Dear Editor,
Several studies of the new outbreak of COVID-19 patients on clinical, epidemiological, and radiological features have now published [1,2]. Apositive RT-PCR result of the discharged patients was reported by Lan et al. [3], we are now reporting the expanded population data of the re-positive patients in Guangzhou City, China.
Collect the discharged COVID-19 patients' information from former epidemiological investigation of Guangzhou Center for Disease Control and Prevention. Data was included the date of onset, date of conformed diagnose, date of discharge, date of first sampling, and the date of the nucleic acid test returned positive. All the discharged patients were followed the criteria of: (a) temperature returned to normal more than 3 days later, (b) Disappearance of respiratory symptoms, (c) substantially improved acute exudative lesions on chest computed tomography (CT) images, and (d) two consecutive negative nucleic acid tests separated by at least 1 day [4].
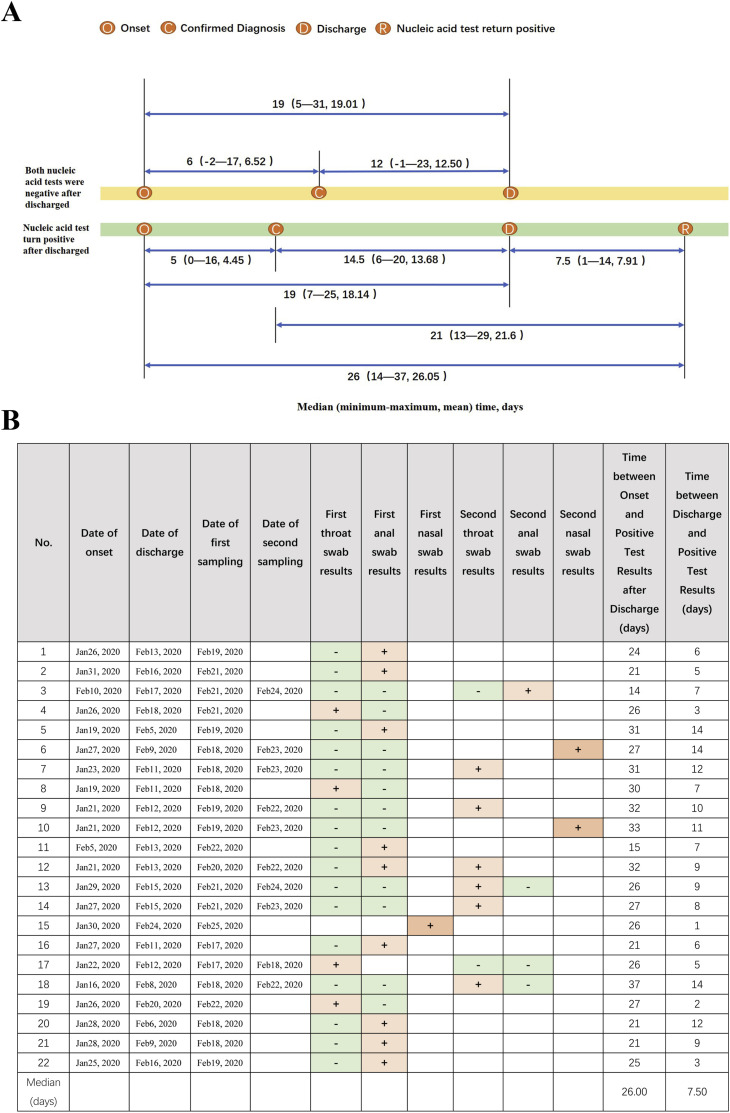
A total of 161 discharged patients of COVID-19 in Guangzhou has retested for SARS-CoV-2, in which 22 patients whose nucleic acid tests were positive accounted for 13.66% (Table S1 in the Supplementary Appendix). As shown in Fig. 1 A, the median time interval between onset of symptom to nucleic acid test return positive after discharge was 26 days (range, 14 to 37; mean, 26.05), in which the longest infection period is 37 days that suggested that the current 14-day medical observation period may be insufficient and needs to be re-evaluated, and the median time interval between the discharge to nucleic acid test return positive was 7.5 days (range, 1 to 14; mean, 7.91) that indicated that the 14-day medical observation period after discharge is an essential measure for controlling epidemic spread.
Fig. 1.
A) Time interval of every two time points among the four time points
Negative group: The median time interval between onset of symptom to diagnosis was 6 days (range,
−
2 to 17; mean, 6.52) and the median time interval between diagnosis to discharge was 12 days (range,
−
1 to 23; mean, 12.50). The median time interval between onset of symptom to discharge was 19 days (range, 5 to 31; mean, 19.01).
Positive group: The median time interval between onset of symptom to diagnosis was 5 days (range, 0 to 16; mean, 4.45). a
nd the median time interval between diagnosis to discharge was 14.5 days (range, 6 to 20; mean, 13.68). The median time interval between onset of symptom to discharge was 19 days (range, 7 to 25; mean, 18.14) and the median time interval between onset of symptom to nucleic acid test turn positive after discharge (the last test was positive) was 26 days (range, 14 to 37; mean, 26.05). The median time interval between the discharge to nucleic acid test turn positive (the last test was positive) was 7.5 days (range, 1 to 14; mean, 7.91) and the median time interval between the diagnosis to nucleic acid test turn positive (the last test was positive) was 21 days (range, 13 to 29; mean, 21.61).
B) RT-PCR results of 22 cases.
The key point that differed the sampling after discharge from the sampling before discharge only including throat swabs and anal swabs is that we increased sampling of nasal swabs. The emergence of 22 discharged patients of return positive suggested that medical institutions should reassess discharge standards and improve sampling methods and types. As listed in Fig. 1B, we selected 3 discharged patients to collect nasal swabs that all were detected to positive of SARS-CoV-2.
Notably, there are two familial clustering cases in the “re-positive” patients.
As previously reported by Zou LR et al. [5], higher viral loads were detected soon after symptom onset, with higher viral loads detected in the nose than in the throat, consequently we suggested that increase nasal swab sampling for SARS-Cov-2 test to reduce false negative rate of nucleic acid test. There were many kinds of specimens collected from one patient, but always only one specimen type was detected for positive of SARS-CoV-2, which indicated that specimen used for nucleic acid test should be collected from multiple body parts before discharge. There are 8 discharged patients tested positive only on the fourth test, including two tests before discharge and two tests after discharge, which shown that relatively high false negative rate was 36.4% (8/22) before the fourth test and suggested that increase the number of tests before discharge. The exact period of infection by far has not been determined, and the knowledge of epidemiological characteristics of COVID-19 were still insufficient so that we need to collect more information to explore.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of. Guangzhou Center for Disease Control and Prevention
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
General Guidance Project of Guangzhou Health and Family Planning Technology 20181A011050(C Xie), Health Industry Scientific Research Project of Gansu Province GSWSKY2018-18(H Zhao), and National Natural Science Foundation of China Grants 81803325(D Wu).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors' contributions
HJ Zhao and ZC Yang designed this study. CJ Xie, JY Lu, D Wu, L Zhang collated the data, and HJ Zhao and ZC Yang discovered and analyzed relevance. ZC Yang and CJ Xie contributed to interpreting the results. HJ Zhao, BQ Rao wrote the manuscript and analyzed the results. ZC Yang revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
None of the authors has any conflict of interest to declare.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2020.101668.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J Am Med Assoc. 2020 doi: 10.1001/jama.2020.1585. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W.J., Ni Z.Y., Hu Y. Clinical characteristics of coronavirus Disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan L., Xu D., Ye G. Positive RT-PCR test results in patients recovered from COVID-19. J Am Med Assoc. 2020 doi: 10.1001/jama.2020.2783. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.China-National-Health-Commission Diagnosis and treatment of 2019-nCoV pneumonia in China (In Chinese) http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml Published February 8, 2020. Accessed March 4, 2020. Web site.
- 5.Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020 doi: 10.1056/NEJMc2001737. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.