Abstract
Antibody fragments for which the sequence is available are suitable for straightforward engineering and expression in both eukaryotic and prokaryotic systems. When produced as fusions with convenient tags, they become reagents which pair their selective binding capacity to an orthogonal function. Several kinds of immunoreagents composed by nanobodies and either large proteins or short sequences have been designed for providing inexpensive ready-to-use biological tools. The possibility to choose among alternative expression strategies is critical because the fusion moieties might require specific conditions for correct folding or post-translational modifications. In the case of nanobody production, the trend is towards simpler but reliable (bacterial) methods that can substitute for more cumbersome processes requiring the use of eukaryotic systems. The use of these will not disappear, but will be restricted to those cases in which the final immunoconstructs must have features that cannot be obtained in prokaryotic cells. At the same time, bacterial expression has evolved from the conventional procedure which considered exclusively the nanobody and nanobody-fusion accumulation in the periplasm. Several reports show the advantage of cytoplasmic expression, surface-display and secretion for at least some applications. Finally, there is an increasing interest to use as a model the short nanobody sequence for the development of in silico methodologies aimed at optimizing the yields, stability and affinity of recombinant antibodies.
Keywords: Nanobodies, Recombinant expression, Fusion immunoreagents, Functionalization strategies, Modeling
Highlights
-
•
There is an increasing request for immunoreagents based on nanobodies.
-
•
The multiplicity of their applications requires constructs with different structural complexity.
-
•
Alternative expression methods are necessary to achieve such structural requirements.
-
•
In silico optimization of nanobody biophysical characteristics becomes more and more reliable.
1. Introduction
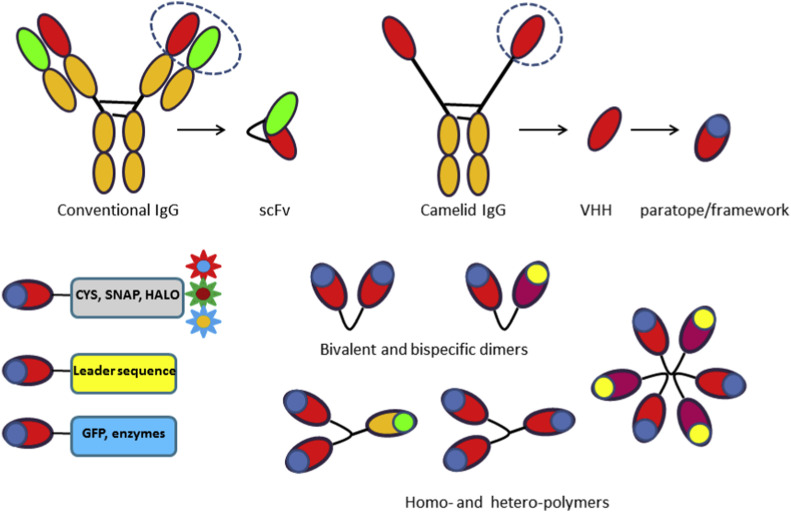
Nanobodies (VHHs) correspond to the heavy-chain variable domain of IgG2 and IgG3 expressed in Camelidae that are devoid of the CH1 domain as well as of the light chain (Fig. 1 ). They represent the smallest antibody fragments (14 kDa) able to preserve the binding affinity and specificity of the original whole antibody and they are appreciated for their structural stability and their simple engineering into reagents suitable for in vitro and in vivo applications [1]. With respect to conventional IgGs (150 kDa), the tiny dimension of nanobody confers the exclusive capacity to bind to cryptic epitopes of viruses [2], a characteristic that at the present could be particularly useful to produce reagents suitable for coronavirus studies. Over the years, it emerged that VHHs are effective crystallography chaperones and molecular tools for protein structural characterization [[3], [4], [5]], convenient carriers for radioisotopes with extremely short half-life to use for in vivo PET/SPECT imaging [[6], [7], [8], [9], [10]], valid immunoreagents for the controlled and oriented functionalization of nanoparticles, nanogels and biosensors [[11], [12], [13], [14]] and that they can be even internalized by mammalian cells when provided with a suitable leader peptide [15]. More recently, their minimal dimension has been particularly appreciated by scientists looking for binders suitable to optimize the performance of super resolution microscopy [[16], [17], [18]] and to create tandem chimeric antigen receptors (CARs) that show higher target specificity due to the possibility of binding simultaneously multiple antigens [19]. Finally, their short sequence makes them the simplest antibody-derived candidate for rational mutagenesis and in silico improvement of biophysical features [[20], [21], [22], [23]]. In parallel to the interest for their technical advantages, the attention for nanobodies has grown exponentially after the expiration of the patents which restricted their use. The consequence has been that lately many new groups explored the field and contributed to its development. The nanobody-related publications, that were only few/year 20 years ago, became tens/year in 2010 and arose to several hundred in 2019, as evidenced by PubMed statistics. This has meant a wider spectrum of VHH applications and the proposal of methodological alternatives at any step of the process that starts from nanobody isolation and proceeds to their production, engineering and development into mature immunoreagents with features useful for their final application [[24], [25], [26], [27]]. For instance, it is the case of nanobodies chosen according to their resistance to physical and chemical conditions or because specific for determined antigen epitopes [28]. Once selected, such clonable molecules can be directly produced as (fusion) immunoreagents with selected functional characteristics, different formats and domain combinations, such as for instance the chromobody with nuclear localization signal used for actin identification by correlative light and electron microscopy [29]. Fusion immunoreagents are either ready-to-use or suitable for controlled custom functionalization with modular chemical partners (Fig. 1) [[30], [31], [32]]. For instance, a tag such as SNAP allows the 1:1 nanobody derivatization with any molecule presenting an O6-benzylguanine group. Advantages and shortcomings of nanobody-based reagents with different structural configurations, binding selectivity and valence have been largely investigated in terms of potency, efficacy and toxicity [[33], [34], [35], [36], [37]]. A paradigmatic example is offered by ALX-0171, an anti-viral therapeutic trimeric nanobody. The reduced mass of the single domain and its recombinant nature enabled the straightforward production of the trivalent reagent which has significantly smaller mass but striking higher neutralization capacity than the monovalent molecule and the commercial humanized monoclonal antibody palivizumab [38]. At the present a large array of vectors is available to produce customized VHHs in both prokaryotic and eukaryotic systems, according to the functional characteristics that they must have to serve their final application. Finally, recombinant nanobodies directed towards different IgG epitopes have been made available recently, providing a valid clonal and simple-to-functionalize alternative to the commercial polyclonal secondary antibodies [39,40].
Fig. 1.
Characteristics of nanobodies and nanobody constructs.
Both heavy- and light-chain variable domains contribute to the antigen-selective binding surface of conventional IgGs, whereas the heavy-chain variable domain of Camelidae IgGs provides alone the complete epitope for the antigen specific recognition. Therefore, the minimal antibody fragments still able to recognize their antigen are the scFv and the VHH, respectively. More exactly, the contact surface of the VHH (paratope) is mostly formed by the amino acids belonging to the three CDRs, whereas the framework residues constitute the conserved structure of the domain, organized in β–sheets. The nanobody single-domain is a recombinant protein that can be fused to minimal (cysteine) or larger tags (SNAP) for controlled, 1:1 functionalization as well as to proteins with their own function, such as fluorescent proteins or enzymes. Such fusion immunoreagents can be coupled to targeting sequences for their selective delivery to desired sub-cellular compartments. Furthermore, nanobodies can be arranged into bi- or multivalent homo- or heteromolecules with other VHHs or other proteins to improve some of their characteristics, such as cell-type specificity, avidity or circulation time, or for combining the target-selectivity of the nanobody with the effector activity of the partners.
2. Nanobody production alternatives
2.1. How to choose between mammalian and bacterial expression systems?
Since VHHs can be usually expressed functional and at high yields in bacteria, their expression in more expensive and demanding systems such as mammalian cells must be justified. It is for instance the case when nanobodies are produced as Fc-fusions to restore the ADCC and CDC effector functions in vivo and to increase their apparent binding affinity by the avidity effect provided by the resulting bivalent molecule. Anti-CXCR4 nanobodies fused to human IgG1 Fc that were expressed and purified from HEK293T cells in suspension specifically induced the ADCC- and CDC-mediate cell death of leukemia cells that overexpressed such receptor but did not affect CXCR4 negative cells [41]. Expi293F cells were used for producing the reconstituted anti-HER1 nanobody-Fc constructs necessary for the fabrication of immunotoxins by means of intein-mediated splicing and assembling with gelonin [42]. Fusions of nanobodies and human IgG1-derived Fc domain have been produced also in CHO cells as immunoreagents suitable for botulinum neurotoxin A protection in vivo. Such constructs were designed to extend the circulation time of the immunoreagents and indeed treated mice remained protected even 14 days after VHH-Fc administration [43]. However, if the fusion of nanobodies with Fc is performed only to provide a tag suitable for in vitro applications, such as binding to Protein A or secondary antibodies, it is possible to produce (not-glycosylated) VHH-Fcs with totally preserved binding capacity in bacteria [44].
A protein that is very often used in combination with antibodies but which is commonly difficult to produce in bacteria is (horseradish) peroxidase. A nanobody-peroxidase fusion would be directly suitable for colorimetric or electrochemical diagnostics. The expression in mammalian cells of such a construct allowed obtaining a chimera in which both moieties were functional and that was successfully used in competitive ELISA [45]. Recently, the search for the production of equivalent immunoreagents in bacteria moved to two promising alternatives. In the first case, an evolved version of soybean ascorbate peroxidase has been fused to nanobodies and the constructs folded successfully into bi-functional reagents when secreted in the bacterial periplasm [46]. In the second, nanobodies preserved their antigen selectivity after being produced in the bacterial cytoplasm fused to the mVirD2 tag. Such tag was then used for binding covalently to a G-quadruplex DNAzyme with peroxidase activity [47].
Nanobodies might require their expression in mammalian cells also when they must act as intrabodies, namely as binders that preserve their functionality even when produced in the reducing environment of the cytoplasm. In this case, intrabody expression in vivo is temporally controlled to induce the binder accumulation at a very specific moment. The applications are multiple. For instance, fluorescent nanobodies can be expressed at a determined physiological phase of the host cells to follow the fate of the corresponding antigens without interfering with their activity and by such a way specifically labeling distinct antigens and subcellular regions [48]. The clear advantage of nanobodies over conventional antibodies is their structural simplicity with a single disulfide bond that in several cases is not obligatory for reaching stable and functional folding. As a consequence, a large share of them can correct fold in the cytoplasm [49] and is not bound to pass through the cellular secretion pathway to achieve their native conformation. This condition has been exploited to design intrabodies that compete for the binding to a specific epitope of their antigens with other molecules present in the cells [50] or immunoprecipitate their target to impair pathological processes of aggregation [51]. The possibility to use intrabodies to selectively modify the activity of a target protein for studying its function in vivo in eukaryotic cells has been achieved by tagging the target proteins and using them in combination with nanobodies that recognize the tag [52]. Anti-tag nanobodies with intrabody features have been successfully exploited to induce visualization, degradation, relocalization, trapping and modification of the protein of interest [53]. The deGradFP system exploits an anti-GFP-Fbox fusion nanobody that promotes the ubiquitination of the bound antigen and its consequent degradation mediated by the proteasome machinery. It was initially optimized for flies [54] but the concept has been recently adapted and extended to other model organisms such as C. elegans and zebrafish and to perform ultrastructural localization of protein interactions [[55], [56], [57], [58]]. It is expected that in the future protein-specific nanobodies will substitute the anti-tag binders because in such a way more flexible multi-dimensional analyses will be possible. However, despite the fact that there is an increasing number of nanobodies specific for human proteins, the community waits for similar reagents suitable for relevant proteins of C. elegans and zebrafish.
Cytoplasmic nanobody expression in mammalian cells has been also exploited for understanding what epitope specificity an antibody must have to neutralize a virus in its physiological environment. Specifically, a nanobody against the Rev protein of HIV was stably expressed in mammalian cell lines and its capacity to block the virus replication was evaluated in the presence of different natural Rev variants [59]. It was therefore possible to map the critical residues involved in the nanobody-antigen contact and predict the potential efficacy of the nanobody for the major HIV-1 subtypes. Recombinant expression of eukaryotic proteins in mammalian cells is also meaningful when it is necessary to obtain optimally folded molecules which underwent all the physiologically relevant post-translational modifications. To monitor their expression and simplify their purification, such proteins were fused to the dual-function tag YFP and then immunopurified by means of anti-YFP nanobodies [60]. Immunopurification of recombinant antigens was successfully performed also by co-expression of their specific nanobodies in the same cells in both, bacterial cytoplasm and periplasm, followed by purification of the complex [61]. There is no report demonstrating the possibility to apply this strategy to mammalian cells but mammalian antigens were purified by immunoaffinity chromatography using nanobodies [62]. Mammalian expression was also instrumental for producing recombinant extracellular vesicles displaying nanobodies specific for tumor biomarkers [63,64]. Such modified vesicles could enable the selective delivery of therapeutic cargos to target cells expressing nanobody-recognized antigens [65].
2.2. Other eukaryotic systems
Pichia pastoris and several plants have been proposed as alternative eukaryotic biofactories for the production of recombinant nanobodies [66,67]. Despite the feasibility demonstration, these efforts did not succeed in convincing the community about the advantage of using such organisms instead of the conventional ones. Apart from the necessity of setting new ad hoc infrastructures, new approaches require protocol optimization and troubleshooting capacity, as underlined by the report indicating that P. pastoris strains are prone to mis-incorporate amino acids under specific fermentation conditions [68]. Pichia has been successfully used to produce a nanobody and its corresponding human Fc-fusion that target the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) receptor-binding domain [69]. Both constructs are characterized by a strong neutralizing activity but, unluckily, the authors’ claim concerning the economic reason for choosing this expression system was not supported by experimental data and comparison with alternative methods. A more relevant example was offered by the recombinant production of the structurally complex hybrid antibody composed by camelid VHH and IgA-Fc domain. Authors looked for alternative to bacteria and Pichia was preferred to plant-based expression as more reliable to yield reagents with good protective effect against gastrointestinal infections in piglets [70].
Innovative strategies exploit nanobodies for modifying in vivo conditions in plants. As already mentioned above, the same therapeutic efficacy would be difficult to obtain by using conventional antibodies that are larger and more prone to fold incorrectly. A nanobody directed against the grapevine fanleaf virus and stably expressed in N. benthamiana and grapevine rootstock, was able to neutralize its target, conferring resistance to the virus-induced degenerative disease [71].
2.3. Bacterial periplasm
The most conventional method for producing recombinant nanobodies is promoting their secretion in the E. coli periplasm. The oxidizing conditions of this sub-cellular compartment favor the formation of the disulfide bond(s) that stabilize(s) the nanobody structure. The folded binders are then usually recovered in the supernatant after a step of osmotic shock performed to permeabilize the bacterial outer membrane. Heat incubation of the supernatant has been successfully applied to purify the relatively thermal-resistant VHHs after precipitation of the more temperature-sensitive E. coli proteins [72] but otherwise affinity purification represents the standard [73]. Classical metal affinity chromatography was paired to maltose binding affinity purification to improve the purity of the recovered nanobodies [74] and streptavidin-mutein affinity chromatography was applied to recover anti-EGFR VHH-Avi tag fusions biotinylated in vivo [75]. This approach enabled also to obtain both mono and tetravalent nanobodies by coupling the mono-biotinylated VHH with wild type streptavidin. The comparison of the functional effect of such binders demonstrated that the tetravalent form was more effective in controlling tumor cell proliferation. Periplasmic produced biotinylated nanobodies were exploited for the functionalization of microbubbles to use as ultrasound contrast agent [76], for the diagnostic of H3N2 and swine influenza, and for the identification of ochratoxin A contaminations in cereals [[77], [78], [79], [80]]. Periplasmic expression was also successfully exploited to produce nanobodies suitable for reacting with maleimide fluorophores [81], fusions between VHHs and nanoluciferase to use as bioluminescent immunoreagents [82], for recovering correct-folded nanobody-intein fusion proteins suitable for thioester reactions and finally for the fabrication of immunomicelles [83]. This expression route was also preferred for producing a hybrid molecule in which an anti-lysozyme nanobody was inserted into a solvent-exposed loop of β–lactamase [84]. Both moieties conserved their function after purification.
Periplasmic accumulation of recombinant proteins has two major drawbacks that can lead to protein aggregation and low yields. The first is due to the presence of incomplete chaperone machinery that can impair correct folding at high expression rate and the second is the consequence of the limited volume of the compartment which can lead to overcrowding. Alternative production of nanobodies and nanobody-fusion constructs has been successfully demonstrated in bacterial cytoplasm [85] and by using secretion routs [86,87] (see sections 5 and 6).
2.4. Cell display
Nanobodies fold into their native and functional structure once secreted at the surface of bacterial or yeast cells and nanobody display have been used for both screening purposes and for preparing whole-cell biosensors [[88], [89], [90], [91]]. The advantages represented by displaying functional macromolecules on cell surface are multiple and have been largely exploited for several classes of proteins, in particular enzymes [92]. There is no necessity to purify the recombinant binders, no engineering is required to link them to the surface and they are already oriented outwards. The bacteria can be directly spotted on a surface and be used as immunocapture elements of biosensors or as both the capture and detection regents of an ELISA test [87,93]. As an alternative, their capacity of agglutinating in solution can be exploited to quantify the antigens present in liquid samples [94]. The system is therefore faster, less expensive and simpler to implement than the conventional protocol that foresees protein purification and functionalization before immobilization. Despite the impossibility to quantify exactly the nanobody concentration in an E. coli sample, the binder amount can be tuned by controlling the level of VHH expression [94]. Nanobody displaying bacteria can co-express cytoplasmic fluorescent proteins for obtaining colored living immunoreagents suitable for light microscopy and flow cytometry applications [87].
Nanobodies were also displayed on bacteria with the aim of facilitating the cell targeting in vivo. Specifically, anti-CD20 nanobodies exposed on Salmonella cells increased their selective accumulation in CD-20 positive tumors [95] with consequent decreased growth of such solid tumors. In a proof-of-principle experiment, nanobodies were anchored on E. coli by means of the SpyTag/SpyCatcher ligation system. In this case bacteria displayed the SpyTag that was available for binding to SpyCatcher-fused anti-GFP antibodies [96]. Of course, the method can be expanded by selecting nanobodies specific for other antigens and could be for instance used to decorate Virus Like-Particles with nanobodies suitable for improving their targeted delivery [97]. Nanobody display was exploited also for preparing Escherichia coli modified as tunable modular platforms for studying cell-cell adhesion and multicellular self-assembly [98]. The approach enabled the inducible and selective adhesion of orthogonal antigen libraries and to design well-defined morphologies useful to study the organization of multicellular structures.
Yeast surface display has been largely used also for panning antibody fragment libraries because enables the sorting of all the selected clones by flow-cytometry. This represents a huge advantage over phage display that requires manual screening of the clones recovered after panning. An interesting recent proposal couples nanobody selection to the possibility to release them directly from the yeast cells after reduction, induced by DTT addition, of the disulfide bond that links the displaying proteins to VHHs. Since in this system nanobodies are expressed fused to a tag compatible with multiple alternative labeling, they can be first biotinylated and then directly adsorbed on streptavidin, for instance for SPR analysis, without necessity to subclone, express and purify them [99].
Also mammalian cell display of nanobodies has found interesting applications, such as their use for the modification of T cells with chimeric antigen receptors (CARs) suitable for cancer immunotherapy. VHHs are preferred to scFvs because are considered less immunogenic, more stable and compact (nanoCARs) [19,100,101].
2.5. Bacterial cytoplasm
The bacterial cytoplasm is a reducing environment that impairs the formation of the nanobody internal disulfide bond(s). However, several VHHs can fold into a functional structure independently on the presence of such disulfide bond(s) [49,102,103] and disulfide bond-independent nanobodies (intrabodies) were successfully produced in E. coli cytoplasm even when fused to large partners such cucurmosin toxin [104]. Since the preliminary results indicated that their anti-Bacilus anthracis nanobodies were stable and functional intrabodies, Anderson et al. [103] expressed them fused to beta galactosidase directly in the cytoplasm of Tuner (DE3) E. coli. Shibuya et al. [105] managed to co-express in bacterial cytoplasm nanobodies fused to complementary split intein moieties for obtaining bispecific VHH constructs by in-cell trans-splicing whereas nanobodies engineered with extra cysteines or ascorbate peroxidase were functionally recovered after production in NEB express F’ cells [39]. Another nanobody with intrabody features has been generated to bind to the ALFA-tag and was effectively used to detect in vivo interactions and pull-down experiments [106]. Intrabodies were also systematically searched to develop therapeutics reagents with the capacity to recognize and block the processes of protein aggregation involved in the progression of neurodegenerative disorders and VHHs with such characteristics and specificity for α–synuclein and prion proteins were identified [107]. For the selective identification of α–synuclein intrabodies, the ability of the E. coli twin-arginine translocation (Tat) system was also exploited to non-covalently secrete protein complexes to the periplasm (FLI-TRAP - functional ligand-binding identification by Tat-based recognition of associating proteins). The resulting nanobodies could be expressed as functional binders in cell cytoplasm [108].
However, the reducing cytoplasmic conditions remain a limiting factor for the folding of the majority of VHHs [109]. An approach to overcome the shortcoming is to express the nanobodies in the cytoplasm of mutant E. coli strains that provide an oxidizing environment. This strategy became available 30 years ago, applied to different recombinant proteins which require disulfide bonds to fold into their native structure and progressively improved with the introduction of more efficient strains such as Rosetta-gami B (DE3) or SHuffle T7 cells. These have been used for the cytoplasmic production of naked nanobodies [110,111] and of their fusion variants containing at their C-terminus an intein-chitin domain suitable for site-specific alkyne functionalization that requires reducing conditions for its functionality [109,112,113]. In one case it was confirmed that the binding properties of the non-modified nanobodies produced in the periplasm were preserved when the same binders were expressed as fusion reagents in the cytoplasm of Shuffle T7 cells [109]. A different approach relies on the use of wild type bacteria with reducing cytoplasm but the parallel overexpression, together with the nanobody of interest, of a sulfhydryl oxidase and DsbC isomerase. Such procedure enabled to produce not only unmodified nanobodies but also more demanding molecules, such as nanobody fusions with GFP and Fc-domains, with no decrease of the binding capacity [44,85].
2.6. Secretion pathway and refolding from inclusion bodies
Secretion of recombinant proteins is a method which enables to recover the targets from the culture media where the amount of contaminants is significantly reduced. While lysis is not necessary, a step of centrifugation or filtration must be considered and the larger volumes require longer loading time during chromatography. Hemolysin secretion pathway was successfully exploited to secrete functional nanobodies directly in the culture media [86]. However, the low obtained yields question the overall advantage of the methodology. Another secretion strategy enabled the recovery of biotinylated nanobodies fused to the AviTag but also in this case the potential advantage of the procedure was not discussed [114]. In contrast, secretion was clearly meaningful when used as an effective pathway to deliver in vivo neutralizing nanobodies produced by Lactobacillus that should reach the target rotavirus in the digestive tract of piglets [115,116].
Inclusion bodies have been recognized for a long time as convenient sources of relatively homogeneous recombinant proteins. The structural characteristics of nanobodies would indicate that refolding protocols should be relatively simple to optimize. Nevertheless, the approach has not been extensively used for nanobodies, probably because a relevant part of them fold correctly as intrabodies and most of the others can be conveniently obtained by means of the procedures described in the previous sections. Elevated refolding yields have been reported for nanobodies expressed as inclusion bodies in bacterial cytoplasm and metal affinity purified after solubilization in urea but their structural and functional integrity was not assessed [117]. A more accurate work compared the periplasmic yields of soluble nanobodies with the amounts recovered by refolding of the same binders from inclusion bodies accumulated in the cytoplasm. In terms of functional binders, the best refolding protocol and the periplasmic expression provided similar yields [118]. Other authors, who tested a higher number of refolding combinations, succeeded in increasing significantly the yield of soluble and functional nanobodies recovered from inclusion bodies [110]. The binding characteristics of the refolded nanobodies were confirmed by comparison with those measured using nanobodies recovered from the soluble fraction.
2.7. In silico optimization
The short sequence of nanobodies (120–130 residues) critically reduces the calculation time necessary for rational in silico optimization of their biophysical characteristics compared to the effort that would be necessary to model larger immunomolecules. Furthermore, nanobodies are suitable for both X-ray crystallography and NMR analyses, therefore several structures are now available for simulating the 3D conformation of new candidates. Recently, the structural information has been also used to understand the specificities of the binding patterns existing between antigens and VHHs [119,120]. This fortunate combination of structural data availability and short sequence stimulated the search for modeling approaches able to suggest mutants performing better than the nanobodies initially isolated by biopanning [121]. Unluckily, a direct comparison between the results obtained with conventional wet-lab approaches (based on random mutagenesis of the initial candidates followed by more stringent in vitro selection steps) and in silico modeling has not yet reported but it would be interesting to evaluate the necessary efforts, resources and achievements specific of the two alternatives. Considering what has been already accomplished with conventional antibodies [122], the available data suggest that a smart conventional approach can still assure results that no in silico system has provided so far [39] but algorithms become progressively more competitive. The following examples show some encouraging results of in silico protocols that follow different strategies to anticipate the potential effect of specific (multiple) single mutations. For instance, a successful computational affinity maturation strategy was based on the combination of Rational Mutation Hotspots Design Protocol and Assisted Design of Antibody and Protein Therapeutics [123] applied to the 3D homology model of the target VHH [124]. Molecular dynamics simulation provided single mutation candidates and the successive combinations of four single mutations resulted in a mutant with 87.4-fold affinity improvement. There are two significant particularities in this result. The first is that the affinity improvement reached the low nanomolar range (K D changed from 278 to 3.2 nM) and the second that such positive effect was associated with an increase of the thermal stability of the mutant with respect to the original nanobody (+7.36 °C), whereas the two parameters were inversely correlated in other reports dealing with similar projects [123,125]. In another case of in silico nanobody affinity maturation, Mahajan and al [108]. Identified a key single mutation by using a combination of replica exchange molecular dynamics (REMD), umbrella sampling, and weighted histogram method together with numerical techniques. This mutation enabled to improve the affinity of the nanobody for its antigen by more than an order of magnitude despite the absence of high-resolution structures of the nanobody, of its α-sin antigen, and of their complex. Simulations could be also exploited to explain the reasons for which specific mutations resulted in unstable nanobodies and in particular indicated that colloidal aggregation can develop even originating from single molecules which fold into stable conformations if their surfaces promote promiscuous interactions [21].
2.8. Practical advice for nanobody expression and purification
Any lab has probably elaborated the nanobody production strategy that is optimal for its necessities. In our lab we constantly modify the protocols trying to update them according to our new observations and the ever changing technical options. The reasons of our choices will be briefly described to serve as a starting point for setting customized methodologies.
When the sequence of a nanobody is available after panning, its expression should be planned considering the final application of the resulting construct. We developed and validated a large array of modular vectors based on pET vector scaffolds that share: i) a conserved cloning site for the nanobody sequence (NcoI/NotI); ii) a further cloning cassette for inserting a “functional tag” such as fluorescent proteins, Avitag, SNAP, free cysteine, SpyTag …; iii) a poly-His tag for affinity purification [13,44,47,62,[126], [127], [128], [129], [130], [131]]. This concept enables to exchange the modules and adapt existing constructs to design new vector versions. Large tags such as SNAP or alkaline phosphatase can slightly affect the binding capacity of the fused nanobodies, whereas small tags (free cysteine, C-tag) usually do not [44,126,128]. Both subcloning from the phagemid and the use of synthetic genes are suitable. This last option is compulsory when the nanobody sequence has been identified by search in a database but can be useful also for the several sequences issued from panning that would require mutagenesis because possess for instance amber codons, as already described elsewhere [132]. However, we noticed that sometimes the yields of the same construct, from “natural source” (i.e. the phage library) and synthetic, were significantly different. In particular, synthetic genes performed worse than original sequences. Unluckily, this is an observation based on few cases and to our knowledge there is no publication on the subject. We can only comment that the codon optimization that the synthesis services implement for adapting the construct to E. coli expression (we have no experience with other expression systems) and to synthesis process could be detrimental for the expression step, maybe because increasing the translation step to a level overwhelming the folding capacity of the cell.
The choice of where and how to express the nanobody fusions has been largely discussed in this review. We opted for the bacterial cytoplasmic accumulation in the presence of sulfhydryl oxidase and DsbC co-expression because it demonstrated being suitable also for large constructs with multiple disulfide bonds such as Fc-VHHs or for proteins that do not fold correctly in the periplasm (several fluorescent proteins) [44]. However, if the lab setting would allow the option, we would suggest parallel small-scale expression from different vectors (also periplasmic) for comparing yields and functionality.
3. Conclusions
The expression of nanobodies and, more and more frequently, nanobodies fused to tags offering orthogonal functions, becomes constantly more diversified [32,130]. This is necessary to obtain reagents that possess the highly differentiated features necessary for fulfilling the always new final application requirements and consequently their production relies on alternative expression conditions [47]. Furthermore, there is an increasing awareness that nanobodies are more differentiated in their structure than initially thought. Specifically, they do not possess a single highly uniform paratope shape but rather may build it combining a large spectrum of different 3D conformations which can involve also the framework residues and confer them several alternative surfaces for interacting with the antigens [119,120,133]. Despite the lack of experimental reports, it can be anticipated that the folding requirements of the different VHH sub-types might be variable as well and, consequently, reference structures used for modeling must be selected with accuracy. Altogether, it is probable that a larger, rather than a smaller, number of expression systems will be necessary to achieve the production of the more and more differentiated nanobody-based immunoreagents necessary to fulfill the future requirements in terms of functional reliability.
Acknowledgements
This work was financially supported by the grants ARRS/N4-0046 and ARRS/J4-9322 provided by the Javna agencija za raziskovalno dejavnost Republike Slovenije.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pep.2020.105645.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 2.Weiss R.A., Verrips C.T. Nanobodies that neutralize HIV. Vaccines. 2019;7:77. doi: 10.3390/vaccines7030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staus D.P., Strachan R.T., Manglik A., Pani B., Kahsai A.W., Kim T.H., Wingler L.M., Ahn S., Chatterjee A., Masoudi A., Kruse A.C., Pardon E., Steyaert J., Weis W.I., Prosser R.S., Kobilka B.K., Costa T., Lefkowitz R.J. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekesi A., Abdellaoui S., Holroyd N., Van Delm W., Pardon E., Pauwels J., Gevaert K., Steyaert J., Derveaux S., Borysik A., Tompa P. Challenges in the structural-functional characterization of multidomain, partially disordered proteins CBP and p300: preparing native proteins and developing nanobody tools. Methods Enzymol. 2018;611:607–675. doi: 10.1016/bs.mie.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 5.Uchański T., Pardon E., Steyaert J. Nanobodies to study protein conformational states. Curr. Opin. Struct. Biol. 2020;60:117–123. doi: 10.1016/j.sbi.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Vaidyanathan G., McDougald D., Choi J., Koumarianou E., Weitzel D., Osada T., Lyerly H.K., Zalutsky M.R. Preclinical evaluation of 18F-labeled anti-HER2 nanobody conjugates for imaging HER2 receptor expression by immuno-PET. J. Nucl. Med. 2016;57:967–973. doi: 10.2967/jnumed.115.171306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krüwel T., Nevoltris D., Bode J., Dullin C., Baty D., Chames P., Alves F. In vivo detection of small tumour lesions by multi-pinhole SPECT applying a (99m)Tc-labelled nanobody targeting the Epidermal Growth Factor Receptor. Sci. Rep. 2016;6 doi: 10.1038/srep21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demine S., Garcia Ribeiro R., Thevenet J., Marselli L., Marchetti P., Pattou F., Kerr-Conte J., Devoogdt N., Eizirik D.L. A nanobody-based nuclear imaging tracer targeting dipeptidyl peptidase 6 to determine the mass of human beta cell grafts in mice. Diabetologia. 2020;63:825–836. doi: 10.1007/s00125-019-05068-5. [DOI] [PubMed] [Google Scholar]
- 9.Lv G., Sun X., Qiu L., Sun Y., Li K., Liu Q., Zhao Q., Qin S., Lin J. PET imaging of tumor PD-L1 expression with a highly specific nonblocking single-domain antibody. J. Nucl. Med. 2020;61:117–122. doi: 10.2967/jnumed.119.226712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krasniqi A., D'Huyvetter M., Devoogdt N., Frejd F.Y., Sörensen J., Orlova A., Keyaerts M., Tolmachev V. Same-day imaging using small proteins: clinical experience and translational prospects in oncology. J. Nucl. Med. 2018;59:885–891. doi: 10.2967/jnumed.117.199901. [DOI] [PubMed] [Google Scholar]
- 11.Nuhn L., Bolli E., Massa S., Vandenberghe I., Movahedi K., Devreese B., Van Ginderachter J.A., De Geest B.G. Targeting protumoral tumor-associated macrophages with nanobody-functionalized nanogels through strain promoted azide alkyne cycloaddition ligation. Bioconjugate Chem. 2018;29:2394–2405. doi: 10.1021/acs.bioconjchem.8b00319. [DOI] [PubMed] [Google Scholar]
- 12.Yong K.W., Yuen D., Chen M.Z., Porter C.J.H., Johnston A.P.R. Pointing in the right direction: controlling the orientation of proteins on nanoparticles improves targeting efficiency. Nano Lett. 2019;19:1827–1831. doi: 10.1021/acs.nanolett.8b04916. [DOI] [PubMed] [Google Scholar]
- 13.Zou T., Dembele F., Beugnet A., Sengmanivong L., Trepout S., Marco S., de Marco A., Li M.H. Nanobody-functionalized PEG-b-PCL polymersomes and their targeting study. J. Biotechnol. 2015;214:147–155. doi: 10.1016/j.jbiotec.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 14.Anderson G.P., Liu J.L., Shriver-Lake L.C., Zabetakis D., Sugiharto V.A., Chen H.W., Lee C.R., Defang G.N., Wu S.L., Venkateswaran N., Goldman E.R. Oriented immobilization of single-domain antibodies using SpyTag/SpyCatcher yields improved limits of detection. Anal. Chem. 2019;91:9424–9429. doi: 10.1021/acs.analchem.9b02096. [DOI] [PubMed] [Google Scholar]
- 15.Tabtimmai L., Suphakun P., Srisook P., Kiriwan D., Phanthong S., Kiatwuthinon P., Chaicumpa W., Choowongkomon K. Cell-penetrable nanobodies (transbodies) that inhibit the tyrosine kinase activity of EGFR leading to the impediment of human lung adenocarcinoma cell motility and survival. J. Cell. Biochem. 2019;120:18077–18087. doi: 10.1002/jcb.29111. [DOI] [PubMed] [Google Scholar]
- 16.Traenkle B., Rothbauer U. Under the microscope: single-domain antibodies for live-cell imaging and super-resolution microscopy. Front. Immunol. 2017;8:1030. doi: 10.3389/fimmu.2017.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cramer K., Bolender A.L., Stockmar I., Jungmann R., Kasper R., Shin J.Y. Visualization of bacterial protein complexes labeled with fluorescent proteins and nanobody binders for STED microscopy. Int. J. Mol. Sci. 2019;20:3376. doi: 10.3390/ijms20143376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virant D., Traenkle B., Maier J., Kaiser P.D., Bodenhöfer M., Schmees C., Vojnovic I., Pisak-Lukáts B., Endesfelder U., Rothbauer U. A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat. Commun. 2018;9:930. doi: 10.1038/s41467-018-03191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Munter S., Ingels J., Goetgeluk G., Bonte S., Pille M., Weening K., Kerre T., Abken H., Vandekerckhove B. Nanobody based dual specific CARs. Int. J. Mol. Sci. 2018;19:403. doi: 10.3390/ijms19020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng X., Wang J., Kang G., Hu M., Yuan B., Zhang Y., Huang H. Homology modeling-based in silico affinity maturation improves the affinity of a nanobody. Int. J. Mol. Sci. 2019;20:4187. doi: 10.3390/ijms20174187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soler M.A., de Marco A., Fortuna S. Molecular dynamics simulations and docking enable to explore the biophysical factors controlling the yields of engineered nanobodies. Sci. Rep. 2016;6:34869. doi: 10.1038/srep34869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soler M.A., Fortuna S., de Marco A., Laio A. Binding affinity prediction of nanobody-protein complexes by scoring of molecular dynamics trajectories. Phys. Chem. Chem. Phys. 2018;20:3438–3444. doi: 10.1039/c7cp08116b. [DOI] [PubMed] [Google Scholar]
- 23.Soler M.A., Medagli B., Semrau M.S., Storici P., Bajc G., de Marco A., Laio A., Fortuna S. A consensus protocol for the in silico optimisation of antibody fragments. Chem. Commun. 2019;55:14043–14046. doi: 10.1039/c9cc06182g. [DOI] [PubMed] [Google Scholar]
- 24.Maaß A., Heiseler T., Maaß F., Fritz J., Hofmeyer T., Glotzbach B., Becker S., Kolmar H. A general strategy for antibody library screening via conversion of transient target binding into permanent reporter deposition. Protein Eng. Des. Sel. 2014;27:41–47. doi: 10.1093/protein/gzt060. [DOI] [PubMed] [Google Scholar]
- 25.Nemoto N., Kumachi S., Arai H. In vitro selection of single-domain antibody (VHH) using cDNA display. Methods Mol. Biol. 2018;1827:269–285. doi: 10.1007/978-1-4939-8648-4_14. [DOI] [PubMed] [Google Scholar]
- 26.Hussack G., Baral T.N., Baardsnes J., van Faassen H., Raphael S., Henry K.A., Zhang J., MacKenzie C.R. A novel affinity tag, ABTAG, and its application to the affinity screening of single-domain antibodies selected by phage display. Front. Immunol. 2017;8:1406. doi: 10.3389/fimmu.2017.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari D., Garrapa V., Locatelli M., Bolchi A. A novel nanobody scaffold optimized for bacterial expression and suitable for the construction of ribosome display libraries. Mol. Biotechnol. 2020;62:43–55. doi: 10.1007/s12033-019-00224-z. [DOI] [PubMed] [Google Scholar]
- 28.de Marco A. Methodologies for the isolation of alternative binders with improved clinical potentiality over conventional antibodies. Crit. Rev. Biotechnol. 2013;33:40–48. doi: 10.3109/07388551.2012.665353. [DOI] [PubMed] [Google Scholar]
- 29.Abdellatif M.E.A., Hipp L., Plessner M., Walther P., Knöll B. Indirect visualization of endogenous nuclear actin by correlative light and electron microscopy (CLEM) using an actin-directed chromobody. Histochem. Cell Biol. 2019;152:133–143. doi: 10.1007/s00418-019-01795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Els Conrath K., Lauwereys M., Wyns L., Muyldermans S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J. Biol. Chem. 2001;276:7346–7350. doi: 10.1074/jbc.M007734200. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y., Li Y., Wu X., Li L., Liu J., Wang Y., Liu Y., Li Q., Wang Z. Identification of anti-CD16a single domain antibodies and their application in bispecific antibodies. Canc. Biol. Ther. 2020;21:72–80. doi: 10.1080/15384047.2019.1665953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Marco A. Nanomaterial bio-activation and macromolecules functionalization: the search for reliable protocols. Protein Expr. Purif. 2018;147:49–54. doi: 10.1016/j.pep.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 33.De Tavernier E., Detalle L., Morizzo E., Roobrouck A., De Taeye S., Rieger M., Verhaeghe T., Correia A., Van Hegelsom R., Figueirido R., Noens J., Steffensen S., Stöhr T., Van de Velde W., Depla E., Dombrecht B. High throughput combinatorial formatting of PcrV nanobodies for efficient potency improvement. J. Biol. Chem. 2016;291:15243–15255. doi: 10.1074/jbc.M115.684241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadopoulos K.P., Isaacs R., Bilic S., Kentsch K., Huet H.A., Hofmann M., Rasco D., Kundamal N., Tang Z., Cooksey J., Mahipal A. Unexpected hepatotoxicity in a phase I study of TAS266, a novel tetravalent agonistic Nanobody® targeting the DR5 receptor. Canc. Chemother. Pharmacol. 2015;75:887–895. doi: 10.1007/s00280-015-2712-0. [DOI] [PubMed] [Google Scholar]
- 35.de Wit R.H., Heukers R., Brink H.J., Arsova A., Maussang D., Cutolo P., Strubbe B., Vischer H.F., Bachelerie F., Smit M.J. CXCR4-Specific nanobodies as potential therapeutics for WHIM syndrome. J. Pharmacol. Exp. Therapeut. 2017;363:35–44. doi: 10.1124/jpet.117.242735. [DOI] [PubMed] [Google Scholar]
- 36.Palomo C., Mas V., Detalle L., Depla E., Cano O., Vázquez M., Stortelers C., Melero J.A. Trivalency of a nanobody specific for the human respiratory syncytial virus fusion glycoprotein drastically enhances virus neutralization and impacts escape mutant selection. Antimicrob. Agents Chemother. 2016;60:6498–6509. doi: 10.1128/AAC.00842-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Compte M., Harwood S.L., Muñoz I.G., Navarro R., Zonca M., Perez-Chacon G., Erce-Llamazares A., Merino N., Tapia-Galisteo A., Cuesta A.M., Mikkelsen K., Caleiras E., Nuñez-Prado N., Aznar M.A., Lykkemark S., Martínez-Torrecuadrada J., Melero I., Blanco F.J., Bernardino de la Serna J., Zapata J.M., Sanz L., Alvarez-Vallina L. A tumor-targeted trimeric 4-1BB-agonistic antibody induces potent anti-tumor immunity without systemic toxicity. Nat. Commun. 2018;9:4809. doi: 10.1038/s41467-018-07195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., Melero J.A., Depla E. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2015;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pleiner T., Bates M., Görlich D. A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J. Cell Biol. 2018;217:1143–1154. doi: 10.1083/jcb.201709115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu J., Li J., Wang W., Wu H., Zhou P., Li Y., He Q., Tu Z. One-step orientated immobilization of nanobodies and its application for immunoglobulin purification. J. Chromatogr. A. 2019;1603:15–22. doi: 10.1016/j.chroma.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 41.Bobkov V., Zarca A.M., Van Hout A., Arimont M., Doijen J., Bialkowska M., Toffoli E., Klarenbeek A., van der Woning B., van der Vliet H.J., Van Loy T., de Haard H., Schols D., Heukers R., Smit M.J. Nanobody-Fc constructs targeting chemokine receptor CXCR4 potently inhibit signaling and CXCR4-mediated HIV-entry and induce antibody effector functions. Biochem. Pharmacol. 2018;158:413–424. doi: 10.1016/j.bcp.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Pirzer T., Becher K.S., Rieker M., Meckel T., Mootz H.D., Kolmar H. Generation of potent anti-HER1/2 immunotoxins by protein ligation using split inteins. ACS Chem. Biol. 2018;13:2058–2066. doi: 10.1021/acschembio.8b00222. [DOI] [PubMed] [Google Scholar]
- 43.Godakova S.A., Noskov A.N., Vinogradova I.D., Ugriumova G.A., Solovyev A.I., Esmagambetov I.B., Tukhvatulin A.I., Logunov D.Y., Naroditsky B.S., Shcheblyakov D.V., Gintsburg A.L. Camelid VHHs fused to human Fc fragments provide long term protection against botulinum neurotoxin a in mice. Toxins. 2019;11:464. doi: 10.3390/toxins11080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Djender S., Schneider A., Beugnet A., Crepin R., Desrumeaux K.E., Romani C., Moutel S., Perez F., de Marco A. Bacterial cytoplasm as an effective cell compartment for producing functional VHH-based affinity reagents and Camelidae IgG-like recombinant antibodies. Microb. Cell Factories. 2014;13:140. doi: 10.1186/s12934-014-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheng Y., Wang K., Lu Q., Ji P., Liu B., Zhu J., Liu Q., Sun Y., Zhang J., Zhou E.M., Zhao Q. Nanobody-horseradish peroxidase fusion protein as an ultrasensitive probe to detect antibodies against Newcastle disease virus in the immunoassay. J. Nanobiotechnol. 2019;17:35. doi: 10.1186/s12951-019-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherwood L.J., Hayhurst A. Periplasmic Nanobody-APEX2 fusions enable facile visualization of Ebola, Marburg, and Mĕnglà virus nucleoproteins, alluding to similar antigenic landscapes among marburgvirus and dianlovirus. Viruses. 2019;11:364. doi: 10.3390/v11040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernardinelli G., Oloketuyi S., Werner S.F., Mazzega E., Högberg B., de Marco A. A compact nanobody-DNAzyme conjugate enables antigen detection and signal amplification. Nat. Biotechnol. 2019;56:1–8. doi: 10.1016/j.nbt.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Dong J.X., Lee Y., Kirmiz M., Palacio S., Dumitras C., Moreno C.M., Sando R., Santana L.F., Südhof T.C., Gong B., Murray K.D., Trimmer J.S. A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. Elife. 2019;8 doi: 10.7554/eLife.48750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olichon A., Surrey T. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J. Biol. Chem. 2007;282:36314–36320. doi: 10.1074/jbc.M704908200. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T., Williams R.L., Rabbitts T.H. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 2007;26:3250–3259. doi: 10.1038/sj.emboj.7601744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paganetti P., Calanca V., Galli C., Stefani M., Molinari M. Beta-site specific intrabodies to decrease and prevent generation of Alzheimer's Abeta peptide. J. Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borg S., Popp F., Hofmann J., Leonhardt H., Rothbauer U., Schüler D. An intracellular nanotrap redirects proteins and organelles in live bacteria. mBio. 2015;6 doi: 10.1128/mBio.02117-14. e02117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguilar G., Matsuda S., Vigano M.A., Affolter M. Using nanobodies to study protein function in developing organisms. Antibodies. 2019;8:16. doi: 10.3390/antib8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caussinus E., Kanca O., Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 55.Shin Y.J., Park S.K., Jung Y.J., Kim Y.N., Kim K.S., Park O.K., Kwon S.H., Jeon S.H., Trinh le A., Fraser S.E., Kee Y., Hwang B.J. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 2015;5 doi: 10.1038/srep14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S., Tang N.H., Lara-Gonzalez P., Zhao Z., Cheerambathur D.K., Prevo B., Chisholm A.D., Desai A., Oegema K. A toolkit for GFP-mediated tissue-specific protein degradation in C. elegans. Development. 2017;144:2694–2701. doi: 10.1242/dev.150094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ariotti N., Rae J., Giles N., Martel N., Sierecki E., Gambin Y., Hall T.E., Parton R.G. Ultrastructural localisation of protein interactions using conditionally stable nanobodies. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamaguchi N., Colak-Champollion T., Knaut H. zGrad is a nanobody-based degron system that inactivates proteins in zebrafish. Elife. 2019;8 doi: 10.7554/eLife.43125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boons E., Li G., Vanstreels E., Vercruysse T., Pannecouque C., Vandamme A.M., Daelemans D. A stably expressed llama single-domain intrabody targeting Rev displays broad-spectrum anti-HIV activity. Antivir. Res. 2014;112:91–102. doi: 10.1016/j.antiviral.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Schellenberg M.J., Petrovich R.M., Malone C.C., Williams R.S. Selectable high-yield recombinant protein production in human cells using a GFP/YFP nanobody affinity support. Protein Sci. 2018;27:1083–1092. doi: 10.1002/pro.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bossi S., Ferranti B., Martinelli C., Capasso P., de Marco A. Antibody-mediated purification of co-expressed antigen-antibody complexes. Protein Expr. Purif. 2010;72:55–58. doi: 10.1016/j.pep.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Aliprandi M., Sparacio E., Pivetta F., Ossolengo G., Maestro R., de Marco A. The availability of a recombinant anti-SNAP antibody in VHH format amplifies the application flexibility of SNAP-tagged proteins. J. Biomed. Biotechnol. 2010 doi: 10.1155/2010/658954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kooijmans S.A., Aleza C.G., Roffler S.R., van Solinge W.W., Vader P., Schiffelers R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles. 2016;5 doi: 10.3402/jev.v5.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kooijmans S.A.A., Gitz-Francois J.J.J.M., Schiffelers R.M., Vader P. Recombinant phosphatidylserine-binding nanobodies for targeting of extracellular vesicles to tumor cells: a plug-and-play approach. Nanoscale. 2018;10:2413–2426. doi: 10.1039/c7nr06966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Jong O.G., Kooijmans S.A.A., Murphy D.E., Jiang L., Evers M.J.W., Sluijter J.P.G., Vader P., Schiffelers R.M. Drug delivery with extracellular vesicles: from imagination to innovation. Acc. Chem. Res. 2019;52:1761–1770. doi: 10.1021/acs.accounts.9b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Marco A. Recombinant antibody production evolves into multiple options aimed at yielding reagents suitable for application-specific needs. Microb. Cell Factories. 2015;14:125. doi: 10.1186/s12934-015-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Q., Zhou Y., Yu J., Liu W., Li F., Xian M., Nian R., Song H., Feng D. An efficient constitutive expression system for anti-CEACAM5 nanobody production in the yeast Pichia pastoris. Protein Expr. Purif. 2019;155:43–47. doi: 10.1016/j.pep.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 68.Schotte P., Dewerte I., De Groeve M., De Keyser S., De Brabandere V., Stanssens P. Pichia pastoris Mut(S) strains are prone to misincorporation of o-methyl-L-homoserine at methionine residues when methanol is used as the sole carbon source. Microb. Cell Factories. 2016;15:98. doi: 10.1186/s12934-016-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao G., He L., Sun S., Qiu H., Tai W., Chen J., Li J., Chen Y., Guo Y., Wang Y., Shang J., Ji K., Fan R., Du E., Jiang S., Li F., Du L., Zhou Y. A novel nanobody targeting middle-east respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J. Virol. 2018;92 doi: 10.1128/JVI.00837-18. e00837-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Virdi V., Palaci J., Laukens B., Ryckaert S., Cox E., Vanderbeke E., Depicker A., Callewaert N. Yeast-secreted, dried and food-admixed monomeric IgA prevents gastrointestinal infection in a piglet model. Nat. Biotechnol. 2019;37:527–530. doi: 10.1038/s41587-019-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hemmer C., Djennane S., Ackerer L., Hleibieh K., Marmonier A., Gersch S., Garcia S., Vigne E., Komar V., Perrin M., Gertz C., Belval L., Berthold F., Monsion B., Schmitt-Keichinger C., Lemaire O., Lorber B., Gutiérrez C., Muyldermans S., Demangeat G., Ritzenthaler C. Nanobody-mediated resistance to Grapevine fanleaf virus in plants. Plant Biotechnol. J. 2018;16:660–671. doi: 10.1111/pbi.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olichon A., Schweizer D., Muyldermans S., de Marco A. Heating as a rapid purification method for recovering correctly-folded thermotolerant VH and VHH domains. BMC Biotechnol. 2007;7:7. doi: 10.1186/1472-6750-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaczmarek J.Z., Skottrup P.D. Selection and characterization of camelid nanobodies towards urokinase-type plasminogen activator. Mol. Immunol. 2015;65:384–390. doi: 10.1016/j.molimm.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 74.Salema V., Fernández L.Á. High yield purification of nanobodies from the periplasm of E. coli as fusions with the maltose binding protein. Protein Expr. Purif. 2013;91:42–48. doi: 10.1016/j.pep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Noor A., Walser G., Wesseling M., Giron P., Laffra A.M., Haddouchi F., De Grève J., Kronenberger P. Production of a mono-biotinylated EGFR nanobody in the E. coli periplasm using the pET22b vector. BMC Res. Notes. 2018;11:751. doi: 10.1186/s13104-018-3852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hernot S., Unnikrishnan S., Du Z., Shevchenko T., Cosyns B., Broisat A., Toczek J., Caveliers V., Muyldermans S., Lahoutte T., Klibanov A.L., Devoogdt N. Nanobody-coupled microbubbles as novel molecular tracer. J. Contr. Release. 2012;158:346–353. doi: 10.1016/j.jconrel.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherwood L.J., Hayhurst A. Hapten mediated display and pairing of recombinant antibodies accelerates assay assembly for biothreat countermeasures. Sci. Rep. 2012;2:807. doi: 10.1038/srep00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu M., Hu Y., Li G., Ou W., Mao P., Xin S., Wan Y. Combining magnetic nanoparticle with biotinylated nanobodies for rapid and sensitive detection of influenza H3N2. Nanoscale Res Lett. 2014;9:528. doi: 10.1186/1556-276X-9-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du T., Zhu G., Wu X., Fang J., Zhou E.M. Biotinylated single-domain antibody-based blocking ELISA for detection of antibodies against swine influenza virus. Int. J. Nanomed. 2019;14:9337–9349. doi: 10.2147/IJN.S218458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun Z., Lv J., Liu X., Tang Z., Wang X., Xu Y., Hammock B.D. Development of a nanobody-AviTag fusion protein and its application in a streptavidin-biotin-amplified enzyme-linked immunosorbent assay for Ochratoxin A in cereal. Anal. Chem. 2018;90:10628–10634. doi: 10.1021/acs.analchem.8b03085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kijanka M.M., van Brussel A.S., van der Wall E., Mali W.P., van Diest P.J., van Bergen En Henegouwen P.M., Oliveira S. Optical imaging of pre-invasive breast cancer with a combination of VHHs targeting CAIX and HER2 increases contrast and facilitates tumour characterization. EJNMMI Res. 2016;6:14. doi: 10.1186/s13550-016-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren W., Li Z., Xu Y., Wan D., Barnych B., Li Y., Tu Z., He Q., Fu J., Hammock B.D. One-step ultrasensitive bioluminescent enzyme immunoassay based on nanobody/nanoluciferase fusion for detection of Aflatoxin B1 in cereal. J. Agric. Food Chem. 2019;67:5221–5229. doi: 10.1021/acs.jafc.9b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reulen S.W., van Baal I., Raats J.M., Merkx M. Efficient, chemoselective synthesis of immunomicelles using single-domain antibodies with a C-terminal thioester. BMC Biotechnol. 2009;9:66. doi: 10.1186/1472-6750-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crasson O., Rhazi N., Jacquin O., Freichels A., Jérôme C., Ruth N., Galleni M., Filée P., Vandevenne M. Enzymatic functionalization of a nanobody using protein insertion technology. Protein Eng. Des. Sel. 2015;28:451–460. doi: 10.1093/protein/gzv020. [DOI] [PubMed] [Google Scholar]
- 85.Veggiani G., de Marco A. Improved quantitative and qualitative production of single-domain intrabodies mediated by the co-expression of Erv1p sulfhydryl oxidase. Protein Expr. Purif. 2011;79:111–114. doi: 10.1016/j.pep.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 86.Ruano-Gallego D., Fraile S., Gutierrez C., Fernández L.Á. Screening and purification of nanobodies from E. coli culture supernatants using the hemolysin secretion system. Microb. Cell Factories. 2019;18:47. doi: 10.1186/s12934-019-1094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oloketuyi S., Dilkaute C., Mazzega E., Jose J., de Marco A. Purification-independent immunoreagents obtained by displaying nanobodies on bacteria surface. Appl. Microbiol. Biotechnol. 2019;103:4443–4453. doi: 10.1007/s00253-019-09823-x. [DOI] [PubMed] [Google Scholar]
- 88.Salema V., Fernández L.Á. Escherichia coli surface display for the selection of nanobodies. Microb Biotechnol. 2017;10:1468–1484. doi: 10.1111/1751-7915.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McMahon C., Baier A.S., Pascolutti R., Wegrecki M., Zheng S., Ong J.X., Erlandson S.C., Hilger D., Rasmussen S.G.F., Ring A.M., Manglik A., Kruse A.C. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 2018;25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roth L., Krah S., Klemm J., Günther R., Toleikis L., Busch M., Becker S., Zielonka S. Isolation of antigen-specific VHH single-domain antibodies by combining animal immunization with yeast surface display. Methods Mol. Biol. 2020;2070:173–189. doi: 10.1007/978-1-4939-9853-1_10. [DOI] [PubMed] [Google Scholar]
- 91.Cavallari M. Rapid antigen and antibody-like molecule discovery by staphylococcal surface display. Methods Mol. Biol. 2020;2070:79. doi: 10.1007/978-1-4939-9853-1_5. 94. [DOI] [PubMed] [Google Scholar]
- 92.Schüürmann J., Quehl P., Festel G., Jose J. Bacterial whole-cell biocatalysts by surface display of enzymes: toward industrial application. Appl. Microbiol. Biotechnol. 2014;98:8031–8046. doi: 10.1007/s00253-014-5897-y. [DOI] [PubMed] [Google Scholar]
- 93.De Marni M.L., Monegal A., Venturini S., Vinati S., Carbone R., de Marco A. Antibody purification-independent microarrays (PIM) by direct bacteria spotting on TiO2-treated slides. Methods. 2012;56:317–325. doi: 10.1016/j.ymeth.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 94.Kylilis N., Riangrungroj P., Lai H.E., Salema V., Fernández L.Á., Stan G.V., Freemont P.S., Polizzi K.M. Whole-cell biosensor with tunable limit of detection enables low-cost agglutination assays for medical diagnostic applications. ACS Sens. 2019;4:370–378. doi: 10.1021/acssensors.8b01163. [DOI] [PubMed] [Google Scholar]
- 95.Massa P.E., Paniccia A., Monegal A., de Marco A., Rescigno M. Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood. 2013;122:705–714. doi: 10.1182/blood-2012-12-474098. [DOI] [PubMed] [Google Scholar]
- 96.van den Berg van Saparoea H.B., Houben D., de Jonge M.I., Jong W.S.P., Luirink J. Display of recombinant proteins on bacterial outer membrane vesicles by using protein ligation. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02567-17. e02567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brune K.D., Howarth M. New routes and opportunities for modular construction of particulate vaccines: stick, click, and glue. Front. Immunol. 2018;9:1432. doi: 10.3389/fimmu.2018.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Glass D.S., Riedel-Kruse I.H. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns. Cell. 2018;174:649–658. doi: 10.1016/j.cell.2018.06.041. e16. [DOI] [PubMed] [Google Scholar]
- 99.Uchański T., Zögg T., Yin J., Yuan D., Wohlkönig A., Fischer B., Rosenbaum D.M., Kobilka B.K., Pardon E., Steyaert J. An improved yeast surface display platform for the screening of nanobody immune libraries. Sci. Rep. 2019;9:382. doi: 10.1038/s41598-018-37212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.An N., Hou Y.N., Zhang Q.X., Li T., Zhang Q.L., Fang C., Chen H., Lee H.C., Zhao Y.J., Du X. Anti-multiple myeloma activity of nanobody-based anti-CD38 chimeric antigen receptor T cells. Mol. Pharm. 2018;15:4577–4588. doi: 10.1021/acs.molpharmaceut.8b00584. [DOI] [PubMed] [Google Scholar]
- 101.Hassani M., Hajari Taheri F., Sharifzadeh Z., Arashkia A., Hadjati J., van Weerden W.M., Abdoli S., Modarressi M.H., Abolhassani M. Engineered Jurkat cells for targeting prostate-specific membrane antigen on prostate cancer cells by nanobody-based chimeric antigen receptor. Iran. Biomed. J. 2020;24:81–88. doi: 10.29252/ibj.24.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Even-Desrumeaux K., Baty D., Chames P. Strong and oriented immobilization of single domain antibodies from crude bacterial lysates for high-throughput compatible cost-effective antibody array generation. Mol. Biosyst. 2010;6:2241–2248. doi: 10.1039/c005279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson G.P., Shriver-Lake L.C., Walper S.A., Ashford L., Zabetakis D., Liu J.L., Breger J.C., Brozozog Lee P.A., Goldman E.R. Genetic fusion of an anti-BclA single-domain antibody with beta galactosidase. Antibodies. 2018;7:36. doi: 10.3390/antib7040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deng C., Xiong J., Gu X., Chen X., Wu S., Wang Z., Wang D., Tu J., Xie J. Novel recombinant immunotoxin of EGFR specific nanobody fused with cucurmosin, construction and antitumor efficiency in vitro. Oncotarget. 2017;8:38568–38580. doi: 10.18632/oncotarget.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shibuya Y., Haga N., Asano R., Nakazawa H., Hattori T., Takeda D., Sugiyama A., Kurotani R., Kumagai I., Umetsu M., Makabe K. Generation of camelid VHH bispecific constructs via in-cell intein-mediated protein trans-splicing. Protein Eng. Des. Sel. 2017;30:15–21. doi: 10.1093/protein/gzw057. [DOI] [PubMed] [Google Scholar]
- 106.Götzke H., Kilisch M., Martínez-Carranza M., Sograte-Idrissi S., Rajavel A., Schlichthaerle T., Engels N., Jungmann R., Stenmark P., Opazo F., Frey S. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 2019;10:4403. doi: 10.1038/s41467-019-12301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Messer A., Butler D.C. Optimizing intracellular antibodies (intrabodies/nanobodies) to treat neurodegenerative disorders. Neurobiol. Dis. 2020;134 doi: 10.1016/j.nbd.2019.104619. [DOI] [PubMed] [Google Scholar]
- 108.Mahajan S.P., Meksiriporn B., Waraho-Zhmayev D., Weyant K.B., Kocer I., Butler D.C., Messer A., Escobedo F.A., DeLisa M.P. Computational affinity maturation of camelid single-domain intrabodies against the nonamyloid component of alpha-synuclein. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-35464-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Billen B., Vincke C., Hansen R., Devoogdt N., Muyldermans S., Adriaensens P., Guedens W. Cytoplasmic versus periplasmic expression of site-specifically and bioorthogonally functionalized nanobodies using expressed protein ligation. Protein Expr. Purif. 2017;133:25–34. doi: 10.1016/j.pep.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 110.Bao X., Xu L., Lu X., Jia L. Optimization of dilution refolding conditions for a camelid single domain antibody against human beta-2-microglobulin. Protein Expr. Purif. 2016;117:59–66. doi: 10.1016/j.pep.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 111.Li D., Ji F., Huang C., Jia L. High expression achievement of active and robust anti-β2 microglobulin nanobodies via E. coli hosts selection. Molecules. 2019;24:2860. doi: 10.3390/molecules24162860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ta D.T., Redeker E.S., Billen B., Reekmans G., Sikulu J., Noben J.P., Guedens W., Adriaensens P. An efficient protocol towards site-specifically clickable nanobodies in high yield: cytoplasmic expression in Escherichia coli combined with intein-mediated protein ligation. Protein Eng. Des. Sel. 2015;28:351–363. doi: 10.1093/protein/gzv032. [DOI] [PubMed] [Google Scholar]
- 113.Graulus G.J., Ta D.T., Tran H., Hansen R., Billen B., Royackers E., Noben J.P., Devoogdt N., Muyldermans S., Guedens W., Adriaensens P. Site-selective functionalization of nanobodies using intein-mediated protein ligation for innovative bioconjugation. Methods Mol. Biol. 2019;2033:117–130. doi: 10.1007/978-1-4939-9654-4_9. [DOI] [PubMed] [Google Scholar]
- 114.Iwaki T., Hara K., Umemura K. Nanobody production can be simplified by direct secretion from Escherichia coli. Protein Expr. Purif. 2020;170 doi: 10.1016/j.pep.2020.105607. [DOI] [PubMed] [Google Scholar]
- 115.Pant N., Marcotte H., Hermans P., Bezemer S., Frenken L., Johansen K., Hammarström L. Lactobacilli producing bispecific llama-derived anti-rotavirus proteins in vivo for rotavirus-induced diarrhea. Future Microbiol. 2011;6:583–593. doi: 10.2217/fmb.11.32. [DOI] [PubMed] [Google Scholar]
- 116.Günaydın G., Alvarez B., Lin Y., Hammarström L., Marcotte H. Co-expression of anti-rotavirus proteins (llama VHH antibody fragments) in Lactobacillus: development and functionality of vectors containing two expression cassettes in tandem. PloS One. 2014;9 doi: 10.1371/journal.pone.0096409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maggi M., Scotti C. Enhanced expression and purification of camelid single domain VHH antibodies from classical inclusion bodies. Protein Expr. Purif. 2017;136:39–44. doi: 10.1016/j.pep.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 118.Noguchi T., Nishida Y., Takizawa K., Cui Y., Tsutsumi K., Hamada T., Nishi Y. Accurate quantitation for in vitro refolding of single domain antibody fragments expressed as inclusion bodies by referring the concomitant expression of a soluble form in the periplasms of Escherichia coli. J. Immunol. Methods. 2017;442:1–11. doi: 10.1016/j.jim.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 119.Mitchell L.S., Colwell L.J. Comparative analysis of nanobody sequence and structure data. Proteins. 2018;86:697–706. doi: 10.1002/prot.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mitchell L.S., Colwell L.J. Analysis of nanobody paratopes reveals greater diversity than classical antibodies. Protein Eng. Des. Sel. 2018;31:267–275. doi: 10.1093/protein/gzy017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Law C.T., Camacho F., Garcia-Alles L.F., Gilleron M., Sarmiento M.E., Norazmi M.N., Acosta A., Choong Y.S. Interactions of domain antibody (dAbκ11) with Mycobacterium tuberculosis Ac2SGL in complex with CD1b. Tuberculosis. 2019;114:9–16. doi: 10.1016/j.tube.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 122.Lippow S.M., Wittrup K.D., Tidor B. Computational design of antibody-affinity improvement beyond in vivo maturation. Nat. Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vivcharuk V., Baardsnes J., Deprez C., Sulea T., Jaramillo M., Corbeil C.R., Mullick A., Magoon J., Marcil A., Durocher Y., O'Connor-McCourt M.D., Purisima E.O. Assisted design of antibody and protein therapeutics (ADAPT) PloS One. 2017;12 doi: 10.1371/journal.pone.0181490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng X., Wang J., Kang G., Hu M., Yuan B., Zhang Y., Huang H. Homology modeling-based in silico affinity maturation improves the affinity of a nanobody. Int. J. Mol. Sci. 2019;20:4187. doi: 10.3390/ijms20174187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sulea T., Hussack G., Ryan S., Tanha J., Purisima E.O. Application of assisted design of antibody and protein therapeutics (ADAPT) improves efficacy of a Clostridium difficile toxin A single-domain antibody. Sci. Rep. 2018;8:2260. doi: 10.1038/s41598-018-20599-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Djender S., Beugnet A., Schneider A., de Marco A. The biotechnological applications of recombinant single-domain antibodies are optimized by the C-terminal fusion to the EPEA sequence (C tag) Antibodies. 2014;3:182–191. [Google Scholar]
- 127.Moutel S., Bery N., Bernard V., Keller L., Lemesre E., de Marco A., Ligat L., Rain G.C., Favre G., Olichon A., Perez F. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife. 2016 doi: 10.7554/eLife.16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ambrosetti E., Paoletti P., Bosco A., Parisse P., Scaini D., Tagliabue E., de Marco A., Casalis L. Quantification of circulating cancer biomarkers via sensitive topographic measurements on single binder nanoarrays. ACS Omega. 2017;2:2618–2629. doi: 10.1021/acsomega.7b00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mazzega E., Beran A., Cabrini M., de Marco A In vitro isolation of nanobodies for selective Alexandrium minutum recognition: a model for convenient development of dedicated immuno-reagents to study and diagnostic toxic unicellular algae. Harmful Algae. 2019;82:44–51. doi: 10.1016/j.hal.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 130.Veggiani G., Giabbai B., Semrau M.S., Medagli B., Riccio V., Bajc G., Storici P., de Marco A. Comparative analysis of fusion tags used to functionalize recombinant antibodies. Protein Expr. Purif. 2020;166 doi: 10.1016/j.pep.2019.105505. [DOI] [PubMed] [Google Scholar]
- 131.Oloketuyi S., Mazzega E., Zavašnik J., Pungjunun K., Kalcher K., de Marco A., Mehmeti E. Electrochemical immunosensor functionalized with nanobodies for the detection of the toxic microalgae Alexandrium minutum using glassy carbon electrode modified with gold nanoparticles. Biosens. Bioelectron. 2020;154 doi: 10.1016/j.bios.2020.112052. [DOI] [PubMed] [Google Scholar]
- 132.de Marco A. Isolation of recombinant antibodies that recognize native and accessible membrane biomarkers. In: Camesano T.A., editor. Nanotechnology To Aid Chemical And Biological Defense, NATO Science for Peace and Security Series A: Chemistry and Biology. Springer; Dordrecht, The Netherlands: 2015. pp. 49–66. [Google Scholar]
- 133.Zimmermann I., Egloff P., Hutter C.A., Arnold F.M., Stohler P., Bocquet N., Hug M.N., Huber S., Siegrist M., Hetemann L., Gera J., Gmür S., Spies P., Gygax D., Geertsma E.R., Dawson R.J., Seeger M.A. Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife. 2018;7 doi: 10.7554/eLife.34317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.