Abstract
Cellular metabolism is central to T cell function and proliferation, with most of the research to date focusing on cancer and autoimmunity. Cellular metabolism is associated with a host of physiological phenomena, from epigenetic changes, to cellular function and fate. For the purpose of this review, we will discuss the metabolism of T cells relating to their differentiation and function. We will cover a variety of metabolic processes, ranging from glycolysis to amino acid metabolism. Understanding how T cell metabolism informs T cell function may be useful to understand alloimmune responses and design novel therapies to improve graft outcome.
Keywords: Immunometabolism, Transplantation, T cell, Tolerance, Rejection
1. Introduction
Human organ availability for transplantation is limited, with too few human donors resulting in a waiting list for potential recipients. As such, it is important to ensure that the organs that are transplanted are accepted for life, to keep patients healthy and from returning to the waiting list. Although allograft survival has improved over the years, the half-life of grafts is still limited, and patients depend on life-long immunosuppressive drugs that increase the risk for infections and cancer and can have graft-damaging side effects. Donor-specific tolerance can be defined as non-responsiveness towards the graft following a brief immunosuppressive regimen that leaves the patient immunocompetent against other pathogens. Although achievable in mice, it remains rare in humans, with many grafts succumbing to long-term failure [1].
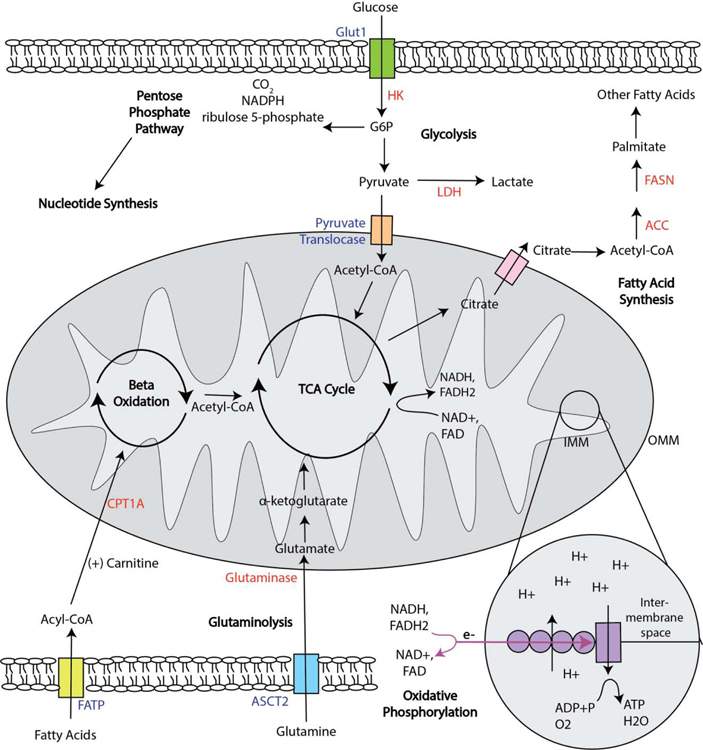
Donor-specific tolerance would require permanent silencing of only the alloreactive T cells, limiting their survival, proliferation, and/or effector functions, while leaving other T cells, including regulatory T cells (Tregs), unaffected. Cellular metabolism is instrumental to T cell survival, function, proliferation, and differentiation [2]. As such, targeting T cell metabolism poses an attractive target for immunotherapies for cancer, autoimmunity, and transplantation. Depending on their cellular needs, T cells will alter their metabolism, either to be catabolic and break down environmental nutrients for energy, or to be anabolic and synthesize necessary components for cellular growth and proliferation. These processes are further described below, and depicted in Figure 1. By taking an in-depth look at T cell metabolism in the context of transplantation, we hope to illustrate how metabolism informs T cell function, and how this knowledge may be harnessed to prevent graft rejection.
Fig. 1.
T Cell Metabolism. Upon uptake, glucose is converted into G6P before further metabolism via glycolysis, the TCA cycle and oxidative phosphorylation, or the pentose phosphate pathway. Metabolism of glutamine is mitochondria-dependent, where glutamine is converted to glutamate, and then the TCA substrate α-ketoglutarate. Fatty acids transported into the cell are converted into acyl-CoA, which combines with carnitine in order to cross the OMM. Once in the mitochondria, acyl-CoA undergoes beta-oxidation, producing acetyl-CoA for the TCA cycle and an acyl-CoA with two fewer carbons for another cycle of beta-oxidation. Importantly, the TCA cycle and pentose phosphate pathway are amphibolic rather than strictly catabolic, and a T cell remains able to undergo both anabolic and catabolic processes at once, such as glycolysis and fatty acid synthesis. A hallmark of both amphibiolic processes is the production of energy rich e- carriers, which are shuttled to the inner mitochondrial membrane and the electron transport chain to undergo oxidative phosphorylation. Transporters are indicated in blue, and enzymes are indicated in red. Abbreviations: acetyl-CoA carboxylase (ACC), alanine-serine-cysteine transporter 2 (ASCT2), fatty acid synthase (FASN), glucose 6 phosphate (G6P), inner mitochondrial membrane (IMM), lactate dehydrogenase (LDH), outer mitochondrial membrane (OMM).
A cell needs energy-rich adenosine triphosphate (ATP) for normal function. ATP can come from a variety of metabolic processes, depending on the nutrient source being catabolized. Glucose is perhaps the most famous example, whereupon being taken up by a cell via the glucose transporter 1 (GLUT1) it is converted into glucose-6-phosphate (G6P) by the rate-limiting enzyme hexokinase [3]. G6P can be serially oxidized into pyruvate, or can enter the pentose phosphate pathway, described below [4]. Additionally, glycolytic metabolites contribute to a variety of cellular processes, such as the biosynthesis of serine from 3-phosphoglycerate, necessary for T cell proliferation [5], or the support of calcium signaling by inhibition of the sarco/ER Ca2+-ATPase, thereby supporting signaling of the nuclear factor of activated T cells (NFAT) [6].
Pyruvate is a substrate for both aerobic glycolysis and the tricarboxylic acid (TCA) cycle. In aerobic glycolysis, pyruvate is kept in the cytoplasm and converted into lactate by the enzyme lactate dehydgrogenase A (LDHA). This differs from oxidative phosphorylation, in which pyruvate is transported into the mitochondria and converted to acetyl coenzyme A (CoA) to join the TCA cycle, an amphibolic pathway that produces energy rich electron (e-) carriers, nicotinamide adenine dinucleotide (NAD) and flavin adenine dinucleotide (FAD), for the electron transport chain (ETC) [7]. Importantly, the TCA cycle is not specific to glucose metabolism, rather, other nutrient sources including fatty acids and amino acids can also generate TCA substrates [2].
Once generated, the e- carriers transport the energy-rich electrons to the ETC, located at the inner mitochondrial membrane, where the energy from the movement of electrons across the ETC is used to generate a hydrogen gradient in the mitochondrial inter-membrane space [8]. Chemiosmosis of hydrogen through the ATP synthase protein, complex V of the ETC, generates the energy needed to synthesize ATP from adenosine diphosphate (ADP) and a free phosphate, consuming oxygen and producing H2O in the process [8]. This entire sequence is referred to as oxidative phosphorylation.
Glycolysis generates a net yield of 2 ATP for every glucose molecule consumed, whereas oxidative phosphorylation generates a net yield of 36 ATP molecules per glucose molecule [3]. Unsurprisingly, at steady state with sufficient oxygen availability, oxidative phosphorylation is favorable for energy production [3]. However, glycolysis generates key biosynthetic intermediates that may be used later for anabolic processes such as cell growth and proliferation, such that there are biological reasons where glycolysis would be preferential, even in aerobic conditions [7].
Similar to glucose, fatty acid β-oxidation (FAO) also involves TCA cycle oxidation. Fatty acids are first brought into the cell via fatty acid transport proteins (FATPs), which can also act as acyl-CoA synthases, thereby creating a concentration deficit and promoting fatty acid transport into the cell [9]. The acyl-CoA is then converted into acylcarnitine by carnitine palmitoyltransferase I (CPT1A) in order to cross the mitochondrial membrane, before being converted back to acyl-CoA [9]. Once across the membrane, the acyl-CoA undergoes 4-step β-oxidation, the products of which are an acyl-CoA shortened by two carbons to undergo another cycle of β-oxidation, an acetyl-CoA to go to the TCA cycle, and two electron carriers to go to the ETC [9]. In this way, FAO is able to generate a lot of energy and is important for cellular homeostasis.
Amino acid metabolism is specific for each individual amino acid; however, generalizations may still be made [10]. The key amino acids in these processes are arginine, cysteine, glutamine, leucine, proline, and tryptophan [10]. Activated T cells rely on glutamine metabolism, which starts with glutamine hydrolysis via glutaminase, producing glutamate [11]. Some of this glutamate undergoes further catabolism to generate the TCA cycle substrate α-ketoglutarate, whereas the rest is used for protein synthesis, the synthesis of glutathione for reactive oxygen species control, or enhanced cysteine uptake [11],[12]. Importantly, α-ketoglutarate can travel to the nucleus as a substrate for demethylases, important for epigenetic changes [13].
Importantly, not all metabolic processes are catabolic; rather, a proliferating cell is highly dependent on synthesis processes as well as nutrients. These anabolic processes include fatty acid synthesis and the pentose phosphate pathway (PPP).
Fatty acid synthesis is important for the generation of cell membranes during proliferation [14]. The main substrate for this process is cytoplasmic acetyl-CoA, which is carboxylated into malonyl-CoA. Seven malonyl-CoA molecules with one acetyl-CoA are combined by fatty acid synthase (FASN) to form palmitate, which can then be elongated and desaturated to form a variety of new fatty acids [14].
The pentose phosphate pathway (PPP) is a cytosolic process involved in nucleotide biosynthesis, amino acid biosynthesis, the production of reducing agents and the prevention of oxidative stress [4]. The oxidative components of the PPP involve the catabolism of glucose derived G6P into carbon dioxide, ribulose 5-phosphate and the electron carrier nicotinamide adenine dinucleotide phosphate (NADPH) [4]. Ribulose 5-phosphate is also a component of the bidirectional, non-oxidative PPP pathway, which forms ribose-5-phosphate, a precursor for nucleotide biosynthesis. This is achieved through the metabolism of glycolytic intermediates fructose 6-phosphate and glyceraldehyde 3-phosphate, as well as sedoheptulose sugar metabolism for amino acid generation [4]. In this way, the pentose phosphate pathway has both anabolic and catabolic components: breaking down metabolites in order to form new ones.
2. T cell differentiation
2.1. Changes in T cell metabolism with activation
Throughout the course of its life, a T cell undergoes drastic metabolic remodeling, necessary for its functional activity. A naïve T cell primarily relies on oxidative phosphorylation for respiration [16]. S1P from endothelial cells, in addition to acting as a lymph node egress signal, stimulates migratory naïve T cells to maintain their mitochondrial ATP production, which is their main source of energy and necessary for their migration [17]. Following TCR-peptide MHC (pMHC) interaction and costimulation, a T cell begins to require increased ATP production and biosynthetic intermediates for productive proliferation and effector function [16]. Activated T cells alter transcription of a variety of metabolic genes, the most notable of which is increased Myc, required for T cell growth and proliferation [18]. The activated T cell increases aerobic glycolysis, necessary for effector cytokine production, such as IFNγ [19], [20]. This was first characterized by Otto Warburg, and is known as the Warburg effect [16], [21], [22]. Increased expression of LDHA facilitates this switch to aerobic glycolysis, promoting the production of acetyl-CoA necessary for histone acetylation and IFNγ production [20]. Aerobic glycolysis is thus associated with effector T cell cytokine production.
Activated T cells also increase their mitochondrial metabolism following a CD28-dependent increase in glutamine uptake [23]. This proves necessary for glutamine oxidation, supporting proliferation and the production of metabolic intermediates that act as building blocks for biosynthesis [24], [25]. Serine is also critical for optimal proliferation by supplying glycine and single carbons to be used in purine nucleotide biosynthesis [5]. Upregulation of additional nutrient transporters including Glut1 (glucose) and Slc7a5 (amino acids) also help support effector function and proliferation [26], [27]. Importantly, the metabolic changes of a T cell are reflective of peptide binding affinity, where peptide-MHC complexes that bind more strongly to the TCR stimulate enhanced glycolysis when compared to weakly binding peptides [28]. As T cell affinity matures over the course of an immune response, with preferential expansion of high affinity clones, it is important to consider the role T cell metabolism may play in the survival of clones of different affinities [29]. Of relevance to transplantation, we have recently shown that acute rejection but not transplantation tolerance is associated with preferential expansion of clones with high avidity for peptide/MHC [30].
Mitochondria are essential for T cell effector responses, and T cells rapidly increase their mitochondrial mass and mitochondrial biogenesis upon stimulation [31], [32]. During chemotaxis, mitochondrial redistribution, fission, and local ATP production are necessary for migration [33], [34]. While mitochondria-derived reactive oxygen species can be detrimental to a resting cell, they are transiently necessary following TCR stimulation for the induction of various activation-associated transcription factors including NF-κB and NFAT [24], [35], [36]. Mitochondrial metabolism also plays a role in both T cell proliferation and effector functions: the malate-aspartate shuttle, complex I of the ETC, is necessary for the aspartate required for nucleotide synthesis and proliferation, whereas succinate dehydrogenase, complex II, suppresses proliferation but is necessary for T cell function [37]. ETC complexes III and IV also contribute to T cell proliferation [32]. In this way, various components of the electron transport chain work independently toward achieving effector T cell function and proliferation, demonstrating that while aerobic glycolysis has always been appreciated as necessary for effector function, mitochondrial metabolism is equally important during an effector response.
Costimulatory and coinhibitory molecules can also have a functional impact on T cell metabolism. For example, CD28 is necessary for increase in glutamine uptake following T cell activation, allowing it to prepare itself for effector function [23]. In contrast, ligation of the coinhibitory receptor programmed cell death protein 1 (PD1) on activated T cells results in decreased glycolysis and amino acid metabolism, instead increasing FAO via greater CPT1A expression [38]. In this way, PD1 prevents the hallmarks of effector metabolism, thereby regulating the effector response. These findings are also important for transplantation, as T cells chronically stimulated in the presence of a persisting graft display high expression of PD1, suggesting that inhibition of glycolysis could prove beneficial for graft outcome [39], [40].
Following an effector response, the T cell population contracts, and T cells that do not die remodel their metabolism in order to form memory. The differences between the metabolism of effector T and memory T cells are extensive. Effector T cells promote an anabolic state, whereas memory T cells are catabolic. As such, effector T cells have low levels of FAO, presumably in order to save citrate for lipid membrane formation, whereas memory cells rely on FAO for differentiation [41]. FAO is supported by AMP-activated protein kinase (AMPK), which is also important for T effector suppression of the mammalian target of rapamycin complex I (mTORC1) under periods of glucose starvation [42]. Retroviral expression of the rate-limiting enzyme of FAO, CPT1A, enhances memory cell survival [43]. Furthermore, the mitochondria of an effector and memory T cell are distinct. Effector mitochondria are punctate and have reduced ETC efficiency, which promotes glycolysis [44]. Mitochondria in memory T cells are larger, with increased overall mass, fusing and forming networks with altered cristae morphology in order to support oxidative phosphorylation [44],[45]. Improved oxidative phosphorylation correlates with the increased spare respiratory capacity, allowing memory T cells to better produce the ATP necessary for their prolonged survival, as well as priming the memory T cells for a strong secondary response.
Upon restimulation, secondary T effectors have greater cytokine and other effector responses compared with primary effectors [46]. Memory T cells remodel their metabolism again, switching back to the anabolic profile of an effector T cell, in order to support effector functions necessary for an immune response. For example, memory T cells are able to promptly switch to glycolysis, necessary for IFNγ production, in a CD28-, protein kinase B (Akt)-, and mTORC2-dependent manner [47]. In the secondary effector cell, this switch is supported through linkage sites between the mitochondria and the endoplasmic reticulum, which facilitates hexokinase I (HK-I) binding the mitochondrial voltage-dependent anion channel (VDAC) and the import of necessary metabolites for respiration [48]. It is also thought that the increased mitochondrial mass found in memory T cells promotes a secondary effector response not only through mitochondrial respiration, but also through mitochondrial associations with HK-I, which promotes glucose accumulation in the cell as glucose-6-phosphate, promoting glycolysis [46]. In this way, memory T cells prepare themselves for a secondary response, linking cellular morphology, metabolism and memory, and underscoring how T cell function is connected to metabolism.
2.2. T effector subsets
Although CD4+ and CD8+ T cells share many hallmarks of T cell activation, they display divergent metabolic phenotypes. While both subsets increase glycolysis and the glucose transporter Glut1 expression upon activation, CD8+ T cells are less glycolytic than CD4+ T cells, due to decreased relative expression of glycolytic enzymes, including hexokinase II (HK-II), whose expression is generally confined to insulin sensitive tissues, as opposed to HK-I, which is ubiquitously expressed [28], [49]. Furthermore, in spite of CD4+ T cells having increased mitochondrial mass compared to CD8+ T cells, CD8+ T cells have an increased reliance on oxidative phosphorylation for cytokine production [28]. The differences in T cell metabolism demonstrated by these two subsets speak to their functional differences. For transplantation, it might be important to understand the metabolic differences between these populations, and other effector subsets, in order to understand and prevent graft rejection.
Glucose has a large impact on T effector cells. Upon stimulation, Th1, Th2, and Th17 cells all increase their Glut1 expression [26], [50]. While Glut1 is not necessary for the survival of a naïve T cell, upon stimulation, Glut1-deficient effector T cells show reduced viability with an inability to grow or proliferate [26], demonstrating the reliance of these cells on glucose utilization, presumably for aerobic glycolysis. This is further supported by evidence from mitochondrial transcription factor A (Tfam)-deficient mice, which show impaired FAO, impaired mitochondrial respiration, increased aerobic glycolysis, and an enhanced inflammatory response following loss of mitochondrial function [31]. While T cells show plasticity in their ability to metabolize the nutrients that surround them, it is clear that glucose is preferred, as this is the key substrate for glycolysis, and thus for effector cytokine production, as described previously.
Metabolism regulates cell fate, and while T cell effector subtypes share many metabolic similarities, they have a few key differences as well. For example, while Th1, Th2, and Th17 cells all rely on mTOR for differentiation, their reliance on the multiprotein complexes mTORC1 and mTORC2 differ [51]. mTORC1 is associated with growth and protein synthesis, whereas mTORC2 is more involved in cell maintenance [52]. Both complexes are necessary for Tfh differentiation, whereas only mTORC1 is necessary for Th17 differentiation and only mTORC2 is necessary for Th2 differentiation [53], [54], [55], [56]. There are conflicting results on whether Th1 differentiation is only dependent upon mTORC1, or whether it also requires mTORC2 [53], [57]. The differences in mTOR requirements among T helper subsets help demonstrate how the environment can impact metabolism and differentiation. T cells respond to antigen stimulation based on their environmental context, including the presence of cytokines, which taken together with the intensity of mTOR activation may modulate the cellular response to activation and ultimately control cell fate [58].
Glutamine metabolism has also been associated with differences in Th1 and Th17 differentiation. Both Th1 and Th17 cells require glutamine for differentiation and proliferation, demonstrated using knockouts of the alanine-serine-cysteine transporter 2 (ASCT2) [59]. Glutaminolysis alters the epigenetic landscape, enhancing IL-2 stimulation of mTORC1 signaling, which in turn promotes Th17 differentiation while inhibiting Th1 differentiation [60]. Interestingly, inhibiting the glutaminase protein (GLS) impaired Th17 cells’ ability to mediate both acute airway inflammation and inflammatory bowel disease, indicating its necessity for Th17 function [60]. Hypoxia Inducible Factor 1 Subunit Alpha (HIF1α) is also important for Th17 differentiation, controlling the glycolytic transcriptional programs that promote Th17 over Treg differentiation [61]. HIF1α directly activates RORγt, the lineage defining transcription factor for Th17, as well as IL-17 via recruitment of RORγt and p300 to its promoter [62]. Expression of HIF1α also suppresses Treg differentiation by promoting a glycolytic phenotype and targeting Foxp3 for degradation [61], [62]. Altogether, the context of stimulation, such as environmental cues, is able to drive unique transcriptional programs in naïve T cells, including unique metabolic profiles, which then dictate T cell differentiation.
2.3. CD4+ regulatory T cells
While Tregs are also CD4+ T cells, it is clear that they have some key metabolic differences when compared to other CD4+ subsets. Tregs share a dependence on FAO with memory T cells, with both subsets using this as their primary input for oxidative phosphorylation [50]. Inhibition of oxidative phosphorylation via rotenone treatment, which suppresses ETC complex I, reduces Treg suppression sufficiently for allograft heart rejection to occur [63]. The electron transport chain, particularly components I and III, is necessary for Treg suppressive capacity, but dispensable for Treg numbers [64], [65]. Glut1-deficient Tregs show normal cell numbers with normal suppressive ability [26], demonstrating that glucose, the key metabolite for aerobic glycolysis, is not required by Tregs. In fact, when Glut1 is transgenically overexpressed, Tregs down-regulate Foxp3, the lineage defining transcription factor of Tregs, and lose suppressor function [66], likely due to the promotion of glycolysis and thus acquisition of T conventional effector functions.
As suggested by glucose regulation of Foxp3 expression, Foxp3 has been linked to programming Treg metabolism to enhance oxidative phosphorylation while suppressing Glut1, Myc, and glycolysis [64], [66]. Foxp3 also inhibits signaling of the PI(3)K, Akt, mTOR axis, thereby reducing glycolysis and promoting Treg suppressive function [66]. In fact, it is necessary to inhibit both mTORC1 and mTORC2 for inducible Treg generation from conventional CD4+ T cells [56], [50]. AMP-activated protein kinase, AMPK, a suppressor of mTORC1, promotes mitochondrial metabolism over glycolysis during times of metabolic stress [67], and Tregs have activated AMPK, which was sufficient for decreasing Glut1 expression and increasing Treg differentiation [50]. Interestingly, while the mTOR pathway negatively regulates Treg differentiation, conditional loss of raptor, an essential component of mTORC1, in Tregs results in loss of suppressive capacity, demonstrating that mTORC1 is necessary for Treg function. In addition to loss of suppressive capacity, T cells deficient in mTORC1 had impaired proliferative capacity [54]. mTORC1 signaling in Tregs is downstream of TCR and IL-2 stimulation and its activity is maintained by amino acids, particularly arginine and leucine, in the absence of continuous TCR signals [68], [54]. Altogether, the mTORC1 axis in Tregs is complex, with a role in cell function rather than differentiation, and is important for metabolic responses to environmental cues. Due to their drastically different functional requirements from effector T cells, Tregs have corresponding changes in their metabolic needs. This knowledge provides an opportunity to differentiate between the two cell types in terms of metabolic drug targeting during transplantation, to be discussed later in this review.
3. T cell metabolism and solid organ transplantation
3.1. T cell dysfunction and environmental impact
Much of our understanding of T cell metabolism comes from models of chronic infection or cancer. These models inform the metabolism of tolerant, alloreactive T cells in transplantation, as tolerant T cells display an antigen-experienced signature (CD44HiCD62LLo) with increased expression of the coinhibitory receptor PD1 and low IL-7Rα (CD127) expression, more similar to chronically stimulated exhausted than memory T cells [39], [69]. In tumor models, exhausted T cells have reduced mitochondrial mass with increased mitochondrial depolarization, indicative of reduced function [70]. The clone 13 model of chronic LCMV infection also generates exhausted T cells with mitochondrial depolarization and reduced glycolysis. In this model, PD1-knockout T cells displayed reduced depolarization and improved glucose uptake than exhausted T cells, suggesting that the metabolic phenotype of exhausted T cells in chronic infection is PD1-dependent [71]. This is not to say that the exhaustion phenotype itself is PD1-dependent, as it has been demonstrated that exhausted CD8+ T cells still develop in the absence of PD1[72]. In contrast to the tumor model, exhausted T cells following clone 13 infection displayed increased mitochondrial mass at day 8 post-infection compared with acute LCMV infection [71]. Further analysis of dysfunctional T cells in a model of murine lupus demonstrated reduced glycolysis as well as decreased mitochondrial mass and function in kidney-infiltrating T cells, similar to tumor-infiltrating T cells [73], [70]. Both the kidney and the tumor microenvironment are known to require high levels of glucose and oxygen, which may promote the dysfunctional T cell phenotype [74].
The nutrient concentrations present in the tumor microenvironment differ from those found in circulation [75], and nutrient availability has been demonstrated to have impacts on corresponding T cell metabolism [42], [76]. CD8+ T cells stimulated in vivo with Ova-expressing Listeria monocytogenes (Lm-OVA) versus in vitro stimulation with OVA and IL-2 demonstrated enhanced mitochondrial metabolism and increased ATP production in vivo compared to in vitro [76]. Using this model, stable isotope tracing of 13C-glucose usage identified that in vitro-stimulated T cells produced more lactate than in vivo-stimulated counterparts, suggesting that the phenotype of Warburg metabolism in vivo may be less extensive than previously believed [76]. Similar results were established in a model of GVHD, where alloreactive T cells stimulated in vitro demonstrated a Warburg metabolism, while those stimulated in vivo showed comparatively less lactate production and Glut1 expression, indicative of a non-Warburg metabolism [77]. There are many differences between the in vivo and in vitro environments, and one prominent difference is nutrient availability, where in vitro culture has a nutrient surplus, potentially altering T cell metabolism and masking the real metabolic phenotypes occurring in vivo.
In addition to nutrient availability, cell signals and interactions can also vary greatly by environment. Recently, lymph node fibroblastic reticular cells have been found to impact the epigenetic landscape of activated T cells by secreting IL-6 [78]. IL-6 alters the chromatin landscape and promotes transcription factor binding of MYC, HIF-1α and HIF-1β, resulting in enhanced glycolytic flux, increased lipogenesis, and an enrichment in oxidative phosphorylation to glycolysis ratio in the FRC-conditioned T cells [78]. These data demonstrate how cell interactions can enhance cellular metabolism, and that the physiological environment of the T cell can have drastic effects on T cell metabolism. For transplantation, these data support the hypothesis that alloreactive T cells in a tolerant environment could display altered T cell metabolic profiles dependent upon their location in the graft versus the periphery, as well type of grafted tissue.
Finally, transplantation must be considered as an environmental insult to the graft, particularly with regards to ischemia/reperfusion, where the graft is initially deprived of oxygen and nutrients, leading to the generation of reactive oxygen species and inflammation, thereby altering organ quality and transplant outcome [79]. When collecting an organ, it is subjected to warm ischemia briefly before a period of cooling, which has recently been associated with dangerously high levels of succinate accumulation and injury in heart grafts [80]. After thirty minutes of warm ischemia, a variety of metabolites were significantly different from those in hearts exposed to thirty minutes of cold ischemia, demonstrating how the metabolic environment of the graft can change during the transplantation process [80]. This is not to say that cold ischemia results in perfect graft outcome, as increased periods of cold ischemia are also associated with long-term graft loss [81], [82]. While this work doesn’t directly investigate T cell metabolism, it provides insight into the graft microenvironment as highly dependent on tissue processing, which can later affect T cell metabolism and potentially attempts at inducing of tolerance.
3.2. Alloreactive T cells
While the exact metabolic phenotype of alloreactive T cells in a tolerant environment has yet to be elucidated, models of transplantation have contributed widely to the field of cellular metabolism. This is particularly true for models of graft versus host disease (GVHD), where alloreactive donor cells attack the host tissue. In GVHD, similar to solid organ transplantation, T cells are responding to an alloantigen that persists. However, in GVHD the antigen is more dispersed, and can be found throughout the body in higher concentrations than in a solid allograft.
In a model of bone marrow transplantation, alloreactive T cells showed enhanced glycolysis as well as FAO when compared with proliferating bone marrow cells [83]. However, this altered metabolic phenotype does not seem to be a specific outcome of alloactivation, as T cells stimulated with alloantigen in vitro only increased aerobic glycolysis, demonstrating a typical metabolism for cells stimulated in vitro [77], [76]. This was corroborated in lung transplantation, where alloreactive CD8+ T cells utilized the highest levels of glucose in the lung tissue during acute rejection [84]. 13C stable isotope tracing following bone marrow transplantation demonstrated that alloreactive T cells in GVHD use glutamine for ribose biosynthesis and the production of TCA cycle intermediates [77]. Additionally, alloreactive cells decrease fatty acid and pyrimidine metabolism in favor of biomass synthesis for cellular proliferation and expansion [85]. In GVHD and acute rejection, alloreactive T cells are activated effectors, indicating that T cell allostimulation results in typical effector T cell responses, and that exposure to alloantigens does not inherently induce distinct metabolic pathways than other antigens.
4. Modifying T cell metabolism
Targeting metabolic functions within cells has become an attractive therapeutic avenue, both in transplantation and cancer. In transplantation, the goal is to limit effector T cell metabolism while maintaining Treg potency. Here we take a look at some common pharmacological approaches to altering cell metabolism in the interest of promoting transplantation tolerance.
4.1. Clinical treatments
Many immunosuppressive therapies alter cellular metabolism. For example, Basiliximab is an IL-2 receptor antagonist used as an induction therapeutic agent in transplantation [86]. Since IL-2 receptor signaling results in downstream activation of mTOR, Basiliximab modulates T cell metabolism [87]. This review will focus on clinically used drugs designed to maintain transplantation tolerance by targeting key aspects of metabolic processes directly, specifically mTOR inhibitor and antimetabolites.
mTOR inhibitors promote transplantation tolerance by preventing downstream metabolism-altering signaling events, thereby blocking T cell proliferation and promoting Treg development [67], [66]. The two direct mTOR inhibitors used in solid organ transplantation are sirolimus and everolimus. Both drugs bind to a 12kd FK-binding protein (FKBP12), which then inhibits mTORC1 via dissociation of the raptor subunit from the complex, hindering downstream phosphorylation [88], [89]. Side effects of these treatments include dyslipidemia, edema, and poor wound healing [89]. Although both drugs inhibit mTOR, everolimus is three-fold weaker at binding FKBP12 than sirolimus [88]. However, everolimus is more potent at preventing mTORC2 activation than sirolimus, which might be why it has been better at preventing antibody-mediated rejection [90]. Additionally, these two drugs have different effects on mitochondrial metabolism when coupled with cyclosporine, a calcineurin inhibitor shown to reduce mitochondrial metabolism in vitro [91]. While sirolimus exacerbated the negative impacts of calcineurin on the mitochondrial metabolism of rat brain tissue, everolimus could distribute into the mitochondria and reverse the effects of calcineurin [91], underscoring the importance of understanding metabolism/drug interactions.
Antimetabolites are drugs interfering with metabolic pathways. In transplantation, mycophenolate mofetil (MMF) and azathioprine (AZA) are antimetabolites used to inhibit T cell proliferation. MMF is a pro-drug, and must be metabolized to mycophenolic acid (MPA) in order to suppress inosine monophosphate dehydrogenase (IMPDH), the rate-limiting step in guanine biosynthesis [92]. IMPDH is important for T cell proliferation and is thus upregulated following T cell stimulation, as lymphocytes lack the ability to salvage purines for synthesis [93], [94]. In contrast, AZA is reduced by glutathione, forming the intermediate 6-mercaptopurine (6-MP) which is further metabolized into a variety of substrates that incorporate into replicating DNA, blocking replication [94]. Similar to MPA, metabolites of AZA also inhibit de novo purine synthesis [94].
The use of antimetabolites and mTOR inhibitors as immunosuppressors following transplantation demonstrates the importance of cellular metabolism for T cell responses. However, neither antimetabolites nor mTOR inhibitors are sufficient to prevent graft rejection alone and other immunotherapies are necessary for targeting T cell cytokine production, such as calcineurin inhibitors or costimulation blockers [95].
4.2. Experimental models
Other forms of metabolic suppression are currently being pursued with the goal of developing donor-specific tolerance. One of the most striking models aimed to induce Treg development by simultaneously blocking glycolysis and glutamine metabolism [96]. Lee and colleagues combined three therapies: 2-deoxyglucose (2-DG), an inhibitor of glycolysis, with 6-diazo-5-oxo-l-norleucine (DON), a glutamine metabolism inhibitor, and metformin, a hypoglycemic agent able to promote fatty acid oxidation. This regimen suppressed CD4+ T cell responses, with the exception of Tregs [96]. Furthermore, this regimen prolonged the survival of both skin and heart allografts compared to untreated controls, demonstrating that targeting both glycolysis and glutamine metabolism sufficiently controlled effector T cell responses.
In cancer, tumors can evade the immune system by competing with T cells for glucose[97]. 2-DG has been clinically tested in a variety of anti-tumor therapies (reviewed in [98]). Side effects associated with the treatment include headaches, drowsiness, and tachycardia [99]. In the tumor microenvironment, tumor cells are responsible for most of the glucose uptake, whereas in transplantation, effector T cells would be the most reliant on glucose, suggesting particular susceptibility to this treatment. This could prove beneficial for transplantation, as a way to control alloreactive T cells. However, in cancer, the treatment may be ceased once the tumor is eliminated. In an alloresponse, new alloreactive T cells may be continuously generated by the host, suggesting the potential long-term necessity of this treatment. Since other tissues, particularly the brain and heart, also utilize high levels of glucose, this could lead to off target effects and limit the practicality of this treatment. This has particularly been demonstrated in rats, where long-term ingestion of 2-DG results in toxicity [100]. Importantly, Lee and colleagues demonstrated that their mice appeared healthy for more than 100 days, suggesting that the combined treatment with 2-DG, DON, and metformin may yet be translatable to the clinic [96].
Glucose deficiency results in the maintained expression of T cell micro-RNAs (miRNAs) specific for the methyltransferase enhancer of zeste homolog 2 (EZH2), which is associated with T cell cytokine production [97]. While reduced glucose availability is detrimental to the anti-tumor T cell response, it provides an interesting avenue of investigation for regulating alloimmune responses. Similar to 2-DG, using micro-RNA to target glycolysis or upstream mediators of T cell proliferation could prove therapeutically beneficial, for example, by preventing the upregulation of Glut1. Another model of interest for informing potential therapies in solid organ transplantation is GVHD. In this model, alloreactive T cells see chronic antigen, and use both glycolysis and oxidative phosphorylation, in contrast to normal effectors, which rely more highly on glycolysis than oxidative phosphorylation for energy [83]. Knowing this, Gatza and colleagues specifically targeted mitochondrial ATPase using the small molecule inhibitor Bz-423, resulting in specific suppression of alloreactive T cells [83]. Finally, it was recently demonstrated that corticosteroids suppress low, but not high affinity memory CD8+ T cell proliferation via inhibition of fatty acid metabolism [101]. While this provides an exciting avenue for the potential control of T cell responses in transplantation, it must also be considered that rejection is linked with the expansion of high affinity T cell clones [30]. Since binding affinity is correlated with T cell metabolic changes, these data suggest that associating to corticosteroids specific therapies targeting high affinity clones will have further potential to prevent this T cell repertoire shift [28], [101], [29]. These models pave the way for future experimental avenues and potentially translational therapeutic strategies to promote solid organ transplantation tolerance.
5. Conclusions and future perspectives
We have compiled evidence detailing the changes in T cell metabolism during cell differentiation, and how research in T cell metabolism has been applied to experimental models and clinical therapies for transplantation. T cell metabolism in transplantation has been historically challenging to study, with research focusing on GVHD and few studies using models of solid organ transplantation. Technologies including 13C stable isotope tracing, metabolic environment profiling, and single cell RNAseq provide potential avenues for further research. Since a caveat of metabolism research is that it has historically been studied in vitro with super-physiological levels of metabolites or oxygen, these techniques utilizable in T cells stimulated in vivo may be more translatable to the clinic for drug testing, as the data might be more representative of physiological processes.
Current metabolism-altering suppressive regimens aim to limit T effector metabolism while leaving Treg metabolism intact, such as by mTOR inhibition. These treatments are insufficient for donor-specific tolerance, with high rates of chronic graft rejection [1]. Glutaminolysis is important for T cell activation [23], [38]. Recent studies in glutamine metabolism offer promising avenues for metabolic therapies, with clinical trials of Dracen Pharma’s glutamine antagonist DRP-104 scheduled to begin in 2020 as a cancer therapeutic [96], [102]. These and similar approaches offer the foundation for future investigations in transplantation.
Finally, in this era of big data, integration of metabolic and transcriptional signatures holds the promise of identifying novel pathways to target to improve transplant outcomes [103]. This approach was recently demonstrated by Hiemer and colleagues, who combined carbon tracing with metabolomic and transcriptomic profiling to investigate human CD4+ T cells following TCR stimulation. While cyclosporine treatment demonstrated broader metabolic impacts than rapamycin, combining the two treatments inhibited T cell proliferation following stimulation with plate bound antibodies [104]. This was an additive effect of the two drugs, demonstrated by network analysis [104]. Studies of this nature provide essential evidence not only for potential drug targets, such as metabolic enzymes, but also for future combinatorial therapies and increasing the efficacy of our current treatment regimens. Additionally, such studies can be used to better understand T cell dysfunction, such as how to promote or prevent dysfunction, for transplantation or cancer, respectively. Altogether, integrative techniques such as network analysis of metabolomics and transcriptomic data provide the potential for translation of findings in immunometabolism to the clinic.
Highlights.
T cells remodel their metabolism for effector function, and again for memory function
Cellular metabolism impacts T cell differentiation and function
Microenvironmental impacts on T cell metabolism include metabolite availability and cellular signaling molecules
Current clinical treatments for transplantation alter T cell metabolism
Acknowledgements
J.B.A. is supported by a NIH predoctoral training grant, award number T32AI7090-41. Additional support comes from NIAID Grant P01AI-97113 (to M.-L.A. and Dr. Anita S. Chong, University of Chicago).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kloc M, Ghobrial RM, Chronic allograft rejection: A significant hurdle to transplant success, Burns Trauma 2 (2014) 3–10. 10.4103/2321-3868.121646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ghesquière B, Wong BW, Kuchnio A, Carmeliet P, Metabolism of stromal and immune cells in health and disease, Nature 511 (2014) 167–176. 10.1038/nature13312. [DOI] [PubMed] [Google Scholar]
- [3].Palmer CS, Ostrowski M, Balderson B, Christian N, Crowe SM, Glucose Metabolism Regulates T Cell Activation, Differentiation, and Functions, Front. Immunol 6 (2015). 10.3389/fimmu.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, Cheung E, Olin-Sandoval V, Grüning N-M, Krüger A, Alam MT, Keller MA, Breitenbach M, Brindle KM, Rabinowitz JD, Ralser M, The return of metabolism: biochemistry and physiology of the pentose phosphate pathway, Biol. Rev. Camb. Philos. Soc 90 (2015) 927–963. 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, Verway MJ, Raissi TC, Tsui H, Boukhaled G, da Costa SH, Frezza C, Krawczyk CM, Friedman A, Manfredi M, Richer MJ, Hess C, Jones RG, Serine Is an Essential Metabolite for Effector T Cell Expansion, Cell Metab 25 (2017) 345–357. 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- [6].Ho P-C, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui Y-C, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM, Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses, Cell 162 (2015) 1217–1228. 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].TeSlaa T, Teitell MA, Techniques to Monitor Glycolysis, Methods Enzymol 542 (2014) 91–114. 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jonckheere AI, Smeitink JAM, Rodenburg RJT, Mitochondrial ATP synthase: architecture, function and pathology, J. Inherit. Metab. Dis 35 (2012) 211–225. 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Houten SM, Wanders RJA, A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation, J. Inherit. Metab. Dis 33 (2010) 469–477. 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu G, Amino acids: metabolism, functions, and nutrition, Amino Acids 37 (2009) 1–17. 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- [11].Le Bourgeois T, Strauss L, Aksoylar H-I, Daneshmandi S, Seth P, Patsoukis N, Boussiotis VA, Targeting T Cell Metabolism for Improvement of Cancer Immunotherapy, Front. Oncol 8 (2018). 10.3389/fonc.2018.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Siska PJ, Kim B, Ji X, Hoeksema MD, Massion PP, Beckermann KE, Wu J, Chi J-T, Hong J, Rathmell JC, Fluorescence-based measurement of cystine uptake through xCT shows requirement for ROS detoxification in activated lymphocytes, J. Immunol. Methods 438 (2016) 51–58. 10.1016/j.jim.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nakajima H, Kunimoto H, TET2 as an epigenetic master regulator for normal and malignant hematopoiesis, Cancer Sci 105 (2014) 1093–1099. 10.1111/cas.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Röhrig F, Schulze A, The multifaceted roles of fatty acid synthesis in cancer, Nat. Rev. Cancer 16 (2016) 732–749. 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- [15].Zhang X, Yang S, Chen J, Su Z, Unraveling the Regulation of Hepatic Gluconeogenesis, Front. Endocrinol 9 (2019). 10.3389/fendo.2018.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Warburg O, Gawehn K, Geissler A, [Metabolism of leukocytes]., Z. Naturforsch. B 13B (1958) 515–516. [PubMed] [Google Scholar]
- [17].Mendoza A, Fang V, Chen C, Serasinghe M, Verma A, Muller J, Chaluvadi VS, Dustin ML, Hla T, Elemento O, Chipuk JE, Schwab SR, Lymphatic endothelial S1P promotes mitochondrial function and survival in naive T cells, Nature 546 (2017) 158–161. 10.1038/nature22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR, The Transcription Factor Myc Controls Metabolic Reprogramming upon T Lymphocyte Activation, Immunity 35 (2011) 871–882. 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chang C-H, Curtis JD, Maggi LB, Faubert B, Villarino AV, O’Sullivan D, Huang SC-C, van der Windt GJW, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL, Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis, Cell 153 (2013) 1239–1251. 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO, Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism, Science 354 (2016) 481–484. 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Warburg O, The Metabolism of Carcinoma Cells, J. Cancer Res 9 (1925) 148–163. 10.1158/jcr.1925.148. [DOI] [Google Scholar]
- [22].Warburg O, On the Origin of Cancer Cells, Science 123 (1956) 309–314. 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- [23].Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA, Glutamine Uptake and Metabolism Are Coordinately Regulated by ERK/MAPK during T Lymphocyte Activation, J. Immunol 185 (2010) 1037–1044. 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang C-R, Schumacker PT, Licht JD, Perlman H, Bryce PJ, Chandel NS, Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling, Immunity 38 (2013) 225–236. 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Newsholme EA, Crabtree B, Ardawi MSM, The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells, Biosci. Rep 5 (1985) 393–400. 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- [26].Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC, The Glucose Transporter Glut1 is Selectively Essential for CD4 T Cell Activation and Effector Function, Cell Metab 20 (2014) 61–72. 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sinclair LV, Rolf J, Emslie E, Shi Y-B, Taylor PM, Cantrell DA, Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation, Nat. Immunol 14 (2013) 500–508. 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jones N, Cronin JG, Dolton G, Panetti S, Schauenburg AJ, Galloway SAE, Sewell AK, Cole DK, Thornton CA, Francis NJ, Metabolic Adaptation of Human CD4+ and CD8+ T-Cells to T-Cell Receptor-Mediated Stimulation, Front. Immunol 8 (2017). 10.3389/fimmu.2017.01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Savage PA, Boniface JJ, Davis MM, A Kinetic Basis For T Cell Receptor Repertoire Selection during an Immune Response, Immunity 10 (1999) 485–492. 10.1016/S1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- [30].Miller ML, McIntosh CM, Williams JB, Wang Y, Hollinger MK, Isaad NJ, Moon JJ, Gajewski TF, Chong AS, Alegre M-L, Distinct Graft-Specific TCR Avidity Profiles during Acute Rejection and Tolerance, Cell Rep 24 (2018) 2112–2126. 10.1016/j.celrep.2018.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Baixauli F, Acín-Pérez R, Villarroya-Beltrí C, Mazzeo C, Nuñez-Andrade N, Gabandé-Rodriguez E, Ledesma MD, Blázquez A, Martin MA, Falcón-Pérez JM, Redondo JM, Enríquez JA, Mittelbrunn M, Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses, Cell Metab 22 (2015) 485–498. 10.1016/j.cmet.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tan H, Yang K, Li Y, Shaw TI, Wang Y, Blanco DB, Wang X, Cho J-H, Wang H, Rankin S, Guy C, Peng J, Chi H, Integrative Proteomics and Phosphoproteomics Profiling Reveals Dynamic Signaling Networks and Bioenergetics Pathways Underlying T Cell Activation, Immunity 46 (2017) 488–503. 10.1016/j.immuni.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Campello S, Lacalle RA, Bettella M, Mañes S, Scorrano L, Viola A, Orchestration of lymphocyte chemotaxis by mitochondrial dynamics, J. Exp. Med 203 (2006) 2879–2886. 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Morlino G, Barreiro O, Baixauli F, Robles-Valero J, González-Granado JM, Villa-Bellosta R, Cuenca J, Sánchez-Sorzano CO, Veiga E, Martín-Cófreces NB, Sánchez-Madrid F, Miro-1 Links Mitochondria and Microtubule Dynein Motors To Control Lymphocyte Migration and Polarity, Mol. Cell. Biol 34 (2014) 1412–1426. 10.1128/MCB.01177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kamiński MM, Sauer SW, Klemke C-D, Süss D, Okun JG, Krammer PH, Gülow K, Mitochondrial Reactive Oxygen Species Control T Cell Activation by Regulating IL-2 and IL-4 Expression: Mechanism of Ciprofloxacin-Mediated Immunosuppression, J. Immunol 184 (2010) 4827–4841. 10.4049/jimmunol.0901662. [DOI] [PubMed] [Google Scholar]
- [36].Kamiński MM, Sauer SW, Kamiński M, Opp S, Ruppert T, Grigaravičius P, Grudnik P, Gröne H-J, Krammer PH, Gülow K, T cell Activation Is Driven by an ADP-Dependent Glucokinase Linking Enhanced Glycolysis with Mitochondrial Reactive Oxygen Species Generation, Cell Rep 2 (2012) 1300–1315. 10.1016/j.celrep.2012.10.009. [DOI] [PubMed] [Google Scholar]
- [37].Bailis W, Shyer JA, Zhao J, Canaveras JCG, Khazal FJA, Qu R, Steach HR, Bielecki P, Khan O, Jackson R, Kluger Y, Maher LJ, Rabinowitz J, Craft J, Flavell RA, Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function, Nature 571 (2019) 403–407. 10.1038/s41586-019-1311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, Li L, Boussiotis VA, PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation, Nat. Commun 6 (2015) 1–13. 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Miller ML, McIntosh CM, Wang Y, Chen L, Wang P, Lei YM, Daniels MD, Watkins E, Solano CM, Chong AS, Alegre M-L, Resilience of T cell-intrinsic dysfunction in transplantation tolerance, Proc. Natl. Acad. Sci (2019). 10.1073/pnas.1910298116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Takahashi T, Hsiao HM, Tanaka S, Li W, Higashikubo R, Scozzi D, Bharat A, Ritter JH, Krupnick AS, Gelman AE, Kreisel D, PD-1 expression on CD8+ T cells regulates their differentiation within lung allografts and is critical for tolerance induction, Am. J. Transplant 18 (2018) 216–225. 10.1111/ajt.14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang L-S, Jones RG, Choi Y, Enhancing CD8 T-cell memory by modulating fatty acid metabolism, Nature 460 (2009) 103–107. 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vázquez G, Yurchenko E, Raissi TC, van der Windt GJW, Viollet B, Pearce EL, Pelletier J, Piccirillo CA, Krawczyk CM, Divangahi M, Jones RG, The Energy Sensor AMPK Regulates T Cell Metabolic Adaptation and Effector Responses In Vivo, Immunity 42 (2015) 41–54. 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- [43].van der Windt GJW, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL, Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development, Immunity 36 (2012) 68–78. 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang C-H, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJW, Chen Q, Huang SC-C, O’Neill CM, Edelson BT, Pearce EJ, Sesaki H, Huber TB, Rambold AS, Pearce EL, Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming, Cell. 166 (2016) 63–76. 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].van der Windt GJW, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL, Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development, Immunity 36 (2012) 68–78. 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].van der Windt GJW, O’Sullivan D, Everts B, Huang SC-C, Buck MD, Curtis JD, Chang C-H, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL, CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability, Proc. Natl. Acad. Sci 110 (2013) 14336–14341. 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gubser PM, Bantug GR, Razik L, Fischer M, Dimeloe S, Hoenger G, Durovic B, Jauch A, Hess C, Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch, Nat. Immunol 14 (2013) 1064–1072. 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- [48].Bantug GR, Fischer M, Grählert J, Balmer ML, Unterstab G, Develioglu L, Steiner R, Zhang L, Costa ASH, Gubser PM, Burgener A-V, Sauder U, Löliger J, Belle R, Dimeloe S, Lötscher J, Jauch A, Recher M, Hönger G, Hall MN, Romero P, Frezza C, Hess C, Mitochondria-Endoplasmic Reticulum Contact Sites Function as Immunometabolic Hubs that Orchestrate the Rapid Recall Response of Memory CD8+ T Cells, Immunity 48 (2018) 542–555.e6. 10.1016/j.immuni.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wilson JE, Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function, J. Exp. Biol 206 (2003) 2049–2057. 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- [50].Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC, Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets, J. Immunol 186 (2011) 3299–3303. 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, Matsuda S, Koyasu S, PI3K-Akt-mTORC1-S6K1/2 Axis Controls Th17 Differentiation by Regulating Gfi1 Expression and Nuclear Translocation of RORγ, Cell Rep 1 (2012) 360–373. 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- [52].Laplante M, Sabatini DM, mTOR signaling at a glance, J. Cell Sci 122 (2009) 3589–3594. 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M, Mammalian Target of Rapamycin Protein Complex 2 Regulates Differentiation of Th1 and Th2 Cell Subsets via Distinct Signaling Pathways, Immunity 32 (2010) 743–753. 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H, mTORC1 couples immune signals and metabolic programming to establish Treg-cell function, Nature 499 (2013) 485–490. 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yang J, Lin X, Pan Y, Wang J, Chen P, Huang H, Xue H-H, Gao J, Zhong X-P, Critical roles of mTOR Complex 1 and 2 for T follicular helper cell differentiation and germinal center responses, eLife. 5 (2016) e17936 10.7554/eLife.17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD, The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2, Nat. Immunol 12 (2011) 295–303. 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Heikamp EB, Patel CH, Collins S, Waickman A, Oh M-H, Sun I-H, Illei P, Sharma A, Naray-Fejes-Toth A, Fejes-Toth G, Misra-Sen J, Horton MR, Powell JD, The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex, Nat. Immunol 15 (2014) 457–464. 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD, The mTOR Kinase Differentially Regulates Effector and Regulatory T Cell Lineage Commitment, Immunity 30 (2009) 832–844. 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nakaya M, Xiao Y, Zhou X, Chang J-H, Chang M, Cheng X, Blonska M, Lin X, Sun S-C, Inflammatory T Cell Responses Rely on Amino Acid Transporter ASCT2 Facilitation of Glutamine Uptake and mTORC1 Kinase Activation, Immunity 40 (2014) 692–705. 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, Johnson ME, de Cubas AA, Wu P, Li G, Zhang Y, Newcomb DC, Wells AD, Restifo NP, Rathmell WK, Locasale JW, Davila ML, Blazar BR, Rathmell JC, Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism, Cell 175 (2018) 1780–1795.e19. 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H, HIF1α–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells, J. Exp. Med 208 (2011) 1367–1376. 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dang EV, Barbi J, Yang H-Y, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen H-R, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F, Control of TH17/Treg Balance by Hypoxia-Inducible Factor 1, Cell 146 (2011) 772–784. 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, Koike MA, Hancock SA, Bhatti TR, Han R, Jiao J, Veasey SC, Sims CA, Baur JA, Wallace DC, Hancock WW, Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival, FASEB J 29 (2015) 2315–2326. 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, Wang Z, Quinn WJ, Kopinski PK, Wang L, Akimova T, Liu Y, Bhatti TR, Han R, Laskin BL, Baur JA, Blair IA, Wallace DC, Hancock WW, Beier UH, Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments, Cell Metab 25 (2017) 1282–1293.e7. 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martínez-Reyes I, Gao P, Helmin KA, Abdala-Valencia H, Sena LA, Schumacker PT, Turka LA, Chandel NS, Mitochondrial complex III is essential for suppressive function of regulatory T cells, Nature 565 (2019) 495–499. 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, Warmoes MO, de Cubas AA, MacIver NJ, Locasale JW, Turka LA, Wells AD, Rathmell JC, Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression, Nat. Immunol 17 (2016) 1459–1466. 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hardie DG, Hawley SA, Scott JW, AMP-activated protein kinase – development of the energy sensor concept, J. Physiol 574 (2006) 7–15. 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shi H, Chapman NM, Wen J, Guy C, Long L, Dhungana Y, Rankin S, Pelletier S, Vogel P, Wang H, Peng J, Guan K-L, Chi H, Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb, Immunity 0 (2019). 10.1016/j.immuni.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R, Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection, Immunity 27 (2007) 670–684. 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- [70].Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, Delgoffe GM, The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction, Immunity 45 (2016) 374–388. 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM, Wherry EJ, Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8+ T Cell Exhaustion, Immunity 45 (2016) 358–373. 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ, Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cellsAbsence of PD-1 dysregulates T cell exhaustion, J. Exp. Med 212 (2015) 1125–1137. 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tilstra JS, Avery L, Menk AV, Gordon RA, Smita S, Kane LP, Chikina M, Delgoffe GM, Shlomchik MJ, Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted, J. Clin. Invest 128 (2018) 4884–4897. 10.1172/JCI120859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chen Y, Fry BC, Layton AT, Modeling Glucose Metabolism in the Kidney, Bull. Math. Biol 78 (2016) 1318–1336. 10.1007/s11538-016-0188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, Dennstedt EA, Vander Heiden MG, Muir A, Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability, eLife 8 (2019) e44235 10.7554/eLife.44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ma EH, Verway MJ, Johnson RM, Roy DG, Steadman M, Hayes S, Williams KS, Sheldon RD, Samborska B, Kosinski PA, Kim H, Griss T, Faubert B, Condotta SA, Krawczyk CM, DeBerardinis RJ, Marsh K, Richer MJ, Chubukov V, Roddy T, Jones RG, Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8+ T Cells, Immunity (2019). 10.1016/j.immuni.2019.09.003. [DOI] [PubMed] [Google Scholar]
- [77].Glick GD, Rossignol R, Lyssiotis CA, Wahl D, Lesch C, Sanchez B, Liu X, Hao L-Y, Taylor C, Hurd A, Ferrara JLM, Tkachev V, Byersdorfer CA, Boros L, Opipari AW, Anaplerotic Metabolism of Alloreactive T Cells Provides a Metabolic Approach To Treat Graft-Versus-Host Disease, J. Pharmacol. Exp. Ther 351 (2014) 298–307. 10.1124/jpet.114.218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Brown FD, Sen DR, LaFleur MW, Godec J, Lukacs-Kornek V, Schildberg FA, Kim H-J, Yates KB, Ricoult SJH, Bi K, Trombley JD, Kapoor VN, Stanley IA, Cremasco V, Danial NN, Manning BD, Sharpe AH, Haining WN, Turley SJ, Fibroblastic reticular cells enhance T cell metabolism and survival via epigenetic remodeling, Nat. Immunol 20 (2019) 1668–1680. 10.1038/s41590-019-0515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Slegtenhorst BR, Dor FJ, Rodriguez H, Voskuil FJ, Tullius SG, Ischemia/reperfusion Injury and its Consequences on Immunity and Inflammation, Curr. Transplant. Rep 1 (2014) 147–154. 10.1007/s40472-014-0017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Martin JL, Costa ASH, Gruszczyk AV, Beach TE, Allen FM, Prag HA, Hinchy EC, Mahbubani K, Hamed M, Tronci L, Nikitopoulou E, James AM, Krieg T, Robinson AJ, Huang MM, Caldwell ST, Logan A, Pala L, Hartley RC, Frezza C, Saeb-Parsy K, Murphy MP, Succinate accumulation drives ischaemia-reperfusion injury during organ transplantation, Nat. Metab 1 (2019) 966–974. 10.1038/s42255-019-0115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Debout A, Foucher Y, Trébern-Launay K, Legendre C, Kreis H, Mourad G, Garrigue V, Morelon E, Buron F, Rostaing L, Kamar N, Kessler M, Ladrière M, Poignas A, Blidi A, Soulillou J-P, Giral M, Dantan E, Each additional hour of cold ischemia time significantly increases the risk of graft failure and mortality following renal transplantation, Kidney Int 87 (2015) 343–349. 10.1038/ki.2014.304. [DOI] [PubMed] [Google Scholar]
- [82].Salahudeen AK, Haider N, May W, Cold ischemia and the reduced long-term survival of cadaveric renal allografts, Kidney Int 65 (2004) 713–718. 10.1111/j.1523-1755.2004.00416.x. [DOI] [PubMed] [Google Scholar]
- [83].Gatza E, Wahl DR, Opipari AW, Sundberg TB, Reddy P, Liu C, Glick GD, Ferrara JLM, Manipulating the Bioenergetics of Alloreactive T Cells Causes Their Selective Apoptosis and Arrests Graft-Versus-Host Disease, Sci. Transl. Med 3 (2011) 67ra8–67ra8. 10.1126/scitranslmed.3001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chen DL, Wang X, Yamamoto S, Carpenter D, Engle JT, Li W, Lin X, Kreisel D, Krupnick AS, Huang HJ, Gelman AE, Increased T Cell Glucose Uptake Reflects Acute Rejection in Lung Grafts, Am. J. Transplant 13 (2013) 2540–2549. 10.1111/ajt.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nguyen HD, Chatterjee S, Haarberg KMK, Wu Y, Bastian D, Heinrichs J, Fu J, Daenthanasanmak A, Schutt S, Shrestha S, Liu C, Wang H, Chi H, Mehrotra S, Yu X-Z, Metabolic reprogramming of alloantigen-activated T cells after hematopoietic cell transplantation, J. Clin. Invest 126 (2016) 1337–1352. 10.1172/JCI82587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ji S-Q, Chen H-R, Yan H-M, Wang H-X, Liu J, Zhu P–, Xiao M–, Xun C-Q, Anti-CD25 monoclonal antibody (basiliximab) for prevention of graft-versus-host disease after haploidentical bone marrow transplantation for hematological malignancies, Bone Marrow Transplant 36 (2005) 349–354. 10.1038/sj.bmt.1705046. [DOI] [PubMed] [Google Scholar]
- [87].Ross SH, Cantrell DA, Signaling and Function of Interleukin-2 in T Lymphocytes, Annu. Rev. Immunol 36 (2018) 411–433. 10.1146/annurev-immunol-042617-053352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Schuler W, Sedrani R, Cottens S, Häberlin B, Schulz M, Schuurman H-J, Zenke G, Zerwes H-G, Schreier MH, SDZ RAD, A NEW RAPAMYCIN DERIVATIVE: Pharmacological Properties In Vitro and In Vivo, Transplantation 64 (1997) 36. [DOI] [PubMed] [Google Scholar]
- [89].Watson CJE, Sirolimus (rapamycin) in clinical transplantation, Transplant. Rev 15 (2001) 165–77. 10.1016/S0955-470X(01)80016-1. [DOI] [Google Scholar]
- [90].Jin Y-P, Valenzuela NM, Ziegler ME, Rozengurt E, Reed EF, Everolimus Inhibits Anti-HLA I Antibody-Mediated Endothelial Cell Signaling, Migration and Proliferation More Potently Than Sirolimus, Am. J. Transplant 14 (2014) 806–819. 10.1111/ajt.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Serkova N, Jacobsen W, Niemann CU, Litt L, Benet LZ, Leibfritz D, Christians U, Sirolimus, but not the structurally related RAD (everolimus), enhances the negative effects of cyclosporine on mitochondrial metabolism in the rat brain, Br. J. Pharmacol 133 (2001) 875–885. 10.1038/sj.bjp.0704142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Allison AC, Eugui EM, Mycophenolate mofetil and its mechanisms of action, Immunopharmacology 47 (2000) 85–118. 10.1016/S0162-3109(00)00188-0. [DOI] [PubMed] [Google Scholar]
- [93].Gu JJ, Stegmann S, Gathy K, Murray R, Laliberte J, Ayscue L, Mitchell BS, Inhibition of T lymphocyte activation in mice heterozygous for loss of the IMPDH II gene, J. Clin. Invest 106 (2000) 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Taylor AL, Watson CJE, Bradley JA, Immunosuppressive agents in solid organ transplantation: Mechanisms of action and therapeutic efficacy, Crit. Rev. Oncol. Hematol 56 (2005) 23–46. 10.1016/j.critrevonc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- [95].Tkachev V, Furlan SN, Watkins B, Hunt DJ, Zheng HB, Panoskaltsis-Mortari A, Betz K, Brown M, Schell JB, Zeleski K, Yu A, Kirby I, Cooley S, Miller JS, Blazar BR, Casson D, Bland-Ward P, Kean LS, Combined OX40L and mTOR blockade controls effector T cell activation while preserving Treg reconstitution after transplant, Sci. Transl. Med 9 (2017). 10.1126/scitranslmed.aan3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lee C-F, Lo Y-C, Cheng C-H, Furtmüller GJ, Oh B, Andrade-Oliveira V, Thomas AG, Bowman CE, Slusher BS, Wolfgang MJ, Brandacher G, Powell JD, Preventing Allograft Rejection by Targeting Immune Metabolism, Cell Rep 13 (2015) 760–770. 10.1016/j.celrep.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhang D, Li J, Wang F, Hu J, Wang S, Sun Y, 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy, Cancer Lett 355 (2014) 176–183. 10.1016/j.canlet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- [98].Landau BR, Laszlo J, Stengle J, Burk D, Certain Metabolic and Pharmacologic Effects in Cancer Patients Given Infusions of 2-Deoxy-D-Glucose, JNCI J. Natl. Cancer Inst 21 (1958) 485–494. 10.1093/jnci/21.3.485. [DOI] [PubMed] [Google Scholar]
- [99].Minor RK, Smith DL, Sossong AM, Kaushik S, Poosala S, Spangler EL, Roth GS, Lane M, Allison DB, de Cabo R, Ingram DK, Mattison JA, Chronic ingestion of 2-deoxy-d-glucose induces cardiac vacuolization and increases mortality in rats, Toxicol. Appl. Pharmacol 243 (2010) 332–339. 10.1016/j.taap.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhao E, Maj T, Kryczek I, Li W, Wu K, Zhao L, Wei S, Crespo J, Wan S, Vatan L, Szeliga W, Shao I, Wang Y, Liu Y, Varambally S, Chinnaiyan AM, Welling TH, Marquez VE, Kotarski J, Wang H, Wang Z, Zhang Y, Liu R, Wang G, Zou W, Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction, Nat. Immunol 17 (2016) 95–103. 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tokunaga A, Sugiyama D, Maeda Y, Warner AB, Panageas KS, Ito S, Togashi Y, Sakai C, Wolchok JD, Nishikawa H, Selective inhibition of low-affinity memory CD8+ T cells by corticosteroids, J. Exp. Med 216 (2019) 2701–2713. 10.1084/jem.20190738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Dracen Pharmaceuticals announces DRP-104 presentations at the 2019 Society of Immunotherapy of Cancer (SITC) Meeting – Dracen Pharmaceutical, (n.d.). https://www.dracenpharma.com/index.php/dracen-pharmaceuticals-announces-drp-104-presentations-at-the-2019-society-of-immunotherapy-of-cancer-sitc-meeting/ (accessed December 13, 2019). [Google Scholar]
- [103].Jha AK, Huang SC-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN, Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization, Immunity 42 (2015) 419–430. 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- [104].Hiemer S, Jatav S, Jussif J, Alley J, Lathwal S, Piotrowski M, Janiszewski J, Kibbey R, Alves T, Dumlao D, Jha A, Bandukwala H, Integrated Metabolomic and Transcriptomic Profiling Reveals Novel Activation-Induced Metabolic Networks in Human T cells, bioRxiv (2019) 635789 10.1101/635789. [DOI] [Google Scholar]