Abstract
Viruses have evolved numerous mechanisms to evade the immune response, including proteins that target the function of cytokines. This article provides an overview of the different strategies used by viruses to block the induction of cytokines and immune signals triggered by cytokines. Examples of virus evasion proteins are presented, such as intracellular proteins that block signal transduction and immune activation mechanisms, secreted proteins that mimic cytokines, or viral decoy receptors that inhibit the binding of cytokines to their cognate receptors. Virus-encoded proteins that target cytokines play a major role in immune modulation, and their contributions to viral pathogenesis, promoting virus replication or preventing immunopathology, are discussed.
Keywords: Cytokine, Cytokine binding protein, Cytokine receptor, Immune evasion, Immunopathology, Inflammation, Interferon, Molecular mimicry, Pattern recognition receptor, Tumor necrosis factor, Viral pathogenesis, Virus
Introduction: Viral Evasion of the Immune Response
Viruses have evolved many different strategies to counteract host defenses, including innate (nonspecific) or adaptive (antigen-specific) immune responses. Important advances have been made in recent years to further understand the complex interaction of viruses with their hosts. Much of the pathology associated with viral diseases may be the result of an unbalanced virus–host interaction, and in some cases it represents uncontrolled attempts of the immune system to destroy the virus leading to damage of the host. A more profound knowledge of the interaction of viruses with the host immune system will help us to understand the molecular mechanisms of viral pathogenesis.
Cytokines are a family of molecules that initiate and orchestrate the immune response and include growth factors, interferons (IFNs), tumor necrosis factor (TNF), interleukins (ILs), and chemokines. Cytokines are normally secreted from cells, although some are expressed at the cell surface, and activate specific responses in cells expressing cognate receptors. The immune system utilizes this complex network of ligands and receptors to orchestrate the immune response. Thus, the activation of a particular set of cytokines as a result of viral infection determines the type of immune response that takes place. In addition, cytokines such as IFN and TNF may restrict virus replication by acting directly on the infected cell.
The importance of cytokines in antiviral defense is highlighted by the numerous mechanisms that viruses encode to block their activity (Alcami and Koszinowski, 2000; Tortorella et al., 2000). The primary purpose of the viral anticytokine strategies is to downregulate the immune responses to infection. However, viruses that replicate in immune cells may take advantage of the effects that cytokines have on cell physiology to enhance the ability of cells to support viral replication, thus promoting viral replication rather than evading the immune system. Viruses may block the synthesis and maturation of cytokines or may modulate cytokine signaling in the target cells. Large DNA viruses such as poxviruses and herpesviruses have captured in their genomes genes encoding host cytokines and cytokine receptors (McFadden and Murphy, 2000; Alcami, 2003; Seet et al., 2003).
Research on virus modulation of the cytokine network has received much attention and expanded enormously in recent years. It is not possible to cover all aspects in depth in an article, and I provide a general view of the different strategies used by viruses to modulate cytokine functions, focusing on some examples that illustrate basic concepts on viral immune evasion. Chemokine modulation by viruses will be discussed elsewhere in this Encyclopedia. More information can be found in review articles (Alcami and Koszinowski, 2000; Tortorella et al., 2000; Katze et al., 2002; Seet et al., 2003; Versteeg and Garcia-Sastre, 2010).
Viral Modulation of Cytokine Expression and Activation
Activation of Pattern Recognition Receptors
The ability of the host to respond to invading pathogens relies on the activation of the innate immune system through a set of germ line–encoded membrane-associated or cytoplasmic receptors, termed pattern recognition receptors (PRRs) that are engaged by virus-derived products named pathogen-associated molecular patterns (PAMPs) (Gurtler and Bowie, 2013; Coccia and Battistini, 2015). Major classes of PRRs include Toll-like receptors, C-type lectin receptors, retinoic acid–inducible gene I-like receptors, nucleotide binding and oligomerization domain-like receptors, and cytosolic DNA-sensing receptors (Blasius and Beutler, 2010; Sancho and Reis e Sousa, 2012; Barbe et al., 2014; Wu and Chen, 2014). Upon engagement, these receptors recruit adaptor proteins that signal downstream and activate several major pathways: the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), the mitogen-activated protein kinases, and the IFN regulatory factor (IRF) pathway (Akira et al., 2006; Honda and Taniguchi, 2006). This pathogen-mediated activation leads to the production of IFN and proinflammatory cytokines. Numerous viruses have been shown to prevent this early activation of cytokines (see below).
Inhibition of the Inflammasome
The inflammasome is a complex that contains caspase-1 and activates the proinflammatory cytokines IL-1β and IL-18 (Lamkanfi and Dixit, 2011; Lupfer et al., 2015; Figure 1 ). The cowpox virus (CPXV) protein cytokine response modifier A (CrmA) and orthologues encoded by vaccinia virus (VACV), ectromelia virus (ECTV), or rabbitpox virus target the enzymatic activity of caspase-1, acting as a pseudosubstrate inhibitor (Ray et al., 1992). CrmA has been shown to contribute to the virulence of CPXV, VACV, and myxoma virus (MYXV) (Thompson et al., 1993; Messud-Petit et al., 1998; MacNeill et al., 2009). Viruses also deploy molecules that prevent inflammasome assembly to enhance virus virulence. This is best illustrated by the action of the Kaposi's sarcoma–associated herpesvirus (KSHV) ORF63, a viral Nlrp1 homologue (Gregory et al., 2011), and the MYXV protein M013L, which resemble the host pyrin-only proteins (Johnston et al., 2005).
Figure 1.

Viral modulation of cytokine synthesis and signal transduction.
Viral Modulation of Cytokine Signal Transduction
There are several examples of viruses that inhibit cytokine-induced intracellular pathways, such as some of the anti-IFN mechanisms discussed below. The interaction of TNF receptors (TNFRs) and Fas with their respective ligands induces the binding of cytoplasmic factors that interact through death domains and death effector domains present in adaptor proteins and in some caspases. These pathways activate NF-κB and other intracellular factors that trigger the activation of innate immune pathways. Molluscum contagiosum virus and KSHV encode inhibitors containing death domains that prevent signal transduction through receptors for TNF and Fas ligand and activation of caspase-8 (Bertin et al., 1997; Thome and Tschopp, 2001; Figure 1). An unrelated protein encoded by the UL36 gene from human cytomegalovirus also associates with caspase-8 and blocks its activation (Skaletskaya et al., 2001). Adenoviruses produce a number of proteins encoded by the E3 region that inhibit TNF- and Fas-mediated apoptosis (Mahr and Gooding, 1999; Wold et al., 1999). The adenovirus receptor internalization and degradation complex, formed by the E3/10.4K and E3/14.5K proteins, induces internalization and subsequent degradation in lysosomes of TNFR and Fas. As a result of this, the infected cell becomes resistant to TNFR- and Fas-mediated apoptosis (Burgert et al., 2002; McNees and Gooding, 2002). African swine fever virus replicates in macrophages, one of the major cytokine producers in the immune system, and encodes a homologue of the inhibitor of kappa-light-chain-enhancer of activated B cells that interacts with the transcription factors NF-κB and nuclear factor–activated T cell and prevents the transcription of cytokine genes (Miskin et al., 1998). Poxviruses encode numerous proteins that inhibit NF-κB activation (Smith et al., 2013; Figure 1). For example, VACV encodes three intracellular proteins (A52, A46, and K7) that block signaling mediated by Toll-like receptors and the IL-1 receptor (IL-1R) by mimicking intracellular adaptor domains of these receptors (Bowie et al., 2000; Harte et al., 2003; Smith et al., 2013). Inhibition of both IL-1- and TNF-mediated NF-κB activation is mediated by other VACV proteins, such as N1 or B14, that act downstream of the convergence of IL-1- and TNF-mediated signaling. Structural studies have shown that some of these VACV proteins (A52 and K7) have a B cell lymphoma-2 fold, a property that is shared with other VACV proteins that function as inhibitors of innate immune signaling pathways (Smith et al., 2013). During virus entry into cells, the interaction of measles virus with its cellular receptor CD46, a complement regulatory protein that protects cells from lysis by complement, blocks the production of IL-12, a potent inducer of IFN-γ and cellular responses (Figure 1).
Lastly, some viruses may subvert cytokine-mediated signaling for their own benefit. The latent membrane protein 1 of Epstein–Barr virus (EBV), a virus that establishes latency in B cells, is a membrane protein that lacks sequence similarity to members of the TNFR superfamily but has a short cytoplasmic sequence that binds components of the TNFR and CD40 transduction machinery. In EBV-infected cells, the multimerization of latent membrane protein 1 in the absence of TNF leads to the recruitment of adaptor molecules and TNF-mediated biological responses such as cell proliferation that may enhance virus replication (Farrell, 1998). In this way, EBV takes advantage of a B cell activation pathway to ensure survival of the infected cell that carries the latent viral genome.
Modulation of IFN Functions
IFNs were discovered because of their ability to protect cells from viral infection. IFNs can be classified as type I (α and β), type II (γ), and type III (λ) and represent one of the first antiviral defense mechanisms to be activated after infection. The finding of anti-IFN strategies encoded by most viruses highlights the key role of IFNs against viral infection. IFNs are produced and secreted in response to viral infection and bind specific receptors which trigger intracellular pathways that activate antiviral mechanisms to protect cells from infection and stimulate leukocytes in the interface of innate and adaptive immunity, and can influence the type of immune response activated after infection (Goodbourn et al., 2000; Katze et al., 2002; Versteeg and Garcia-Sastre, 2010; Coccia and Battistini, 2015).
More than 170 viral proteins have been shown to block the activity of IFN at different levels: (1) inhibition of IFN production; (2) blockade of IFN binding to specific receptors; (3) interference with signal transduction pathways induced by IFN; and (4) inhibition of antiviral effector functions induced by IFN.
Viral Inhibition of IFN Synthesis
Recognition of viruses is mediated by membrane-associated or cytoplasmic PRRs, which initiate the innate immune response and trigger the production of IFNs (Coccia and Battistini, 2015; Figure 2 ). Coronaviruses, such as severe acute respiratory syndrome coronavirus, encode an exoribonuclease that is involved in RNA-proofreading but also degrades viral PAMPs to avoid detection from the immune system (Kindler and Thiel, 2014). These viruses replicate their genome within double membrane vesicles, protecting the genome from being recognized by the host (Knoops et al., 2008). Dengue virus NS2B/3 protease binds and cleaves the adaptor protein simulator of IFN genes and prevents IFN induction (Aguirre et al., 2012).
Figure 2.

Activation of pattern recognition receptors (PRRs) by viruses, induction of interferon (IFN), and antiviral effects of IFN.
Secreted Viral IFN Receptors and IFN Binding Proteins
Poxviruses encode proteins that are secreted from infected cells and bind with high-affinity type I or type II IFN and prevent their interaction with interferon receptors (IFNRs) (McFadden and Murphy, 2000; Alcami, 2003; Epperson et al., 2012; Smith et al., 2013; Figure 3 ). This mechanism prevents IFN-mediated signaling and the initiation of biological effects.
Figure 3.
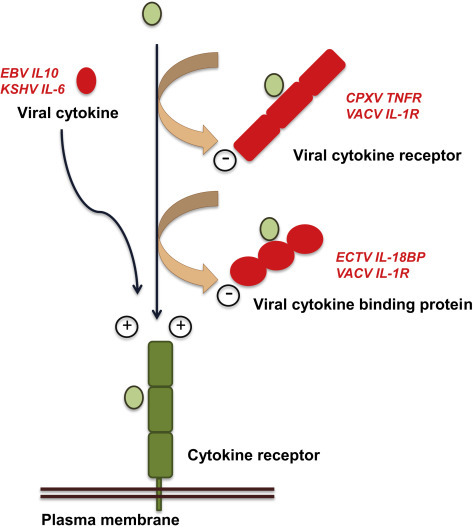
Cytokines, cytokine receptors, and cytokine-binding proteins encoded by viruses.
The viral type II IFNR is encoded by MYXV and the orthopoxviruses VACV, CPXV, and ECTV (Upton et al., 1992; Alcami and Smith, 1995; Mossman et al., 1995). This viral decoy receptor has sequence similarity to the IFN binding domain of the cellular type II IFNR. The crystal structure of the ECTV type II IFNR showed a novel C-terminal region required for the tetramerization of the decoy receptor bound to two IFN-γ dimers (Nuara et al., 2008). A role during viral infection has been demonstrated by the attenuated phenotype of a MYXV mutant lacking this IFN inhibitor (Mossman et al., 1996).
VACV, CPXV, and highly virulent orthopoxviruses (variola virus, monkeypox virus, and ECTV) encode a secreted type I interferon binding protein (IFNBP) that has very limited sequence similarity to the cellular counterparts (Colamonici et al., 1995; Symons et al., 1995; Figure 2). The orthologue encoded by yaba-like disease virus was shown to inhibit also human type III IFN (Huang et al., 2007). The viral type I IFNBP is secreted initially but binds to the cell surface through the interaction with glycosaminoglycans to be retained in the vicinity of the infected tissues and presumably to increase its efficacy (Alcami et al., 2000; Montanuy et al., 2011). VACV and ECTV mutants lacking the type I IFNBP gene are attenuated in mice and their ability to grow in tissues is impaired (Symons et al., 1995; Xu et al., 2008).
Interference with Signal Transduction Induced by IFNs
The interaction of IFNs with specific receptors at the cell surface triggers a complex cascade of intracellular pathways through Janus kinase (JAK) and signal transducers and activators of transcription (STAT) pathways (Goodbourn et al., 2000; Katze et al., 2002; Versteeg and Garcia-Sastre, 2010; Coccia and Battistini, 2015; Figure 2). These signaling events cause the translocation of IFN-stimulated gene (ISG) factor 3 to the nucleus, which binds to IFN-stimulated response elements and activates the transcription of ISGs. These genes act to promote viral clearance, establish an antiviral state, and stimulate cells at the interface of innate and adaptive immunity. Genes upregulated by IFN include the low-molecular-weight protein 2 and 7 subunits of the proteasome, involved in the generation of viral peptides for major histocompatibility complex (MHC) class I and class II molecules, the molecules that present viral peptides to T cells. Other genes upregulated by IFNs are the double-stranded RNA (dsRNA)-dependent protein kinase (PKR) and the 2′-5′-oligoadenylate synthetase (2′5′OAS) that activates ribonuclease L (RNase L).
Many viral proteins have been shown to interfere with the IFN signaling cascade (Goodbourn et al., 2000; Katze et al., 2002; Versteeg and Garcia-Sastre, 2010; Coccia and Battistini, 2015; Figure 2). For example, the T antigen of murine polyoma virus binds to and inactivates JAK1 (Weihua et al., 1998), whereas the protein V from simian virus 5 interacts with STAT1 and induces its degradation by the proteasome (Didcock et al., 1999). Other paramyxovirus V proteins target different components of the IFN signaling pathway. Type II human parainfluenza virus V protein targets STAT2, mumps virus V protein targets both STAT1 and STAT3, and measles virus V protein causes a defect in STAT nuclear accumulation and STAT-inducible transcription (Parisien et al., 2001; Nishio et al., 2002; Palosaari et al., 2003; Ulane et al., 2003). KSHV uses a different mechanism by encoding a homologue of IRF that represses the IFN-mediated transcriptional activation of host genes (Zimring et al., 1998).
Viral Modulation of IFN Effector Functions
One of the best-characterized antiviral effects of IFN is the induction of the PKR and 2′5′OAS pathways that lead to the blockade of viral gene expression in the cell (Gale and Katze, 1998; Goodbourn et al., 2000; Katze et al., 2002; Versteeg and Garcia-Sastre, 2010; Coccia and Battistini, 2015). The replication and transcription of viral genomes leads to the formation of dsRNA that activates PKR, which phosphorylates the translation initiation factor 2α (eIF-2α) causing its inactivation and the subsequent arrest of protein synthesis in infected cells (Figure 2). Viral mechanisms that prevent the activation of PKR are varied: (1) proteins encoded by reovirus (σ3) and VACV (E3) that bind dsRNA; (2) RNAs encoded by adenovirus (VAI) and HIV (TAR) that bind PKR but do not activate the enzyme; and (3) proteins encoded by herpes simplex virus (HSV) (US11), hepatitis C virus (US5A), and human immunodeficiency virus (HIV) (Tat) that bind directly to PKR and inactivate the enzyme. An alternative strategy to block the PKR pathway is to prevent the phosphorylation of eIF-2α. Poxviruses encode an eIF-2α homologue, the K3 protein in VACV, which functions as a substrate of PKR and prevents phosphorylation of eIF-2α. The glycoprotein E2 from hepatitis C virus acts as a competitive substrate with eIF-2α for PKR binding. Alternatively, the HSV IC34.5 protein activates protein phosphatase 1α that removes the phosphate groups from eIF-2α.
The formation of dsRNA during viral replication also activates the enzyme 2′5′OAS, producing 2′5′OA which in turn activates RNase L and the degradation of RNA within the infected cell, causing the blockade of viral replication. HSV produces analogs of 2′5′OA which bind RNase L and inhibit this pathway. The proteins σ3 from reovirus and E3L from VACV bind dsRNA and thus inhibit the activation of 2′5′OAS as well as PKR (see above).
Molecular Mimicry: Viral Cytokines and Cytokine Receptors
Sequencing of the genome of large DNA viruses (poxviruses and herpesviruses) uncovered the presence of viral proteins that mimic cytokines and their receptors (McFadden and Murphy, 2000; Alcami, 2003; Seet et al., 2003; Epperson et al., 2012; Smith et al., 2013; Figure 3). The sequence similarity suggests that, during evolution, viruses may have ‘stolen’ host genes that initiate or coordinate the immune response and incorporated them into their genomes. Functional studies have shown that the biological proteins of the viral proteins differ from their cellular counterparts suggesting that viruses may have altered the function of these host proteins for their own benefit.
Viral Cytokines
Viral cytokines act as agonists inducing specific immune responses that are beneficial for the virus. The VACV epidermal growth factor homologue activates cell growth and promotes viral replication, but an immune-related function has not been defined (McFadden et al., 1994). The EBV-encoded IL-10 homologue was the first viral cytokine identified with immunomodulatory activity (Hsu et al., 1990; Figure 3). EBV IL-10 inhibits the induction of IFN-γ and suppresses cellular immunity but lacks the immunostimulatory properties of the host counterpart. Other examples of viral cytokines are the IL-6 homologue encoded by KSHV and the IL-17 homologue encoded by herpesvirus saimiri (Yao et al., 1995; Moore et al., 1996). Virus-encoded IL-6 and IL-17 also promote the proliferation of B and T cells, respectively, increasing the presence of target host cells in the infected tissues that may favor viral replication.
Secreted Viral Cytokine Receptors
Viral genes encoding soluble cytokine receptors have been identified in the poxvirus and herpesvirus families. Viral cytokine receptors are secreted versions of cellular cytokine receptors that lack transmembrane and cytoplasmic domains. These viral proteins function as decoy receptors binding cytokines with high affinity and blocking their activity. A second class of cytokine receptors, known as binding proteins, has very limited sequence similarity to the host cytokine receptors (Figure 3).
The first virus-encoded cytokine receptor identified was the MYXV TNFR (Smith et al., 1991). Four different genes encoding viral TNFRs were subsequently identified in other poxviruses such as CPXV, VACV, and ECTV and were designated CrmB, CrmC, CrmD, and CrmE (Hu et al., 1994; Smith et al., 1996; Loparev et al., 1998; Saraiva and Alcami, 2001; Figures 1 and 3). A variety of TNFRs in poxvirus genomes provide distinct binding properties and may have been optimized to inhibit the immune response in different hosts or tissues (Pontejo et al., 2015). CrmB and CrmD contain a C-terminal domain, named smallpox virus–encoded chemokine receptor domain, that binds chemokines (Alejo et al., 2006). A secreted protein encoded by tanapox virus and yaba-like disease virus that binds TNF but adopts a MHC class I-like fold adds complexity to this family of viral proteins (Brunetti et al., 2003; Yang et al., 2009). Clinical isolates of human cytomegalovirus, but not laboratory strains, encode a membrane TNFR homologue (UL144) related to herpesvirus entry mediator (Smith et al., 1991). UL144 binds B lymphocyte and T lymphocyte attenuator, a natural ligand of herpesvirus entry mediator, and inhibits T cell proliferation (Cheung et al., 2005). In addition, UL144 is a potent activator of NF-κB, upregulates the chemokine CCL22, and may help immune evasion by attracting Th2 and regulatory T cells (Poole et al., 2006).
CPXV and ECTV encode a secreted version of CD30, another member of the TNFR superfamily (Panus et al., 2002; Saraiva et al., 2002). Viral CD30 not only works as a decoy receptor blocking the interaction of CD30 to CD30 ligand but also induces reverse signaling in CD30 ligand–expressing cells. Viral CD30 downregulates type 1 cytokine-mediated inflammatory responses in mice (Saraiva et al., 2002), but this protein is not a virulent factor in a mouse model of ECTV infection (mousepox) (Alejo et al., 2009).
The viral IL-1R acts as a decoy receptor that, in contrast to the membrane IL-1Rs encoded by the host, binds exclusively IL-1β and not IL-1α or IL-1R antagonist (Alcami and Smith, 1992; Spriggs et al., 1992; Figures 1 and 3). Deletion of this gene in VACV leads to enhanced virulence associated with fever and accelerated death in mice infected through the respiratory route, a natural route of poxvirus transmission (Alcami and Smith, 1992, Alcami and Smith, 1996). This illustrates that the function of the viral IL-1R and other virus-encoded cytokine inhibitors may be to reduce immunopathology caused by excess production of cytokines rather than to evade the immune response.
Viral Cytokine Binding Proteins
IL-18 is a proinflammatory cytokine required, together with IL-12, for the induction of IFN-γ and the generation of an efficient cellular response against viral infections. The viral IL-18 binding protein (IL-18BP) is unrelated to cellular membrane IL-18 receptors but shows a structure similar to that of the human IL-18BP, a secreted protein that downregulates the immune response (Krumm et al., 2008; Figures 1 and 3). The viral IL-18BP is expressed by a variety of poxviruses including M. contagiosum virus, a poxvirus that causes benign skin tumors and persists in the skin for months without inducing an inflammatory response (Xiang and Moss, 1999; Smith et al., 2000). The poxvirus IL-18BP downregulates IFN-γ production and natural killer cell responses in ECTV-infected mice (Born et al., 2000; Smith et al., 2000). The EBV-encoded colony stimulating factor 1 binding protein has limited sequence similarity to the cellular counterpart and it has been proposed to modulate the response of macrophages during EBV infection (Strockbine et al., 1998).
Modulation of Chemokine Networks
Chemokines are chemotactic cytokines that induce cell migration and other biological effects, such as cell differentiation and angiogenesis (Baggiolini, 1998). Herpesviruses and poxviruses modulate chemokine activity through the expression of chemokine homologues, chemokine receptor homologues, and secreted chemokine-binding proteins (Lalani et al., 2000; McFadden and Murphy, 2000; Murphy, 2001; Alcami, 2003). The function of these viral proteins during the infectious cycle is diverse and will be discussed elsewhere in this Encyclopedia.
Conclusions and Future Perspectives
The reasons why a virus blocks a particular immune mechanism may depend on the replication and transmission mechanisms adopted by the virus. It will be of interest to determine which viral genes are required for the initial steps of the infection or for efficient transmission to another host in the presence of an active immune system. The apparent complexity and redundancy of viral proteins that evade immunity may only be understood in the context of viral infection in the host. The lack of good animal models of infection for many viruses is limiting our understanding of the function of viral immune evasion strategies in vivo.
A better knowledge of the interaction of viruses with the host defense mechanisms will help us to understand the mechanisms of viral pathogenesis. Very often, the pathology observed in viral infections is not due to viral replication but to the damage that the immune response to infection may cause in the host. Some human inflammatory and autoimmune diseases are the result of an uncontrolled production of immune mediators such as TNF and IL-1. We are finding examples of viral proteins that may enhance these immunopathological responses in the infected host.
The deletion of viral immune evasion proteins from the viral genome causes viral attenuation in most cases due to increased host defenses. However, enhanced viral virulence has been observed in VACV and CPXV when some viral functions that block IL-1β, complement factors, or chemokines are inactivated (Alcami and Smith, 1992, Alcami and Smith, 1996; Miller et al., 1997; Reading et al., 2003). These surprising observations suggest that the function of some of the viral immune evasion proteins is not to promote viral replication but to ‘protect’ the host from immunopathology, mediated in these examples by IL-1β, complement, or chemokines, respectively. Increased host survival will be beneficial since the virus fully depends on the host for its replication. The complex interaction of each virus with the immune system is unique, and a better understanding of this interaction will lead to new strategies to control viral replication.
See also
IMMUNITY TO VIRAL INFECTIONS | Chemokines and Viral Infections; IMMUNITY TO VIRAL INFECTIONS | Innate Cytokine Responses and Their Functions during Viral Infections; IMMUNITY TO VIRAL INFECTIONS | Sensors of Viral Infection.
Acknowledgments
Work in the author's laboratory is funded by the Spanish National Research Council and the Spanish Ministry of Economy and Competitiveness.
References
- Aguirre S., Maestre A.M., Pagni S. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- Alcami A., Koszinowski U.H. Viral mechanisms of immune evasion. Immunol. Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A., Smith G.L. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell. 1992;71:153–167. doi: 10.1016/0092-8674(92)90274-g. [DOI] [PubMed] [Google Scholar]
- Alcami A., Smith G.L. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J. Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A., Smith G.L. A mechanism for the inhibition of fever by a virus. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11029–11034. doi: 10.1073/pnas.93.20.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A., Symons J.A., Smith G.L. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 2000;74:11230–11239. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejo A., Ruiz-Arguello M.B., Ho Y. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5995–6000. doi: 10.1073/pnas.0510462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejo A., Saraiva M., Ruiz-Arguello M.B. A method for the generation of ectromelia virus (ECTV) recombinants: in vivo analysis of ECTV vCD30 deletion mutants. PLoS One. 2009;4:e5175. doi: 10.1371/journal.pone.0005175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- Barbe F., Douglas T., Saleh M. Advances in Nod-like receptors (NLR) biology. Cytokine Growth Factor Rev. 2014;25:681–697. doi: 10.1016/j.cytogfr.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Bertin J., Armstrong R.C., Ottilie S. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A.L., Beutler B. Intracellular Toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Born T.L., Morrison L.A., Esteban D.J. A poxvirus protein that binds to and inactivates IL-18, and inhibits NK cell response. J. Immunol. 2000;164:3246–3254. doi: 10.4049/jimmunol.164.6.3246. [DOI] [PubMed] [Google Scholar]
- Bowie A., Kiss-Toth E., Symons J.A. A46R and A52R from vaccinia virus are antagonists of host IL-1 and Toll-like receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti C.R., Paulose-Murphy M., Singh R. A secreted high-affinity inhibitor of human TNF from Tanapox virus. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4831–4836. doi: 10.1073/pnas.0737244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgert H.G., Ruzsics Z., Obermeier S. Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol. 2002;269:273–318. doi: 10.1007/978-3-642-59421-2_16. [DOI] [PubMed] [Google Scholar]
- Cheung T.C., Humphreys I.R., Potter K.G. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia E.M., Battistini A. Early IFN type I response: learning from microbial evasion strategies. Semin. Immunol. 2015;27:85–101. doi: 10.1016/j.smim.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamonici O.R., Domanski P., Sweitzer S.M. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J. Biol. Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- Didcock L., Young D.F., Goodbourn S., Randall R.E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson M.L., Lee C.A., Fremont D.H. Subversion of cytokine networks by virally encoded decoy receptors. Immunol. Rev. 2012;250:199–215. doi: 10.1111/imr.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P.J. Signal transduction from the Epstein–Barr virus LMP-1 transforming protein. Trends Microbiol. 1998;6:175–177. doi: 10.1016/s0966-842x(98)01262-1. discussion 177–178. [DOI] [PubMed] [Google Scholar]
- Gale M., Jr., Katze M.G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Didcock L., Randall R.E. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Gregory S.M., Davis B.K., West J.A. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 2011;331:330–334. doi: 10.1126/science.1199478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte M.T., Haga I.R., Maloney G. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 2003;197:343–351. doi: 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Hsu D.H., de Waal Malefyt R., Fiorentino D.F. Expression of interleukin-10 activity by Epstein–Barr virus protein BCRF1. Science. 1990;250:830–832. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- Hu F., Smith C.A., Pickup D.J. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- Huang J., Smirnov S.V., Lewis-Antes A. Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9822–9827. doi: 10.1073/pnas.0610352104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.B., Barrett J.W., Nazarian S.H. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Katze M.G., He Y., Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Kindler E., Thiel V. To sense or not to sense viral RNA – essentials of coronavirus innate immune evasion. Curr. Opin. Microbiol. 2014;20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm B., Meng X., Li Y. Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20711–20715. doi: 10.1073/pnas.0809086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani A.S., Barrett J.W., McFadden G. Modulating chemokines: more lessons from viruses. Immunol. Today. 2000;21:100–106. doi: 10.1016/s0167-5699(99)01556-x. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M., Dixit V.M. Modulation of inflammasome pathways by bacterial and viral pathogens. J. Immunol. 2011;187:597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- Loparev V.N., Parsons J.M., Knight J.C. A third distinct tumor necrosis factor receptor of orthopoxviruses. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3786–3791. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C., Malik A., Kanneganti T.D. Inflammasome control of viral infection. Curr. Opin. Virol. 2015;12:38–46. doi: 10.1016/j.coviro.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill A.L., Moldawer L.L., Moyer R.W. The role of the cowpox virus crmA gene during intratracheal and intradermal infection of C57BL/6 mice. Virology. 2009;384:151–160. doi: 10.1016/j.virol.2008.10.041. [DOI] [PubMed] [Google Scholar]
- Mahr J.A., Gooding L.R. Immune evasion by adenoviruses. Immunol. Rev. 1999;168:121–130. doi: 10.1111/j.1600-065x.1999.tb01287.x. [DOI] [PubMed] [Google Scholar]
- McFadden G., Graham K., Opgenorth A. Poxvirus growth factors. In: McFadden G., editor. Viroceptors, Virokines and Related Immune Modulators Encoded by DNA Viruses. R.G. Landes Company; Georgetown, Texas: 1994. pp. 1–15. [Google Scholar]
- McFadden G., Murphy P.M. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr. Opin. Microbiol. 2000;3:371–378. doi: 10.1016/s1369-5274(00)00107-7. [DOI] [PubMed] [Google Scholar]
- McNees A.L., Gooding L.R. Adenoviral inhibitors of apoptotic cell death. Virus Res. 2002;88:87–101. doi: 10.1016/s0168-1702(02)00122-3. [DOI] [PubMed] [Google Scholar]
- Messud-Petit F., Gelfi J., Delverdier M. Serp2, an inhibitor of the interleukin-1beta-converting enzyme, is critical in the pathobiology of myxoma virus. J. Virol. 1998;72:7830–7839. doi: 10.1128/jvi.72.10.7830-7839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.G., Shchelkunov S.N., Kotwal G.J. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology. 1997;229:126–133. doi: 10.1006/viro.1996.8396. [DOI] [PubMed] [Google Scholar]
- Miskin J.E., Abrams C.C., Goatley L.C., Dixon L.K. A viral mechanism for inhibition of the cellular phosphatase calcineurin. Science. 1998;281:562–565. doi: 10.1126/science.281.5376.562. [DOI] [PubMed] [Google Scholar]
- Montanuy I., Alejo A., Alcami A. Glycosaminoglycans mediate retention of the poxvirus type I interferon binding protein at the cell surface to locally block interferon antiviral responses. FASEB J. 2011;25:1960–1971. doi: 10.1096/fj.10-177188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.S., Boshoff C., Weiss R.A., Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- Mossman K., Nation P., Macen J. Myxoma virus M-T7, a secreted homolog of the interferon-gamma receptor, is a critical virulence factor for the development of myxomatosis in European rabbits. Virology. 1996;215:17–30. doi: 10.1006/viro.1996.0003. [DOI] [PubMed] [Google Scholar]
- Mossman K., Upton C., Buller R.M.L., McFadden G. Species specificity of ectromelia virus and vaccinia virus interferon-g binding proteins. Virology. 1995;208:762–769. doi: 10.1006/viro.1995.1208. [DOI] [PubMed] [Google Scholar]
- Murphy P.M. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2001;2:116–122. doi: 10.1038/84214. [DOI] [PubMed] [Google Scholar]
- Nishio M., Garcin D., Simonet V., Kolakofsky D. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology. 2002;300:92–99. doi: 10.1006/viro.2002.1509. [DOI] [PubMed] [Google Scholar]
- Nuara A.A., Walter L.J., Logsdon N.J. Structure and mechanism of IFN-gamma antagonism by an orthopoxvirus IFN-gamma-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1861–1866. doi: 10.1073/pnas.0705753105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palosaari H., Parisien J.P., Rodriguez J.J. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panus J.F., Smith C.A., Ray C.A. Cowpox virus encodes a fifth member of the tumor necrosis factor receptor family: a soluble, secreted CD30 homologue. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8348–8353. doi: 10.1073/pnas.122238599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien J.P., Lau J.F., Rodriguez J.J. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology. 2001;283:230–239. doi: 10.1006/viro.2001.0856. [DOI] [PubMed] [Google Scholar]
- Pontejo S.M., Alejo A., Alcami A. Comparative biochemical and functional analysis of viral and human secreted TNF decoy receptors. J. Biol. Chem. 2015;290:15973–15984. doi: 10.1074/jbc.M115.650119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E., King C.A., Sinclair J.H., Alcami A. The UL144 gene product of human cytomegalovirus activates NFkappaB via a TRAF6-dependent mechanism. EMBO J. 2006;25:4390–4399. doi: 10.1038/sj.emboj.7601287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray C.A., Black R.A., Kronheim S.R. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- Reading P.C., Symons J.A., Smith G.L. A soluble chemokine-binding protein from vaccinia virus reduces virus virulence and the inflammatory response to infection. J. Immunol. 2003;170:1435–1442. doi: 10.4049/jimmunol.170.3.1435. [DOI] [PubMed] [Google Scholar]
- Sancho D., Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M., Alcami A. CrmE, a novel soluble tumor necrosis factor receptor encoded by poxviruses. J. Virol. 2001;75:226–233. doi: 10.1128/JVI.75.1.226-233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M., Smith P., Fallon P.G., Alcami A. Inhibition of type 1 cytokine-mediated inflammation by a soluble CD30 homologue encoded by ectromelia (mousepox) virus. J. Exp. Med. 2002;196:829–839. doi: 10.1084/jem.20020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet B.T., Johnston J.B., Brunetti C.R. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- Skaletskaya A., Bartle L.M., Chittenden T. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., Davis T., Wignall J.M. T2 open reading frame from Shope fibroma virus encodes a soluble form of the TNF receptor. Biochem. Biophys. Res. Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- Smith C.A., Hu F.Q., Smith T.D. Cowpox virus genome encodes a second soluble homologue of cellular TNF receptors, distinct from CrmB, that binds TNF but not LT alpha. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- Smith G.L., Benfield C.T., Maluquer de Motes C. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J. Gen. Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- Smith V.P., Bryant N.A., Alcami A. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. J. Gen. Virol. 2000;81:1223–1230. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- Spriggs M.K., Hruby D.E., Maliszewski C.R. Vaccinia and cowpox viruses encode a novel secreted interleukin-1-binding protein. Cell. 1992;71:145–152. doi: 10.1016/0092-8674(92)90273-f. [DOI] [PubMed] [Google Scholar]
- Strockbine L.D., Cohen J.I., Farrah T. The Epstein–Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J. Virol. 1998;72:4015–4021. doi: 10.1128/jvi.72.5.4015-4021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons J.A., Alcami A., Smith G.L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Thome M., Tschopp J. Regulation of lymphocyte proliferation and death by FLIP. Nat. Rev. Immunol. 2001;1:50–58. doi: 10.1038/35095508. [DOI] [PubMed] [Google Scholar]
- Thompson J.P., Turner P.C., Ali A.N. The effects of serpin gene mutations on the distinctive pathobiology of cowpox and rabbitpox virus following intranasal inoculation of Balb/c mice. Virology. 1993;197:328–338. doi: 10.1006/viro.1993.1594. [DOI] [PubMed] [Google Scholar]
- Tortorella D., Gewurz B.E., Furman M.H. Viral subversion of the immune system. Annu. Rev. Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- Ulane C.M., Rodriguez J.J., Parisien J.P., Horvath C.M. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 2003;77:6385–6393. doi: 10.1128/JVI.77.11.6385-6393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C., Mossman K., McFadden G. Encoding of a homolog of IFN-g receptor by myxoma virus. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- Versteeg G.A., Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihua X., Ramanujam S., Lindner D.J. The polyoma virus T antigen interferes with interferon-inducible gene expression. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1085–1090. doi: 10.1073/pnas.95.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W.S., Doronin K., Toth K. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr. Opin. Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Moss B. IL-18 binding and inhibition of interferon gamma induction by human poxvirus-encoded proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11537–11542. doi: 10.1073/pnas.96.20.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., Cohen M., Tang Y. The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. J. Exp. Med. 2008;205:981–992. doi: 10.1084/jem.20071854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., West A.P., Jr., Bjorkman P.J. Crystal structure of TNFalpha complexed with a poxvirus MHC-related TNF binding protein. Nat. Struct. Mol. Biol. 2009;16:1189–1191. doi: 10.1038/nsmb.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Fanslow W.C., Seldin M.F. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- Zimring J.C., Goodbourn S., Offermann M.K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J. Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


