Abstract
The rapid global emergence of SARS-CoV-2 has been the cause of significant health concern, highlighting the immediate need for antivirals. Viral RNA-dependent RNA polymerases (RdRp) play essential roles in viral RNA synthesis, and thus remains the target of choice for the prophylactic or curative treatment of several viral diseases, due to high sequence and structural conservation. To date, the most promising broad-spectrum class of viral RdRp inhibitors are nucleoside analogues (NAs), with over 25 approved for the treatment of several medically important viral diseases. However, Coronaviruses stand out as a particularly challenging case for NA drug design due to the presence of an exonuclease (ExoN) domain capable of excising incorporated NAs and thus providing resistance to many of these available antivirals. Here we use the available structures of the SARS-CoV RdRp and ExoN proteins, as well as Lassa virus N exonuclease to derive models of catalytically competent SARS-CoV-2 enzymes. We then map a promising NA candidate, GS-441524 (the active metabolite of Remdesivir) to the nucleoside active site of both proteins, identifying the residues important for nucleotide recognition, discrimination, and excision. Interestingly, GS-441524 addresses both enzyme active sites in a manner consistent with significant incorporation, delayed chain termination, and altered excision due to the ribose 1′-CN group, which may account for the increased antiviral effect compared to other available analogues. Additionally, we propose structural and function implications of two previously identified RdRp resistance mutations in relation to resistance against Remdesivir. This study highlights the importance of considering the balance between incorporation and excision properties of NAs between the RdRp and ExoN.
Keywords: RNA-Dependent RNA polymerase, Exonuclease, Nucleotide analogue, Remdesivir, Resistance, Mutation, Coronavirus, COVID-19
Highlights
-
•
Sequence comparison of SARS-CoV and SARS-CoV-2 for nsp12 (RdRp) and nsp14 (ExoN) show near structural & functional identity.
-
•
Base & ribose modifications of GS-441524 5′-TP vs ATP point to specific discrimination mechanisms & active site residues.
-
•
Lassa virus N & SARS-CoV nsp14 ExoN domains allow modeling catalytically competent SARS-CoV-2 nsp14 Exonuclease.
-
•
RNA terminated with GS-441524 5′-MP at the 3′-end indicate a problematic accommodation at the nsp14 ExoN active site.
-
•
Anti-SARS-CoV-2 nucleoside analog design must consider balance between incorporation & excision by nsp12 & nsp14.
1. Introduction
The recent emergence of a new SARS-like coronavirus in December 2019 (now named SARS-CoV-2) likely from the Huanan seafood market in Wuhan China, is the cause of significant global health concern. SARS-CoV-2 has been shown to be closest related (~88%) to two bat-derived SARS-like CoVs (bat-SL-CoVZC45 and bat-SL-CoVZXC21), with ~79% overall sequence identity to SARS-CoV and ~50% to MERS-CoV. At the present time, any mention of the number of cases, affected countries, and case/fatality ratio will be outdated at print time.
Coronaviruses (CoV) are enveloped, positive-sense RNA viruses of the order Nidovirales, with genome sizes ranging from 26 to 32 kilobases (Gorbalenya et al., 2006). They have been identified across a range of avian and mammalian hosts, but did not attract much attention until November 2002, with the emergence of severe acute respiratory syndrome CoV (SARS-CoV) from Guangdong in southern China (Peiris et al., 2003), resulting in over 8000 human infections and 774 deaths across 37 countries. In September 2012, a second human pathogen: middle-east respiratory syndrome CoV (MERS-CoV), emerged in Saudi Arabia (Zaki et al., 2012), causing 2494 confirmed cases of infection with 858 deaths, a case fatality rate of >34%.
To date, neither prophylactic nor therapeutic options are available for the control or treatment of any human CoV. Recent compassionate clinical trials of the anti-malaria drug hydroxychloroquine as well as Remdesivir (see below) have been conducted, and await publication in scientific journals. The rapid global emergence of SARS-CoV-2 outlines the importance of and immediate need for antivirals. Potential broad-spectrum targets include viral gene products that are widely conserved and do not exist in the host cell, or that are structurally and functionally different enough from cellular homologous to achieve selective inhibition. For RNA viruses, the RNA-dependent RNA polymerase (RdRp) presents an optimal target due to its crucial role in RNA synthesis, lack of host homolog and high sequence and structural conservation. The RdRp remains the target of choice for the treatment of several viral diseases, including chronic liver disease caused by hepatitis C virus infection. Arguably the most promising, broad-spectrum class of viral RdRp inhibitors are nucleos(t)ide analogues (NAs). Upon delivery into the host cell, nucleoside/nucleotide prodrugs are metabolized into an active 5′-triphosphate form (5′-TP) which competes with endogenous nucleotides as substrates for the viral RdRp. The NAs are subsequently incorporated into the nascent viral RNA by the RdRp, accounting for the antiviral effect through several mechanisms of action (MoA).
Firstly, NA incorporation may cause termination of RNA synthesis. This can be either obligate, i.e. the analogue lacks a 3′-OH required for RNA chain extension, or non-obligate, i.e. the analogue perturbs the product RNA structure enough to stop further synthesis by the polymerase. Sofosbuvir is a pro-drug of 2′F-2′C-ME-UMP that has a 3′-OH but acts as a chain-terminator nonetheless (Fung et al., 2014). In combination with other drugs, Sofosbuvir is widely and successfully used to cure HCV infections, but no data are available regarding activity against coronaviruses.
A second MoA exists without termination or slow-down of RNA synthesis, but rather through high level incorporation of NA-TPs throughout the nascent RNA. These NAs are unable to be recognized as ‘regular’ Watson-Crick nucleobases during subsequent rounds of RNA synthesis from the NA-containing template. This results in an increase in mutations and ultimately leads to non-viable genomes, a process known as ‘lethal mutagenesis’ (Crotty et al., 2000). Ribavirin (Rbv) and Favipiravir (Furuta et al., 2002) belong to this class of antivirals, and are active against a variety of viruses (eg., HCV, influenza virus, Ebola virus). In the case of SARS-CoV, Rbv shows some efficacy in infected cells (Chen et al., 2004), Rbv-5′-monophosphate (Rbv-MP) is incorporated into RNA in vitro (Ferron et al., 2018), however the drug does not control coronavirus replication in patients (Booth et al., 2003; Gross and Bryson, 2015). Likewise, the mutagenic effect of β-D-N4 hydroxycytidine (EIDD-1931) and its isopropyl ester prodrug EIDD-2801 against a number of viruses has been demonstrated including coronaviruses (Agostini et al., 2019), and very recently SARS-CoV-2 (biorxiv.org/content/10.1101/2020.03.19.997890v1.full.pdf).
The two MoAs outlined above are not mutually exclusive. The domination of one mechanism over the other, as well as variations and/or intermediate effects are dictated by the structural and functional properties of the viral RdRp. In any case, the viral RdRp's low fidelity, ie., its inability to distinguish NA-TPs from endogenous NTPs, is largely responsible for both antiviral effects.
Coronaviruses stand out as a particularly challenging case for NAs drug design due to the presence of an additional, CoV-specific mechanism which impairs NAs potency. NAs incorporated into RNA can be removed by the CoV exonuclease (ExoN) residing in the N-terminal domain of nsp14 (Bouvet et al., 2012, Minskaia et al., 2006, Snijder et al., 2003). The CoV ExoN interacts with the very processive trimeric RNA polymerase complex consisting of the viral RdRp (nsp12) and associated cofactors (nsp7 and nsp8) to perform proofreading activity (Ferron et al., 2018; Subissi et al., 2014). This mechanism was demonstrated for Rbv-TP, which is readily incorporated as a purine nucleotide in biochemical assays. However, 3′-terminal Rbv-MP is readily excised from RNA by the SARS-CoV ExoN, a removal that is thought to jeopardize NA potency against CoVs (Ferron et al., 2018; Smith et al., 2013). Rbv is thus ineffective at doses regularly used to treat other viral infections such as hepatitis C virus (McHutchison et al., 1998), respiratory syncytial virus (Hall et al., 1983) and Lassa fever virus (McCormick et al., 1986).
Despite this, NAs remain a good option for the treatment of CoV infections based on the high level of structural conservation of the viral RdRps, particularly regarding nucleotide binding sites. Additionally, NAs display a relatively high barrier to resistance relative to other antivirals, as escape mutations likely come at a high cost to viral replication. In support of this, the broad-spectrum AMP analogue Remdesivir (GS-5734) which is currently used for the treatment of several diverse viral infections has shown promising results, inhibiting viral replication of both SARS-CoV and MERS-CoV in various in vitro systems, including primary human airway epithelial cultures, and reducing disease severity in a mouse model (Sheahan et al., 2017; Agostini et al., 2018). Recently, Remdesivir was shown to inhibit SARS-CoV-2 viral replication in cell culture, supporting the potential of this NA produg to be used for the broad-spectrum treatment of CoV infections (Wang et al., 2020). Importantly, passaging of the model β-CoV murine hepatitis virus (MHV) in the presence of the parent nucleoside GS-441524 yielded two resistance mutations in the viral RdRp (F476L and V553L), which were also shown to confer resistance in SARS-CoV (F480L and V557L in SARS-CoV) (Agostini et al., 2018). However, resistance came at a cost to viral replication in vitro, and furthermore attenuated SARS-CoV in vivo. As is the case for Rbv-TP, the ExoN of CoV may be able to excise incorporated GS 441524-MP, as shown by the increased potency against ExoN deficient viruses (Agostini et al., 2018). This raises interesting questions as to the balance between incorporation of NA-TPs by the RdRp and removal by the ExoN of the NA-MPs, whereby the NA-TPs must be incorporated by the polymerase faster than the excision rate of the ExoN.
Here we perform structural-based sequence alignments of the RdRp and ExoN domains of the SARS-CoV to those of the SARS-CoV-2. We report the high level of sequence conservation in key motifs within these enzymes supporting the conjecture that NAs can be used as broad-spectrum antivirals to treat different CoVs. We review the molecular mechanism of resistance against Remdesivir brought by the F480L and V557L mutations (Agostini et al., 2018; Sexton et al., 2016), as well as possible nucleoside structural determinants (modifications at the ribose and nucleobase) for optimal NA efficiency. Likewise, using the crystal structure of SARS-CoV nsp14 Exo as well as structural alignment with other homologous DE (D/E)Eh ExoN enzymes, such as that of the N protein of Lassa virus (Jiang et al., 2013), we map the contacts of the RNA terminated with GS-441524-MP at the 3′-end with the SARS-CoV ExoN active site. Coupling NAs with ExoN inhibitors may be an attractive option, and may further reduce viral escape potential.
2. Results
2.1. Genetic diversity in the nsp12 gene
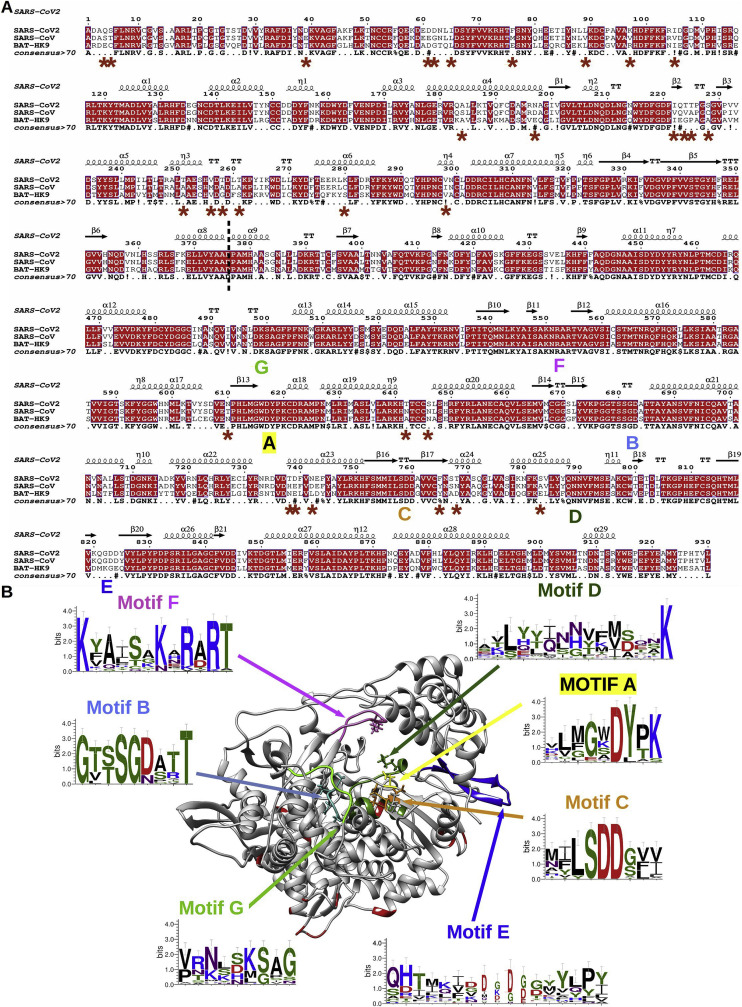
As of Feb 17, 2020, 90 complete SARS-CoV-2 genome sequences have been published and analyzed in the Nexstrain repository (Nextstrain.org), with the first sequence deposited December 2019. Alignment of nsp12 for the whole CoV family indicates that the SARS-CoV-2 nsp12 is almost identical to that of the SARS-CoV (96% identity 98% similarity): a total of 31 amino acid (aa) changes are present along the protein of 804 amino acids (Fig. 1 A). Of these, twenty two map to the nucleotidyltransferase (NiRAN) (Lehmann et al., 2015); a large domain located N-terminal of the RdRp core that does not appear to play a structural nor functional role in polymerase activity (Shannon & Canard, unpublished).. While the NiRAN is a genetic marker for the Nidovirales order for which no viral or cellular homologs have been identified, it characteristically displays a low level of sequence conservation throughout the order. Furthermore, while a nucleotidylation activity has been defined for the small-genome arterivirus equine arteritis virus (EAV) (Lehmann et al., 2015), the exact role of the NiRAN domain in the Nidovirus viral life-cycle is unknown. The remaining nine mutations are located in the C-terminal RdRp domain of nsp12 (Fig. 1A), and only one of them (S783A) is a non-conservative mutation.
Fig. 1.
(A) Amino acid sequence alignment of selected coronavirus nsp12 protein, including: SARS-CoV2 (YP_009724389), SARS-CoV (NC_004718); BatHK9 (NC_009021). SARS-CoV2 nsp12 secondary structure (derived from PDB: 6NUR) is indicated on top. Fully conserved residues are in red font and boxed, whereas partially conserved residues are displayed in red font (above 70% conservation). A consensus sequence is generated using same criteria uppercase is identity, “.” is no consensus, ! is any of I,V; $ is any of L,M, % is any of F,Y; # is any of N,D,Q,E,B,Z. Red star marked the mutation between SARS-CoV2 and SARS-CoV. The NiRAN and the polymerase domains are separated by a dashed line. The polymerase conserved motifs (A–G) are shown under the consensus sequence. Alignments were made using Seaview (Gouy et al., 2010) and Espript (Gouet et al., 2003) (B) Cartoon representation of nsp12 SARS-CoV (PDB: 6NUR). The motifs are colored as follows: A, yellow; B, cyan; C, orange; D, forest green; E, blue; F, pink; G, light green. Around the structure are WebLogo (Crooks et al., 2004) highlighting the A-G motifs. All structural models were made using Chimera (Pettersen et al., 2004).
We subsequently performed a structure-based alignment using the available Cryo-EM structure of SARS-CoV nsp12 to evaluate the potential structural and functional impact of the SARS-CoV-2 amino acid changes (Fig. 1B). The SARS-CoV RdRp domain adopts a classical right hand fold with “Fingers”, “Thumb”, and “Palm” subdomains (Kirchdoerfer and Ward, 2019). At both the structural and sequence levels, the conserved, canonical A to G motifs can be readily identified, and none of the mutations map within these motifs. We conclude that the SARS-CoV-2 RdRp structure and function is unlikely to differ significantly from that of SARS-CoV, which allows the latter to be considered as a faithful surrogate for structure/function analysis.
2.2. A model of RNA and incoming NTP at the nsp12 polymerase active-site
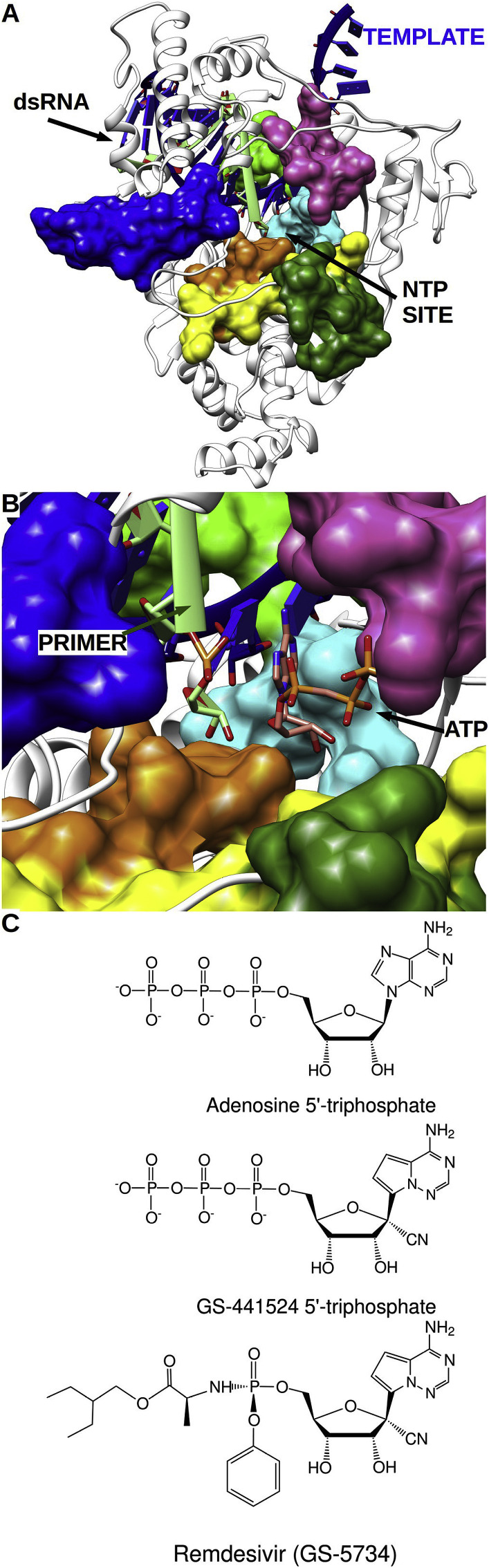
In the absence of an X-ray or Cryo-EM structure of a ternary complex of a CoV RdRp bound to RNA template-primer and incoming nucleotide triphosphate (NTP), we took advantage of the structural and phylogenetic relatedness to picornavirus RdRps, for which the structures of several ternary RdRp-RNA-NTP elongation complexes exist (Kirchdoerfer and Ward, 2019). Using the SARS-CoV nsp12 cryo-EM structure (PDB 6NUR), we built a model by stacking nsp12 onto a set of related RdRps described by Peersen (2019) and used the T7 RNA polymerase ternary complex (Yin and Steitz, 2004) to position both RNA and the incoming NTP. RNA and ATP were subsequently modelled at the SARS-CoV nsp12 active site. Overall, the structure is similar to that of the poliovirus polymerase: Motifs A-G encircle and constitute the polymerase active site, with an open NTP entry tunnel leading to the catalytic center (Fig. 2 A). In CoV nsp12, motifs A,B,C,D and F the most conserved, and this conservation extends to Picornaviridae and Flaviviridae families (Campagnola et al., 2015): it is thus reasonable to extrapolate that the mechanism of action is similar to that of other viral RdRps, and in particular to Picornaviridae with which CoVs share the “small thumb” feature (Peersen, 2019). The ribose moiety of the NTP poised to be incorporated is properly positioned by a conserved serine located in motif B and probably by residues of motif F (Fig. 2B, light blue and purple, see below).
Fig. 2.
(A) Cartoon representation of nsp12 SARS-CoV-2 with spatial arrangement of RdRp motifs complexed with a modelled RNA template (blue) and neo synthesised (light green). Models are derived from either PDB model 6NUR or 6M71; the latter has a 2.9 Å resolution (biorxiv.org/content/10.1101/2020.03.16.993386v1.full.pdf) and they exhibit a RMSD of 0.612 Å over 790 amino acids, which means that the model is valid in both. Color code is the same as in Fig. 1. (B) Close up of the NTP entry and catalytic sites with NTP in position to be incorporated. Position of RNA and NTP was obtained by superimposition of references crystal structures of poliovirus and T7 RNA polymerases elongation complexes (PDB 4K4S and 1S76). (C) Chemical structures of nucleotides discussed throughout the manuscript including: (a) ATP (b) GS-441524-TP (c) Remdesivir.
2.3. GS-441524-TP at the nsp12 active-site
The spectrum of NAs active against human pathogenic CoVs has been recently reviewed (Pruijssers and Denison, 2019). Five NAs; Remdesivir, β-D-N4 hydroxycytidine, Gemcitabine, 6-Azauridine, Mizoribine and Acyclovir-Fleximer, show moderate-to-potent activity against three human pathogenic CoVs (SARS-CoV, MERS- CoV, and HCoV-NL63). One striking feature of this activity spectrum is the structural peculiarity of analogues relative to their potency towards CoVs as well as other RNA viruses. Obviously, the activity spectrum can be linked to either a modified ribose, a modified base, of both.
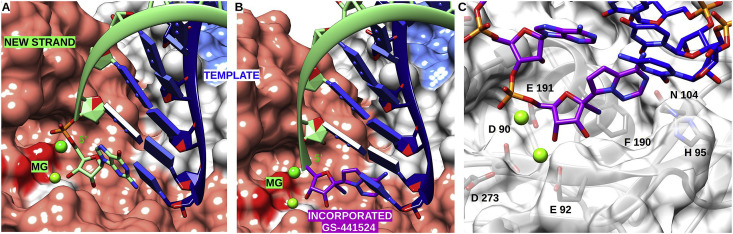
Remdesivir (GS-5734) is one of the few nucleotide analogues reported to be active against SARS-CoV (reviewed in (Pruijssers and Denison, 2019)). It carries a cyano group on the 1′ position of the ribose (1′-CN) and a 4-aza-7,9-dideazaadenosine nucleobase (pseudo-adenine) linked to the ribose by a C–C bond (Fig. 2C). Fig. 3 A shows the natural substrate ATP poised to receive the catalytic nucleophile attack on its α-phosphate. Modelling GS 441524-TP at the nsp12 active site shows that the 1′-CN group is freely accommodated in the close vicinity of motif B (Fig. 3B), a motif that has been reported to act as a fidelity check-point during active site closure with a conserved serine (here S682, see below) (Campagnola et al., 2015).
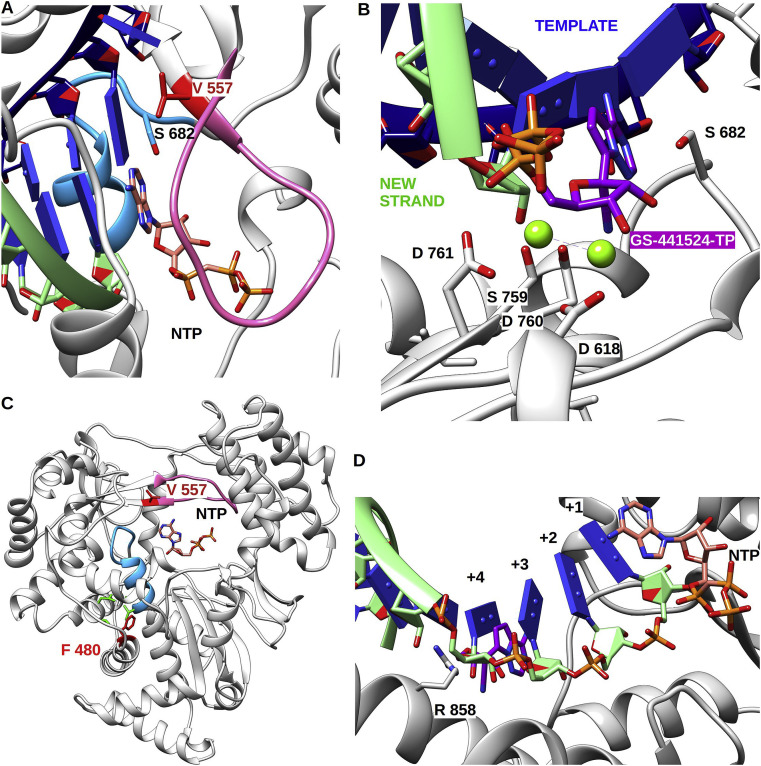
Fig. 3.
Cartoon representation of nsp12 SARS-CoV polymerase domain (PDB: 6NUR) pre-incorporation of GS-441524-TP and mutation mapping. (A) Top view of V 557 with RNA strands and NTP in pre-incorporation position. V 557 forms a hydrophobic wall upon which the template base is stacked. (B) Close up of RNA strands with GS-441524-TP in a pre-incorporation position at the catalytic site. Template strand is in blue, newly synthesised strand is in light green, and GS-441524-TP in purple. Key residues of active site (from motif A, B and C) are shown in stick and labelled. (C) Remdesivir resistance mutations highlighted in red on the cartoon representation of nsp12 SARS-CoV polymerase domain. F 480 faces a patch of hydrophobic residues shown in green that indirectly impact motif B (in cyan). V 557 is located on the side of the template groove and is at the end of motif F (in pink). NTP in pre-incorporation position is shown in stick. (D) View of the newly synthesised RNA in mix representation stick from position 1 to 4 and ribbon for position five and above, for clarity reason template RNA is not shown. Shown is a NTP in a pre-incorporation position and newly nascent RNA. In position +4 the conserved R 858 is poised to make a steric clash with the 1′-CN group of GS-441524 once it has been incorporated and translocated.
2.4. Structural analysis of nsp12 with GS 441524-TP and related resistance mutations of Remdesivir (GS 5734)
In a recent study, Agostini et al. evaluated the mode of action (MoA) of Remdesivir (GS-5734)21. Passage of wild-type (WT) MHV with the Remdesivir (GS-5734) and/or parent nucleoside GS-441524 resulted in phenotypic resistance associated with two nonsynonymous mutations in the predicted fingers subdomain of the nsp12 RdRp (F480L and V557L SARS-CoV numbering). Given the potential of NAs as broad-spectrum antivirals for the treatment of CoV infections, it is imperative to understand the structural and functional implications of these mutations. The SARS-CoV nsp12 structure (Kirchdoerfer and Ward, 2019) allows us to generate a reliable prediction by extending on prior knowledge gathered from other viral RdRps.
Neither of the resistance mutations directly impact the catalytic site nor substrate-binding pocket, but rather cause minor structural alterations which likely impact an NTP ‘checking step’ performed by the polymerase before catalysis (Fig. 3A and B): Residue F480 is located in the hydrophobic core of the protein, at the interface between the fingers and palm subdomains (Fig. 3C). The phenyl ring is orientated towards hydrophobic residues from the palm (V637, L638) and the fingers (I579, V693), and plays a structural role in tightening the two domains by a strong mesh of hydrophobic interactions. In contrast, residue V557 is located at the end of motif F on the second strand forming the side of the RNA template entry channel (Fig. 3A). Based on the theoretical model derived from the poliovirus polymerase elongation complex (4K4S) (Gong et al., 2013), it can be predicted that the valine side chain faces the template RNA and interacts with the base to be paired with the incoming NTP. F480 is completely buried while V557 is partially exposed to solvent with solvent accessibility of 4.2 and 35.2% respectively. Nevertheless, the server of protein thermodynamic stability changes upon single-site mutations (Dehouck et al., 2009) and predicts for both mutations a slight increase in Free energy (ΔG) of the protein, corresponding to a structural relaxation.
The conserved F480L mutation reduces the amount of interactions within the hydrophobic core, thereby retaining the overall conformation while lowering the structural rigidity. The resulting increase of free energy should allow a greater degree of freedom of the secondary structure elements which harbors those hydrophobic residues. In related viral RdRps, the serine of motif B is thought to be responsible for allowing the proper positioning of the ribose moiety of the incoming NTP via the establishment of a hydrogen bond network with the 2’ OH group of the ribose (Fig. 3A). The structure of motif B is considered loose, but its conformational change upon binding the NTP is considered as a fidelity check point of viral RdRps (Peersen, 2017). Therefore, one possible interpretation is that phenylalanine to leucine mutation allows a relaxation in the positioning of the serine, leading to an altered fidelity check. Only detailed experimental work using, eg., pre-steady state kinetics, would determine how a better incorporation of ATP relative to that of GS 441524-TP might be achieved.
Valine 557 is also located in the vicinity of motif B (Fig. 3A and C). The consequence of a valine to leucine mutation is an extended hydrophobic side chain, generating a steric hindrance with the template RNA. As a result, it is expected that the RNA would be deviated from the grove away from the serine of motif B. Again, it is possible to hypothesize that this deviation would alter the serine fidelity check, allowing easier incorporation of ATP relative to GS 441524-TP thus resulting in discrimination against the latter. Again, validation of this hypothesis requires detailed kinetics investigating nucleotide incorporation at the nsp12 active site.
It is interesting to note that the nucleotide at the 3′-end shows some possible room at its 1′-position, hinting that GS 441524-TP, once incorporated, could potentially translocate (Fig. 3D). However, after about 4 translocation events, we anticipate a major steric clash of the 1′-CN group with R858, suggesting that GS 441524-TP might act as a delayed chain terminator. Delayed chain termination mediated by GS 441524-MP has been demonstrated for Respiratory Syncitial virus (Warren et al., 2016), Nipah virus (Jordan et al., 2018) and Ebola virus (Gordon et al., 2020) RdRps, and more relevant to this discussion for the MERS-CoV RdRp complex (Gordon et al., 2020). Likewise, the action of the ExoN remains to be determined once GS 441524-MP has been incorporated and buried in the nascent RNA chain. Future biochemical experiments should address these issues which have implication not only for Remdesivir MoA but also for any up-coming therapeutic NA.
2.5. Genetic diversity in the nsp14 ExoN
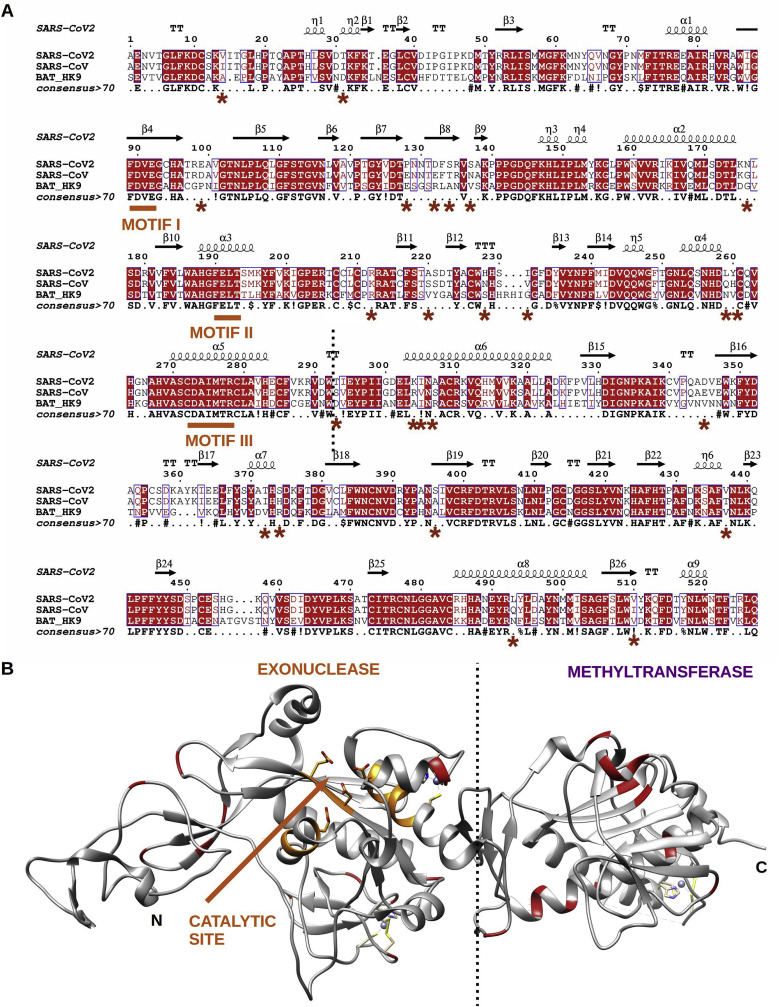
The overall identity of nsp14 sequences from the SARS-CoV Frankfurt isolate and the SARS-CoV-2 Wuhan-1 isolate is 95% regarding both the entire nsp14 gene and the ExoN domain alone (Fig. 4 A). Out of the 14 amino-acid changes in the ExoN domain, four involve non-conserved amino-acids. The ExoN active site has been structurally defined with three conserved motifs, DXE, W(X)4EL, and DAIMTR (Ogando et al., 2019). None of the amino-acid changes map to these motifs, which is confirmed by a structure based alignment performed in a similar way as that of Fig. 1. Amino-acid changes can thus be considered as polymorphisms, and the SARS-CoV nsp14 ExoN domain can be considered a valid model for that of SARS-CoV-2.
Fig. 4.
(A) Amino acid sequence alignment of selected coronavirus nsp14 proteins: SARS-CoV-2 (YP_009724389), SARS-CoV (NC_004718); BatHK9 (NC_009021). SARS-CoV nsp14 secondary structure (derived from PDB: 5NFY) is indicated on top. Fully conserved residues are in red and boxed, whereas partially conserved residues are displayed in red only (above 70% conservation). A consensus sequence is generated using same criteria as in Fig. 1. Residues marked with a red star show polymorphisms between SARS-CoV-2 and SARS-CoV. The N-terminal exonuclease (ExoN) and C-terminal methyltransferase domains are separated by a dashed vertical line. The ExoN catalytic residues (motifs I, II, and III) are underlined in orange. (B) Cartoon representation of the crystal structure of the nsp14 SARS-CoV (PDB: 5NFY). The nsp14 ExoN domain, catalytic site is highlighted in orange; and star residues are shown in red. None of the mutations are in the vicinity of the catalytic site.
2.6. A model of RNA at the nsp14 exonuclease active-site
We took advantage of the SARS-CoV nsp14 crystal structure and the exonuclease structure of Lassa N protein (Jiang et al., 2013). Both proteins belong to the same DE (D/E)Dh family, and the Lassa N exoN domain has been crystallized with dsRNA, which thus allows modelling of the dsRNA into the nsp14 ExoN active site (Fig. 5 ).
Fig. 5.
The excision step catalyzed by nsp14. Close-up of nsp14 ExoN active site (PDB: 5NFY). Modelling of active site ions and RNA was done using a superimposition of the related Lassa virus ExoN structure complexed with RNA and ions (PDB: 4FVU and 4GV9). (A) Reference position of RNA and ions in the excision of an RNA 3′-end nucleotide. (B) Position of RNA and ions in the case of GS-441524-MP excision. GS-441524-MP is structurally superimposed to the ribose of nucleotide in position of A. The distorted base of GS-441524-MP would force the ribose to move in place of the ions preventing the proper distances for an efficient two metal ion catalysis to happen. (C) Same view as B detailed with catalytic (DEED) and binding residues (FHN) in stick and transparent surface.
Fig. 5A shows that the modeling a regular 3′-terminal nucleotide in the active site is possible without any major structural distortion, as expected. When this model is built with GS 441524-TP, it shows that the cyano group at the 1′-ribose position of GS 441524-TP would fit easily (Fig. 5B). However, no proper positioning of the pseudo-adenine can be easily achieved. Superimposition of the ribose so as to position the metal ions for proper catalysis implicates a potential steric clash of the GS 441524 pseudo-adenine with surrounding nucleotides (Fig. 5C). In other words, if the pseudo-adenine base-pairs properly, the α-phosphate would be pushed away and unaligned for catalysis with the metal ions. It is thus likely that excision of GS 441524-MP is significantly less efficient than that of a regular nucleotide. Alternately, the GS 441524-MP might remain stuck in the polymerase active site as described above for delayed chain termination, and be inaccessible for excision.
Taken together, the structural features of GS 441524-TP are consistent with the latter acting as a NA-TP substrate at the nsp12 active site, and a somehow disabled substrate for excision by the nsp14 ExoN, which could account for its anti-CoV effect.
3. Discussion
As proven by the recent SARS-CoV-2 outbreak, the emergence of new, significant human pathogens from zoonotic CoVs is a threat, and the development of broad-spectrum antivirals remains a priority. The RdRp domain remains an extremely attractive target for antivirals due to the high level of structural-conservation across diverse RNA viruses; denoting the potential for broad-spectrum applicability coupled with the advantage of having a relatively high barrier for resistance mutations. NAs constitute an attractive option, with success against several diverse RNA viruses. Nevertheless, CoVs are a particularly challenging case due to the additional presence of a unique exonuclease activity in their nsp14 gene product, which confers resistance to several commonly used NAs including Rbv via an error correcting mechanism.
Remdesivir (GS-5734) remains one of the most promising NA candidates for the treatment of CoV infections, and has recently been advanced to phase 3 trials clinical trials for SARS-CoV-2 based on encouraging pre-clinical results for SARS-CoV (Sheahan et al., 2017; Agostini et al., 2018) and MERS-CoV (de Wit et al., 2020). Studies with MHV and SARS-CoV revealed a partial resistance phenotype could be provided by two mutations in the RdRp domain, although these mutations came at a cost to overall replication (Agostini et al., 2018). Here, we have mapped the two resistance mutants, F480L and V557L, on the SARS-CoV nsp12 structure and describe the structural and functional implications of these changes. While neither mutant directly impacts the catalytic site nor substrate-binding pocket, both result in an increase in free-energy and consequentially a structural relaxation which likely affects the polymerase fidelity-check performed prior to nucleotide incorporation.
Interestingly, MHV V553 had been previously identified as a potential fidelity-regulating residue in CoVs based on comparison with known fidelity-altering mutants in the coxsackie B3 polymerase (Sexton et al., 2016). A V553I mutation engineered into murine hepatitis virus CoV was associated with a decrease in accumulation of mutations over time (a.k.a increased fidelity) and was shown to confer resistance to the mutagens 5-fluorouridine (5-FU) and 5-azacytidine (5-AZC). The conservation of fidelity-regulating residues between distantly related RNA viruses indicates that these RdRps function in an analogous manner due to a high level of structural-conservation, despite limited sequence-similarity.
To discover and design novel potent NAs against CoVs, an additional level of complexity has to be addressed relative to other RNA pathogenic viruses. NAs incorporated into coronaviral RNA have to escape the proofreading ExoN brought by the nsp14 N-terminus domain. In the absence nsp14 structural data at reasonable resolution for the drug-designer (ie., <2.5 Å) (Ferron et al., 2018; Ma et al., 2015), and in complex with relevant substrates, we show here that the related Lassa virus N protein ExoN domain for which this type of data exist (Jiang et al., 2013) can guide NA MoA studies. As shown in Fig. 5 the replacement of a standard nucleotide by GS-441524 generate a structural distortion and a clash between its non-planar nucleobase part and the template base. To accommodate this structurally, the ribose would have to move away from its canonical position and towards the catalytic ions. Due to the small size of the catalytic cavity, this would generate in turn a displacement on the catalytic ions from the active site, as shown for a similar two-metal ion catalysis ExoN reaction where any slight modification in ion position or distortion leads to inhibition of the reaction (Yekwa et al., 2017). Better structurally defined nsp14:RNA complexes are needed to fully understand the structural requirements of 3′-terminal nucleotides for ExoN-mediated removal. Of note, the engineered MHV V553I RdRp mutant was found to be inextricably linked with ExoN activity, i.e. the fidelity effect could only be detected in ExoN deficient virus (Sexton et al., 2016). Importantly, as is the case for Rbv, the ExoN of CoV may be able to recognise and excise incorporated GS 441524-MP, as shown by the increased potency against ExoN deficient viruses (Agostini et al., 2018). However, unlike other Rbv and 5-FU, Remdesivir is active on WT, and ExoN (−) mutants show only modest 4.5 fold increase in sensitivity to Remdesivir, indicating that inhibition likely originates by a mixed effect balanced between incorporation and excision.
The increased efficiency of Remdesivir relative to other NAs indicates that its metabolite GS 441524-TP must be incorporated by the polymerase at a level superior to that of the excision rate of the GS 441524-MP by the ExoN, and/or be less accessible or removed less-efficiently by the ExoN. In contrast, while Rbv-TP also appears to be efficiently incorporated into the nascent RNA by the RdRp, it is likely to be excised just as efficiently by the ExoN.
CoV RdRps are able to accommodate a wide variety of different chemical modifications of NAs (e.g., on the ribose, on the base, on both) (Pruijssers and Denison, 2019). The ability of the ExoN domain to recognise these diverse modifications should therefore be taken into consideration, and represents a potential avenue which could be exploited for NA drug development. Studies which analyse the difference in recognition, incorporation of GS 441524 5′-TP and excision of GS 441524-MP compared to other NA-TPs by the CoV RdRp, ExoN and other replication machinery will provide crucial insights into proper design of novel NAs.
Funding
This work was performed with the support of REACTing, a multi-disciplinary collaborative network of French research institutions working on emerging infectious diseases, which aims to prepare and respond to epidemics, and the Fondation pour la Recherche Médicale (Aide aux équipes).
Acknowledgements
The authors would like to thank Veronique Fattorini for excellent technical assistance at various stages of this project.
Contributor Information
Francois Ferron, Email: francois.ferron@afmb.univ-mrs.fr.
Bruno Canard, Email: bruno.canard@afmb.univ-mrs.fr.
References
- Agostini M.L. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L. Small-Molecule antiviral β-d-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C.M. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. J. Am. Med. Assoc. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- Bouvet M. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., Peersen O.B. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015;89:275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- de Wit E. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020 doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinforma. Oxf. Engl. 2009;25:2537–2543. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- Ferron F. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung A. Efficiency of incorporation and chain termination determines the inhibition potency of 2’-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob. Agents Chemother. 2014;58:3636–3645. doi: 10.1128/AAC.02666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Kortus M.G., Nix J.C., Davis R.E., Peersen O.B. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PloS One. 2013;8 doi: 10.1371/journal.pone.0060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020 doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Robert X., Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Gross A.E., Bryson M.L. Oral ribavirin for the treatment of noninfluenza respiratory viral infections: a systematic review. Ann. Pharmacother. 2015;49:1125–1135. doi: 10.1177/1060028015597449. [DOI] [PubMed] [Google Scholar]
- Hall C.B., Walsh E.E., Hruska J.F., Betts R.F., Hall W.J. Ribavirin treatment of experimental respiratory syncytial viral infection. A controlled double-blind study in young adults. J. Am. Med. Assoc. 1983;249:2666–2670. [PubMed] [Google Scholar]
- Jiang X. Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression. J. Biol. Chem. 2013;288:16949–16959. doi: 10.1074/jbc.M112.420521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.C. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J.B. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 1986;314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- McHutchison J.G. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- Minskaia E. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S. The curious case of the Nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peersen O.B. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017;234:4–20. doi: 10.1016/j.virusres.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peersen O.B. A comprehensive superposition of viral polymerase structures. Viruses. 2019;11 doi: 10.3390/v11080745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet Lond. Engl. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pruijssers A.J., Denison M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton N.R. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016;90:7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekwa E., Khourieh J., Canard B., Papageorgiou N., Ferron F. Activity inhibition and crystal polymorphism induced by active-site metal swapping. Acta Crystallogr. Sect. Struct. Biol. 2017;73:641–649. doi: 10.1107/S205979831700866X. [DOI] [PubMed] [Google Scholar]
- Yin Y.W., Steitz T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]