Abstract
Background
Circ-ZNF609 has been shown to modulate cancer progression in several kinds of cancer. However, the role of circ-ZNF609 played in lung adenocarcinoma (LAUD) remains unclear. In this study, we investigated the role of circ-ZNF609 in regulating LAUD cancer progression.
Materials and Methods
Quantitative reverse transcription polymerase chain reaction was conducted to evaluate circ‐ZNF609 expression in 52 LAUD tissues and 52 matched adjacent normal tissues, LAUD cell lines and bronchial epithelial cell line (HBE). The direct interaction between miR-1224-3p and circ-ZNF609 or EVT1 was verified using luciferase reporter assay and RNA immunoprecipitation assay. Cell Counting Kit-8, colony formation assay, cell-cycle analysis were utilized to examine the effect of circ-ZNF609 or miR-1224-3p on cell proliferation.
Results
Circ-ZNF609 was significantly upregulated in LAUD tissues and cell lines, and its inhibition induced reduced cell proliferation of LAUD cells. Mechanistically, we demonstrated that circ-ZNF609 sponged miR-1224-3p to promote EVT1 expression. More importantly, miR-1224-3p overexpression strongly suppressed circZNF609-induced malignant phenotype of LAUD cells.
Conclusion
Circ-ZNF609 enhances LAUD progression by increasing oncogenic EVT1 expression via sponging miR-1224-3p, revealing that circ-ZNF609/miR-1224-3p/ETV1 axis may be a promising therapeutic target for LAUD treatment.
Keywords: circ-ZNF609, lung adenocarcinoma, miR-1224-3p, EVT1, ceRNA
Introduction
Lung cancer is a deadly disease that is responsible for the largest number of cancer-related deaths annually.1 Lung adenocarcinoma (LAUD), the most common subtype of lung cancer, accounts for approximately 40% of lung malignancies.2 The difficulty of early diagnosis and the lack of effective therapies lead to the prognosis of LAUD remaining dismal.3 Therefore, the impetus for investigating the molecular mechanisms regulating malignant progression and identifying potential therapeutic targets is strong needed.
Circular RNAs (circRNAs) are known as a class of endogenous noncoding RNAs that lacking 5ʹand 3ʹends and shape a covalently closed loop.4 Because of the stable structure, circRNAs are inherently resistant to RNAse activity.5 Furthermore, there are selectively conserved microRNA (miRNA) target sites inside circRNAs, which enable circRNAs to act as miRNA sponges for modulating gene expression.6 Currently, a broad interest pointed to circRNAs due to its important biological function “miRNA sponge” in mediating cancer progression.7 For instance, circ-ADAMTS13 inhibits the growth of hepatocellular carcinoma by sponging miR-484.3 Another study demonstrated that circ-PRMT5 triggered Epithelial-Mesenchymal Transition of bladder cancer cells by interacting with miR-30c8. Han et al showed that circMTO1 impaired the cell proliferation of liver cancer via sponging miR-9.9 Therefore, circRNAs are considered as a promising class of molecules to better elucidate the mechanism underpinning cancer progression.
Emerging evidence indicates a critical role of circ-ZNF609 (ID: hsa_circ_0000615 in circBase) in human cancers, including breast cancer,9 renal carcinoma,10 colorectal cancer,11 and rhabdomyosarcoma.12 Importantly, higher expression of circ-ZNF609 was detected in all above cancer biopsies compared with paracancerous tissues, and its depletion could efficiently hamper the aggressive biological behavior of cancer cells. These observations suggest that circ-ZNF609 may serve as a predictive biomarker for cancer prognosis and a potential target for cancer therapy. However, the role of circ-ZNF609 in LAUD remains largely unexplored.
In this study, we confirmed that circ-ZNF609 is strongly up-regulated in LAUD tissues and LAUD cell lines. We further verified that circ-ZNF609 enhances LAUD cell proliferation by increasing ETV1 expression via sponging miR-1224-3p, indicating that circ-ZNF609 may represent a promising therapeutic target for the treatment of LAUD.
Materials and Methods
Clinical Samples
Fifty-two LAUD tumor tissues and paired noncancer tissues were collected from primary LAUD patients who never received preoperative radiotherapy or chemotherapy at Lianshui people’s hospital between 2009 and 2019. LAUD tissues were histologically confirmed by three independent pathologists. The methodologies utilized in this study were under the supervision of the ethics committee at Lianshui people’s hospital and were conducted in accordance with the Declaration of Helsinki. The patients participated in this study had provided their written informed consent.
CircZNF609 and ETV1 Plasmid Construction and Stable Transfection
We cloned the human cDNA sequences of circZNF609 and ETV1 into pcircRNA 2.2 hsa vector and pcDNA3.1 vector, respectively. The vectors were both bought from BersinBio Co., Ltd. (Guangzhou, China). Then we transfected cells with the overexpression vector by using Lipofectamine 2000 (Invitrogen), and selected cells by G418.
Cell Culture and Transfection
Human LAUD cell lines (HCC827, NCI-H23, SPC-A1, H1975, H1299 and A549) and a normal human bronchial epithelial cell line (HBE) were all obtained from American Type Culture Collection (ATCC; Shanghai, China) and cultured in RPMI-1640 complete medium supplemented with 10% fetal bovine serum in the incubator at 37°C.
A549 and SPC-A1 cells were transfected with circ-ZNF609 small interfering RNAs, circ-ZNF609 expression vector, miR-1224-3p mimics and their matched negative controls (GenePharma, Shanghai, China) by using Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocols. The sequences of si-circZNF609 was 5ʹ-GTCAAGTCTGAAAAGCAA TGA-3ʹ.
Quantitative RT-PCR
Total RNA of LAUD tumor tissues and paired noncancer tissues or LAUD cell lines and HBE cells was extracted by using RNAsimple kit (Tiangen, Beijing, China), followed by reverse-transcribed to cDNA via a Reverse Transcription kit (Takara Biotechnology Co., Ltd.). Quantitative RT-PCR was conducted with the SYBR Green qPCR kit (Tiangen, Beijing, China) and 2−ΔΔCq method was employed to calculate the relative RNA expression. GAPDH and U6 were used as an internal control. The sequences of the primers used in this study were as follows:
circ-ZNF609:
Forward: 5ʹ-GCCACACTACCAGACAACATC-3ʹ
Reverse: 5ʹ-TAATACGACTCACTATAGGGAACCGGCTACACTGCGG-3ʹ
hsa-miR-1224-3p:
Forward: 5ʹ- CCCCACCTCCTCTCTCCTCAG-3ʹ
ETV1:
Forward: 5ʹ-TGGCAGTTTTTGGTAGCTCTTC-3ʹ
Reverse: 5ʹ-CGGAGTGAACGGCTAAGTTTATC-3ʹ
GAPDH:
Forward: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ
Reverse: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ
U6:
Forward: 5ʹ- GCTTCGGCAG CACATATACTAAAAT-3ʹ
Reverse: 5ʹ- CGCTTCACGAATT TGCGTGTCAT-3ʹ
Cell Counting Kit-8 (CCK-8)
A549 and SPC-A1 cells were seeded into 96-well plates at a density of 1×104 cells/well. After being incubated at 37°C overnight, the cells were transfected with si- circ-ZNF609, circ-ZNF609 or circ-ZNF609 in combination with miR-1224-3p and their matched controls. Ten microliter CCK-8 solution (Apexbio, HOU, USA) was added into each well after cells were incubated for 24, 48, 72 and 96 h. Following 2 h incubation, the absorbance value of each well was measured at 450 nm via a microplate reader.
Colony Formation Assay
A549 and SPC-A1 cells were seeded in 6-well plates at a density of 200 cells/well and incubated at 37°C overnight. Then the cells were transfected with si- circ-ZNF609, circ-ZNF609 or circ-ZNF609 in combination with miR-1224-3p and their matched controls for 14 days. After being fixed with 100% methanol and stained with 0.1% violet, cell colonies were counted using a light microscope.
Cell Cycle Analysis
Cell cycle was analyzed according to manufacturer’s protocols by using a cell cycle detection kit (KeyGENBioTECH, Nanjing, China). Briefly, the concentration of A549 and SPC-A1 cells was adjusted to 1×106 cells/mL, and the cells were fixed with 70% cold ethanol at 4ºC overnight. Next day, the cells were washed with PBS and RNase A was added to the cells. After the cells were stained with 400 μL of propidium iodide, the cell cycle was examined via flow cytometry (BD Biosciences).
Luciferase Reporter Assay
A fragment of circ-ZNF609 and ETV1 3`-UTR with or without mutant binding site of miR-1224-3p was inserted into FL luciferase reporter vector (Obio, Shanghai, China), respectively. A549 and SPC-A1 cells were transfected with miR-1224-3p or control mimics in combination with the above luciferase vectors using Lipofectamine 2000 (Invitrogen). The luciferase activity of cells was determined after 2 days via a Luciferase Reporter Assay Kit (BioVision, SFO, USA). The relative Luciferase activity was normalized to that of the groups transfected with the luciferase reporter vector containing mutant binding site of miR-1224-3p.
Target Prediction Tools
The Circular RNA Interactome online tool (https://circinteractome.nia.nih.gov/) was used to predict the miRNA-binding sites of circ-ZNF609. TargetScan, a freely available online software, was used to predict the possible target gene of miR-1224-3p.
Western Blot
A549 and SPC-A1 cells were lysed by RIPA buffer containing a protease inhibitor (KeyGENBioTECH, Nanjing, China). Then the proteins were separated on 8–10% SDS-PAGE gels, followed by being transferred to PVDF membranes (Merck Millipore, Billerica, MA, USA). First, the membranes were blocked with 5% nonfat milk dissolved in TBST (a mixture of Tris-buffered saline and Tween 20) at room temperature for 1 h. Then the membranes were incubated with primary antibodies against ETV1 (1:500, #27381-1-AP, proteintech, Rosemont, USA) and GAPDH (1:5000, #10494-1-AP, Proteintech, Rosemont, USA) at 4ºC overnight. Next day, the membranes were incubated with the secondary antibodies for 1 h at room temperature. The signals were assessed by chemiluminescence (Millipore, Billerica, MA, USA).
Statistical Analysis
All experiments were repeated at least three times. Results were expressed as the mean ± standard deviation. Student’s t-test and one-way ANOVA were used to analyze the statistical significance for comparisons of two groups and multiple groups, respectively. All analysis was conducted via SPSS 21.0 software (International Business Machines Corp, NY, USA). P<0.05 was considered statistically significant.
Results
Circ-ZNF609 Expression Is Up-Regulated in LUAD
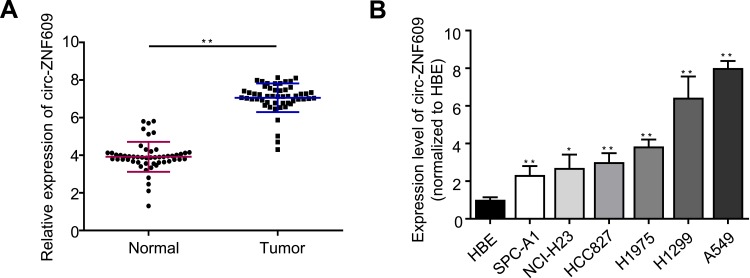
First, in an attempt to investigate whether the expression of circ-ZNF609 was modulated during cancer progression, we used qRT-PCR to examine circ-ZNF609 expression in 52 pairs of LAUD biopsies (52 LAUD tissues and 52 matched adjacent normal tissues). We observed that circ-ZNF609 was profoundly overexpressed in LAUD tissue samples as compared to that in normal tissues (Figure 1A). Consistently, circ-ZNF609 was significantly up-regulated in all LAUD cell lines compared with in bronchial epithelial cell line (HBE) as detected by qRT-PCR (Figure 1B). Of note, A549 and SPC-A1 cells expressed the highest and lowest level of circ-ZNF609, respectively. Hence, A549 and SPC-A1 cell lines were utilized as cell models for the following experiments. Taken together these data reveal that circ-ZNF609 participates in the development and progression of LAUD.
Figure 1.
Circ-ZNF609 expression is up-regulated in LUAD. (A) qRT-PCR quantification of circ-ZNF609 expression in LAUD and adjacent normal tissues. (n=52/group). (B) qRT-PCR analysis for circ-ZNF609 expression in six LAUD cell lines. The levels of circ-ZNF609 expression were normalized to normal human bronchial epithelial HBE cells. Values are expressed in mean ± SD. *p<0.05, **p<0.01.
Downregulation of Circ-ZNF609 Impairs Growth of LUAD Cells
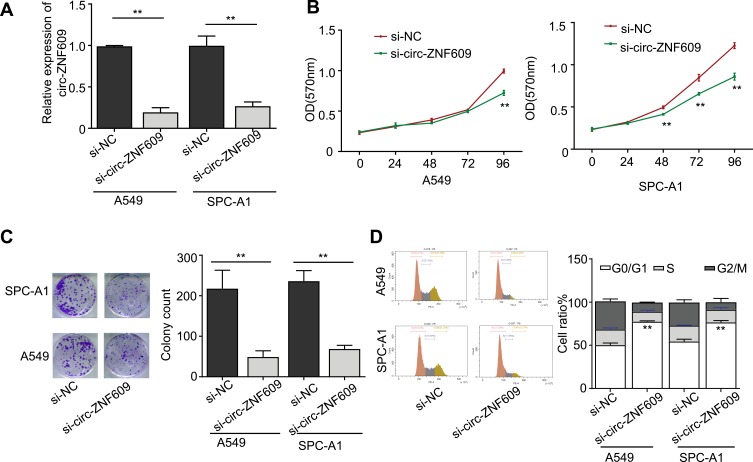
To understand the oncogenic role of circ-ZNF609 in LAUD, we next silenced its expression by utilizing siRNA that against circ-ZNF609. We first confirmed that the siRNA efficiently knocked down circ-ZNF609 expression in both A549 and SPC-A1 cell lines via qRT-PCR (Figure 2A). The CCK-8 assays demonstrated that circ-ZNF609 depletion significantly inhibited the proliferation of A549 cells on day 4 and suppressed SPC-A1 cell proliferation all the time (Figure 2B). Similarly, circ-ZNF609 silencing obviously decreased the colony numbers of both A549 and SPC-A1 cells (Figure 2C). Furthermore, we observed that more A549 and SPC-A1 cells remained in G0/G1 phase after transfection with circ-ZNF609 siRNA as analyzed by flow cytometry (Figure 2D). Collectively, these results suggest that circ-ZNF609 plays an important role in enhancing the proliferation of LAUD cells.
Figure 2.
Downregulation of circ-ZNF609 impairs the growth of LUAD cells. (A) qRT-PCR analysis for circ-ZNF609 expression in si-NC or si-circ-ZNF609 treated A549 and SPC-A1 cells. (B) Effect of si-NC or si-circ-ZNF609 on the cell proliferation of A549 and SPC-A1 cells for 24, 48, 72, 96 h was detected by MTT assay. (C) The proliferation of A549 and SPC-A1 cells treated with si-NC or si-circ-ZNF609 was measured by colony formation assay. (D) The cell-cycle distribution of A549 and SPC-A1 cells treated with si-NC or si-circ-ZNF609 was assessed by flow cytometry. Values are expressed in mean ± SD. **p<0.01.
Circ-ZNF609 Can Sponge miR-1224-3p in LUAD Cells
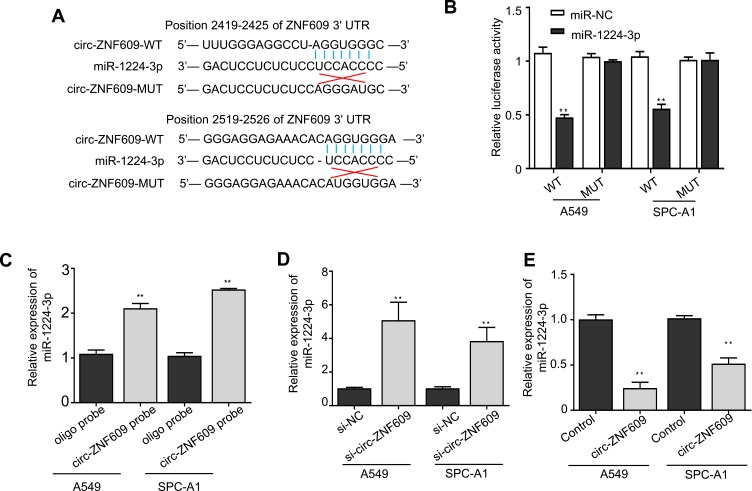
It has been illustrated that circ-ZNF609 can serve as sponges for miRNAs.9 Indeed, we found that circ-ZNF609 have two putative binding sites with miR-1224-3p through searching a web tool named CircInteractome (Figure 3A). Accordingly, we performed luciferase reporter and RNA pull-down assays to confirm the hypothesis. We transfected A549 and SPC-A1 cells with reported plasmids containing the wild-type or mutant circ-ZNF609 sequence in combination with miR-1224-3p mimics or negative controls, and found that the luciferase activity of the circ-ZNF609 WT was remarkably declined by miR-1224-3p overexpression (Figure 3B). In contrast, miR-1224-3p overexpression did not have any effect on the luciferase activity in the cells transfected with circ-ZNF609 MUT (Figure 3B). The results of RNA pull-down verified that circ-ZNF609 did interact with miR-1224-3p (Figure 3C). Consequently, miR-1224-3p expression in A549 and SPC-A1 cells was strongly up-regulated via silencing circ-ZNF609 (Figure 3D). In addition, when circ-ZNF609 was overexpressed in A549 and SPC-A1 cells, miR-1224-3p expression was reduced by at least 45% as determined by qRT-PCR (Figure 3E). In summary, these findings indicate that circ-ZNF609 promotes LAUD cell proliferation possibly via sponging miR-1224-3p.
Figure 3.
Circ-ZNF609 can sponge miR-1224-3p in LUAD cells. (A) Schematic representation of the base pair match between circ-ZNF609-wide-type and miR-1224-3p, and base-pair mismatch between circ-ZNF609-mutant and miR-1224-3p. (B) The luciferase activity of circ-ZNF609-wide-type or -mutant luciferase vector in A549 and SPC-A1 cells transfected with miR-NC or miR-1224-3p. (C) RNA pull down assay in A549 and SPC-A1 cells, and qRT-PCR analysis was used to determine the expression levels of miR-1224-3p pulled down by circ-ZNF609 or oligo probe. (D) qRT-PCR analysis for miR-1224-3p expression in A549 and SPC-A1 cells that were treated with si-NC or si-circ-ZNF609. (E) qRT-PCR analysis for miR-1224-3p expression in control or circ-ZNF609-overexpressing A549 and SPC-A1 cells. Values are expressed in mean ± SD. **p<0.01.
ETV1 Is Identified as a Target of miR-1224-3p in LUAD Cells
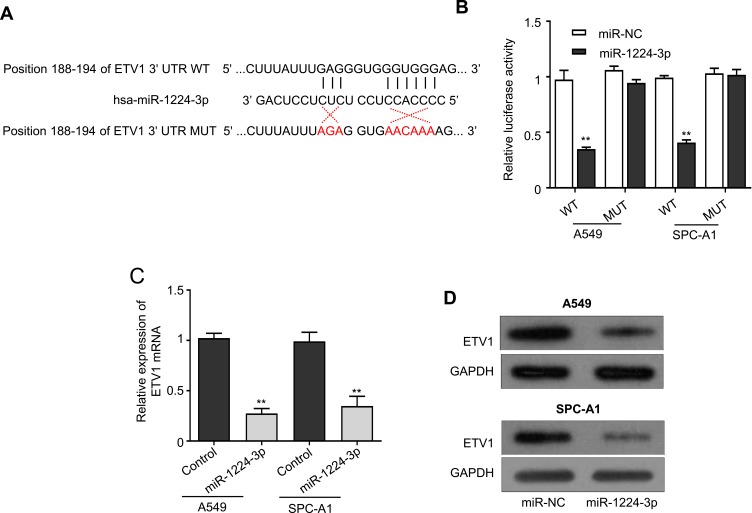
Based on the evidence that circ-ZNF609 sponges miR-1224-3p, thus, we searched for potential target genes of miR-1224-3p by using a web tool named TargetScan. We discovered that ETV1 might be the target of miR-1224-3p (Figure 4A), and then we conducted luciferase reporter assay to confirm this finding. A549 and SPC-A1 cells were transfected with reported plasmids containing the wild-type or mutant ETV1 sequence in combination with miR-1224-3p mimics or negative controls. As expected, the cells cotransfected with ETV1 WT and miR-1224-3p mimics had obviously reduced luciferase activity compared to the control (Figure 4B). Additionally, miR-1224-3p overexpression did not alter the luciferase activity in the cells transfected with ETV1 MUT (Figure 4B). Using qRT-PCR, we showed that ETV1 expression in A549 and SPC-A1 cells was significantly decreased by miR-1224-3p overexpression (Figure 4C). The same results were obtained via Western blot analysis (Figure 4D). All these results demonstrate that ETV1 is the target gene of miR-1224-3p.
Figure 4.
ETV1 is identified as a target of miR-1224-3p in LUAD cells. (A) Schematic representation of the base pair match between miR-1224-3p and EVT1, and base pair mismatch between miR-1224-3p and EVT1. (B) The luciferase activity of EVT1-wide-type or -mutant luciferase vector in A549 and SPC-A1 cells transfected with miR-NC or miR-1224-3p. (C) qRT-PCR analysis for ETV1 expression in control or miR-1224-3p-overexpressing A549 and SPC-A1 cells. (D) The protein expression of ETV1 in control or miR-1224-3p-overexpressing A549 and SPC-A1 cells was evaluated by Western blot. Values are expressed in mean ± SD. **p<0.01.
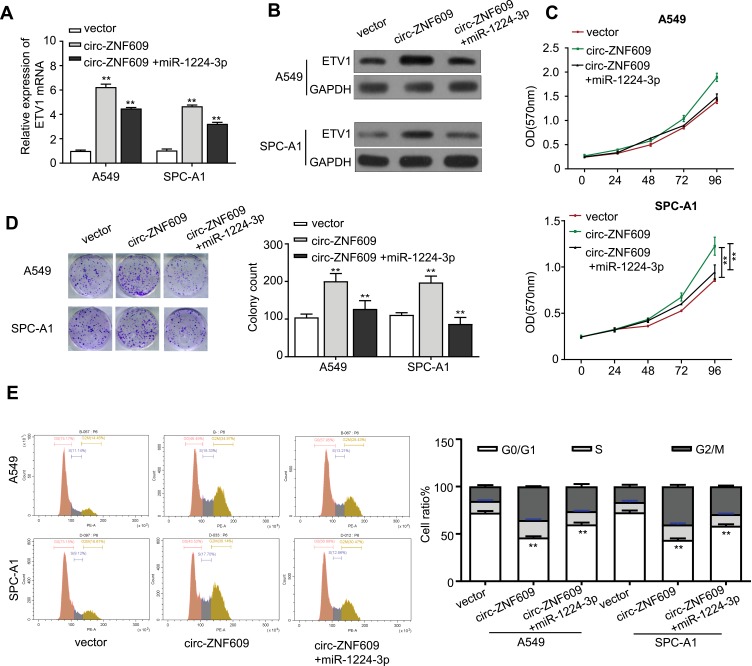
Circ-ZNF609 Mobilizes miR-1224-3p/ETV1 Axis to Promote the Growth of LUAD Cells
ETV1 has been demonstrated to accelerate prostate and gastric cancer progression.13,14 We were wondering whether circ-ZNF609 is capable to regulate EVT1 expression through interacting with miR-1224-3p. We first constructed ETV1 overexpression vector in order to identify the relationship between miR-1224-3p and ETV1. As expected, ETV1 expression in ETV1 overexpressing A549 cells was strongly inhibited by treating with miR-1224-3p mimics, illustrating that miR-1224-3p inhibits ETV1 expression (Supplement Figure 1A). We next constructed circ-ZNF609 overexpression vector, which worked efficiently in A549 and SPC-A1 cells as shown in Supplement Figure 1B, for understanding if circ-ZNF609 mediate ETV1 expression via miR-1224-3p. Both qRT-PCR and Western blot analysis illustrated that EVT1 expression was increased in A549 and SPC-A1 cells via overexpressing circ-ZNF609, and this effect was further neutralized when the cells were transfected with miR-1224-3p mimics (Figure 5A and B). In addition, qRT-PCR and Western blot analysis also demonstrated that EVT1 expression was suppressed in A549 and SPC-A1 cells transfected with si-circ-ZNF609, and this inhibition was abolished by adding miR-1224-3p inhibitor (Supplement Figure 2A and B). Next, we started to clarify whether circ-ZNF609/miR-1224-3p/ETV1 axis influence LAUD growth. Of note, we observed that miR-1224-3p mimics could abolish the increase in the proliferation of A549 and SPC-A1 cells that induced by overexpressing circ-ZNF609 (Figure 5C). The same results were acquired in colony formation assays (Figure 5D). Consistently, flow cytometry analysis showed that the cell population remained in G0/G1 phase was rescued by miR-1224-3p mimics in circ-ZNF609 overexpressing A549 and SPC-A1 cells (Figure 5E). Moreover, miR-1224-3p inhibitor statistically increased the proliferation and colony formation of A549 and SPC-A1 cells transfected with si-circ-ZNF609 (Supplement Figure 2C and D). These data confirmed that circ-ZNF609 acts as a sponge of miR-1224-3p to elevate ETV1 expression, leading to LAUD progression.
Figure 5.
Circ-ZNF609 mobilizes miR-1224-3p/ETV1 axis to promote the growth of LUAD cells. (A) qRT-PCR analysis for EVT1 expression in control or circ-ZNF609-overexpressing A549 and SPC-A1 cells after transfection with miR-1224-3p. (B) Western blot analysis for EVT1 protein expression in control or circ-ZNF609-overexpressing A549 and SPC-A1 cells that were transfected with miR-1224-3p. (C) The cell proliferation of control or circ-ZNF609-overexpressing A549 and SPC-A1 cells after being transfected with control or miR-1224-3p-overexpressing vector for 24, 48, 72, 96 h. (D) Colony formation assay for control or circ-ZNF609-overexpressing A549 and SPC-A1 cells transfected with control or miR-1224-3p-overexpressing vector. (E) Flow cytometry analysis revealed the cell-cycle distribution of control or circ-ZNF609-overexpressing A549 and SPC-A1 cells transfected with control or miR-1224-3p. Values are expressed in mean ± SD. **p<0.01.
Discussion
The accumulating evidence that circRNA serves as sponges for miRNA to modulate cancer progression raised interest in identifying the potential targets for circRNA aimed at developing novel anticancer therapies. Currently, exciting reports have verified that circ-ZNF609 contributes to several kinds of tumor progression and its inhibition effectively impairs the growth of tumor cells.10 However, little is known concerning the role of circ-ZNF609 played in LAUD cancer. In this study, we evaluated whether circ-ZNF609 affects LAUD cancer progression and elucidated the underlying mechanisms of circ-ZNF609 function.
The up-regulated expression of circ-ZNF609 was detected in human breast cancer,9 renal carcinoma,10 colorectal cancer,11 and rhabdomyosarcoma12 in contrast with the corresponding normal tissues. Our experiments were in line with the previous findings that circ-ZNF609 was strongly increased in human LAUD tissue and cell lines compared to normal controls, which provides the evidence that circ-ZNF609 may participate in regulating LAUD progression.
Nowadays, it has not been well clarified the role of circRNA in carcinogenesis. Numerous studies suggest that circRNAs mainly function as miRNA sponges and mediate cell behavior through circRNA-miRNA-mRNA axis.15 For instance, circ-ZNF609 has been shown to enhance malignant behavior of breast cancer cells by increasing p70S6K expression via sponging miR-145-5p.9 In our experiments, we discovered that circ-ZNF609 could sponge miR-1224-3p in LAUD cells. The previous study demonstrated that the expression of miR-1224-3p is closely correlated with the prognosis of bladder cancer.16 Indeed, we illustrated that miR-1224-3p was capable to inhibit ETV1 expression in LAUD cells, which is identified to play an oncogenic role in melanoma,17 prostate cancer18–20 and breast cancer.21 In agreement with previous findings, we verified that circ-ZNF609 promoted ETV1 expression by sponging miR-1224-3p in LAUD cells, and this up-regulation could be retarded when the cells were transfected with miR-1224-3p mimics. More importantly, ectopic expression of miR-1224-3p efficiently reduced the enhanced cell proliferation of LAUD cells induced by circ-ZNF609. Our results demonstrated that circ-ZNF609/miR-1224-3p/ETV1 axis play a critical role in modulating LAUD cell proliferation.
Conclusions
In summary, this study shows that circ-ZNF609 exerts its oncogenic effects in LAUD cells by up-regulating ETV1 expression through act as miR-1224-3p sponge. Furthermore, these data offer a better understanding of the mechanisms of circ-ZNF609 modulating LAUD cancer progression and reveals circ-ZNF609/miR-1224-3p/ETV1 axis can be a potential therapeutic target for LAUD.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wong MCS, Lao XQ, Ho KF, Goggins WB, Tse SLA. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7(1):14300. doi: 10.1038/s41598-017-14513-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myers DJ, Wallen JM. Cancer, Lung Adenocarcinoma. Treasure Island (FL): StatPearlsed; 2019. [Google Scholar]
- 3.Zhang B, Chen M, Jiang N, Shi K, Qian R. A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the proliferation of lung adenocarcinoma. Cancer Biol Ther. 2019;20(8):1127–1135. doi: 10.1080/15384047.2019.1598762 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 5.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 7.Cui X, Wang J, Guo Z, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17(1):123. doi: 10.1186/s12943-018-0877-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Chen RX, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–6330. doi: 10.1158/1078-0432.CCR-18-1270 [DOI] [PubMed] [Google Scholar]
- 9.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression[J]. Hepatology, 2017, 66(4):1151–1164. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 10.Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654. doi: 10.1002/jcp.v234.7 [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Xia J, Yang J, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J BUON. 2018;23(5):1343–1349. [PubMed] [Google Scholar]
- 12.Rossi F, Legnini I, Megiorni F, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38(20):3843–3854. doi: 10.1038/s41388-019-0699-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Zhang L, Ma Z, et al. ETV1 induces epithelial to mesenchymal transition in human gastric cancer cells through the upregulation of Snail expression. Oncol Rep. 2013;30(6):2859–2863. doi: 10.3892/or.2013.2776 [DOI] [PubMed] [Google Scholar]
- 14.Baena E, Shao Z, Linn DE, et al. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev. 2013;27(6):683–698. doi: 10.1101/gad.211011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 16.Miah S, Dudziec E, Drayton RM, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012;107(1):123–128. doi: 10.1038/bjc.2012.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jane-Valbuena J, Widlund HR, Perner S, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70(5):2075–2084. doi: 10.1158/0008-5472.CAN-09-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–599. doi: 10.1038/nature06024 [DOI] [PubMed] [Google Scholar]
- 19.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–624. doi: 10.1038/ng.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41(5):524–526. doi: 10.1038/ng.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Netzer S, Leenders F, Dumont P, Baert JL, de Launoit Y. Ectopic expression of the ets transcription factor ER81 in transgenic mouse mammary gland enhances both urokinase plasminogen activator and stromelysin-1 transcription. Transgenic Res. 2002;11(2):123–131. doi: 10.1023/A:1015248525364 [DOI] [PubMed] [Google Scholar]