Abstract
The liver contains heterogeneous cell types that exhibit distinct spatial, molecular, and functional properties. The cells in the liver communicate with each other via a network of ligands and receptors and undergo cell type-specific transcriptomic reprogramming in disease. The advance of single-cell genomics has provided a powerful tool for unraveling the complexity of liver cells in health and disease with unprecedented resolution. In this review, we discuss insights gained from the recent single-cell RNA sequencing studies that shed new light on the molecular nature of liver cell heterogeneity, intercellular crosstalk, and disease-associated reprogramming.
Keywords: liver, single-cell, scRNA-seq, NASH, HCC, zonation, immune
The liver is the largest internal organ that comprises hepatocytes and non-parenchymal cells (NPCs), including liver sinusoidal endothelial cells (LSECs), the resident macrophage Kupffer cells (KCs), hepatic stellate cells (HSCs), biliary epithelial cells (BECs, or cholangiocytes), and diverse immune cell types. A unique anatomical feature of the liver is its dual blood supply from the portal vein and hepatic artery (Figure 1A). Blood flow from portal triad to the central vein creates gradients of nutrients, oxygen, hormones, and gut-derived endotoxins that collectively shape the molecular and functional heterogeneity of hepatocytes. While the concept of liver zonation was supported by early work using histological approaches, unraveling the molecular nature of liver cell heterogeneity remains a challenging problem. The liver plays a central role in glucose (gluconeogenesis and glycogen storage) and lipid metabolism (lipogenesis, fatty acid β-oxidation, and lipoprotein metabolism). Elevated hepatic glucose production and excess fat accumulation are hallmarks of diabetes and non-alcoholic fatty liver disease (NAFLD), respectively. The latter may progress to non-alcoholic steatohepatitis (NASH), a more severe form of metabolic liver disease characterized by persistent liver injury, inflammation, and liver fibrosis. In addition to hepatocytes, NPCs are known to regulate diverse aspects of liver disease pathogenesis. However, their heterogeneity and dynamic regulation in disease have not been fully elucidated.
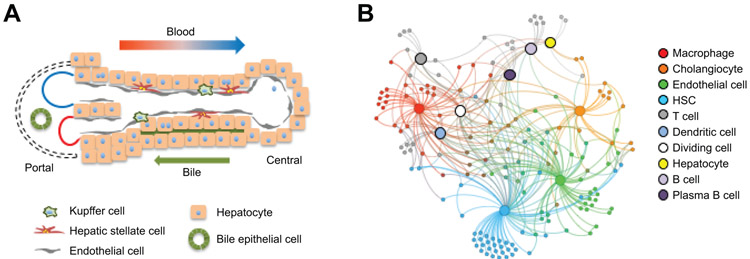
Figure 1. Liver cell heterogeneity and crosstalk.
(A) A diagram illustrating liver cell heterogeneity and zonation. Portal vein and hepatic artery are indicated in blue and red, respectively. (B) A ligand-receptor signaling network among different cell types revealed by single-cell secretome gene analysis.
The emerging single-cell genomics technologies have transformed research on complex organs and tissues (1). In particular, single-cell RNA sequencing (scRNA-seq) provides a powerful tool for mapping the transcriptomic landscape of the cells with unprecedented resolution and delineating the dynamic nature of cellular events in development, homeostasis, and disease. This review covers recent single-cell studies on mouse and human liver, and provides perspectives on the new insights into the nature of liver cell heterogeneity, intrahepatic ligand-receptor crosstalk, and disease-associated cellular reprogramming.
Liver cell heterogeneity and zonation at single-cell resolution
Liver zonation is a prominent aspect of liver cell heterogeneity (2). Using scRNA-seq, Halpern et al. obtained transcriptomic data for over 1,500 FACS-sorted liver cells (3). To reconstruct their spatial heterogeneity, the authors developed a probability-based strategy to assign hepatocytes along the portal-central lobule axis based on mRNA expression of a panel of zone-specific liver genes. Analysis of the single-cell dataset revealed pervasive and distinct patterns of hepatocyte gene expression that can be mapped to different virtual lobule layers. A technical limitation of scRNA-seq is that it is best suited for quantitative analysis of highly expressed genes. To address this challenge and more comprehensively characterize the molecular nature of liver zonation, Ben-Moshe et al. combined cell sorting using zone-specific cell surface markers (CD73 and E-cadherin) and gating strategies to obtain hepatocytes corresponding to different zones (4). RNAseq and proteomic analyses of these pooled hepatocytes revealed greater details of spatial gene expression in the liver at mRNA, miRNA and protein levels. In a separate study, scRNA-seq of human liver cells revealed remarkably conserved features of liver zonation between mouse and human (5).
Several principles of metabolic and signaling zonation have emerged from these studies. For example, periportal hepatocytes assume several energetically demanding tasks, such as secretion of plasma proteins, gluconeogenesis, and urea cycle, that are coupled to their access to oxygen-rich blood supply and high capacity for oxidative metabolism (4, 6). As such, periportal hepatocytes exhibit abundant expression of Albumin and key enzymes including PCK1 (gluconeogenesis), ARG1 (urea cycle), and SDHD (mitochondrial oxidation) to carry out these metabolic functions. Pericentral hepatocytes are specialized in glycolytic metabolism, de novo lipogenesis, triglyceride synthesis, and detoxification. This restriction of hepatocyte metabolism to specific zones has important disease implications. As such, hepatic steatosis is most conspicuously observed in pericentral hepatocytes in NAFLD patients (7), likely reflecting their role in lipogenesis and triglyceride synthesis. Hepatocytes are known to harbor polyploid genomes contained within one or two nuclei in the cell (8). This heterogeneity of hepatocyte polyploidization and its associated transcriptomic signature represent yet another dimension of liver cell heterogeneity that is primed for exploration using single-cell technologies.
Endothelial cells lining arterial, venous and lymphatic vessels are known to exhibit distinct molecular and functional properties (9). Similar to hepatocytes, different endothelial compartments in the liver are exposed to varying concentrations of nutrients, oxygen and hormones and exhibit unique patterns of gene expression. By taking advantage of the knowledge on zonated hepatocyte transcriptomes, Halpern et al. developed a paired-cell sequencing approach to probe heterogeneity of liver endothelial cells and bioinformatically inferred their spatial information (10). Similar molecular signatures of LSEC zonation were observed in independent scRNA-seq studies on NPCs isolated from healthy and NASH mouse liver (11) and in human liver (5). Comparative analysis of LSEC transcriptomes obtained from healthy and NASH livers revealed profound defects in endothelial gene expression and vascular signaling that may contribute to disease pathogenesis (11). These studies provided an important first step in unraveling the molecular and functional heterogeneity of liver endothelial cells. Interestingly, liver macrophages also exhibited features of zonation, as illustrated by gradient expression of the scavenger receptor MARCO (12). Future work is needed to dissect the signaling mechanisms underlying liver cell zonation and its role in liver physiology and disease.
Landscape of intercellular crosstalk among liver cell types
Intercellular crosstalk via ligands and receptors is a fundamental feature of organ biology. Hepatocytes and NPCs release an array of secreted factors that exert diverse biological effects in the liver and beyond. As such, several hepatokines have emerged as important regulators of nutrient metabolism and energy balance by acting on the central nervous system and other peripheral tissues (13, 14). Despite the expanding role of intercellular signaling in metabolic physiology and tissue homeostasis, a comprehensive survey of ligand and receptor gene expression and the landscape of intercellular crosstalk in the liver have not been fully elucidated. Xiong et al. performed single-cell secretome gene analysis on NPCs and hepatocytes isolated from mouse liver (11). The secretome gene set includes those predicted to encode secreted factors (1,272) and membrane receptors (755) in the mouse genome (15). A notable feature of secretome gene expression is that many ligands and receptors exhibit cell type-restricted patterns of expression that are highly conserved, as revealed by comparative analysis of the mouse and human scRNA-seq datasets. By integrating single-cell expression data with a ligand-receptor interaction database, a highly connected signaling network among ten major liver cell types was generated (Figure 1B). This work provides a roadmap for dissecting the complexity and biological role of intercellular crosstalk within the mammalian liver.
HSCs express numerous genes encoding the structural proteins of extracellular matrix (ECM) and those contributing to ECM modeling (16). Analysis of the HSC secretome genes revealed two unexpected features (11). First, HSCs appear to function as a signaling hub by secreting growth factors, cytokines, and chemokines, which were collectively termed “stellakines”. Based on ligand-receptor pairing analysis, many of these stellakines were predicted to act on endothelial cells and different immune cell types. The expression of several stellakines was elevated following diet-induced NASH, suggesting that paracrine signaling mediated by HSC-derived secreted factors may contribute to disease progression. In parallel, HSCs appeared to express a large number of membrane receptors including regulators of HSC proliferation, activation, and apoptosis. Unexpectedly, several G-protein coupled receptors responsive to vasoactive hormones exhibited abundant mRNA expression in HSCs, underscoring a potentially important role of HSC contractile regulation in liver biology. Compared to bulk RNA sequencing, single-cell transcriptomic analysis provides a sensitive method for delineating the precise cellular sources of ligands and receptors and revealing the landscape of intercellular crosstalk in the liver. Future challenges include functional assessments of the predicted signaling modalities and their biological functions in health and disease.
A single-cell perspective of liver development and regeneration
Development of the mammalian liver entails sequential specification of the hepatocyte lineage and the formation three-dimensional (3D) tissue architecture involving multiple cell types. Using scRNA-seq, Camp et al. reconstructed hepatocyte-like lineage progression from induced pluripotent stem cells in two-dimensional (2D) monoculture and 3D multilineage organoid systems (17). The latter is uniquely suited for deconstructing liver cell heterogeneity during development and complex intercellular interactions between the hepatic, stromal, and endothelial lineages. Compared to 2D culture, the liver bud hepatoblasts in organoids acquired an epithelial migration gene signature characteristic of organ budding. Further, the authors observed striking concordance of gene signatures between the 3D liver bud and fetal liver cells. Analysis of the single-cell transcriptomic data from the organoid cultures revealed a network of potential ligand-receptor crosstalk among different lineages. Vascular endothelial growth factor (VEGF) was identified as a key molecular signal that potentiates endothelial network formation and hepatoblast differentiation (17).
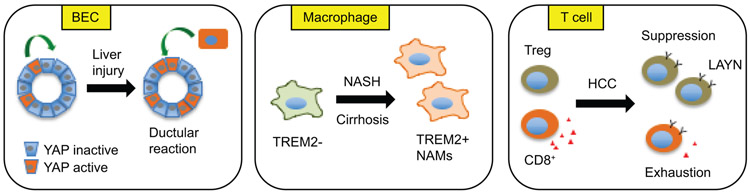
The adult liver retains robust regenerative capacity that supports organ homeostasis and normal liver functions. In response to injury, hepatocytes can re-enter the cell cycle and undergo proliferation to replace dead hepatocytes (18). In addition, biliary epithelial cells (BECs) also provide a source of progenitor cells that can differentiate into hepatocytes and repopulate the liver parenchyma following injury. While BECs have been established as facultative liver stem cells that exhibit a high degree of plasticity (18), the intrinsic heterogeneity of BECs and the mechanisms that govern their progenitor function have not been fully elucidated. Recent work using single-cell transcriptomics from several groups shed light on cellular heterogeneity of BECs in healthy liver and following liver injury (Figure 2). Pepe-Mooney et al. performed scRNA-seq on Epithelial Cell Adhesion Molecule positive (EpCAM+) BECs and identified two subtypes that were distinguished by their differential expression of the Hippo/YAP gene signature (19). The YAP-high cluster exhibited correlated expression of YAP target genes and was enriched for mechanical and adhesion signaling. This heterogeneity of BECs was confirmed using single-molecule RNA FISH and in a YAP reporter mouse strain. The YAP transcriptional program appeared to be highly responsive to DDC-induced injury and is required for BEC homeostasis in adult liver. Interestingly, hepatocyte-specific YAP inactivation revealed a crucial role for this signaling pathway in injury-induced reprogramming of hepatocytes toward a progenitor, biliary-like cell fate. In a separate CRISPR-based screen for regulators of ductular reaction using BEC-like organoids, Planas-Paz et al. also uncovered YAP as a factor that serves a critical role in triggering ductular reaction following DDC-induced liver injury (20). In contrast, Wnt/β catenin signaling appeared dispensable for injury-induced BEC expansion and ductular reaction. Single-cell transcriptomic analysis revealed a molecular signature consistent with robust YAP signaling in EpCAM+ BECs.
Figure 2. Heterogeneity and reprogramming of liver cells in disease.
BECs are heterogeneous and serve as a progenitor pool during liver injury (left). NASH-associated macrophages (NAMs) are marked by TREM2 expression and emerge during diet-induced NASH in mice and in human NASH and liver cirrhosis (middle). Accumulation of LAYN-positive Tregs and exhausted CD8+ T cells are a hallmark of the HCC tumor microenvironment (right).
The profound heterogeneity of BECs was also observed in two single-cell studies on human fetal and adult liver cells. Segal et al. identified a hepatobiliary hybrid progenitor (HHyP) cell population in human fetal liver that exhibited a molecular profile distinct from fetal hepatocytes, mature hepatocytes and mature BECs (21). These cells were localized to the ductal plate of fetal liver and expressed both hepatocyte and BEC markers, a transcriptional signature characteristic of bipotent mouse progenitors. In contrast, adult BECs in human liver were marked by expression of TROP2, Similar to mouse BECs, EpCAM+ cells isolated from fetal and adult human livers also contained heterogeneous cell populations. Single-cell analysis of EpCAM+ human BECs revealed the existence of three transcriptionally heterogeneous populations that correspond to hepatocytes, bipotent progenitor cells, and cholangiocytes with low-to-high gradient expression of TROP2 (5). The putative progenitor cells with intermediate levels of TROP2 expression exhibited the highest organoid forming capacity in culture. These observations further support the notion that a subset of bile epithelial cells serves as a source for liver progenitors.
The immune landscape of NASH, liver cirrhosis, and liver cancer
During NASH pathogenesis, persistent liver injury triggers the recruitment and reprogramming of various immune cells (22). Recent single-cell analysis of sorted myeloid cells (23) and total NPCs (11) isolated from healthy and diet-induced NASH mouse liver revealed new insights into the dynamic nature of liver macrophages. Diet-induced NASH was associated with a pronounced expansion of both liver resident KCs and monocyte-derived macrophages (MDM). In Xiong et al. study (11), three subpopulations of macrophages were uncovered that correspond to MDM and KC expressing low or high levels of Trem2, a putative scavenger receptor implicated in Alzheimer’s Disease. Because Trem2+ macrophages were largely absent in healthy liver and strongly induced during NASH, this population of macrophages was named NASH-associated macrophages (NAM) (Figure 2). NAM exhibited a transcriptional signature enriched for genes involved in lysosomal degradation, phagocytosis and antigen presentation, suggesting that they may play a role in the clearance of dead hepatocytes. Ramachandran et al. generated single-cell atlas of human liver NPCs using different pools of sorted cells from healthy and cirrhotic livers (24). Interestingly, TREM2-positive scar-associated macrophages (SAMacs) were also found to be present in the fibrotic niche of human liver cirrhosis and exhibit a pro-fibrogenic phenotype. Krenkel et al. also observed macrophage heterogeneity in NASH liver (11); however, whether any of the three clusters identified corresponds to NAM was not analyzed. It is important to note that liver macrophages did not appear to subdivide into the canonical M1 and M2 populations in these studies, illustrating the necessity for reassessing the nature of molecular and functional polarization of macrophages in liver disease.
Altered immune landscape is a feature of tumor microenvironment associated with liver cancer (25). Several immune cell types including regulatory T cells (Tregs), exhausted CD8+ effector T cells, tumor associated macrophages, and myeloid-derived suppressor cells have been shown to accumulate in the hepatocellular carcinoma (HCC) microenvironment (26). To better understand the mechanisms and regulatory pathways leading to T cell exhaustion and accumulation of Tregs in liver cancer, Zheng et al. performed scRNA-seq on sorted T cells from peripheral blood, normal liver tissue adjacent to tumor, and tumor tissues from six HCC patients (27). Comparative analysis of single-cell transcriptomic data revealed gene signatures unique for tumor-associated Tregs and exhausted CD8+ T cells (Figure 2). In addition, effector CD8+ T cells were less abundant and displayed more pronounced exhaustion phenotype and clonal expansion in the tumor environment. Layilin (LAYN) was identified as a gene that exhibited highly specific expression in tumor-derived Tregs and exhausted CD8+ T cells. Functional analysis revealed an inhibitory role of LAYN in CD8+ T cell function.
More recently, Zhang et al. used a combination of full-length and 3’ scRNA-seq technologies to generate a high-resolution map of the immune landscape of HCC (28). By surveying multiple immune-relevant sites (tumor, adjacent liver, hepatic lymph node, blood, and ascites), the authors identified LAMP3+ dendritic cells as a key cell type that originated from tumor, migrated to hepatic lymph nodes, and shaped lymphocyte function via ligand-receptor signaling. These studies provided unprecedented insights into cellular heterogeneity, molecular nature, and functional implications of the immune landscape associated with human HCC. Additional single-cell studies that focus on stromal cells are needed to provide a more comprehensive picture of the HCC microenvironment and its role in tumorigenesis and therapeutic response.
Future perspectives
Single-cell genomics offers powerful tools for unraveling cell heterogeneity in complex organs, the nature of intercellular crosstalk, and disease-associated cellular reprogramming with unprecedented resolution. New technologies are being developed to further expand single-cell analysis to include spatial information of cells in their native tissue context , such as sequential fluorescence in situ hybridization (seqFISH) and spatially barcoded transcriptomic analysis (29, 30). In addition, multimodal and integrated methods are available for high-resolution mapping of chromatin state (DNA methylation, chromatin accessibility, histone modifications), genome sequence, and cell surface protein expression in combination with single-cell transcriptomic analysis, as discussed in a recent review (31). These emerging tools are expected to reveal multi-dimensional details of the liver microenvironment in physiological and disease states. The construction of a comprehensive cell atlas for healthy and disease tissues will open up heretofore inaccessible avenues for biological exploration, disease modeling, and therapeutic discovery.
Acknowledgements
This work was supported by NIH (DK102456, DK112800 and DK118731 to J.L.). H.K. was supported by an NRSA fellowship (FDK117615) and MSTP training grant (T32GM007863).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Birnbaum KD. Power in Numbers: Single-Cell RNA-Seq Strategies to Dissect Complex Tissues. Annu Rev Genet 2018;52:203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Moshe S, Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol 2019;16:395–410. [DOI] [PubMed] [Google Scholar]
- 3.Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017;542:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Moshe S, Shapira Y, Moor AE, Manco R, Veg T, Bahar Halpern K, Itzkovitz S. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab 2019;1:899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, Pessaux P, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jungermann K, Katz N. Functional specialization of different hepatocyte populations. Physiol Rev 1989;69:708–764. [DOI] [PubMed] [Google Scholar]
- 7.Chalasani N, Wilson L, Kleiner DE, Cummings OW, Brunt EM, Unalp A, Network NCR. Relationship of steatosis grade and zonal location to histological features of steatohepatitis in adult patients with non-alcoholic fatty liver disease. J Hepatol 2008;48:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang MJ, Chen F, Lau JTY, Hu YP. Hepatocyte polyploidization and its association with pathophysiological processes. Cell Death Dis 2017;8:e2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augustin HG, Koh GY. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 2017;357. [DOI] [PubMed] [Google Scholar]
- 10.Halpern KB, Shenhav R, Massalha H, Toth B, Egozi A, Massasa EE, Medgalia C, et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol 2018;36:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, et al. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol Cell 2019;75:644–660 e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meex RCR, Watt MJ. Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat Rev Endocrinol 2017;13:509–520. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Sharma VP, Shen H, Xiao Y, Zhu Q, Xiong X, Guo L, et al. The hepatokine Tsukushi gates energy expenditure via brown fat sympathetic innervation. Nature Metabolism 2019;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang GX, Zhao XY, Meng ZX, Kern M, Dietrich A, Chen Z, Cozacov Z, et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med 2014;20:1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 2008;88:125–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017;546:533–538. [DOI] [PubMed] [Google Scholar]
- 18.Shin S, Kaestner KH. The origin, biology, and therapeutic potential of facultative adult hepatic progenitor cells. Curr Top Dev Biol 2014;107:269–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pepe-Mooney BJ, Dill MT, Alemany A, Ordovas-Montanes J, Matsushita Y, Rao A, Sen A, et al. Single-Cell Analysis of the Liver Epithelium Reveals Dynamic Heterogeneity and an Essential Role for YAP in Homeostasis and Regeneration. Cell Stem Cell 2019;25:23–38 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas-Paz L, Sun T, Pikiolek M, Cochran NR, Bergling S, Orsini V, Yang Z, et al. YAP, but Not RSPO-LGR4/5, Signaling in Biliary Epithelial Cells Promotes a Ductular Reaction in Response to Liver Injury. Cell Stem Cell 2019;25:39–53 e10. [DOI] [PubMed] [Google Scholar]
- 21.Segal JM, Kent D, Wesche DJ, Ng SS, Serra M, Oules B, Kar G, et al. Single cell analysis of human foetal liver captures the transcriptional profile of hepatobiliary hybrid progenitors. Nat Commun 2019;10:3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017;17:306–321. [DOI] [PubMed] [Google Scholar]
- 23.Krenkel O, Hundertmark J, Abdallah AT, Kohlhepp M, Puengel T, Roth T, Branco DPP, et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 2019. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cariani E, Missale G. Immune landscape of hepatocellular carcinoma microenvironment: Implications for prognosis and therapeutic applications. Liver Int 2019;39:1608–1621. [DOI] [PubMed] [Google Scholar]
- 26.Wan S, Kuo N, Kryczek I, Zou W, Welling TH. Myeloid cells in hepatocellular carcinoma. Hepatology 2015;62:1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169:1342–1356 e1316. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019;179:829–845 e820. [DOI] [PubMed] [Google Scholar]
- 29.Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 2019;568:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353:78–82. [DOI] [PubMed] [Google Scholar]
- 31.Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet 2019;20:257–272. [DOI] [PubMed] [Google Scholar]