Highlights
-
•
This study is currently the largest case series to date on the RT-PCR findings of COVID-19 throughout disease course.
-
•
The average contagious period of infected patients was 20 days.
-
•
Longer observation period and more than 2 series of negative viral test are necessary for patients ≥65 years.
Keywords: COVID-19, SARS-CoV-2, RT-PCR, Dynamic profile
Abstract
Background
With the spread of Coronavirus Disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection, its effect on society is amplified. We aimed to describe the viral detection results across different timepoints throughout the disease course.
Methods
A retrospective study of 301 confirmed COVID-19 patients hospitalized at Tongji Hospital in Wuhan, China, were included. Demographic characteristics of the patients were collected. Upper respiratory specimens (throat and/or nasal swabs) were obtained and analyzed by real-time RT-PCR for SARS-CoV-2 infection. Period of viral infection and the contagious stage were analyzed.
Results
Of 301 hospitalized COVID-19 patients, the median age was 58 years and 51.2 % were male. The median period between symptoms presence and positive SARS-CoV-2 RT-PCR results was 16 days (IQR, 10–23, N = 301). The median period between symptoms presence and an effective negative SARS-CoV-2 RT-PCR result was 20 days (IQR, 17–24; N = 216). Infected patient ≥65 years old stayed contagious longer (22 days vs 19 days, p = 0.015). Although two consecutive negative results were confirmed in 70 patients, 30 % of them had positive viral test results for the third time. Using specimens from nasal swabs to run the RT-PCR test showed a higher positive rate than using specimens from throat swabs.
Conclusions
This large-scale investigation with 1113 RT-PCR test results from 301 COVID-19 patients showed that the average contagious period of SARS-CoV-2 infected patients was 20 days. Longer observation period and more than 2 series of negative viral test are necessary for patients ≥65 years.
1. Introduction
To date, an outbreak of infectious diseases--coronavirus disease 2019 (COVID-19) associated with Severe Acute Respiratory Syndrome Coronavirus -2 (SARS-CoV-2) continues in Wuhan, Hubei Province, China [1,2]. The clinical manifestations of patients with COVID-19 included fever, dyspnea, fatigue, dry cough, myalgia, lymphopenia and radiographic findings of pneumonia. For severe and critical cases, patients suffered from acute respiratory distress syndrome (ARDS), acute respiratory failure, other serious complications and even death [3,4]. The spread of virus has struck Wuhan, and even the whole country, and has caused unmeasurable losses in every aspect. Over 50 countries are fighting against the disease. As of Feb 26th, 2020, 81,015 cases were diagnosed worldwide, and 2762 patients died from this disease (death rate 3.41 %). 1
Genomic studies have shown that SARS-CoV-2 shared around 80 % identity sequencing with SARS-CoV, which caused a global epidemic with 8096 confirmed cases worldwide in 2002–2003 [5]. Presumed person-to-person transmission of SARS-CoV-2 was suggested based on epidemiology and clinical evidences [[6], [7], [8]]. Recent reports suggested that asymptomatic COVID-19 infected individuals could also be the source of transmission [9,10]. To successfully contained the epidemic of SARS-CoV-2, public health interventions such as disease detection and isolation were critical.
Currently, the dynamic profile of SARS-CoV-2 viral load after onset of symptoms is not clear. Recent report by Zou et al. from 18 patients with COVID-19 suggested that the viral nucleic acid shedding pattern of SARS-CoV-2 appears different from that in patients with SARS-CoV [9]. Therefore, the aim of this large-scale retrospective study was to analyze the dynamic profile of SARS-CoV-2 and to explore the impact of demographic parameters on it.
2. Methods
2.1. Study design and participants
A total of 301 hospitalized patients (admission date from Jan 21st to Feb 11th, 2020) with confirmed SARS-CoV-2 infection in three branches (Hankou, Sino-French new city and Optical Valley) of Tongji Hospital at Tongji Hospital of Huazhong University of Science and Technology in Wuhan, China were included in this study. Tongji hospital is one of the major and largest hospital for COVID-19 treatment in Wuhan. All enrolled patients were confirmed diagnosed of COVID-19 according to the diagnosis and treatment guideline for SARS-CoV-2 from Chinese National Health Committee (Version 5) and the interim guidance from Centers for Disease Control and Prevention [11,12]. All data (test dates and results of RT-PCR assay) were collected up to the final follow-up date (February 26th, 2020).
2.2. Data collection and definitions
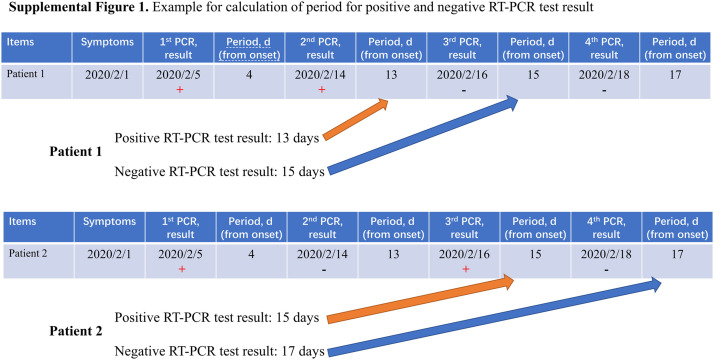
Data (clinical information and results of RT-PCR for SARS-CoV-2 viral nucleic acid detection) were collected from the electronic medical record system. The following information were collected for analysis: 1. Demographic characteristics such as age and gender; 2. Clinical characteristics such as date of onset (defined as the first date when the symptoms were reported), date of admission and date of discharge; 3. SARS-CoV-2 RT-PCR characteristics. Throat and/or nasal swabs were collected for the SARS-CoV-2 viral nucleic acid detection in sequential time-points. Positive SARS-CoV-2 RT-PCR assay is defined as the period from the date of onset to the date of last positive RT-PCR test result. Effective negative SARS-CoV-2 RT-PCR assay is defined as the period from the date of onset to the date of first negative RT-PCR test result (In some cases, patients got positive RT-PCR results following false negative results, which were considered non-effective negative results in our study).
2.3. Real-time reverse transcription polymerase chain reaction assay for SARS-CoV-2
Throat swab samples or deep nasal cavity swab samples were collected for extracting COVID-19 RNA from patients suspected of having COVID-19 infection. The collected swabs were placed into a collection tube with 200 μL of virus preservation solution, and total RNA was extracted within 2 h using magnetic beads (Tianlong, Xi’an, China). The extracting solution was used for one step RT-PCR assay of COVID-19 RNA. Two target genes, including open reading frame 1ab (ORF1ab) and nucleocapsid protein (N), were simultaneously amplified and tested during the real-time RT-PCR assay. Target 1 (ORF1ab): forward primer CCCTGTGGGTTTTACACTTAA; reverse primer ACGATTGTGCATCAGCTGA; and the probe 5′-VIC-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3’. Target 2 (N): forward primer GGGGAACTTCTCCTGCTAGAAT; reverse primer CAGACATTTTGCTCTCAAGCTG; and the probe 5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′. The real-time RT-PCR assay was performed using a COVID-19 nucleic acid detection kit according to the manufacturer’s protocol (Shanghai Huirui Biotechnology Co., Ltd). Reaction mixture contains 7.5 μL of RT-PCR reaction buffer, 5 μL of ORF 1ab/N gene reaction solution, 1.5 μL of enzyme mixture, 11 μL of RNA specimen. RT-PCR assay was performed under the following conditions: reverse transcriptional reaction at 50 °C for 15 min, initial denaturation at 95 °C for 5 min, 45 cycles of denaturation at 95 °C for 10 s and extending and collecting fluorescence signal at 55 °C for 45 s. A cycle threshold value (Ct-value) less than 35 was defined as a positive test result, and a Ct-value of 39.2 or more was defined as a negative test. A medium load, defined as a Ct-value of 35 to less than 39.2, required confirmation by retesting.
2.4. Statistical analysis
All continuous variables were presented as medians (interquartile range, IQR) and analyzed with Mann-Whitney U test Categorical variables were reported as whole numbers and percentages. All p values were reported as two-sided with a significance level of 0.05. All statistical tests were performed in SPSS version 24.0 (IBM, NY, USA).
3. Results
3.1. Demographic and clinical characteristics
A total of 301 patients diagnosed as COVID-19 were included in this study. According to the guideline [12], all included patients were mild to moderate. No patient was transferred to ICU. The median age was 58 years (IQR, 44–68; range, 10–92 years), comprising 154 (51.2 %) men and 147 (48.8 %) women. The median period from symptom onset to admission was 9 days (IQR, 7−12; range, 0–31). On the last follow-up day (February 26th, 2020), 246 (81.7 %) patients were still hospitalized while 55 (18.3 %) patients were discharged. (Table 1 )
Table 1.
The demographic and clinical characteristics of all patients.
| Variables | All Patients |
|---|---|
| Clinical petameters (N = 301) | |
| Age, median (IQR), y | 58.0 (44.0−68.0) |
| <65 | 63.5% (191/301) |
| ≥65 | 36.5% (110/301) |
| Gender | |
| Male | 51.2 % (154/301) |
| Female | 48.8 % (147/301) |
| Onset of symptom to admission, median (IQR), d | 9 (7−12) |
| Status | |
| In-hospital | 81.7 % (246/462) |
| Discharge | 18.3 % (55/301) |
| SARS-CoV-2 RT-PCR assay | |
| Total tests | 1113 |
| Tests/patient | 3.7 tests/patient |
| Throat swabs | 92.7 % (1028/1113) |
| Nasal swabs | 7.6 % (85/1113) |
| Onset of symptom to, median (IQR), d | |
| First SARS-CoV-2 RT-PCR assay | 8.0 (5.0−12.0) |
| Last SARS-CoV-2 RT-PCR assay | 24.0 (20.0−28.0) |
| Positive SARS-CoV-2 RT-PCR assay | 16.0 (10.0−23.0) |
| Negative SARS-CoV-2 RT-PCR assay | 20.0 (17.0−24.0) (N = 216) |
| Positive rate of SARS-CoV-2 RT-PCR, d | |
| Day 0−7 | 97.9 % (137/140) |
| Day 8−14 | 68.8 % (152/221) |
| Day 15−21 | 36.3 % (127/350) |
| Day 22−28 | 30.0 % (92/307) |
| >28 days | 26.3 % (25/95) |
Abbreviations: IQRinterquartile range; RT-PCRreal-time reverse transcription polymerase chain reaction.
3.2. Dynamic profile of SARS-CoV-2 infection
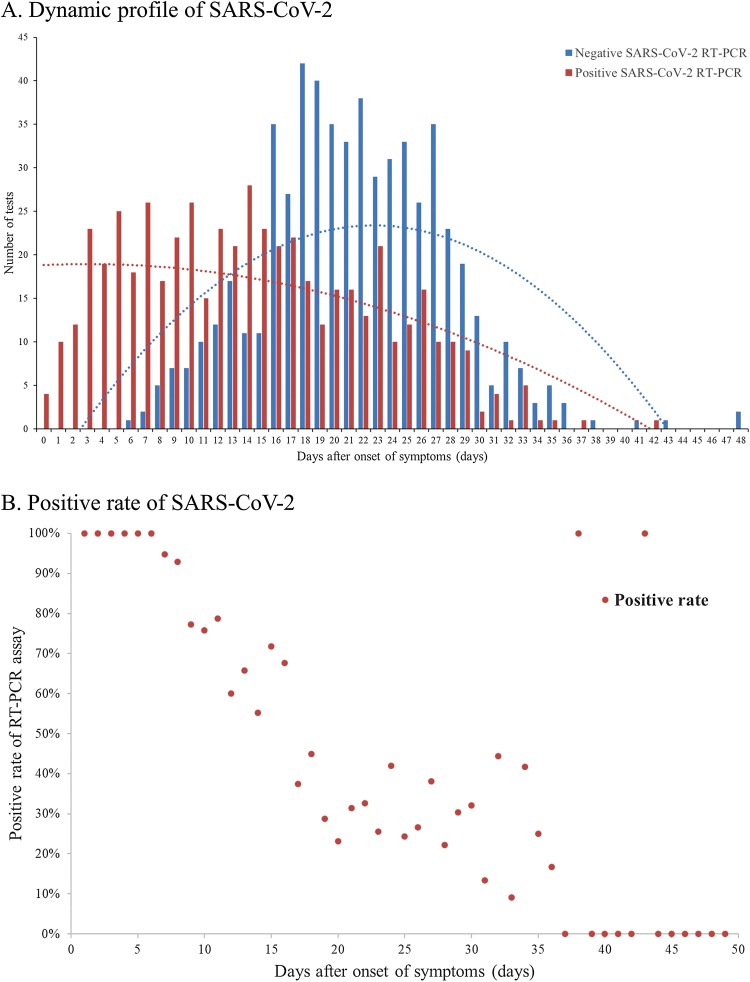
The total number of SARS-CoV-2 RT-PCR assay from 301 included COVID-19 patients was 1113, with 3.7 tests per patient. The median period from symptoms onset to the first RT-PCR assay was 8 days (IQR, 5–12; range, 0–32). (Table 1) At the last follow-up, 85 (28.2 %) patients still got positive results of RT-PCR assay for SARS-CoV-2. As shown in Fig. 1 A, the median period from symptoms onset to positive SARS-CoV-2 RT-PCR test result was 16 days (IQR, 10–23; range, 0–42, N = 301). The median period from symptoms onset to negative SARS-CoV-2 RT-PCR test result was 20 days (IQR, 17–24; range, 7–44, N = 216). The positive rate of RT-PCR assay was highest at day 0−7 (97.9 %), followed by 68.8 %, 36.3 %, 30.0 % and 26.3 % at day 8−14, day 15−21, day 22−28 and >28 days respectively. (Fig. 1B) The median period for last RT-PCR assay was 24 days (IQR, 20–28; range, 9–48).
Fig. 1.
Dynamic Profile of SARS-CoV-2 Infection.
(A) Dynamic Profile of SARS-CoV-2 Detected by RT-PCR from 301 COVID-19 Patients (N = 1113). Numbers of the positive (red bar) and negative (blue bar) results of SARS-CoV-2 RT-PCR were sum on every day based on the days after the onset. (B) Positive rate of SARS-CoV-2 Detected by RT-PCR from 301 COVID-19 Patients (N = 1113). Percentage of positive results of SARS-CoV-2 RT-PCR were calculated on every day based on the days after the onset.
3.3. Impact of demographic factors on dynamic profile of SARS-CoV-2
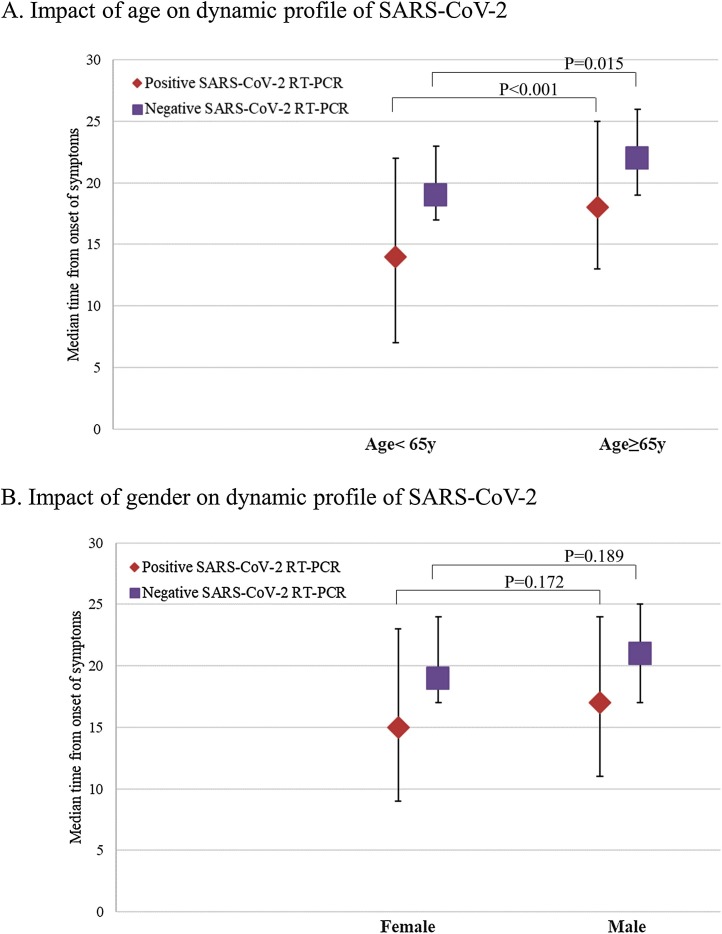
We investigated the impact of age and gender on the dynamic profile of SARS-CoV-2 by RT-PCR assay. As shown in Fig. 2 A, the median period of positive SARS-CoV-2 RT-PCR test result was significantly longer in older (≥65 years) patients (18 days, IQR, 13–25 vs 14 days, IQR, 7–22, p < 0.001). The median period of negative SARS-CoV-2 RT-PCR test result was also significantly longer in older (≥65 years) patients (22 days, IQR, 19–26 vs 19 days, IQR, 17–23, p = 0.015). As shown in Fig. 2B, female patients had a shorter median period of positive and negative SARS-CoV-2 RT-PCR test result than male patients (15 days, IQR, 9–23 vs 17 days, IQR, 11–24, p = 0.172) and (19 days, IQR, 17–24 vs 21 days, IQR, 17–25, p = 0.189), respectively. However, the difference is not statistically significant.
Fig. 2.
Impact of Age and Gender on Dynamic Profile of SARS-CoV-2.
(A) The median time of SARS-CoV-2 infection (positive, red triangle) and SARS-CoV-2 conversion (negative, purple square) grouped by age. (B) The median time of SARS-CoV-2 infection (positive, red triangle) and SARS-CoV-2 conversion (negative, purple square) grouped by gender. The I bars indicate the 25th and 75th percentiles. P < .05 was considered statistically significant.
3.4. Estimation of false negative of SARS-CoV-2 RT-PCR assay
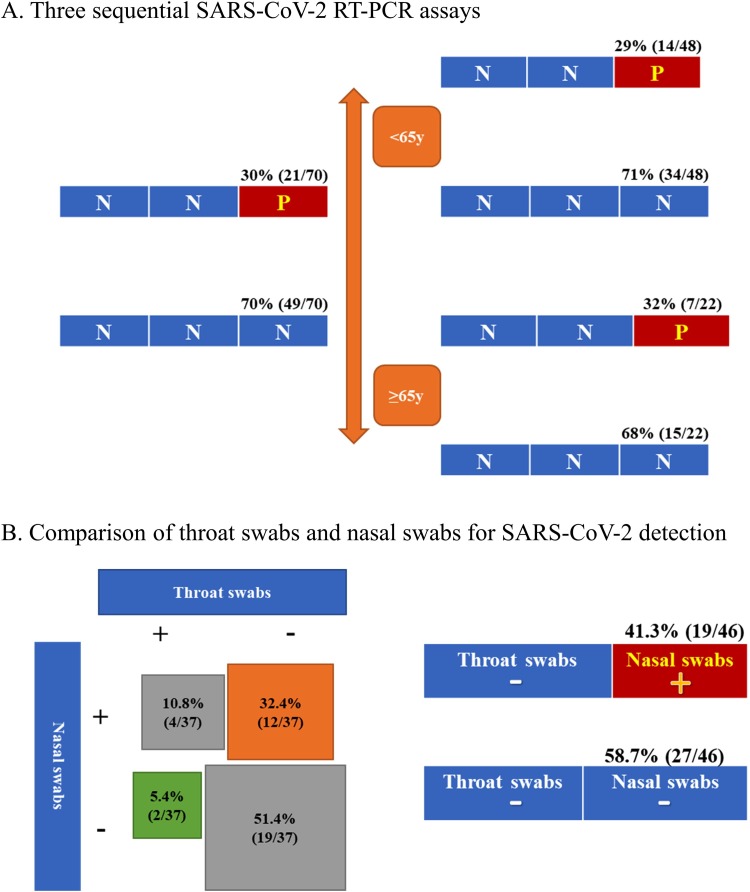
Of the 301 patients, 70 (23.3 %) had the records of three consecutive SARS-CoV-2 RT-PCR assays with the negative results for the first two tests. Among these 70 patients, 21 (30 %) patients had positive SARS-CoV-2 RT-PCR results for the third time. As illustrated in Fig. 3 A, older (≥65 years) patients had a higher third-time positive rate (32 %, 7/22) than younger (29 %, 14/48) patients had, although the difference is not significant (p = 0.82).
Fig. 3.
Estimation of false negative result of SARS-CoV-2 RT-PCR assay and comparison of throat swabs and nasal swabs.
(A). Three sequential SARS-CoV-2 RT-PCR assays. Red square indicates positive results for the third time and blue square indicate negative results. (B) Comparison of SARS-CoV-2 RT-PCR assay from paired specimens obtained by throat swabs and nasal swabs.
3.5. Throat swabs and Nasal swabs for SARS-CoV-2 RT-PCR assay
Of the 1113 tests for SARS-CoV-2 by RT-PCR assay, 74 tests (37 pairs) were obtained from both throat swabs and nasal swabs at the same time. As shown in Fig. 3B, results of positive nasal swabs and negative throat swabs were found in 12 pairs of tests (32.4 %), while results of positive throat swabs and negative nasal swabs were found in only 2 pairs of tests (5.4 %). Same results of throat and nasal swabs were found in 23 pairs (62.2 %), with 4 pairs (10.8 %) for both positive and 19 pairs (51.4 %) for both negative. 46 pairs (92 tests) were obtain from throat swabs at one certain time point followed by nasal swabs at the next time-point. As shown in Fig. 3B, negative throat swabs followed by a positive nasal swab were found in 41.3 % (19/46) of samples, while negative throat swabs followed by a negative nasal swab were found in 58.7 % (27/46) of samples. According to the viral infection confirmed by nasal swabs, the estimated false negative rate of SARS-CoV-2 RT-PCR assay with specimens from throat swabs is 41.3 %.
4. Discussion
This is a large-scale report from 301 COVID-19 patients with 1113 samples of RT-PCR tests for SARS-CoV-2 detection. The data and results from this study is notable for providing solid evidence of SARS-CoV-2 dynamic profile in patients infected.
Family of human coronavirus is the main pathogens of respiratory infection. Global epidemics caused by two highly pathogenic coronavirus, SARS-CoV and MERS-CoV, were reported during 2002–2003 and 2012 (ongoing) [5,13]. Although SARS-CoV-2 share similar sequencing characteristics with SARS-CoV and MERS-CoV, study of case series suggested the viral nucleic acid shedding pattern of patients infected with SARS-CoV-2 is different from SARS-CoV, which had a modest viral loads in the early stage and peaked approximately 10 days after symptoms onset [14]. Our study analyzed the dynamic profile of SARS-CoV-2 in hospitalized COVID-19 patients. The median period between onset of symptoms and positive SARS-CoV-2 RT-PCR test result was 16 days (IQR, 10–23). (Fig. 1A) Positive rate of SARS-CoV-2 RT-PCR was detected soon after the onset of symptoms, with a gradually decreasing trend thereafter. We showed that after 4 weeks (>28 days), 26.3 % samples are still positive for SARS-CoV-2. (Fig. 1B) The above findings suggested that SARS-CoV-2 viral replication has a relatively long period in infected patients compared to SARS-CoV [14]. Although viral replication is not necessarily associated with severity of immunopathological damage, it implied that, to limit transmission of the disease, infected patients need exclusive monitoring until at least two consecutive negative RT-PCR results are obtained. It should be noted that one patient had a positive result of SARS-CoV-2 RT-PCR assay on day 42 after the onset of symptoms. This, currently, is the longest period had been reported.
The impact of demographic factors on dynamic profile of SARS-CoV-2 has not been studied before. The previous studies suggested that coronavirus is more likely to infect older individuals with weaker immune functions [3,4,15]. We collected the test results of SARS-CoV-2 RT-PCR assay and performed the subgroup analysis by age. Older patients (≥65 years) had a significant longer time interval (18 days) of positive SARS-CoV-2 RT-PCR test result than patients <65 years (14 days). (Fig. 2A) One possible explanation is that the dysfunction of immune system in older patients resulted in a prolonged elimination of the virus. Male patients had a slightly longer time interval (17 days) of positive SARS-CoV-2 RT-PCR assay than female patients (15 days). Smoking and alcohol history may relate to the above finding.
As a result of errors in sampling and testing, false negative result of RT-PCR for SARS-CoV-2 is very common in clinical settings. Meanwhile, it is recommended by the current diagnosis and treatment guideline for SARS-CoV-2 from Chinese National Health Committee that the criteria to discharge a patient included the relief of symptoms, improvement in radiography and two consecutive negative RT-PCR results for SARS-CoV-2 [12]. We analyzed results from 70 patients with three consecutive viral tests. We found that 21 patients (30 %) had a positive third-time RT-PCR test, even though the results were negative for the previous two tests. (Fig. 3A) This evidence revealed the defect (high false positive rate) of current SARS-CoV-2 detection method and questioned the criteria for discharge and discontinuation of quarantine mentioned above. Releasing potential contagions to public will have substantial influence in disease control and transmission, especially when patients were asymptomatic [10,16,17].
Recent report of 18 COVID-19 patients by Zou et al. suggested that higher viral loads were detected in the nose than in the throat [9]. We compared the results of SARS-CoV-2 RT-PCR assay for 37 pairs of specimens from nasal and throat swabs. Positive nasal swabs and negative throat swabs were found in 12 pairs (32.4 %) of samples, compared to only 2 pairs of samples were positive throat swabs and negative nasal swabs. (Fig. 3B). 46 pairs of samples are an initial negative SARS-CoV-2 RT-PCR assay from throat swabs followed by a sequential SARS-CoV-2 RT-PCR assay from nasal swabs. We found that 19 pairs (41.3 %) of samples from nasal swabs were positive regardless of the initial negative throat swabs. These findings suggested that upper respiratory specimens obtained from nasal swabs were more sensitive and reliable for SARS-CoV-2 RT-PCR assay.
To the date of this manuscript drafted (February 27th, 2020), COVID-19 rapidly spreads from Wuhan to the entire country. The epidemic of COVID-19 also threatened countries such as Korea, Italy, Iraq and Japan, etc. [2]. We have acquired some knowledge of the virus from series case reports and laboratory findings, while much work remains to be done and many questions remain unanswered. Evidence suggested that the outbreaks of COVID-19 may be correlated to its rapid person-to-person transmission ability. Since specific treatment had not been validated for COVID-19, traditional public health tactics—isolation, quarantine and community containment are critical to control the spread [[17], [18], [19], [20]]. This study has found valuable and solid evidence of the dynamic profile of SARS-CoV-2 and offered suggestion to improve current detection method and criteria for discharge. The overall period of patient infected by SARS-CoV-2 was relatively long especially in old individuals. Therefore, we suggested prolonged observation and repeat confirmation of RT-PCR assay from nasal swabs specimens for safe discharges.
The present study has several limitations that should be taken into consideration. First, in this retrospective setting, the accuracy of SARS-CoV-2 RT-PCR may vary (improvement of the detection protocol and gain of experience in sampling). Second, this study only investigated the impact of age and gender on dynamic profile of SARS-CoV-2, while other factors were not included. Third, some information of such as RT-PCR test after discharge, period of negative SARS-CoV-2 RT-PCR test result in some included patients were incomplete. Therefore, a prospective study with a strict inclusion criteria and more clinical-pathological measurements are needed to validate the findings.
5. Conclusions
In summary, in this study we for the first time provided information of SARS-CoV-2 dynamic profile, with the median duration for viral conversion of 20 days after onset of symptom. Old age is the risk factor for prolonged virus replication (22 days). Throat swabs should be replaced by nasal swabs to obtain the specimens for viral detection and the current criteria for discharge should be adjusted.
Funding
No funding resources to declare for this study.
Ethical approval
This study was approved by the Ethics committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All procedures followed in this study were in accordance with the 1964 Helsinki Declaration and later versions.
Informed consent
Oral consent was obtained from patients involved before enrollment when data were collected retrospectively.
Transparency document
CRediT authorship contribution statement
Ai Tang Xiao: Conceptualization, Writing - original draft. Yi Xin Tong: Formal analysis, Project administration. Chun Gao: Formal analysis, Project administration. Li Zhu: Data curation, Methodology, Software. Yu Jie Zhang: Data curation, Methodology, Software. Sheng Zhang: Conceptualization, Writing - original draft.
Declaration of Competing Interest
All authors declare that there are no conflicts of interest.
Acknowledgement
The authors declare that they have no competing interests. No financial support to declare. We thank Ms. Cheng Chen for English grammatic correction of this manuscript.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104346.
Contributor Information
Ai Tang Xiao, Email: pekjean@126.com.
Yi Xin Tong, Email: yx_tong@126.com.
Chun Gao, Email: gaochun75@hotmail.com.
Li Zhu, Email: 125683844@qq.com.
Yu Jie Zhang, Email: 1179204979@qq.com.
Sheng Zhang, Email: aloof3737@126.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J. Med. Virol. 2020 doi: 10.1002/jmv.25678. Published online Jan 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zunyou W., Jennifer M.G. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. Published online February 24. [DOI] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia inWuhan, China: a descriptive study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30211-7. Published January 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. Published January 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization; Geneva: 2004. Summary of Probable SARS Cases with Onset of Illness from 1 November 2002 to 31 July 2003. (https://www. who. int/ csr/ sars/ country/ table2004_04_21/en/) [Google Scholar]
- 6.Ping Y., Jiang Z., Zhengdong Z. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa077. Published February 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F., Yuan S., Kok K. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. Published online January 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan. China. JAMA. 2020 doi: 10.1001/jama.2020.1585. Published online February 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2001737. Published online February 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothe C., Schunk M., Sothmann P. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2001468. Published online January 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.2020. Interim Infection Prevention and Control Recommendations for Patients with Confirmed Coronavirus Disease 2019 (COVID-19) or Persons under Investigation for COVID-19 in Healthcare Settings.https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control.html Update: February 21. [Google Scholar]
- 12.China National Health Commission . 2020. Diagnosis and treatment of 2019-nCoV pneumonia in China. (Version 5)http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a/files/ab6bec7f93e64e7f998d802991203cd6.pdf In Chinese. Published February 8, 2020. Accessed February 26. [Google Scholar]
- 13.Haagmans B.L., Al Dhahiry S.H., Reusken C.B. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiris J.S., Chu C.M., Cheng V.C. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badawi A., Ryoo S.G. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): a systematic review and meta-analysis. Int. J. Infect. Dis. 2016;49:129–133. doi: 10.1016/j.ijid.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan L.T., Nguyen T.V., Luong Q.C. Importation and human-to-human transmission of a novel coronavirus in Vietnam. Published online January 28. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., Guan X., Wu P. Early transmission dynamics inWuhan, China, of novel coronavirus-infected pneumonia. Published on January 29, 2020. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCloskey B., Heymann D.L. SARS to novel coronavirus: old lessons and new lessons. Epidemiol. Infect. 2020;148:e22. doi: 10.1017/S0950268820000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Z., Wang L., Cauchemez S. Risk for transportation of 2019 novel coronavirus disease from Wuhan to other cities in China. Emerg Infect Dis. 2020;26(5) doi: 10.3201/eid2605.200146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan L., Dan X., Guangming Y. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2783. Published online February 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.