Abstract
This work reports on a rapid diagnostic platform for the detection of Plasmodium falciparum lactate dehydrogenase (PfLDH), a representative malaria biomarker, using a microfluidic microplate-based immunoassay. In this study, the microfluidic microplate made it possible to diagnose PfLDH with a small volume of sample (only 5 μL) and short time (< 90 min) compared to conventional immunoassays such as enzyme-linked immunosorbent assay (ELISA). Moreover, the diagnostic performance of PfLDH showed high sensitivity, specificity, and selectivity (i.e., 0.025 pg/μL in phosphate-buffered saline and 1 pg/μL in human serum). The microfluidic-based microplate sensing platform has the potential to adapt simple, rapid, and accurate diagnoses to the practical detection of malaria.
Keywords: Microfluidic microplate, Immunoassay, Malaria, PfLDH, Diagnosis
Introduction
Malaria is a serious infectious disease that is transmitted from mosquitoes to humans causing 219 million infections and 435,000 deaths worldwide in 2017 according to the World Health Organization (WHO) [1, 2]. Despite vast efforts to reduce the risk of malaria, it still has high mortality and morbidity due to inaccurate diagnosis and increased drug resistance [3]. Malaria infection is caused by Plasmodium parasites that are transmitted by the bite of Anopheles spp. mosquitoes such as Plasmodium falciparum (P. falciparum) and Plasmodium vivax (P. vivax) [1]. In particular, P. falciparum is an important target of malaria diagnosis because it accounts for 90% of worldwide malaria mortality [4]. P. falciparum has various biomarkers including lactate dehydrogenase (LDH), histidine rich protein2 (HRP2), aldolase, and hypoxanthine phosphoribosyl transferase [5]. Conventional diagnosis of P. falciparum is mainly dependent on HRP2 detection by immunoassay [6]. However, HRP2 deletion mutants have been reported in several countries [7–12]. Therefore, it is necessary to develop an alternative diagnostic biomarker instead of HRP2. In this study, we chose P. falciparum lactate dehydrogenase (PfLDH) as an alternative biomarker for the diagnosis of malaria (P. falciparum), which is a water-soluble enzyme that converts pyruvate to lactate in glycolysis in P. falciparum infection [5, 13, 14].
Conventional malaria diagnosis methods include a microscopic examination and antibody-based rapid diagnostic tests (RDTs) [5, 15–19]. Although the microscopic examination is the gold standard for malaria diagnosis and is rapid and cost-effective, it requires highly trained personnel. Meanwhile, the RDT, a lateral flow immunoassay, is a useful method in low-resource environments because it is possible to provide cost-effective, rapid (< 30 min) and simple detection methods using the nitrocellulose strip [5, 20]. However, RDTs have sensitivity limitations, such as the inability to detect < 200 parasites/µL or < 1 ng/mL [16, 19, 21, 22]. To overcome the limitations of existing diagnostics, several studies demonstrated improved detection efficiency by introducing nanoparticles such as magnetic beads. Markwalter et al. successfully detected PfLDH up to 21.1 ± 0.4 parasites/mL within 45 min using antibody-immobilized magnetic beads as a colorimetric assay [14]. Additionally, they developed a simultaneous capture and sequential detection of two malarial biomarkers (PfLDH and HRP2) on magnetic microparticles [23]. Kim et al. detected HRP2 up to 0.1 ng/mL using antibody-immobilized magnetic beads and quantum dots using an automated droplet-based microfluidic device [20]. Although these studies were developed to overcome the limitations of conventional enzyme-linked immunosorbent assay (ELISA), nanoparticle-based immunoassays still require a relatively complex surface functionalization process and a large amount of antibody to immobilize the antibody.
To overcome these limitations of conventional ELISA-based diagnosis and to improve the diagnosis of PfLDH, we present a microfluidic microplate-based immunoassay. This microfluidic microplate is an Optimiser™ microplate, which is one of the next-generation immunoassay platforms, and the existing 96-well plate was developed in the form of a microfluidic channel [24–26]. In this study, we demonstrated that the microfluidic microplate-based immunoassay provides an ultrafast, simple, and precise immunoassay for PfLDH diagnosis. The microfluidic microplate-based immunoassay significantly reduced the amount of reagents (5 μL) and diagnosis time (< 90 min) compared to conventional ELISA, as well as enabling high-sensitivity diagnostics (0.025 pg/µL). Additionally, we confirmed that PfLDH in human serum can be diagnosed up to 1 pg/µL. Based on the results of our study, we expect that the microfluidic microplate-based immunoassay platform will be widely used for infectious disease diagnosis as well as in malaria.
Results and discussion
Microfluidic microplate-based immunoassay for PfLDH diagnosis
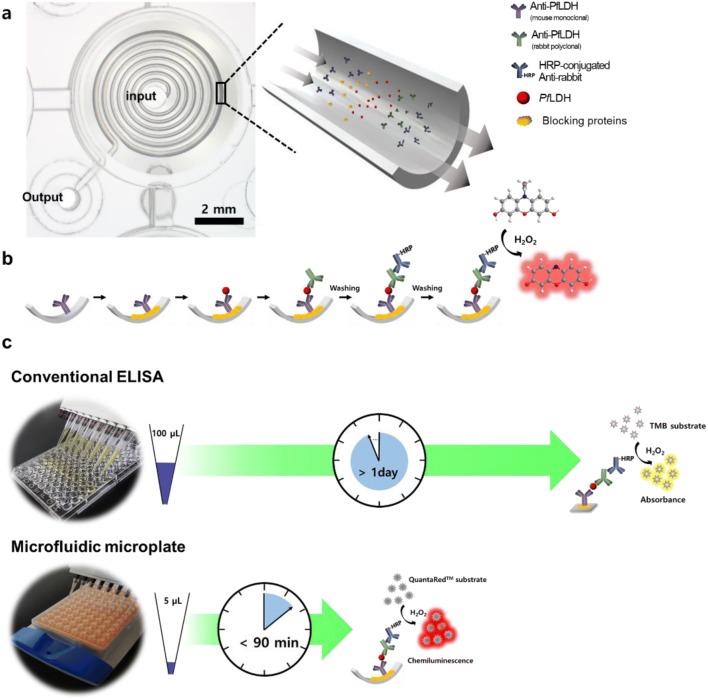
Figure 1a illustrates the construction of a microfluidic microplate and the procedure for detecting PfLDH. The whole body of this microplate consisted of a conventional 96-well plate with an inlet for pipette injection, an outlet that is open toward the absorbent pad, and a microfluidic channel between them. The microfluidic microplate is a spiral microfluidic channel and has a 1.5-fold larger surface area and a 50-fold surface-area-to-volume ratio than conventional ELISA plates [24]. The microfluidic microplate is operated by capillary action between the microchannel and adsorbent pad. The process is passive flow regulation, which can lead to accurate and rapid immunoassay results. This flow system involves the sequential addition of reagents, such as antibodies and antigens, to the microfluidic channel (Fig. 1b). The diagnostic procedure in the microfluidic channel is similar to that of conventional ELISA. In general, conventional ELISA was used to target the absorbance signal from the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate; however, the microfluidic microplate-based immunoassay used the chemiluminescence signal from the 10-acetyl-3,7-dihydroxyphenoxazine (ADHP)-based chemiluminescent substrate due to the microfluidic channel structural properties. ADHP is not normally a fluorescent molecule; instead, it is converted to a fluorescent form (resorufin) in the presence of horseradish peroxidase (HRP) and hydroxide peroxide [27]. Consequently, microfluidic microplates have the advantage of using a highly accessible microfluidic surface, capillary design, and highly sensitive substrate compared to ELISA. Therefore, the volume required to diagnose the targets can be significantly reduced, and the overall diagnosis time can be much faster than that of conventional ELISA (Fig. 1c). This immunoassay platform shows that it is appropriate to diagnose PfLDH with a small amount of antibody and fast diagnosis time.
Fig. 1.
Schematic illustration for diagnosis of PfLDH using microfluidic microplate. a Optical image of the microfluidic microplate. The microfluidic microplate allows for low volume and rapid immunoassay due to microfluidic channel. b Flow sequence for detection of PfLDH. c Comparison of PfLDH detection method using conventional ELISA and microfluidic microplate
Performance of the microfluidic microplate for PfLDH immunoassay
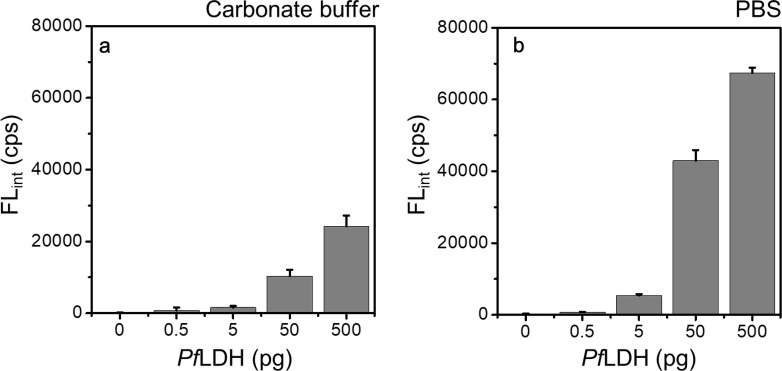
To optimize the microfluidic microplate-based immunoassay for PfLDH, we selected an antibody pair through a sandwich ELISA (data not shown). Based on the results of the ELISA, monoclonal mouse PfLDH antibody was selected as the capture antibody (Cap-Ab), and polyclonal rabbit PfLDH antibody was selected as the primary antibody (1st-Ab). In this condition, we confirmed that PfLDH was diagnosed up to 0.1 pg/µL (10 pg in 100 μL) by ELISA (Fig. 2). To use selected antibody pairs in the microfluidic microplate, the optimization of Cap-Ab adsorption was significantly important to the whole immunoassay process. We compared the diagnosis efficiency in carbonate-bicarbonate buffer (pH 9.6, Fig. 3a) and phosphate-buffered saline (PBS, pH 7.4, Fig. 3b) to optimize Cap-Ab adsorption. In general, carbonate-bicarbonate buffer was used as a coating buffer in ELISA because the high pH was attributed to the better dissolution of proteins into the buffer and improved adsorption to the positive charged plate. However, Cap-Ab in PBS showed better diagnosis efficiency than Cap-Ab in carbonate-bicarbonate buffer (Fig. 3). This indicates that PBS (pH 7.4) was appropriate substance to adsorb Cap-Ab on the microfluidic microplate wall.
Fig. 2.
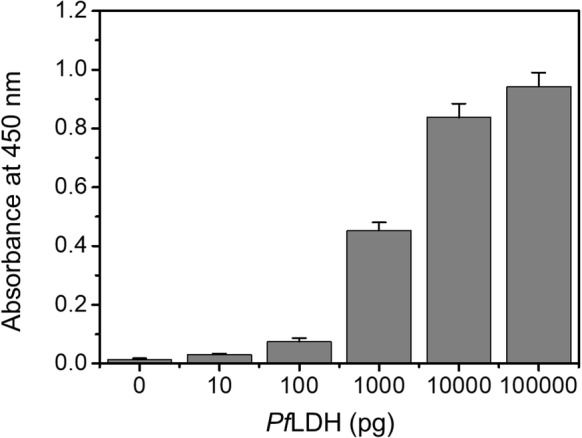
Diagnosis of PfLDH using the ELISA. The optical density were measured at 450 nm wavelength with 0 to 100 ng/100 μL (n = 3)
Fig. 3.
Optimization of the Cap-Ab adsorption. The Cap-Ab coating efficiency was compared two coating buffers. To detect the PfLDH in the microfluidic microplate, the Cap-Ab was prepared in a carbonate-bicarbonate buffer (pH 9.6) and b PBS (pH 7.4). The chemiluminescence were measured by BioTek multimode reader (Excitation: 530 nm, Emission: 590 nm, n = 3)
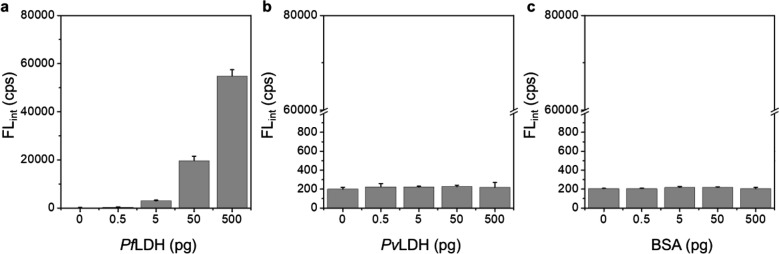
To evaluate the PfLDH diagnostics capability in the microfluidic microplate, we diagnosed PfLDH in the range from 0.05 to 500 pg, including the negative control (PfLDH 0 pg). As shown in Fig. 4, PfLDH showed a limit of detection (LOD) of 0.025 pg/µL (0.125 pg in 5 μL), and the diagnosis of PfLDH was visually confirmed through the chemiluminescence image. The LOD was calculated as the minimum detectable signal (FLPfLDH=0 + 3SDPfLDH=0). Additionally, we compared PfLDH with Plasmodium vivax lactate dehydrogenase (PvLDH) and BSA for selectivity and specificity at concentrations of 0, 0.5, 5, 50, and 500 pg/5 µL. As shown in Fig. 5, this platform showed high selectivity and specificity for PfLDH. Based on the results of experiments, it was clearly shown that PfLDH can be ultrafast and precise in our sensing platform.
Fig. 4.
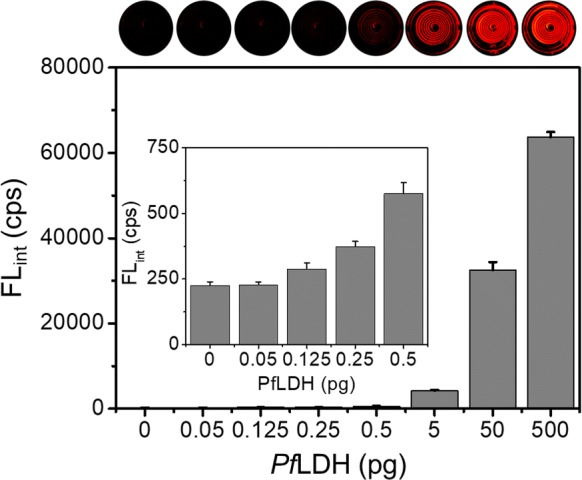
Diagnosis of PfLDH using the microfluidic microplate-based immunoassay. Chemiluminescence intensity (n = 3) and chemiluminescence images of the microfluidic microplate to the detection of PfLDH
Fig. 5.
Selectivity and specificity of microfluidic microplate-based immunoassay for detection of PfLDH. Diagnosis of aPfLDH, bPvLDH, and c BSA in PBS-based blocking solution (n = 3)
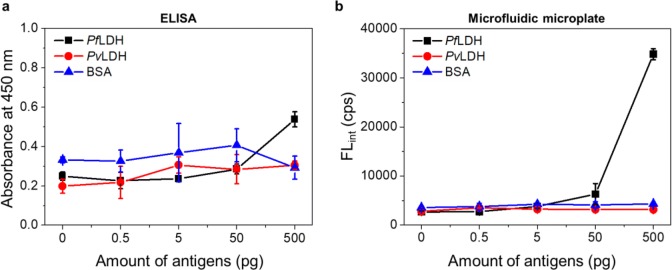
To confirm its applicability in clinical diagnosis, we performed the diagnosis of PfLDH in human serum. PfLDH, PvLDH, and BSA were prepared in commercial human serum at concentrations of 0, 0.5, 5, 50, and 500 pg/5 µL. Figure 6a shows the diagnosis results of the PfLDH in human serum using ELISA. The results show that the non-specific binding is high, even in the absence of PfLDH. It means that PfLDH in human serum can be detected by ELISA but is not suitable for highly sensitive diagnosis due to its high non-specific binding. In general, one of the drawbacks of ELISA is the false-positive result due to non-specific binding in the presence of various proteins, such as human serum, it must be improved for application in clinical diagnosis [28]. In contrast to the ELISA results, Fig. 6b shows that PfLDH in human serum can be diagnosed up to 1 pg/µL (5 pg in 5 μL) with high selectivity and specificity using microfluidic microplate. This LOD is an ideal sensitivity at which to adapt the clinical application because PfLDH-infected patients typically show PfLDH plasma levels of approximately 3–15 pg/μL [13, 29]. Recently, Tonigold et al. demonstrated that the conditions of antibody adsorption (i.e., pH and isoelectric point of antibody) were significantly affected by the orientation of the antibody [30]. They validated that adsorbed antibodies on the polystyrene (PS) beads showed superior targeting properties compared with covalently coupled antibodies with the PS beads. The microfluidic microplate is also composed of PS; therefore, the Cap-Ab, which is well oriented on the microfluidic channel, led to the high sensing capability of PfLDH, including PfLDH spiked in human serum. Additionally, the large surface area and capillary forces of the microfluidic channel also contributed to the sensing performance. Based on the results of PfLDH diagnosis in human serum, we expect that our immunoassay platform can be widely used for the clinical diagnosis of infectious diseases such as malaria.
Fig. 6.
Diagnosis of PfLDH, PvLDH, and BSA in human serum using a ELISA and b microfluidic microplate (n = 3). All antigens were prepared in the range of 0–500 pg in human serum
Diagnostics capability of the microfluidic microplate-based immunoassay
Table 1 summarizes and compares recent studies on the diagnosis of PfLDH. Recently, many studies have developed high-sensitivity, high-selectivity, and cost-effective diagnostics based on aptamers or nanoparticles [31–35]. Nanoparticles, such as novel-metal nanoparticles and magnetic nanoparticles, are applied for lateral flow immunoassay or combined with aptamers to improved diagnostic performance. However, these sensors require the complicated surface functionalization and stabilization of nanoparticles or require a relatively large amount of sample volume. On the other hand, the microfluidic microplate-based immunoassay improved the conventional 96-well plate to a microfluidic channel, significantly reducing the time and amount of reagents required for the diagnosis and showing that high-sensitive diagnosis to the PfLDH. Based on the results, the microfluidic microplate-based immunoassay shows that it not only brings cost-effective for antibody use but also provides easier, simpler, faster, and superior sensitive diagnosis than recently developed sensors.
Table 1.
Comparison of recent PfLDH detection studies
| Diagnosis | Target | Bio-recognition | Output | LOD | Volume | References |
|---|---|---|---|---|---|---|
| Microfluidic microplate | PfLDH in buffer | Antibody | Fluorescence intensity | 0.025 pg/μL | 5 μL | This study |
| Microfluidic microplate | PfLDH in human serum | Antibody | Fluorescence intensity | 1 pg/μL | 5 μL | This study |
| ELISA | PfLDH in buffer | Antibody | Absorbance | 0.1 pg/μL | 100 μL | This study |
| Lateral flow immunoassay | PfLDH in buffer | Antibody | Colorimetric assay | 10 pg/μL | 30 μL | [31] |
| AuNPs based aptasensor | PfLDH in lysed RBCsa solution (Mimic real sample) | Aptamer | Colorimetric assay | 38 pg/μL | 10 μL | [32] |
| MNP-Qdot aptasensor | PfLDH in buffer | Aptamer | Fluorescence intensity | 0.0066 pg/μL | 100 μL | [33] |
| Aptamer-tethered enzyme capture (APTEC) assay | PfLDH in buffer | Aptamer | Colorimetric assay | 4.9 pg/μL | 10 μL | [34] |
| Aptamer-tethered enzyme capture (APTEC) assay | PfLDH in serum | Aptamer | Colorimetric assay | 50 pg/μL | 40 μL | [35] |
aRBC red blood cells
Conclusions
In conclusion, we demonstrated that simple, ultrafast, and accurate diagnosis of PfLDH using microfluidic microplates is possible. We reduced the amount of reagent required for diagnosis to 5 μL and reduced the diagnosis time to 90 min. The microfluidic microplate-based sensing platform provides a user-friendly method and a cost-effective approach for an immunoassay-based method for PfLDH diagnosis. Our sensing platform with optimized antibody pairs of capture and detection antibodies showed highly sensitive, selective, and specific diagnosis of PfLDH. We expect that microfluidic microplates will be used for malaria diagnosis rapidly and accurately in hospitals.
Experimental section
Materials
The microfluidic microplate plate (Optimiser™ microplate), holder, and absorbent pad were provided from MiCo BioMed Co., Ltd. (Seoul, Korea). Mouse monoclonal PfLDH antibody was purchased from Fapon Biotech (BRCMALS212, Guangdong, China), and rabbit polyclonal PfLDH antibody was purchased from LifeSpan BioScience, Inc. (LS-C488775, Seattle, USA). PfLDH and PvLDH were provided by BioNano Health Guard Research Center (H-GUARD). Polyclonal anti-rabbit HRP-tagged antibody (HRP-Ab) was obtained from Cell Signaling Technology, Inc. (Danvers, USA). Phosphate-buffered saline (PBS), blocking solution (StartingBlock™) and HRP substrate kit (QuantaRed™ Enhanced Chemifluorescent HRP substrate kit) were purchased from Thermo Fisher Scientific Ltd. (Waltham, USA). Bovine serum albumin (BSA) and sodium carbonate were purchased from Sigma-Aldrich (Louis, USA). Human serum (from human male AB plasma, USA origin, sterile-filtered) was purchased from Sigma-Aldrich (Louis, USA).
Expression and purification of PfLDH
As malaria biomarkers, PfLDH and PvLDH were prepared for malaria diagnostics. In order to clone PfLDH and PvLDH, the full length gene encoding for the LDH was amplified via the polymerase chain reaction (PCR). The PCR product was inserted into the pET 21a vector using to generate pET-PfLDH and pET-PvLDH. The gene was all verified by DNA sequencing, and transformed into the expression host, E. coli BL21 (DE3) (Stratagene, CA) for the expression of the recombinant proteins. The transformed cells were grown at 37 °C with shaking to an OD600 of 0.6. The cells were induced with 1 mM isopropyl-2-D-thiogalactopyranoside (IPTG, GibcoBRL, MD), and grown for an additional 14 h at 25 °C. The cells were then harvested and disrupted via sonication, after which the soluble and insoluble fractions were separated by centrifugation. The soluble fractions were loaded onto a IDA-miniexcellose affinity column (Bioprogen Co., Republic of Korea) and washed three times with equilibration buffer (50 mM Tris–Cl, 0.5 N NaCl, pH 8.0), respectively. The recombinant proteins were then eluted with 0.5 M imidazole in the same buffer (50 mM Tris–Cl, 0.5 N NaCl, pH 8.0), and dialyzed against phosphate-buffered saline (PBS, pH7.4). The purified PfLDH and PvLDH were resolved on 12% Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and the gels were stained with Coomassie’s Brilliant Blue R250. Protein concentrations were determined by the Bradford method, using bovine serum albumin as a standard.
Validation of antibody pairs using ELISA
To select the antibody pairs for detection of PfLDH, Cap-Ab and 1st-Ab were selected by ELISA. As a Cap-Ab, mouse monoclonal PfLDH antibody (200 μL, 2 μg/mL) dissolved in sodium bicarbonate 1 M (pH 9.6), is incubated in a 96-well plate (Corning® 96 Well EIA/RIA Assay Microplate) at 4 °C for overnight. After the reaction, BSA 1% in PBS buffer (100 μL) was added to the wells and reacted at 37 °C for 30 min. After the removal of BSA solution, PfLDH (100 μL dissolved in PBS) was added to the wells in a concentration range of 1 μg/mL to 100 μg/mL and reacted at 37 °C for 120 min. After PfLDH treatment, rabbit polyclonal PfLDH antibody as a 1st-Ab (100 μL, 1/2000 dilution) was immediately added onto the PfLDH solution. After 120 min incubation at 37 °C, the wells were washed 2 times with PBS buffer. Then, HRP-Ab was added on the wells and reacted at 37 °C for 120 min. Once again, the wells were washed 4 times with PBS buffer. Finally, TMB substrate reagent (BD Biosciences, USA) composed of TMB and hydrogen peroxide are mixed at a ratio of 1: 1, and 100 μL was added each well. After 5 min, stop solution was added onto the wells and measured absorbance at 450 nm in microplate reader (Multiskan™ FC Microplate Photometer, Thermo Fisher Scientific, USA).
Immunoassay of PfLDH using the microfluidic microplate
The immunoassay of PfLDH in the microfluidic microplate was performed in the same order as conventional ELISA. Cap-Ab (5 μL, 10 μg/mL) dispersed in coating buffer (PBS and carbonate-bicarbonate buffer) was first immobilized on the microfluidic channel for 10 min. Five microliters of blocking solution were added to the channels and incubated for 10 min. Next, PfLDH (each 5 μL, 0–100 ng/mL) and 1st-Ab (1/100 dilution) were treated and incubated for 10 min in order. The microplates were then incubated with 5 μL PBS for 10 min to remove unbound substances and were treated with HRP-tagged Ab (1/2500 dilution) for 10 min. Then, all channels were washed with PBS (30 μL) twice. After washing, Quantared™ Enhanced Chemifluorescent HRP Substrate (Enhancer solution: Stable peroxide solution: ADHP = 50:50:1) was added to the channel for chemiluminescence. After 10 min, the chemiluminescent signal was measured using a BioTek multimode reader (Cytation5, USA). Additionally, the chemiluminescence image was observed by stereomicroscope (SMZ18, Nikon, Japan) with a fluorescence filter (excitation 530 nm). To confirm the diagnostic ability of PfLDH in human serum, PfLDH, PvLDH and BSA were prepared from human serum. Three antigens were prepared in PBS 10 times higher than the target concentration before diagnosis. These antigens were then spiked in human serum at a volume ratio of 1:9. The immunoassay procedure was the same as described above.
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by Ministry of Science and ICT (MSIT) (NRF-2019R1C1C1006084 and NRF-2019R1C1C1006867), The Center for BioNano Health-Guard funded by the Ministry of Science and ICT as Global Frontier Project (H-GUARD_2013M3A6B2078950) and KRIBB Research Initiative Program.
Abbreviations
- WHO
World Health Organization
- P. falciparum
Plasmodium falciparum
- P. vivax
Plasmodium vivax
- LDH
Lactate dehydrogenase
- HRP2
Histidine rich protein2
- PfLDH
Plasmodium falciparum lactate dehydrogenase
- PvLDH
Plasmodium vivax lactate dehydrogenase
- RDTs
Rapid diagnostic tests
- ELISA
Enzyme-linked immunosorbent assay
- TMB
3,3′,5,5′-tetramethylbenzidine
- ADHD
10-acetyl-3,7-dihydroxyphenoxazine
- HRP
Horseradish peroxidase
- Cap-Ab
Capture antibody
- 1st-Ab
Primary antibody
- HRP-Ab
HRP-tagged antibody
- PBS
Phosphate-buffered saline
- BSA
Bovine serum albumin
- LOD
Limit of detection
- PCR
Polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- APTEC
Aptamer-tethered enzyme capture
- H-GUARD
BioNano Health Guard Research Center
Authors’ contributions
WSL takes the lead through the whole experiments. WSL and KJK performed the detection of PfLDH by using microfluidic microplate. KP, SYL, UJL, and YBS performed the ELISA for selection of antibody pairs and prepared malaria biomarkers (PfLDH and PvLDH). JJ conceived of the idea and supervised the project. WSL, TK and JJ mainly wrote the manuscript and all authors provided feedback and helped the completion of the final manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by Ministry of Science and ICT (MSIT) (NRF-2019R1C1C1006084 and NRF-2019R1C1C1006867), The Center for BioNano Health-Guard funded by the Ministry of Science and ICT as Global Frontier Project (H-GUARD_2013M3A6B2078950) and KRIBB Research Initiative Program.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Phillips MA, Burrows JN, Manyando C, Van Huijsduinen RH, Van Voorhis WC, Wells TNCC. Nat. Rev. Dis. Prim. 2017;3:17. doi: 10.1038/nrdp.2017.50. [DOI] [PubMed] [Google Scholar]
- 2.WHO . World Malaria Report 2018. Geneva: World Health Organization; 2018. [Google Scholar]
- 3.World Health Organization . World Malaria Report 2018. Geneva: World Health Organization; 2015. [Google Scholar]
- 4.Snow RW. BMC Med. 2015;13:14. doi: 10.1186/s12916-014-0254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragavan KV, Kumar S, Swaraj S, Neethirajan S. Biosens. Bioelectron. 2018;105:188. doi: 10.1016/j.bios.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 6.Cheung YW, Dirkzwager RM, Wong WC, Cardoso J, Costa JDN, Tanner JA. Biochimie. 2018;145:131. doi: 10.1016/j.biochi.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Molina-Cruz A, Zilversmit MM, Neafsey DE, Hartl DL, Barillas-Mury C. Annu. Rev. Genet. 2016;50:447. doi: 10.1146/annurev-genet-120215-035211. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Xing H, Zhao Z, Yang Z, Cao Y, Li W, Yan G, Sattabongkot J, Cui L, Fan Q. Acta Trop. 2015;152:26. doi: 10.1016/j.actatropica.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solano CM, Okoth SA, Abdallah JF, Pava Z, Dorado E, Incardona S, Huber CS, Oliveira AM, Bell D, Udhayakumar V, Barnwell JW. PLoS One. 2015;10:e0131576. doi: 10.1371/journal.pone.0131576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. PLoS ONE. 2016;11:1. doi: 10.1371/journal.pone.0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viana GMR, Okoth SA, Flannery LS, Barbosa DRL, de Oliveira AM, Goldman IF, Morton LC, Huber C, Anez A, Machado RLD, Camargo LMA, Do Valle SCN, Povoa MM, Udhayakumar V, Barnwell JW. PLoS One. 2017;12:1. [Google Scholar]
- 12.Beshir KB, Sepúlveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, De Boer JG, Sutherland C, Cunningham J, Hopkins H. Sci. Rep. 2017;7:1. doi: 10.1038/s41598-017-15031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung Y-W, Kwok J, Law AWL, Watt RM, Kotaka M, Tanner JA. Proc. Natl. Acad. Sci. 2013;110:15967. doi: 10.1073/pnas.1309538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markwalter CF, Davis KM, Wright DW. Anal. Biochem. 2016;493:30. doi: 10.1016/j.ab.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 15.McKenzie FE, Sirichaisinthop J, Miller RS, Gasser RA, Wongsrichanalai C. Am. J. Trop. Med. Hyg. 2003;69:372. doi: 10.4269/ajtmh.2003.69.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. Am. J. Trop. Med. Hyg. 2007;77:119. doi: 10.4269/ajtmh.2007.77.119. [DOI] [PubMed] [Google Scholar]
- 17.Kilian AHD, Metzger WG, Mutschelknauss EJ, Kabagambe G, Langi P, Korte R, Von Sonnenburg F. Trop. Med. Int. Heal. 2000;5:3. doi: 10.1046/j.1365-3156.2000.00509.x. [DOI] [PubMed] [Google Scholar]
- 18.Gillet P, Maltha J, Hermans V, Ravinetto R, Bruggeman C, Jacobs J. Malar. J. 2011;10:1. doi: 10.1186/1475-2875-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray CK, Gasser RA, Magill AJ, Miller RS. Clin. Microbiol. Rev. 2008;21:97. doi: 10.1128/CMR.00035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim C, Hoffmann G, Searson PC. ACS Sensors. 2017;2:766. doi: 10.1021/acssensors.7b00119. [DOI] [PubMed] [Google Scholar]
- 21.Bell D, Wongsrichanalai C, Barnwell JW. Nat. Rev. Microbiol. 2006;4:682. doi: 10.1038/nrmicro1474. [DOI] [PubMed] [Google Scholar]
- 22.Bell D, Peeling RW. Nat. Rev. Microbiol. 2006;4:S34. doi: 10.1038/nrmicro1524. [DOI] [PubMed] [Google Scholar]
- 23.Markwalter CF, Ricks KM, Bitting AL, Mudenda L, Wright DW. Talanta. 2016;161:443. doi: 10.1016/j.talanta.2016.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kai J, Puntambekar A, Santiago N, Lee SH, Sehy DW, Moore V, Han J, Ahn CH. Lab Chip. 2012;12:4257. doi: 10.1039/c2lc40585g. [DOI] [PubMed] [Google Scholar]
- 25.Vashist SK, Luppa PB, Yeo LY, Ozcan A, Luong JHT. Trends Biotechnol. 2015;33:692. doi: 10.1016/j.tibtech.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Jung W, Han J, Choi JW, Ahn CH. Microelectron. Eng. 2014;132:46. doi: 10.1016/j.mee.2014.09.024. [DOI] [Google Scholar]
- 27.Lü JM, Lin PH, Yao Q, Chen C. J. Cell Mol. Med. 2010;14:840. doi: 10.1111/j.1582-4934.2009.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Güven E, Duus K, Lydolph MC, Jørgensen CS, Laursen I, Houen G. J. Immunol. Methods. 2014;403:26. doi: 10.1016/j.jim.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Martin SK, Rajasekariah GH, Awinda G, Waitumbi J, Kifude C. Am. J. Trop. Med. Hyg. 2009;80:516. doi: 10.4269/ajtmh.2009.80.516. [DOI] [PubMed] [Google Scholar]
- 30.Tonigold M, Simon J, Estupiñán D, Kokkinopoulou M, Reinholz J, Kintzel U, Kaltbeitzel A, Renz P, Domogalla MP, Steinbrink K, Lieberwirth I, Crespy D, Landfester K, Mailänder V. Nat. Nanotechnol. 2018;13:862. doi: 10.1038/s41565-018-0171-6. [DOI] [PubMed] [Google Scholar]
- 31.Mthembu CL, Sabela MI, Mlambo M, Madikizela LM, Kanchi S, Gumede H, Mdluli PS. Anal. Methods. 2017;9:5943. doi: 10.1039/C7AY01645J. [DOI] [Google Scholar]
- 32.Jain P, Chakma B, Singh NK, Patra S, Goswami P. Mol. Biotechnol. 2016;58:497. doi: 10.1007/s12033-016-9946-x. [DOI] [PubMed] [Google Scholar]
- 33.Kim C, Searson PC. Bioconjug. Chem. 2017;28:2230. doi: 10.1021/acs.bioconjchem.7b00328. [DOI] [PubMed] [Google Scholar]
- 34.Dirkzwager RM, Kinghorn AB, Richards JS, Tanner JA. Chem. Commun. 2015;51:4697. doi: 10.1039/C5CC00438A. [DOI] [PubMed] [Google Scholar]
- 35.Dirkzwager RM, Liang S, Tanner JA. ACS Sensors. 2016;1:420. doi: 10.1021/acssensors.5b00175. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.