Highlights
-
•
Dendrimer-aptamer conjugates.
-
•
Polymer-aptamer conjugates.
-
•
Delivery aptamers.
-
•
Cancer.
Abstract
Aptamers are RNA or DNA oligonucleotides interacting to form unique 3D target conformations with high affinity and specificity, and are emerging as a powerful class of ligands for therapeutic applications. In addition, dendrimers are well-defined nano-sized symmetric polymeric molecules. In this review, we provide an analysis of the use of dendrimers modified with aptamers as nonviral vectors to specifically target tumor cells. Various anticancer agents have been encapsulated with dendrimers complexing with aptamers, including epirubicin, camptothecin, Bcl-xL short hairpin (sh)RNA, and 5-fluorouracil rhodamine-labeled dextran. Other types of polymeric nanoparticle (NP)–aptamer bioconjugates have also been developed and loaded with Pt(IV) derivatives, to target specific tumor cells.
Aptamer-based tools for therapeutic monitoring for precision therapy
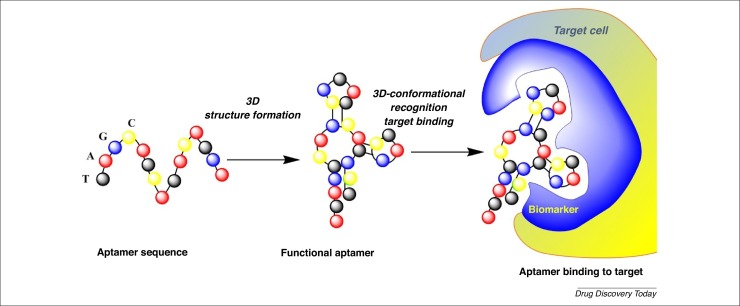
Aptamer comes from the Latin ‘aptare’ meaning to fit together or join, and from the Greek ‘meros’ meaning part. A nucleic acid aptamer is a short chain of artificial oligonucleotides of RNA or single-stranded DNA with a variable region of ∼40–100 nucleotide bases [1]. Their unique, flexible 3D structure, and their high affinity and specificity with ligand-binding capabilities, are strongly related to their sequence. These important characteristics increase their ability to differentiate between targets. In contrast to antibodies, aptamers can bind to functional domains of their cognate target protein (e.g., substrate binding pockets and allosteric sites) (Fig. 1 ) [2]. Aptamers are considered to be ‘smart ligands’ and are often named ‘nucleic acid antibodies’. The binding between aptamers and user-defined targets occurs through electrostatic interactions, which result in their versatility. The strong binding affinity and exceptional specificity induces a nano- or pico-molar dissociation constant (Kd). Their targets vary from small ions, such as Zn2+, and large proteins, such as coagulation factor VIII, to whole cells, viruses, and tissues [3]. It is well documented that the molecular recognition of target molecules by aptamers makes use of their secondary or tertiary structures. Interaction with small molecules (e.g., amino acids) induces a structural switch, changing the internal loop structure of the aptamer, compared with the arrangement when binding with large molecules (e.g., enzymes, regulatory proteins, growth factors, or monoclonal antibodies) [1].
Figure 1.
2D schematic representation of aptamer function.
The most important advantages of aptamers over traditional antibodies in clinical applications include: (i) non-immunogenicity; (ii) high cell/tissue selectivity and penetration; and (iii) numerous potential targets. From a drug discovery point of view, aptamers also have the following advantages over traditional antibodies: (i) thermally stability; (ii) less batch variability; (iii) short production time; and (iv) low cost. Importantly, large quantities of aptamers can be obtained using biochemical synthesis, such as the systematic evolution of ligands by exponential enrichment (SELEX), high-throughput aptamer identification screens (HAPIscreen), and nonequilibrium capillary electrophoresis of equilibrium mixture (NECEEM). One area of interest for the purified protein-based SELEX is the selection of aptamers that recognize cell surface receptors.
Aptamers are rapidly eliminated from the body by renal clearance. The short half-lives of unmodified aptamers in vivo remains one of the major challenges for the development of therapeutic aptamers. Indeed, RNA-based aptamers are prone to hydrolytic breakdown degradation by nucleases. To avoid this degradation, several modifications of RNA aptamers have been performed to improve their bioavailability and the pharmacokinetic parameters: (i) 2-fluoro pyrimidine modifications; (ii) 2-O-methyl nucleotides; (iii) 3-end cap; and (iv) introduction of cholesterol or polyethylene glycol (PEG) as anchor groups [4].
A variety of applications for aptamers have also been developed, including: (i) new drugs; (ii) therapeutic tools; (iii) drug delivery; (iv) diagnosis of disease based on aptamer-based tests, using, for example, ELISAs ; (v) bioimaging; (vi) analytical reagents; (vii) food inspection; and (viii) detection, such as by using optical aptasensors (fluorescence and colorimetric sensors), for the identification and quantification of environmental pollutants, such as heavy metal ions, piezoelectric (mass-dependent) aptasensors for the detection of viruses, bacteria, and toxins, and electrochemical aptasensors for the detection of low-mass molecules and ions [5].
Thus far, only one aptamer-based drug has been approved by the US Food and Drug Administration (FDA): Macugen® (a VEGF165 aptamer or pegaptanib) in 2004 [6]. This aptamer, which strong inhibits angiogenesis by inhibiting the extracellular form of endothelial growth factor, is used to treat adults with wet-form age-related macular degeneration and has been developed by Eyetech.IN/Pfizer. Macugen® is a modified RNA derived from a 2′fluoro pyrimidine aptamer and contains 2′O-methyl purine modifications to enhance its stability against endonucleases. A second type of modification has been performed, including the introduction of 5′-PEG moiety and a 3′dT attached via a 3′–3′ linkage, to induce a good pharmacokinetic profile and strong protection against exonucleases. Macugen® has also been encapsulated in poly(lactic-co-glycolic acid) (PLGA) microspheres as a transscleral delivery nanodevice [7] as well the antivascular endothelial growth factor (VEGF) aptamer EYE001 [8].
The medical applications of aptamers focus on RNA bases, which are more structurally diverse than DNA-based aptamers. For example, on the cytoplasmic membranes of tumor cells, specific aptamers interact with A30, A9, A10, TTA1, and MUC1-specific factors that are intensively overexpressed only on the surface of tumor cell membranes [9] and are used for in vivo molecular imaging and theranostics [10]. Aptamers can also be used as carriers of polymeric NPs, such as polylactides, conjugating antiproliferative drugs, such as Taxol [11]. In addition, aptamers can deliver cisplatin from aptamer-functionalized Pt (IV) prodrug-PLGA–PEG NPs to prostate tumor cells [12]. Of particular interest is the preclinical and clinical development of aptamers for thrombotic events and anticoagulation monitoring [13]. It is well known that PEGylated polymeric NPs have significantly reduced systemic clearance compared with similar NPs without PEG chains [14].
Interestingly, aptamers can be used as specific carriers of therapeutic oligonucleotides, such as small interfering (si)RNAs, miRNAs, shRNAs, and antisense oligonucleotides (ASOs), for diagnosis, precision medicine, and personalized medicine in, for example, cancer treatment [15]. The main aim is to prepare and develop chimera aptamers in which one part is the aptamer, for targeting purposes, linked to a second part that is the therapeutic oligodeoxynucleotides (ODNs), transported by the aptamer.
Dendrimers in nanomedicine
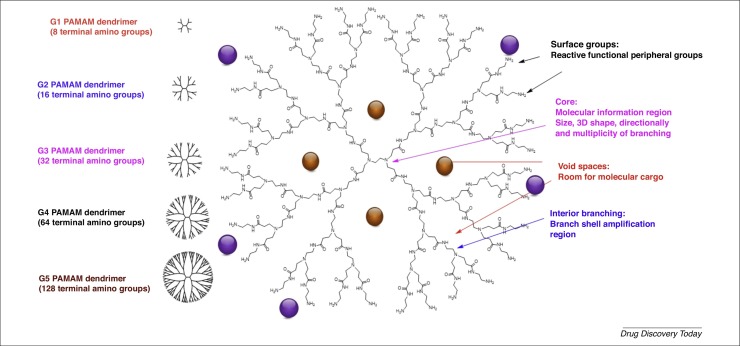
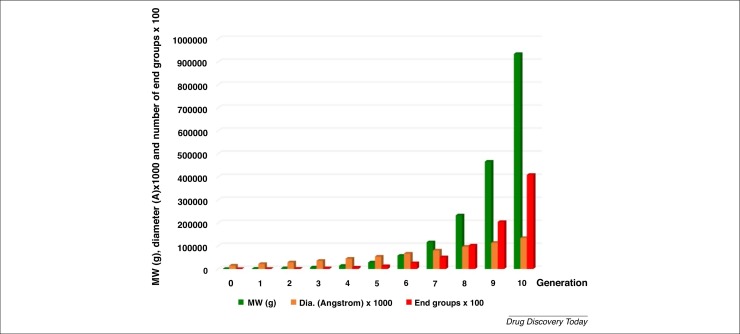
The chemistry of dendrimers was first introduced in 1978 by Fritz Vogtle and coworkers, as well as by Tomalia et al. Numerous biological studies using dendrimers have been carried out, as reviewed elsewhere 16, 17, 18, 19. Dendrimers represent a unique class of polymers, with highly branched 3D architectures, the shape and size of which can be precisely controlled. Size is one of the most important parameters to be evaluated, because it strongly influences clearance kinetics, biodistribution, and in vivo efficacy. NPs >200 nm in size generally induce immune responses and are absorbed by Kupffer cells, which are an important component of the mononuclear phagocytic system. By contrast, smaller NPs (<150 nm) induced rapid clearance from circulation, with NPs <10 nm being easily cleared through the kidney or lymph nodes [14]. Dendrimers display an exponential number of dendritic branches (hydrophobic and hydrophilic moieties) radiating out from a central core unit. Fig. 2 shows the structural units of the well-known poly(amidoamine) (PAMAM) dendrimers, such as core, surface groups, void spaces, and interior branching. Interior layers (generations, Gn, where n is generally from 1 to 6, and more exceptionally up to 12 and recently even 13) comprise regularly repeating branching units attached to the core [20]. As shown in Fig. 2, the dendrimers can encapsulate biologically active compounds (yellow balls) as well as conjugate molecules (violet balls). Dendrimers can also be used as drugs (active per se) [21]. The dendrimer diameter increases linearly whereas the number of surface groups and the molecular weight increases exponentially for each generation (Fig. 3 ).
Figure 2.
Evolution of molecular weight (MW), diameter (Dia.), and number of functional groups on the surface of poly(amidoamine) (PAMAM) dendrimers related to generation as an example.
Figure 3.
2D schematic representation of poly(amidoamine) (PAMAM) dendrimers.
Importantly, the dendrimer collection, based on multiple variations of their chemical structure, allows the monitoring of the chemical properties of core, branches, and surface groups by the introduction of varied chemical groups (basic, acidic, hydrophobic, hydrogen-bonding capability, charges, etc.) [22]. These fine-tuning chemical transformations allow modification of the composition, architecture, and properties of dendrimers, and are key physicochemical properties to improve their in vitro [23] and in vivo [24] behaviors.
Dendrimer–aptamer bioconjugates in nanomedicine
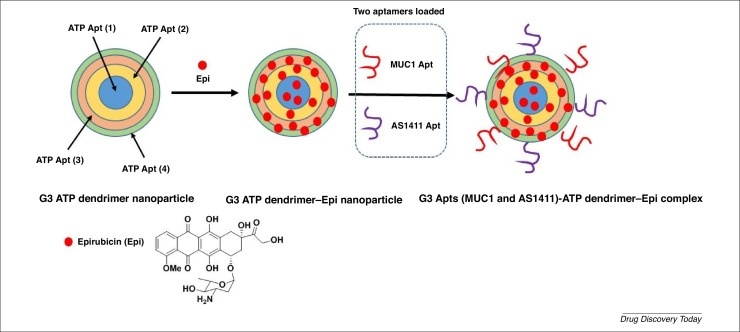
An interesting study by Taghdisi et al. involved the development of aptamer-based dendrimers with a double targeting strategy (two aptamers: MUC1 and AS1411) and encapsulating epirubicin (Epi), an anthracycline drug, acting against cancer cells in vitro and in vivo (Fig. 4 ) [25]. Clinical administration of Epi (e.g., to breast and colon cancers) has been limited by nonspecific interactions inducing cardiotoxicity and bone marrow suppression. Consequently, the development of a new nanosystem, avoiding these adverse effects, was needed. The new strategy is based on the preferential accumulation of chemotherapeutic agents at the cancer site, consequently decreasing the adverse effects. The MUC1 aptamer selectively binds to the transmembrane glycosylated mucin-1 (glycoprotein), which is overexpressed in many tumors, whereas the single-stranded AS1411 aptamer (ATPAS1411, a 26-mer DNA aptamer with the sequence: 5’-Thiol-C6 linker- GGTGGTGGTGGTTGTGGTGGTGGTGG 3’) specifically binds to the nucleus membrane protein nucleolin, which is overexpressed on the plasma membrane of tumor cells. The authors designed and developed an original nanosystem, based on an aptamer–dendrimer conjugate (generation 3), to deliver Epi to MCF-7 breast cancer cells and murine C26 colon carcinoma cells in the presence of two aptamers (MUC1 and AS1411) as targeting agents on the DNA nanostructure surface. The size of the chemically stable G3 aptamer–dendrimer was 36.4 nm. Based on fluorescence intensity and gel retardation assays, the formation of the dendriplex was analyzed, and the maximum quenching ratio was ∼1:25 dendrimer/Epi, corresponding to 25 mol Epi loaded in 1 mol G3 aptamer-dendrimer. The release of Epi from the aptamer-dendrimer-Epi complex was pH dependent, with the more rapid release of Epi in acidic pH (pH 5.5, as in cancerous tissues and lysosomes) versus physiological pH: 63% of Epi was released at pH 5.5 and 15% at pH 7.4 (72 h, 37 °C). The probable protonation of Epi leads to a decrease in the interactions between Epi and dendrimer. Nevertheless, this process is not unique because catalytic enzymes from lysosomes should participate in the disassembly of the dendrimer, inducing the release of Epi.
Figure 4.
2D schematic representation of G3 aptamer(MUC1 and AS1411)–dendrimer–epirubicin (Epi) complex preparation.
Flow cytometry analysis clearly demonstrated that the dendriplex was specifically internalized into tumor cells (MCF-7 and C26) by receptor-mediated endocytosis using the MUC1 aptamer, and by using the AS1411 aptamer nonstandard mechanism, leading to accumulation of the complex in the nucleus. No internalization was observed with the nontargeted Chinese hamster ovary (CHO) cells. Internalization effects were also validated by the results of cell viability assays (MTT) with MCF-7, C26, and CHO cells. The aptamer–dendrimer–Epi dendriplex showed lower toxicity in CHO cells versus Epi alone, with a cell viability of 100% and ∼40%, respectively, whereas the aptamer–dendrimer–Epi complex was more cytotoxic in MCF-7 and C26 tumor cells compared with Epi alone, with a cell viability of ∼30% and ∼80%, respectively (72 h post treatment).
Interestingly, the aptamer–dendrimer–Epi dendriplex showed better in vivo antitumor activity against mice bearing C26 cells versus both Epi alone and MUC1-dendrimer-Epi, and similar in vivo activities compared with a AS1411–dendrimer–Epi complex. These data confirmed that the aptamer–dendrimer–Epi dendriplex modified with two types of aptamer with different targeting sites resulted in good internalization into target cancer cells.
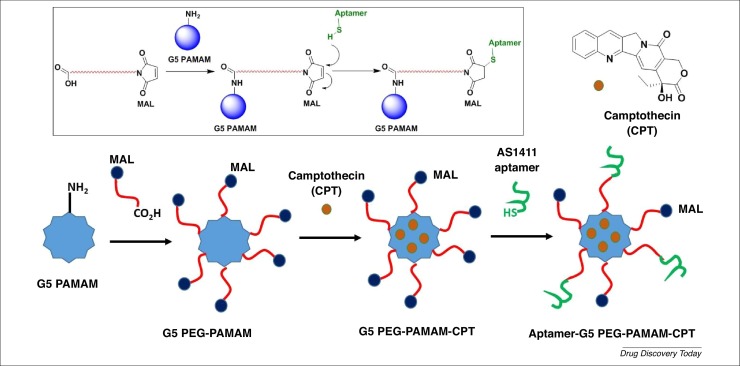
The same authors designed and developed PEGylated G5 PAMAM dendrimers loaded with camptothecin (CPT; a topoisomerase inhibitor used as an anticancer agent) and surface grafted with AS1411 anti-nucleolin aptamers for site-specific targeting to colorectal cancer cells overexpressing nucleolin receptors (Fig. 5 ) [26]. One of the problems of the drug delivery system is the inability to discriminate target cells from nontarget cells, which could result in off-target delivery. Specific drug delivery approaches used cell surface receptors that are overexpressed on the surface of tumor cells. After binding the aptamers to the cell surface receptors, internalization of the aptamer–receptor complexes occurs via a receptor-mediated endocytic pathway process, finally allowing delivery of the loaded drugs. The targeted delivery of chemotherapeutic agents, for instance, could increase their efficacy and reduce their off-target effects. The efficiency of the AS1411-loading and camptothecin-encapsulating content was 8.1% and 93.67%, respectively. The in vitro release assay of camptothecin demonstrated a sustained release of the anticancer agent over 4 days in both phosphate buffer saline (PBS, pH 7.4) and citrate buffer (pH 5.5). Compared with PEG-PAMAM-CPT, the targeted Apt-PEG-PAMAM-CPT formulation displayed a slower CPT release profile because of the decreased diffusion rate of CPT from cavities of dendrimers after the dendrimer periphery was further modified with the aptamer.
Figure 5.
2D schematic representation of the preparation of the aptamer–G5 polyethylene glycol (PEG)-poly(amidoamine) (PAMAM)-camptothecin (CPT) dendrimer.
The biocompatibility of PEG-PAMAM and Apt-PEG-PAMAM dendrimers versus plain PAMAM dendrimers was investigated via hemolytic assays. The PAMAM dendrimer showed significant hemolytic toxicity, whereas PEG-PAMAM and Apt-PEG-PAMAM caused lower hemolytic activities (<5% at 5 mg/mL concentration).
Two nucleolin-positive cell lines (HT29 and C26) and a nucleolin-negative cell line (CHO) were used to evaluate the anticancer activity of Apt-PEG-PAMAM-CPT, PEG-PAMAM-CPT, and free CPT. MTT assays showed that the Apt-PEG-PAMAM-CPT nanodevice displayed higher antiproliferation activity towards nucleolin-positive HT29 and C26 colorectal cancer cells compared with PEG-PAMAM-CPT. Apt-PEG-PAMAM-CPT was internalized after binding to nucleolin protein. The IC50s of CPT, PEG-PAMAM-CPT, and Apt- PEG-PAMAM-CPT against the HT29 cell line were 15 mg/mL, 10 mg/mL, and 3 mg/mL, respectively, whereas, against C26 cell lines, IC50s were 18 mg/mL, 8 mg/mL, and 1.5 mg/mL, respectively. The improvement in the cellular uptake of HT29 and C26 cells with Apt-PEG-PAMAM-CPT was validated using fluorescence microscopy and flow cytometry techniques. In vivo studies in colorectal carcinoma cell line C26 subcutaneously allografting BALB/C mice showed that the Apt- PEG-PAMAM-CPT nanodevice displayed potent antitumor activity and improved mouse survival rates (3 mg/kg i.v. injection twice weekly for 3 weeks). The tumor growth inhibition effect of PEG-PAMAM-CPT was better than that of free CPT, but the most potent effect was obtained with Apt-PEG-PAMAM-CPT. The enhanced tumor-inhibition effects of Apt-PEG-PAMAM-CPT compared with PEG-PAMAM-CPT might result from the targeting effect of the conjugated AS1411 aptamer that can bind to nucleolin proteins on the C26 cells, thus possibly delaying clearance from the tumor site
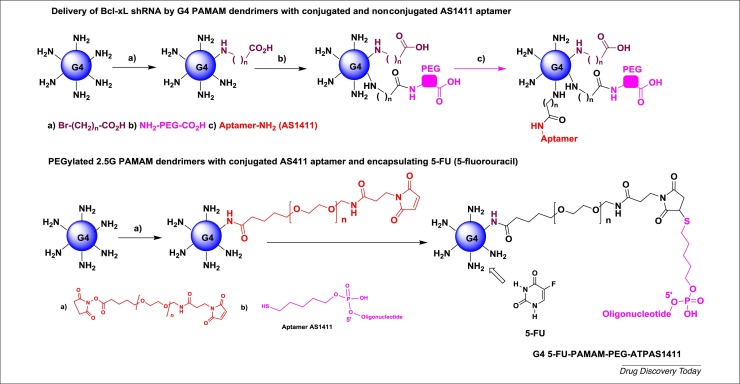
The same authors also reported the delivery of Bcl-xL shRNA by G4 PAMAM dendrimers, which are grafted by aptamers as a targeting ligand (Fig. 6 ) [27]. The G4 PAMAM dendrimer was modified with: (i) 10-bromodecanoic acid (10C) and 10C-PEG chains; (ii) AS1411 aptamer (covalently or noncovalently) as a nucleolin-targeting ligand on target cancer cells, which is overexpressed in various cancer cells; and (iii) a shRNA plasmid for specific knock down of antiapoptotic Bcl-xL protein expression in nucleolin-positive cancer cells. Bcl-xL is expressed in several solid tumors, such as neuronal tumors, adenocarcinoma, bladder cancers, and gastric cancer. The prepared polyplexes showed typical particle sizes of 128–230 nm, with an increased dimension after aptamer conjugations. The charge of all dendriplexes after aptamer conjugations were positive (12.76–19.13 mV). Immunocytochemistry analysis showed the presence of nucleolin receptors on the surface of A549 cells (adenocarcinomic human alveolar basal epithelial cells) but not on L929 cells (negative control). The transfection efficiency of complexes was evaluated in nucleolin-expressing A549 cells, using L929 cells as negative control, and clearly showed strong transfection efficacy with polyplexes bearing covalent or noncovalent aptamers versus nontargeted vectors in A549 cells; noncovalent conjugation also exhibited more efficiency compared with covalent binding. Notably, all the polyplexes prepared showed >70% viability with or without the AS1411 aptamer. Based on Western blot analyses of the expression level of Bcl-xL in A549 cells, its expression was reduced by ∼55% with polyplexes bearing shRNA versus polyplexes bearing scrambled shRNA. The complexes with aptamer only, or with scrambled shRNA, did not significantly change the protein expression level. These results showed the high silencing of Bcl-xL expression by shRNA using G4 PAMAM dendrimers bearing the AS411 aptamer compared with the PAMAM dendrimer without an aptamer. The active polyplex also induced apoptosis in A549 target cells, as validated by flow cytometric analysis.
Figure 6.
2D schematic representation of G4 polyethylene glycol (PEG)-poly(amidoamine) (PAMAM) dendrimers modified with AS1411 aptamer and G4 5-fluorouracil (5-FU)-poly(amidoamine) (PAMAM)- polyethylene glycol (PEG)-ATPAS1411.
Further work by the same team concerned the design and development of functionalized PEGylated 2.5 G PAMAM dendrimers with a conjugated AS411 aptamer and encapsulating 5-fluorouracil (5-FU), a common chemotherapeutic drug (Fig. 6) [28]. This nanoparticle (named PAMAM-PEG-ATPAS1411) increased 5-FU accumulation in cancerous tumors (MKN45 gastric cancer cells) at an effective concentration. The highest encapsulation efficiency was observed with a 1:1 formulation (5-FU/PAMAM ratio). Atomic force microscopy (AFM) topographic images of surface structures confirmed the chemical structure of the PAMAM NPs. The surface charge (zeta potential) of the PAMAM-PEG-ATPAS1411 dendriplex was –13.6 mV. An anti-proliferation assay (MTT test) against HERK293 and MKN45 cells, at a dose of 100 mg/mL, showed that both 5-FU and the PAMAM-PEG-ATPAS1411 complex displayed a percentage viability of ∼30% against HERK293 cells, and of ∼40% and ∼20%, respectively, against MKN45 cells. Good linearity was also observed between the effect and dose. IC50s were ∼10−8 g/mL for both 5-FU and the PAMAM-PEG-ATPAS1411 dendriplex against HERK293 cells, and ∼10−8 g/mL for 5-FU and ∼10−9 g/mL for the PAMAM-PEG-ATPAS1411 complex against MKN45 cells. In vivo imaging studies, using BODIPY fluorescent dye, were performed in mice, and confirmed that the PAMAM-PEG-ATPAS1411 complex was internalized in MKN45 cells as a result of specific nucleolin receptor expression. The linkage of the AS1411 aptamer on the surface of the functionalized PAMAM dendrimer increased the quantity of the conjugated BODIPY dye complex in the vicinity of the tumors.
Polymer–aptamer bioconjugates in nanomedicine
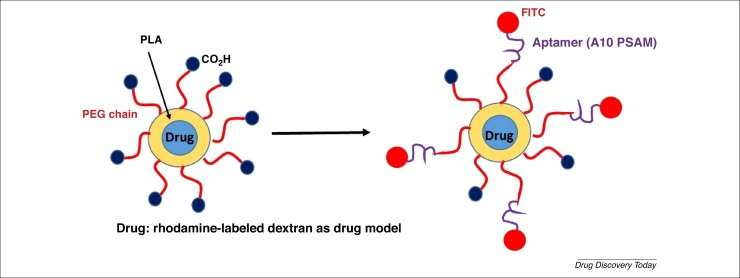
Of particular interest is the design and development of polymeric NP–aptamer bioconjugates targeting prostate-specific membrane antigen (PSMA), which is overexpressed on prostate epithelial cancer cells. These conjugates are based on a two-polymer system: a poly(lactic acid)-block-PEG copolymer with a terminal carboxylic acid functional group (PLAPEG-COOH) (Fig. 7 ) [29]. This NP encapsulated rhodamine-labeled dextran as a model of drug, and conjugating aptamer, A10 PSAM, as a targeting agent. A 5′-FITC-label compound was linked to the end of the aptamers to analyze the specific binding of membrane surface interactions using light transmission or fluorescent microscopy techniques. These negatively surface-charged NPs (−50 mV) minimized the nonspecific interactions with the negatively charged nucleic acid RNA aptamers, whereas the PEG chains enhanced the circulating half-life and contributed to the decreased uptake in nontargeted cells. These microNP–aptamer bioconjugates showed a narrow size distribution. The PEGylated NPs efficiently targeted prostate LNCaP (PSMA+) epithelial cells [with a 77-fold increase in binding versus PC3 (PSMA–) cells as a control]. More importantly, these NPs did not targeted cells where no expression of PSA occurred. The binding of the NPs to LNCaP epithelial cells was easily detectable in an early-stage evaluation. Based on fluorescent microscopy and 3D image reconstruction, the NP–aptamer bioconjugates were strongly internalized into LNCaP epithelial cells compared with PC3 cells, as were the rhodamine-dextran encapsulated NP-aptamer bioconjugates.
Figure 7.
2D schematic representation of polymeric nanoparticle–aptamer bioconjugates encapsulating rhodamine-labeled dextran (as a drug model) bearing A10 PSAM aptamer and FITC.
The same team developed an interesting construction based on Pt(IV)-encapsulated PSMA targeted NPs of biodegradable and noncytotoxic PLGA-PEG-functionalized material [12]. This construction bears aptamer-targeting PSMA (a PSMA aptamer) on its surface, with FITC as a fluorescent probe. The polydispersity of the Pt(IV) cargo (Pt-NP-Apt) increased with the percentage loading of Pt(IV): a percentage loading of 5%, 20%, 50%, and 70% resulted in sizes of 132 nm, 135 nm, 167 nm, and 172 nm, and a polydispersity index of 0.171, 0.205, 0.444, and 0.479, respectively. The controlled release kinetics of the Pt(IV) complex in PBS (pH = 7.4) reached 66% after 24 h, and ∼90% after 45 h. The targeted uptake of Pt-NP-Apt occurred via endocytosis in PSMA-expressing prostate cancer cells, such as LNCaP (PSMA+) cells, through receptor-mediated endocytosis, inducing its accumulation in this type of cell. No accumulation of the Pt(IV) complex was observed in PC3 (PSMA–) cells. Only 12% of the total Pt(IV) was released after 2 h in PBS medium. In vitro cellular cytotoxicity assays showed the high cytotoxicity of Pt-NP-Apt to LNCaP cells (IC50 = 0.03 μM) versus nontargeted nanoparticles (Pt-NP) (IC50 = 0.13 μM), and cisplatin (IC50 = 2.4 μM). The IC50s of Pt-NP and Pt-NP-Apt against PC3 were both ∼0.12 μM, whereas cisplatin showed an IC50 of 0.18 μM. Using the GFP monoclonal antibody R-C18, which is used in the detection of cisplatin-induced Pt adducts in DNA, crosslinks between Pt(IV) and DNA were observed in LNCaP cells.
An additional example was described by Tong et al., who used several techniques concerning one-step co-precipitation synthesis, and the development of tunable polymeric NPs based on A10 aptamer-coated paclitaxel-polylactide (LA) nanoconjugates targeting specific tumor cell lines [30]. The A10 aptamer was identified using the SELEX technique, and targeted PSMA in vitro and in vivo. The fluorescence dye Cy5 was also used to initiate LA polymerization, resulting in Cy5-PLA NPs (Cy5-PLA-NCs), which were then co-precipitated with a PLA-PEG-CO2H polymer, resulting in PLA-PEG-CO2H/Cy5-PLA-NCs NPs. These NPs had a size of 132.8 nm and a polydispersity of 0.031. Condensation of the A10 aptamer with the carboxylic functions of PLA-PEG-CO2H/Cy5-PLA-NCs resulted in aptamer/PLA-PEG-CO2H/Cy5-PLA-NCs (size: 157.1 nm, polydispersity: 0.144). Confocal images showed that the uptake of NPs coated with the aptamer was significantly enhanced versus the uncoated NPs in LNCaP (PSMA+). In the same assay, no difference between coated and uncoated nanoparticles was observed in PC3 cells (PSMA–). The binding of coated NPs were higher than that of uncoated NPs in LNCaP cells, but no difference was observed in PC3 cells.
Multivalent polymer–aptamer versus dendrimer–aptamer conjugates
Compared with peptides (small sizes and molecular weight), a major advantage of using both polymer–aptamer conjugates and dendrimer–aptamer conjugates is to increase the contact area between the ligand and the target receptor, which can enhance the potency and interactions with the target receptor [31]. To increase the biostability, for example against nuclease hydrolysis in human serum, several strategies have been developed based on polymer–aptamer conjugates [32]: (i) 3′ and 5′-biotin–aptamer conjugates [33]; (ii) cholesterol and lipid moiety conjugates [34]; (iii) 5′-end PEGylation conjugates [35]; (iv) modifications of the sugar ring of the aptamers [36]; (v) 4′-oxygen replacement of the aptamers [37]; (vi) modification of the phosphodiester linkage and bases of the aptamers [38]; and (vii) direct conjugation of aptamers to NPs, generally via a spacer or a linker to maintain aptamer-binding activity. Examples of such approaches include: (i) an antinucleolin-specific DNA aptamer (AS1411) linked to DOX-loaded liposomes [39] and a AS1411 aptamer conjugated to liposomes with cisplatin [40]; (ii) an aptamer functionalized on the surface of PLGA-b-PEG-CO2H-based NPs [41]; (iii) quantum dot-MUC1 aptamers [42]; and (iv) thiolated nucleic acids functionalized on gold NPs (AuNPs) [43]. There appear to be some similarities in morphology between multivalent polymer–aptamer and dendrimer–aptamer conjugates, although further research is required to confirm these. A 3′-biotin-modified–DNA aptamer targeting severe acute respiratory syndrome (SARS) coronavirus helicase has also been reported [33].
Concluding remarks and future perspectives
The characteristic features of aptamers enable their development for many applications and in any instance where an antibody is traditionally used. In addition, aptamers can be used in biomarker discovery, drug discovery, diagnostics, and therapeutics, where traditional antibodies cannot. Specialized delivery vehicles, such as dendrimers, coating specific aptamers, represent a novel approach to tackle tumor cells that overexpress the corresponding antigens, such as prostate LNCaP (PSMA+) epithelial cells. Here, we have highlighted and analyzed the use of engineered dendrimers and polymers as nonviral carriers, targeting specific tumors based on the binding of aptamers to their respective biomarkers on the surface of the tumor cells, and the subsequent, controlled release of chemotherapeutic drugs. However, few in vivo studies have been performed using dendrimer–aptamer bioconjugates. Thus, there is a need for additional in vivo studies. Such studies are required to evaluate the biodistribution of these vehicles after systemic administration, as well as their anticancer properties. The selective delivery of a therapeutic dose of a drug is a major advantage of this type of NP. Lastly, more broadly, these powerful nanodrugs could be developed for the specific targeted drug delivery to treat myriad diseases.
Acknowledgments
S.M. and X.S. acknowledge the support of Fundacao para a Ciencia e a Tecnologia (FCT) with Portuguese Government funds through the CQM Strategic Project PEst-OE/ QUI/UI0674/2013, and ARDITI-Agencia Regional parao Desenvolvimento da Investigacao Tecnologia through the project M1420-01-0145-FEDER-000005 – Centro de Quımica da Madeira – CQM+ (Madeira 14-20 Program). J-P.M. thanks the CNRS for financial support. X.S. acknowledges financial support from Shanghai Education Commission through the Shanghai Leading Talents Program, and National Natural Science Foundation of China (81761148028 and21773026). X.S., S.M., and J-P.M. thank the PRC NSFC-CNRS 2019. V.C. acknowledges the support of the Spanish Ministerio de Economía y Competitividad (project SAF2017-89288-R from MINECO/AEI/FEDER/UE) and from JCCM (project SBPLY/19/180501/000067). S.M., V.C., and J-P.M. acknowledge transnational EuroNanoMed III-funded projects including ERANET EuroNanoMed III and the respective funding partner organizations.
Contributor Information
Serge Mignani, Email: serge.mignani@parisdescartes.fr, serge.mignani@staff.uma.pt.
Xangyang Shi, Email: xshi@dhu.edu.cn.
Jean-Pierre Majoral, Email: majoral@lcc-toulouse.fr.
References
- 1.Kudłak B., Wieczerzak M. Aptamer based tools for environmental and therapeutic monitoring: a review of developments, applications, future perspectives. Crit. Rev. Env. Sci. Technol. 2019;7:1064–3389. [Google Scholar]
- 2.Toh S.Y. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015;64:392–403. doi: 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Vahidi H. Challenges to design and develop of DNA aptamers for protein targets. II. Development of the aptameric affinity ligands specific to human plasma coagulation factor VIII using SEC-SELEX. Iran J. Pharm. Res. 2017;16:737–744. [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacevic K.D. Pharmacokinetics, pharmacodynamics and safety of aptamers. Adv. Drug Deliv. Rev. 2018;134:36–50. doi: 10.1016/j.addr.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y. Recent advances in aptamer discovery and applications. Molecules. 2019;24:941. doi: 10.3390/molecules24050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ew N.G., Adamis A.P. Anti-VEGF aptamer (pegaptanib) therapy for ocular vascular diseases. Ann. N. Y. Acad. Sci. 2006;1082:151–171. doi: 10.1196/annals.1348.062. [DOI] [PubMed] [Google Scholar]
- 7.Sapino S. Ocular drug delivery: a special focus on the thermosensitive approach. Nanomaterials. 2019;9:884. doi: 10.3390/nano9060884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karen K.G. Controled delivery of the anti-VEGF aptamer EYE001 with poly(lactic-co-glycolic)acid microspheres. Invest. Ophthalmol. Visual Sci. 2003;44:290–299. doi: 10.1167/iovs.01-1156. [DOI] [PubMed] [Google Scholar]
- 9.Cerchia L., de Franciscis V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010;28:517–525. doi: 10.1016/j.tibtech.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Bouvier-Müller A., Duconge F. Application of aptamers for in vivo molecular imaging and theranostics. Adv. Drug Deliv. Rev. 2018;134:94–106. doi: 10.1016/j.addr.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Tong R. The formulation of aptamer-coated paclitaxel-polylactide nanoconjugates and their targeting to cancer cells. Biomaterials. 2010;31:3043–3053. doi: 10.1016/j.biomaterials.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhara S. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponce A.T., Hong K.L. A mini-review: clinical development and potential of aptamers for thrombotic events treatment and monitoring. Biomedicines. 2019;7:55. doi: 10.3390/biomedicines7030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexis F. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nuzzo S. Potential and challenges of aptamers as specific carriers of therapeutic oligonucleotides for precision medicine in cancer. Cancers. 2019;11:1521. doi: 10.3390/cancers11101521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomalia D.A. Special issue: functional dendrimers. Molecules. 2016;21:1035. doi: 10.3390/molecules21081035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kannan R. Emerging concepts in dendrimer based nanomedicine: from design principles to clinical application. J. Intern. Med. 2014;276:576–617. doi: 10.1111/joim.12280. [DOI] [PubMed] [Google Scholar]
- 18.Chauhan A., Kulhari H., editors. Pharmaceutical Applications of Dendrimers. Elsevier; 2020. [Google Scholar]
- 19.Mignani S. Can dendrimers based nanoparticles fight neurodegenerative diseases? Current situation versus other established approaches. Progr. Polym. Sci. 2017;64:23–51. [Google Scholar]
- 20.Sowinska M., Urbanczyk-Lipkowska Z. Advances in the chemistry of dendrimers. New J. Chem. 2014;38:2168–2203. [Google Scholar]
- 21.Brahmi N.E. Original multivalent copper(II)-conjugated phosphorus dendrimers and corresponding mononuclear copper(II) complexes with antitumoral activities. Mol. Pharm. 2013;10:1459–1464. doi: 10.1021/mp4000184. [DOI] [PubMed] [Google Scholar]
- 22.Mignani S., Majoral J.-P. Dendrimers as macromolecular tools to tackle from colon to brain tumor types: a concise overview. New J. Chem. 2013;37:3337–3357. [Google Scholar]
- 23.Mignani S. Exploration of biomedical dendrimer space based on in vitro physicochemical parameters: key factor analysis (Part 1) Drug Discov. Today. 2019;24:1176–1183. doi: 10.1016/j.drudis.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Mignani S. Exploration of biomedical dendrimer space based on in vivo physicochemical parameters: key factor analysis (Part 2) Drug Discov. Today. 2019;24:1184–1192. doi: 10.1016/j.drudis.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Taghdisi S.M. Double targeting and aptamer-assisted controlled release delivery of epirubicin to cancer cells by aptamers-based dendrimer in vitro and in vivo. Eur. J. Pharm. Biopharm. 2016;102:152–158. doi: 10.1016/j.ejpb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Mona Alibolandia M. Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int. J. Pharm. 2017;519:352–364. doi: 10.1016/j.ijpharm.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 27.Ayatollahia S. Aptamer-targeted delivery of Bcl-xL shRNA using alkyl modified PAMAM dendrimers into lung cancer cells. Int. J. Biochem. Cell Biol. 2017;92:210–217. doi: 10.1016/j.biocel.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Behrooz A.B. Smart Bomb AS1411 aptamer-functionalized/PAMAM dendrimer nanocarriers for targeted drug delivery in the treatment of gastric cancer. Clin. Exp. Pharmacol. Physiol. 2017;44:41–51. doi: 10.1111/1440-1681.12670. [DOI] [PubMed] [Google Scholar]
- 29.Farokhzad O.C. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 30.Rong Tong R. The formulation of aptamer-coated paclitaxel–polylactide nanoconjugates and their targeting to cancer cells. Biomaterials. 2010;31:3043–3053. doi: 10.1016/j.biomaterials.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin J.T. Synthesis of multivalent polymer-aptamer conjugates with enhanced inhibitory potency. Int. J. Nanomed. 2018;13:5249–5253. doi: 10.2147/IJN.S174673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fadwa Odeh F. Aptamers chemistry: chemical modifications and conjugation strategies. Molecules. 2020;25:3. doi: 10.3390/molecules25010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shum K.T. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C.H. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis c virus (HCV) NS5B. Protein. Mol. Ther. Nucleic Acids. 2015;4:e254. doi: 10.1038/mtna.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prodeus A. Targeting the PD-1/PD-L1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers. Mol. Ther. Nucleic Acids. 2015;4:e237. doi: 10.1038/mtna.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruckman J. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165) Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 37.Minakawa N. Investigations toward the selection of fully-modified 40-thioRNA aptamers: Optimization of in vitro transcription steps in the presence of 40-thioNTPs. Bioorg. Med. Chem. 2008;16:9450–9456. doi: 10.1016/j.bmc.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 38.Diafa S. Generation of aptamers with an expanded chemical repertoire. Molecules. 2015;20:16643–16671. doi: 10.3390/molecules200916643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing H. Selective delivery of an anticancer drug with aptamer-functionalized liposomes to breast cancer cells in vitro and in vivo. J. Mater. Chem. B. 2013;1:5288–5297. doi: 10.1039/C3TB20412J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Z. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew. Chem. Int. Ed. Engl. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 41.Alshaer W. Aptamer-guided nanomedicines for anticancer drug delivery. Adv. Drug Deliv. Rev. 2018;134:122–137. doi: 10.1016/j.addr.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Savla R. Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin conjugate for imaging and treatment of cancer. J. Control. Release. 2011;153:16–22. doi: 10.1016/j.jconrel.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 2012;134:7266–7269. doi: 10.1021/ja3014055. [DOI] [PubMed] [Google Scholar]