Abstract
Purpose:
Image-guided prostate biopsies are routinely acquired in the diagnosis and treatment monitoring of prostate cancer, yielding useful tissue for identifying metabolic biomarkers and therapeutic targets. We developed an optimized biopsy tissue culture protocol in combination with [1,6-13C2]glucose labeling and quantitative high-resolution NMR to measure glycolysis and tricarboxcylic acid (TCA) cycle activity in freshly acquired living human prostate biopsies.
Methods:
We acquired 34 MRI-ultrasound fusion-guided prostate biopsies in vials on ice from 22 previously untreated patients. Within 15 min, biopsies were transferred to rotary tissue culture in 37°C prostate medium containing [1,6-13C2]glucose. Following 24 h of culture, tissue lactate and glutamate pool sizes and fractional enrichments were quantified using quantitative 1H high resolution magic angle spinning Carr-Purcell-Meiboom-Gill (CPMG) spectroscopy at 1°C with and without 13C decoupling. Lactate effluxed from the biopsy tissue was quantified in the culture medium using quantitative solution-state high-resolution NMR.
Results:
Lactate concentration in low-grade cancer (1.15 ± 0.78 nmol/mg) and benign (0.74 ± 0.15 nmol/mg) biopsies agreed with prior published measurements of snap-frozen biopsies. There was substantial fractional enrichment of [3-13C]lactate (≈70%) and [4-13C]glutamate (≈24%) in both low-grade cancer and benign biopsies. Although a significant difference in tissue [3-13C]lactate fractional enrichment was not observed, lactate efflux was significantly higher (P < 0.05) in low-grade cancer biopsies (0.55 ± 0.14 nmol/min/mg) versus benign biopsies (0.31 ± 0.04 nmol/min/mg).
Conclusion:
A protocol was developed for quantification of lactate production–efflux and TCA cycle activity in single living human prostate biopsies, allowing metabolic labeling on a wide spectrum of human tissues (e.g., metastatic, post-non-surgical therapy) from patients not receiving surgery.
Keywords: biopsy, efflux, lactate, metabolism, prostate, tracer
1 |. INTRODUCTION
Prostate cancer metabolism is characterized by a shift to aerobic glycolysis and lactate production, known as the Warburg effect.1–5 Yet, it is not currently known how tumor metabolism changes in the setting of human castration-resistant and/or neuroendocrine prostate cancer (CRPC/NEPC). Recent studies have identified genetic alterations with significant putative metabolic effects in human CRPC.6,7 To investigate these effects, it is essential to measure metabolism in living human prostate tissue. Our laboratory has previously used an optimized rotary tissue culture system to perform 13C NMR metabolic labeling studies in slices of human prostate tissue acquired from radical prostatectomy (RP).8–10 Yet, RP is not standard of care for CRPC or NEPC, so the tissue slice technique cannot be applied in this clinically significant cohort of patients to yield a better insight into their respective metabolic adaptations. Although it is clinically feasible to obtain biopsy samples from patients with CRPC and/or NEPC, a novel metabolic profiling protocol that is optimized for living human biopsies is needed to study these specimens.
In the current work, we demonstrate that the combination of rotary biopsy culture, 13C metabolic labeling, and NMR can quantify metabolism in freshly acquired living human prostate biopsies. The current research focuses on the development of the techniques necessary to acquire and measure the Warburg effect in living patient-derived biopsies, acquired before treatment for the determination of cancer presence, Gleason grade, and extent for personalized therapeutic selection. By definition, this was an early stage cancer patient population, resulting in the acquisition of mainly benign and low grade (primary Gleason pattern 3) prostate cancer biopsies in this study. However, the ultimate goal is to use the optimized techniques developed in this study to assess the Warburg effect and other changes in metabolism in biopsies from patients with advanced disease who would not normally receive surgical therapy.
A particular motivation for studying the metabolism of prostate cancer comes from the preclinical and clinical development of hyperpolarized 13C MRI techniques,11–14 with clinical trials of hyperpolarized [1-13C]pyruvate in prostate cancer now underway at multiple sites.15 In prostate cancer, there is considerable heterogeneity of prognosis for patients, particularly for those with a cancer of Gleason 7 histopathological score or lower.16 Therefore, there is significant potential for hyperpolarized 13C metabolic imaging of primary prostate cancer to provide additional information to the clinician, in combination with Gleason scoring and clinical risk scores such as the Cancer of the Prostate Risk Assessment (CAPRA) score.17 In the metastatic setting, it is widely recognized that metabolic responses to therapy can significantly precede anatomic responses,18 which means that hyperpolarized 13C metabolic imaging may be able to better inform in real-time spatially resolved treatment monitoring for the clinician and patient in conjunction with serum tumor markers such as prostate-specific antigen (PSA).19
In hyperpolarized 13C MRI, the measured signal is proportional to the product of (1) probe polarization, (2) the fractional enrichment of the precursor being converted to the produced metabolite, and (3) the pool size of the produced metabolite.20 In the ex vivo setting, it becomes possible to disentangle these factors, ideally leading to a better interpretation and understanding of the hyperpolarized 13C signal10 and enabling the development of new hyperpolarized probes and potential therapeutic targets.
A significant challenge in the metabolic profiling of specific biochemical pathways using stable isotopes in individual living prostate biopsies by NMR is the small tissue mass ranging from 1–6 mg, which limits the achievable SNR. Therefore, in the present work, we have focused on optimizing the experimental parameters such as the duration of time in rotary biopsy culture and the parameters of the NMR acquisition so as to achieve sufficient metabolite signals from a small mass of tissue incubated with 13C-labeled metabolic substrates.
2 |. METHODS
2.1 |. Biopsy acquisition
Transrectal 18-gauge prostate biopsies from 22 previously untreated patients (34 total biopsies, 27 normal prostate biopsies, 5 Gleason 6(3 + 3) cancer biopsies, and 2 Gleason 7(3 + 4) cancer biopsies) were acquired with Institutional Review Board approval. Biopsies were acquired using an ultrasound-MRI fusion system that provided registration of transrectal ultrasound images to prior prostate MRI (UroNav, InVivo, Gainesville, FL). The clinical efficacy of this system has been evaluated previously.21 Up to 2 research biopsies per patient were acquired from any visible multiparametric 1H MRI or ultrasound lesions once the clinical biopsy acquisition had been completed. In cases with fewer than 2 visible lesions, a biopsy was sometimes taken from a normal-appearing area of the peripheral zone. Immediately following acquisition, biopsies were stored in vials on ice and were transferred to rotary culture within 15 min.
2.2 |. Biopsy culture medium
A formulation of PFMR-4A medium with supplements optimized for prostate tissue culture was previously described.8,9 For this study, it was found that HEPES and threonine in this medium posed a potential challenge for the quantification of NMR spectra. Consequently, we developed a formulation of bicarbonate-buffered, HEPES-free, threonine-free, pyruvate-free F-12 medium containing all of the previously described supplements.8,9 (The complete medium recipe is provided in Supporting Information Tables S1 and S2). For labeling studies, the medium contained exclusively [1,6-13C2]glucose at 1.0 g/L.
2.3 |. Biopsy rotary tissue culture
Techniques previously described for the culture of thin, precision-cut prostate tissue slices8 were adapted for culture of biopsies. Each biopsy was loaded onto a titanium mesh inside of 1 well of a 6-well plate containing 2.5 mL per well of pre-warmed F-12 prostate medium with [1,6-13C2]glucose. The plates were loaded onto a rotary tissue culture system (Alabama R&D) (Figure 1B) within a 37°C, 5% CO2 incubator, and the biopsies were cultured for either 24 or 26 h. For a subset of samples, the biopsy culture medium was collected and replaced with fresh pre-warmed medium at the 2-h time point to quantify the initial rate of lactate efflux, in addition to the lactate efflux after 24 h of culture. Each 6-well plate also contained a control well containing medium but without any biopsy tissue. At the end of the culture period, the medium was collected and stored at −80°C.
FIGURE 1.
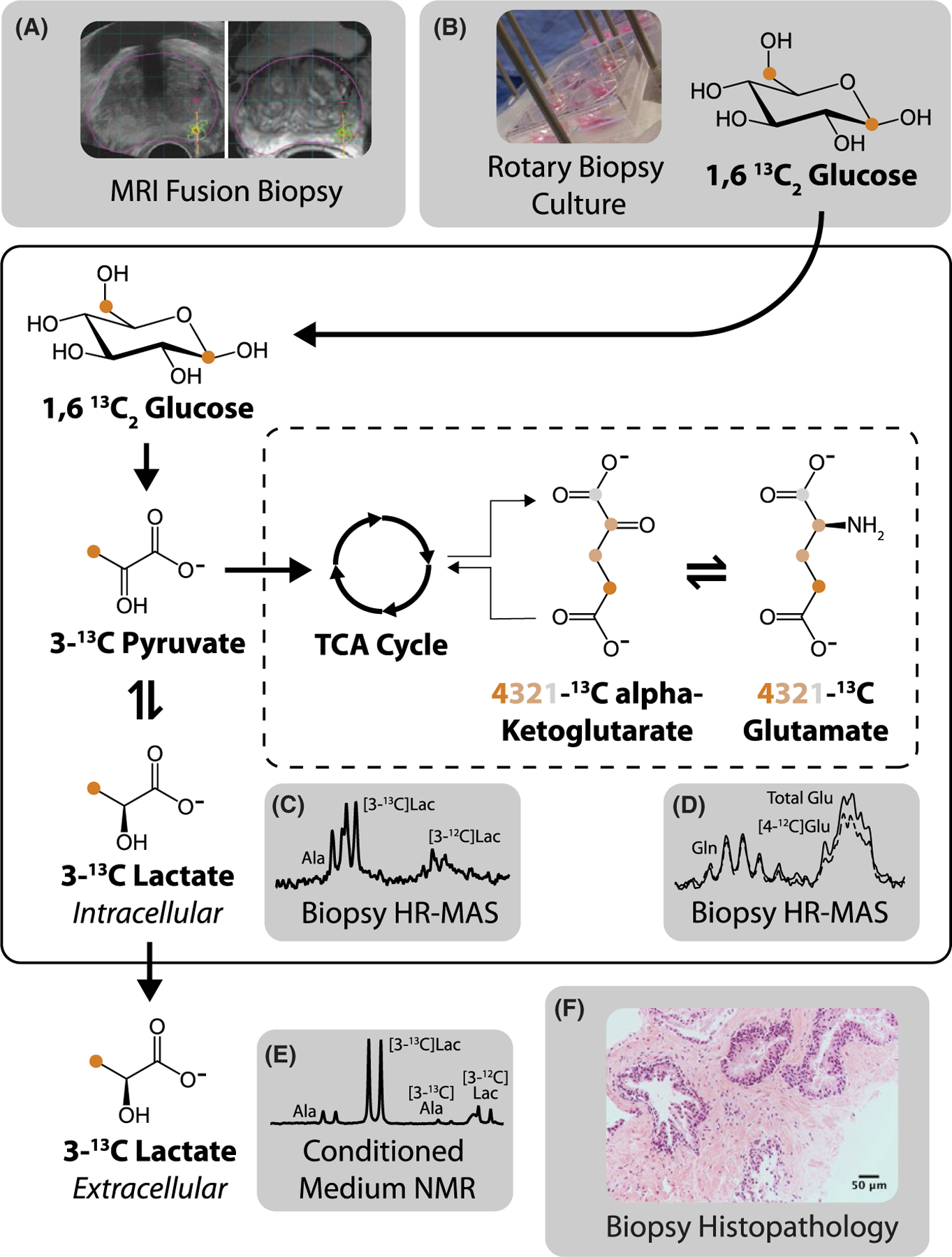
Study protocol. (A) MRI fusion biopsy was followed by (B) rotary tissue culture with [1,6-13C2]glucose. (C) Tissue lactate and (D) glutamate concentration and fractional enrichment were quantified using HR-MAS of biopsy samples, whereas (E) lactate efflux in media was quantified using NMR. (F) The biopsy histopathology was obtained after culture
2.4 |. Biopsy rinsing
After culture, the biopsies were placed in ice-cold phosphate-buffered saline for <1 min, blotted with a delicate task wipe, and stored at −80°C. This procedure was intended to remove medium that could otherwise contaminate the post-culture biopsy high resolution-magic angle spinning (HR-MAS) NMR spectra as we have shown previously.22
2.5 |. Biopsy tissue NMR measurements
Frozen biopsies were loaded without thawing into a sealed zirconium HR-MAS rotor22,23 on a tared digital balance to measure the wet weight of the biopsy tissue. Six μL of D2O/0.05% trimethylsilylpropanoic acid (TSP) solution was further added to the rotor and weighed to enable deuterium locking as well as a chemical shift reference via TSP. The HR-MAS rotor was sealed and loaded into a 500 MHz Varian gHX Nano (proton-carbon double resonance) 4 mm indirect detection HR-MAS probe maintained at 1°C. Acquisition was performed at a spin rate of 2.25 kHz using a Varian Inova console with VNMRJ 4 after locking on the D2O signal. The probe tuning, 90° pulse width and water saturation frequency were calibrated for each sample. Each sample was shimmed using an automated FID shimming routine for 20 min (see Supplemental Information). 1H Carr-Purcell-Meiboom-Gill (CPMG) spectra were acquired with 60,000 complex points, 3-s acquisition time, 2-s water suppression pulse, 6-s overall repetition time, and 1024 scans. It was previously established that this repetition time yielded fully relaxed spectra in human prostate tissue.24 The CPMG TE was set to 288 ms. The spacings of 180° pulses within the CPMG sequence were synchronized to the spinning period of the rotor.24 To quantify metabolite total pools in addition to 13C-labeled fractions, CPMG data sets were acquired both with and without heteronuclear 13C decoupling using a GARP pulse22,25 for a total CPMG scan time of 3 h, 25 min.
2.6 |. Biopsy histopathology
Following the HR-MAS data acquisition, the biopsies were fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) and high-molecular weight cytokeratin for interpretation by a board-certified genitourinary pathologist. For each sample the pathologist determined the primary Gleason pattern, secondary Gleason pattern, Gleason score, percentage of cancer, percentage of glandular tissue, and percentage of stromal tissue.
2.7 |. Biopsy media NMR measurements
The samples of biopsy medium were thawed from storage at −80°C. NMR samples were prepared by adding 65 mg of D2O/0.05% TSP to 540 mg of conditioned medium and placed in 5 mm NMR tubes. The sample data were acquired at 600 MHz using a Bruker Avance III console equipped with a 5 mm Bruker BBFO (broadband double resonance) direct detection probe. The 90° pulse was calibrated, and automated z-axis shimming was performed up to 8th order in addition to tuning of all first-order shims (see Supporting Information Tables S3 and S4). Water-suppressed data were acquired using a fully relaxed ZGCPPR pulse sequence with a 12-s acquisition time (114,942 points) and 3-s relaxation delay and 32 scans (8 min per sample).
2.8 |. NMR data processing
All NMR data sets were processed in MestreNova 12 (Mestrelab Research S.L.). The data sets were zero-padded by a factor of 2, apodized with a 0.25 Hz exponential filter, and automatically phased and baselined using a Whittaker smoother algorithm26 and manually adjusted as needed. Peaks of interest were automatically picked and fitted using a Lorentzian-Gaussian shape function with a simulated annealing algorithm (500 coarse iterations, 100 fine iterations, and a local minima filter of 25 were used). All automated peak fits were tweaked after visual assessment for fit quality based on the residual signal and adjusted when necessary.
2.9 |. NMR quantification
In the biopsy HR-MAS CPMG data, peak areas were quantified relative to the calibrated amplitude of the electronic reference to access in vivo concentrations (ERETIC) signal, corrected for the number of metabolite protons.23 Appropriate T2 corrections were applied based on HR-MAS measurements in human prostate tissue at 1°C and 500 MHz (lactate C3 T2 = 251 ms, glutamate C4 T2 = 275 ms).4,10 Metabolite amounts were further standardized by the wet tissue weight of the sample, measured after culture, to yield a biopsy metabolite concentration in (nmol/mg). Fractional enrichments were computed as follows.
The glutamate fractional enrichment at the 4-position was computed by differencing the quantification of the 2.34 ppm C4 multiplet in the 13C decoupled versus non-decoupled CPMG spectrum. This obviated the need to quantify the [4-13C]glutamate satellite peaks, which are known to underlie other peaks such as the [4-12C]glutamine peak and additionally could be further split by the presence of [3-13C]glutamate labeling on the same molecule (Figure 1D).
In the fully relaxed media data, the lactate peak area was quantified relative to the known peak area and measured mass of TSP and corrected for the relative number of protons. The lactate amount was further standardized by the wet tissue weight of the biopsy, as well as the duration of the biopsy tissue culture, to yield a lactate efflux rate in (nmol/min)/(mg tissue).
2.10 |. LIVE/DEAD assays and imaging
At the end of the culture experiment, a limited number of biopsies were transferred to a new well containing medium with ethidium homodimer-1 at 0.5 μmol/L and calcein-AM at 2.5 μmol/L.27 The biopsies were maintained in culture for a further 2 h and then rinsed for 10 min in phosphate-buffered saline under dark conditions before blotting with a delicate task wipe and imaging on a laser scanning confocal microscope (Zeiss 780). A two-photon excitation was used at a 759 nm wavelength with a 25 × water immersion objective, a 1024 × 1024 image size, and 2 averages. The detection windows were set as 493–556 nm for green and 615–741 nm for red. Z-stack images were acquired using overlapping optical sectioning with thickness determined by 1 Airy unit, with the maximum possible Z-stack height to assess tissue viability throughout the sample. We found that our extended staining protocol allowed the stain to diffuse throughout the biopsy, which made it possible to acquire a Z-stack covering the entire biopsy thickness (~400 μm). Because of the time spent at room temperature on the microscope, the samples analyzed in this manner were not used for any HR-MAS data acquisition after imaging.
3 |. RESULTS
3.1 |. Overview of study protocol
The MRI fusion biopsy procedure provided registration of ultrasound to multiparametric prostate MRI, with the MRI lesion highlighted for biopsy targeting (Figure 1A). The freshly acquired biopsies were cultured with [1,6-13C2] glucose that was converted into [3-13C]pyruvate via glycolysis (Figure 1B). The pyruvate interconverted with the intracellular lactate pool, which was quantified after culture using HR-MAS. In Figure 1C, the 500 MHz HR-MAS spectrum shows doublets of [3-12C]alanine, [3-13C]lactate, and [3-12C]lactate going from left to right (total scan time for this spectrum: 1 h, 42 min). The pyruvate also entered the TCA cycle via pyruvate dehydrogenase, which led to labeling at the 4-position of alpha-ketoglutarate following 1 turn of the cycle. This interconverted with glutamate to produce labeling at the 4-position of glutamate. Figure 1D shows the higher amplitude of the multiplets corresponding to the C4 protons of glutamate when decoupling is turned on (total scan time for this spectrum, with decoupling both off and on: 3 h, 25 min). Figure 1E shows a representative spectrum of the medium signifying the efflux of lactate and alanine from the biopsy tissue as evidenced by the 13C satellite peaks of the respective methyl protons. The biopsy histopathology after culture demonstrated well-defined glands with preserved tissue microarchitecture (Figure 1F: H&E staining, 20× magnification). The representative biopsy was benign with 30% glandular tissue and 70% stromal tissue (a detail of a glandular region is shown).
3.2 |. Study data set
We obtained a total of 34 biopsies from 22 previously untreated patients. Of these biopsies, 16 were targeted to a radiologist-identified MRI lesion using the MRI-ultrasound fusion biopsy device. Twenty-seven benign biopsies and 7 cancer biopsies were acquired. The mean ± SE wet tissue weights were 3.65 ± 0.28 mg for benign biopsies and 4.40 ± 0.49 mg for cancer biopsies. Among the benign biopsies, the average percentage of benign glandular tissue was 17% as determined by a board-certified genitourinary pathologist. Of the 7 cancer biopsies, 5 had a Gleason score of 6(3 + 3) and 2 had a Gleason score of 7(3 + 4). The average percentage of cancer within the cancer biopsies was 12%, and the average percentage of benign glandular tissue was 9%. Of the 7 cancer biopsies, 4 were acquired from MRI-fusion targeted regions and 3 were acquired from non-MRI targeted regions.
3.3 |. Biopsy media NMR measurements
We observed considerable efflux of [3-13C]lactate into the medium during culture. In the representative NMR spectra normalized to tissue mass shown in Figure 2A, the cancer (yellow) medium after 24 h of culture demonstrated a higher level of [3-13C]lactate efflux compared to the benign (blue) medium.
FIGURE 2.
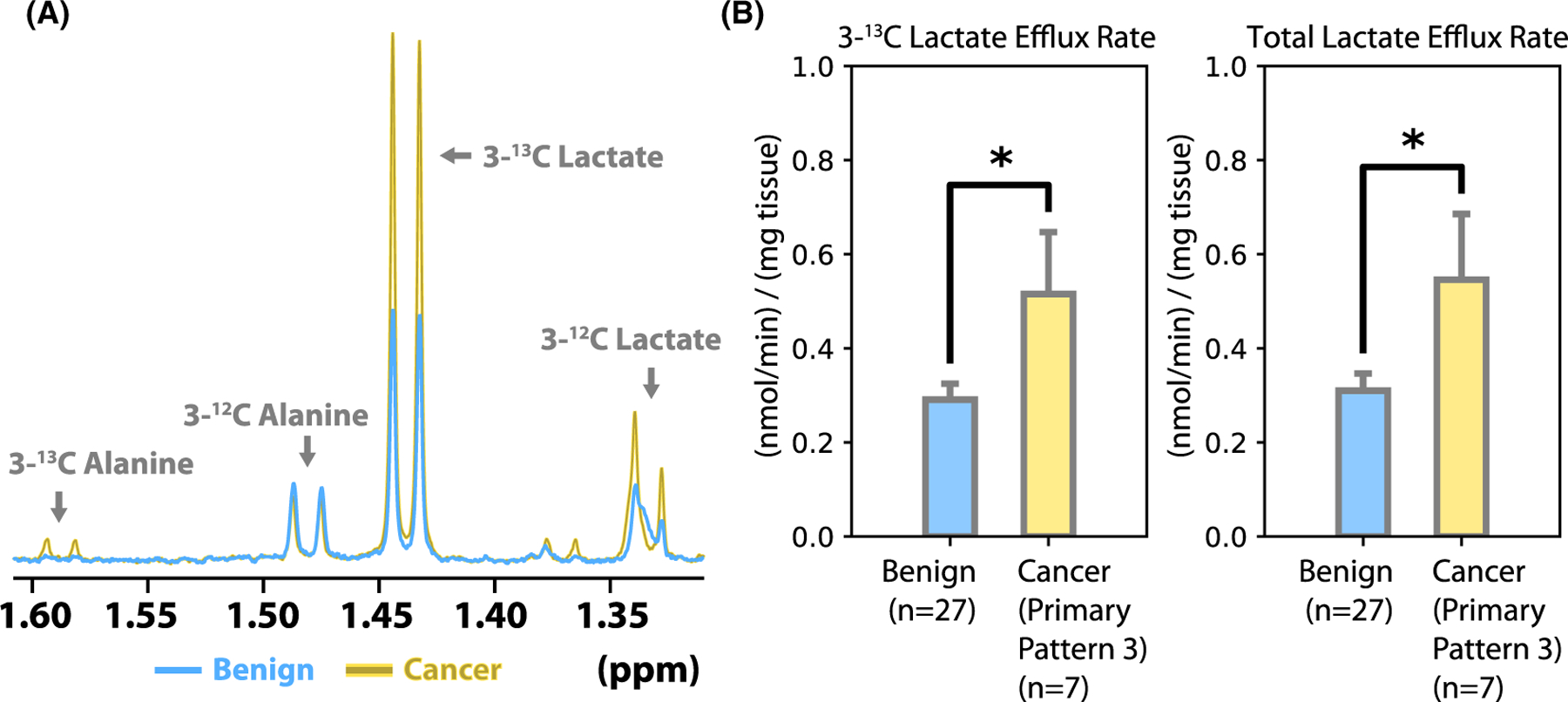
Biopsy lactate efflux and concentration. (A) Representative NMR spectra from benign (blue) and cancer (yellow) media. The broad peak that is superimposed with the 3-12C lactate doublet at 1.33 ppm is a medium additive that was deconvolved during the quantification process. (B) Quantification of [3-13C]lactate efflux and total lactate efflux in media. There was significantly greater [3-13C]lactate and total lactate efflux in cancer biopsies compared to benign biopsies (P < 0.05)
As shown in Figure 2B, benign biopsies had a [3-13C] lactate efflux rate of 0.29 ± 0.04 nmol/min/mg (mean ± SE), whereas cancer biopsies had a [3-13C]lactate efflux rate of 0.52 ± 0.14 nmol/min/mg measured over 24 h in culture. Benign biopsies had a total lactate efflux rate of 0.31 ± 0.04 nmol/min/mg (mean ± SE), whereas cancer biopsies had a total lactate efflux rate of 0.55 ± 0.14 nmol/min/mg measured over 24 h in culture. Both the 3-13C and total lactate efflux rates were significantly higher for the cancer biopsies (P < 0.05, two-tailed Student’s t-test for independent samples). The fractional enrichment of the effluxed lactate was 95 ± 1% for benign biopsies and 94 ± 1% for cancer biopsies, with no significant fractional enrichment differences observed between benign and cancer biopsies.
In an effort to assess the stability of lactate efflux over time in culture, for the majority of biopsies the lactate efflux rate per minute was measured from media collected after 2 h of biopsy culture (“0–2 h”) and again from media collected after a further 24 h of biopsy culture (“2–26 h”). We did not observe significant differences between the lactate efflux rates measured over these 2 time points (Figure 3A: benign 0–2 h: 0.33 ± 0.05 nmol/min/mg, benign 2–26 h: 0.28 ± 0.03 nmol/min/mg; and 3B: cancer 0–2 h: 0.49 ± 0.10 nmol/min/mg, cancer 2–26 h: 0.53 ± 0.23 nmol/min/mg).
FIGURE 3.
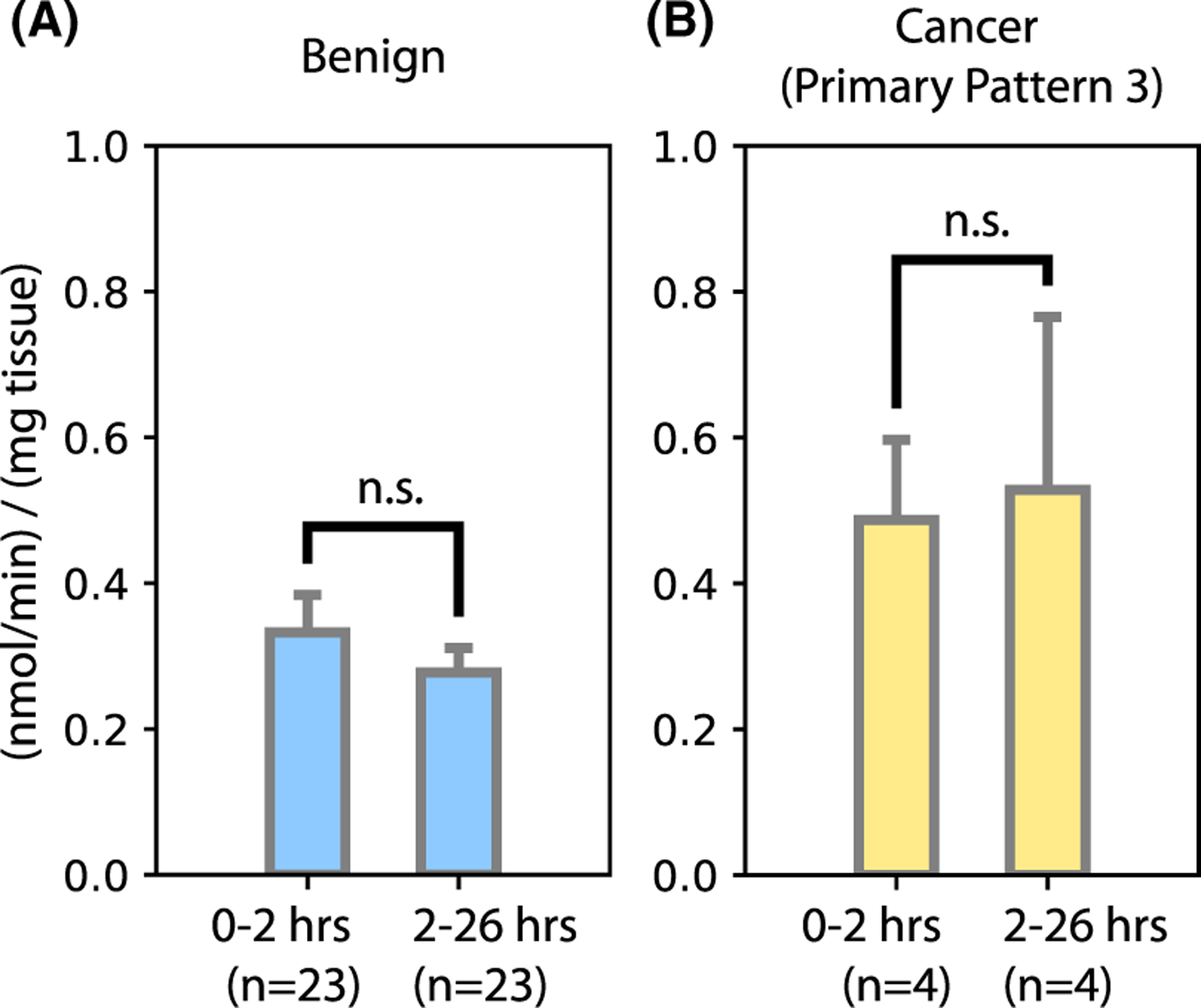
Biopsy lactate efflux over time. The biopsy lactate efflux rate per minute was measured from media collected after 2 h in culture (“0–2 h”) and again collected after a further 24 h in culture with the same biopsy (“2–26 h”)
3.4 |. Biopsy tissue NMR measurements
Of the 34 biopsies cultured during this study, 7 were used for non-HR-MAS experiments after culture and 6 were not measured by HR-MAS because of technical issues. We obtained high-quality HR-MAS data from 21 biopsies suitable for metabolite quantification. Of these HR-MAS samples, 16 were benign, 4 were Gleason score 6(3 + 3), and 1 was Gleason score 7(3 + 4). It was possible to observe a number of metabolites in the HR-MAS CPMG spectra from biopsy tissues after culture, including metabolites that we have previously quantified in prostate tissue such as phospholipids28 (Figure 4A). However, the focus of the present study was lactate production and efflux in culture. Despite a trend toward higher lactate levels in the cancer biopsies, we did not observe significant differences between benign and cancer biopsies after culture (Figure 4B). The [3-13C]lactate concentration after culture was 0.56 ± 0.12 nmol/mg (mean ± SE) in benign biopsies and 0.73 ± 0.49 nmol/mg in cancer biopsies. The total lactate concentration after culture was 0.74 ± 0.15 nmol/mg (mean ± SE) in benign biopsies and 1.15 ± 0.78 nmol/mg in cancer biopsies. The fractional enrichment was 72 ± 6% in benign biopsies and 71 ± 6% in cancer biopsies.
FIGURE 4.
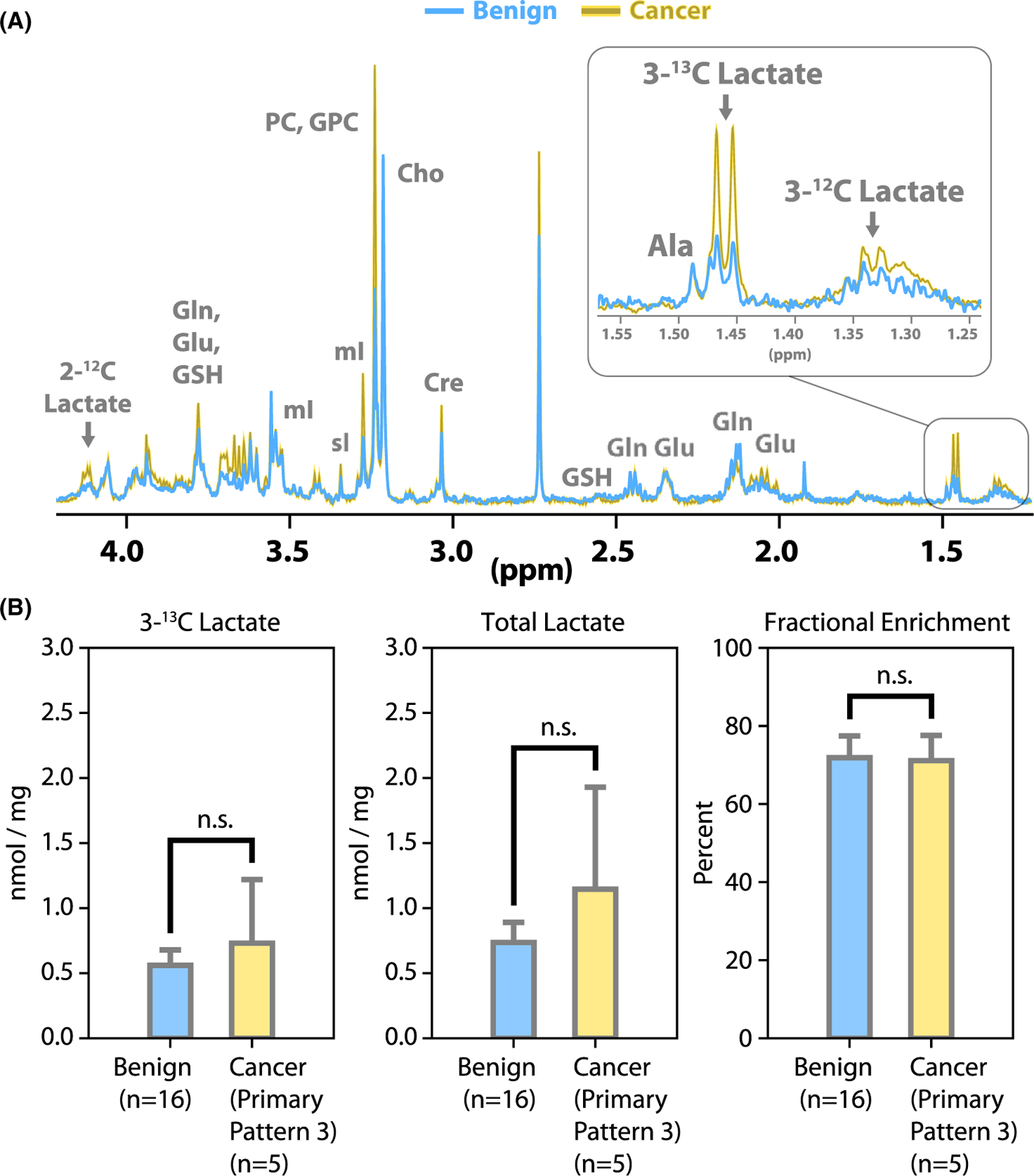
Biopsy tissue HR-MAS spectra. (A) Examples of CPMG spectra from benign (blue) and cancer (yellow) samples after culture. The [2-12C]lactate peak is visible at left, as a quartet split by the long-range coupling (~4 Hz) to [3-13C]lactate. Ala, alanine; Glu, glutamate; Gln, glutamine; GSH, glutathione; Cre, creatine; Cho, choline; PC, phosphocholine; GPC, glycerophosphocholine; mI, myo-inositol; sI, scyllo-inositol. (B) Quantified CPMG data: [3-13C]lactate concentration, total lactate concentration and fractional enrichment of lactate at the 3-position
3.5 |. Labeling of glutamate from [1,6-13C2] glucose
We were able to detect an increase of the glutamate C4 multiplet amplitude at 2.34 ppm with 13C decoupling turned on, indicating fractional enrichment (representative spectrum, Figure 5A). However, no significant differences were observed between the benign and cancer (primary pattern 3) biopsies in terms of the glutamate concentration and/or fractional enrichment (Figure 5B). As shown in Figure 5B, the [4-13C]glutamate concentration after culture was 0.32 ± 0.10 nmol/mg (mean ± SE) in benign biopsies and 0.27 ± 0.15 nmol/mg in cancer biopsies. The total glutamate concentration after culture was 1.18 ± 0.23 nmol/mg in benign biopsies and 0.92 ± 0.47 nmol/mg in cancer biopsies. The fractional enrichment of C4 glutamate after culture was 24 ± 3% in benign biopsies and 24 ± 7% in cancer biopsies.
FIGURE 5.
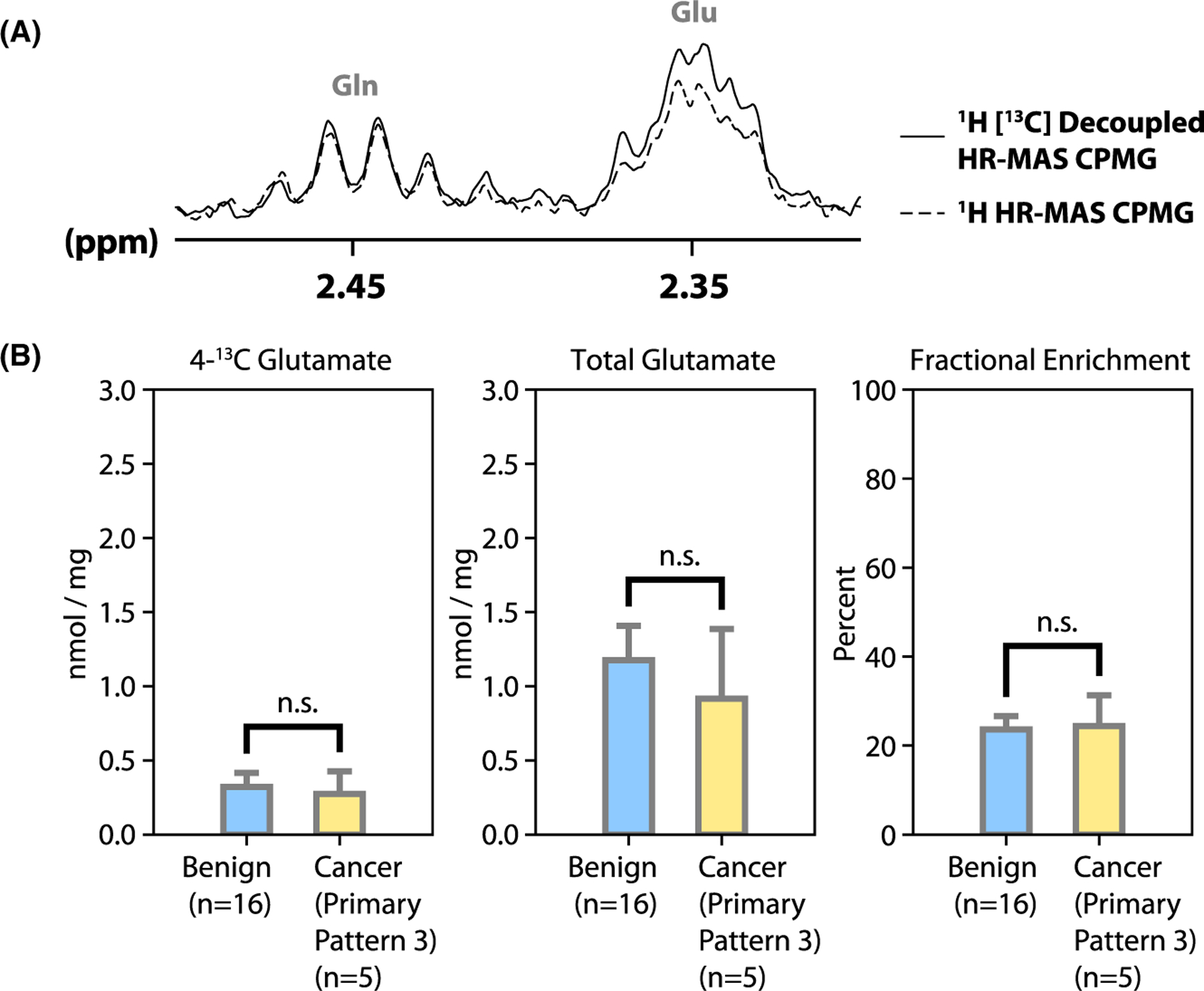
Labeling of glutamate from glucose. (A) There was an increase of the glutamate C4 multiplet amplitude at 2.34 ppm with 13C decoupling turned on, indicating fractional enrichment. (B) Quantified CPMG data: 4-13C glutamate concentration, total glutamate concentration, and fractional enrichment of glutamate at the 4-position
3.6 |. Biopsy LIVE/DEAD staining
We assessed the biopsy viability after 26 h in rotary culture using a fluorescent vital stain that is trapped in living cells (Calcein-AM) combined with a nuclear stain that enters the nuclei of dead cells (ethidium homodimer-1). Scattered cells demonstrated staining with ethidium homodimer-1, but the biopsies were primarily stained by the calcein-AM (Figure 6A), indicating that the majority of cells remained viable in biopsies after culture. The corresponding histopathology slides further demonstrated intact glandular architecture after culture (Figure 6B).
FIGURE 6.
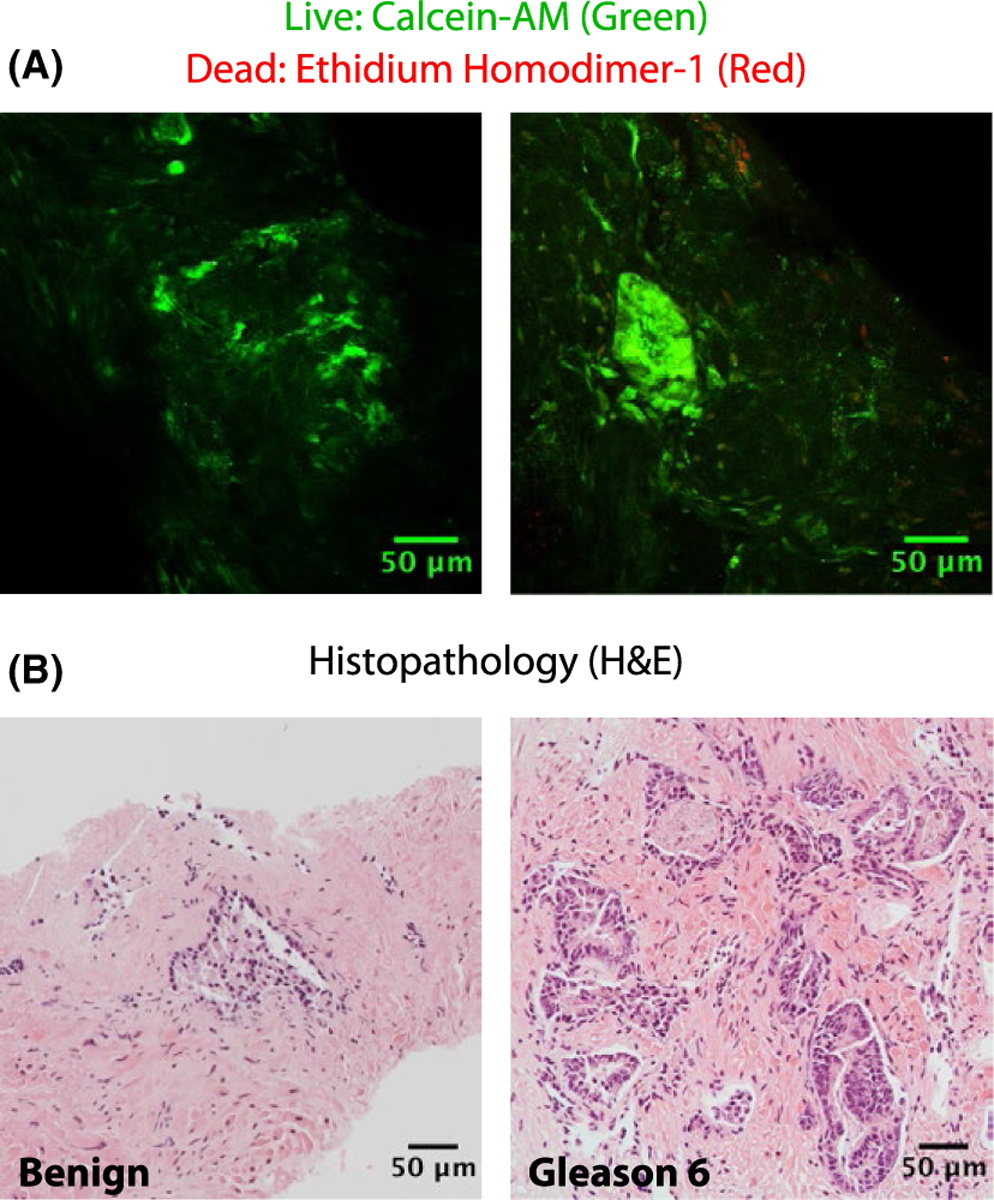
Biopsy LIVE/DEAD staining. (A) Confocal 2-photon microscopy from biopsy samples stained with calcein-AM and ethidium homodimer-1 following 26 h in culture. (B) Biopsy histopathology obtained after confocal microscopy. Each hematoxylin and eosin (H&E) stained image originates from the same sample as the confocal image directly above but not from the exact same location in the tissue
4 |. DISCUSSION
In this study, we found that it was possible to perform NMR measurements of metabolism in individual living human prostate biopsies. Furthermore, the 13C-labeled and overall rates of lactate efflux in culture were significantly higher for low-grade (primary Gleason pattern 3) cancer biopsies as compared to benign biopsies (Figure 2). In the HR-MAS data (Figure 4), for benign biopsies, we observed a tissue lactate concentration of 0.74 ± 0.15 nmol/mg, in good agreement with a prior published measurement of 0.61 ± 0.28 nmol/mg in snap-frozen biopsy samples by Tessem et al.4 For cancer biopsies, we observed a tissue lactate concentration of 1.15 ± 0.78 nmol/mg, as compared to a prior measurement of 1.59 ± 0.61 nmol/mg in snap-frozen cancer biopsies.4 This discrepancy can perhaps be explained by the fact that the present data set contained only low-grade prostate cancer biopsies (primary Gleason pattern 3), whereas the data set from Tessem et al.4 contained specimens of low- and high-grade prostate cancer (primary Gleason patterns 3 and 4). These findings are consistent with the notion that there is a more pronounced Warburg effect in high-grade prostate cancer such as primary Gleason pattern 4.29
Interestingly, although a significant difference in tissue lactate fractional enrichment coming from [1,6-13C2]glucose was not observed, the overall rate of lactate efflux in culture was significantly higher for primary Gleason pattern 3 cancer biopsies as compared to benign biopsies. This was presumably because of not only the trend toward higher lactate pool size (Figure 4), but also the propensity of the cancer tissue to export the lactate (Figure 2). This is consistent with prior data showing overexpression of the lactate dehydrogenase-A (LDHA) isoform (favoring lactate production from pyruvate) as well as overexpression of the monocarboxylate transporter MCT4, favoring lactate efflux, in human prostate cancer tissue compared to benign prostatic tissue.10 Moreover, prior studies have shown that both increasing expression of MCT4 mRNA as well as MCT4 protein (quantified via immunohistochemistry) correlate with higher Gleason score, suggesting that lactate efflux from prostate biopsies would provide a sensitive assessment of aggressive disease.30–33
In this study, a longer duration of [1,6-13C2]glucose labeling was used in culture to yield a medium [3-13C]lactate peak with higher SNR. It was therefore important to demonstrate that the rate of lactate efflux did not change over time, and in this study the lactate efflux rate was not significantly different at late (2–26 h) versus early (0–2 h) time points (Figure 3). Theoretically, it would also be possible to increase the lactate concentration (and hence lactate SNR) effluxed into the media by decreasing the medium volume that is used during culture. However, we used a rotary tissue culture system that relies on a 2.5 mL medium volume within a 6-well plate to provide optimal exposure to medium and 5% CO2/21% O2 air. In our experience (data not shown), alternatives such as 24-well plates do not yield the same level of tissue viability and would need to be further optimized to provide the correct ratio of medium to air exposure over time. It is also possible to increase the SNR of dilute NMR metabolites via lyophilization34; however, using the presented protocol, we achieved sufficient media lactate SNR without additional sample processing.
The fractional enrichment of C4 glutamate coming from [1,6-13C2]glucose in the biopsies studied is an indication of TCA cycle activity during rotary biopsy culture (Figures 1 and 5). In the current work, we obtained a C4 glutamate fractional enrichment of 24%. This value should be viewed as a balance between TCA cycle metabolism of [1,6-13C2]glucose as well as uptake of compounds that can contribute to the unlabeled tissue glutamate pool such as glutamine and glutamate. The culture medium used for this study contained 1 mM glutamine and 0.1 mM glutamate (Supporting Information).
In this study, the biopsy metabolite levels and lactate efflux rates were normalized to wet tissue weight, which could be suboptimal in studies of highly cellular tumors. In our data set, we observed a range of tissue cellularity for both benign samples (Figures 1F and 6, left) and cancer samples (Figure 6, right). As determined by a board-certified genitourinary pathologist, in benign samples, the average percentage of glandular tissue was 17%, whereas in cancer samples, the average percentage of glandular tissue was 9%, and the average percentage of cancer was 12%. In future studies with a higher number of samples, we intend to fit a multivariate model of lactate production as a function of the predictor variables of cancer percentage and benign glandular percentage.
The choice of an appropriate metabolite normalization standard is an ongoing challenge in metabolic studies of heterogeneous tissues, as opposed to cells. In studies of cultured cells, it is common to normalize metabolites to the number of cells or to a metric of cell mass such as total protein. In tissues, automated cell counting and/or protein quantification requires tissue digestion, making it impossible to obtain high-quality histopathology. It is possible to estimate tissue cellularity using automated image analysis of histopathology slides35; however, given the heterogeneity of human prostate tissue, this approach might yield inaccurate results unless the fixed, embedded, and stained tissue is comprehensively sliced to yield a 3D image stack. Another possibility is to analyze the metabolites of interest in terms of their ratios to a reference metabolite.10,36,37 This can be a robust method, but it does yield a metric that is sensitive to the reference metabolite as well as the metabolite of interest. Therefore, each normalization technique has strengths and limitations to practical implementation.
This study had a number of limitations. Our goal was to develop the biopsy culturing and metabolic profiling protocol, so we focused on biopsies routinely acquired from patients undergoing active surveillance or workup for prostate-related symptoms who had not been previously treated. Because of the low prevalence of aggressive cancer in this population, we only obtained biopsies containing benign glands or primary Gleason pattern 3 cancer, which limited our ability to further characterize the metabolic phenotype of aggressive prostate cancer.
5 |. CONCLUSION
In summary, a protocol was developed that allowed the quantification of lactate production and efflux in single living human prostate biopsies (1–6 mg of tissue), as well as the potential to quantify TCA metabolism through the quantification of fractional enrichment of glutamate. This was accomplished by optimizing experimental parameters such as the duration of time in rotary tissue culture (24 h) and the parameters of the NMR acquisition to achieve sufficient SNR from small metabolite signals within a 3 h and 25 min HR-MAS acquisition time. Validation studies demonstrated that this optimized protocol provided measurements of lactate production and/or fractional enrichment and efflux consistent with prior data describing lactate pool size, LDHA, and MCT4 expression in benign human prostatic tissue versus prostate cancer.4,10 Having established this protocol, similar measurements can then be performed on treatment-resistant and/or metastatic prostate cancer biopsies for a better understanding of metabolic perturbations in the setting of aggressive prostate cancer.
Supplementary Material
TABLE S1 Culture medium formulation
TABLE S2 Sources of culture medium ingredients
TABLE S3 Biopsy tissue NMR measurements: automated HR-MAS shimming routine
TABLE S4 Biopsy media NMR measurements: automated shimming routine
ACKNOWLEDGMENTS
The authors wish to thank Rosalie Nolley of Stanford University for preparing components of the culture medium and Professor Matthew Merritt of the University of Florida for useful discussions. Additionally, we wish to thank the MRI technologists and nursing staff of our institution as well as the patients who donated their tissue for this study.
Funding information
National Cancer Institute, Grant/Award Number: 5R01CA215694; National Institute of Biomedical Imaging and Bioengineering - P41 EB013598; Department of Defense - W81XWH-17-1-0471.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Warburg O, Posener K, Negelein E. Über den stoffwechsel der carcinomzelle. Biochem Zeitschr. 1924;152:309–344. [Google Scholar]
- 2.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. [DOI] [PubMed] [Google Scholar]
- 3.Ros S, Santos CR, Moco S, et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–343. [DOI] [PubMed] [Google Scholar]
- 4.Tessem MB, Swanson MG, Keshari KR, et al. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn Reson Med. 2008;60:510–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurhanewicz J, Vigneron DB, Brindle K, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson D, Van Allen E, Wu Y-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quigley DA, Dang HX, Zhao SG, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174(758–769):e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maund SL, Nolley R, Peehl DM. Optimization and comprehensive characterization of a faithful tissue culture model of the benign and malignant human prostate. Lab Invest. 2013;94:208–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peehl DM, Stamey TA. Growth responses of normal, benign hyperplastic, and malignant human prostatic epithelial cells in vitro to cholera toxin, pituitary extract, and hydrocortisone. Prostate. 1986;8:51–61. [DOI] [PubMed] [Google Scholar]
- 10.Keshari KR, Sriram R, VanCriekinge M, et al. Metabolic reprogramming and validation of hyperpolarized 13C lactate as a prostate cancer biomarker using a human prostate tissue slice culture bioreactor. Prostate. 2013;73:1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen AP, Albers MJ, Cunningham CH, et al. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T—initial experience. Magn Reson Med. 2007;58:1099–1106. [DOI] [PubMed] [Google Scholar]
- 12.Larson PEZ, Kerr AB, Chen AP, et al. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008;194:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albers MJ, Bok R, Chen AP, et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68:8607–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson PEZ, Hu S, Lustig M, et al. Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies. Magn Reson Med. 2011;65:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1–13C]pyruvate. Sci Transl Med. 2013;5:198ra108–198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–560. [DOI] [PubMed] [Google Scholar]
- 17.Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco cancer of the prostate risk assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173:1938–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. 2007;13:1382–1387. [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal R, Vigneron DB, Kurhanewicz J. Hyperpolarized 1-[13C]-pyruvate magnetic resonance imaging detects an early metabolic response to androgen ablation therapy in prostate cancer. Eur Urol. 2017;72:1028–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comment A, Merritt ME. Hyperpolarized magnetic resonance as a sensitive detector of metabolic function. Biochemistry. 2014;53:7333–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion–guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313: 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin YS, Albers MJ, Butler TN, Spielman D, Peehl DM, Kurhanewicz J. Methods for metabolic evaluation of prostate cancer cells using proton and 13C HR-MAS spectroscopy and [3–13C] pyruvate as a metabolic substrate. Magn Reson Med. 2009;62:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albers MJ, Butler TN, Rahwa I, et al. Evaluation of the ERETIC method as an improved quantitative reference for 1H HR-MAS spectroscopy of prostate tissue. Magn Reson Med. 2009;61:525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson MG, Zektzer AS, Tabatabai ZL, et al. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55:1257–1264. [DOI] [PubMed] [Google Scholar]
- 25.Shaka AJ, Barker PB, Freeman R. Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson. 1969;1985(64):547–552. [Google Scholar]
- 26.Carlos Cobas J, Bernstein MA, Martín-Pastor M, Tahoces PG. A new general-purpose fully automatic baseline-correction procedure for 1D and 2D NMR data. J Magn Reson. 2006;183:145–151. [DOI] [PubMed] [Google Scholar]
- 27.Johnson S, Rabinovitch P. Ex vivo imaging of excised tissue using vital dyes and confocal microscopy. Curr Protoc Cytom. 2012;Chapter 9:Unit 9.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keshari KR, Tsachres H, Iman R, et al. Correlation of phospholipid metabolites with prostate cancer pathologic grade, proliferative status and surgical stage – impact of tissue environment. NMR Biomed. 2011;24:691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eidelman E, Twum-Ampofo J, Ansari J, Siddiqui MM. The metabolic phenotype of prostate cancer. Front Oncol. 2017;7:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi S, Xue H, Wu R, et al. The MCT4 gene: a novel, potential target for therapy of advanced prostate cancer. Clin Cancer Res. 2016;22:2721–2733. [DOI] [PubMed] [Google Scholar]
- 31.Pértega-Gomes N, Vizcaíno JR, Miranda-Gonçalves V, et al. Monocarboxylate transporter 4 (MCT4) and CD147 overexpression is associated with poor prognosis in prostate cancer. BMC Cancer. 2011;11:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertega-Gomes N, Felisbino S, Massie CE, et al. A glycolytic phenotype is associated with prostate cancer progression and aggressiveness: a role for monocarboxylate transporters as metabolic targets for therapy. J Pathol. 2015;236:517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao J, Chen H, Madigan Mc, et al. Co-expression of CD147 (EMMPRIN), CD44v3–10, MDR1 and monocarboxylate transporters is associated with prostate cancer drug resistance and progression. Br J Cancer. 2010;103:1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiebler ML, Miyamoto KK, White M, Maygarden SJ, Mohler JL. In vitro high resolution 1H-spectroscopy of the human prostate: benign prostatic hyperplasia, normal peripheral zone and adenocarcinoma. Magn Reson Med. 1993;29:285–291. [DOI] [PubMed] [Google Scholar]
- 35.Sriram R, Gordon J, Baligand C, et al. Non-invasive assessment of lactate production and compartmentalization in renal cell carcinomas using hyperpolarized 13C pyruvate MRI. Cancers (Basel). 2018;10:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson MG, Keshari KR, Tabatabai ZL, et al. Quantification of choline- and ethanolamine-containing metabolites in human prostate tissues using 1H HR-MAS total correlation spectroscopy. Magn Reson Med. 2008;60:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elkhaled A, Jalbert LE, Phillips JJ, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4:116ra5–116ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Culture medium formulation
TABLE S2 Sources of culture medium ingredients
TABLE S3 Biopsy tissue NMR measurements: automated HR-MAS shimming routine
TABLE S4 Biopsy media NMR measurements: automated shimming routine


