Abstract
The recognition of DNA as an immune stimulatory molecule is an evolutionarily conserved mechanism to initiate rapid innate immune response against microbial pathogens. Recently, the cGAS-STING pathway has been discovered as an important DNA sensing machinery in innate immunity and viral defense. Recent advances have now expanded the roles of cGAS-STING to cancer. Highly aggressive, unstable tumors have evolved to co-opt this program to drive tumorigenic behaviors. In this review, we will discuss the link between the cGAS-STING DNA sensing pathway with anti-tumor immunity as well as cancer progression, genomic instability, the tumor microenvironment, and pharmacologic strategies for cancer therapy.
Statement of significance
The cGAS-STING pathway is an evolutionary conserved defense mechanism against viral infections. Given its role in activating immune surveillance, it has been assumed that this pathway primarily functions as a tumor suppressor. Yet, mounting evidence now suggests that depending on the context, cGAS-STING signaling can also have tumor and metastasis-promoting functions and its chronic activation can paradoxically induce an immune suppressive tumor microenvironment.
Introduction
Cancer has long been described as a wound that does not heal. This important paradigm was first proposed by Rudolph Virchow and highlights the parallels between cancer formation and inflammation (1,2). In normal physiologic response to injury, several mechanisms regulate the timely termination of wound-healing processes (3). However, mounting evidence now paint a complex landscape in which unresolved inflammation is a potent driver of tumorigenesis (4–6). The cytosolic DNA sensing cGAS-STING pathway has emerged as an important mechanism to drive inflammation-driven tumor growth (7). Indeed, chronic activation of cGAS-STING and its downstream effector programs, such as TBK1, has been linked with persistent inflammation and cancer progression (7,8).
cGAS (cyclic GMP-AMP synthase) is a cytosolic DNA sensor that serves to mount an immune response against the invasion of microbial pathogens such as viruses (9). Activation of cGAS, in turn, stimulates the adapter protein STING (stimulator of interferon genes) to trigger interferon signaling (10). The existence of homologs for cGAS and STING across both eukaryotes and prokaryotes suggests that DNA sensing is an evolutionarily conserved mechanism against pathogenic infections (11–15). Beyond the antimicrobial function of cGAS and STING, recent evidence has expanded their roles to cancer, including other cellular functions such as DNA repair and autophagy. In this review, we discuss the dichotomous roles of cGAS-STING in tumorigenesis and the profound implications of this pathway for novel therapeutic approaches against cancer.
Overview of cGAS-STING signaling
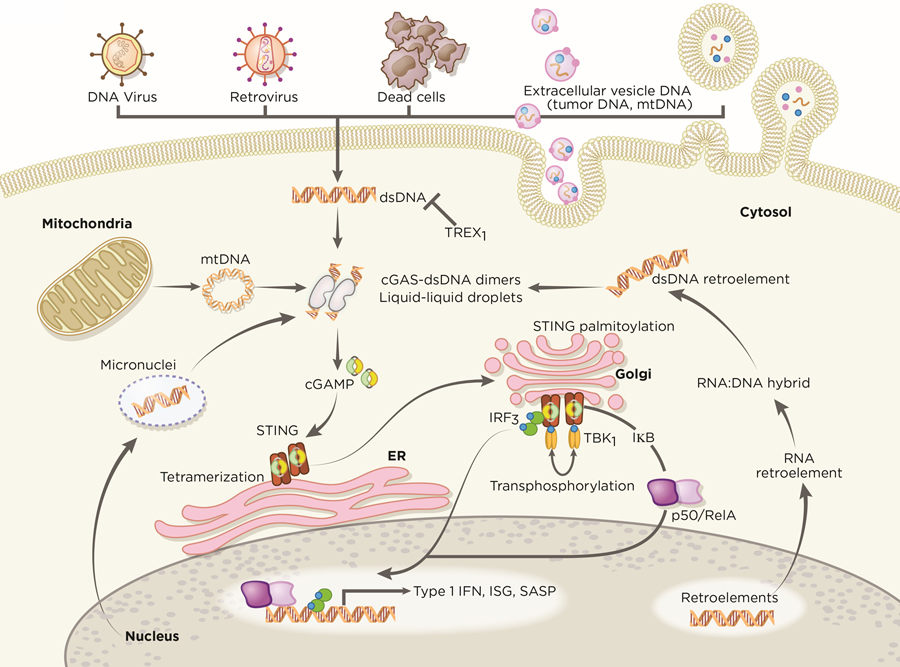
cGAS is activated by interacting with double-stranded (dsDNA) in a sequence-independent manner (9,14,16). The DNA ligands bind with cGAS in a minimal 2:2 complex to induce conformational changes that allow cGAS to catalyze ATP and GTP into 2’,3’-cyclic GMP-AMP (cGAMP), a cyclic dinucleotide comprising of both 2’−5’ and 3’−5’ phosphodiester linkages (17–19). Interestingly, longer DNA is more potent in activating cGAS and promotes liquid-like droplet formations, in which cGAS and dsDNA are spatially concentrated for efficient cGAMP synthesis (20–22). This liquid-liquid phase transition depends on the concentrations of cGAS and DNA, suggesting that a minimal threshold of DNA content must be surpassed to activate cGAS – such as in viral infections. The second messenger cGAMP then activates STING at the endoplasmic reticulum (ER), in which STING undergoes a higher-order oligomerization to form tetramers (23,24) and translocate from the ER to ER-Golgi intermediate compartments (Figure 1). At the Golgi, palymitolyation of STING has been proposed to recruit TANK binding kinase-1 (TBK1) and interferon regulatory factor 3 (IRF3) (25,26). Recent structural studies revealed that tetramerization of STING serves as a signaling platform to recruit and activate TBK1 dimers (26). In turn, TBK1 trans-phosphorylates the C-terminal domains of STING to recruit IRF3 for activation (26,27) at which point, IRF3 translocates to the nucleus and exerts its transcriptional function in expressing immune-stimulated genes (ISG) and type 1 interferons (IFN) (9,19). In parallel, STING also activates IKK to mediate the induction of NF-κB-driven inflammatory genes. Following activation, STING is trafficked to endolysosomes for degradation (28).
Figure 1. cGAS-STING signaling in immunity.
cGAS is an innate immune sensor that recognizes a diverse array of cytosolic dsDNA, which includes DNA with viral, apoptotic, exosomal, mitochondrial, micronuclei, and retroelement origins. cGAS oligomerizes with dsDNA in a 2:2 complex. The interaction of cGAS with DNA induces the formation of liquid droplets through phase transition, in which cGAS exerts its catalytic role to generate the second messenger 2’,3’-cGAMP. The presence of cGAMP stimulates STING at the ER, which undergoes higher-ordered tetramerization. STING translocates from the ER to Golgi compartments and is palmitoylated. STING serves as a signaling platform for TBK1 and IKK. TBK1 phosphorylates STING, which in turn recruits IRF3 for TBK1-mediated phosphorylation. Activated IRF3 dimerizes and translocates to the nucleus. IKK-mediated phosphorylation of the inhibitory IκB protein licenses nuclear entry of p50-RelA. Together with IRF3 in the nucleus, p50-RelA dimers stimulate the transcriptional expression of interferon and other immune-stimulatory genes. Artwork by Terry Helms, Graphics Department, MSKCC.
cGAS-STING in Antiviral Defense
The cGAS-STING pathway serves as a crucial bridge between detection of viral pathogens and immune host defense mechanisms. Growing evidence support the notion that cGAS is an indiscriminate, immune sensor for virus-derived dsDNA, which in turn stimulates downstream interferon signaling. Several reports have demonstrated the protective role of cGAS against a broad range of DNA viruses, which includes vaccinia virus, Kaposi’s sarcoma-associated herpesvirus (KSHV), murine gammaherpesvirus 68, and herpes simplex 1 (HSV-1) (29–31). Furthermore, cGAS also participates in the restriction of retroviruses, such as HIV (32). DNA intermediates generated from reverse transcription of the HIV genome may be recognized by cGAS to stimulate downstream STING-TBK1 signaling (33). This unique property of cGAS-STING has important therapeutic implications as cancers frequently exhibit de-repression of endogenous retroviral elements, which is accentuated by epigenetic modifying therapies (34–36).
Beyond canonical RNA sensors that are extensively reviewed elsewhere (37–39), the cGAS-STING DNA sensing pathway may have indirect roles in facilitating immune responses against RNA viruses. Indeed, cGAS-deficient mice were more vulnerable to West Nile viral infections (30). However, the molecular details in how cGAS-STING restricts RNA viruses remain poorly understood. One plausible mechanism may involve the inadvertent release of organelle-enclosed DNA as a pathologic consequence of viral infections. Indeed, Dengue RNA viruses were shown to induce the cytoplasmic exposure of mitochondrial DNA within infected cells to elicit cGAS-STING dependent IFN signaling, illustrating a mechanism by which cGAS can act as an indirect immune sensor against the presence of dsRNA (40). In cancer, such a mechanism may further sensitize responses to dsRNA as ISG-expressing tumors were revealed to upregulate RIG-I in the setting of chronic STING activation (41).
Despite the crucial role of the cGAS-STING-IFN pathway in antiviral defense, viral pathogens have acquired evasive strategies to escape host immune surveillance to drive pathologic states, such as chronic infection and cancer (42,43). For instance, the HPV18 and human adenovirus 5 DNA tumor viruses encode viral oncoproteins E7 and E1A, respectively, that antagonize STING (44). The ability of viral oncoproteins to suppress the DNA sensing pathway suggests that such oncogenic viruses have evolved dual functions to both promote malignancy and suppress innate immune signaling. Other cancer-related viruses such as KSHV and Hepatitis B express interferon regulatory factor 1, tegument protein ORF52, and viral polymerases that potently disrupt the cGAS-STING pathway (29,45,46). Interestingly, Eaglesham et al. revealed that viral poxins modulate STING signaling by degrading 2’3-cGAMP (42). It remains to be determined if such a mechanism exists in tumor-causing viruses to evade immune recognition. This wide repertoire of viral antagonists against cGAS-STING highlights the importance of evolutionary pressures for DNA tumor viruses to counter this DNA sensing pathway and downstream antitumor programs, which will be described in later sections.
Mechanisms of cGAS-STING activation in cancer
Unlike normal cells, cancer cells are often replete with cytosolic dsDNA. This DNA can be derived from multiple sources including genomic, mitochondrial as well as exogenous origins. The effects of ectopic cytosolic dsDNA in cancer are still poorly understood yet evidence suggests that it may play both anti-tumorigenic and pro-tumorigenic effects in a manner that is dependent on the specific context as well as stage of tumor progression. It is likely that during the early steps of transformation, cytosolic DNA leads to immune surveillance as well as cancer cell-intrinsic senescence. On the other hand, the loss of key cell-cycle and immune checkpoint effectors may enable cytosolic dsDNA to activate chronic inflammatory signaling associated with pro-survival and metastatic programs. Recent evidence suggests that chromosomal instability (CIN) is a primary source of cytosolic dsDNA. CIN is a hallmark of human cancer and it is often associated with tumor progression, therapeutic resistance, and distant metastasis. Cancer cells harboring unstable genomes are prone to chromosome missegregation during mitosis. One consequence of such segregation defects is the generation of micronuclei, a reservoir of nuclear-derived genomic substrates, in a cell-cycle dependent manner (47). Micronuclear envelopes are prone to rupture and expose their genomic contents into the cytosol, which in turn triggers the cGAS-cGAMP-STING pathway (48,49) (Figure 1). The regulators of cGAS-STING activation, such as nucleases (DNAses) and the process of cellular compartmentalization itself, are reviewed elsewhere (50). STING mediates the transcriptional activity of a broad repertoire of molecular programs, which include inflammation, senescence, autophagy, and metastasis. Following a similar mechanism, acute genomic stressors induced by radiation, cisplatin, and intrinsic DNA damage generates cytosolic DNA to activate cGAS-STING in cancer cells (7,48,49,51). It is possible that the ability of CIN to promote cGAS-STING activation explains why complex aneuploidy patterns that result from chromosome missegregation are relatively uncommon during the early steps of tumorigenesis and expand once the tumor develops tolerance for a chronic inflammatory milieu (48,49,52–55).
Apart from the nuclear compartment, the mitochondria may serve as another genomic source to stimulate the cytosolic DNA sensing pathway in cancer (Figure 1). Malignant cells undergoing oxidative stress and mitochondrial dysfunction release mtDNA in the cytosol (56). The molecular details that allow mtDNA to escape from the mitochondria require further investigation, but may involve permeabilization of the outer and inner mitochondrial membranes (57). These findings are congruent with the rationale behind several anticancer therapies targeting permeabilization of mitochondria, which results in the release of potent biomolecules to trigger cell death (58,59). It is possible that these pharmacologic compounds induce the cytoplasmic leakage of mtDNA to promote cGAS-STING mediated antitumor responses. As a strategy to avoid the DNA sensing machinery, tumors harboring mitochondrial dysfunction have been shown to actively silence STING, which effectively ablates downstream IFN signaling (56).
On the other hand, tumors also acquire mtDNA from the extracellular milieu to engage the cGAS dsDNA sensing cascade. Indeed, tumors deficient in mtDNA reconstituted their mtDNA pool by exosomal transfer from host cells to restore mitochondrial function and enhance metastatic potential (60,61). Together with recent reports of the cGAS-STING pathway in driving malignant behaviors (62), these observations raise an interesting possibility that cancer cells with scarce cytosolic DNA substrates may seek extratumoral mtDNA to sustain cGAS-STING driven tumorigenesis in addition to supplementing mitochondrial functions.
Interestingly, mtDNA and genomic DNA may not be synonymous with respect to their ability to activate cGAS. Recent evidence suggests that nucleosome-bound chromatin has a higher binding affinity for cGAS, but reduces the catalytic activity of cGAS in comparison to unchromatinized DNA (49,63). These findings suggest that stearic hindrance and constrained architecture of cytosolic chromatin may limit cGAS-catalyzed generation of 2’3-cGAMP, which aligns with the possibility that genetic structures devoid of histones such as mtDNA are more efficient in activating cGAS. However, this concept remains controversial as a separate study suggests that cytoplasmic chromatin increases cGAS activity (51).
Other sources of genomic substrates, such as apoptotic-derived DNA, exosomes, and transposable elements, may also elicit cGAS-STING activation in tumors (Figure 1). For instance, horizontal transfer of DNA from dead HRASV12;c-MYC rat cancer cells engendered tumorigenic phenotypes in p53-deficient mouse fibroblasts (64). In addition, the uptake of stromal-derived and tumor-derived exosomes by cancer cells can also promote malignant behaviors (65,66). The precise composition of the biomolecules within exosomes that are responsible for driving such effects is poorly understood. However, recent reports have linked interferon signaling with exosomal DNA detection by cGAS, as well as chronic cGAS-STING stimulation with metastasis (62,67,68). These observations raise an interesting prospect that delivery of exosome-derived DNA to cancer cells may trigger the cGAS-STING pathway to upregulate pro-tumorigenic programs. Finally, dysregulation of chromatin remodeling genes and interferon signaling has been shown to induce the transcriptional de-repression of retroviral coding sequences residing in 3’ untranslated regions of interferon-stimulated genes in cancer cells (34). The resulting accumulation of dsRNA and reverse-transcribed dsDNA stimulates the RIG-I RNA and cGAS DNA sensing pathways, respectively. Importantly, these immune pathways may further promote interferon responses, which in turn, maintain transcriptional de-repression, facilitating a positive feedback loop to enhance RNA and DNA recognition. With the advent of pharmacologic inhibitors targeting epigenetic modulators for cancer therapy, these observations raise an important question if long-term treatment with such agents might exacerbate tumor progression through chronic innate immune stimulation.
Noncanonical Activation of cGAS and STING in Cancer
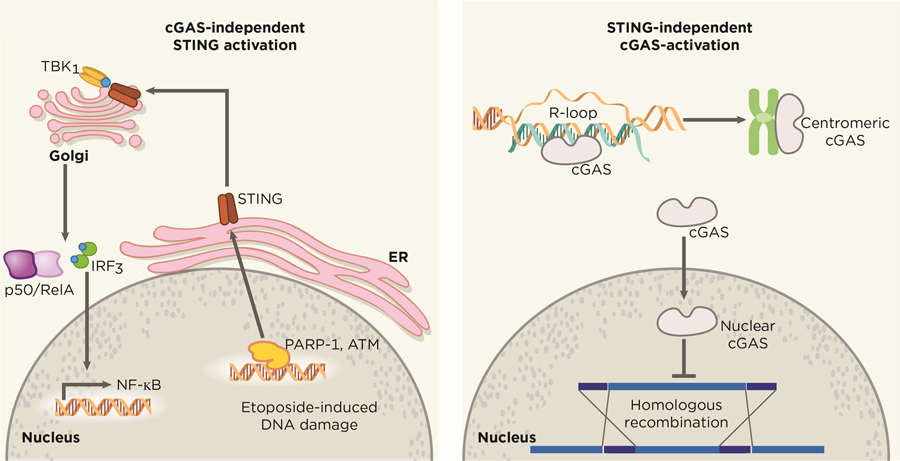
In particular cellular contexts, STING activation is not fully dependent on the cytosolic DNA sensor cGAS (Figure 2). Beyond the classical cGAS-cGAMP-STING axis, Dunphy et al. have revealed an alternative mechanism in which the DNA repair proteins ATM (ataxia telangiectasia mutated) and PARP-1 cooperate with the DNA binding protein IFI16 to promote noncanonical STING signaling in response to etoposide-induced DNA damage (69). In a similar vein, STING regulates cell-cycle progression in a cGAS-independent manner in specific tumor models, such as in HCT116 colorectal carcinomas (70). These results have important implications as tumors lacking cGAS expression may still sustain active STING through other DNA binding partners. These observations open an opportunity to explore how other DNA sensors and adapter proteins may converge on the STING signaling platform and the molecular properties that dictate such engagements in cancer.
Figure 2. Independent functions cGAS and STING.
cGAS and STING possess alternative functions that diverge from the uniaxial and canonical cGAS-cGAMP-STING pathway, exerting effects that are independent from each other. Nuclear translocation of cytosolic cGAS has been shown to drive tumorigenesis through the blockade of HR repair in a STING-independent manner. Breakdown of the nuclear membrane in mitosis has been suggested to allow nuclear entry of cytosolic cGAS, which remains catalytically active and binds to centromeric satellites and LINE DNA repeats. On the other hand, STING can also be activated in a cGAS-independent manner. Etoposide-induced DNA damage recruits the DNA-damage response factors PARP-1 and ATM, which in turn activates STING to drive canonical NF-κB and IRF3-mediated interferon signaling. Artwork by Terry Helms, Graphics Department, MSKCC.
Beyond its function as an innate cytosolic sensor for DNA damage, cGAS may have noncanonical roles in the nucleus (48,49). Contrary to its dominant 2’3-cGAMP production in the cytosol, cGAS is highly enriched in the nuclear compartment on long interspersed nuclear elements and centromeric satellites (71). The mechanism in which cGAS gains nuclear entry remains unknown, but has been proposed to occur as a result of nuclear membrane disassembly in mitosis. It is worth noting that nuclear-localized cGAS exhibit limited responses to endogenous DNA compared to exogenous DNA, suggesting unknown regulators exist to dampen immune responses against self-DNA. Perhaps, the inhibitor effect of chromatin on cGAS catalytic activity is an evolutionary strategy to avoid immune recognition of self-DNA by preventing mitotic cells from ectopically activating cGAS after nuclear-envelope breakdown. This observation also raises the exciting possibility for cGAS to adopt alternative functions in the nucleus. For instance, the unique behavior of cGAS in binding nuclear elements highlights an interesting hypothesis that nuclear cGAS may exert functional roles in epigenetic or chromatin architecture modulation. Furthermore, nuclear cGAS has been shown to facilitate tumorigenesis through the inhibition of homologous-recombination (HR) DNA repair in response to genotoxic-stress induced DNA damage (72,73) (Figure 2). In normal cells, the ability of nuclear cGAS to impair homologous recombination might derive its evolutionary origin from selective advantage for such cells to counter DNA integration by proviruses (74–76).
The nuclear function of cGAS has important implications in tumor formation and cancer therapy. Several cellular safeguards exist to inhibit the propagation of pre-neoplastic cells harboring unresolved DNA damage (77,78). In this setting, it is likely that nuclear cGAS will trigger cell-cycle arrest and apoptotic programs. Indeed, radiation exposure to bone marrow-derived macrophages with functional cGAS exhibited increased mitotic cell death compared to cGAS−/− cells (72). On the other hand, tumors with high tolerance to DNA damage can provide a permissive environment for nuclear cGAS to optimally exert its tumorigenic effects in response to genotoxic stress. For example, chronic radiation exposure in such cells generates double-strand breaks and induces nuclear translocation of cGAS. Subsequently, nuclear cGAS suppresses the HR-DNA repair machinery to license further accumulation of DNA damage in the cancer genome, establishing a feed forward cycle of chromosome instability. The extent of DNA damage is limited as increasing chromosome instability beyond a tolerable threshold reduces tumor fitness (79). This notion highlights an intriguing role for nuclear cGAS in sensitizing tumors to DNA-damaging agents by facilitating lethal accumulations of genetic lesions.
cGAS-STING and Immune Activation and Immune Evasion in Cancer
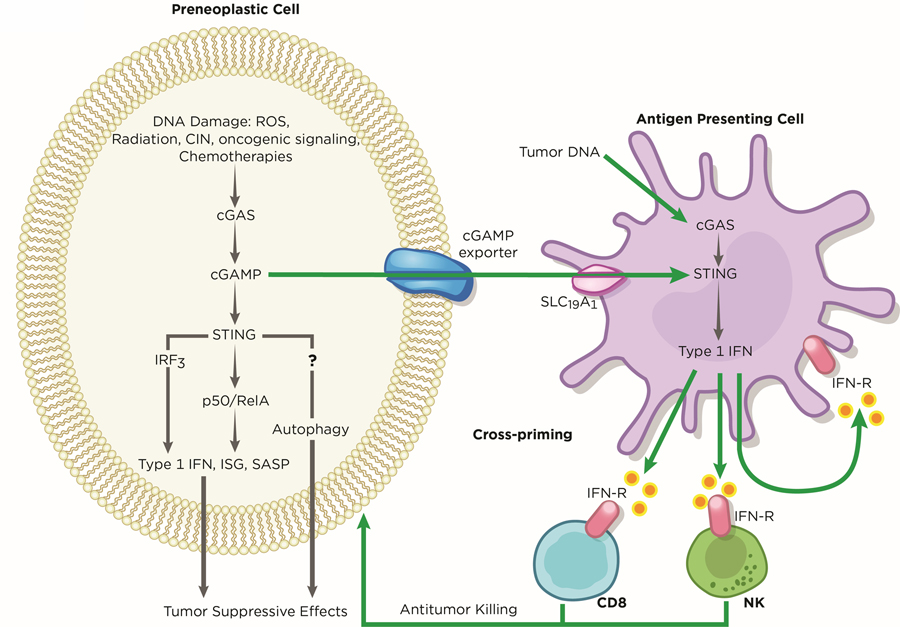
Innate cytosolic DNA sensing plays a crucial role in mounting anti-tumor responses in both a tumor cell-autonomous and non-cell autonomous manner. This tumor surveillance mechanism has been well-characterized and is mediated by infiltrating immune cells, such as natural killer (NK) cells and T cells, through IFN signaling (80,81). Activation of cGAS-STING in cancer cells may serve as a barrier to early neoplastic progression through the upregulation of a battery of inflammatory genes, such as Type 1 IFNs (Figure 3). Importantly, activation of this pathway also mediates the secretion of pro-inflammatory cytokines, chemokines, proteases, and growth factors that are collectively termed SASP (senescence-associated secretory phenotype), which has roles in restricting tumorigenesis (51,53,82). The resulting immune-stimulatory factors from this cascade can either attenuate tumor growth in a cancer cell-autonomous manner or recruit immune cells for tumor clearance (51,52,70). Indeed, suppression of STING in melanoma and pancreatic cancer cells led to reduced immune infiltration, which allowed increased tumor growth in vivo (83,84).
Figure 3. Tumor Suppressive Roles of cGAS-STING.
In early preneoplastic cells, the cGAS-STING pathway exerts its function as a tumor suppressor against oncogenic effects induced by DNA damage. Several sources of DNA damage, such as oxidative stress, radiation, low chromosome instability, hyper-activation of oncogene signaling, and chemotherapies, generates cytosolic DNA, which is sensed by cGAS within tumors. cGAS, in turn stimulates STING to upregulate the expression of type 1 IFN, ISG, and SASP genes. Through an unknown mechanism, STING-mediated autophagy may cooperate with IRF3 and canonical NF-κB signaling to inhibit or delay cancer progression. In addition, the cGAS-STING pathway allows crosstalk between tumors and neighboring immune cells to regulate antitumor immunity. Tumor-derived cGAMP and tumor DNA licenses antigen presenting cells, such as dendritic cells, to activate cGAS-STING signaling, triggering immune cells to mediate tumor clearance. Artwork by Terry Helms, Graphics Department, MSKCC.
Apart from cancer specific cGAS-STING activation, the host may also harness this inflammatory pathway for tumor surveillance (Figure 3). Antigen-presenting cells (APCs), such as dendritic cells (DCs) and macrophages, are thought to clear necrotic malignant cells (85,86). Through an unestablished mechanism, tumor DNA is thought to be transferred and released into the cytosol of DCs and macrophages (87). The accumulation of tumor DNA, in turn, activates STING-IRF3-induced IFN signaling to enforce tumor-antigen presentation on DCs and, as such, cross-prime CD8 T cells for anti-tumor immunity (87–89). Alternatively, tumors may secrete cGAMP into the extracellular space, which is imported into host immune cells through the folate transporter SLC19A1 (90). Tumor-derived cGAMP subsequently activates host STING-IRF3 and induce NK-mediated tumor killing (81). Although DCs are suggested as primary responders to tumor cGAMP (88), it is possible that other cell types such as fibroblasts also possess such function, which may depend on tumor types and the microenvironment. Further studies will be required to unveil the full repertoire of immune cells that detect tumor-cGAMP and how the tumor and host cGAS-STING axis cooperates to facilitate tumor suppression.
Tumors with high levels of CIN face a unique challenge in which their chronic release of cytosolic DNA poses a risk to activate cGAS-STING mediated IFN signaling. To circumvent such suppressive effects, cancer cells have adopted strategies to inhibit this pathway and drive tumorigenic programs. Cancer cells may silence the cytosolic DNA sensing pathway to evade immune surveillance. Decreased protein expression of cGAS and STING has been shown in a small number of late-stage tumors (91–93). This is congruent with a pan-cancer analysis that revealed a small subset of tumors (~1–25%) harboring increased methylations in the promoters of cGAS and STING compared to matched normal tissues (94). In this same report, reduced methylation patterns were observed in other tumors such as pancreatic and thyroid cancers, suggesting that loss of cGAS and STING to facilitate immune evasion is not a universal feature of cancer and is likely context-dependent (95).
The majority of tumors, however, retain some level of cGAS and STING protein expression, which suggests that direct silencing of these genes is not the dominant mechanism for immune evasion. Tumors may escape immune detection by co-opting STING-dependent DNA sensing. For example, HER2-AKT activation in melanoma and colorectal adenocarcinoma cells was shown to selectively abrogate the appropriate activation of the TBK1-IRF3 signaling downstream of STING (96). Furthermore, cancer cells treated with STING agonist markedly increased PD-L1 expression and pro-inflammatory cytokines (97,98). Although the mechanism remains to be determined, Lim et al. have demonstrated that RelA/NF-κB signaling stabilizes PD-L1 protein expression to coordinate evasion of T cell immune surveillance and promote tumor growth (99), which raises a unique prospect for the STING-driven RelA/NF-κB pathway in cancer cells to modulate co-inhibitory checkpoint molecules for immune evasion. In addition, STING-dependent DNA sensing may play a crucial role in shaping an immunosuppressive tumor microenvironment. In human squamous cell carcinomas, STING signaling abrogated tumor immunogenicity by recruiting regulatory T cells (100). In this same vein, exposure to radiation in MC38 colon tumors altered the immune landscape by mobilizing myeloid suppressor cells in a host STING-dependent manner (101). Additional studies will be required to further delineate the stepwise mechanism in how tumor and host STING activity facilitates a potent immunosuppressive environment. Collectively, these observations highlight the importance of cGAS-STING activation not only in eliciting tumor immunogenicity, but also driving malignant behaviors, such as immune evasion. Such dichotomous roles for this DNA sensing pathway will be further explored below.
cGAS-STING and Senescence – a double-edged sword
Recent studies have revealed an important link between the cGAS-STING cascade and cellular senescence, a state of irreversible cell-cycle arrest. In response to senescence-inducing genotoxic stress, such as radiation and oncogene activation, growth-arrested cells generate cytosolic DNA substrates, which alerts the cytoplasmic DNA sensor cGAS (51,52,70). This, in turn, activates STING-mediated induction of SASP in a NF-κB dependent manner (51,53,82) (Figure 3). Components of the SASP promote inflammation and reinforce cell-cycle arrest (102,103). Indeed, oncogenic RAS activation has been shown to induce senescence and SASP in a cGAS-STING dependent manner (51,52). The fact that SASP is driven by NF-κB and not by the IRF3-IFN pathway, suggests non-redundant roles of downstream STING effectors in regulating senescence (51).
Transcriptional data of a subset of human tumors revealed that poor patient survival associated with reduced cGAS and STING expression (53,93). In line with these findings, experimental evidence suggests that loss of cGAS or STING exhibits compromised senescence and SASP responses, accelerated spontaneous immortalization, and increased tumor growth (51–53,70). It is important to note that high expression of STING has also been shown to correlate with poor prognosis in a subset of colorectal cancer patients (104), reflecting the potential role for STING in promoting tumor growth and immune evasion. The incongruent relationship between cGAS-STING RNA levels and patient outcome across different cancers may reflect technical limitations of bulk RNA sequencing in these studies, i.e. actual cGAS-STING expression state is masked by contaminating stromal cells, or represent biological differences in which cGAS-STING exerts tumor-promoting or suppressing roles in particular tumor types.
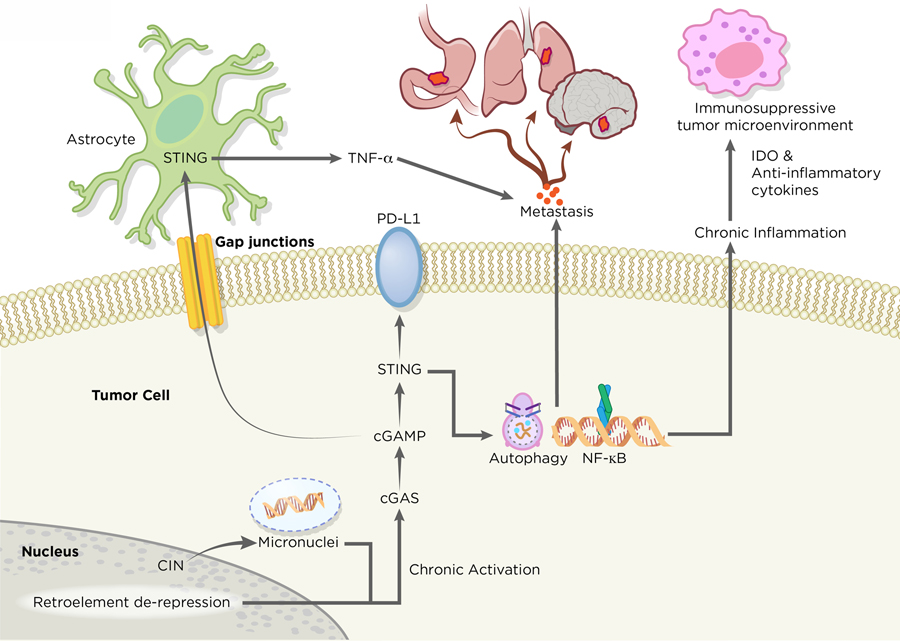
Paradoxically, and in the absence of functional downstream cell cycle effectors such as p53 and p21 among others, chronic stimulation of cGAS-STING signaling exerts tumorigenic effects by establishing an immune suppressive tumor microenvironment leading to metastasis and resistance to chemotherapeutic agents (5). Unlike acute STING-driven SASP, chronic SASP-related inflammation is correlated with malignant behaviors, such as evasion of oncogene-induced senescence and immune-suppression (51,105). Similarly, chromosomally unstable tumors chronically activate STING-dependent noncanonical NF-κB signaling to drive secretion of proinflammatory cytokines and metastasis (62) (Figure 4). The molecular basis underlying the pro-tumorigenic SASP remains to be fully elucidated, yet it may involve particular oncogenic triggers and alternative splicing events that change the composition of SASP to include immune-suppressive and pro-inflammatory cytokines (105,106). Importantly, the release of oncogenic SASP-related factors is not limited to cancer cells. Neighboring growth-arrested fibroblasts can release soluble factors that elicit proliferation of pre-malignant human keratinocytes. This interesting crosstalk between normal tissue and cancer cells raises the possibility that the host-STING mediated SASP phenotypes may have non-cell autonomous roles in driving tumorigenesis (107). Taken together, these observations suggest that the consequences of cGAS-STING inflammatory signaling is context dependent. Acute activation of STING in early neoplastic cells reinforce cell-cycle arrest through SASP. As cancer cells tolerate long-term cGAS-STING signaling and lose downstream cell cycle regulators, the inflammatory processes adopt pro-tumorigenic roles that enable senescence evasion and aggressive tumor growth.
Figure 4. Tumor Promoting Roles of cGAS-STING.
In metastatic tumor cells, the cGAS-STING pathway exerts its role in promoting tumorigenesis. Tumors with high chromosome instability generate micronuclei, which ruptures and releases DNA to the cytosol. STING serves as a signaling platform for a diverse array of tumorigenic programs. Chronic activation of STING signaling leads to suppression of type 1 IFN production and concurrent upregulation of noncanonical NF-κB signaling, pro-survival programs and metastasis. The release of immune-stimulating molecules, such as indoleamine 2,3-dioxygenase (IDO) and anti-inflammatory cytokines, from tumors allows the formation of an immunosuppressive microenvironment. STING may have additional roles in promoting immune evasion and metastasis through regulation of PD-L1 expression and induction of autophagy. Apart from cancer cell-autonomous roles of cGAS-STING, tumors can directly communicate with their external environment through the transfer of cGAMP to neighboring cells through gap junctions, which accelerates neoplastic progression. Artwork by Terry Helms, Graphics Department, MSKC.
It is worth noting that precise spatial control may exist for cGAS activation in cancer. Using human monocyte cell lines, Barnett et al. have revealed that the N-terminus of inactive cGAS anchors to phosphatidylinositol 4,5-bisphosphate PI(4,5)2 at the plasma membrane, which restricts cGAS access to self-DNA (108). Upon activation, cGAS is released into the cytosol to recognize endogenous dsDNA and initiate interferon responses. The interaction of cGAS with PI(4,5)2 raises an interesting prospect for phosphoinositide 3 kinase(PI3K) in regulating cGAS activation, and if high PI3k/AKT signaling licenses chronic activation of cGAS-STING signaling for tumorigenesis. Further work will be required to delineate the molecular requirements that determine the fate of downstream STING signaling to either promote or suppress tumor formation.
cGAS-STING and Autophagy in Cancer
Emerging evidence now supports a link between autophagy and the DNA sensing adapter STING. Autophagy is a primordial function of STING that likely preceded STING’s role in interferon signaling (109,110). Gui et al. have shown that autophagy constitutes an IFN-independent pathway in response to viral infection (110). The paradoxical role of autophagy in tumor progression has been widely studied. In normal cells, autophagy modulates cytoprotective effects to maintain homeostasis by mediating functions that include clearance of damaged organelles, misfolded proteins, and oxygen radicals (111). The removal of such cytotoxic elements impairs the acquisition of genetic abnormalities and mutations that would lead to cancer formation. For instance, mouse hepatocytes with defects in ATG5 and ATG7 exhibit higher levels of oxidative stress and genomic DNA damage that favor their transformation to liver adenomas (112). However, aggressive tumors can co-opt autophagy to drive pro-survival and metastatic programs (111,113,114). The increased metabolic and energy demand to sustain malignancy may force cancer cells to adopt autophagy as a protective mechanism for survival. It is likely that the dual role of autophagy in cancer may be context-dependent based on factors such as genotype, tumor type, and microenvironment.
Similar to autophagy, STING has been shown to adopt dichotomous roles in tumor progression (Figures 3 and 4). It is tempting to postulate that such duality of STING functions emerged as a consequence of its role as a regulator of autophagy. In line with this notion, Nassour et al. demonstrated STING-driven macroautophagy, a subtype of autophagy involving autophagolysosomes, is a critical step in preventing the proliferation of cells undergoing replicative crisis (109). Attenuation of either cGAS-STING or autophagy licensed RB-p53 deficient cells to bypass telomeric crisis and tolerate acquisition of both chromosomal copy number and structural alterations. It is worth noting that this consequence of cGAS-STING activation may depend on the genetic context as functional p53, alternatively, triggers autophagy-independent senescent programs (70,115). The molecular mechanism underlying the STING-autophagy-cell death axis remains to be determined but potential pathways may involve regulation of mitophagy or calcium signaling (116,117). Beyond cellular senescence, these observations illuminate a novel role for STING-mediated autophagy as an additional barrier against early neoplastic progression in normal cells.
Alternatively, the endoplasmic reticulum (ER) stress response and autophagy can contribute to the development of advanced cancers by enabling cancer cells to survive in stressful environments (118). Disseminating cancer cells (DCCs) were shown to exert their dormant and immune evasive potentials through the ER stress response, in which this imbalance of ER homeostasis may help fuel metastasis (113,114) (Figure 4). Taken together with the pro-tumorigenic role for cGAS-STING in chromosomally unstable tumors (62), it is possible that, in certain cellular contexts, the self-DNA sensing pathway may act in concert with autophagy-ER stress programs to potentiate tumor-promoting behaviors. In line with this hypothesis, ER stress induced the upregulation of STING expression and resistance to EGFR tyrosine kinase inhibitors in non-small cell lung cancers (119). Apart from the tumor-cell intrinsic role of ER stress in driving malignancy, the tumor microenvironment may exploit such metabolic programs to facilitate immune suppression (118). The tumor milieu was proposed to restrict proper uptake of nutrients such as glucose in intratumoral T cells, which leads to ER stress and subsequent activation of the IRE1α–XBP1 unfolded protein response pathway. Chronic activation of IRE1α–XBP1 reprograms T cells to adopt a dysfunctional state with decreased antitumor functions. Future studies will be required to define both the molecular hierarchy between cGAS-STING and ER stress and how these pathways interact in driving tumorigenesis.
cGAS-STING and Metastasis
Recent work has demonstrated an important link between chromosome instability (CIN) and tumor metastasis (62). Ongoing chromosome segregation errors generate cytosolic dsDNA that is sensed by the cGAS-STING pathway (49,62). Tumors harboring unstable genomes engage in STING-dependent noncanonical NF-κB and inflammatory responses that favor invasion and metastasis (Figure 4). In parallel, tumors with CIN also suppress anti-viral type 1 IFN production, which may explain the susceptibility of IFN-defective malignant cells to viral oncolysis (120,121). Apart from the role of CIN in driving malignant behaviors by intrinsic cGAS-STING activation in tumors, there is growing evidence that metastasis may be driven in a tumor cell non-autonomous manner. In metastatic human breast tumors, cancer cells communicate with adjacent astrocytes through cGAMP signaling (122). cGAMP generated by tumor cGAS is exported to astrocytes via gap junctions, which in turn activate astrocyte STING and initiate the release of inflammatory cytokines to promote tumor progression and metastatic cancer cell survival in the brain. This bidirectional crosstalk mediated by cGAMP transfer between tumors and normal tissues is reminiscent of mechanisms observed in anti-viral defense, suggesting a potential role in which tumor-derived cGAMP may allow cancer cells to interact with their environment and exert their tumorigenic effects (123).
A crucial unanswered question is how tumors alter the downstream circuitry of STING to adopt metastatic behaviors. One possible mechanism may involve precise control of STING expression levels. In support of this hypothesis, a recent study revealed that the magnitude of STING signaling determined the induction of apoptotic programs in macrophages and T lymphocytes (124), suggesting that modulation of STING activity may select for distinct downstream effector programs. Further investigation is warranted to uncover the molecular requirements and context that dictate metastatic promoting or suppressive outcomes downstream of the cGAS-STING cascade.
cGAS-STING and Response to DNA damaging therapies
The crucial role of the cGAS-STING pathway as a cytosolic DNA sensor and activator of both adaptive and innate immune responses highlights its clinical relevance of DNA-damaging therapies. A growing body of evidence have shown that therapeutic agents such as radiation, poly-(ADP-ribose) polymerase inhibitors (PARPi), and etoposide help generate cytosolic DNA and invoke STING-dependent interferon production for anti-tumor immunity (49,69,125,126). For example, radiation treatment has been linked with cGAS-STING to suppress tumor growth (48,88). Furthermore, PARP inhibition has been shown to elicit cGAS-STING signaling in tumors and host immune cells to promote tumor clearance, proinflammatory signaling, and increased immune infiltration (125–128). It is important to note that the particular stimulus of DNA damage may elicit markedly different programs to activate STING signaling. In contrast to PARP inhibition and radiation, cytosolic DNA induced by etoposide can trigger STING-mediated secretion of IFNs in a cGAS independent manner (69). Taken together, these observations align with the notion that tumors with inherent defects in DNA repair pathways are permissive to cytoplasmic exposure of DNA, which in turn may induce immunogenicity in a cGAS-STING-IRF3 dependent manner (129). Such synergy between DNA damage and cGAS-STING activation raises an interesting prospect for combinatorial strategies targeting these pathways to improve immunologic clearance of tumors.
Persistent stimulation of the cGAS-STING pathway can lead to therapeutic resistance. STING-induced IFN activation by radiation promotes an immunosuppressive environment by recruiting myeloid-derived suppressor cells to MC38 adenocarcinoma cells (101). However, other immune cell types may also help diminish the therapeutic efficacy of radiation. Contrary to the classical role of DCs in IR-tumor control, these cells may concurrently adopt pro-tumorigenic behaviors. An intricate balance between STING-dependent canonical and noncanonical NF-κB pathways co-exists in DCs to modulate antitumor immunity after radiation (130). Defective RelB in DCs with functional STING-IRF3-RelA signaling led to increased IFN production and regression of transplanted murine adenocarcinoma tumors. These results suggest that concurrent activation of the STING-noncanonical NF-κB pathway mitigates optimal radiotherapy effects. As described before, chronic stimulation of cGAS-STING favors noncanonical NF-κB to mediate metastatic behaviors within breast tumors (62). Therefore, it is possible that prolonged activation of STING enables immune cells to participate in a similar mechanism to adopt pro-tumorigenic functions. Taken together, the role of noncanonical NF-κB in mediating malignant phenotypes in both cancer and immune cells provides a rationale for this pathway as a candidate pharmacologic target. Future work will be required to refine our understanding of how the STING-noncanonical NF-κB cascade may drive metastasis and cancer drug resistance.
STING agonists and antagonists – a personalized approach
The ability to harness the host’s immune system in targeting cancer has unleashed a paradigm shift in cancer treatment. Therapeutic antibodies that block immune checkpoint proteins, such as CTLA-4 and PD-1/PD-L1, have led to durable responses in a subset of cancer patients (131–133). However, a substantial proportion of patients either do not receive durable clinical benefit from initial therapy with either CTLA-4 or PD-1 inhibitors or relapse after a partial response (134–140). The limitations of immune-checkpoint therapy highlight the need to search for other modulators that may enhance tumor immunogenicity.
The role of cGAS-STING signaling in modulating antitumor responses has sparked the development of pharmacologic agonists for STING. Inspired by the discovery that 2’3’-cGAMP that activates human STING to initiates robust downstream interferon signaling (9,17–19,141,142), the majority of STING-activating agents are synthetic analogs of 2’3’-cGAMP, which includes chemical modifications that increases STING-induced antitumor efficacy by engendering synthetic cGAMP resistant to hydrolysis (142,143). Direct pharmacologic activation of STING has been shown to restrict tumor growth and enhance immunogenicity in several cancer-bearing mouse models. In immune-competent mouse models of colorectal cancer and melanoma, intra-tumoral injection of cGAMP activates STING-driven interferon responses in dendritic cells, which in turn presents tumor-associated antigens on major histocompatibility complexes to activate CD8 T cells for antitumor killing (143–145). In a subset of tumors such as B cell malignancies, STING may serve as a therapeutic vulnerability as STING agonists have been shown to induce apoptosis, allowing the release of tumor antigens to further cross-prime antitumor T-cells (146). Beyond T-cell driven tumor regression, STING agonists have also been shown to exert critical roles in ensuring the full execution of cell-mediated immunity. Autologous tumor re-challenge in mice previously treated with DMXAA, a mouse-selective STING agonist, impaired reformation of primary tumors (143). Most importantly, STING agonists may also serve as promising adjuvants. cGAMP administration in tumor-bearing mice potentiated the therapeutic effects of immune checkpoint inhibitors and radiation therapy (87,88,145). This synergy may be a consequence of STING activation-induced expression of immunosuppressive molecules, such as PD-L1, in tumors (97). These therapeutic benefits were absent in STING-deficient mice, which highlights the crucial role of host STING signaling in enforcing tumor immunogenicity in non-immunogenic tumors.
The promising avenue of STING as a pharmacologic target for immunotherapy has accelerated the development of human STING agonists, in the form of synthetic cGAMP analogs, for clinical trials. Careful design of such analogs is necessary for clinical efficacy as allelic variants of human STING are poorly responsive to 3’−5’ phosphodiester-linked cGAMP (98,141). Two clinical phase 1 trials are ongoing for intratumoral delivery of STING agonists (ADU-S100 and MK-1454) in solid human tumors and lymphomas (147,148). Dose escalations were well-tolerated and evidence of CD8 tumor infiltrations at injected tumor lesions were observed, including patients with prior treatment with checkpoint inhibitors (148). However, preliminary results have revealed only a modest clinical response with no significant activity seen upon single-agent administration and only partial responses when STING agonists were administered concurrently with anti-PD1 therapy in advanced tumors. It remains to be determined why such limited responses were observed, but the intratumoral approach with the ADU-S100 and MK-1454 STING agonists may have restricted optimal antitumor activity at non-injected tumor lesions. In an attempt to overcome this challenge and expand the pharmacologic toolbox for immunotherapy, Ramanjulu et al. have recently discovered amidobenzimidazole as a novel STING agonist, in which intravenous delivery of this small-molecule elicited robust tumor regression in immune-competent mice (149). The safety profile of systemic administration of STING agonists will remain an important clinical consideration and the effect of such drugs on the patient’s hemodynamic status will need to be closely monitored.
Are chromosomally unstable tumors inherently resistant to STING agonists?
Despite the excitement in harnessing STING agonists, the recent link between cGAS-STING and metastasis provides a rationale for STING inhibition in late-stage cancers (62). Although acute STING activation poses a barrier to early tumorigenesis, prolonged activation of the cGAS-STING signaling axis may inadvertently suppress innate anti-tumor immunity and drive aggressive behaviors. The chronic release of genomic DNA into the cytosol licenses persistent activation of cGAS-STING to suppress antitumor phenotypes and drive metastasis, which may explain the lack of durable clinical benefit from STING agonists in late-stage human tumors. It is likely that constitutive cGAS-STING activation in chromosomally unstable tumors induces pre-existing resistance to STING agonists as such tumors already evolved to eschew the deleterious effects of cytosolic DNA. Pre-existing resistance to constitutive cGAS activity in cancer might also explain the lack of clinical response to STING agonists and raise the promise for STING antagonists, such as the recent discovery of a covalent inhibitor of STING palmitoylation, in therapeutic interventions in attenuating metastasis (150). Furthermore, the role of cGAS in activating STING and inhibiting HDR to drive tumorigenesis highlights the unique prospect for cGAS inhibitors as an orthogonal approach to abrogate cGAS-STING signaling in late-stage cancers (151–153).
The clinical and molecular guidelines for STING agonists and antagonists remain to be determined. Differences in CIN state between primary and metastatic lesions suggest that active CIN may be one factor that helps predict personalized treatments. Indeed, increased CIN, chronic cGAS-STING activation, and poor patient survival, were correlated with metastatic and not primary tumors (62), which reinforces the exciting prospect for STING inhibitors in advanced cancers. Moreover, precise modulation of STING may be required for optimal therapeutic benefit. A recent preclinical study demonstrated that persistent STING activation only mildly reduced tumor growth and abrogated the development of T-cell driven adaptive immunity (154). In a similar vein, chronic engagement of cGAS-STING led to increased carcinogen-induced tumor formation (7). These results suggest that a delicate balance must be maintained for effective anti-tumor responses, in which surpassing a particular threshold of STING activity may license progression of malignancy. Therefore, careful patient selection based on genomic and phenotypic markers of CIN might provide insight into which patients might benefit from STING pathway inhibition or activation.
Concluding Remarks
Beyond the canonical role of cGAS-STING in antiviral immunity, recent evidence has emerged that expands the functional roles of cGAS-STING to cancer. Tumors are able to co-opt the cGAS-STING pathway to either suppress or promote malignancy by regulating a diverse array of molecular programs such as immune activation and evasion, senescence, metastasis, and autophagy.
However, important questions remain unanswered in our understanding of the molecular requirements and context that define how tumors regulate each pathway to drive tumorigenesis in a cGAS-STING dependent manner. For instance, it remains unclear how aggressive tumors with a functional cGAS-STING axis suppress type 1 IFN signaling and upregulate alternative programs, such as the canonical and noncanonical NF-κB pathways, to adopt metastatic behaviors. One possibility may involve modulation of the intensity of cGAS-STING activation in determining the switch between tumor suppressive or promoting functions (124). Yet, the underlying mechanisms that define such outcomes remain poorly understood and require further studies.
The rapid progress in further resolving the molecular details of the cGAS-STING pathway have led to the discoveries of novel roles for cGAS and STING. Recent work has revealed that cGAS and STING may act independently from one another. Indeed, STING activation in response to etoposide-induced DNA damage was independent from the catalytic function of cGAS (69). Apart from its localization in the cytosol, cGAS also exists in the nuclear compartment (49,51,71–73). The presence of nuclear cGAS raises an important question in how the innate DNA sensor tolerates self-DNA (53), which likely involves suppression of nuclear cGAS activity through an undetermined mechanism (71). Further work will be required to define the molecular context in which cGAS and STING act in concert or independently.
Finally, an appreciation of the dichotomous roles of cGAS-STING in cancer progression has great importance for advancing cancer therapeutic strategies. A thorough understanding of the temporal and spatial controls in which cGAS-STING exerts a tumor promoting or suppressive role will improve our ability to appropriately target this pathway an ensuring adequate patient selection. In normal or preneoplastic cells, cGAS-STING activation may sensitize tumors to immune cell clearance. On the other hand, inhibition of this pathway may elicit a vulnerability in late-stage aggressive cancers and as such, pose as a barrier to metastatic spread. Currently, clinical trials are harnessing STING agonists to induce tumor immunogenicity. However, hyper-activation of STING signaling may inadvertently worsen clinical outcomes if tumors have already co-opted the STING pathway to drive malignant programs and to suppress its antitumor functions. It is likely that the particular tumor stage, genotype, CIN state, basal level of cGAS-STING activation, and microenvironment will dictate therapeutic responses to STING agonists or antagonists. Therefore, careful selection of patients will be required to identify the subset of patients who will benefit from pharmacologic therapy that either activate or inhibit STING.
Acknowledgements
SFB is supported by the Office Of The Director, National Institutes Of Health of the National Institutes of Health under Award Number DP5OD026395 High-Risk High-Reward Program, the Department of Defense Breast Cancer Research Breakthrough Award W81XWH- 16-1-0315 (Project: BC151244), the Burroughs Wellcome Fund Career Award for Medical Scientists, the Parker Institute for Immunotherapy at MSKCC, the Josie Robertson Foundation, and the MSKCC core grant P30-CA008748.
Footnotes
Disclosure of Potential Conflicts of Interest
SFB holds a patent related to some of the work described targeting CIN and the cGAS-STING pathway in advanced cancer. He owns equity in, receives compensation from, and serves as a consultant and the Scientific Advisory Board and Board of Directors of Volastra Pharmaceuticals Inc. He has also consulted for Sanofi. JK declares no conflicts of interest.
References
- 1.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet 2001;357(9255):539–45 doi 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 2.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 1986;315(26):1650–9 doi 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 3.Henson PM. Dampening inflammation. Nat Immunol 2005;6(12):1179–81 doi 10.1038/ni1205-1179. [DOI] [PubMed] [Google Scholar]
- 4.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 2009;30(7):1073–81 doi 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 5.Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420(6917):860–7 doi 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140(6):883–99 doi 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun 2014;5:5166 doi 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462(7269):108–12 doi 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (New York, NY) 2013;339(6121):786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009;461(7265):788–92 doi 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kranzusch PJ, Lee ASY, Wilson SC, Solovykh MS, Vance RE, Berger JM, et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 2014;158(5):1011–21 doi 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin M, Hiroyasu A, Guzman RM, Roberts SA, Goodman AG. Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep 2018;23(12):3537–50 e6 doi 10.1016/j.celrep.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol 2015;15(12):760–70 doi 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Wu FH, Wang X, Wang L, Siedow JN, Zhang W, et al. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res 2014;42(13):8243–57 doi 10.1093/nar/gku569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 2012;149(2):358–70 doi 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013;498(7454):332–7 doi 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature 2013;498(7454):380–4 doi 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al. Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 2013;153(5):1094–107 doi 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 2013;341(6152):1390–4 doi 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreeva L, Hiller B, Kostrewa D, Lassig C, de Oliveira Mann CC, Jan Drexler D, et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 2017;549(7672):394–8 doi 10.1038/nature23890. [DOI] [PubMed] [Google Scholar]
- 21.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018;361(6403):704–9 doi 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, et al. Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 2018;174(2):300–11.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang G, Zhang C, Chen ZJ, Bai XC, Zhang X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 2019;567(7748):389–93 doi 10.1038/s41586-019-0998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B, Du F, Xu P, Shu C, Sankaran B, Bell SL, et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 2019;569(7758):718–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, et al. Activation of STING requires palmitoylation at the Golgi. Nat Commun 2016;7:11932 doi 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019;567(7748):394–8 doi 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015;347(6227):aaa2630 doi 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 28.Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu J, Dobbs N, et al. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Rep 2017;21(11):3234–42 doi 10.1016/j.celrep.2017.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, et al. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 2015;112(31):E4306–15 doi 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014;505(7485):691–5 doi 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015;520(7548):553–7 doi 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013;341(6148):903–6 doi 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, et al. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J 2014;33(24):2937–46 doi 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med 2018;24(8):1143–50 doi 10.1038/s41591-018-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassiotis G Endogenous retroviruses and the development of cancer. J Immunol 2014;192(4):1343–9 doi 10.4049/jimmunol.1302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishak CA, Classon M, De Carvalho DD. Deregulation of Retroelements as an Emerging Therapeutic Opportunity in Cancer. Trends Cancer 2018;4(8):583–97 doi 10.1016/j.trecan.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell 2011;147(2):409–22 doi 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 2012;86(6):2900–10 doi 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chow KT, Gale M Jr., Loo YM. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu Rev Immunol 2018;36:667–94 doi 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 40.Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2017;2:17037 doi 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Golji J, Brodeur LK, Chung FS, Chen JT, deBeaumont RS, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med 2019;25(1):95–102 doi 10.1038/s41591-018-0302-5. [DOI] [PubMed] [Google Scholar]
- 42.Eaglesham JB, Pan Y, Kupper TS, Kranzusch PJ. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 2019;566(7743):259–63 doi 10.1038/s41586-019-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Z, Damania B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016;19(2):150–8 doi 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015;350(6260):568–71 doi 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Li J, Chen J, Li Y, Wang W, Du X, et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol 2015;89(4):2287–300 doi 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J-j, Li W, Shao Y, Avey D, Fu B, Gillen J, et al. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell host & microbe 2015;18(3):333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012;482(7383):53–8 doi 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017;548(7668):466–70 doi 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017;548(7668):461–5 doi 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science 2019;363(6431) doi 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 51.Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017;550(7676):402–6 doi 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gluck S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 2017;19(9):1061–70 doi 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A 2017;114(23):E4612–E20 doi 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santaguida S, Richardson A, Iyer DR, M’Saad O, Zasadil L, Knouse KA, et al. Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Dev Cell 2017;41(6):638–51 e5 doi 10.1016/j.devcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012;337(6102):1678–84 doi 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- 56.Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov 2019;9(1):34–45 doi 10.1158/2159-8290.CD-18-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riley JS, Quarato G, Cloix C, Lopez J, O’Prey J, Pearson M, et al. Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J 2018;37(17) doi 10.15252/embj.201899238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov 2010;9(6):447–64 doi 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 59.Galluzzi L, Larochette N, Zamzami N, Kroemer G. Mitochondria as therapeutic targets for cancer chemotherapy. Oncogene 2006;25(34):4812–30 doi 10.1038/sj.onc.1209598. [DOI] [PubMed] [Google Scholar]
- 60.Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 2015;21(1):81–94 doi 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A 2017;114(43):E9066–E75 doi 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553(7689):467–72 doi 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zierhut C, Yamaguchi N, Paredes M, Luo JD, Carroll T, Funabiki H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019;178(2):302–15 e23 doi 10.1016/j.cell.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci U S A 2001;98(11):6407–11 doi 10.1073/pnas.101129998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014;159(3):499–513 doi 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schafer R, Beerling E, et al. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015;161(5):1046–57 doi 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitai Y, Kawasaki T, Sueyoshi T, Kobiyama K, Ishii KJ, Zou J, et al. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J Immunol 2017;198(4):1649–59 doi 10.4049/jimmunol.1601694. [DOI] [PubMed] [Google Scholar]
- 68.Diamond JM, Vanpouille-Box C, Spada S, Rudqvist NP, Chapman JR, Ueberheide BM, et al. Exosomes Shuttle TREX1-Sensitive IFN-Stimulatory dsDNA from Irradiated Cancer Cells to DCs. Cancer Immunol Res 2018;6(8):910–20 doi 10.1158/2326-6066.CIR-17-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, et al. Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-kappaB Signaling after Nuclear DNA Damage. Mol Cell 2018;71(5):745–60 e5 doi 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ranoa DRE, Widau RC, Mallon S, Parekh AD, Nicolae CM, Huang X, et al. STING Promotes Homeostasis via Regulation of Cell Proliferation and Chromosomal Stability. Cancer Res 2019;79(7):1465–79 doi 10.1158/0008-5472.CAN-18-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gentili M, Lahaye X, Nadalin F, Nader GPF, Lombardi EP, Herve S, et al. The N-Terminal Domain of cGAS Determines Preferential Association with Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Rep 2019;26(13):3798 doi 10.1016/j.celrep.2019.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang H, Panda S, Xue X, Liang F, Sung P, & Gekara NOO The innate immune DNA sensor cGAS is a negative regulator of DNA repair hence promotes genome instability and cell death. bioRxiv 2018;465401.
- 73.Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018;563(7729):131–6 doi 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 74.Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol 1999;80 ( Pt 10):2535–43 doi 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 75.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 2003;55(8):451–8 doi 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 76.He CQ, Xie ZX, Han GZ, Dong JB, Wang D, Liu JB, et al. Homologous recombination as an evolutionary force in the avian influenza A virus. Mol Biol Evol 2009;26(1):177–87 doi 10.1093/molbev/msn238. [DOI] [PubMed] [Google Scholar]
- 77.Sherr CJ. Principles of tumor suppression. Cell 2004;116(2):235–46 doi 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 78.Sun W, Yang J. Functional mechanisms for human tumor suppressors. J Cancer 2010;1:136–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andor N, Maley CC, Ji HP. Genomic Instability in Cancer: Teetering on the Limit of Tolerance. Cancer Res 2017;77(9):2179–85 doi 10.1158/0008-5472.CAN-16-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res 2009;69(7):3077–85 doi 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, Raulet DH. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 2018;49(4):754–63 e4 doi 10.1016/j.immuni.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 2008;6(12):2853–68 doi 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takashima K, Takeda Y, Oshiumi H, Shime H, Okabe M, Ikawa M, et al. STING in tumor and host cells cooperatively work for NK cell-mediated tumor growth retardation. Biochem Biophys Res Commun 2016;478(4):1764–71 doi 10.1016/j.bbrc.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 84.Ho SS, Zhang WY, Tan NY, Khatoo M, Suter MA, Tripathi S, et al. The DNA Structure-Specific Endonuclease MUS81 Mediates DNA Sensor STING-Dependent Host Rejection of Prostate Cancer Cells. Immunity 2016;44(5):1177–89 doi 10.1016/j.immuni.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 85.Sauter B, Albert ML, Francisco L, Larsson M, Somersan S, Bhardwaj N. Consequences of cell death: exposure to necrotic tumor cells, but not primary tissue cells or apoptotic cells, induces the maturation of immunostimulatory dendritic cells. J Exp Med 2000;191(3):423–34 doi 10.1084/jem.191.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smyth MJ, Godfrey DI, Trapani JA. A fresh look at tumor immunosurveillance and immunotherapy. Nat Immunol 2001;2(4):293–9 doi 10.1038/86297. [DOI] [PubMed] [Google Scholar]
- 87.Woo S-R, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MYK, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014;41(5):830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014;41(5):843–52 doi 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med 2011;208(10):1989–2003 doi 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ritchie C, Cordova AF, Hess GT, Bassik MC, Li L. SLC19A1 Is an Importer of the Immunotransmitter cGAMP. Mol Cell 2019;75(2):372–81 e5 doi 10.1016/j.molcel.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia T, Konno H, Barber GN. Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res 2016;76(22):6747–59 doi 10.1158/0008-5472.CAN-16-1404. [DOI] [PubMed] [Google Scholar]
- 92.Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep 2016;14(2):282–97 doi 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song S, Peng P, Tang Z, Zhao J, Wu W, Li H, et al. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep 2017;7:39858 doi 10.1038/srep39858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Konno H, Yamauchi S, Berglund A, Putney RM, Mule JJ, Barber GN. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 2018;37(15):2037–51 doi 10.1038/s41388-017-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bakhoum SF, Cantley LC. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018;174(6):1347–60 doi 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu S, Zhang Q, Zhang F, Meng F, Liu S, Zhou R, et al. HER2 recruits AKT1 to disrupt STING signalling and suppress antiviral defence and antitumour immunity. Nat Cell Biol 2019;21(8):1027–40 doi 10.1038/s41556-019-0352-z. [DOI] [PubMed] [Google Scholar]
- 97.Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med 2015;7(283):283ra52 doi 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Corrales L, McWhirter SM, Dubensky TW Jr., Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest 2016;126(7):2404–11 doi 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016;30(6):925–39 doi 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang D, Xiao-Feng H, Guan-Jun D, Er-Ling H, Sheng C, Ting-Ting W, et al. Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim Biophys Acta 2015;1852(11):2494–503 doi 10.1016/j.bbadis.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 101.Liang H, Deng L, Hou Y, Meng X, Huang X, Rao E, et al. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat Commun 2017;8(1):1736 doi 10.1038/s41467-017-01566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 2008;133(6):1019–31 doi 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 103.Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 2008;133(6):1006–18 doi 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 104.An X, Zhu Y, Zheng T, Wang G, Zhang M, Li J, et al. An Analysis of the Expression and Association with Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer. Mol Ther Nucleic Acids 2019;14:80–9 doi 10.1016/j.omtn.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toso A, Revandkar A, Di Mitri D, Guccini I, Proietti M, Sarti M, et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep 2014;9(1):75–89 doi 10.1016/j.celrep.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 106.Georgilis A, Klotz S, Hanley CJ, Herranz N, Weirich B, Morancho B, et al. PTBP1-Mediated Alternative Splicing Regulates the Inflammatory Secretome and the Pro-tumorigenic Effects of Senescent Cells. Cancer Cell 2018;34(1):85–102 e9 doi 10.1016/j.ccell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A 2001;98(21):12072–7 doi 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, et al. Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction between Self- and Viral DNA. Cell 2019;176(6):1432–46 e11 doi 10.1016/j.cell.2019.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nassour J, Radford R, Correia A, Fuste JM, Schoell B, Jauch A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 2019;565(7741):659–63 doi 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019;567(7747):262–6 doi 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Curr Opin Cell Biol 2010;22(2):206–11 doi 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011;25(8):795–800 doi 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pommier A, Anaparthy N, Memos N, Kelley ZL, Gouronnec A, Yan R, et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science 2018;360(6394) doi 10.1126/science.aao4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018;9(1):1944 doi 10.1038/s41467-018-04070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J, Bakhoum SF. Expanding the Role of STING in Cellular Homeostasis and Transformation. Trends Cancer 2019;5(4):195–7 doi 10.1016/j.trecan.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu J, Chen YJ, Dobbs N, Sakai T, Liou J, Miner JJ, et al. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J Exp Med 2019;216(4):867–83 doi 10.1084/jem.20182192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bhatelia K, Singh K, Prajapati P, Sripada L, Roy M, Singh R. MITA modulated autophagy flux promotes cell death in breast cancer cells. Cell Signal 2017;35:73–83 doi 10.1016/j.cellsig.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 118.Song M, Sandoval TA, Chae CS, Chopra S, Tan C, Rutkowski MR, et al. IRE1alpha-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 2018;562(7727):423–8 doi 10.1038/s41586-018-0597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Terai H, Kitajima S, Potter DS, Matsui Y, Quiceno LG, Chen T, et al. ER Stress Signaling Promotes the Survival of Cancer “Persister Cells” Tolerant to EGFR Tyrosine Kinase Inhibitors. Cancer Res 2018;78(4):1044–57 doi 10.1158/0008-5472.CAN-17-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chiocca EA. Oncolytic viruses. Nat Rev Cancer 2002;2(12):938–50 doi 10.1038/nrc948. [DOI] [PubMed] [Google Scholar]
- 121.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med 2000;6(7):821–5 doi 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 122.Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016;533(7604):493–8 doi 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 2015;349(6253):1232–6 doi 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 124.Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, Villunger A, et al. Signalling strength determines proapoptotic functions of STING. Nat Commun 2017;8(1):427 doi 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ding L, Kim HJ, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep 2018;25(11):2972–80 e5 doi 10.1016/j.celrep.2018.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, et al. PARPi Triggers the STING-Dependent Immune Response and Enhances the Therapeutic Efficacy of Immune Checkpoint Blockade Independent of BRCAness. Cancer Res 2019;79(2):311–9 doi 10.1158/0008-5472.CAN-18-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pantelidou C, Sonzogni O, De Oliveria Taveira M, Mehta AK, Kothari A, Wang D, et al. PARP Inhibitor Efficacy Depends on CD8(+) T-cell Recruitment via Intratumoral STING Pathway Activation in BRCA-Deficient Models of Triple-Negative Breast Cancer. Cancer Discov 2019;9(6):722–37 doi 10.1158/2159-8290.CD-18-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov 2019;9(5):646–61 doi 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst 2017;109(1) doi 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hou Y, Liang H, Rao E, Zheng W, Huang X, Deng L, et al. Non-canonical NF-kappaB Antagonizes STING Sensor-Mediated DNA Sensing in Radiotherapy. Immunity 2018;49(3):490–503 e4 doi 10.1016/j.immuni.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest 2015;125(9):3384–91 doi 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang JC, Hughes M, Kammula U, Royal R, Sherry RM, Topalian SL, et al. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. J Immunother 2007;30(8):825–30 doi 10.1097/CJI.0b013e318156e47e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res 2013;19(19):5300–9 doi 10.1158/1078-0432.CCR-13-0143. [DOI] [PubMed] [Google Scholar]
- 134.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017;168(4):707–23 doi 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 2015;373(19):1803–13 doi 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med 2016;375(19):1856–67 doi 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ferris RL, Licitra L, Fayette J, Even C, Blumenschein G Jr., Harrington KJ, et al. Nivolumab in Patients with Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck: Efficacy and Safety in CheckMate 141 by Prior Cetuximab Use. Clin Cancer Res 2019. doi 10.1158/1078-0432.CCR-18-3944. [DOI] [PMC free article] [PubMed]
- 138.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372(21):2018–28 doi 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 139.Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2015;33(18):2004–12 doi 10.1200/JCO.2014.58.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol 2015;33(17):1889–94 doi 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 2013;3(5):1355–61 doi 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 2013;39(6):1019–31 doi 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep 2015;11(7):1018–30 doi 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li T, Cheng H, Yuan H, Xu Q, Shu C, Zhang Y, et al. Antitumor Activity of cGAMP via Stimulation of cGAS-cGAMP-STING-IRF3 Mediated Innate Immune Response. Sci Rep 2016;6:19049 doi 10.1038/srep19049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Demaria O, De Gassart A, Coso S, Gestermann N, Di Domizio J, Flatz L, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A 2015;112(50):15408–13 doi 10.1073/pnas.1512832112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tang CH, Zundell JA, Ranatunga S, Lin C, Nefedova Y, Del Valle JR, et al. Agonist-Mediated Activation of STING Induces Apoptosis in Malignant B Cells. Cancer Res 2016;76(8):2137–52 doi 10.1158/0008-5472.CAN-15-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harrington KJ, Brody J, Ingham M, Strauss J, Cemerski S, Wang M LBA15 Preliminary results of the first-in-human (FIH) study of MK-1454, an agonist of stimulator of interferon genes (STING), as monotherapy or in combination with pembrolizumab (pembro) in patients with advanced solid tumors or lymphomas ESMO Congress. Volume 29 Munich, Germany Annals of Oncology; 2018. [Google Scholar]
- 148.Meric-Bernstam Funda TLW, Hodi F. Stephen, Messersmith Wells, Lewis Nancy, Talluto Craig, Dostalek Mirek, Tao Aiyang, McWhirter Sarah M., Trujillo Damian, Luke Jason J. Phase I Dose-finding Study of MIW815 (ADU-S100), an Intratumoral STING Agonist, in Patients With Advanced Solid Tumors or Lymphomas Society for Immunotherapy for Cancer. Washington D.C: 2018. [Google Scholar]
- 149.Ramanjulu JM, Pesiridis GS, Yang J, Concha N, Singhaus R, Zhang SY, et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 2018;564(7736):439–43 doi 10.1038/s41586-018-0705-y. [DOI] [PubMed] [Google Scholar]
- 150.Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, et al. Targeting STING with covalent small-molecule inhibitors. Nature 2018;559(7713):269–73 doi 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 151.Lama L, Adura C, Xie W, Tomita D, Kamei T, Kuryavyi V, et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat Commun 2019;10(1):2261 doi 10.1038/s41467-019-08620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hall J, Brault A, Vincent F, Weng S, Wang H, Dumlao D, et al. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS One 2017;12(9):e0184843 doi 10.1371/journal.pone.0184843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vincent J, Adura C, Gao P, Luz A, Lama L, Asano Y, et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat Commun 2017;8(1):750 doi 10.1038/s41467-017-00833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sivick KE, Desbien AL, Glickman LH, Reiner GL, Corrales L, Surh NH, et al. Magnitude of Therapeutic STING Activation Determines CD8+ T Cell-Mediated Anti-tumor Immunity. Cell reports 2018;25(11):3074–85.e5. [DOI] [PubMed] [Google Scholar]