Abstract
The large dsDNA virus herpes simplex virus 1 (HSV-1) is considered to be genetically stable, yet it can rapidly evolve in response to strong selective pressures such as antiviral treatment. Deep sequencing has revealed that clinical and laboratory isolates of this virus exist as populations that contain a mixture of minor alleles or variants, similar to many RNA viruses. The classic virology approach of plaque purifying virus creates a genetically homogenous population, but it is not clear how closely this represents the mixed virus populations found in nature. We sought to study the evolution of mixed versus highly purified HSV-1 populations in controlled cell culture conditions, to examine the impact of this genetic diversity on evolution. We found that a mixed population of HSV-1 acquired more genetic diversity and underwent a more dramatic phenotypic shift than a plaque-purified population, producing a viral population that was almost entirely syncytial after just ten passages. At the genomic level, adaptation and genetic diversification occurred at the level of minor alleles or variants in the viral population. Certain genetic variants in the mixed viral population appeared to be positively selected in cell culture, and this shift was also observed in clinical samples during their first passages in vitro. In contrast, the plaque-purified viral population did not appear to change substantially in phenotype or overall quantity of minor allele diversity. These data indicate that HSV-1 is capable of evolving rapidly in a given environment, and that this evolution is facilitated by diversity in the viral population.
Keywords: HSV-1, evolution, minor variant, viral fitness, syncytia
1. Introduction
HSV-1 is a widespread pathogen in the alphaherpesvirus family (Roizman et al. 2013). It is prevalent across the world, infecting an estimated 3.7 billion people, who experience a spectrum of disease outcomes from asymptomatic infections to recurring skin lesions or rarely, encephalitis (Looker et al. 2015). HSV-1 productively replicates in epithelial cells and then latently infects neuronal cells, leading to lifelong persistence in the host. The virus can periodically reactivate from latency to productively replicate in epithelial cells, with the potential for multiple cycles of latency and reactivation from the neuronal reservoir over the life of an infected individual (Roizman et al. 2013). This replication strategy has great advantages for viral persistence, and it also provides multiple opportunities for the virus to evolve within a host. During each active replication event, the virus may undergo selection in response to the cellular and immunological selective pressures that it faces (Hall and Almy 1982; Bower et al. 1999; Drake and Hwang 2005; Sanjuan et al. 2010; McCrone and Lauring 2018). Following productive replication, the progeny viruses can either be transmitted to a new host or may re-enter the nervous system of the current host, potentially expanding the latent reservoir of virus for future reactivation cycles (Kennedy et al. 2015; Lieberman 2016). As the selective pressures between epithelial and neuronal cellular environments are likely different, each cycle of latency and reactivation may create new genetic diversity in the viral population, which may be followed by a bottleneck upon the conversion to latency.
While cycles of latency and reactivation are a well-established and accepted aspect of herpesvirus biology (Pellett and Roizman 2013; Roizman et al. 2013), the concept of HSV-1 exhibiting substantial genetic diversity and evolutionary potential within an infected individual has only recently been established (Bower et al. 1999; Minaya et al. 2017; Pandey et al. 2017; Greninger et al. 2018; Renner and Szpara 2018; Shipley et al. 2018). As with other herpesviruses, HSV-1 has a large double-stranded DNA (dsDNA) genome, and these viruses have generally been considered to be genetically stable in comparison to viruses with an RNA genome (Sanjuan et al. 2010; Sanjuán and Domingo-Calap 2016). However, an increasing body of evidence supports the idea that herpesviruses, including HSV-1, exist as a diverse genetic population in vivo (Depledge et al. 2014; Hage et al. 2017; Renzette et al. 2014, 2016). A similar level of diversity may exist in vitro as well, depending on the method of preparation (reviewed in Renner and Szpara 2018). This genetic diversity can be generated through multiple mechanisms, including polymerase error, copy number variation, and recombination (Hall and Almy 1982; Drake and Hwang 2005; Lee et al. 2015). The HSV-1 polymerase has been previously demonstrated to have a low mutation rate (1 × 10−7 to 1 × 10−8 mutations per base per infectious cycle), although these studies were performed on a single gene in a unique coding region of the HSV-1 genome (Hall and Almy 1982; Brown 2004; Drake and Hwang 2005). The HSV-1 genome consists of unique long (UL) and unique short (US) coding regions, which are flanked by large structural repeats [Internal Repeat Long/Short (IRL/S), Terminal Repeat Long/Short (TRL/S)]. Tandem repeats (TRs) occur frequently in the HSV-1 genome but are especially enriched in the IRL/S and TRL/S regions. Copy number variation or length fluctuations of tandem repeats and homopolymer tracts are a frequent source of genetic variation in strains of HSV-1 (Szpara et al. 2014). Repetitive regions of the HSV-1 genome also have very high G + C content, which favors recombination (Lee et al. 2015). Recombination allows for increased genetic diversity in the absence of polymerase error, which may be especially relevant for herpesviruses (Bloom and Stevens 1994; Lee et al. 2015; Lassalle et al. 2016). These mechanisms contribute to a high level of variation in the large terminal and internal repeats, which contain genes that are critical to HSV-1 replication (ICP0, ICP4, y34.5) (Szpara et al. 2010; Roizman et al. 2013; Parsons et al. 2015). Crucially, HSV-1 is known to respond quite rapidly in response to strong selective pressures such as antiviral drug treatment, whether through de novo mutation or selection of existing variants in the population (Hall and Almy 1982; Burrel et al. 2010; Sauerbrei et al. 2010; Houldcroft et al. 2017).
The genetic diversity of a population of HSV-1 can undergo genetic drift (e.g., single nucleotide polymorphisms (SNPs), insertions/deletions (Indels), or TR length fluctuation) or even more dramatic genetic shifts (e.g., recombination) both in vivo and in vitro. There have been few studies that have followed a population of HSV-1 through longitudinal sampling of one person (Shipley et al. 2018), or studied known transmission events (Pandey et al. 2017). These studies imply that viral population diversity can be generally maintained through transmission (Pandey et al. 2017), but that the viral population can drift or adapt during subsequent rounds of latency and reactivation (Shipley et al. 2018). However, as with most viruses, HSV-1 is quite often studied in cell culture or animal models. Each of these approaches have limitations and differences from authentic replication in the natural host in vivo, with concomitant impacts on viral evolution and genetic diversity. As an example, HSV-1 is routinely propagated in monolayers of African green monkey kidney (Vero) cells. These cells support robust viral replication and are easily manipulated in a laboratory environment. However, these cells are not human in origin and they represent only one cell type (described as fibroblast-like) (Wathelet et al. 1992) of the multiple epithelial and mucosal cell types that HSV-1 encounters in a normal human infection (Roizman et al. 2013). In addition to the lack of adaptive immune control in cell culture, Vero cells lack innate immune defenses, due to a defect in interferon signaling (Emeny and Morgan 1979; Mosca and Pitha 1986; Wathelet et al. 1992). Finally, HSV-1 stocks are often passaged multiple times in Vero cells to amplify the virus, without considering the potential for genetic drift from the source stock. One classic approach used by virologists to reduce the potential for genetic drift was to isolate virus from individual plaques on a monolayer, which is referred to as plaque-purification. This practice constitutes a severe genetic bottleneck, which could lead to unexpected consequences if a rare or deleterious genetic variant is unintentionally selected (Szpara et al. 2010; Jaramillo et al. 2013; Parsons et al. 2015).
In this study, we utilized serial passage of two HSV-1 populations to begin to address several outstanding questions about the ability of this virus to evolve in cell culture. We utilized plaque morphology as an initial marker for genetic diversity, based on our prior observation that a frequently used HSV-1 strain (F) contained a mixture of syncytial and non-syncytial plaque phenotypes (Szpara et al. 2010; Parsons et al. 2015). We also used a ‘purified’ clonal population of the same strain of HSV-1, which was generated through plaque purification of a non-syncytial representative of strain F. By subjecting each of these viral populations to serial passage in Vero cells, we investigated the contribution of genetic diversity to viral fitness, detected the frequency and distribution within the genome of new genetic variants, and gained insight into the mechanisms underlying the generation and maintenance of genetic diversity. These results have ramifications for our understanding of the outcome of viral mutagenesis and generation of new recombinants in vitro, as well as shedding new light on the genetic diversity of HSV-1.
2. Materials and methods
2.1 Cells and viruses
Vero (African Green Monkey Kidney) cells (ATCC, CCL-81) were maintained at 37°C with 5 per cent CO2. Cells were cultured in Dulbecco’s minimal essential medium (DMEM) supplemented with 10 per cent Fetal Bovine Serum (FBS), penicillin-streptomycin (Life Technologies—Gibco) and l-Glutamine (Thermo Fischer Scientific-HyClone). HSV-1 strain F has been previously described (Szpara et al. 2010; Parsons et al. 2015). In our previous description of mixed plaque morphologies in this stock of HSV-1 F, we referred to this stock as ‘FOriginal’, and here we refer to it as F-Mixed or Mixed throughout the paper. The Purified population used here is the same as the previously described FLarge (Parsons et al. 2015), which was obtained following three sequential plaque purifications. For Fig. 7, the purified Syncytial population (F-Syncytial) is the same as the previously described FSyncytial (Parsons et al. 2015). The consensus genome sequence of the Purified (FLarge) population is GenBank accession KM222724 and that of the purified Syncytial population (FSyncytial) is GenBank accession KM222725. Clinical isolates shown in Fig. 5 were collected at University of Washington from participants within one year of their first episode of genital HSV-1 infection. Isolates were collected from genital lesions, placed into viral transport media, and cultured once on Vero cells (Passage 0, P0) before transfer of samples to the Pennsylvania State University for further studies. The University of Washington Human Subjects Division reviewed and approved the collection of clinical isolates, and all participants provided written informed consent.
Figure 7.
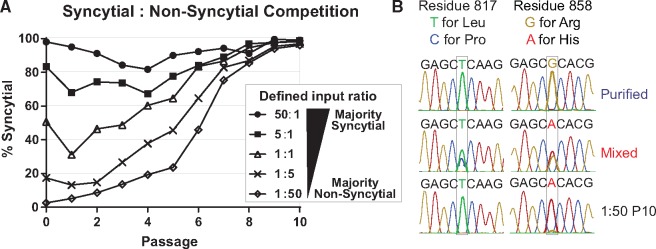
Defined mixtures of purified syncytial and non-syncytial HSV-1 populations reveal a universal advantage of the syncytial phenotype during sequential passage in Vero cells. (A) Purified populations of HSV-1 that displayed a uniformly syncytial or non-syncytial phenotype were mixed in defined ratios (as shown in legend) and allowed to compete during sequential passages of infection on Vero cells. At each passage, the plaque morphology of the resulting viral population was counted and plotted as the percentage of total plaques that were syncytial. MOI (0.01) and time to harvest (72 hpi) of each passage are the same as the initial in vitro evolution experiment described in Figure 1. (B) The tail region of gB (encoded by UL27) that contains the L817P and R858H variants described in Figure 6 was Sanger sequenced from each purified starting population, the Mixed P10 population, and the 1:5 and 1:50 passage 10 (P10) populations. Representative traces are shown with the key nucleic acids highlighted, for the non-syncytial stocks (e.g., Purified), for the mixed population (e.g., Mixed), and for the syncytial stocks (shown here is 1:50 P10; similar data for purified syncytial and 1:5 P10 are not shown). Note that these reads are the reverse-complement of those shown in Figure 6.
Figure 5.
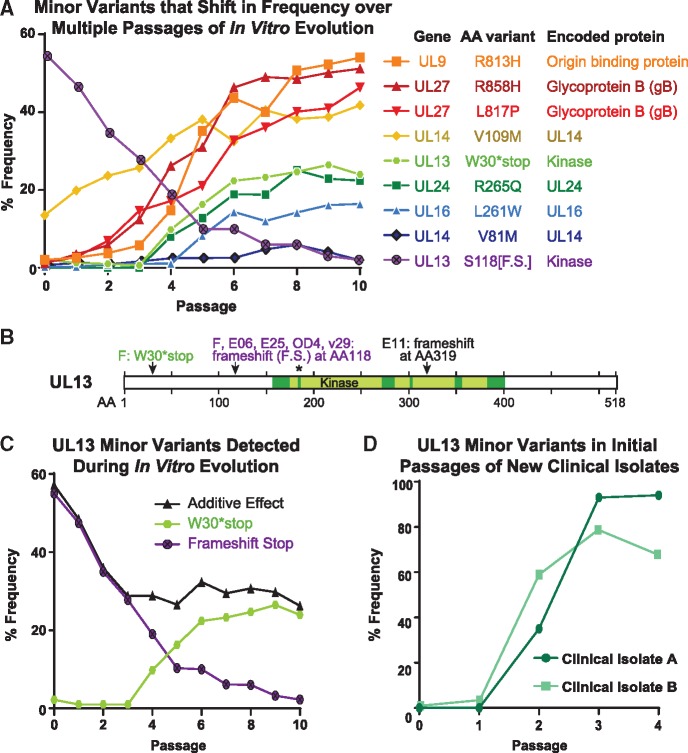
Minor variant dynamics shift over sequential passages in vitro. (A) A subset of the observed high-confidence minor variants in the Mixed population were plotted by their frequency in the viral population over passage (see Table 1 for full list of minor variants). Each variant would cause a change in the translated protein, whether through premature stop (UL13 S118[F.S.], W30*stop) or a missense variant (all others). Variants and their encoded proteins are listed in the legend according to their frequency at passage 10 (P10). (B) A diagram of the UL13 protein (518 AA in length) depicts the location of the six previously described catalytic domains of this kinase (Smith & Smith 1989), with an asterisk (*) denoting the catalytic lysine where a single point mutation can disrupt kinase activity (Cano-Monreal et al. 2008; Kawaguchi et al. 2003). The minor variants observed in this study, and consensus-level mutations observed in other virus strains in prior studies, are indicated by arrows and labels. GenBank accessions: strain F, GU734771 (Szpara et al. 2010); E25, HM585506; E06, HM585496; E11, HM585500 [all three from (Szpara et al. 2014)]; OD4, JN420342 (Lee et al. 2015). (C) The minor variants observed in the UL13 gene were separated for specific analysis. Each variant is predicted to produce a catalytically-inactive UL13 kinase. Additive Effect refers to the arithmetic addition of the frequency of each of these two individual variants. (D) New clinical isolates of HSV-1 were passaged in Vero cells, genome sequenced, and examined for minor variants in the UL13 gene at each passage. The frequency of genetic variants that would produce an inactive UL13 kinase are plotted over each passage.
2.2 In vitro evolution experiments
Each viral population was used to infect a T-150 flask of Vero cells at an MOI of 0.01 in DMEM with 2 per cent FBS, penicillin–streptomycin, and l-Glutamine. Seventy-two hours post-infection (hpi) virus was harvested by scraping, followed by three cycles of freezing and thawing. Each cycle of infection and harvest was considered a passage. The harvested virus was then titered on Vero cells using DMEM with 2 per cent FBS, penicillin–streptomycin, l-Glutamine, and methylcellulose. At 72 h post-infection, cell monolayers were fixed and stained with methanol and methylene blue. Following a determination of titer, plaque morphologies were identified by counting at least 200 plaques per passage. Plaques were counted as standard cytopathic effect (CPE) if they displayed rounding of individual cells after infection (Parsons et al. 2015). Plaques were counted as syncytial if they had a fused center, with the appearance of multiple nuclei within a shared membrane (Parsons et al. 2015). This process then continued through each of 10 passages to create a ‘lineage’. For the Mixed population, three independent lineages were created.
2.3 Other viral infections
For the competition experiments in Fig. 7, Vero cells were infected at an MOI of 0.01, at the indicated ratios of F-Purified: F-Syncytial. For the growth curves in Fig. 8, Vero cell monolayers were infected at a MOI of 10 (single-step) or 0.01 (multiple-step) and harvested by scraping at the indicated time-points. Following three rounds of freezing and thawing, the resulting virus stock was titered on Vero cells as above. These experiments were performed in triplicate.
Figure 8.
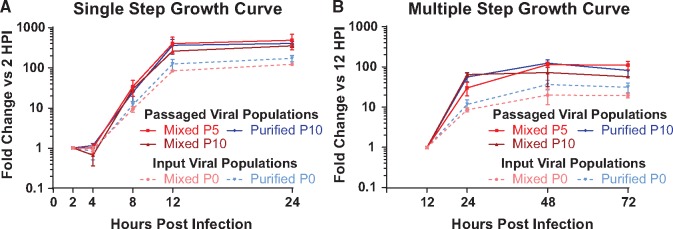
Passaged virus populations replicate more than input viral populations. Viral populations from the indicated passages of the Mixed or Purified stocks were used to infect Vero cells in (A) a single step growth curve (MOI = 10), or in (B) a multiple step growth curve (MOI = 0.01). Each assay was done in triplicate, with viral harvest and quantification by titering at the indicated timepoints. Data are plotted as titer compared to the first time-point for each experiment (2 h for A, 12 h for B). Error bars indicate standard deviation. In both growth curves, comparisons of passaged viral populations versus the input viral populations were significantly different at the final two time-points (two-way ANOVA, P < 0.05).
2.4 Next-generation sequencing
Next-generation sequencing was performed as previously described (Parsons et al. 2015). Briefly, nucleocapsid DNA was prepared by a high (MOI = 5) infection of Vero cells, followed by collection of cells and media. Nucleocapsids were isolated with Freon and pelleted through a glycerol gradient. Nucleocapsids were then lysed using SDS and Proteinase K, and viral DNA was extracted using phenol-chloroform. The viral nucleocapsid DNA was then used to prepare barcoded sequencing libraries, according to the Illumina TruSeq DNA Sample prep kit instructions. Libraries were quantified and assessed by Qubit (Invitrogen, CA), Bioanalyzer (Agilent), and library adapter qPCR (KAPA Biosystems). Illumina MiSeq paired-end sequencing (2 × 300 bp) was completed according to manufacturer’s recommendations, using 17 pM library concentration as input to the MiSeq. A consensus viral genome for each strain was assembled using a de novo viral genome assembly (VirGA) workflow (Parsons et al. 2015). Consensus genomes for each population were aligned and visualized using Geneious (Kearse et al. 2012) (Biomatters). A multi-line fasta file of these consensus genomes, as well as the alignments used for Fig. 2, are available in a ScholarSphere data repository at this DOI: https://doi.org/10.26207/t2vg-jd33.
Figure 2.
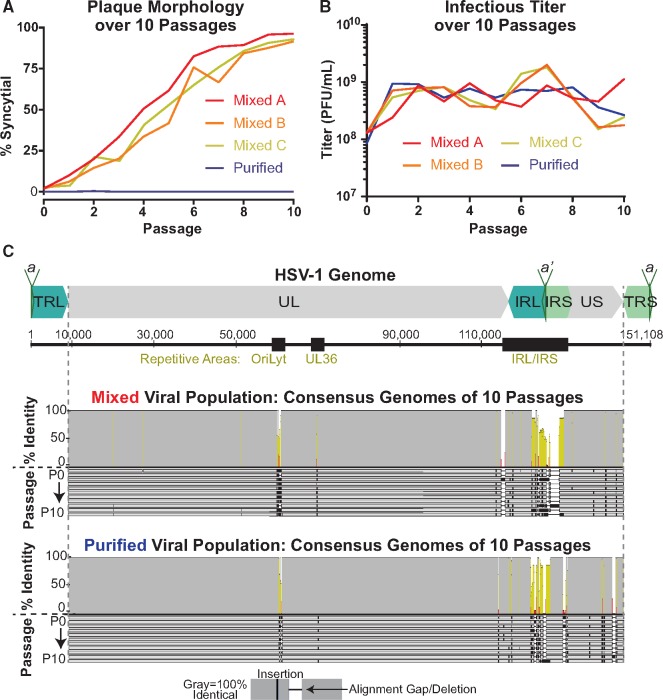
Gross changes in plaque morphology occur over sequential passages, while consensus-level genome changes are limited. (A) Plaque morphology was quantified for each passage of the Mixed and Purified viral populations. The Mixed population was passaged for three independent lineages (A, B, and C). (B) The viral titer was measured at each passage of the Mixed and Purified lineages. (C) A diagram of the full-length HSV-1 genome, depicts the structural repeats and major sites of tandem repeats (black boxes). An alignment of the consensus genome for each of ten passages of the Mixed and Purified viral populations are shown here. Each genome is represented by a gray bar and passages are displayed in order (P0–P10). Identical sequences are noted by gray in the identity plot above the alignment, with yellow and red indicating lesser levels of identity across the alignment. The terminal repeats (TRL/TRS) are trimmed from the consensus genomes as to not over-represent diversity in these duplicated regions. (UL, Unique Long; US, Unique Short; TRL, Terminal Repeat Long; IRL, Internal Repeat Long; IRS, Internal Repeat Short; TRS—Terminal Repeat Short, a/a’, repeat-containing site involved in genome cleavage and packaging).
2.5 Minor variant analysis
Minor variants (MVs) within each genome were detected using VarScan 2.4.2 (Koboldt et al. 2012), in a similar manner as previously described (Pandey et al. 2017; Bowen et al. 2018; Shipley et al. 2018). We used parameters intended to eliminate sequencing-induced errors from the initial calling of MVs. Minimum variant allele frequencies ≥0.02; base call quality ≥20; read depth at the position ≥100; independent reads supporting minor allele ≥5. Minor variants with directional strand bias ≥90 per cent were excluded. MVs that passed these filters were then annotated onto the genome using SnpEff (Cingolani et al. 2012) to determine their mutational effects. Finally, we used the alignment of the sequentially passaged consensus genomes to detect large gaps and/or insertions that were observed in single genomes, and to locally reassemble these suspected errors. Once curated, these genomes were re-analyzed for minor variants as before. See Supplementary File 1 (Mixed viral populations) and Supplementary File 2 (Purified viral populations) for full list of minor variants detected at each passage from P0 to P10. High confidence minor variants (Fig. 5 and Table 1) were defined by the presence of a given variant in 3 or more consecutive passages in a lineage for Lineage 1 or noted for presence at P10 for Lineages 2 and 3.
Table 1.
High confidence minor variants in F Mixed population (* indicates variants highlighted in Figure 5)
| Gene | Nucleotide Change | Variant Type | AA Change | Allele frequency (Percent) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P0 | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | L2 ^ P10 | L3 ^ P10 | ||||
| UL7 | C to T | Synonymous | Phe209Phe | 9.4 | 14.0 | 23.9 | 21.8 | 23.4 | 25.7 | 22.3 | ||||||
| Between UL8/9 | G to C | Non-Coding | 7.2 | 6.4 | 8.1 | 5.9 | 9.2 | 10.6 | ||||||||
| Between UL8/9 | G Deletion (Homopolymer) | Non-Coding | 14.3 | 13.4 | 12.9 | 17.0 | 13.8 | 6.5 | 12.8 | 11.1 | ||||||
| UL9* | G to A | Missense | Arg813His | 2.0 | 3.7 | 5.7 | 14.8 | 35.1 | 43.6 | 40.1 | 10.1 | 10.9 | ||||
| UL9* | A to G | Missense | His813Arg | 49.3 | 47.8 | 46.0 | ||||||||||
| UL9 | C to T | Missense | Arg203Cys | 6.9 | 7.9 | 6.8 | 7.2 | 6.1 | 6.2 | 8.4 | 25.9 | 17.5 | ||||
| UL9 | G to A | Missense | Arg200His | 2.2 | 3.6 | 3.6 | 7.2 | 6.0 | 6.4 | 8.4 | 8.7 | 9.2 | 22.3 | 13.0 | ||
| UL9 | G to A | Missense | Ser105Asn | 2.6 | 2.4 | 2.2 | 3.1 | 5.1 | 4.1 | 3.6 | 10.2 | |||||
| UL9 | A to G | Missense | Ser105Gly | 2.3 | 3.8 | 5.6 | 5.4 | 8.0 | 10.3 | 12.5 | 15.0 | 14.4 | 14.7 | |||
| Between UL10/11 | C Insertion (Homopolymer) | Non-Coding | 4.3 | 4.1 | 3.6 | 2.1 | 3.8 | 3.6 | 3.8 | 4.7 | 5.0 | 3.6 | 3.7 | 7.4 | 6.0 | |
| UL12 | C to T | Synonymous | Leu287Leu | 11.2 | 16.4 | 22.9 | 20.1 | 28.5 | 23.8 | 23.9 | ||||||
| UL12 | C to T | Missense | Ala204Val | 7.3 | 9.2 | 11.3 | 16.8 | 17.1 | 18.9 | 3.3 | ||||||
| UL13 | C to T | Missense | Arg432Cys | 12.5 | 17.7 | 24.4 | 26.7 | 17.9 | 21.8 | 23.0 | 15.6 | 12.4 | 13.6 | 26.4 | 28.9 | |
| UL13* | G to A | Premature Stop | Trp30Stop | 2.2 | 9.9 | 16.3 | 22.4 | 23.3 | 24.7 | 26.5 | 24.0 | |||||
| UL13* | G Insertion (Homopolymer) | Premature Stop | Frameshift 118, Stop 161 | 45.0 | ||||||||||||
| UL13* | G Deletion (Homopolymer) | Premature Stop | Frameshift 118, Stop 161 | 47.5 | 35.0 | 27.8 | 19.0 | 10.3 | 10.0 | 6.2 | 6.0 | 3.3 | 2.3 | 4.7 | 3.2 | |
| UL14 | C to A | Missense | Leu135Met | 2.5 | 2.3 | 3.9 | 4.0 | 3.4 | 3.8 | 3.9 | 3.4 | 2.6 | 3.6 | 3.1 | 3.3 | |
| UL14 | C to T | Synonymous | Asp132Asp | 2.0 | 2.2 | 5.4 | 6.2 | 8.5 | 6.8 | 9.4 | 11.9 | 14.4 | 13.6 | 18.2 | 25.6 | |
| UL14* | G to A | Missense | Val109Met | 13.5 | 19.9 | 23.7 | 25.7 | 33.3 | 38.1 | 32.5 | 40.5 | 38.3 | 38.8 | 41.8 | 41.3 | 38.1 |
| UL14* | G to A | Missense | Val81Met | 2.5 | 2.6 | 2.7 | 4.8 | 6.0 | 4.1 | |||||||
| UL16* | C to T | Missense | Leu261Phe | 8.2 | 14.4 | 12.0 | 14.2 | 16.1 | 16.4 | |||||||
| UL17 | C to A | Missense | Ala227Glu | 2.1 | 2.0 | 3.4 | 3.7 | 4.3 | 4.2 | 2.7 | ||||||
| UL17 | G to A | Missense | Ala3Thr | 9.9 | 15.0 | 24.6 | 24.5 | 26.7 | 24.8 | 23.2 | ||||||
| Between UL17/18 | C Insertion (Homopolymer) | Non-Coding | 3.1 | 5.8 | 5.0 | 5.4 | 4.5 | 6.9 | 4.9 | 4.7 | ||||||
| UL18 | C to T | Synonymous | Leu275Leu | 2.4 | 3.3 | 3.1 | 2.5 | 3.1 | 3.4 | 10.6 | 12.0 | |||||
| UL19 | C to T | Missense | Pro486Leu | 2.5 | 2.5 | 3.0 | 3.7 | 3.0 | 3.5 | 4.2 | ||||||
| Between UL20/21 | C to T | Non-Coding | 3.6 | 3.3 | 2.2 | 2.6 | 2.9 | 2.2 | 2.3 | 3.1 | 2.7 | |||||
| Between UL22/23 | C to G | Non-Coding | 10.4 | 20.7 | 23.7 | 20.4 | 26.4 | 28.5 | 29.7 | |||||||
| UL22 | C to T | Synonymous | Leu480Leu | 2.5 | 2.9 | 3.5 | 3.4 | 4.4 | 4.1 | 4.6 | 3.1 | 13.3 | 14.5 | |||
| UL24 | C to T | Missense | Arg63Cys | 2.8 | 3.6 | 2.5 | 2.8 | 2.3 | 2.7 | |||||||
| UL24* | G to A | Missense | Arg265Gln | 8.1 | 12.8 | 18.9 | 18.8 | 25.1 | 22.9 | 22.4 | ||||||
| UL25 | G to A | Synonymous | Pro146Pro | 5.5 | 12.6 | 13.4 | 12.0 | 14.1 | 15.4 | |||||||
| UL25 | G to A | Missense | Ala357Thr | 3.0 | 2.9 | 4.3 | 3.5 | 2.7 | 2.1 | |||||||
| Between UL25/26 | T Insertion | Non-Coding | 16.0 | 15.3 | 12.5 | 15.5 | 18.9 | 19.8 | 26.2 | 26.6 | 23.2 | 27.2 | 25.7 | 14.5 | 16.8 | |
| UL26 | G to A | Synonymous | Ala263Ala | 6.3 | 10.1 | 13.6 | 15.3 | 15.8 | 18.4 | 4.2 | ||||||
| UL26 | C to A | Synonymous | Ala362Ala | 3.2 | 2.7 | 3.5 | 4.0 | 3.8 | 4.4 | 2.8 | ||||||
| Between UL26/27 | T Insertion | Non-Coding | 4.8 | 3.4 | 3.4 | 4.2 | 3.5 | 5.8 | 4.4 | 5.5 | 19.5 | 7.1 | ||||
| Between UL26/27 | C to T | Non-Coding | 2.5 | 12.7 | 20.4 | 30.4 | 26.3 | 35.3 | 32.9 | 28.5 | 4.9 | 5.7 | ||||
| UL27 | G to A | Missense | Arg885His | 2.9 | 3.5 | 4.1 | 5.4 | 4.8 | 4.3 | 5.2 | 8.5 | 4.7 | ||||
| UL27* | G to A | Missense | Arg858His | 3.5 | 5.8 | 12.4 | 26.2 | 31.0 | 46.3 | 49.0 | 48.6 | 35.9 | ||||
| UL27* | A to G | Missense | His858Arg | 50.1 | 48.9 | 47.3 | ||||||||||
| UL27* | T to C | Missense | Leu817Pro | 2.8 | 6.9 | 14.7 | 17.2 | 21.1 | 32.8 | 36.1 | 40.1 | 41.0 | 46.3 | 30.9 | 47.6 | |
| UL27 | G to A | Synonymous | Lys544Lys | 7.9 | 10.5 | 13.4 | 17.7 | 15.9 | 16.0 | |||||||
| UL30 | C to T | Missense | Ala25Val | 2.8 | 3.3 | 2.9 | 2.9 | 2.5 | 4.6 | 13.5 | 14.5 | |||||
| UL30 | G to A | Synonymous | Lys938Lys | 8.6 | 15.3 | 20.8 | 20.2 | 20.7 | 21.8 | 23.6 | ||||||
| UL30 | G to A | Missense | Arg181His | 2.2 | 2.9 | 3.2 | 2.1 | 3.3 | 2.4 | |||||||
| UL34 | C to T | Synonymous | Gly109Gly | 6.1 | 8.8 | 10.5 | 12.7 | 14.9 | 13.4 | |||||||
| UL36 | G to A | Missense | Ala981Thr | 2.5 | 2.2 | 3.0 | 2.6 | 2.8 | 11.4 | 7.8 | ||||||
| UL36 | C to T | Missense | Thr860Met | 3.2 | 2.6 | 3.4 | 2.3 | 2.5 | ||||||||
| UL37 | C to T | Missense | Ser890Leu | 2.6 | 2.2 | 3.2 | 3.5 | 2.4 | 3.2 | |||||||
| UL39 | G to A | Synonymous | Gln736Gln | 3.1 | 3.2 | 3.7 | 2.8 | 2.8 | ||||||||
| UL40 | C to T | Synonymous | Leu301Leu | 3.9 | 2.7 | 3.0 | 2.5 | 3.4 | 2.3 | 2.2 | ||||||
| UL42 | C to T | Missense | Thr135Met | 2.8 | 3.5 | 2.6 | 2.6 | |||||||||
| UL42 | C to T | Synonymous | Leu325Leu | 10.2 | 12.5 | 23.3 | 23.2 | 22.2 | 21.0 | 21.8 | ||||||
| UL46 | G to A | Missense | Ala185Thr | 2.0 | 2.4 | 3.7 | 2.1 | 2.2 | 2.9 | |||||||
| Between UL49/50 | G to A | Non-Coding | 2.1 | 3.2 | 2.6 | 2.2 | 2.7 | |||||||||
| Between UL53/54 | G to A | Non-Coding | 2.9 | 3.4 | 3.6 | 3.6 | 3.2 | |||||||||
| UL52 | G to A | Synonymous | Gly2Gly | 2.1 | 2.7 | 2.9 | 2.5 | |||||||||
| Between UL56/IRL | C to T | Non-Coding | 9.5 | 14.2 | 24.0 | 23.4 | 21.1 | 18.4 | ||||||||
| US6 | G to A | Synonymous | Gln52Gln | 2.0 | 2.0 | 3.3 | 3.0 | |||||||||
| US6 | C to T | Synonymous | Val356Val | 2.7 | 2.7 | 2.4 | ||||||||||
| US8A | A to G | Missense | Thr109Ala | 2.4 | 2.5 | 2.4 | 2.7 | 2.1 | ||||||||
| US8A | C to T | Missense | Ala149Val | 2.6 | 2.4 | 2.9 | 2.5 | 2.2 | 2.3 | 3.1 | 2.6 | 3.8 | 5.0 | |||
| US9 | G to A | Missense | Val65Ile | 11.4 | 12.4 | 8.7 | 11.8 | |||||||||
indicates variants highlighted in Figure 5. See Supplementary Files 1 (Mixed) and 2 (Purified) for full lists of Minor Variants in all passages of Lineage 1.
Minor variants also detected via deep-sequencing of Passage 10 (P10) of Lineages 2 or 3 (L2/L3) are listed as well.
2.6 Data availability
Consensus genomes for starting viral populations have been deposited in GenBank as follows: Purified (FLarge), KM222724; purified Syncytial (FSyncytial), KM222725; Mixed (FOriginal), GU734771. The alignments used for Fig. 2, which include the consensus genomes of all of the passaged viral populations, have been deposited at ScholarSphere: https://doi.org/10.26207/t2vg-jd33. Raw sequence reads are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) as Bioproject ID PRJNA565399.
Results
3.1 Viral phenotypes can evolve over sequential passage in culture
While viruses with a DNA genome are often considered genetically stable (Sanjuan et al. 2010; Sanjuán and Domingo-Calap 2016), they are able to evolve rapidly in response to strong selective pressures, such as antiviral drug selection (Burrel et al. 2010; Sauerbrei et al. 2010; Houldcroft et al. 2017). It is becoming increasingly evident that these viruses, including HSV-1, can exist as populations that contain significant genetic diversity (Renzette et al. 2014; Renner and Szpara 2018). We sought to determine the effects of this genetic diversity on the in vitro evolution of HSV-1, in the context of the selective pressures present in Vero cell culture. To conduct these experiments, we used two related starting populations of HSV-1 strain F. The F-Mixed population (subsequently ‘Mixed’) is representative of the original HSV-1 F strain that was previously described to contain ∼3 per cent syncytial plaques, as well as an unexpected UL13 frameshift mutation (Fig. 1) (Szpara et al. 2010; Parsons et al. 2015). This variability in plaque phenotype was an early indication that the Mixed population was genetically heterogeneous. The F-Purified population (subsequently ‘Purified’) was derived from the Mixed population by three rounds of sequential plaque purification of a non-syncytial variant (Szpara et al. 2010; Parsons et al. 2015). It displays a homogenous, non-syncytial plaque morphology (Fig. 1). Each viral population was used to infect a separate monolayer of Vero cells at a MOI of 0.01. Following lysis of the cell monolayer, the resulting sample was harvested, titered, and examined for plaque morphology. This procedure is referred to as a passage for the purposes of these experiments. The titered viral stock from the first passage was used to infect the next monolayer of Vero cells at the same low MOI, and this process was repeated for 10 sequential passages. The viral population at each passage was also deep-sequenced (Fig. 1). Low MOI conditions were chosen to allow for multiple rounds of replication in each passage, enabling the most fit viral genotypes to expand in the population.
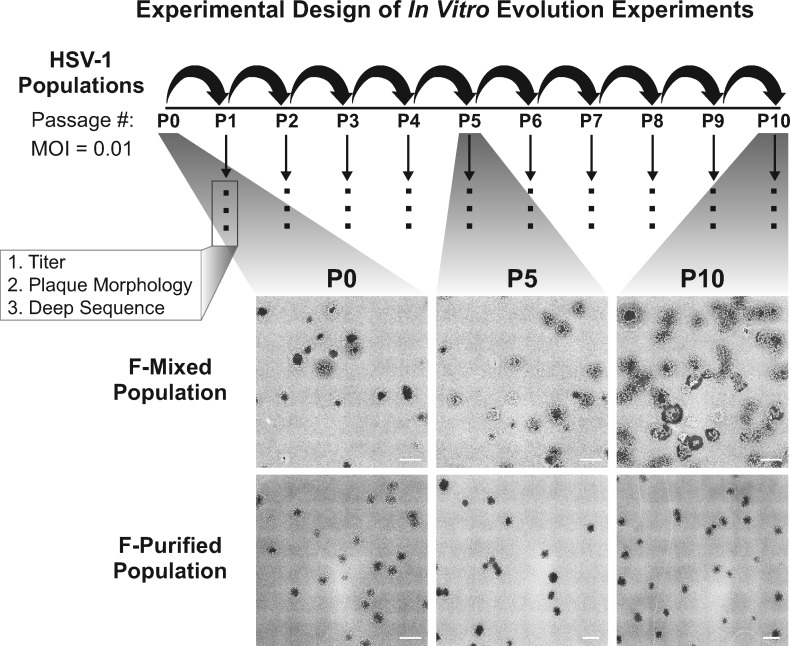
Figure 1.
Schematic of the in vitro evolution experiment approach and observed changes in plaque morphology: two populations of HSV-1 strain F were used to infect Vero cells at a multiplicity of infection (MOI) of 0.01. After infection was allowed to proceed for 72 h, progeny viral populations were harvested. This infectious cycle is referred to as a passage, and each viral population was carried through ten passages. Following each passage, each viral population was titered, visually examined for plaque morphology (see ‘Materials and Methods’ section for details), and prepared for sequencing. The entire series of 10 passages was performed in triplicate for the Mixed population and once for the Purified population. All replicates were titered and quantified for plaque morphology. Deep sequencing was performed on one lineage for each starting viral population. Plaque images were obtained by plating virus at limiting dilution on monolayers of Vero cells, then fixing and staining with methylene blue at 72 hpi. Tiled images (6 × 6) were exported from Nikon NIS-Elements software, and contrast inverted using Adobe Photoshop to show plaques more clearly. Scale bars indicate 2 mm.
Over the course of ten sequential passages, we noticed dramatic changes in plaque morphology in the Mixed population (Figs 1 and 2A). However, we did not observe substantial changes in viral titer across the ten passages in either viral population (Fig. 2B). At passage 0 (P0), we observed ∼3 per cent syncytial plaques in the Mixed population. The frequency of syncytial plaque-forming viruses in the population increased steadily with each passage, until nearly 100 per cent syncytial plaques were observed at passage 10 (P10). The conversion from predominantly non-syncytial to syncytial plaques in the population was observed in three independent lineages of the passaging experiment (Fig. 2A). In contrast, the Purified population did not contain any syncytial plaques at P0, and the syncytial plaque phenotype was not acquired over the course of ten passages. Using plaque morphology as a visual marker of genetic diversity, these results suggested that the more genetically diverse Mixed population was better able to adapt or evolve over the course of this experiment.
3.2 Few changes in consensus genomes were observed over ten passages
Because each independent Mixed lineage displayed similar changes in plaque morphology, we chose one lineage each for Mixed and Purified viral populations and used high-throughput sequencing (HTS) to examine the viral genetic diversity at each passage. Full-length, consensus genomes were assembled for each passage using a de novo assembly pipeline (Parsons et al. 2015). We describe these as consensus genomes because they condense the sequencing results from the entire viral population into the most common nucleotide observed at each location in the genome (Renner and Szpara 2018). Therefore, these genomes represent the consensus of the sequencing reads that were aligned to each position. We found that the consensus genomes were similar overall from passage to passage within the same lineage (i.e. Mixed versus Purified), which enabled curation and improvement of the assembled sequences. The high percent identity of a full-genome alignment derived from each set of ten passages is presented in Fig. 2C. While the consensus genomes were generally similar, sequence variation was observed in highly repetitive regions of these consensus genomes, where assembly of the HSV genome from short-read sequencing is technically challenging (Renner and Szpara 2018). These regions include the PQ amino acid repeats in the gene encoding the tegument protein VP1/2 (UL36), the inverted repeats surrounding the lytic origin of replication (OriLyt), and the large internal and terminal copies of the repeats flanking UL and US (IRL/S and TRL/S) (Szpara et al. 2010). Surprisingly, the Mixed viral population did not display differences in genes associated with syncytia formation in the consensus genomes until passage 8 (P8). This genetic data differs from the timing of our visual detection of the syncytial plaque phenotype, which represented a majority of the observed plaques by P4. The asynchrony of consensus-level genomic data with the visually observable phenotype motivated a deeper analysis of the sequencing results, to analyze how minor alleles in the viral population may be contributing to the observed diversity in phenotypes.
3.3 Minor variants reveal evolution in the viral population
Because the analysis of consensus genomes from the passaged viral populations was insufficient to explain the observed phenotypic differences, we analyzed the sequencing data for evidence of genotypes that were detected as minor alleles in the viral population. These minor variants, which we define as nucleotide alleles present in <50 per cent of the sequencing reads at a given locus, are not otherwise represented in the consensus genomes. With sufficiently deep sequencing coverage, these variants can generally be identified with confidence. The assembled genomes across all ten passages had an average coverage depth of 563 reads/position for the Mixed viral population and 517 reads/position for the Purified viral population. We identified nucleotide substitutions and insertions/deletions (indels) that were present in greater than 2 per cent but <50 per cent of the sequencing reads for a given sample (2 per cent cutoff as the threshold of detection; see ‘Materials and methods’ for additional criteria.) Since the frequency of sequencing reads is correlated with the frequency of alleles in the population, this analysis revealed the genetic diversity present in each starting population, as well as the specific changes that occurred over passage, and their relative proportion in each sequential passage.
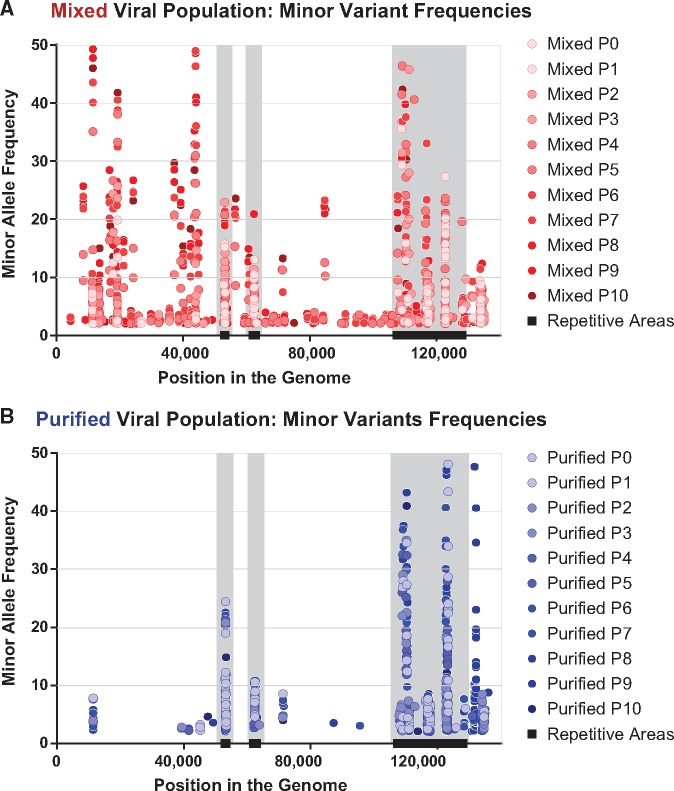
For each minor variant, we examined its position in the genome as well as its frequency in the population. We detected minor variants in each viral population, albeit at different sites and frequencies in each passage and population (Figs 3 and 4; see Supplementary Files 1 and 2 for full list). Both viral populations had minor variants in highly repetitive areas of the genome, but the Mixed viral population also had multiple minor variants in the unique (e.g., UL) coding regions (Figs 3A and 4A). As each virus population was passaged, the pattern of observed minor variants became more distinct. The Mixed population became more diverse, with new minor variants detected throughout the genome, and in a substantial percentage of the overall virus population (Fig. 4A). In contrast, the vast majority of detected minor variants in the Purified population were in the highly repetitive regions of the HSV-1 genome (Renner and Szpara 2018) (Figs 3B and 4B). These results indicated that the initially higher level of genetic diversity in the Mixed population (i.e., Mixed P0 in Fig. 3A versus Purified P0 in Fig. 3B) likely seeded the future evolution of that viral population.
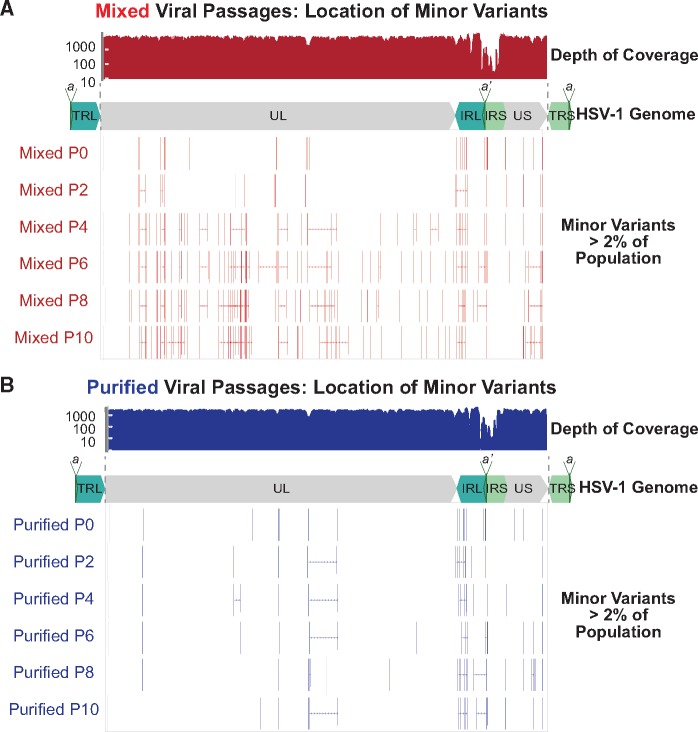
Figure 3.
Minor variants are widespread in the mixed, but not the purified, viral populations. Each passage of the (A) Mixed and (B) Purified viral populations were sequenced and analyzed for minor variants. A subset of passages are shown here for space considerations. Each minor variant present in 2% or more of the viral popiulation is shown as a vertical bar, and multiple variants within one gene are connected with a horizontal line. A plot of sequence read depth across the HSV-1 genome is shown for P0 of each virus population (results are representative of all passages). Across all passages, the average coverage depth across the genome was 563 reads/position for the Mixed viral populations and 517 reads/position for the Purified viral populations. A diagram of the HSV-1 genome is shown in each panel for spatial orientation.
Figure 4.
Minor variants in the mixed viral populations occur at more diverse locations and at higher frequencies than in the purified viral populations. Minor variants were plotted according to their location in the genome (x-axis), as well as by their observed frequency in each passage of these viral populations (y-axis). Variants that occurred in the same genomic location are seen as vertical columns of dots. Lighter colors indicate variants observed in earlier passages, whereas darker colors indicate later passages. Overall, a larger number and higher frequency of minor variants are observed in (A) the Mixed viral population than in (B) the Purified viral population. Highly repetitive regions of the HSV-1 genome are denoted by black bars (OriLyt, UL36 PQ repeats, and the internal repeat region) and gray vertical columns.
While the repetitive regions of herpesvirus genomes have previously been demonstrated to be areas of high variability (Szpara et al. 2010, 2014; Lee et al. 2015; Parsons et al. 2015), it is difficult to measure length fluctuations and other variations using short-read HTS technology in these regions (Renner and Szpara 2018). To surmount this challenge, we used the sequential nature of these passages to identify minor variants with high confidence, due to their appearance in multiple sequential passages. We identified a number of these high-confidence minor variants (Table 1) in the Mixed population lineage. In contrast, we were unable to identify any such high-confidence variants of this type in the Purified population lineage. While we cannot disregard the minor variants that sporadically appear in both of these viral populations, the lack of observed consistency from passage to passage makes these sporadic minor variants more difficult to conclusively identify, especially when they occurred in the aforementioned repetitive regions. This suggests that if these variants do arise, they are not increasing in frequency (e.g., due to positive selection), or that the low number of virions transferred at each passage is not allowing them to continue in the time course (i.e., an MOI = 0.01 uses <0.01 per cent of the >109 PFU in each stock). These data suggest that when HSV-1 has a more diverse starting genetic population (i.e., the Mixed population), it is more able to change in response to the pressures of a given environment.
3.4 Frequency of minor variants may reveal selective pressures in vitro
We next investigated the types of minor variants that were present and how their frequencies changed over sequential passages of the viral population. We found that most of the high-confidence minor variants were missense mutations, though examples of non-coding and synonymous or silent mutations were also identified (Table 1). Many minor variants increased as a percentage of the population over sequential passage, while others seemed to reach an equilibrium level of standing variation (Fig. 5A and Table 1). We did not observe any minor variants that were clearly linked as a haplotype, in that their frequencies did not change in parallel. Accordingly, we treated each minor variant an individual contributor to the population. Minor variants that significantly increased in the viral population potentially conferred a selective advantage under these experimental conditions. This outcome is most clearly illustrated by the syncytia-inducing variants in UL27 (glycoprotein B, gB), which match previously described spontaneous mutations observed in this and other strains of HSV-1 (Bzik et al. 1984; Engel et al. 1993; Diakidi-Kosta et al. 2003; Rogalin and Heldwein 2015).
Other minor variants appeared to reach an equilibrium in frequency within the viral population, which is a more difficult pattern to interpret. We found a particularly intriguing example of equilibration within the population with minor variants present in the UL13 gene. UL13 is the HSV-1 encoded representative of the Conserved Herpesvirus-encoded Protein Kinases (CHPK). UL13 kinase activity is dispensable in vitro (Gershburg et al. 2015), but it is required for axonal transport in vivo (Coller and Smith 2008). The kinase domain of this enzyme is located in the C-terminal end of the protein, with the crucial catalytic lysine located at amino acid 176 (Fig. 5B) (Kawaguchi et al. 2003; Cano-Monreal et al. 2008; Gershburg and Pagano 2008). We identified two distinct minor variants that would cause premature termination of UL13 and would not be anticipated to have any kinase activity. We refer to these variants as W30*stop, a missense mutation, and Frameshift Stop (or S118[F.S.]), an indel in a homopolymer (Fig. 5B and 5C). The Frameshift Stop insertion was present in a slight majority of the initial population (51 per cent) and it rapidly declined in frequency over sequential passages. However, the W30*stop variant increased in frequency in the population over the same passages. In combination, these two presumably defective variants of the UL13 gene added up to a stable level of ∼30 per cent of the viral population. Thus, we observed a tolerance and/or a selection of UL13 variants that would presumably produce inactive kinases (Fig. 5B and 5C).
The presence of defective UL13 variants in HSV-1 populations is not unique to the F strain. In a recent publication by our group that compared a cultured clinical isolate to direct-from-patient swab-based sequencing of a genital HSV-1 lesion, we detected UL13 minor variants only in the cultured material (Shipley et al. 2018). We extended this investigation to two newly isolated clinical genital HSV-1 isolates, and likewise detected UL13-inactivating minor variants in these cultured populations. These variants rose in frequency over the initial passages in vitro, reaching substantially higher levels, in fewer passages, than the rate of change observed in the Mixed population (Fig. 5D). It is as yet unknown why these viral populations would maintain standing variation in the UL13 gene, and/or favor the inactivation of UL13 in Vero cell culture.
3.5 Syncytial phenotype is favored in Vero cells
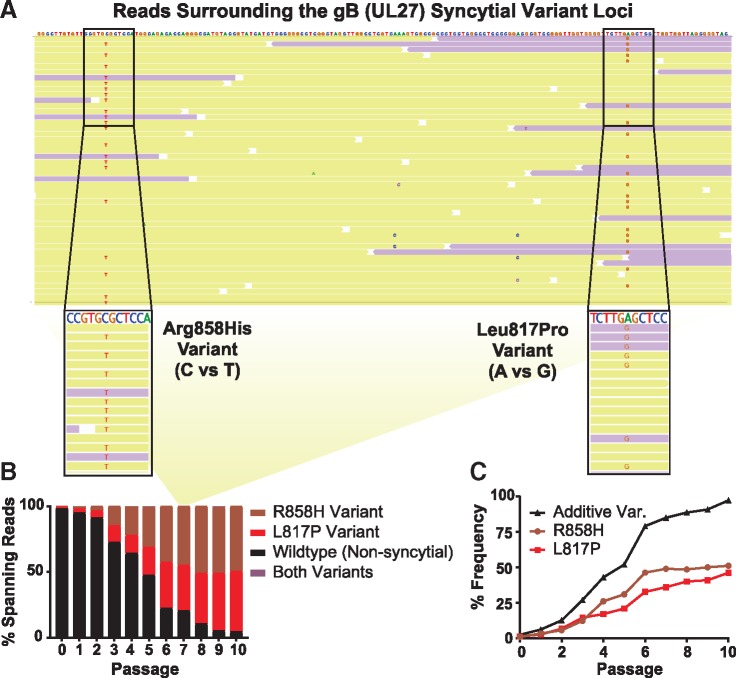
While the role of UL13 in Vero cell culture remains unclear, it is clear that the syncytial plaque phenotype is favored under these conditions. Three independent lineages of Mixed viral populations became dominated by syncytial variants (Fig. 2) under these experimental conditions, and the selective advantage of syncytia formation in Vero cell culture has been previously reported (Padilla et al. 1997). However, it is known that a syncytial phenotype can be caused by multiple independent genetic variants, and we can infer that each syncytial variant could have a different effect on viral fitness. We were able to identify two previously described syncytial variants in the Mixed viral population, both of which affect the gene encoding gB (UL27), at amino acids 817 (L to P) and 858 (R to H) (Bzik et al. 1984; Engel et al. 1993; Diakidi-Kosta et al. 2003; Rogalin and Heldwein 2015) (Fig. 6). Each of these variants steadily increased in frequency within the viral population over subsequent passages, but neither variant reached a majority of the population (and therefore became represented in the consensus genomes) until P8 (Fig. 6C). As noted above, even when the visually observed phenotype indicated that the viral population was 88 per cent syncytial (P7, Fig. 2A), there was no genetic evidence of this at the level of the consensus genome. Intriguingly, while neither variant alone was sufficient to account for the entirety of this widespread syncytial phenotype, simply adding the frequency of each variant in the population neatly mirrored the frequency of syncytial plaques (Figs 6C and 2A).
Figure 6.
The combination of individual variants in gB (UL27) explained the syncytial phenotype observed in the Mixed viral population. The UL27 gene encoding gB includes two syncytia-inducing minor variants that encode an Arginine to Histidine change at amino acid 858 or a Leucine to Proline change at amino acid 817. Sequencing reads that span the nucleotides encoding both sites were identified for each passage. (A) Shown here is a random subset of the passage 7 (P7) sequencing reads surrounding this region, of which a total of 366 reads span both minor variants. The consensus genome is displayed at the top, with individual sequencing reads aligned below (yellow indicates forward-aligning sequencing reads, while purple indicates the reverse). Nucleotides matching the consensus genome are not shown, such that minor alleles that do not match the consensus sequence are the only ones highlighted. The relevant stretches of these sequencing reads are magnified, with any nucleotide that varies from the consensus genome noted (C–T for Arg858His or A–G for Leu817Pro). Note that the UL27 gene is on the reverse strand of the HSV-1 genome, so that the depicted nucleotides are in the reverse complement of what would be transcribed to produce the UL27 mRNA. (B) Across all ten passages, the percent of sequencing reads spanning this region that contained one, both, or neither variant is summarized here. Data for ‘both’ are included but are not visible on the graph, since only six sequence reads ever included both variants (three at P5, and three at P9). The yellow background panel in (B) connects the subset of P7 sequence read data shown in (A) to the relevant overall P7 percentage data graphed in (B). (C) The frequency of each variant in gB (UL27) was plotted alone, or as the sum of their frequency (Additive Variation (Var.)).
These data suggest that each of these syncytial variants in gB occurred and proliferated independently. Therefore, we sought to address whether or not these variants ever co-occurred on the same stretch of DNA. The proximity of these variants in the genome enabled us to identify a number of sequencing reads, each corresponding to one physical piece of DNA, that spanned both minor variant loci in gB (UL27) (Fig. 6A). We detected each minor variant within this subset of reads at a frequency that was comparable to the overall frequency of the variant in the population (Fig. 6B–6C). We found that each variant occurred independently. Across all reads spanning these loci in ten passages, we found only six instances of these minor variants co-occurring in a single sequence read. We interpret these results as an indication that the syncytial phenotype, rather than any individual genetic variant, is being selected for under these conditions of Vero cell culture.
To test this hypothesis, we conducted a competition experiment in which two purified populations of HSV-1 were mixed in defined ratios and allowed to replicate under the same conditions as the original in vitro evolution experiment. We made use of a previously described syncytial clone of the F-Mixed population (Szpara et al. 2010), which carries the R858H syncytial variant in gB, as well as the same F-Purified population as was used in Fig. 1. These purified populations of uniformly syncytial or uniformly non-syncytial viruses were competed in defined ratios, where one or the other plaque phenotype was dominant at the start (50:1, 5:1, 1:5, and 1:50, Syncytial: Non-Syncytial). We also included one case of equal starting input (1:1, Syncytial: Non-Syncytial). As in the original in vitro evolution experiment, we found that the viral populations eventually displayed a syncytial plaque phenotype (Fig. 7A). The starting ratio influenced the rate of syncytial virus takeover, as shown by the slopes of each curve in Fig. 7. However, all ratio mixtures were predominantly syncytial by P10. Sanger sequencing of the gB (UL27) tail region (containing residues 817 and 858) in the tenth passage of the 1:5 and 1:50 mixes (Fig. 7B) suggests that the initial F-Syn population expanded to dominate in the mixed populations, rather than the syncytial phenotype occurring due to de novo mutations over passage. These results are similar to those obtained in the initial in vitro evolution experiment, where the R858H and L817P variants in gB were observed at very low levels in the initial Mixed population (R858H, 1.1 per cent; L817P, 1.4 per cent—below our threshold for reporting minor variants), before expanding over passage. Combined with the results observed in the Purified population passages, the data imply that in these experimental conditions we predominantly observed an expansion of more fit genotypes in the population, as opposed to de novo genetic variation.
3.6 Genetic variation outside of syncytia formation facilitates viral fitness
Finally, we examined whether or not each evolved viral population was more fit for replication than the starting population. To do this, we compared the single-step and multiple-step growth kinetics for representative populations of the passaged viruses. We chose P0, P5, and P10 as representative of the Mixed viral population at the starting, midpoint, and endpoint of the in vitro evolution experiment. For the Purified viral population, we examined only the P0 and P10 populations, due to the limited genetic changes observed in that viral population. Under both single-step growth conditions (MOI = 10), and multiple-step growth conditions (MOI = 0.01), we found that the passaged viral populations increased in titer to a greater degree than their original starting populations (Fig. 8). The differences in replication between passaged and input viral populations were statistically significant (P < 0.05) at the final two time points in both growth curves (Two-way ANOVA), although the final titers of these stocks were overall quite similar (Fig. 8 and Fig. 2B). Despite the continued genetic changes between P5 and P10 for the Mixed viral population, we did not detect any additional increases in replication by P10 in these assays. It is possible that the effect of these genetic changes is more subtle than can be detected by the assays and/or time-points chosen in these experiments.
4. Discussion
In this study, we examined the evolution of HSV-1 during viral replication in vitro. We sequentially passaged either a mixed or a purified population of HSV-1 in Vero cell culture, and we quantified the outcome both in terms of observed phenotypes and in genomic changes over time. These approaches enabled us to monitor how the viral population changed from passage to passage, to detect potentially advantageous viral genotypes and/or phenotypes under these conditions, and to examine how a diverse versus a homogeneous pool of viral genomes impacted the subsequent genetic drift of each viral population. We found that the more diverse (‘Mixed’) viral population was much more prone to change during passaging, with a syncytial plaque phenotype emerging as an advantageous trait under these Vero cell culture conditions. While the selection for syncytial plaques in Vero cell culture has been observed previously (Padilla et al. 1997), the precise mechanisms for this selection remain unclear. It is possible that syncytia allow the virus to spread to uninfected cells more rapidly than the non-syncytial phenotype and expand in the viral population (Yamada et al. 1986).
In addition to the genetic variants associated with syncytia formation, we observed the proliferation of a multitude of other genetic variants that may also confer a selective advantage in Vero cell culture conditions. Because Vero cell culture is the main methodology behind HSV-1 and HSV-2 propagation in most virology labs, both in experimental and diagnostic lab settings, these data provide key insights on the need to detect and understand viral evolution and selection in vitro. This is especially critical if the goal is to use laboratory observations to improve our understanding of viral population dynamics in actual human infections.
From these experiments, we were able to gain a number of useful insights for HSV-1, which may well extend to other herpesviruses and large DNA viruses. First, these experiments emphasized the contributions of standing variation and genetic diversity to HSV-1 evolution. While few would suggest that viruses with a large dsDNA genome do not evolve, the prevailing sentiment is that DNA viruses have a substantially slower rate of evolution than RNA viruses (Sanjuan et al. 2010; Renzette et al. 2011). Thus, the concept of quasispecies or of viruses existing as populations with a mixture of genotypes has generally been limited to the RNA virus world. However, recent experiments that take advantage of modern deep-sequencing technology have shown that dsDNA viruses possess substantial genetic diversity in vivo as well as in laboratory conditions (Spatz et al. 2008; Dargan et al. 2010; Depledge et al. 2011; Elde et al. 2012; Renzette et al. 2014, 2016; Parsons et al. 2015; Palser et al. 2015; Murrell et al. 2016; Risso-Ballester et al. 2016; Pandey et al. 2017; Weiss et al. 2018; Trimpert et al. 2019). The diversity of genomes in an RNA virus population has been linked to viral fitness (Acevedo et al. 2014; Korboukh et al. 2014), and there is no reason to expect a different result with DNA virus populations. Indeed, in these experiments, the genetically diverse (‘Mixed’) viral population was able to adapt more readily to replication in Vero cells, while the more homogeneous (‘Purified’) population appeared to be much more stable.
From a technical standpoint, these experiments highlighted the usefulness of deep sequencing for the analysis of viral populations. If we were to assume that the consensus genomes of each Mixed passage were indicative of the entire viral population, we could not have explained why there were more syncytial plaques observed over sequential passages, because the consensus genome did not change until passage 8 (P8). A focus solely on consensus-level genome analysis would also have missed the dynamics of the inactivating variants of UL13, not to mention the array of other genetic variants that fluctuated in the Mixed viral population (Fig. 5A, Table 1). Limiting the genetic analysis to the consensus level, particularly when using deep-sequencing approaches, eliminates a great deal of useful information about the viral population (Renner and Szpara 2018). When performing similar evolution experiments to those performed here, but with different viruses, other researchers have found the most compelling evidence of evolution at the minority variant level (Elde et al. 2012; Hildebrandt et al. 2014; Grubaugh et al. 2015; Morley et al. 2016; Morley and Turner 2017).
Several of the specific minor variants observed in this study are ones that have been previously noted to occur spontaneously in HSV-1 and related herpesvirus strains during propagation in vitro. For instance, the occurrence of spontaneous syncytia-inducing mutations in the gene encoding gB (UL27) has been well-documented in the literature (Bzik et al. 1984; Engel et al. 1993; Diakidi-Kosta et al. 2003; Rogalin and Heldwein 2015). Likewise, the appearance of mutations that truncate the UL13 kinase, whether through an early stop codon or a frameshift, have been previously noted in multiple strain of HSV-1 by our lab and others (Szpara et al. 2010, 2014; Lee et al. 2015) (Fig. 5B). Despite prior observation in the literature, earlier works have not connected these variants to a fitness advantage in Vero cell culture per se. For instance, early studies noting the selective advantage of HSV-1 syncytial phenotypes in cell culture did not have knowledge of the genetic basis of these phenotypes, and thus could not track these genetic variants over time (Yamada et al. 1986; Padilla et al. 1997). Future work is now warranted to investigate the potential positive selection of these and the other minor variants detected here that rise in frequency over sequential passages (e.g., Fig. 5A). The rapid rise in frequency of UL13 variants observed in early passages of two clinical isolates (Fig. 5D) suggests that the initial passages of new clinical isolates may serve as a useful screening approach to detect which of these minor variants are most impactful for viral fitness in vitro—i.e., those most frequently observed and/or rapidly selected in newly cultured clinical isolates. Intriguingly, both the UL13 and UL14 variants observed here were also detected in the initial culture passages of the genital HSV-1 isolate v29 (Shipley et al. 2018).
These data lay the foundation for a number of subsequent explorations using similar approaches. First, as mentioned above, it is a goal of our lab and others to model the dynamics of HSV-1 infection that occur in human patients in a controlled setting of laboratory culture. Ideally, this controlled setting would minimize the genetic drift observed here in Vero cell culture. To minimize HSV-1 genetic drift, it may be useful to explore using alternative cell lines (e.g., species-matched or with intact interferon signaling, both of which are missing in Vero cells) or by using a different multiplicity of infection, or by changing the duration of infection. The in vitro evolution approach could also be used to investigate HSV-1’s response to specific selective pressures, such as exposure to antibodies, complement, or antiviral drugs. This type of in vitro evolution experiment has been performed with other viruses (Elde et al. 2012; Hildebrandt et al. 2014; Morley et al. 2016; Cone et al. 2017), though more frequently with RNA viruses rather than DNA viruses. While conventional reverse genetic approaches allow for detailed examination of the function of a specific gene product, forward genetic approaches such as in vitro evolution passaging experiments will allow us to study how the entirety of the viral genome responds to selective pressures. In addition, the unbiased approach of forward genetics enables the exploration of individual missense variants, as well as epistatic interactions of variants, and even copy number variants, as seen as prior studies (Elde et al. 2012; Hildebrandt et al. 2014; Morley et al. 2016; Cone et al. 2017). These experiments may be especially fruitful in the case of large DNA viruses like HSV-1, where multiple genes have pleiotropic and/or unknown functions, or undefined networks of interactions.
Supplementary Material
Acknowledgements
The authors would also like to thank Nathan Arnett for his contributions to the passaging experiments, and the other members of the Szpara lab for their critical reading of the article and figures.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
The authors acknowledge support from the Eberly College of Science and the Huck Institutes of the Life Sciences at Pennsylvania State University, as well as NIH grant support from R21 AI130676 and R01 AI132692 to M.L.S., and P01 AI030731 to C.M.J. This project was funded, in part, under a grant with the Pennsylvania Department of Health using Commonwealth Universal Research Enhancement Program (CURE) funds (M.L.S.).
Conflict of interest: None declared.
References
- Acevedo A., Brodsky L., Andino R. (2014) ‘Mutational and Fitness Landscapes of an RNA Virus Revealed through Population Sequencing’, Nature, 505: 686–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D. C., Stevens J. G. (1994) ‘Neuron-Specific Restriction of a Herpes Simplex Virus Recombinant Maps to the UL5 Gene’, Journal of Virology, 68: 3761–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen C. D. et al. (2018) ‘HSV-1 Strains Circulating in Finland Demonstrate Uncoupling of Geographic and Phenotypic Variation’. Journal of Virology, 93: e01824–18. [DOI] [PMC free article] [PubMed]
- Bower J. R. et al. (1999) ‘Intrastrain Variants of Herpes Simplex Virus Type 1 Isolated from a Neonate with Fatal Disseminated Infection Differ in the ICP34.5 Gene, Glycoprotein Processing, and Neuroinvasiveness’, Journal of Virology, 73: 3843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. (2004) ‘Effect of Gene Location on the Evolutionary Rate of Amino Acid Substitutions in Herpes Simplex Virus Proteins’, Virology, 330: 209–20. [DOI] [PubMed] [Google Scholar]
- Burrel S. et al. (2010) ‘Genotypic Characterization of UL23 Thymidine Kinase and UL30 DNA Polymerase of Clinical Isolates of Herpes Simplex Virus: Natural Polymorphism and Mutations Associated with Resistance to Antivirals’, Antimicrobial Agents and Chemotherapy, 54: 4833–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzik D. J. et al. (1984) ‘Nucleotide Sequence of a Region of the Herpes Simplex Virus Type 1 gB Glycoprotein Gene: Mutations Affecting Rate of Virus Entry and Cell Fusion’, Virology, 137: 185–90. [DOI] [PubMed] [Google Scholar]
- Cano-Monreal G. L., Tavis J. E., Morrison L. A. (2008) ‘Substrate Specificity of the Herpes Simplex Virus Type 2 UL13 Protein Kinase’, Virology, 374: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P. et al. (2012) ‘A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff’, Fly, 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller K. E., Smith G. A. (2008) ‘Two Viral Kinases Are Required for Sustained Long Distance Axon Transport of a Neuroinvasive Herpesvirus’, Traffic, 9: 1458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K. R. et al. (2017) ‘Emergence of a Viral RNA Polymerase Variant during Gene Copy Number Amplification Promotes Rapid Evolution of Vaccinia Virus’, (G. McFadden, Ed’) Journal of Virology, 91: e01428–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargan D. J. et al. (2010) ‘Sequential Mutations Associated with Adaptation of Human Cytomegalovirus to Growth in Cell Culture’, The Journal of General Virology, 91: 1535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge D. P. et al. (2014) ‘Evolution of Cocirculating Varicella-Zoster Virus Genotypes during a Chickenpox Outbreak in Guinea-Bissau’, Journal of Virology, 88: 13936–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge D. P. et al. (2011) ‘Specific Capture and Whole-Genome Sequencing of Viruses from Clinical Samples’, PLoS One, 6: e27805. (R. Jhaveri, Ed.) /DOI: 10.1371/journal.pone.0027805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakidi-Kosta A. et al. (2003) ‘A Single Amino Acid Substitution in the Cytoplasmic Tail of the Glycoprotein B of Herpes Simplex Virus 1 Affects Both Syncytium Formation and Binding to Intracellular Heparan Sulfate’, Virus Research, 93: /99–108. [DOI] [PubMed] [Google Scholar]
- Drake J. W., Hwang C. B. C. (2005) ‘On the Mutation Rate of Herpes Simplex Virus Type 1’, Genetics, 170: 969–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N. C. et al. (2012) ‘Poxviruses Deploy Genomic Accordions to Adapt Rapidly against Host Antiviral Defenses’, Cell, 150: 831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny J. M., Morgan M. J. (1979) ‘Susceptibility of Various Cells Treated with Interferon to the Toxic Effect of Poly(rI).Poly(rC) Treatment’, Journal of General Virology, 43: 253–5. [DOI] [PubMed] [Google Scholar]
- Engel J. P., Boyer E. P., Goodman J. L. (1993) ‘Two Novel Single Amino Acid Syncytial Mutations in the Carboxy Terminus of Glycoprotein B of Herpes Simplex Virus Type 1 Confer a Unique Pathogenic Phenotype’, Virology, 192: /112–20.DOI: 10.1006/viro.1993.1013 [DOI] [PubMed] [Google Scholar]
- Gershburg E., Pagano J. S. (2008) ‘Conserved Herpesvirus Protein Kinases’, Biochimica et Biophysica Acta (Bba) - Proteins and Proteomics, 1784: 203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershburg S. et al. (2015) ‘The UL13 and US3 Protein Kinases of Herpes Simplex Virus 1 Cooperate to Promote the Assembly and Release of Mature, Infectious Virions’, PLoS One, 10: /e0131420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A. L. et al. (2018) ‘Ultrasensitive Capture of Human Herpes Simplex Virus Genomes Directly from Clinical Samples Reveals Extraordinarily Limited Evolution in Cell Culture’, mSphere, 3: e00283–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N. D. et al. (2015) ‘Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness’, PLOS Pathogens, 11: /e1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage E. et al. (2017) ‘Characterization of Human Cytomegalovirus Genome Diversity in Immunocompromised Hosts by Whole-Genome Sequencing Directly from Clinical Specimens’, The Journal of Infectious Diseases, 215: 1673–83. [DOI] [PubMed] [Google Scholar]
- Hall J. D., Almy R. E. (1982) ‘Evidence for Control of Herpes Simplex Virus Mutagenesis by the Viral DNA Polymerase’, Virology, 116: 535–43. [DOI] [PubMed] [Google Scholar]
- Hildebrandt E. et al. (2014) ‘Characterizing the Molecular Basis of Attenuation of Marek’s Disease Virus via in Vitro Serial Passage Identifies de Novo Mutations in the Helicase-Primase Subunit Gene UL5 and Other Candidates Associated with Reduced Virulence’, Journal of Virology, 88: 6232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houldcroft C. J., Beale M. A., Breuer J. (2017) ‘Clinical and Biological Insights from Viral Genome Sequencing’, Nature Reviews Microbiology, 15: 183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo N. et al. (2013) ‘Evidence of Muller’s Ratchet in Herpes Simplex Virus Type 1’, Journal of General Virology, 94: 366–75. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y. et al. (2003) ‘Conserved Protein Kinases Encoded by Herpesviruses and Cellular Protein Kinase cdc2 Target the Same Phosphorylation Site in Eukaryotic Elongation Factor 1delta’, Journal of Virology, 77: 2359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. (2012) ‘Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data’, Bioinformatics, 28: 1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P. G. E. et al. (2015) ‘A Comparison of Herpes Simplex Virus Type 1 and Varicella-Zoster Virus Latency and Reactivation’, Journal of General Virology, 96: 1581–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C. et al. (2012) ‘VarScan 2: Somatic Mutation and Copy Number Alteration Discovery in Cancer by Exome Sequencing’, Genome Research, 22: 568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korboukh V. K. et al. (2014) ‘RNA Virus Population Diversity, an Optimum for Maximal Fitness and Virulence’, Journal of Biological Chemistry, 289: 29531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F. et al. (2016) ‘Islands of Linkage in an Ocean of Pervasive Recombination Reveals Two-Speed Evolution of Human Cytomegalovirus Genomes’, Virus Evolution, 2: DOI: 10.1093/ve/vew017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. et al. (2015) ‘Recombination Analysis of Herpes Simplex Virus 1 Reveals a Bias toward GC Content and the Inverted Repeat Regions’, Journal of Virology, 89: 7214–23. (R. M. Longnecker, Ed.) /DOI: 10.1128/JVI.00880-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P. M. (2016) ‘Epigenetics and Genetics of Viral Latency’, Cell Host & Microbe, 19: /619–28.DOI: 10.1016/j.chom.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker K. J. et al. (2015) ‘Global and Regional Estimates of Prevalent and Incident Herpes Simplex Virus Type 1 Infections in 2012’, PLoS One, 10: e0140765.DOI: 10.1371/journal.pone.0140765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone J. T., Lauring A. S. (2018) ‘Genetic Bottlenecks in Intraspecies Virus Transmission’, Current Opinion in Virology, 28: 20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaya M. A. et al. (2017) ‘Molecular Evolution of Herpes Simplex Virus 2 Complete Genomes: Comparison between Primary and Recurrent Infections’, Journal of Virology, 91: 17. /e00942– [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley V. J. et al. (2016) ‘Evolution in Spatially Mixed Host Environments Increases Divergence for Evolved Fitness and Intrapopulation Genetic Diversity in RNA Viruses’, Virus Evolution, 2: /vev022.DOI: 10.1093/ve/vev022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley V. J., Turner P. E. (2017) ‘Dynamics of Molecular Evolution in RNA Virus Populations Depend on Sudden versus Gradual Environmental Change’, Evolution, 71: /872–83.DOI: 10.1111/evo.13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca J. D., Pitha P. M. (1986) ‘Transcriptional and Posttranscriptional Regulation of Exogenous Human Beta Interferon Gene in Simian Cells Defective in Interferon Synthesis’, Molecular and Cellular Biology, 6: 2279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell I. et al. (2016) ‘Genetic Stability of Bacterial Artificial Chromosome-Derived Human Cytomegalovirus during Culture in Vitro’, Journal of Virology, 90: 3929–43.DOI: 10.1128/JVI.02858-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla J. et al. (1997) ‘In Vitro Selection of Variants of Herpes Simplex Virus Type 1 Which Differ in Cytopathic Changes’, Microbiology and Immunology, 41: /203–7. [DOI] [PubMed] [Google Scholar]
- Palser A. L. et al. (2015) ‘Genome Diversity of Epstein-Barr Virus from Multiple Tumor Types and Normal Infection’, Journal of Virology, 89: 5222–37.DOI: 10.1128/JVI.03614-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey U. et al. (2017) ‘Inferred Father-to-Son Transmission of Herpes Simplex Virus Results in near-Perfect Preservation of Viral Genome Identity and in Vivo Phenotypes’, Scientific Reports, 7: /13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L. R. et al. (2015) ‘Rapid Genome Assembly and Comparison Decode Intrastrain Variation in Human Alphaherpesviruses’, mBio, 6: e02213–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Roizman B. (2013). ‘Herpesviridae’, in Knipe D. M., Howley P. M. (eds.) Fields Virology, 6th ed., Vols 1-2, Vol. 2, pp. 1802–22. Lippincott Williams & Wilkins: Philadelphia, PA. [Google Scholar]
- Renner D. W., Szpara M. L. (2018) ‘The Impacts of Genome-Wide Analyses on Our Understanding of Human Herpesvirus Diversity and Evolution’, Journal of Virology, 92: e00908–17.DOI: 10.1128/JVI.00908-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N. et al. (2011) ‘Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants’, PLoS Pathogens, 7: e1001344–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N. et al. (2014) ‘Human Cytomegalovirus Intrahost Evolution—a New Avenue for Understanding and Controlling Herpesvirus Infections’, Current Opinion in Virology, 8: 109–15.DOI: 10.1016/j.coviro.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N., Kowalik T. F., Jensen J. D. (2016) ‘On the Relative Roles of Background Selection and Genetic Hitchhiking in Shaping Human Cytomegalovirus Genetic Diversity’, Molecular Ecology, 25: /403–13.DOI: 10.1111/mec.13331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso-Ballester J., Cuevas J. M., Sanjuán R. (2016) ‘Genome-Wide Estimation of the Spontaneous Mutation Rate of Human Adenovirus 5 by High-Fidelity Deep Sequencing’, PLoS Pathogens, 12: /e1006013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalin H. B., Heldwein E. E. (2015) ‘Interplay between the Herpes Simplex Virus 1 gB Cytodomain and the gH Cytotail during Cell-Cell Fusion’, Journal of Virology, 89: /12262–72.DOI: 10.1128/JVI.02391-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Knipe D. M., Whitley R. (2013). ‘Herpes Simplex Viruses’, in Knipe D. M., Howley P. M. (eds) Fields Virology, 6th ed., Vols 1-2, Vol. 2, pp. 1823–97. Lippincott Williams & Wilkins: Philadelphia, PA. [Google Scholar]
- Sanjuán R., Domingo-Calap P. (2016) ‘Mechanisms of Viral Mutation’, Cellular and Molecular Life Sciences, 73: /4433–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R. et al. (2010) ‘Viral Mutation Rates’, Journal of Virology, 84: 9733–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerbrei A. et al. (2010) ‘Phenotypic and Genotypic Characterization of Acyclovir-Resistant Clinical Isolates of Herpes Simplex Virus’, Antiviral Research, 86: 246–52. [DOI] [PubMed] [Google Scholar]
- Shipley M. M. et al. (2018) ‘Genome-Wide Surveillance of Genital Herpes Simplex Virus Type 1 from Multiple Anatomic Sites over Time’, The Journal of Infectious Diseases, 218: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. (1989) ‘Identification of New Protein Kinase-Related Genes in Three Herpesviruses, Herpes Simplex Virus, Varicella-Zoster Virus, and Epstein-Barr Virus’, Journal of Virology, 63: /450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatz S. J. et al. (2008) ‘Clustering of Mutations within the Inverted Repeat Regions of a Serially Passaged Attenuated Gallid Herpesvirus Type 2 Strain’, Virus Genes, 37: /69–80.DOI: 10.1007/s11262-008-0242-0 [DOI] [PubMed] [Google Scholar]
- Szpara M. L. et al. (2014) ‘Evolution and Diversity in Human Herpes Simplex Virus Genomes’, Journal of Virology, 88: /1209–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpara M. L., Parsons L., Enquist L. W. (2010) ‘Sequence Variability in Clinical and Laboratory Isolates of Herpes Simplex Virus 1 Reveals New Mutations’, Journal of Virology, 84: 5303–13.DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpert J. et al. (2019) ‘A Proofreading-Impaired Herpesvirus Generates Populations with Quasispecies-like Structure’, Nature Microbiology, DOI: 10.1038/s41564-019-0547-x [DOI] [PubMed] [Google Scholar]
- Wathelet M. G., Berr P. M., Huez G. A. (1992) ‘Regulation of Gene Expression by Cytokines and Virus in Human Cells Lacking the type-I Interferon Locus’, European Journal of Biochemistry, 206: /901–10.DOI: 10.1111/j.1432-1033.1992.tb16999.x [DOI] [PubMed] [Google Scholar]
- Weiss E. R. et al. (2018) ‘Early Epstein-Barr Virus Genomic Diversity and Convergence toward the B95.8 Genome in Primary Infection’, Journal of Virology, 92: DOI: 10.1128/JVI.01466-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Uno F., Nii S. (1986) ‘In Vitro Cytopathology and Pathogenicity to Inbred Mice Shown by Five Variants of a Laboratory Strain of Type 1 Herpes Simplex Virus’, Archives of Virology, 90: 183–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.