ABSTRACT
Extracellular DNA (eDNA) plays an important role in both the aggregation of bacteria and in the interaction of the resulting biofilms with polymorphonuclear leukocytes (PMNs) during an inflammatory response. Here, transmission electron and confocal scanning laser microscopy were used to examine the interaction between biofilms of Pseudomonas aeruginosa and PMNs in a murine implant model and in lung tissue from chronically infected cystic fibrosis patients. PNA FISH, DNA staining, labeling of PMN DNA with a thymidine analogue and immunohistochemistry were applied to localize bacteria, eDNA, PMN-derived eDNA, PMN-derived histone H3 (H3), neutrophil elastase (NE) and citrullinated H3 (citH3). Host-derived eDNA was observed surrounding bacterial biofilms but not within the biofilms. H3 localized to the lining of biofilms while NE was found throughout biofilms. CitH3, a marker for neutrophil extracellular traps (NETs) was detected only sporadically indicating that most host-derived eDNA in vivo was not a result of NETosis. Together these observations show that, in these in vivo biofilm infections with P. aeruginosa, the majority of eDNA is found external to the biofilm and derives from the host.
Keywords: polymorphonuclear leukocyte, neutrophil extracellular traps, NETosis, histone, elastase
eDNA is not observed within in vivo biofilms, but surrounding the biofilms. A PMN-derived layer of eDNA may provide a passive physical shield for the biofilm against antibiotics and phagocytes.
INTRODUCTION
Persistent bacterial infections present an increasing challenge to health care practitioners worldwide. For Pseudomonas aeruginosa bacterial aggregation shows consistency with biofilm formation in both the tolerance and resistance to both antibiotics and the host immune system (Alhede et al. 2011; Bjarnsholt et al. 2013). Most studies of bacteria are based on planktonic cultures consisting primarily of single cells grown in vitro where both aggregation and host factors are missing (Moser et al. 2017). To understand the underlying mechanism(s) causing persistence of chronic bacterial infections, it is important to investigate the interplay between bacterial biofilms and the host immune system. A common denominator of both bacterial biofilms and cells of the innate host defense is extracellular DNA (eDNA). In vitro studies have shown eDNA, produced by the bacteria themselves, to be important primarily for initial biofilm formation as a major structural component of bacterial biofilms (Whitchurch et al. 2002) and as a stabilizer that contributes to increased tolerance of biofilms to antibiotics in vitro (Allesen-Holm et al. 2006; Izano et al. 2008; Chiang et al. 2013; Das, Sehar and Manefield 2013). The role of eDNA in vivo has still not been established, but during the last decade the use of animal model to study eDNA has been published (Jurcisek et al. 2007; Conover, Mishra and Deora 2011; Jung et al. 2017; Soavelomandroso et al. 2017). However, most of the studies only focus on the eDNA as a component originating from the bacteria and being a part of the biofilm, because this has been shown in vitro. Unfortunately, the host response and/or the in vivo environment is not taken into account. The common feature of chronic bacterial infections is persistent inflammation dominated by polymorphonuclear leukocytes (PMNs), which border bacterial biofilms but are unable to eradicate them (Bjarnsholt et al. 2009). This type of inflammation is classified as acute inflammation. Successful resolution of acute inflammation involves apoptosis and engulfment of demised PMNs by macrophages (Cox, Crossley and Xing 1995). However, during chronic bacterial infections, PMNs are thought to undergo necrosis, during which release of toxic compounds such as NE, oxidants and eDNA actually increase inflammation (Cox, Crossley and Xing 1995). Furthermore, components such as NE contribute to the destruction of tissue in chronic wounds and at sites of inflammation in the lungs of cystic fibrosis (CF) patients (Ohlsson and Olsson 1977; Yager and Nwomeh 1999; Voynow, Fischer and Zheng 2008; McCarty et al. 2012; Suleman 2016). PMNs actively secrete neutrophil elastase (NE) and histones attached to DNA, via a process termed NETosis (Steinberg and Grinstein 2007), both in vitro and in vivo in acute infections (Brinkmann et al. 2004; Yipp et al. 2012). This process is collectively referred to as neutrophil extracellular traps (NETs), which capture and kill bacteria. Since 2004, formation of NETs has been proposed as a function of neutrophils and other immune cells (Goldmann and Medina 2012) activated in vitro and in vivo in response to various bacterial components (Yipp et al. 2012; Kenny et al. 2017). However, a recent review questioned whether NETs play a role in innate immunity and thereby trapping of bacteria (Sørensen and Borregaard 2016). Release of eDNA by NETosis has, to the best of our knowledge, never been demonstrated in association with inflammation caused by chronic bacterial infections, despite evidence of close contact between PMNs and bacteria.
At present, our knowledge about the interaction between in vivo biofilms and the host immune system during chronic infections is limited and we do not know the role or localization of eDNA in relation to the biofilms in vivo. Unsuccessful eradication of infectious biofilms despite accumulation of PMNs has not been explained fully. Thus, to obtain more detailed information about the interplay between bacterial biofilms and PMNs during chronic infection and to increase our understanding of the role of eDNA, we studied these phenomena for the first time, directly in a murine implant model of biofilm infection and directly in human samples of chronic-infected CF patients. By using transmission electron microscopy (TEM) we were able to study the interaction between PMNs and biofilm of Pseudomonas aeruginosa formed on a silicone implant inserted in the peritoneal cavity of mice. Furthermore, labeling the DNA of PMNs during their S-phase using the Click-iT technology in vivo and confocal scanning laser microscopy (CSLM), showed PMN-derived DNA did not localize with biofilms. To establish the localization of specific components originating from PMNs we used immunohistochemistry and found that PMN-derived histone H3 (H3) and NE co-localized with biofilms, but citrullinated H3 (citH3) and PMN-derived DNA did not. The results obtained using the mouse model could be correlated directly to chronic bacterial infections in human CF lungs.
RESULTS
TEM-based examination of the interaction between biofilms and PMNs in a murine implant model shows damaged PMNs
Previously, we used scanning electron microscopy (SEM) (van Gennip et al. 2012) to examine the interaction between immune cells and P. aeruginosa biofilms in a murine implant model and found a marked influx of PMNs toward the biofilms. We then used CSLM to identify PMNs according to their segmented lobular nucleus. Here, we used TEM to examine the in-depth additional detailed interaction between bacteria and PMNs, and to visualize the matrix of the biofilm. Implants were inspected at three different time points (6, 24, and 48 h post-insertion) to illustrate progression of infection and the immune response.
At 6 h post-insertion, we noted intact PMNs containing internalized bacteria within vacuoles, which indicates active phagocytosis (Fig. 1A). PMNs with internalized bacteria were confined to areas with a low bacterial density. Areas with a higher bacterial density decreased the presence of intact PMNs (Fig. 1B). At 24 h post-insertion, bacterial aggregation resulted in the formation of large biofilms lining the surface of the inside of the implants (Fig. 1C). PMNs frequently adjoined the bacteria. These PMNs were enlarged and had damaged cell membranes (Fig. 1C). Intact PMNs at this time point were rare. Matrix material surrounding the bacteria within the biofilm was evident (Fig. 1 E–G). At 48 h post-insertion, the biofilm appeared denser and the PMNs were tightly interfaced with the bacteria (Fig. 1D). Matrix material was clearly visible (Fig. S1, Supporting Information).
Figure 1.
(A), Pseudomonas aeruginosa is internalized by PMNs in the murine implant model. Top row: TEM images showing intact and active PMNs containing internalized P. aeruginosa (arrows) 6 h post-insertion of a pre-coated silicone implant (Bar: 2 μm). Bottom row: internalized bacteria are magnified (Bar: 500 nm). Images represents sections obtained from two biological samples (two implants). (B–D), Interaction between PMNs and P. aeruginosa in the murine implant model. Pre-coated silicone implants were inserted into the peritoneal cavity of BALB/c mice. The interaction between PMNs and bacteria was imaged by TEM at 6 h (B), 24 h (C) and 48 h (D) post-insertion. In (B) black arrows points to bacteria and in (C) to a P. aeruginosa biofilm. Arrow heads: PMNs (Bar: 5 μm). Images represents sections obtained from two biological samples (two implants). (E-G), Matrix material surrounding P. aeruginosa on a silicone implant at 24 h post-insertion. TEM images showing matrix material surrounding P. aeruginosa bacteria at different magnifications. Matrix material (black arrows) can be seen between the bacteria. (E) Bar: 2 μm; (F) Bar: 1 μm; and (G) Bar: 200 nm. Images represents sections obtained from two biological samples (two implants).
In vitro interaction of P. aeruginosa and PMNs results in lysis of PMNs
When we examine the in vitro interaction between PMNs and aggregated P. aeruginosa microscopically, time series reveal that PMNs lyse resulting in release, expansion and dissolution of DNA (Movie 1 and Fig. S5, Supporting Information). The staining of the nuclear content, DNA, with propidium iodide (PI), increases over time as the membrane of the PMNs becomes damaged. At the end the DNA disappears (the red color from PI fades) (Movie 1 and Fig. S5, Supporting Information).
PMN-derived eDNA surrounds in vivo biofilms in a murine implant model
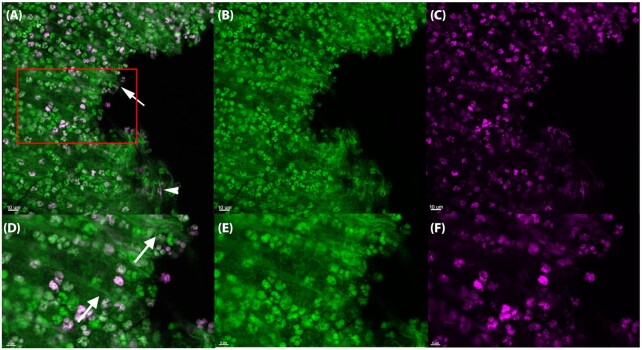
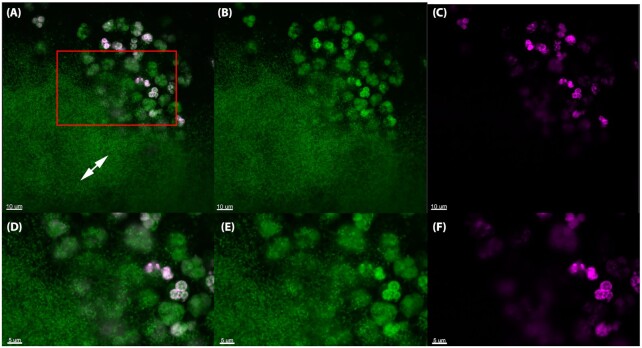
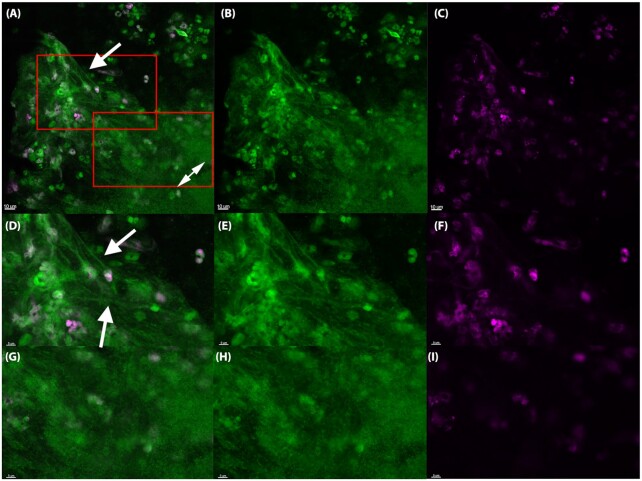
To examine eDNA in vivo, we stained silicone implants ex vivo from the murine implant model with SYTO9 (green) which binds to DNA together with the Click-iT® DNA labeling technology kit to determine the origin (i.e. murine or bacterial) of the eDNA in biofilms in vivo. This method utilizes a modified thymidine analogue, 5-ethynyl-2'-deoxyuridine (EdU), which is incorporated into DNA during active DNA synthesis in the S-phase of the eukaryotic cell cycle. Ex vivo, PMN DNA was detected by labeling with a fluorescent Alexa Fluor® dye (pink). Treatment of healthy mice showed the presence of EdU-labeled PMNs in the blood 2–5 days after treatment. Since the release of PMNs into the blood increases during an infection (Hansen, Karle and Valerius 1978), we decided to treat the mice two days pre-infection to increase the probability of EdU-labeled PMNs would reach the implant and biofilm post-infection. The percentage of EdU-labeled PMNs in the images presented in Figs 2–4 indicated 45% (165 pink/370 green), 42% (28 pink/66 green) and 89% (32 pink/36 green), respectively.
Figure 2.
In vivo DNA labeling of PMNs in the murine implant model. PMNs in the murine implant model were labeled with Click-iT®, in which a modified thymidine analogue, EdU (5-ethynyl-2'-deoxyuridine), is incorporated into DNA during active DNA synthesis in murine immune cells in vivo, and hence will label only DNA originating from murine PMNs. DNA is labeled ex vivo with Alexa Fluor® 647 (pink) and counterstained with SYTO9 (green) on an implant 24 h post-insertion in the peritoneal cavity. The number of PMNs stained with SYTO9 were estimated to 370 whereas the EdU labeled (pink) PMNs were estimated to 165. Thereby 45% PMNs in the images were EdU labeled. In the PMN accumulations SYTO9 (green)-stained DNA strings (white arrows) were observed as well as a few pink DNA strings (white arrowheads). (A, D), Merged images showing both EDU (pink) and SYTO9 (green) staining. (B, E), Images showing only the SYTO9 staining. (C, F), images showing only the EDU staining. Red square in A indicates magnified area in D–F. Images represents staining obtained from four biological samples (four implants).
Figure 4.
The in vivo biofilm lack PMN-derived DNA 48 h post-insertion in the murine implant model. PMNs in the murine implant model were labeled with Click-iT®, in which a modified thymidine analogue, EdU (5-ethynyl-2'-deoxyuridine), is incorporated into DNA during active DNA synthesis in murine immune cells in vivo, and hence will label only DNA originating from murine PMNs. DNA is labeled ex vivo with Alexa Fluor® 647 (pink) and counterstained with SYTO9 (green) on an implant 48 h post-insertion in the peritoneal cavity. The number of PMNs stained with SYTO9 were estimated to 36 whereas the EdU labeled (pink) PMNs were estimated to 32. Thereby 89% PMNs in the images were EdU labeled. Labeling was observed in PMNs (pink) but was absent from biofilms (double-headed arrows), suggesting that PMNs are not a source of eDNA in biofilms. (A, D), Merged images showing both EDU (pink) and SYTO9 (green) staining. (B, E), Images showing only the SYTO9 staining. (C, F), images showing only the EDU staining. Red square in A indicates magnified area in D-F. Images represents staining obtained from four biological samples (four implants).
Long strings of eDNA (green) were clearly visible in the PMN accumulations with interacting bacteria (Fig. 2) and surrounding the biofilms (Fig. 3), but not as part of the biofilm itself (Figs 3 and 4).
Figure 3.
The in vivo biofilm lack PMN-derived DNA 24 h post-insertion in the murine implant model. PMNs in the murine implant model were labeled with Click-iT®, in which a modified thymidine analogue, EdU (5-ethynyl-2'-deoxyuridine), is incorporated into DNA during active DNA synthesis in murine immune cells in vivo, and hence will label only DNA originating from murine PMNs. DNA is labeled ex vivo with Alexa Fluor® 647 (pink) and counterstained with SYTO9 (green) on an implant 24 h post-insertion in the peritoneal cavity. The number of PMNs stained with SYTO9 were estimated to 66 whereas the EdU-labeled (pink) PMNs were estimated to 28. Thereby 42% PMNs in the images were EdU labeled. In the PMN accumulations SYTO9 (green)-stained DNA strings (white arrows) were observed. EdU labeling was observed in PMNs (pink), but was absent from biofilms (double headed arrows), suggesting that PMNs are not a source of eDNA in biofilms. (A, D), merged images showing both EDU (pink) and SYTO9 (green) staining. (B, E), Images showing only the SYTO9 staining. (C, F), images showing only the EDU staining. Red squares in A indicates magnified area in D-F and G-I. Images represents staining obtained from four biological samples (four implants).
As seen in Fig. 2, eDNA strings labeled with pink dye was clearly visible in close proximity to PMNs 24 h post-insertion of the implant. However, no pink eDNA was detected in areas occupied by bacterial biofilms (Figs 3 and 4) at 24 h or 48 h post-insertion, indicating that PMN-derived eDNA is not present internally within these structures.
eDNA surrounds in vivo biofilms in lung tissue of chronically infected CF patients
Previously, peptide nuclear acid (PNA) fluorescence in situ hybridization (FISH) and 4’,6-diamidino-2-phenylindole (DAPI) have been used to identify bacterial biofilms surrounded by PMNs in deparaffinated sections of CF lung tissue (Bjarnsholt et al. 2009). The PNA probes bind to bacterial ribosomal 16S RNA (Stender et al. 2002) and DAPI binds to the A-T regions of double-stranded DNA. Here, we used PNA FISH (red) in combination with DAPI (blue) to show that eDNA was localized external to the biofilm in CF lung tissue (Fig. 5A–C). The biofilm stained red by a Texas Red-labeled P. aeruginosa-specific PNA probe, and the nuclei of PMNs were clearly stained blue, as were the strings of DNA outside the biofilm (Fig. 5A). Furthermore, we used the DNA stain propidium iodide (PI), which stain all cells red due to the fixation of the tissue, whereas only strings of DNA were observed outside the biofilms.
Figure 5.
PNA FISH and DAPI staining of CF lung tissue. Deparaffinated CF lung tissue section stained with (A), PNA FISH to show a P. aeruginosa (red) and (B), DAPI (blue) to show DNA of PMNs and eDNA. (C), Shows an overlay of A and B. (D), A deparaffinated CF lung tissue section stained with the DNA stain propidium iodide (PI) to illustrate both biofilm and eDNA. White arrows point to P. aeruginosa biofilm and black arrows point to eDNA as strings. (Bars: 9 μm).
Localization of histone H3 (H3), citrullinated H3 (citH3) and NE in a murine implant model
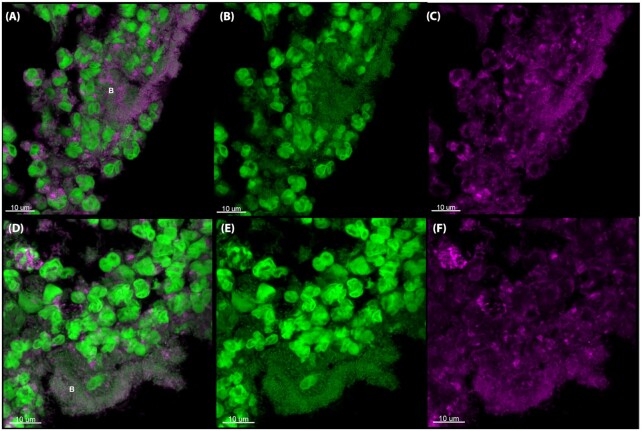
To localize components originating from murine PMNs during a bacterial biofilm infection, we stained deparaffinated sections of silicone implants isolated 24 h post-insertion in the peritoneal cavity of mice with antibodies specific for citH3 (Fig. 6), H3 (Fig. 7) or NE (Fig. 8). All of these components, especially citH3, have also been associated with NETs. Hence, we used citH3 as a marker for NETs (Neeli, Khan and Radic 2008; Wang et al. 2009; Wong et al. 2015; Barliya et al. 2017).
Figure 6.
citH3 antibody staining of the murine implant model. CSLM images of deparaffinized sections of silicone implants from the murine implant model 24 h post-insertion. The sections were stained with primary antibodies specific for citrullinated histone H3 (citH3). The secondary antibody was conjugated to Alexa Fluor 647 (pink). SYTO9 (green) was used as a counterstain. SYTO9 stains DNA in bacteria and eukaryotic cells. (A, D), Merged images of both the antibody (pink) and SYTO9 (green) staining. (B, E), Are only the SYTO9 staining. (C, F), are only the antibody staining. The images represent two implants. The letter B indicates biofilm. Images represents staining of 2–4 sections obtained from two biological samples (two implants).
Figure 7.
H3 antibody staining of the murine implant model. CSLM images of deparaffinized sections of silicone implants from the murine implant model 24 h post-insertion. The sections were stained with primary antibodies specific for histone H3 (H3). The secondary antibody was conjugated to Alexa Fluor 647 (pink). SYTO9 (green) was used as a counterstain. SYTO9 stains DNA in bacteria and eukaryotic cells. (A, D), Merged images of both the antibody (pink) and SYTO9 (green) staining. (B, E), Is only the SYTO9 staining. (C, F), is only the antibody staining. The images represent two implants. The letter B indicates biofilm. Images represents staining of 2–4 sections obtained from two biological samples (two implants).
Figure 8.
NE antibody staining of the murine implant model. CSLM images of deparaffinized sections of silicone implants from the murine implant model 24 h post-insertion. The sections were stained with primary antibodies specific for NE. The secondary antibody was conjugated to Alexa Fluor 647 (pink). SYTO9 (green) was used as a counterstain. SYTO9 stains DNA in bacteria and eukaryotic cells. (A, D), Merged images of both the antibody (pink) and SYTO9 (green) staining. (B, E), Are only the SYTO9 staining. (C, F), Are only the antibody staining. The images represent two implants. The letter B indicates biofilm. Images represents staining of 2–4 sections obtained from two biological samples (two implants).
H3 localized to areas surrounding bacterial biofilms. Only weak staining was observed inside the biofilm; these findings are in agreement with those described above (i.e. no eDNA originating from PMNs was observed within biofilms). citH3 localized with some PMNs, but the lack of citH3 signifies that the eDNA is not generated by NETosis.
In contrast staining with specific antibodies against NE revealed distinct co-localization of biofilms and NE, indicating that NE is incorporated into biofilms (Fig. 8). Control images are presented in Fig. S4A. For specific identification of P. aeruginosa a PNA FISH image is presented in Fig. S4C (Supporting Information).
Localization of H3, citH3 and NE in lung tissue of chronically infected CF patients
Co-localization of NE and H3 with P. aeruginosa biofilms in the murine implant model led us to examine deparaffinated lung sections from explanted CF patients with chronic bacterial infections. To verify function and correct binding of the three antibodies when using human PMNs, we stained in vitro phorbol myristate acetate (PMA) generated NETs from human PMNs. We were able to show co-localization of NE and citH3 with NETs, but not with H3 (Fig. S2, Supporting Information). The lack of H3 co-localization correlates well with the fact that NETosis results in a partial degradation of H3 and post-translational histone modifications and when using PMA as stimulation a total loss of full length H3 is observed (Papayannopoulos et al. 2010; Kenny et al. 2017). However, we did find co-localization of H3 with the nucleus of unstimulated PMNs, which indicates a functional antibody (Fig. S3, Supporting Information). NE also co-localized with the cytoplasma of unstimulated PMNs of deparaffinated sections (Fig. S3, Supporting Information).
In CF lungs the distribution of citH3, H3 and NE was similar to that observed in the murine implant model. CitH3 was observed in a few PMNs but did not co-localize with bacterial biofilms (Fig. 9A–C). H3 was observed at the periphery of biofilms but not co-localized with the biofilm (Fig. 9D–F). NE co-localized with bacteria within the biofilm (Fig. 9G–I). Control images are shown in Fig. S4B (Supporting Information). To estimate the co-localization of NE, H3 and citH3 with in vivo biofilms we used Manders’ co-localization coefficient as seen in Fig. 9J. A value of 1, means that the green pixels (SYTO9) are perfectly co-localized with the pink pixels (antibody). The co-localization coefficient confirmed what we visually observed, H3 localized outside of biofilms (P < 0.0001), citH3 sometimes to outside of biofilms (P < 0.0007) and NE localized primarily to biofilms (P < 0.25). There was not a significant difference between the outside and biofilm areas for NE. Some of the biofilms seemed to be located in a substance material, presumably alginate as previously shown (Bjarnsholt et al. 2009) due to mucoid P. aeruginosa. These biofilms did not co-localize with NE. Instead NE was co-localized with the surrounding PMNs.
Figure 9.
Immunostaining of CF lung tissue. CSLM images of deparaffinized sections of CF lung tissue. The sections were stained with primary antibodies specific for citrullinated histone H3 (citH3) (A-C). Histone H3 (D-F) or NE (G-I). The secondary antibody was conjugated to Alexa Fluor 647 (pink). SYTO9 (green) was used as a counterstain. SYTO9 stains DNA in bacteria and eukaryotic cells. (A, D, G), Merged images of both the antibody (pink) and SYTO9 (green) staining. (B, E, H), Are only the SYTO9 staining. (C, F, I), Are only the antibody staining. The images represent staining of 2–4 sections from three different CF lungs. B indicates biofilm. J: Co-localization of biofilm with antibodies in CF lungs shown as Manders’ co-localization coefficient. ‘Outside biofilm’ are areas containing PMNs and ‘biofilms’ are ROI defined biofilms. Sample size (n): NE outside biofilms (16), NE biofilms (20), H3 outside biofilms (18), H3 biofilms (17), citH3 outside biofilms (15), citH3 biofilms (18). There was significant difference between H3 outside biofilms and H3 biofilms (P < 0.0001) and citH3 outside biofilms and biofilms (P < 0.0007). No significant difference was found between NE outside biofilms and biofilms (P < 0.25). P < 0.05 was considered significant.
DISCUSSION
Surprisingly, our visual findings show that, in vivo, eDNA is concentrated external to bacterial biofilms rather than inside as shown previously in vitro by us and others (Whitchurch et al. 2002; Alhede et al. 2011). In fact, the lack of SYTO9 and DAPI staining within the bacterial biofilms in chronic bacterial infections truly questions the significance of the, otherwise in vitro important, role of eDNA in biofilms in vivo as a scaffold of the biofilm. Instead it highlights the important and detrimental feature of host-derived eDNA as a protective shell surrounding the bacteria. This barrier of host eDNA surrounding a biofilm in vivo, which could both limit dissemination of bacteria and shield the biofilm from phagocytosis, may be manifested by necrotic lysis of PMNs rather than through NETosis. Thus, our results are in support of deposition of PMN content in a zone between the PMNs and the biofilm as recently demonstrated in experimental P. aeruginosa ocular biofilm infection where the deposition of NETosis derived material prevented dissemination (Thanabalasuriar et al. 2019). In our study, we found only scarce amount of host-derived eDNA and citH3 suggesting a modest contribution by NETosis to the material in the zone between PMNs and biofilm. However, dissemination of the bacteria may be prevented by the NE localized to the biofilm and therefore we propose other mechanisms than NETosis to contribute to the stalemate and chronicity of biofilm infection as illustrated in Fig. 10.
Figure 10.
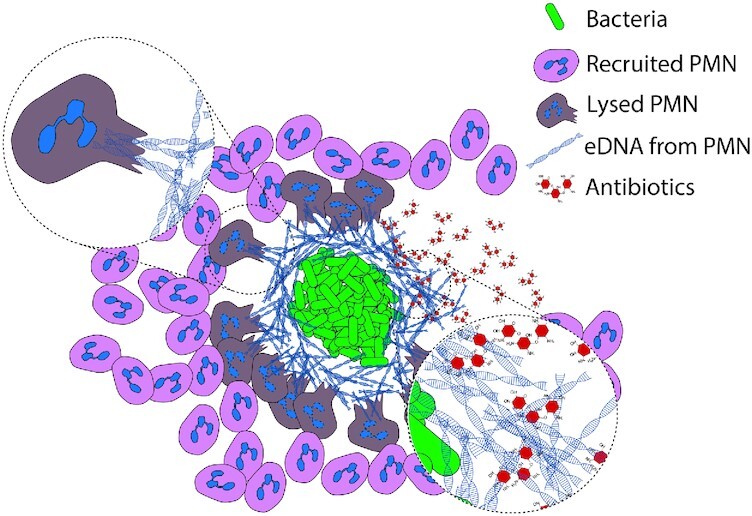
Hypothesis for formation of an eDNA shield in chronic bacterial infections in vivo. Early during biofilm formation, PMNs are able to phagocytose and destroy single bacteria or very small particles of bacterial cells. As the biofilm develops, bacterial aggregates evade phagocytosis and induce a necrotic cell death of PMNs. In chronic bacterial infections PMNs are continuously recruited to the site of biofilms where they release eDNA via necrosis. This released eDNA does not become incorporated into the biofilm itself. The PMN-derived layer of eDNA, which constitutes a secondary matrix, may provide a passive physical shield for the biofilm against cationic antibiotics such as tobramycin and additional phagocytes.
Using TEM we observed biofilm formation and swollen PMNs with damaged membranes, which is in agreement with our former SEM observations (van Gennip et al. 2012) showing PMNs failing to eradicate bacterial biofilms. The matrix material seen between the bacteria in the biofilm has not been fully characterized, but in vitro studies show the importance of eDNA for initial formation of young surface biofilms in vitro (Whitchurch et al. 2002); accordingly, it is believed to be an important structural component (Allesen-Holm et al. 2006; Izano et al. 2008). Furthermore, P. aeruginosa has been shown to be attached to eDNA in CF sputum (Walker et al. 2005) and incorporate external eDNA into the biofilm matrix in vitro (Chiang et al. 2013). PMNs were enlarged and had damaged cell membranes, presumably due to exposure to the bacterial virulence factor, rhamnolipid, which is detrimental to PMNs (Alhede et al. 2009; van Gennip et al. 2009). It has previously been shown that P. aeruginosa produce rhamnolipids which cause PMNs to swell and burst, thereby releasing their cytoplasmic and nuclear content into the surrounding area (Jensen et al. 2007). In an unpublished study, we observed that when PMNs were exposed to aggregates of P. aeruginosa in vitro approx. 60% PMNs were killed within 8 h (unpublished Alhede et al.). The cellular components including eDNA from necrotic PMNs are believed to serve as a biological matrix for biofilm formation and a protective shield within the biofilm increasing tolerance to treatment to tobramycin in vitro (Walker et al. 2005; Chiang et al. 2013). This was confirmed in a mouse model where a rhlA mutant (unable to excrete rhamnolipid) did not cause any PMN lysis and no release of cytoplasmic and nuclear content, thus consistent with a mechanism of lysis by the biosurfactant rather than a programmed NET pathway for eDNA release (van Gennip et al. 2009).
The lack of strings in in vivo biofilms indicate that unlike in vitro biofilms, eDNA is not a direct part of the biofilm. Visually eDNA from P. aeruginosa has been shown to form a net both in a stationary grown biofilm and in the flow cell biofilm (Allesen-Holm et al. 2006; Alhede et al. 2011). Furthermore, eDNA from P. aeruginosa is believed to be double-stranded DNA (Allesen-Holm et al. 2006) and since DAPI bind to double-stranded DNA the lack of DAPI staining signifies the lack of eDNA in the in vivo biofilm. Also, the measurement of double-stranded DNA has been shown to decrease when P. aeruginosa change from the planktonic state to the aggregating state (Das et al. 2014). However, the eDNA of a plate grown colony of P. aeruginosa have been shown to be 5 μm long (Chiba et al. 2015) which mean that visually the eDNA fragment would be almost as small as a single P. aeruginosa bacterium. Visually this does not correlate to our in vivo and others in vitro findings. However, we might have to rethink the need for a structural matrix depending on the environment of the biofilm outside the laboratory. Maybe when the biofilm is exposed to high forces such as fluid it needs to establish a solid structure, but in the CF lung another type of structure is established within the mucus to withstand the immune cells. Accordingly, this indicates that neither bacterial nor PMN-derived eDNA is incorporated into biofilms in vivo, but eDNA from necrotic PMNs is more likely accumulating outside the biofilms.
Since we only found host-derived eDNA outside infectious biofilms in vivo, we speculated whether in vivo biofilm matrix structures comprised other components originating from PMNs (e.g. H3, citH3 or NE). PMNs undergoing apoptosis, necrotic cell death or NETosis release different histones (i.e. H3 during necrotic cell death and citH3 during NETosis), either into the surrounding area or attached to NETs (Brinkmann et al. 2004). Release of these extracellular histones promotes secretion of pro-inflammatory cytokines and recruitment of immune cells to sites of infection (Westman et al. 2015). Histones bind to lipopolysaccharide (LPS) derived from P. aeruginosa, which neutralizes the positive charge of the histone molecule (Rose-Martel and Hincke 2014); with the outcome of antimicrobial activity (Rose-Martel et al. 2017). The fact that histones are antimicrobial (Hirsch 1958; Brinkmann et al. 2004), might not be the case in this study. Since H3 is rich in arginine it may rather play a role in the arginine pathway in P. aeruginosa, which breaks down arginine to ornithine via the arginine deiminase pathway to sustain growth under anoxic conditions. Ornithine is a carbon source used to maintain growth during periods in which nitrate is either limited or lacking (Vander Wauven et al. 1984; Stalon et al. 1987; Eschbach et al. 2004). Arginine has also been shown to contribute to biofilm formation (Bernier et al. 2011).
Here, we showed that H3 is mostly present along the periphery of bacterial biofilms, but not within the biofilms. This could indicate that H3 is still bound to eDNA. By contrast, citH3 did not localize with biofilms in either the mouse model or CF lung samples. CitH3 is a post-translational modification of H3. Conversion of arginine to citrulline is accomplished by peptidylarginine deiminase 4 (PAD4) and is a prerequisite for in vitro generated NETosis since the modification of arginine residues to citrulline changes the charge of the histones, leading to massive chromatin de-condensation, NETs (Wang et al. 2009; Lewis et al. 2015; Wong et al. 2015). NETs are highly de-condensed chromatin structures that comprise mainly DNA, although histones and antimicrobial granular substances such as NE are also present (Brinkmann et al. 2004). Histone citrullination in vitro depends on stimulation of PMNs (Neeli and Radic 2013; Konig and Andrade 2016); indeed, an increase in histone citrullination is associated with chromatin de-condensation during NETs formation and LPS-induced early responses to inflammatory stimuli of PMNs (Neeli, Khan and Radic 2008; Wang et al. 2009). However, one widely used stimulus, PMA, has resulted in conflicting results in the literature and has been shown to induce NETs with and without co-localized citH3 and H3 (Brinkmann et al. 2004; Konig and Andrade 2016; Neeli and Radic 2016). In addition citrullination of H3 is strongly reduced when PMNs are stimulated during anoxic conditions (Lopes et al. 2018), which is present in the endobronchial secretion in infected CF lungs (Worlitzsch et al. 2002; Kolpen et al. 2014).
The lack of host-derived eDNA and citH3 associated with bacterial biofilms in the present study demonstrates that NETosis probably does not occur in chronic biofilm infections. Release of DNA under these conditions is not an active process as in NETosis; rather, it is more likely due to lysis of PMNs (Jensen et al. 2007) which correlates to the presence of H3 and absence of citH3. In addition, our findings that NETosis seemingly plays no role in chronic P. aeruginosa infections correlates to the depletion of molecular oxygen at the site of chronic biofilm infections (Kolpen et al. 2010), thereby preventing NADPH oxidase from generating the reactive oxygen species (ROS) needed to release NETs (Fuchs et al. 2007). Thus, PAD4-dependent NET formation might not be responsible for DNA release in CF airways and we speculate whether citH3 may serve as a biomarker for discriminating chronic from acute infections. However, NET generation independent of ROS generated by NADPH oxidase has been shown in vivo (Kolaczkowska et al. 2015) therefore further work is needed to confirm this. Nevertheless, eDNA in sputum samples from CF patients have been shown to originate from PMNs (Lethem et al. 1990) with NETs as the major component (Manzenreiter et al. 2012). However, since NETs are antimicrobial this should benefit the CF patients since this should lead to eradication of the present bacteria. On the contrary eDNA has been shown to increase the viscosity of the mucus and bind antibiotics such as tobramycin (Potter et al. 1965; Ramphal et al. 1988). Furthermore, by treating the CF patients with DNase an improved lung function in CF patients and an increased effect of antibiotics have been reported (Shak et al. 1990; Hunt et al. 1995).
We did observe NE within biofilms, although this may have been derived from a necrotic cell death of PMNs external to the biofilm, followed by passive diffusion into the biofilm structure. NE is stored in primary (azurophilic) granules and released upon degranulation, phagocytosis and cell death. NE cleaves a wide variety of proteins, including bacterial virulence factors (Weinrauch et al. 2002). The purpose of NE is to degrade phagocytosed proteins; however, in CF patients NE damages the airways (Janoff et al. 1979). Furthermore, NE is an important component of NETs (Brinkmann et al. 2004). However, since NE was found within the biofilms and eDNA was not supports the lack of NETs, since in these structures NE is bound to NETs. Furthermore, unlike in acute infections caused by Gram-negative bacteria P. aeruginosa is not killed by NE following aggregation and biofilm formation (Belaaouaj et al. 1998). The absence of NE-mediated killing is likely attributed to production of the protease inhibitor ecotin by bacteria (Eggers et al. 2004). In addition, NET formation induced by P. aeruginosa depends strongly on flagella motility (Floyd et al. 2016), which may further explain the absence of NETs in CF lungs in which the flagella of P. aeruginosa are down-regulated (Ahmed et al. 1993) and in which flagellin, which is the major component of flagella, may be cleaved by NE (Lopez-Boado et al. 2004). Since a decrease and/or absence of flagella motility is associated with formation of bacterial biofilms (Chaban, Hughes and Beeby 2015), a lack of motile flagella may lead to a lack of detectable NETs.
Limitations: Our study applies different microscopic techniques for visual inspection of different model systems and patient samples. An image-based study as this has limitations as to showing representing images of the investigated question. However, as in this case appropriate experimental setups are not available and since the imaging technique continual improves, we are now able to study small details such as DNA. We think that the visual information obtained significantly contributes to new information on the in vivo setting of chronic bacterial infections. The in vitro models available also have limitations when studying the interaction between immune cells and biofilms since most often the immune cells are isolated from their natural environment and the bacteria are grown in a media not resembling the in vivo environment.
In conclusion, this study is the first to examine the direct distribution of host and bacterial eDNA during chronic bacterial infections in vivo. Unlike after stimulation in vitro, stimulation of PMNs by bacterial biofilms during chronic infections in vivo does not seem to trigger active release of NETs; however, necrotic PMNs do release eDNA, histone H3 and antibacterial enzymes such as NE.
We did not find evidence of eDNA being part of in vivo biofilms, instead the PMN-derived layer of eDNA, which constitutes a secondary matrix, may provide a passive physical shield for the biofilm against cationic antibiotics such as tobramycin and additional phagocytes. A similar spatial arrangement has recently been described in a murine model of P. aeruginosa infection of the cornea in which the bacterial biofilm and neutrophil layers are separated by a ‘dead zone’ rich in eDNA and NE (Thanabalasuriar et al. 2019).
Thus, we believe that eDNA released by PMNs due to a necrotic cell death contribute to the protection afforded to bacteria by biofilms (Fig. 10). These novel findings shed new light on the origin and role of eDNA in in vivo biofilms and unexpectedly question the role of extracellular host DNA during inflammation associated with chronic bacterial infections and of NET activation.
MATERIALS AND METHODS
Murine implant model
All experiments were performed using a wild-type (WT) P. aeruginosa strain obtained from Professor Barbara Iglewski (University of Rochester Medical Center, NY, USA). The strain is QS proficient, except for the reduced production of C4-HSL previously noted for this P. aeruginosa variant (Köhler et al. 2001). Bacteria obtained from freezer stocks were plated onto blue agar plates (State Serum Institute, Denmark) and incubated overnight at 37°C. Blue agar plates are used to select Gram-negative bacilli (Høiby 1974). One colony was used to inoculate overnight cultures grown in Luria-Bertani (LB) medium at 37°C with shaking at 180 rpm. Female BALB/c mice (6 weeks-of-age) were purchased from Taconic M&B A/S (Ry, Denmark) and maintained on standard mouse chow and water ad libitum for 2 weeks before challenge. The murine implant model was generated as described previously (Bjarnsholt et al. 2010; van Gennip et al. 2012), with some modifications. Briefly, the silicon tubes (Ole Dich Instrumentmakers Aps. Silicone Pumpeslange 60 Shore A (ID: 4.0 mm, OD: 6.0 mm, wall: 1.0 mm) were cut to a length of 4 mm, and the bacterial pellet from a centrifuged overnight culture was resuspended in 0.9% NaCl (OD600nm = 0.1). Mice were anesthetized by subcutaneous (s.c.) injection of hypnorm/midazolam (Roche) [hypnorm (0.315 mg ml−1 fentanyl citrate; 10 mg ml−1 fluanisone); midazolam (5 mg ml−1) and sterile water; 1:1:2 v/v. The pre-coated silicone implants were inserted in the peritoneal cavity of the mice. The mice were treated with bupivacaine and Temgesic® as post-operative pain relief. Experiments were terminated with euthanization of the mice with intraperitoneal injection of pentobarbital (DAK; 10.0 ml kg−1 body weight).
Preparation of samples for TEM
The silicone implants were fixed in 2% glutaraldehyde in 0.05 M sodium phosphate buffer pH 7.4. Growing biofilms on a silicone tube presents a challenge with respect to sample preparation prior to the different microscopy techniques. First, a traditional preparation method was used for TEM to get an indication of the interaction between PMNs and the biofilm. However, some of the steps involved in the embedding process were shortened. Samples were observed with a CM 100 TEM (Philips, the Nederlands) operated at 80 kV. Imaged were recorded with a side-mounted Olympus Veleta camera (resolution, 2048 × 2048 pixels (2 K × 2 K)) and the iTEM (Olympus, Germany) software package.
In vitro experiments with PMNs and P. aeruginosa
Wild-type P. aeruginosa (strain PAO1) was obtained from the Pseudomonas Genetic Stock Center (www.pseudomonas.med.ecu.edu; strain PAO0001). Bacteria were tagged with a stable green fluorescent protein (GFP) constitutively expressed by plasmid pMRP9 (Davies et al. 1998). PAO1 cultures were grown for 24 h at 37°C in 12 well microtiter plates containing LB medium. Human blood was collected from healthy volunteers with the approval of the Danish Scientific Ethical Board (H-3–2011-117). PMNs were isolated as described (Bjarnsholt et al. 2005). Isolated PMNs were resuspended in krebs ringer buffer containing 10 mM glucose at 37°C (final density, 2.5 × 107 PMNs/ml). Propidium iodide (2.5 ug/ml) was used as an indicator of cell death. PAO1 (350 μl) and PMNs (50 μl) were mixed in a 12 well microtiter plate and observed under a confocal microscope for 35 min.
Click-iT™
Mice were treated with EdU (Click-iT™ EdU Alexa Fluor™ 647 Imaging Kit) (C10640, ThermoFisher) for 2 days pre-infection and until termination of the experiment. Each mouse was injected i.p. twice a day with 25 mg/g EdU dissolved in 0.9% NaCl (Zeng et al. 2010). Control mice received 0.9% NaCl without EdU. Ex vivo implants were fluorescently labeled using the Click-iT kit, according to the manufacturer's instructions, and stained with SYTO9 (S34934, ThermoFisher) prior to observation under a microscope.
The percentage of SYTO9 stained PMN nuclei and EdU labeled cells were estimated using the Measuring PRO add on to Imaris 8.5 (Bitplane, Switzerland). Individual nuclei were identified by applying isomeric surfaces and separated with the build in object separator as spheres with a 5–10 µm diameter in green (SYTO9) spectra. Similarly, antibody stained cells were separated and counted as ∼10 µm diameter spheres in pink (EdU) spectra. The results were returned as simple numeric counts.
In vitro generated NETs
In vitro NETs were generated according to Vong et al. (Vong, Sherman and Glogauer 2013) and Brinkmann et al. (Brinkmann et al. 2012) with modifications. PMNs were isolated according to Hu et al. (Hu 2012) using polymorphprep with modifications. Isolated PMNs were suspended in RPMI 1640 with glutamine and were seated in an ibidi slide coated with poly-L-lysine (80 604, ibidi) for 30 min before stimulation with PMA for 3 h. The PMNs were incubated at 37 degrees and 5% CO2. PMNs was washed with PBS and fixed in 4% PFA pH 7.2 for 15 min, washed in 0.025% Triton X-100 twice before proceeding with antibody staining as described below for antibody staining of deparaffinated tissue sections. Before imaging the channels were filled with Ibidi mounting medium (50001, ibidi).
Ex vivo paraffin samples
Samples from the mouse model, the lungs of CF patients and unactivated human PMNs were fixed in 4% formalin and paraffin-embedded according to standard protocols. Sections (4 μm thick) were cut using a standard microtome, fixed on glass slides and stored at 4°C until required.
Lung tissue from chronically infected CF patients
Explanted lungs tissue from three contemporary intensively treated P. aeruginosa infected CF patients were examined (Bjarnsholt et al. 2009).
PNA FISH
PNA FISH staining was performed as previously described (Kirketerp-Møller et al. 2008; Bjarnsholt et al. 2009). Texas Red-labeled P. aeruginosa-specific PNA probe both in hybridization solution (AdvanDx, Inc., Woburn, MA), was added to each section and hybridized in a PNA FISH workstation covered by a lid at 55°C for 90 min. The slides were washed for 30 min at 55°C in wash solution (AdvanDx) and counterstained with DAPI (D1306, ThermoFisher).
Antibody staining of deparaffinated tissue sections
Sections were deparaffinized in 99.9% xylene (twice for 5 min each time) and subsequently hydrolyzed in 99.9% ethanol (twice for 5 min each time), followed by 96.5% ethanol (twice for 3 min each time). Finally, sections were rinsed in water (twice for 5 min each time). Heat-induced antigen retrieval was performed in citrate buffer, pH 6.0 (Sigma C9999) for 15 min in a domestic microwave oven. For unstimulated human PMNs a permabilization step was included using 0.1% Triton X-100 for 20 min. Samples were washed twice (5 min each time) in 0.025% triton X-100, drained and blocked for 2 h at room temp in 10% goat serum in 1% BSA/PBS. Samples were drained and incubated for 16–18 h at 4°C with a primary antibody (Abcam Cat# ab1791, RRID:AB_302613, Abcam Cat# ab5103, RRID:AB_304752 or Abcam Cat# ab21595, RRID:AB_446409), diluted 1:200 in 2% goat serum/1% BSA in PBS). Negative controls were isotype IgG (Abcam Cat# ab27478, RRID:AB_2616600) or absence of primary antibody. Samples were drained and washed in 0.025% Triton X-100 (twice for 5 min each) with gentle agitation. Samples were then incubated (1 h at room temp in the dark) with a secondary antibody (Abcam Cat# ab150083, RRID:AB_2714032) diluted 1:1000 in 1% BSA/PBS. Finally, samples were washed three times in PBS (5 min each time) and counterstained with SYTO9 (S34934, ThermoFisher). ProLong™ gold antifade mountant (Thermofisher, P36934) was applied before adding the coverslip.
Co-localization coefficient
Co-localization refers to the geometric co-distribution of two fluorescent labels or color channels. Co-localization was acquired in Zen (version 2.1 SP3 FP2 (black)) as described by Zeiss ‘Acquiring and analyzing data for co-localization experiments in AIM or Zen software’. The Manders’ co-localization coefficient (Manders, Verbeek and Aten 1993) for SYTO9 (green channel) was used to show the co-localization of the antibody (pink channel) using 2D images. Perfect co-localization will give the value 1. The crosshair was set to 1003 for the green channel and 2500 for the pink channel based on single stained deparaffinated sections according to the manual by Zeiss. Sections from three different patients were used. Biofilm and PMN areas were selected by region of interest (ROIs) in each deparaffinated section. All biofilm areas in one image were included. GraphPad Prism was used for statistical analysis using unpaired t-test. P < 0.05 was considered significant.
Confocal scanning laser microscopy
All observations and image acquisition were performed using a confocal scanning laser microscope (LSM510, LSM 710 or LSM 880; Carl Zeiss GmbH, Germany). Images were obtained using 40 × 63 × or 100 × oil objective lenses. Images were scanned at 405 nm (blue), 488 nm (green) and 633 nm (far red). Images of the biofilms were generated using Imaris software including the magnifications (version 8.4, Bitplane AG). Images were processed for display using Photoshop software (Adobe). Presence of biofilms were defined by presence of bacterial cluster larger than 5 µm in diameter
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen, Zhila Nikrozi for preparing samples for TEM, Heidi Marie Paulsen for preparing paraffin sections and Steen Seier Poulsen for helpful discussions regarding antibody staining procedures. M.A., M.A. and T.B. were funded by the Lundbeck Foundation. M.A. was also founded by Hørslev-Fonden and Torben and Alice Frimodt Fonden.
ETHICS STATEMENT
All animal studies were carried out in accordance with the European convention and Directive for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes, and with Danish law on animal experimentation. All experiments were authorized and approved by the National Animal Ethics Committee, Denmark (The Animal Experiments Inspectorate, dyreforsoegstilsynet.dk; permit numbers: 2012−15−2934−00677 and 2014−15−0201−00259).
Human blood was collected from healthy volunteers with the approval of the Danish Scientific Ethical Board (H-3–2011-117). All donors provided written informed consent.
Infected tissue was collected following approval by the Danish Scientific Ethical Board (KF465 01278432). The samples used were collected with written informed consent for a previous study [9], and only from adults. The samples were anonymized for this study.
AUTHOR CONTRIBUTIONS
M.A. conducted the mouse experiments and antibody staining experiments and wrote and edited the manuscript.
M.A. undertook microscopic analysis of Click-It and edited the manuscript.
K.Q. assisted with TEM preparation and image recording and edited the manuscript.
P.Ø.J. performed the in vitro PMN experiments, contributed to data interpretation and edited the manuscript.
K.N.K estimated EdU labeled cells with Imaris, performed PNA FISH, helped with the co-localization, illustrated Fig. 10 and edited the manuscript.
P.S. contributed to data interpretation and edited the manuscript.
T.B. performed the in vitro PMN experiments and PNA FISH, contributed to data interpretation and edited the manuscript.
Conflicts of interest. None declared.
REFERENCES
- Ahmed K, Dai TC, Ichinose Aet al.. Neutrophil response to Pseudomonas aeruginosa in respiratory infection. Microbiol Immunol. 1993;37:523–9. [DOI] [PubMed] [Google Scholar]
- Alhede M, Bjarnsholt T, Jensen PØet al.. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–8. [DOI] [PubMed] [Google Scholar]
- Alhede M, Kragh KN, Qvortrup Ket al.. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One. 2011;6:e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang Let al.. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–28. [DOI] [PubMed] [Google Scholar]
- Barliya T, Dardik R, Nisgav Yet al.. Possible involvement of NETosis in inflammatory processes in the eye: evidence from a small cohort of patients. Mol Vis. 2017;23:922–32. [PMC free article] [PubMed] [Google Scholar]
- Belaaouaj A, McCarthy R, Baumann Met al.. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4:615–8. [DOI] [PubMed] [Google Scholar]
- Bernier SP, Ha DG, Khan Wet al.. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res Microbiol. 2011;162:680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, van Gennip M, Jakobsen THet al.. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. Nat Protoc. 2010;5:282–93. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Alhede M, Alhede Met al.. The in vivo biofilm. Trends Microbiol. 2013;21:466–74. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PØ, Fiandaca MJet al.. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–58. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PØ, Burmølle Met al.. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–83. [DOI] [PubMed] [Google Scholar]
- Brinkmann V, Goosmann C, Kuhn LIet al.. Automatic quantification of in vitro NET formation. Front Immunol. 2012;3:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann Cet al.. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–5. [DOI] [PubMed] [Google Scholar]
- Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: For motility and a whole lot more. Semin Cell Dev Biol. 2015;46:91–103. [DOI] [PubMed] [Google Scholar]
- Chiang WC, Nilsson M, Jensen PØet al.. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57:2352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba A, Sugimoto S, Sato Fet al.. A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb Biotechnol. 2015;8:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover MS, Mishra M, Deora R. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS One. 2011;6:e16861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G, Crossley J, Xing Z. Macrophage engulfment of apoptotic neutrophils contributes to the resolution of acute pulmonary inflammation in vivo. Am J Respir Cell Mol Biol. 1995;12:232–7. [DOI] [PubMed] [Google Scholar]
- Das T, Sehar S, Manefield M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep. 2013;5:778–86. [DOI] [PubMed] [Google Scholar]
- Das T, Sehar S, Koop Let al.. Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS One. 2014;9:e91935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JPet al.. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–8. [DOI] [PubMed] [Google Scholar]
- Eggers CT, Murray IA, Delmar VAet al.. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem J. 2004;379:107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach M, Schreiber K, Trunk Ket al.. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol. 2004;186:4596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd M, Winn M, Cullen Cet al.. Swimming motility mediates the formation of neutrophil extracellular traps induced by flagellated pseudomonas aeruginosa. PLoS Pathog. 2016;12:e1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs TA, Abed U, Goosmann Cet al.. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann O, Medina E. The expanding world of extracellular traps: not only neutrophils but much more. Front Immunol. 2012;3:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen NE, Karle H, Valerius NH. Neutrophil kinetics in acute bacterial infection. A clinical study. Acta Med Scand. 1978;204:407–12. [DOI] [PubMed] [Google Scholar]
- Hirsch JG. Bactericidal action of histone. J Exp Med. 1958;108:925–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N. Pseudomonas aeruginosa infection in cystic fibrosis. Relationship between mucoid strains of Pseudomonas aeruginosa and the humoral immune response. Acta Pathol Microbiol Scand [B] Microbiol Immunol. 1974;82:551–8. [PubMed] [Google Scholar]
- Hu Y. Isolation of human and mouse neutrophils ex vivo and in vitro. Methods Mol Biol. 2012;844:101–13. [DOI] [PubMed] [Google Scholar]
- Hunt BE, Weber A, Berger Aet al.. Macromolecular mechanisms of sputum inhibition of tobramycin activity. Antimicrob Agents Chemother. 1995;39:34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WBet al.. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoff A, White R, Carp Het al.. Lung injury induced by leukocytic proteases. Am J Pathol. 1979;97:111–36. [PMC free article] [PubMed] [Google Scholar]
- Jensen PØ, Bjarnsholt T, Phipps Ret al.. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153:1329–38. [DOI] [PubMed] [Google Scholar]
- Jung CJ, Hsu RB, Shun CTet al.. AtlA mediates extracellular DNA release, which contributes to streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun. 2017;85:e00252–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bookwalter JE, Baker BDet al.. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–99. [DOI] [PubMed] [Google Scholar]
- Kenny EF, Herzig A, Kruger Ret al.. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. 2017;6:e24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirketerp-Møller K, Jensen PØ, Fazli Met al.. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46:2717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, van Delden C, Curty LKet al.. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E, Jenne CN, Surewaard BGet al.. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat Commun. 2015;6:6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpen M, Kuhl M, Bjarnsholt Tet al.. Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. PLoS One. 2014;9:e84353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpen M, Hansen CR, Bjarnsholt Tet al.. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax. 2010;65:57–62. [DOI] [PubMed] [Google Scholar]
- Konig MF, Andrade F. A critical reappraisal of neutrophil extracellular traps and NETosis mimics based on differential requirements for protein citrullination. Front Immunol. 2016;7:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lethem MI, James SL, Marriott Cet al.. The origin of DNA associated with mucus glycoproteins in cystic fibrosis sputum. Eur Respir J. 1990;3:19–23. [PubMed] [Google Scholar]
- Lewis HD, Liddle J, Coote JEet al.. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol. 2015;11:189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JP, Stylianou M, Backman Eet al.. Evasion of immune surveillance in low oxygen environments enhances candida albicans virulence. MBio. 2018;9:e02120–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Boado YS, Espinola M, Bahr Set al.. Neutrophil serine proteinases cleave bacterial flagellin, abrogating its host response-inducing activity. J Immunol. 2004;172:509–15. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169:375–82. [DOI] [PubMed] [Google Scholar]
- Manzenreiter R, Kienberger F, Marcos Vet al.. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros. 2012;11:84–92. [DOI] [PubMed] [Google Scholar]
- McCarty SM, Cochrane CA, Clegg PDet al.. The role of endogenous and exogenous enzymes in chronic wounds: a focus on the implications of aberrant levels of both host and bacterial proteases in wound healing. Wound Repair Regen. 2012;20:125–36. [DOI] [PubMed] [Google Scholar]
- Moser C, Pedersen HT, Lerche CJet al.. Biofilms and host response - helpful or harmful. APMIS. 2017;125:320–38. [DOI] [PubMed] [Google Scholar]
- Neeli I, Radic M. Opposition between PKC isoforms regulates histone deimination and neutrophil extracellular chromatin release. Front Immunol. 2013;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeli I, Radic M. Current challenges and limitations in antibody-based detection of citrullinated histones. Front Immunol. 2016;7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol. 2008;180:1895–902. [DOI] [PubMed] [Google Scholar]
- Ohlsson K, Olsson I. The extracellular release of granulocyte collagenase and elastase during phagocytosis and inflammatory processes. Scand J Haematol. 1977;19:145–52. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Metzler KD, Hakkim Aet al.. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter JL, Matthews LW, Spector Set al.. Complex formation between basic antibiotics and deoxyribonucleic acid in human pulmonary secretions. Pediatrics. 1965;36:714–20. [PubMed] [Google Scholar]
- Ramphal R, Lhermitte M, Filliat Met al.. The binding of anti-pseudomonal antibiotics to macromolecules from cystic fibrosis sputum. J Antimicrob Chemother. 1988;22:483–90. [DOI] [PubMed] [Google Scholar]
- Rose-Martel M, Hincke MT. Antimicrobial histones from chicken erythrocytes bind bacterial cell wall lipopolysaccharides and lipoteichoic acids. Int J Antimicrob Agents. 2014;44:470–2. [DOI] [PubMed] [Google Scholar]
- Rose-Martel M, Kulshreshtha G, Ahferom Berhane Net al.. Histones from Avian Erythrocytes Exhibit Antibiofilm activity against methicillin-sensitive and methicillin-resistant Staphylococcus aureus. Sci Rep. 2017;7:45980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shak S, Capon DJ, Hellmiss Ret al.. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci USA. 1990;87:9188–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soavelomandroso AP, Gaudin F, Hoys Set al.. Biofilm structures in a mono-associated mouse model of clostridium difficile infection. Front Microbiol. 2017;8:2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen OE, Borregaard N. Neutrophil extracellular traps - the dark side of neutrophils. J Clin Invest. 2016;126:1612–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalon V, Vander Wauven C, Momin Pet al.. Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J Gen Microbiol. 1987;133:2487–95. [DOI] [PubMed] [Google Scholar]
- Steinberg BE, Grinstein S. Unconventional roles of the NADPH oxidase: signaling, ion homeostasis, and cell death. Science's STKE: Signal Transduction Knowledge Environment. 2007;2007:pe11. [DOI] [PubMed] [Google Scholar]
- Stender H, Fiandaca M, Hyldig-Nielsen JJet al.. PNA for rapid microbiology. J Microbiol Methods. 2002;48:1–17. [DOI] [PubMed] [Google Scholar]
- Suleman L. Extracellular bacterial proteases in chronic wounds: a potential therapeutic target? Adv Wound Care (New Rochelle). 2016;5:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanabalasuriar A, Scott BNV, Peiseler Met al.. Neutrophil extracellular traps confine Pseudomonas aeruginosa ocular biofilms and restrict brain invasion. Cell Host & Microbe. 2019;25:526–36. e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gennip M, Christensen LD, Alhede Met al.. Interactions between Polymorphonuclear Leukocytes and Pseudomonas aeruginosa Biofilms on Silicone Implants In Vivo. Infect Immun. 2012;80:2601–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gennip M, Christensen LD, Alhede Met al.. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117:537–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C, Pierard A, Kley-Raymann Met al.. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984;160:928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong L, Sherman PM, Glogauer M. Quantification and visualization of neutrophil extracellular traps (NETs) from murine bone marrow-derived neutrophils. Methods Mol Biol. 2013;1031:41–50. [DOI] [PubMed] [Google Scholar]
- Voynow JA, Fischer BM, Zheng S. Proteases and cystic fibrosis. Int J Biochem Cell Biol. 2008;40:1238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TS, Tomlin KL, Worthen GSet al.. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun. 2005;73:3693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li M, Stadler Set al.. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y, Drujan D, Shapiro SDet al.. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. [DOI] [PubMed] [Google Scholar]
- Westman J, Papareddy P, Dahlgren MWet al.. Extracellular histones induce chemokine production in whole blood ex vivo and leukocyte recruitment in vivo. PLoS Pathog. 2015;11:e1005319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PCet al.. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. [DOI] [PubMed] [Google Scholar]
- Wong SL, Demers M, Martinod Ket al.. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med. 2015;21:815–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlitzsch D, Tarran R, Ulrich Met al.. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager DR, Nwomeh BC. The proteolytic environment of chronic wounds. Wound Repair Regen. 1999;7:433–41. [DOI] [PubMed] [Google Scholar]
- Yipp BG, Petri B, Salina Det al.. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med. 2012;18:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Pan F, Jones LAet al.. Evaluation of 5-ethynyl-2'-deoxyuridine staining as a sensitive and reliable method for studying cell proliferation in the adult nervous system. Brain Res. 2010;1319:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.