Dear Editor,
Corona Virus Disease 2019 (COVID-19) has caused a global pandemic in over 100 countries. Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) was identified as the pathogen of COVID-19. Both SARS-CoV-2 and SARS-CoV belong to the subgenus sarbecvirus of Coronaviridae. One major structural protein Spike (S) has attracted great attention because of its function in recognizing receptor, angiotensin-converting enzyme 2 (ACE2). ACE2 binds to the receptor-binding motif (RBM) in the receptor-binding domain (RBD) of SARS-CoV-2 and SARS-CoV. Although ACE2 is distributed in many organs, lung is the major target of both SARS-CoV-2 and SARS-CoV. SARS-CoV-2 tends to infect fewer organs than SARS-CoV. For instance, COVID-19 patients showed less diarrhea than SARS patients.1 The underlying mechanism remains inconclusive.
Integrins are heterodimeric proteins comprising α and β subunits in cell surface. Many integrins recognize Arg-Gly-Asp (RGD) and Lys-Gly-Asp (KGD) motifs which are displayed on the exposed loops of proteins. RGD could associate broader types of integrins such as αVβ1, αVβ3, αVβ5, αVβ6, αVβ8, αⅡbβ3, αMβ2, αLβ2 and α3β1, while KGD-recognizing integrins are restricted to αⅡbβ3, αVβ5, αVβ6 and αVβ8.2 A recent study proposed a proviral role for integrins in SARS-CoV-2 entry.3 In current study, we found RGD/KGD motif presents not only in S protein but also in its receptor ACE2. We suggested inhibitory roles for integrins in the entry of both SARS-CoV-2 and SARS-CoV.
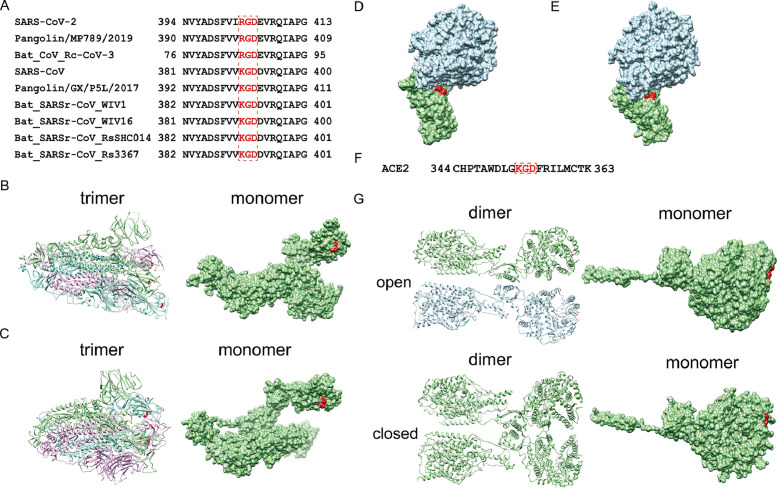
RGD and KGD are two classical integrin-binding motifs. First, we performed sequence analysis of S proteins from SARS-CoV-2 (YP_009724390.1) and SARS-CoV (NP_828851.1). An RGD motif (403–405) was identified in SARS-CoV-2 S protein (Fig. 1 A). In SARS-CoV S protein, there is a KGD motif (390-392) (Fig. 1A). By searching NCBI databases and literatures, we identified similar RGD/KGD motifs in several coronaviruses including Pangolin/MP789/2019,4 Pangolin/GX/P5L/2017,4 Bat_CoV_Rc-CoV-3 (BBJ35999.1), Bat_SARSr-CoV_WIV1 (AGZ48828.1), Bat_SARSr-CoV_WIV16 (ALK02457.1), Bat_SARSr-CoV_RsSHC014 (AGZ48806.1), Bat_SARSr-CoV_Rs3367 (AGZ48818.1). These results suggested that RGD/KGD integrin-binding motif is conserved in several coronaviruses including SARS-CoV-2 and SARS-CoV.
Fig. 1.
Identification of integrin-binding motifs in S proteins from SARS-CoV-2/ SARS-CoV and ACE2 protein. (A) RGD/KGD is present in S protein from SARS-CoV-2, SARS-CoV, pangolin coronaviruses, and some bat coronaviruses. RGD/KGD motif is in red box. (B) RGD is in an exposed loop of SARS-CoV-2 S protein. Left: trimer of SARS-CoV-2 S protein (PDB: 6VSB). Right: monomer of SARS-CoV-2 S protein. RGD motif is colored red. (C) KGD is in an exposed loop of SARS-CoV S protein. Left: trimer of SARS-CoV S protein (PDB: 5XLR). Right: monomer of SARS-CoV S protein. KGD motif is colored red. (D) RGD of SARS-CoV-2 S protein locates near the groove of the complex of ACE2 and SARS-CoV-2 RBD (PDB: 6LZG). Red: RGD motif. Grey: ACE2, Green: SARS-CoV-2 RBD. (E) KGD of SARS-CoV S protein locates near the groove of the complex of ACE2 and SARS-CoV RBD (PDB: 2AJF). Red: KGD motif. Grey: ACE2, Green: SARS-CoV RBD. (F) KGD presents in human ACE2. KGD motif is in red box. (G) KGD locates in an exposed loop of ACE2. Left: dimer of full-length human ACE2 in complex with B0AT1. Right: monomer of human ACE2. Upper: open conformation (PDB: 6M1D). Lower: closed conformation (PDB: 6M18). KGD motif is colored red.
In order to associate with integrin, RGD/KGD motif should be exposed to the surface of the protein. To investigate whether RGD/KGD motif in S proteins from SARS-CoV-2 and SARS-CoV is on the surface, we analyzed the structures of SARS-CoV-2 S trimer (PDB: 6VSB)5 and SARS-CoV S trimer (PDB: 5XLR)6 by Chimera software Ver 1.14. RGD located at the top of protrusions in SARS-CoV-2 S, and KGD is displayed on exposed loop in SARS-CoV S (Fig. 1B and 1C). These results suggested that S proteins are accessible to integrins.
RGD/KGD motif in S protein is close to RBM of S protein. Next, we checked the structure of ACE2 with SARS-CoV-2 S RBD (PDB: 6LZG) and ACE2 with SARS-CoV S RBD (PDB: 2AJF)7 by Chimera software Ver 1.14. RGD/KGD in S protein is near the groove of complex of RBD and ACE2 (Fig. 1D and 1E). If S protein associates with integrin, there would be no space for ACE2 to contact with S.
ACE2 is known to associate with integrin. An RGD sequence (203–205) in ACE2 is partially buried inside the protein and thus not responsible for integrin association.8 We identified a KGD motif in 353–355 of ACE2 (BAB40370.1), which is a key region for binding S protein (Fig. 1F). This KGD motif is on the exposed small loop in the structure of open ACE2 dimer (PDB: 6M1D) and closed ACE2 dimer (PDB: 6M18),9 which indicated that integrin could interact with ACE2 through this motif (Fig. 1G).
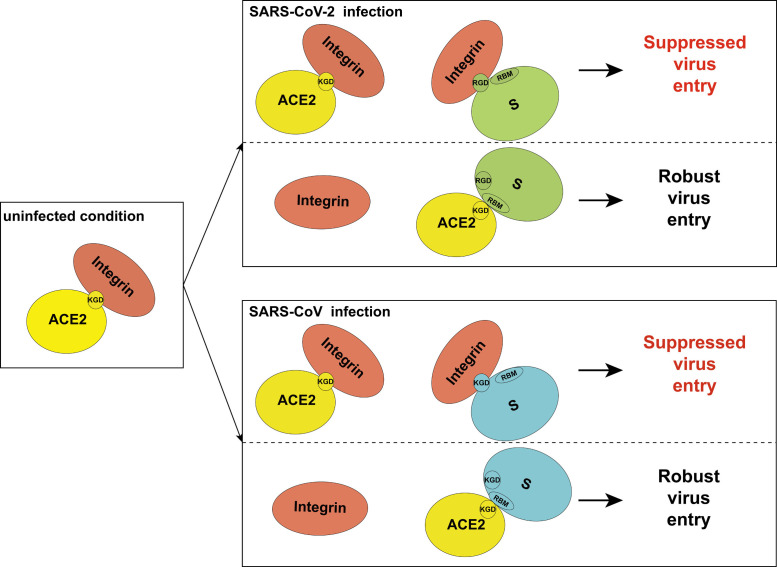
We proposed the following model for the role of integrin in receptor recognition of S protein (Fig. 2 ). Upon SARS-CoV-2 or SARS-CoV infection, integrin could interact with ACE2 and S protein individually. Integrin associates with ACE2 through its KGD motif including K353, which is one of key AAs for S protein recognition. Integrin associates with S protein by its RGD/KGD motif, which would shield the space of RBM for contacting with ACE2. In those scenarios, both ACE2 K353 (inside KGD) and S protein RBM (near RGD/KGD) are shielded by integrin. Thus, ACE2 could not recognize S protein, leading to a suppressed viral entry. In the condition of ACE2 targeting of S protein, integrin no longer blocks ACE2-S interaction, resulting in a robust virus entry.
Fig. 2.
Proposed model for the inhibitory role of integrin in SARS-CoV-2 and SARS-CoV.
In uninfected condition, integrin could associate with ACE2 through KGD motif. Upon SARS-CoV-2 or SARS-CoV infection, integrin could interact with ACE2 (through KGD) and S protein (through RGD/KGD) individually, which will mask the interface between S protein and ACE2. Thus, ACE2 could not recognize S protein, leading to a suppressed viral entry. When ACE2 associates with S protein, a robust virus entry will occur.
Several host proteins were predicted to associate with SARS-CoV-2 S protein.3 , 10 Recently, people reported that the SARS-CoV-2 S protein acquired an RGD integrin-binding motif and claimed that this motif was absent from other coronaviruses.3 They further speculated that RGD acquired by SARS-CoV-2 would promote virus entry by association of integrins, thus enhance the transmission ability. We disagree with that because both RGD and KGD could recognize integrins. The difference is that RGD recognizes more types of integrins than KGD. When integrins bind to S protein, the stereo-hindrance effect will prevent ACE2 targeting by S protein. Moreover, ACE2 contains a KGD integrin-binding motif inside the S-binding region. When integrins bind to S protein, ACE2 targeting by S protein will also be inhibited. Taken together, our model favors an inhibitory role for integrins in virus entry by associating with both S protein and ACE2.
Potential association of S protein and integrins provide mechanistic insights for the pathogenesis of SARS-CoV-2 and SARS-CoV. Because RGD recognized a broader spectrum of integrins than KGD, more integrins could block receptor binding of SARS-CoV-2 S than that of SARS-CoV S. Consequently, SARS-CoV-2 would infect fewer organs than SARS-CoV, which might partially explain why SARS-CoV-2 caused less mortality than SARS-CoV.
In conclusion, we identified an RGD/KGD integrin-binding motif in S proteins from SARS-CoV-2 and SARS-CoV. We also discovered a KGD integrin-binding motif in ACE2. Integrins were predicted to inhibit receptor targeting of S proteins from SARS-CoV-2 and SARS-CoV by shielding both S protein and ACE2. We proposed a previous unappreciated inhibitory role for integrin in virus entry, which will improve our understanding on the virus entry for both SARS-CoV-2 and SARS-CoV.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by grants from National Key Plan for Research and Development of China [2016YFD0500300], National Natural Science Foundation of China [81871663 and 81672035], the Innovation Project of Shandong Academy of Medical Sciences and Academic promotion programme of Shandong First Medical University [2019LJ001]. We thank Dr. Jianxun Qi for sharing the structure of 6LZG.
References
- 1.He Y., Wang Z., Li F., Shi Y. Public health might be endangered by possible prolonged discharge of SARS-CoV-2 in stool. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo Y.J., Chung C.H., Huang T.F. From discovery of snake venom disintegrins to a safer therapeutic antithrombotic agent. Toxins (Basel) 2019;11 doi: 10.3390/toxins11070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177 doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y. 2020. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. doi: 10.1101/2020.02.13.945485 bioRxiv:2020.02.13.945485. [DOI]
- 5.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Clarke N.E., Fisher M.J., Porter K.E., Lambert D.W., Turner A.J. Angiotensin converting enzyme (ACE) and ACE2 bind integrins and ACE2 regulates integrin signalling. PLoS One. 2012;7:e34747. doi: 10.1371/journal.pone.0034747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]