transduced cells after transplantation in vivo. Thus, a tightly regulated erythroid-specific knockdown of BCL11A mediated by the transcriptional control elements present in BCH-BB694 and LCR-shRNAmiR LVVs is essential to allow for stem cell engraftment, long-term reconstitution, and therapeutic efficacy. To confirm the lack of any negative effect of LCR-shRNAmiR transduction on engraftment, we performed competitive transplantation experiments. Lineage-negative BM cells from CD45.1+ B6 mice were transduced with the LCR-shRNAmiR LVV (which contains the Venus fluorescent reporter) and mixed with an equal number of cells transduced with an LVV encoding only the dTomato fluorochrome under transcriptional control of the LCR as a neutral competitor (Figure S5). Transduction rates of both competitor populations were similar as assessed by colony assays (Figure S6), with an average VCN of ∼1 c/dg across both groups. The relative frequency of different colony types and the absolute number of colonies were also comparable between groups. The cell mixture was transplanted into congenic CD45.2 B6 recipients, and the relative ratio of the two populations in peripheral blood (PB) was monitored in 4-week intervals over a 16-week reconstitution period (Figure 4). Two independent experiments were performed that differ in their irradiation dose used for conditioning. For the first experiment, recipient animals received 10 Gy total body irradiation, which led to incomplete and heterogeneous engraftment of both groups of donor cells (Figure 4A, open circles). Consequently, in a second independent experiment, the dose of irradiation was increased to 11.5 Gy, which led to near-complete donor chimerism (Figure 4A, closed circles). We were able to assess the frequency of Venus- and dTomato-positive cells in all animals from both experiments and calculate log2-transformed relative ratios to obtain Gaussian distribution for statistical analysis. As shown in Figure 4B, the competitor populations showed equal engraftment, suggesting that LVV-mediated erythroid lineage-specific knockdown of BCL11A is compatible with normal HSPC engraftment and function in this assay. Furthermore, there were no signs of skewing in hematopoietic lineages in the BM, PB, or spleen of transplanted animals (Figures 4C–4E).
Figure 4.
Competitive Transplantation of LCR-shRNAmiR versus an Empty Control Vector
Lineage-negative donor cells from CD45.1 mice were transduced with LCR-shRNAmiR co-expressing Venus or with empty control vector expressing the dTomato fluorescent reporter. Cells were mixed in equal proportions and transplanted into CD45.2 recipient animals. (A) The donor CD45 engraftment was assessed in peripheral blood (PB) in two separate cohorts (open and closed circles represent first and second cohorts) at various time points. Each dot represents one animal. (B) The relative ratio of transduced cells from the two competitor populations over time. Each dot represents one animal. (C) Total donor cell engraftment and lineage distribution in individual animals in bone marrow (BM), in PB (D), and in spleen (E).
Xenotransplantation of Gene-Modified CD34+ Cells into NSG Mice
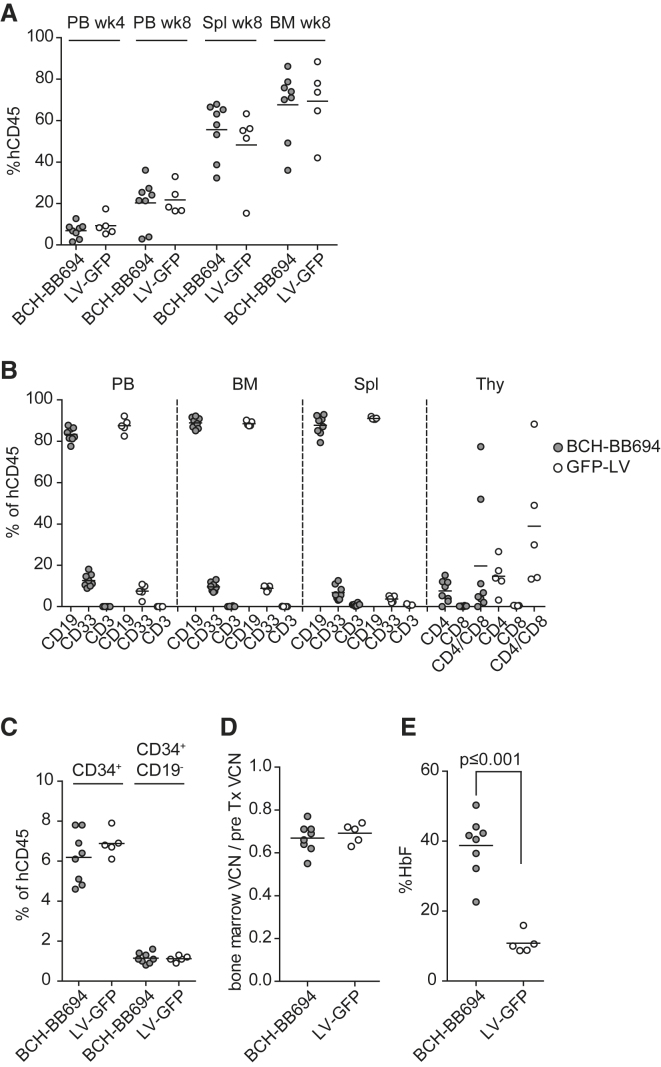
With the aim of confirming these findings in a model system with primary human cells, a xenograft transplantation experiment was performed using HD mPB CD34+ cells transduced with BCH-BB694 LVV or a neutral control vector expressing only GFP. Similar engraftment of human CD45+ cells was observed in PB 4 and 8 weeks post-transplantation and in spleen and BM at 8 weeks post-transplantation for both groups (Figure 5A). The lineage distribution in PB, BM, spleen, and thymus (Figure 5B) and the frequency of BM CD34+ and CD34+/CD19− cells (Figure 5C) were comparable between groups. The frequency of B cells was similar between groups, indicating normal BCL11A function in lymphoid cells. To determine any toxicity in the engrafting HSPC population, we divided the VCN determined in BM cells 8 weeks post-transplantation by the pre-transplant VCN, determined from a cell product that was kept in culture for 14 days. The VCNs from harvested total BM (post-transplant) (Figure 5D) were similar to the VCN of the cells transplanted (pre-transplant) in both groups. Similar results for VCNs were obtained with samples from PB, human CD45+ (hCD45+) BM, hCD34+ BM, spleen, and thymus (Table S1). These data demonstrate the absence of engraftment defects or toxicity mediated by the LVV. Because human erythropoiesis is poorly supported in NSG mice, we isolated hCD34+ cells from BM of transplanted animals and induced erythroid differentiation in vitro. HbF levels in erythroid cells derived from the BCH-BB694 LVV treatment group demonstrated 39% ± 8% HbF compared with 10% ± 3% in the control GFP LVV group (Figure 5E) as determined by HPLC, with the amount of HbF closely correlating with the VCN in differentiated samples (Figure S7).
Figure 5.
Comparative Performance of BCH-BB694 in a Xenotransplant Model
(A) Engraftment of human cells in PB 4 and 8 weeks post-transplantation, and in spleen (Spl) and BM 8 weeks after transplantation. (B) The relative contribution of human CD45+ cells to the B cell lineage (CD19), myeloid lineage (CD33), and T cells (CD3) was assessed by flow cytometry 8 weeks post-transplantation. PB, BM, Spl, and thymus (Thy) were analyzed. (C) Frequency of human hematopoietic stem and progenitor cells (HSPCs) was assessed in the BM of transplanted NSG mice. (D) Recovery of VCN from total BM relative to the initial VCN. (E) Induction of HbF after erythroid in vitro differentiation of hCD34+ cells isolated from the BM of transplanted animals. HbF was significantly induced in BCH-BB694-transduced cells; differences in all other panels are not significant (two-sided unpaired t test). Each data point represents one animal: BCH-BB694, n = 8; LV-GFP, n = 5. pre-Tx VCN, pre-transplant VCN.
Large-Scale Validations Using SCD Patient CD34+ Cells and GMP-Grade LVV
Lastly, we performed large-scale transduction experiments using the manufacturing protocol to be utilized for the generation of the cell products for the planned clinical trial (ClinicalTrial.org: NCT03282656). The goal of these large-scale transductions was to establish the appropriate MOI to achieve a VCN in the desired range (0.7–5 c/dg). Four independent engineering runs were performed on plerixafor mPB CD34+ cells from SCD subjects.44 The purity of CD34+ cells was between 93.9% and 99.2%. VCNs were assessed after 7 days in culture (Table 1). The first engineering run was performed on previously cryopreserved cells with research-grade BCH-BB694 LVV at an MOI of 6 or 25, leading to VCNs of 3.7 and 11.1 c/dg, respectively. After this initial pilot experiment, three other engineering runs were performed under GMP conditions with clinical-grade BCH-BB694 LVV using MOIs ranging from 15 to 37, which resulted in VCNs between 3.5 and 7.5 c/dg. The last two engineering runs used an MOI of 15, resulting in a VCN within the desired range. The cell products generated at the end of each run successfully passed sterility, endotoxin, and mycoplasma tests.
Table 1.
Clinical-Scale Transductions Performed under Good Manufacturing Practices Conditions
| Donor 1 | Donor 2 | Donor 3 | Donor 4 | |||||
|---|---|---|---|---|---|---|---|---|
| Fresh versus frozen cells | frozen | frozen | fresh | fresh | ||||
| % CD34+ cells | 99.2 | 93.9 | 99.1 | 96.3 | ||||
| No. of cells used for transduction | 3.8E+7 | 3.65E+7 | 1.18E+8 | 2.03E+8 | ||||
| Vector grade | research | clinical | clinical | clinical | ||||
| Prestimulation (hours:minutes) | 44:55 | 42:00 | 42:00 | 42:00 | ||||
| Transduction (hours:minutes) | 19:00 | 19:42 | 19:42 | 20:00 | ||||
| MOI | 6 | 25 | 25 | 37 | 15 | 25 | 37 | 15 |
| VCN (c/dg) | 3.7 | 11.1 | 5.2 | 7.5 | 3.7 | 6.0 | 7.1 | 3.5 |
Mobilized CD34+ cells pre-stimulated and transduced as described in Materials and Methods. The first validation was performed with research-grade BCH-BB694, and the subsequent three validations were performed with clinical-grade vector. Multiplicity of infection (MOI), duration of pre-stimulation, and transduction are indicated. After 7 days in culture, cells were harvested for DNA extraction, and the average vector copy number (VCN) per cell was assessed by qPCR. Tests for sterility (BAC T Alert), endotoxin, mycoplasma, and recombinant competent lentivirus (RCL) were also performed.
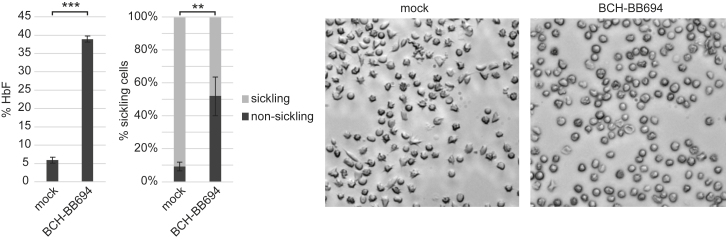
Transduced SCD CD34+ cells from one donor were erythroid differentiated in vitro to quantify HbF and physiological analyses using a sickling assay. After 18 days of differentiation, the average VCN was 2.8 c/dg, resulting in 39% ± 1% HbF of total hemoglobin (Figure 6). Enucleated RBCs derived from this sample were exposed to the potent sickling-inducing agent metabisulfite. Although >90% of mock-treated cells demonstrated sickling or dysmorphic appearance caused by hemoglobin polymerization, this number was significantly reduced to 48% in cells treated with BCH-BBB694 LVV (Figure 6) coincident with the presence of increased levels of HbF.
Figure 6.
In Vitro HbF Induction and Sickling Assay with BCH-BB694-Transduced SCD Cells Manufactured under GMP Conditions
CD34 cells transduced with BCH-BB694 under GMP conditions were differentiated into erythrocytes. The VCN and HbF induction were determined, and enucleated erythrocytes were enriched by fluorescence-activated cell sorting (FACS) to assess metabisulfite-induced sickling. The relative proportion of dysmorphic versus normally shaped cells was determined by light microscopy. n = 3 technical replicates. Average VCN of total cells prior to FACS: 2.8. Error bars: SD. Statistical test: two-sided t test, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
Discussion
We previously developed an LVV that mediates downregulation of BCL11A via RNAi selectively in erythroid cells using a microRNA-adapted short hairpin RNA (shRNAmiR). Significant downregulation of BCL11A in erythroid cells leads to sustained reactivation of γ-globin, the production of HbF, reduced polymerization of sickle-containing hemoglobin, and significant mitigation of the hematologic effects of SCD.35 Our strategy and vector configuration underwent several stages of development to address a variety of problems limiting clinical applicability. First, a polymerase II-driven shRNAmiR replaced the polymerase III-driven shRNAs to reduce non-specific cytotoxicity related to excessive shRNA expression and allowing expression via more physiological promoters.41 Second, the shRNAmiR was placed under control of erythroid-specific promoter/enhancer elements to avoid downregulation of BCL11A in HSCs and B cell progenitors, cells in which BCL11A expression is essential.34, 35, 36 As described here, we also changed the vector backbone in order to improve viral titers to a level that is required for the efficient genetic modification of patient primary HSCs at large scale, an essential step to bring this therapy to the clinic. Low viral titers are a practical bottleneck particularly for LVVs containing the globin LCR regulatory elements and β-globin sequences due to their large size,48, 49, 50 and LVV-expressing miRNA hairpins have reduced titers because of the partial degradation of vector genomes through the cellular miRNA-processing complex DROSHA during virus production.51, 52, 53 Low titers are problematic because they limit the achievable transduction rate and VCN, particularly within the rare long-term HSC compartment, and ultimately the therapeutic efficacy for the patient.
We performed preclinical experiments showing that clinical-grade vector can be produced for the BCH-BBB694 LVV vector at high titers in GMP production at large scale. In colony assays, we provide evidence that we consistently obtained transduction of >80% of hematopoietic progenitor-derived colony-forming units, and that almost all transduced erythroid colonies show high levels of HbF expression in vitro. These parameters are particularly important in SCD gene therapy because a too-large fraction of unmodified cells may lead to a less than complete reversal of the clinical phenotype. RBCs without sufficient HbF will remain prone to intracellular HbS polymerization and sickling.54 Although the therapeutic efficacy of gene therapy approaches for SCD is augmented by a substantial selection advantage of high HbF-containing mature RBCs due to an increased lifespan,55, 56, 57, 58, 59, 60 and potentially also due to reduced ineffective erythropoiesis,61 it is difficult to extrapolate the fraction of gene-modified HSCs that is required for the prevention of specific clinical manifestations. Two somewhat related parameters are the main determinants of therapeutic success. First, PB in gene therapy patients treated with BCH-BB694 LVV and other gene therapy approaches, including methods of gene editing, will consist of a mixture of gene-modified and unmodified cells, the latter of which contain mainly HbS and are still prone to sickling. The number of RBCs containing mostly HbS depends in large part on the efficiency of genetic modification of long-term reconstituting stem cells and the effectiveness of the conditioning regimen to prevent endogenous HSC recovery. In our and others’ previous work, it has been estimated that 20% gene-corrected HSCs would lead to 80% non-sickling erythrocytes in the periphery.55,58,60,62 Second, the gene-modified cells will contain varying levels of HbF depending on the efficacy of the vector in expressing the heterologous gene, in this case the shRNAmiR, while in other cases a hemoglobin gene. Although the level of intracellular HbF required to prevent HbS polymerization in vivo is not currently known, the use of a physiological switch to both increase HbF and concurrently reduce HbS has clear theoretical advantages. HbF has potent anti-sickling characteristics, as shown in previous work that has suggested a level of HbF of approximately one-third of the total cellular content of hemoglobin would prevent HbS polymerization, while the concurrent reduction in intracellular HbS further attenuates the tendency for polymer formation.39,40,63 These estimates are also informed by the phenotype of patients co-inheriting a HbS mutation with genetic mutations leading to the persistence of HbF production into adulthood and by the experience with patients who have varying responses to hydroxyurea treatment.54,64
Also complicating predictions of the extent of HbF production needed to attenuate SCD is that the multiple pathophysiological manifestations of SCD may require different levels of gene modification and HbF per RBC for phenotypic correction.54,64, 65, 66, 67 For instance, the attenuation of hemolysis may require different levels of HbF compared with the vaso-occlusive and other vascular manifestations of the disease. For example, stroke is one of the life-threatening conditions of SCD, and the current consensus for secondary stroke prevention in SCD patients is to lower HbS to 30% or less by erythrocyte transfusion.68, 69, 70 This assumes a clear bimodal distribution of pure HbA or HbS RBCs, which is not the case for gene therapy settings, but still is informative in that a similar level of RBCs protected from sickling needs to be reached in gene therapy to achieve comparable positive clinical outcomes for patients. Thus, both the pancellular nature of the response to effective HSC gene therapy and the potent anti-polymerization effect of HbF as reported here may be important in the overall phenotype correction to be anticipated in a clinical trial.
In viral vector-mediated gene therapy there is a well-established connection between the fraction of gene-modified cells, the VCN, and the distribution of different VCNs in individual cells.71 In order to maximize the percentage of gene-modified HSCs and the level of HbF per cell, it is desirable to achieve high VCNs in the infused cell product. However, this has to be balanced against safety concerns associated with high VCNs due to the potential risk of insertional oncogenesis,26,72,73 and also against economic considerations of using high volumes of costly vector. We demonstrate at VCNs of 4–11 c/dg the safety of the BCH-BB694 LVV vector using the IVIM assay.46 This confirms findings by others who used very similar vector configurations,42,74, 75, 76, 77 although the predictive value of available genotoxicity assays is limited.
An additional concern of lineage-targeted BCL11A knockdown is that any residual expression of the shRNAmiR in HSC or B cells may negatively affect engraftment or B cell reconstitution because BCL11A is essential in those cells.35,36,78 We addressed this concern by stringent competitive transplantation experiments, where cells transduced with the BCL11A knockdown vector competed with cells transduced with a neutral LVV encoding only a fluorescent reporter. Equivalent frequencies of both competitor populations were detected in transplanted animals, suggesting the absence of biologically relevant levels of leaky expression in HSCs. To account for potential species-specific differences, we performed xenotransplantation experiments using human CD34+ cells that were transduced to high VCNs (up to 9.35 c/dg in vivo) with BCH-BB694 LVV or a neutral GFP LVV. Even minimal leakiness of shRNAmiR expression by the vector at this high VCN would be predicted to lead to loss of gene-modified cells as previously demonstrated.35 However, cells transduced with BCH-BB694 LVV showed similar engraftment characteristics as cells transduced with the neutral control vector. Overall, in the studies reported here, human cell engraftment, lineage distribution, and specifically B cell reconstitution were similar, as was the recovery rate of VCN pre- versus post-transplantation in both groups. No evidence of transduced B or stem cell impairment has been observed. These data are consistent with a previous report that showed therapeutic efficacy and the complete absence of detectable expression in CD34+ BM cells isolated from transplanted NSG animals.35 In summary, all experimental evidence indicates that the BCH-BB694 LVV is safe for use in HSC gene therapy.
One hurdle in the clinical translation of gene therapies is the scale-up of cell manufacturing protocols, which may lead to lower than desired gene modification rates. We performed a series of large-scale validation runs that identified suitable conditions for transducing SCD CD34+ cells to generate cell products within a desired target VCN range and confirmed the compatibility of the procedures with therapeutic efficacy in an in vitro sickling assay, underscoring the potency of our vector.
In conclusion, the data presented here form the basis for an ongoing pilot and feasibility clinical trial for the treatment of SCD (ClinicalTrials.org: NCT03282656) using autologous HSCs transduced with an LVV expressing a shRNAmiR against BCL11A under the control of an erythroid-specific promoter. This is also the first example of a clinical application of RNAi for the gene therapy of a monogenic blood disorder and may pave the way toward gene therapy of other severe diseases using a shRNAmiR approach.79
Materials and Methods
Lentiviral Transduction of Human CD34+ Cells
Cells were obtained from patients at Boston Children’s Hospital after informed consent under a protocol approved by the Institutional Review Board. CD34+ cells were enriched from mPB apheresis products from healthy and SCD donors using magnetic CD34 beads (Miltenyi, Germany).44 Purified CD34+ cells were prestimulated at 1 × 106 cells/mL for 44 ± 4 h in CellGro Serum-free Media (CellGenix, Portsmouth, NH, USA) supplemented with human stem cell factor (hSCF) 100 ng/mL, human thrombopoietin (hTPO) 100 ng/mL, and hFlt-3L 100 ng/mL (all PeproTech, Rocky Hill, NJ, USA) in a standard humidified tissue culture incubator (5% CO2). Then cells were enumerated and transduced with the LVV at an MOI as indicated for 22 ± 4 h before downstream processing or cryopreservation in Cryostor5 (BioLife Solutions, Bothell, WA, USA). MOIs were calculated based on titrations on human osteosarcoma cells (HOS) cells using standard procedures.
Erythroid Differentiation in Liquid Culture
Following transduction, a subset of cells was cultured in erythroid differentiation media in a standard humidified tissue culture incubator for 2 weeks at 37°C and 5% CO2. The erythroid differentiation media consisted of Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 1% penicillin/streptomycin (Pen/Strep), 20 ng/mL hSCF, 1 ng/mL hIL-3 (all PeproTech, Rocky Hill, NJ, USA), 2 IU/mL erythropoietin (R&D Systems, Minneapolis, MN, USA), and 20% fetal bovine serum. After 14 days, cells were centrifuged (∼300 × g, 10 min), washed in PBS, and lysed in HPLC-grade water. After high-speed centrifugation (20,000 × g, 30 min at 4°C), supernatants were analyzed by IE-HPLC. Alternatively, the three-stage erythroid differentiation protocol developed by Giarratana et al.45 was followed.
Hemoglobin Analysis by IE-HPLC
After erythroid differentiation, cells were washed in PBS (Sigma-Aldrich, St. Louis, MO, USA) and analyzed by IE-HPLC on a PolyCATA 200 × 2.1 mm 5 μm 1,000 Å (PolyC#202CT0510; PolyLC, Columbia, MD, USA) using the mobile phases: phase A, Tris 40 mM, KCN 3 mM, in HPLC-grade water adjusted to a pH 6.5 with acetic acid (reagents from Sigma-Aldrich, St. Louis, MO, USA); and phase B, Tris 40 mM, KCN 3 mM in HPLC-grade water, NaCl 0.2M adjusted to a pH 6.5 with acetic acid (reagents from Sigma-Aldrich, St. Louis, MO, USA). A timed 24-min program was used to create a 2%–100% B gradient with a flow rate of 0.3 mL/min using a Shimadzu Prominence Chromatograph (Shimadzu Scientific Instruments, Columbia, MD, USA). The column oven temperature was 30°C, and the sample tray was at kept at 4°C. The peaks were detected at 418 nm. Retention times were determined using the AFSC hemoglobin standard (Helena Laboratories, Beaumont, TX, USA).
Clonogenic Cultures
Approximately 500 CD34+ cells in total were seeded into MethoCult Classic H4434 (STEMCELL Technologies, Vancouver, BC, Canada) for triplicates in 3.5-cm plates following the manufacturer’s protocol. After 2 weeks of culture, colonies were scored by morphology, enumerated, and either plucked as individual colonies or pooled and subjected to qPCR for assessment of VCNs (c/dg).
Mouse Transplantations
B6 (C57BL/6J), BoyJ (B6.SJL-Ptprca Pepcb/BoyJ), and NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) aged 4–8 weeks were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). B6 animals were conditioned with 10 or 11.5 Gy and NSG mice with 2.7 Gy, and transplanted retro-orbitally with 1 × 106 cells per animal. Blood samples were obtained via retro-orbital bleeds. For final analysis, mice were sacrificed using CO2. Spleen and thymus were removed and ground over a 100-μm mesh (Merck Millipore, Danvers, MA, USA) to obtain cell suspensions; femur and tibia were flushed out and filtered through a 100-μm mesh to obtain BM cells. The procedures were approved under Boston Children's Hospital Institutional Animal Care and Use Committee (BCH IACUC) #17-01-3364R.
Flow Cytometry and Fluorescence-Assisted Cell Sorting
Cells suspensions from cultured cells or from transplanted mice were stained with combinations of surface antibodies for mCD45-Vioblue, hCD3-allophycocyanin (APC) (both Miltenyi, Germany), hCD45-PerCP-Cy5.5, hCD235a-phycoerythrin (PE) (both BioLegend, San Diego, CA, USA), hCD4-PE-Cy7, hCD71-APC, hCD19-PE-Cy7, hCD33-PE, hCD71-APC (all BD Pharmingen, San Jose, CA, USA), and hCD34-APC (BD Biosciences, San Jose, CA, USA). Mouse and human Fc-Block were added when staining cells isolated from mice. Analyses were performed on BD Fortessa SOP or BD LSRII SOP equipped with UV, 405-, 488-, 561-, and 633-nm lasers, and cell sorting was performed on a BD AriaII machine. Analysis of flow cytometry data was performed using the BD Diva and Tree Star FlowJo software.
PCR and VCN Assay
Genomic DNA was extracted using the QIAGEN DNeasy protocol (QIAGEN, Hilden, Germany). VCN was assessed by real-time polymerase chain reaction, performed using TaqMan Fast Master Mix (Invitrogen, Carlsbad, CA, USA) and 0.9 mM GAG forward (5′-GGAGCTAGAACGATTCGCAGTTA-3′) and reverse (5′-GGTTGTAGCTGTCCCAGTATTTGTC-3′) primers, GAG FAM probe (5′-[FAM]-ACAGCCTTCTGATGTCTCTAAAAGGCCAGG-[TAMRA]-3′), and RNASE-P-VIC control TaqMan assay (Invitrogen). PCR was run using Fast program on Applied Biosystems StepOnePlus real-time thermocycler (Invitrogen, Carlsbad, CA, USA). VCN was assessed relative to a reference known to contain one copy of integrated viral DNA per haploid genome.
Assessment of VCN and HbF on Individual BFU-E Colonies
Erythroid colonies were plucked individually under a microscope. Each colony was washed in PBS (∼300 × g for 10 min) and resuspended in 100 μL of HPLC-grade water. A total of 20 μL was used for VCN assessment by qPCR, and 80 μL was used for hemoglobin analysis by IE-HPLC. Genomic DNA was column purified using the DNA Extract All Reagents Kit (Thermo Fisher, Waltham, MA, USA).
IVIM
The IVIM assay was conducted as previously described.46,47 Lineage-negative cells from B6 mice were isolated using the murine lineage depletion kit (Miltenyi, Germany) according to the manufacturer’s instructions and transduced in StemSpan-medium (STEMCELL Technologies, Cologne, Germany) supplemented with 1% Pen/Strep, 50 ng/mL mSCF, 100 ng/ml hFLT-3L, 100 ng/mL hIL-11, and 20 ng/mL m-IL-3 (all PeproTech, Hamburg, Germany). Cells were further expanded in IMDM supplemented with the above-mentioned antibiotics and cytokines, as well as 10% fetal calf serum and 2 mmol/L glutamine for 15 days. A total of 100 cells/well was seeded on 96-well plates and incubated for 14 days in IMDM conditions. Mock cells usually do not grow under these conditions. Wells containing proliferated insertional mutants were detected by microscopic evaluation and an enzymatic assay (MTT).80 The positive wells were used to calculate the replating frequencies (RFs) using the R package limdil.81 According to metadata, plates with insertional mutants in three or more wells (Q1 level) were counted as positive assays. The incidence of positive assays was compared by Fisher’s exact test with Benjamini-Hochberg correction for multiple comparisons.
Large-Scale Engineering Runs of CD34+ Cell Transduction under Good Manufacturing Practices
For engineering runs, large-scale vector preparations were produced by transient transfection of HEK293T cells with LVV construct and packaging plasmids, followed by microfiltration, ion exchange chromatography, concentration, and diafiltration by tangential flow filtration and fill-finish. All vector preparations were titrated on HOS cells. Four large-scale validations were performed at Connell and O’Reilly Families Cell Manipulation Core Facility at Dana-Farber Cancer Institute (Boston, USA). Plerixafor mobilized CD34+ cells44 enriched by CliniMACS device (Miltenyi Biotec, Auburn, CA, USA) were pre-stimulated in CellGenix SCGM media supplemented with recombinant human (rh)TPO, rhSCF, and rhFLT3-L (all 100 ng/mL from CellGenix, Portsmouth, NH, USA) at a density of 1 × 106 cells/mL for 44 ± 4 h at 37°C and 5% CO2. Cells were then transduced at MOIs indicated in Table 1 at 5 × 106 cells/mL with BCH-BB694 LVV for 22 ± 4 h as previously described.82, 83, 84 The final product was cryopreserved in Cryostor5 (BioLife Solutions, Bothell, WA, USA) prior to downstream analyses.
In Vitro Sickling Assay
At the end of in vitro erythroid differentiation,45 enucleated RBCs were sorted using Hoechst 33342 (5 μg/mL; Invitrogen, Carlsbad, CA, USA). Sorted cells were resuspended in 50 μL PBS, mixed with an equal volume of 2% metabisulfite in PBS, and incubated at 37°C for 20 min in a chamber slide (Nunc/Life Technologies, Grand Island, NY, USA) prior to recording and analysis. Cells with irregular structure, protruding spikes, or sickle shape were counted as sickling cells.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8.3 software (GraphPad Software, San Diego, CA, USA). The standard statistical test used in most cases is a two-sided unpaired t test unless indicated otherwise. Statistical significance is indicated with a p value; N.S. denotes p > 0.05. Statistical significance differences are indicated with asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005. For Figures 2E and 2F we applied a Gompertz-model,85 and the three parameters characterizing the Gompertz model have been estimated by means of the easynls package86 available in R.
Author Contributions
C.B., O.N., and D.A.W. wrote the manuscript; C.B., O.N., M.R., M. Bonner, A.S., and D.A.W. designed experiments; C.B., O.N., M.R., S.G., G.P., C.H., M.M., M.A., D.A., A.T., D.K., M. Bentler, H.N., L.C., and M. Bonner performed experiments; A.S., J.M., G.V., E.E., and M.A. provided technical help and advice; D.P. performed statistical analyses.
Conflicts of Interest
O.N., G.P., L.C., M. Bonner, and G.V are employees and shareholders of bluebird bio, Inc. DAW licensed IP relevant to BCL11A vector to bluebird bio and has the potential to receive future royalty/milestone payments. DAW has received payment in the past through BCH institutional licensing agreement with bluebird bio.
Acknowledgments
D.A.W. received research funding from bluebird bio, Inc. The work was supported by NHLBI grant 5U01HL117720 (to D.A.W. and C.B.), a sponsored research agreement with bluebird bio, Inc. (D.A.W.), and the German Research Foundation (cluster of excellence REBIRTH [Exc 62/2] and SFB738) (M.R. and A.S.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.03.015.
Supplemental Information
References
- 1.Rees D.C., Williams T.N., Gladwin M.T. Sickle-cell disease. Lancet. 2010;376:2018–2031. doi: 10.1016/S0140-6736(10)61029-X. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization Management of haemoglobin disorders: report of a joint WHO-TIF meeting, Nicosia, Cyprus, November 16–18, 2007. 2008. https://www.who.int/genomics/WHO-TIF_genetics_final.pdf
- 3.Hassell K.L. Population estimates of sickle cell disease in the U.S. Am. J. Prev. Med. 2010;38(Suppl 4):S512–S521. doi: 10.1016/j.amepre.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Neel J.V. The Inheritance of Sickle Cell Anemia. Science. 1949;110:64–66. doi: 10.1126/science.110.2846.64. [DOI] [PubMed] [Google Scholar]
- 5.Piel F.B., Steinberg M.H., Rees D.C. Sickle Cell Disease. N. Engl. J. Med. 2017;377:305. doi: 10.1056/NEJMc1706325. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg M., Nagel R. HbSC disease and HbC disorders. In: Steinberg M.H., Forget B.G., Higgs D.R., editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Second Edition. Cambridge University Press; 2009. pp. 525–548. [Google Scholar]
- 7.Wayne A.S., Kevy S.V., Nathan D.G. Transfusion management of sickle cell disease. Blood. 1993;81:1109–1123. [PubMed] [Google Scholar]
- 8.Kato G.J., Piel F.B., Reid C.D., Gaston M.H., Ohene-Frempong K., Krishnamurti L., Smith W.R., Panepinto J.A., Weatherall D.J., Costa F.F., Vichinsky E.P. Sickle cell disease. Nat. Rev. Dis. Primers. 2018;4:18010. doi: 10.1038/nrdp.2018.10. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh M.M., Fitzhugh C.D., Tisdale J.F. Allogeneic hematopoietic stem cell transplantation for sickle cell disease: the time is now. Blood. 2011;118:1197–1207. doi: 10.1182/blood-2011-01-332510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters M.C., De Castro L.M., Sullivan K.M., Krishnamurti L., Kamani N., Bredeson C., Neuberg D., Hassell K.L., Farnia S., Campbell A., Petersdorf E. Indications and Results of HLA-Identical Sibling Hematopoietic Cell Transplantation for Sickle Cell Disease. Biol. Blood Marrow Transplant. 2016;22:207–211. doi: 10.1016/j.bbmt.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gluckman E., Cappelli B., Bernaudin F., Labopin M., Volt F., Carreras J., Pinto Simões B., Ferster A., Dupont S., de la Fuente J., Eurocord, the Pediatric Working Party of the European Society for Blood and Marrow Transplantation, and the Center for International Blood and Marrow Transplant Research Sickle cell disease: an international survey of results of HLA-identical sibling hematopoietic stem cell transplantation. Blood. 2017;129:1548–1556. doi: 10.1182/blood-2016-10-745711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters M.C., Patience M., Leisenring W., Eckman J.R., Buchanan G.R., Rogers Z.R., Olivieri N.E., Vichinsky E., Davies S.C., Mentzer W.C. Barriers to bone marrow transplantation for sickle cell anemia. Biol. Blood Marrow Transplant. 1996;2:100–104. [PubMed] [Google Scholar]
- 13.Krishnamurti L., Abel S., Maiers M., Flesch S. Availability of unrelated donors for hematopoietic stem cell transplantation for hemoglobinopathies. Bone Marrow Transplant. 2003;31:547–550. doi: 10.1038/sj.bmt.1703887. [DOI] [PubMed] [Google Scholar]
- 14.Gragert L., Eapen M., Williams E., Freeman J., Spellman S., Baitty R., Hartzman R., Rizzo J.D., Horowitz M., Confer D., Maiers M. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N. Engl. J. Med. 2014;371:339–348. doi: 10.1056/NEJMsa1311707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiuti A., Cattaneo F., Galimberti S., Benninghoff U., Cassani B., Callegaro L., Scaramuzza S., Andolfi G., Mirolo M., Brigida I. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 16.Hacein-Bey-Abina S., Le Deist F., Carlier F., Bouneaud C., Hue C., De Villartay J.P., Thrasher A.J., Wulffraat N., Sorensen R., Dupuis-Girod S. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 17.Hacein-Bey-Abina S., Hauer J., Lim A., Picard C., Wang G.P., Berry C.C., Martinache C., Rieux-Laucat F., Latour S., Belohradsky B.H. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott M.G., Schmidt M., Schwarzwaelder K., Stein S., Siler U., Koehl U., Glimm H., Kühlcke K., Schilz A., Kunkel H. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 20.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boztug K., Schmidt M., Schwarzer A., Banerjee P.P., Díez I.A., Dewey R.A., Böhm M., Nowrouzi A., Ball C.R., Glimm H. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 23.Eichler F., Duncan C., Musolino P.L., Orchard P.J., De Oliveira S., Thrasher A.J., Armant M., Dansereau C., Lund T.C., Miller W.P. Hematopoietic Stem-Cell Gene Therapy for Cerebral Adrenoleukodystrophy. N. Engl. J. Med. 2017;377:1630–1638. doi: 10.1056/NEJMoa1700554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 25.Malik P., Grimley M., Quinn C.T., Shova A., Courtney L., Lutzko C., Kalfa T.A., Niss O., Mehta P.A., Chandra s. Gene Therapy for Sickle Cell Anemia Using a Modified Gamma Globin Lentivirus Vector and Reduced Intensity Conditioning Transplant Shows Promising Correction of the Disease Phenotype. Blood. 2018;132(Suppl 1):1021. [Google Scholar]
- 26.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson A.A., Walters M.C., Kwiatkowski J., Rasko J.E.J., Ribeil J.A., Hongeng S., Magrin E., Schiller G.J., Payen E., Semeraro M. Gene Therapy in Patients with Transfusion-Dependent β-Thalassemia. N. Engl. J. Med. 2018;378:1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- 28.Marktel S., Scaramuzza S., Cicalese M.P., Giglio F., Galimberti S., Lidonnici M.R., Calbi V., Assanelli A., Bernardo M.E., Rossi C. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ß-thalassemia. Nat. Med. 2019;25:234–241. doi: 10.1038/s41591-018-0301-6. [DOI] [PubMed] [Google Scholar]
- 29.Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W. Gene Therapy in a Patient with Sickle Cell Disease. N. Engl. J. Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 30.Lettre G., Sankaran V.G., Bezerra M.A., Araújo A.S., Uda M., Sanna S., Cao A., Schlessinger D., Costa F.F., Hirschhorn J.N., Orkin S.H. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. USA. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uda M., Galanello R., Sanna S., Lettre G., Sankaran V.G., Chen W., Usala G., Busonero F., Maschio A., Albai G. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. USA. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menzel S., Garner C., Gut I., Matsuda F., Yamaguchi M., Heath S., Foglio M., Zelenika D., Boland A., Rooks H. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 33.Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 34.Liu P., Keller J.R., Ortiz M., Tessarollo L., Rachel R.A., Nakamura T., Jenkins N.A., Copeland N.G. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 35.Brendel C., Guda S., Renella R., Bauer D.E., Canver M.C., Kim Y.J., Heeney M.M., Klatt D., Fogel J., Milsom M.D. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J. Clin. Invest. 2016;126:3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luc S., Huang J., McEldoon J.L., Somuncular E., Li D., Rhodes C., Mamoor S., Hou S., Xu J., Orkin S.H. Bcl11a Deficiency Leads to Hematopoietic Stem Cell Defects with an Aging-like Phenotype. Cell Rep. 2016;16:3181–3194. doi: 10.1016/j.celrep.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J., Peng C., Sankaran V.G., Shao Z., Esrick E.B., Chong B.G., Ippolito G.C., Fujiwara Y., Ebert B.L., Tucker P.W., Orkin S.H. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer D.E., Kamran S.C., Orkin S.H. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood. 2012;120:2945–2953. doi: 10.1182/blood-2012-06-292078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brittenham G.M., Schechter A.N., Noguchi C.T. Hemoglobin S polymerization: primary determinant of the hemolytic and clinical severity of the sickling syndromes. Blood. 1985;65:183–189. [PubMed] [Google Scholar]
- 40.Poillon W.N., Kim B.C., Rodgers G.P., Noguchi C.T., Schechter A.N. Sparing effect of hemoglobin F and hemoglobin A2 on the polymerization of hemoglobin S at physiologic ligand saturations. Proc. Natl. Acad. Sci. USA. 1993;90:5039–5043. doi: 10.1073/pnas.90.11.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guda S., Brendel C., Renella R., Du P., Bauer D.E., Canver M.C., Grenier J.K., Grimson A.W., Kamran S.C., Thornton J. miRNA-embedded shRNAs for Lineage-specific BCL11A Knockdown and Hemoglobin F Induction. Mol. Ther. 2015;23:1465–1474. doi: 10.1038/mt.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Negre O., Bartholomae C., Beuzard Y., Cavazzana M., Christiansen L., Courne C., Deichmann A., Denaro M., de Dreuzy E., Finer M. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of β-thalassemia and sickle cell disease. Curr. Gene Ther. 2015;15:64–81. doi: 10.2174/1566523214666141127095336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atweh G.F., Loukopoulos D. Pharmacological induction of fetal hemoglobin in sickle cell disease and beta-thalassemia. Semin. Hematol. 2001;38:367–373. doi: 10.1016/s0037-1963(01)90031-9. [DOI] [PubMed] [Google Scholar]
- 44.Esrick E.B., Manis J.P., Daley H., Baricordi C., Trébéden-Negre H., Pierciey F.J., Armant M., Nikiforow S., Heeney M.M., London W.B. Successful hematopoietic stem cell mobilization and apheresis collection using plerixafor alone in sickle cell patients. Blood Adv. 2018;2:2505–2512. doi: 10.1182/bloodadvances.2018016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giarratana M.C., Rouard H., Dumont A., Kiger L., Safeukui I., Le Pennec P.Y., François S., Trugnan G., Peyrard T., Marie T. Proof of principle for transfusion of in vitro-generated red blood cells. Blood. 2011;118:5071–5079. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modlich U., Bohne J., Schmidt M., von Kalle C., Knöss S., Schambach A., Baum C. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Modlich U., Navarro S., Zychlinski D., Maetzig T., Knoess S., Brugman M.H., Schambach A., Charrier S., Galy A., Thrasher A.J. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar M., Keller B., Makalou N., Sutton R.E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 49.Morgan R.A., Unti M.J., Aleshe B., Brown D., Osborne K.S., Koziol C., Ayoub P.G., Smith O.B., O’Brien R., Tam C. Improved Titer and Gene Transfer by Lentiviral Vectors Using Novel, Small β-Globin Locus Control Region Elements. Mol. Ther. 2020;28:328–340. doi: 10.1016/j.ymthe.2019.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbinati F., Campo Fernandez B., Masiuk K.E., Poletti V., Hollis R.P., Koziol C., Kaufman M.L., Brown D., Mavilio F., Kohn D.B. Gene Therapy for Sickle Cell Disease: A Lentiviral Vector Comparison Study. Hum. Gene Ther. 2018;29:1153–1166. doi: 10.1089/hum.2018.061. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y.P., Vink M.A., Westerink J.T., Ramirez de Arellano E., Konstantinova P., Ter Brake O., Berkhout B. Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies. RNA. 2010;16:1328–1339. doi: 10.1261/rna.1887910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poluri A., Sutton R.E. Titers of HIV-based vectors encoding shRNAs are reduced by a dicer-dependent mechanism. Mol. Ther. 2008;16:378–386. doi: 10.1038/sj.mt.6300370. [DOI] [PubMed] [Google Scholar]
- 53.Park H.H., Triboulet R., Bentler M., Guda S., Du P., Xu H., Gregory R.I., Brendel C., Williams D.A. DROSHA Knockout Leads to Enhancement of Viral Titers for Vectors Encoding miRNA-Adapted shRNAs. Mol. Ther. Nucleic Acids. 2018;12:591–599. doi: 10.1016/j.omtn.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinberg M.H., Chui D.H., Dover G.J., Sebastiani P., Alsultan A. Fetal hemoglobin in sickle cell anemia: a glass half full? Blood. 2014;123:481–485. doi: 10.1182/blood-2013-09-528067. [DOI] [PubMed] [Google Scholar]
- 55.Walters M.C., Patience M., Leisenring W., Rogers Z.R., Aquino V.M., Buchanan G.R., Roberts I.A., Yeager A.M., Hsu L., Adamkiewicz T., Multicenter Investigation of Bone Marrow Transplantation for Sickle Cell Disease Stable mixed hematopoietic chimerism after bone marrow transplantation for sickle cell anemia. Biol. Blood Marrow Transplant. 2001;7:665–673. doi: 10.1053/bbmt.2001.v7.pm11787529. [DOI] [PubMed] [Google Scholar]
- 56.Franco R.S., Lohmann J., Silberstein E.B., Mayfield-Pratt G., Palascak M., Nemeth T.A., Joiner C.H., Weiner M., Rucknagel D.L. Time-dependent changes in the density and hemoglobin F content of biotin-labeled sickle cells. J. Clin. Invest. 1998;101:2730–2740. doi: 10.1172/JCI2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franco R.S., Yasin Z., Palascak M.B., Ciraolo P., Joiner C.H., Rucknagel D.L. The effect of fetal hemoglobin on the survival characteristics of sickle cells. Blood. 2006;108:1073–1076. doi: 10.1182/blood-2005-09-008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altrock P.M., Brendel C., Renella R., Orkin S.H., Williams D.A., Michor F. Mathematical modeling of erythrocyte chimerism informs genetic intervention strategies for sickle cell disease. Am. J. Hematol. 2016;91:931–937. doi: 10.1002/ajh.24449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kean L.S., Manci E.A., Perry J., Balkan C., Coley S., Holtzclaw D., Adams A.B., Larsen C.P., Hsu L.L., Archer D.R. Chimerism and cure: hematologic and pathologic correction of murine sickle cell disease. Blood. 2003;102:4582–4593. doi: 10.1182/blood-2003-03-0712. [DOI] [PubMed] [Google Scholar]
- 60.Andreani M., Testi M., Gaziev J., Condello R., Bontadini A., Tazzari P.L., Ricci F., De Felice L., Agostini F., Fraboni D. Quantitatively different red cell/nucleated cell chimerism in patients with long-term, persistent hematopoietic mixed chimerism after bone marrow transplantation for thalassemia major or sickle cell disease. Haematologica. 2011;96:128–133. doi: 10.3324/haematol.2010.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu C.J., Krishnamurti L., Kutok J.L., Biernacki M., Rogers S., Zhang W., Antin J.H., Ritz J. Evidence for ineffective erythropoiesis in severe sickle cell disease. Blood. 2005;106:3639–3645. doi: 10.1182/blood-2005-04-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu C.J., Gladwin M., Tisdale J., Hsieh M., Law T., Biernacki M., Rogers S., Wang X., Walters M., Zahrieh D. Mixed haematopoietic chimerism for sickle cell disease prevents intravascular haemolysis. Br. J. Haematol. 2007;139:504–507. doi: 10.1111/j.1365-2141.2007.06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maier-Redelsperger M., Noguchi C.T., de Montalembert M., Rodgers G.P., Schechter A.N., Gourbil A., Blanchard D., Jais J.P., Ducrocq R., Peltier J.Y. Variation in fetal hemoglobin parameters and predicted hemoglobin S polymerization in sickle cell children in the first two years of life: Parisian Prospective Study on Sickle Cell Disease. Blood. 1994;84:3182–3188. [PubMed] [Google Scholar]
- 64.Akinsheye I., Alsultan A., Solovieff N., Ngo D., Baldwin C.T., Sebastiani P., Chui D.H., Steinberg M.H. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118:19–27. doi: 10.1182/blood-2011-03-325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marcus S.J., Kinney T.R., Schultz W.H., O’Branski E.E., Ware R.E. Quantitative analysis of erythrocytes containing fetal hemoglobin (F cells) in children with sickle cell disease. Am. J. Hematol. 1997;54:40–46. doi: 10.1002/(sici)1096-8652(199701)54:1<40::aid-ajh6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 66.Platt O.S., Brambilla D.J., Rosse W.F., Milner P.F., Castro O., Steinberg M.H., Klug P.P. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 67.Odenheimer D.J., Sarnaik S.A., Whitten C.F., Rucknagel D.L., Sing C.F. The relationship between fetal hemoglobin and disease severity in children with sickle cell anemia. Am. J. Med. Genet. 1987;27:525–535. doi: 10.1002/ajmg.1320270305. [DOI] [PubMed] [Google Scholar]
- 68.Yawn B.P., Buchanan G.R., Afenyi-Annan A.N., Ballas S.K., Hassell K.L., James A.H., Jordan L., Lanzkron S.M., Lottenberg R., Savage W.J. Management of sickle cell disease: summary of the 2014 evidence-based report by expert panel members. JAMA. 2014;312:1033–1048. doi: 10.1001/jama.2014.10517. [DOI] [PubMed] [Google Scholar]
- 69.Kassim A.A., Galadanci N.A., Pruthi S., DeBaun M.R. How I treat and manage strokes in sickle cell disease. Blood. 2015;125:3401–3410. doi: 10.1182/blood-2014-09-551564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pegelow C.H., Adams R.J., McKie V., Abboud M., Berman B., Miller S.T., Olivieri N., Vichinsky E., Wang W., Brambilla D. Risk of recurrent stroke in patients with sickle cell disease treated with erythrocyte transfusions. J. Pediatr. 1995;126:896–899. doi: 10.1016/s0022-3476(95)70204-0. [DOI] [PubMed] [Google Scholar]
- 71.Fehse B., Kustikova O.S., Bubenheim M., Baum C. Pois(s)on—it’s a question of dose. Gene Ther. 2004;11:879–881. doi: 10.1038/sj.gt.3302270. [DOI] [PubMed] [Google Scholar]
- 72.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Braun C.J., Boztug K., Paruzynski A., Witzel M., Schwarzer A., Rothe M., Modlich U., Beier R., Göhring G., Steinemann D. Gene therapy for Wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci. Transl. Med. 2014;6:227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 74.Roselli E.A., Mezzadra R., Frittoli M.C., Maruggi G., Biral E., Mavilio F., Mastropietro F., Amato A., Tonon G., Refaldi C. Correction of beta-thalassemia major by gene transfer in haematopoietic progenitors of pediatric patients. EMBO Mol. Med. 2010;2:315–328. doi: 10.1002/emmm.201000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lidonnici M.R., Paleari Y., Tiboni F., Mandelli G., Rossi C., Vezzoli M., Aprile A., Lederer C.W., Ambrosi A., Chanut F. Multiple Integrated Non-clinical Studies Predict the Safety of Lentivirus-Mediated Gene Therapy for β-Thalassemia. Mol. Ther. Methods Clin. Dev. 2018;11:9–28. doi: 10.1016/j.omtm.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arumugam P.I., Higashimoto T., Urbinati F., Modlich U., Nestheide S., Xia P., Fox C., Corsinotti A., Baum C., Malik P. Genotoxic potential of lineage-specific lentivirus vectors carrying the beta-globin locus control region. Mol. Ther. 2009;17:1929–1937. doi: 10.1038/mt.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poletti V., Urbinati F., Charrier S., Corre G., Hollis R.P., Campo Fernandez B., Martin S., Rothe M., Schambach A., Kohn D.B., Mavilio F. Pre-clinical Development of a Lentiviral Vector Expressing the Anti-sickling βAS3 Globin for Gene Therapy for Sickle Cell Disease. Mol. Ther. Methods Clin. Dev. 2018;11:167–179. doi: 10.1016/j.omtm.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu Y., Wang J., Khaled W., Burke S., Li P., Chen X., Yang W., Jenkins N.A., Copeland N.G., Zhang S., Liu P. Bcl11a is essential for lymphoid development and negatively regulates p53. J. Exp. Med. 2012;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levitt N., Briggs D., Gil A., Proudfoot N.J. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 80.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 81.Bonnefoix T., Bonnefoix P., Verdiel P., Sotto J.J. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. J. Immunol. Methods. 1996;194:113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
- 82.Uchida N., Nassehi T., Drysdale C.M., Gamer J., Yapundich M., Demirci S., Haro-Mora J.J., Leonard A., Hsieh M.M., Tisdale J.F. High-Efficiency Lentiviral Transduction of Human CD34+ Cells in High-Density Culture with Poloxamer and Prostaglandin E2. Mol. Ther. Methods Clin. Dev. 2019;13:187–196. doi: 10.1016/j.omtm.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masiuk K.E., Zhang R., Osborne K., Hollis R.P., Campo-Fernandez B., Kohn D.B. PGE2 and Poloxamer Synperonic F108 Enhance Transduction of Human HSPCs with a β-Globin Lentiviral Vector. Mol. Ther. Methods Clin. Dev. 2019;13:390–398. doi: 10.1016/j.omtm.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tisdale J.F., Kanter J., Mapara M.Y., Kwiatkowski J.L., Krishnamurti L., Schmidt M., Miller A., Pierciey F., Shi W., Ribeil J. Current results of lentiglobin gene therapy in patients with severe sickle cell disease treated under a refined protocol in the phase 1 Hgb-206 study. Blood. 2018;132(Suppl 1) 1026–1026. [Google Scholar]
- 85.Gompertz B. XXIV. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. In a letter to Francis Baily, Esq. FRS &c. Philos. Trans. R. Soc. Lond. 1825;115:513–583. doi: 10.1098/rstb.2014.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaps M., Lamberson W.R. Cabi; 2017. Biostatistics for Animal Science. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.