Abstract
The oxytocin receptor (OXTR) is a key regulator of stress and anxiety and may be regulated by both psychosocial risk factors and gonadal hormones,making it an attractive candidate for study in postpartum depression (PPD). The objective of this study was to investigate both serum hormone and PPD specific DNA methylation variation in the OXTR. Illumina HM450 microarray data generated in a prospective PPD cohort identified significant associations (P=0.014) with PPD in an intronic region in the OXTR located 4bp proximal to an estrogen receptor (ER) binding region. Pyrosequencing confirmed moderate evidence for an interaction of CpGs in the region with childhood abuse status to mediate PPD. These CpGs located on chr3 at positions 8810078 and 8810069 exhibited significant associations with postpartum depression scores from an independent cohort of 240 women with no prior psychiatric history. Hormone analysis suggested a PPD specific negative correlation of DNA methylation in the region with serum estradiol levels. Estradiol levels and OXTR DNA methylation exhibited a significant interaction to associate with the ratio of allopregnanolone to progesterone. Cumulatively, the data corroborate our previous hypotheses of a PPD specific increased sensitivity of epigenetic reprogramming at estrogen target genes and suggests that OXTR epigenetic variation may be an important mediator of mood relevant neuroactive steroid production.
Keywords: Postpartum depression, Trauma, DNA methylation, OXTR, Allopregnanolone, Estradiol
1. Introduction
Postpartum depression (PPD) affects between 10 and 20% of women (Josefsson et al., 2001; Miller, 2002; Pearlstein et al., 2009) and has significant adverse effects on both mother and child (Breese McCoy, 2011; Cuijpers et al., 2008; Field, 2011; Hirst and Moutier, 2010; O’Hara, 2009; Soufia et al., 2010). PPD afflicts some populations at even higher rates, for example,30%of women with a history of depression and 52% of women with bipolar disorder (Viguera et al., 2011). A growing body of evidence indicates that an increased sensitivity to change in gonadal hormone levels may mediate a biological vulnerability to PPD, with much of the available evidence implicating the estrogens (Bloch et al., 2000; Guintivano et al.,2014; Mehta et al., 2014). Critically, it is not the levels of estrogens so much as differences in the downstream responses and physiological consequences to them that may confer risk onto vulnerable women.
Importantly, PPD has also been associated with differences in levels of other hormones, including corticotrophin releasing hormone (Magiakou et al., 1996), triiodothyronine (Bunevicius et al., 2009; Pedersen et al., 2007), testosterone (Aswathi et al., 2015), and oxytocin (Skrundz et al., 2011), among others. Some of these associations may represent the complex interplay between dysregulated hormone systems and their downstream consequences. Of particular interest are hormones linked with estrogen signaling including progesterone, its metabolites, and oxytocin.
Progesterone withdrawal and progesterone receptor antagonists lead to depressive phenotypes. Metabolites of progesterone in the brain, specifically allopregnanolone, modulate GABA(A) receptors in a concentration dependent manner, resulting in sedation and anxiolytic effects in some cases and anxiogenic, aggressive, and irritable effects in others (Andreen et al., 2009; Backstrom et al., 2011; Studd, 2011). Levels of allopregnanolone may also be closely tied to estradiol signaling, as estradiol administration to ovariectomized (OVX) rats restored deficits in both this progesterone metabolite and beta-endorphin (Yim et al., 2010) and resulted in anxiolytic effects in another progesterone withdrawal model rat (Windle et al., 2006).
There is growing interest in the oxytocin (OXT) system in PPD. It is a key hormone in the initiation of maternal behavior after parturition (Stuebe et al., 2012). Low prenatal plasma OXT levels have been observed in PPD (Skrundz et al., 2011). We propose the oxytocin receptor (OXTR) is an attractive candidate for study of epigenetic variation associated with PPD as modulation of the OXT system occurs with estrogen at the OXTR. Estradiol increases OXTR gene transcription (Mamrut et al., 2013) resulting in elevations in the uterus (Franczak et al., 2002) and numerous brain regions including the hippocampus, amygdala, and arcuate nucleus (Quinones-Jenab et al., 1997). Recently, Bell et al. (2015) demonstrated DNA methylation associations of the OXTR with PPD. The association involved an interaction of OXTR genotype at rs53576 and epigenetic factors such that antenatally euthymic women who developed PPD had increased OXTR DNA methylation in GG homozygotes.
In addition to biological evidence, epidemiological evidence points to a different series of risk factors for perinatal depression. Antenatal depression and PPD both exhibit psychosocial risk factors such as low levels of partner and social support (Jeong et al., 2013; Robertson et al., 2004), while trauma history strongly predicts PPD (Meltzer-Brody et al., 2013). In a recent review and synthesis of studies from 2000 to 2013, Yim et al. (2015) identified strong risk for PPD based on chronic strain, severe life events, relationship quality, and partner and maternal support. In light of the biological and epidemiological data, an attractive hypothesis is that stressors associated with childbirth and caring for a new born in the postpartum period may interact with underlying biological vulnerabilities to result in the onset of depressive symptoms. Furthermore, this system may be moderated by protective factors such as social support or risk factors such as early life trauma or severe life events. In this way, the physiological consequences of estrogen sensitivity and a lack of social support may result in physiological changes ultimately resulting in a depressive phenotype.
Given the importance of the OXT system for social bonding and buffering stress and anxiety, the oxytocin receptor (OXTR) represents an attractive target for bridging the gap between psychosocial stressors and estrogen sensitivity in PPD and antenatal depression. Blocking the Oxtr in progesterone withdrawal model rats led to increased hypothalamic pituitary adrenal (HPA) axis activation (Windle et al., 2006), while OXT administration inhibited stress activation of the HPA axis through recruitment of GABAergic neurons in another study (Smith et al., 2015). In humans, there is evidence that implicates a moderating effect of childhood trauma on OXT administration induced mood (Ammerman et al., 2012; Bakermans-Kranenburg et al., 2012; Huffmeijer et al., 2011). Furthermore, genetic variations in the OXTR along with early adversity have been found to be predictive of the development of depressive and anxiety symptoms (Myers et al., 2014; Thompson et al., 2011).
Recently, we published an association of PPD biomarker loci with serum hormone levels of estradiol and allopregnanolone, two mood relevant reproductive hormones (Osborne et al., 2015). This study suggested that variations in DNA methylation at earlier time points during pregnancy were associated with changes in hormone levels at later time points, suggesting that a PPD specific sensitivity to estrogen observed at PPD biomarker loci may translate into similar observations in estrogen responsive loci such as OXTR. In light of this as well as the gene expression modulating effects of estradiol on the OXTR, we sought to test the hypothesis that OXTR DNA methylation is associated with PPD at functionally relevant CpGs and that PPD will interact with traumatic experiences in early life to influence OXTR DNA methylation. We sought to test the hypothesis that gene expression differences would be observed in 1st or 3rd trimester antenatal blood in women who developed PPD. We hypothesize that OXTR DNA methylation will be negatively associated with serum estradiol levels and that this relationship may be specific to PPD cases. Furthermore, we hypothesize OXTR DNA methylation will be associated with the ratio of allopregnanolone to progesterone, gonadal hormones downstream of estradiol with potential implications for regulating mood.
2. Material and methods
2.1. Human samples
Subjects were derived from two prospective cohorts designed to study PPD and one cohort where DNA was collected long after pregnancy. The prospective cohorts included the Johns Hopkins Prospective PPD sample reported on previously by our group (Guintivano et al., 2014) and publicly available gene expression data collected by Mehta et al. (2014), who investigated gene expression in pregnant women prospectively followed until a PPD outcome could be determined. Both prospective cohorts comprised women with previous diagnoses of mood disorder (Guintivano et al., 2014; Mehta et al., 2014). The third cohort, the Franconian Maternal Health Evaluation Studies (FRAMES) cohort, consisted of psychiatrically healthy women, who were prospectively evaluated for depression and who had blood drawn for DNA at 1–3 years postpartum. Detailed information on study subjects is available below and in Table 1.
Table 1.
Sample demographics.
| Cohort | Johns Hopkins Prospective PPD Cohort | Prospective Gene Expression PPD Cohort | FRAMES cohort |
|---|---|---|---|
| Total | 51 | 61 | 240 |
| PPD: Antenatal Euthymic | 10 | 1st:15; 3rd: 15 | 5 |
| PPD: Antenatal Depressed | 12 | 1st:14; 3rd: 18 | 0 |
| No PPD: Antenatal Euthymic | 22 | 1st:22; 3rd: 28 | 235 |
| No PPD: Antenatal Depressed | 7 | 0 | 0 |
| PPD Assessment | Prospective Clinical | Prospective HDRS, BD1, EPDSa | Prospective HDRS |
| Age | 30.68 ±6.32 | 33 | 32.7 ± 0.018 |
| 1st trimester | 9 | 51 | 0 |
| 2nd trimester | 22 | 0 | 0 |
| 3rd trimester | 20 | 61 | 0 |
| Postpartum | 0 | 0 | 240 |
| % Caucasian: African American: Other | 30:70:0% | 85:15:0% | 100:0:0% |
| Childhood Sexual Abuse | 19 | NA | NA |
| No Childhood Sexual Abuse | 29 | NA | NA |
| Procedures | |||
| Illumina HM450 beadchip | 51 | 0 | 0 |
| OXTR Targeted Pyrosequencing | 51 | 0 | 240 |
| Illumina HumanHT-12 V4.0 expression beadchip | 0 | 61 | 0 |
| Hormone Analysis | 31 | 0 | 0 |
Depression diagnoses as reported by Mehta et al. (2014) for the various time points were based on satisfying all of the following criteria: 1st trimester: Hamilton Depression Rating Scale (HDRS)>14, Edinburgh Postnatal Depression Scale (EPDS)>12, Beck Depression Inventory (BDI)>15; 3rd Trimester: HDRS>15, EPDS>15, BDI>12; 3 months postpartum: HDRS>14, EPDS>11, BDI>16.
2.1.1. Johns Hopkins prospective PPD cohort
The Johns Hopkins Prospective PPD cohort derives from 93 pregnant women recruited at the Women’s Mood Disorders Center at Johns Hopkins. Subjects were prospectively followed during pregnancy and after delivery in order to identify genetic and clinical characteristics that precede the development of a postpartum depressive episode. Of this cohort, 51 subjects provided a blood sample for genetic and epigenetic analysis. The average age of the participants was 30.6 and 70% of the sample was Caucasian. Approximately 66% of participants had a history of Major Depression and 33% had Bipolar Disorder (I, II or NOS). Participants were managed by their treating psychiatrist as clinically indicated and were evaluated during each trimester of pregnancy and then 1 week, 1 month and 3 months postpartum. Women were classified as being depressed if they met DSM-IV criteria for a Major Depressive Episode (MDE) based on a psychiatric interview at each time point (first, second, and third trimester and 1 week and 1 month postpartum). Diagnosis of PPD was based on DSM-IV criteria and was confined to individuals experiencing an MDE within the first 4 weeks following parturition. Childhood sexual abuse status was binarily coded as a response to the question “Were you sexually assaulted as a child?¨during a clinical interview. The trimester of blood draw is depicted in Table S1. Prospective human subjects research at Johns Hopkins was conducted under IRB protocol # 00008149.
2.1.2. Prospective gene expression PPD cohort
Data from this cohort were generated by Mehta et al. (2014) and were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE45603. Subjects reportedly derived from a longitudinal cohort recruited at the Emory Women’s Mental Health Program. The sample was 33 years old on average and approximately 85% Caucasian. Approximately 59.6% were diagnosed with MDD and 40.4% with bipolar disorder, as defined by the Structured Clinical Interview for DSM-IV Axis I Disorders. Depressive symptoms were assessed in the 1st and 3rd trimester and within the first 7 weeks following pregnancy using the Beck Depression Inventory (BDI), the 17-item Hamilton Depression Rating Scale (HDRS), and the Edinburgh Postnatal Depression Scale (EPDS). Depression diagnoses as reported by Mehta et al., for the 1st trimester and 3rd trimester time points were based on satisfying all of the following criteria: 1st trimester: HDRS>14,EPDS>12,BDI>15;3rdTrimester:HDRS>15, EPDS>15, BDI>12; 3 months postpartum: HDRS>14, EPDS>11, BDI>16 such that the sample consisted of N=18 always depressed women, N=28 antenatally euthymic, and N=17 postpartum onset depression cases (Table 1).
2.1.3. Franconian maternal health evaluation studies (FRAMES) cohort
DNA was obtained from a subset of 421 women enrolled in the Franconian Maternal Health Evaluation Studies (FRAMES) and who were evaluated for genetic associations with postpartum outcomes in previous studies (Mehta et al., 2012). Of these samples a total of N=240 passed quality control for pyrosequencing following subsequent analyses. The mean age of the sample was 32.7±0.018years and 100% of the sample was Caucasian. Women did not have a previous psychiatric diagnosis. Women responded to the HDRS prospectively during the third trimester, between 48 and 72h, and between 6 and 8 months after parturition and PPD was classified as scores greater than 14 at the 6–8 month time point. All blood drawn for this study ranged in age between 1 and 3 years after parturition. The study was approved by the Ethics Committee at Erlangen University Hospital, Nuremberg, Germany. All participants received detailed information and provided written consent. Human subjects research was performed under IRB protocol # 00049309.
2.2. Illumina HM450 microarray data
Genome-wide DNA methylation data generated on the Illumina Human Methylation 450 (HM450) bead array were generated in 51 women from the Johns Hopkins Prospective sample previously (Guintivano et al., 2014). Data can be found on the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE44132.
2.3. Sodium bisulfite pyrosequencing
Bisulfite conversion was carried out using EZ DNA Methylation Gold Kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions on N=51 subjects from the Johns Hopkins Prospective cohort and N=240 subjects from the FRAMES cohort. Nested PCR amplifications were performed with a standard PCR protocol in 25 μl volume reactions containing 3–4μl of sodium-bisulfite-treated DNA, 0.2μM primers, and master mix containing Taq DNA polymerase (Sigma Aldrich, St. Louis, MO). Primer sequences can be found in Table S1. PCR amplicons were processed for pyrosequencing analysis according to the manufacturer’s standard protocol (Qiagen) using a PyroMark MD system (QIAGEN, Germantown, MD) with Pyro Q-CpG 1.0.9 software (QIA-GEN) for CpG methylation quantification. Only data passing internal quality checks for sodium bisulfite conversion efficiency, signal to noise ratio, and the observed vs. expected match of the predicted pyrogram peak profile using reference peaks were incorporated in subsequent analyses. Data generated derive from one technical replicate.
2.4. rs53576 genotyping
Genotyping of the rs53576 SNP was performed for N = 51 women from the Johns Hopkins Prospective cohort. The allele frequency of the A allele in European Caucasians, from which the FRAMES cohort derives, is reported as zero based on dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). As such, this cohort was not genotyped. Genotyping was performed on an Applied Biosystems 7900HT Fast-Real Time PCR system using TaqMan assay C___3290335_10 using cycling conditions recommended by the manufacturer. Genotype calls were made using the Allelic Discrimination function of the SDS 2.4 software.
2.5. Gene expression data
Gene expression data generated on the Illumina HumanHT-12 V4.0 expression beadchip by Mehta et al. (2014) was downloaded from GEO accession GSE45603. Raw data was normalized with the variance stabilizing transformation method (Huber et al., 2002) using the ‘justvsn’ function from the vsn package in R. OXTR gene expression (Probe: ILMN 1804929) values were extracted from the normalized dataset for subsequent analysis.
Investigation of cortical gene expression associations with DNA methylation was accomplished using the BrainCloud tool (Colantuoni et al., 2011). BrainCloud is a publically available dataset of prefrontal cortical gene expression levels and DNA methylation levels generated on the Illumina HM27 microarray, an earlier variant of the HM450 microarray used in the Johns Hopkins Prospective PPD cohort and containing a subset of overlapping probes.
2.6. Serum hormone analysis
Participant blood was collected at each visit in four 10 ml EDTA tubes and were immediately centrifuged at 4◦C for 30 min. The serum was then aliquoted into 2 ml microcentrifuge tubes, snap frozen on dry ice, and immediately stored in a −80◦C freezer. The blood samples we chose for analysis were from women from the Johns Hopkins Prospective cohort who had data for at least one antepartum blood draw and one postpartum visit with psychological data (N = 31). Participants whose blood was chosen for analysis did not differ by demographic characteristic or psychiatric diagnosis from those whose blood was not chosen. Blood was analyzed with the following kits: Allopregnanolone EIA kit from Arbor Assays LLC (Ann Arbor, MI, USA) Cat 3 KC44-H1, Progesterone EIA kit from Alpco (Salem, NH, USA) catalogue # PROHU-E01, Estradiol Alpco Elisa kit catalogue number 11ESPHU-E 1. All sample runs passed quality control by generating low coefficient of variance percentages (CV%) among replicate control samples. Data values represent the average of two technical replicates per individual.
2.7. Statistical analysis
All statistical tests were performed in R (http://www.r-project.org/). Using an Anderson-Darling test from the nortest package, all distributions of data that rejected the null hypothesis of normality were subsequently evaluated with non-parametric tests. All statistical tests performed were two tailed and a p < 0.05 is considered significant. Unless otherwise specified ± denotes the standard error of the mean. Correlation analyses were performed with Pearson’s correlation. Interaction analyses or those requiring covariate adjustment were performed with linear regression. For linear regression modeling, non-standardized coefficients are presented. Bonferroni adjustment was used to correct for multiple tests in the discovery analysis used to identify key CpGs to investigate further. Student’s t tests and Wilcoxon Rank Sum tests were used to test group mean differences in post hoc analyses in samples with Gaussian and non-Gaussian distributions, respectively. Estradiol levels were log transformed to generate a Gaussian distribution for downstream parametric tests. Hormone levels were adjusted by taking the residuals of a linear model of hormone level as a function of trimester to adjust for pregnancy related increases in hormone levels that will not necessarily be observed for DNA methylation.
3. Results
3.1. Associations between OXTR DNA methylation and PDD
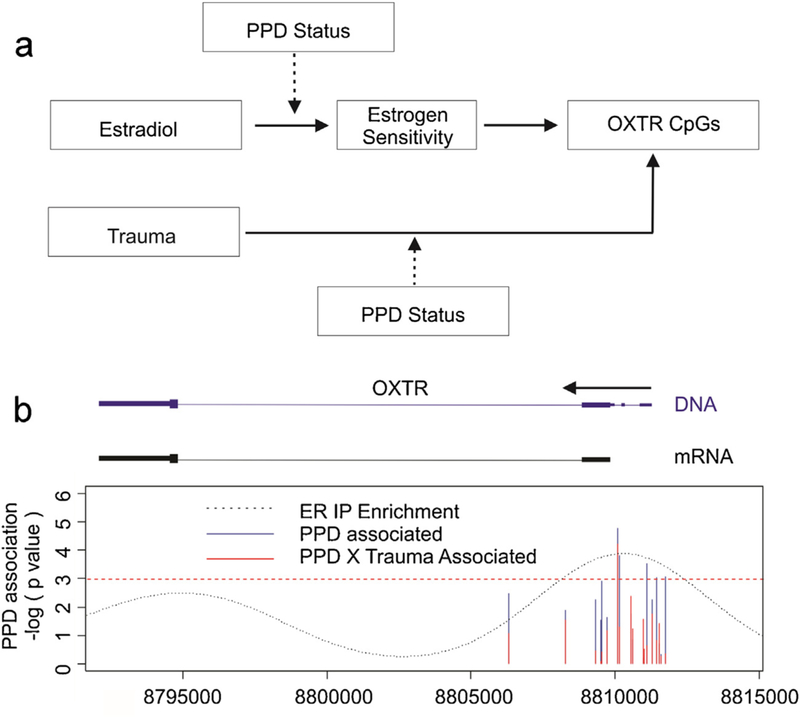
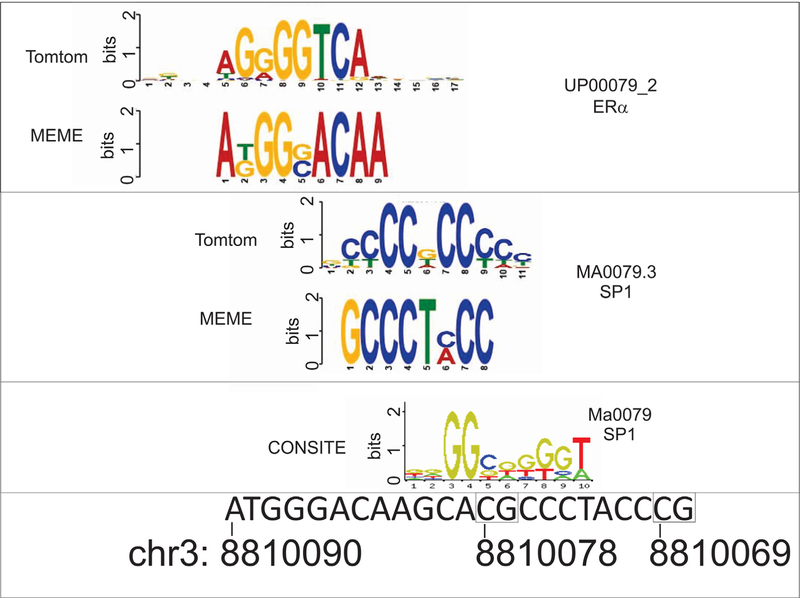
We sought to address the hypothesis that OXTR DNA methylation is associated with PPD at functionally relevant loci. As a first step, we limited our investigation to only those CpGs within the OXTR gene located on the Illumina HM450 array with potential functional relevance to brain gene expression levels using the BrainCloud tool. Of the 18 HM450 microarray probes spanning the OXTR gene, only cg25140571 was present in the BrainCloud application and exhibited a significant positive correlation with OXTR gene expression levels derived from prefrontal cortical tissue (Fig. S1). Of the 18 loci, 7 exhibited significant correlation with cg25140571 after Bonferroni correction for multiple testing, suggesting they had potential for functional relevance in the brain (Table 2). These 7 CpGs were subsequently tested for association to PPD and while 3 of them, including the brain gene expression relevant cg25140571 exhibited nominal significance, only cg12695586 exhibited a Bonferroni corrected p value below 10% (Table 2, Fig. 1). The association remained significant after adjusting for the estimated cell proportions (Linear regression: 0.0062, p = 0.026). Notably, the next most significant CpG, cg19619174, was directly adjacent to cg12695586. Intriguingly, the genomic location of cg12695586 was enriched for estrogen receptor a binding based on ENCODE data (Fig. 1) suggesting the presence of an estrogen response element (ERE) at this location. Further analysis of the sequence in the region corroborated this interpretation identifying that cg12695586 is proximal to an estrogen receptor alpha (ER) motif and within a specificity protein 1 (SP1) transcription factor binding site (Result S1, Fig. 2). Notably, these ERE proximal CpGs are directly upstream of an mRNA splice variant E05109 as denoted by the UCSC genome browser.
Table 2.
OXTR CpG association with cg25140571 and PPD.
| HM450 ID | CHR | Coordinate | cg2514057 Association |
PPD Association |
Bonferroni P | ||||
|---|---|---|---|---|---|---|---|---|---|
| Ra | P | Bonferroni P | βb | Error | P | ||||
| cg12695586 | 3 | 8810077 | 0.45 | 0.00083 | 0.015 | −1.6 | 0.63 | 0.014 | 0.08 |
| cg19619174 | 3 | 8810139 | 0.43 | 0.0015 | 0.027 | −1.09 | 0.51 | 0.039 | 0.23 |
| cg25140571 | 3 | 8811437 | 1 | NA | NA | −2.55 | 1.34 | 0.062 | 0.37 |
| cg02192228 | 3 | 8809536 | 0.39 | 0.0044 | 0.079 | −2.63 | 1.49 | 0.084 | 0.50 |
| cg15317815 | 3 | 8809306 | 0.45 | 0.00098 | 0.018 | −3.1 | 2.09 | 0.14 | 0.84 |
| cg04523291 | 3 | 8809501 | 0.41 | 0.0028 | 0.05 | −2.25 | 2.01 | 0.27 | 1.00 |
| cg23391006 | 3 | 8811279 | 0.44 | 0.0014 | 0.025 | −0.46 | 0.34 | 0.17 | 1.00 |
| cg08535600 | 3 | 8810980 | 0.47 | 0.00046 | 0.0083 | 0.03 | 0.57 | 0.96 | 5.76 |
| cg11589699 | 3 | 8806317 | 0.4 | 0.0041 | 0.074 | ||||
| cg00385883 | 3 | 8808259 | 0.32 | 0.02 | 0.36 | ||||
| cg27501759 | 3 | 8809715 | 0.25 | 0.072 | 1 | ||||
| cg03987506 | 3 | 8810549 | 0.29 | 0.037 | 0.67 | ||||
| cg00078085 | 3 | 8810592 | 0.34 | 0.016 | 0.29 | ||||
| cg17285225 | 3 | 8811004 | 0.04 | 0.75 | 1 | ||||
| cg09353063 | 3 | 8811092 | 0.36 | 0.0097 | 0.17 | ||||
| cg00247334 | 3 | 8811543 | 0.33 | 0.019 | 0.34 | ||||
| cg17036624 | 3 | 8811601 | 0.44 | 0.0013 | 0.023 | ||||
| cg14483142 | 3 | 8811758 | 0.39 | 0.005 | 0.09 | ||||
Pearson’s Correlation: N=51 subjects.
Linear Regression: N=51 subjects, non-standardized coefficients.
Fig. 1.
Location of model associated OXTR DNA methylation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
(a) A schematic representation of the model being tested in the manuscript. Estrogen sensitivity was not directly tested in our models, but represents a hypothesized explanatory variable mediating the effects of estradiol on OXTR DNA methylation. (b) A plot of the nominal negative natural log of the p values for loci associated with postpartum depression (PPD) (y axis) as a function of genomic coordinates on chr 3 in the region of the OXTR gene (hg 19). Only those 7CpGs exhibiting significant Bonferroni corrected associations to the cg25140571 probe found in BrainCloud are depicted. The horizontal red dashed line denotes a p value of 5%. ENCODE data downloaded from Gene Expression Omnibus accession GSE32465 was used to evaluate empirically determined binding sites for ER binding in ECC1 and T-47D cells. Data was downloaded from Gene Expression Omnibus accession GSE32465 and the frequency of sequences aligning to genomic coordinates following chromatin immunoprecipitation with antibodies for estrogen receptor alpha (ER) are depicted by the dotted black line. A schematic of the genomic DNA OXTR gene as depicted on the UCSC genome browser (https://genome.ucsc.edu/) (blue) and mRNA for splice variant E05109 (black) is depicted to scale with the horizontal black arrow denoting the direction of transcription.
Fig. 2.
In silico transcription factor binding predictions in OXTR.
Output from the MEME, Tomtom, and CONSITE software identifying an estrogen receptor alpha (ER) and specificity protein 1 (SP1) transcription factor binding sites in the region of cg1269556 at chr3: 8810078 (hg19). The region of chr3 is depicted to scale below the identified motifs. Boxes surround CpGs at positions chr3: 8810078 and 8810069 interrogated by pyrosequencing.
Previous groups have reported an interaction of OXTR genotype at rs53576 with OXTR DNA methylation that was specific to women who were antenatally euthymic and became depressed in the postpartum period, as opposed to those antenatally depressed women who remained depressed (Bell et al., 2015). No significant genotype effects were observed in our cohort (Result S2). However, independent of rs53576 genotype, antenatally depressed PPD cases had lower levels of OXTR DNA methylation than antenatally euthymic PPD cases (Student’s t test: N = 12, Antenatally Depressed PPD = 9.8 ± 0.11%, N = 10, Antenatally Euthymic PPD = 11 ± 0.17%, p = 0.038). We subsequently attempted to validate this association using sodium bisulfite pyrosequencing at CpGs proximal to the implicated site of ER binding including chr3: 8810078 (hg19) corresponding to the CpG represented in probe cg12695586 (chr3:8810078) and an adjacent CpG at chr3:8810069. Pyrosequencing based DNA methylation was significantly correlated with the microarray probe cg12695586 derived values at both chr3:8810078 (Pearson’s Correlation: R=0.53, p=9.1×10−5) (Fig. S2) and chr3:8810069 (Pearson’s Correlation: R = 0.39, p = 0.0092). For subsequent analyses as justified in Result S3, we averaged methylation across these two CpG positions to reduce technical variation and refer to these CpGs as the ERE proximal CpGs. Similar to the microarray findings, DNA methylation derived by pyrosequencing at ERE proximal CpGs demonstrated a significantly lower level in DNA methylation in PPD cases arising from women who were antenatally depressed and stayed depressed relative to those that developed depression postpartum (Student’s t test: N=12, Antenatally Depressed PPD=5.59±0.06%, N=9, Antenatally Euthymic PPD=6.46±0.1%, p=0.035).
3.2. Association of trauma and PPD status on OXTR DNA methylation at ERE proximal CpGs
We next addressed the hypothesis that trauma history is associated with OXTR ERE proximal CpG methylation and that this effect may be moderated by PPD status. Childhood abuse status and PPD significantly interacted to modulate DNA methylation levels (Linear Regression: Abuse β = 0.78±0.34, p=0.027; PPD β = 0.32±0.33, p=0.33; interaction β = −1.2±0.53, p=0.03, F=2.46, df = 3/42, Model p=0.1) and were similar to those observed using microarray derived values (Result S4). Post hoc analysis demonstrated that childhood abuse significantly increases DNA methylation in women that did not develop PPD (Student’s t test: N=11, Abuse = 6.62±0.09%, N=16, No Abuse = 5.84±0.05%, p=0.045) but does not exhibit an effect in women who developed PPD (Student’s t test: N=7, Abuse = 5.73±0.08%, N=12, No Abuse = 6.16±0.08%, p = 0.25), suggesting that trauma history has independent effects on OXTR ERE proximal DNA methylation in non-PPD women.
In light of our above observations in the microarray data, we assessed this relationship in the context of cases of PPD that derived from antenatally depressed vs. antenatally euthymic women. Controlling for antenatal depression status as an additive covariate results in a general improvement of the model fit as well as significance of the interactive factors (Linear Regression: Abuse β = 0.80±0.32, p=0.016; PPD β = 0.31±0.32, p=0.11; Antenatal Depression Status β = −0.72±0.25, p=0.008, Abuse X PPD interaction β = −1.2±0.53, p=0.024, F=3.86, df = 4/41, Model p=0.009).
3.3. Independent replication of PPD predictive model in women with no previous psychiatric history
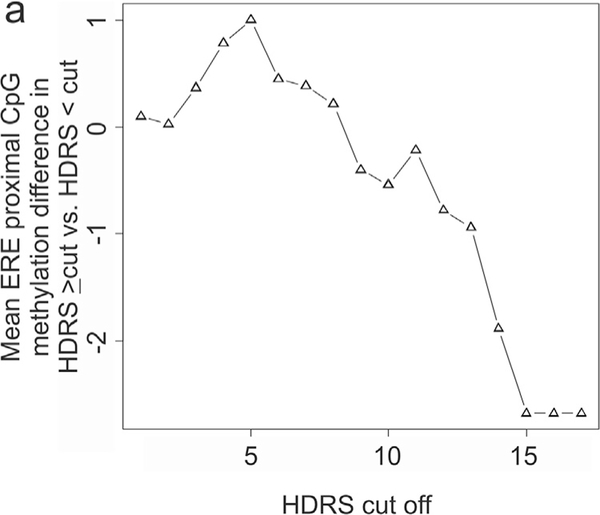
We assessed for DNA methylation changes at OXTR ERE proximal CpGs in an independent cohort of women without a prior history of psychiatric diagnosis. OXTR DNA methylation levels were significantly lower in blood from women with 6–8 month HDRS scores of 14 or greater (Wilcoxon Rank Sum test: N=5, HDRS≥14=3.96±0.27%, N=235, HDRS<14=5.84±0.01%, p=0.034). In general, we observed a linear relationship between mean OXTR DNA methylation at CpGs chr3:8810078 and 8810069 and HDRS cut off (Pearson’s Correlation: R = −0.87, p = 5.18×10−6) (Fig. 3). It is possible that some women reporting high HDRS scores in the third trimester may remain high into the postpartum period and meet DSM-V criteria for PPD. As such, we assessed all women reporting an HDRS of greater or equal to 14 at any of the assessed time points and observed a significantly lower DNA methylation in this group of women (Wilcoxon Rank Sum test: N=16, HDRS≥14=4.91±0.16%, N=224, HDRS<14=5.87±0.01%, p=0.035).
Fig. 3.
OXTR DNA methylation decrease with HDRS severity.
(a) A plot of the mean DNA methylation levels at OXTR CpGs ch3:8810078 and 8810069 (y axis) in women from the Franconian Maternal Health Evaluation Studies (FRAMES) cohort with Hamilton Depression Rating Scale (HDRS) values greater and less than the cut off value (x axis).
3.4. OXTR gene expression is lower in PPD
Using a Prospective Gene Expression cohort generated by Mehta et al. (2014), we addressed the hypothesis that OXTR gene expression levels will be associated with PPD. A significantly lower OXTR gene expression was observed in 3rd trimester in blood of PPD cases (Wilcoxon Rank Sum, N = 33, PPD = 6 ± 0.012, N = 28, non-PPD = 6.2 ± 0.015, p = 0.031) but not 1st trimester blood (Wilcoxon Rank Sum, N = 29, PPD = 6 ± 0.013, N = 22, non-PPD = 6.1 ± 0.022, p = 0.39). Like the DNA methylation observations, this 3rd trimester relationship held true for antenatally depressed women who remained depressed in the postpartum period Wilcoxon Rank Sum, (N = 18, PPD = 5.9 ± 0.021, N = 28, non-PPD = 6.2 ± 0.015, p = 0.036) but not in antenatally euthymic women who became depressed postpartum (Wilcoxon Rank Sum, N = 15, PPD = 6 ± 0.028, N = 28, non-PPD = 6.2 ± 0.015, p = 0.16). As above, no significant differences were observed for 1st trimester derived OXTR expression levels in separate analyses for antenatally depressed (N = 14, PPD = 6 ± 0.03, N = 22, non-PPD = 6.1 ± 0.022, p = 0.49) or antenatally euthymic subgroups (N = 15, PPD = 6 ± 0.023, N = 22, non-PPD = 6.1 ± 0.022, p = 0.47).
3.5. Evaluation of serum hormone levels
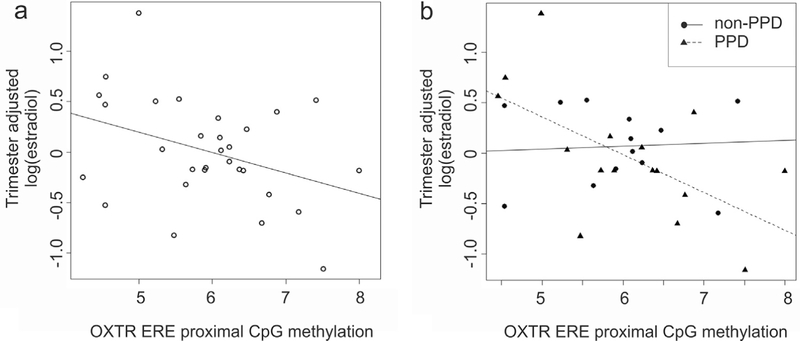
Mean DNA methylation levels at OXTR ERE proximal CpGs were evaluated against serum hormone concentrations in the Johns Hopkins Prospective cohort. A significant association was observed for levels of estradiol on OXTR DNA methylation (Pearson’s Correlation: R = −0.37, p = 0.043) (Fig. 4). Removal of one outlier located outside the interquartile range for estradiol levels results in a non-significant correlation of R = −0.32, p = 0.083. We excluded the outlier and then randomly selected data points to exclude from the remaining set (permuting 10,000 times) in order to assess if the lack of significance is a result of a loss of statistical power or if the removed data point is driving the observed relationship. The result was not significant (P = 0.06), suggesting that the removal of the specific outlier is not the only probe that will contribute to nonsignificant findings and that removal of others can result in the same effect. A closer investigation revealed that PPD status influenced the direction of the observed association. After adjusting for trimester as a covariate, a significant interaction between PPD status and OXTR DNA methylation on estradiol levels was observed (Linear Regression: OXTR DNA methylation β = 0.027 ± 0.14, p = 0.84; PPD β = 2.39 ± 1.13, p = 0.044; interaction β =−0.41±0.18, p=0.038, F=8.32, df=4/25, Model p=2.0×10−4). Post hoc analysis demonstrated a significant negative correlation of OXTR ERE proximal CpG methylation with estradiol in women who developed PPD (Pearson’s Correlation: R = −0.59, p = 0.015) but not in those who did not develop PPD (Pearson’s Correlation: R = 0.07, p = 0.81). This relationship remained significant in N = 8 antenatally depressed cases that developed PPD (Pearson’s Correlation: R = −0.80, p = 0.017) but was not observed in N = 9 antenatally euthymic cases that developed PPD (Pearson’s Correlation: R = −0.10, p = 0.81). The significant association in PPD cases was not influenced by removal of outlier estradiol values.
Fig. 4.
OXTR DNA methylation association with serum hormone levels.
(a) A scatter plot of the trimester adjusted natural log of estradiol (y axis) as a function of the mean OXTR DNA methylation at ERE proximal CpGs located at chr3:8810078 and 8810069 (x axis) (Pearson’s Correlation: R=−0.37, p=0.043). Removal of one outlier located outside the interquartile range for estradiol levels results in a non-significant correlation of R=−0.32, p=0.083. We excluded the outlier and then randomly selected data points to exclude from the remaining set (permuting 10,000 times) in order to assess if the lack of significance is a result of a loss of statistical power or if the removed data point is driving the observed relationship. The result was not significant (P=0.06), suggesting that the removal of the specific outlier is not the only probe that will contribute to nonsignificant findings and that removal of others can result in the same effect. (b) A scatterplot of the trimester adjusted natural log of estradiol levels (y axis) as a function of OXTR DNA methylation levels at ERE proximal CpGs for women who did (Pearson’s Correlation: R=−0.59, p=0.015) and did not develop PPD (Pearson’s Correlation: R=0.07, p=0.81). The significant association in PPD cases was not influenced by removal of outlier estradiol values.
After adjusting for trimester, estradiol levels interacted with OXTR DNA methylation to strongly and significantly associate with the ratio of allopregnanolone to progesterone (Linear Regression: OXTR DNA methylation β = 0.23 ± 0.068, p = 0.0018; log(estradiol)β = 0.18 ± 0.059, p = 0.0042; interaction β = 0.030± 0.0097,p=0.003, F=13.3, df =4/25, Model p=6.1×10−6) (Fig. S3). The interaction suggests that the ratio of allopregnanolone to progesterone will be low when OXTR ERE proximal CpG methylation is high and estradiol is high or conversely when both estrogen and OXTR DNA methylation are low. Conversely, when OXTR ERE proximal CpG methylation and estradiol levels are low or high relative to each other, the ratio of allopregnanolone to progesterone will be high. Cumulatively, these data suggest that OXTR ERE proximal CpG methylation in conjunction with estradiol exposure may be important for determining levels of neuroactive steroid production.
4. Discussion
Using a candidate approach, we investigated DNA methylation variation in HM450 microarray probes spanning the OXTR gene for association to PPD in a prospective PPD cohort. By first focusing on only those CpGs capable of marking functionally relevant changes in brain gene expression, we were able to target our search to a select group of CpGs located at a region demonstrating the maximal enrichment for ER binding in response to estradiol treatment. While the location of these CpGs may function as a promoter specific region for the mRNA splice variant E05109, they may also drive transcription of the full transcript as they are correlated with cortical expression associated CpGs.
Intriguingly, the only CpG demonstrating a Bonferroni corrected p value below 8% for a negative association of DNA methylation with PPD was identified at the peak for ER binding. An analysis of the sequence demonstrated that the CpG at microarray probe cg12695586 appeared to be positioned in the middle of an SP1 transcription factor binding site. Interestingly, estrogen receptors (ER) have been shown to interact with SP1 transcription factors and result in gene silencing through the recruitment of co-repressor complexes (Bartella et al., 2012; Vivar et al., 2010). Mutation analysis in the ovine Oxtr demonstrate that estradiol driven Oxtr gene expression is mediated by CG rich SP1 transcription factor binding sites (Fleming et al., 2006). Our previous work identified an enrichment for SP1 transcription factor binding motifs among murine hippocampal loci that exhibited estradiol induced DNA methylation changes (Guintivano et al., 2014). More recently, next generation sequencing analysis has demonstrated that DNA methylation at SP1 transcription factor binding sites in the murine Oxtr gene are positively correlated with gene expression variation in various brain regions (Harony-Nicolas et al., 2014). Notably, the genomic region under study was in the promoter; however, the directionality of SP1 DNA methylation and Oxtr gene expression is consistent with the positive correlation implicated by the BrainCloud tool in our study. Cumulatively, the data suggest the implicated CpG (cg12695586) at chr3 position 8810078 and proximal CpGs in the region may have functional relevance to human OXTR gene expression. While use of the BrainCloud tool limited the assessment of the gene expression relevance of OXTR DNA methylation to the cortex and not the hippocampus, cross tissue hormonal effects may facilitate similar neuronal gene expression changes in other structures or peripheral tissues and should be studied in future work. Indeed, when assessing OXTR gene expression in independent prospective study data generated in blood by Mehta et al. (2014), we observed a significantly lower level in OXTR gene expression with PPD in third trimester blood. It is important to keep in mind that the reported associations occur in peripheral tissues and so an extrapolation to brain relevant changes in structures such as the cortex, hippocampus, amygdala, or other relevant brain structures that may be responsible for mood symptoms in PPD must be made with caution. One hypothesis for the detectability of PPD associated epigenetic changes in blood is that disease specific hormonal reprogramming of the epigenome occurs in the brain and affects mood, and that systematic circulation of the same hormones results in concomitant marking of the epigenome in peripheral cells.
We identified a negative association of pregnancy estradiol levels with DNA methylation in the studied region suggesting the observed PPD association may be the downstream consequence of estrogen mediated epigenetic reprogramming. Interestingly, the effect was confined to antenatal blood from the women who would later develop PPD. These observations are consistent with previous work out of our laboratory, which suggested that PPD may arise due to an increased sensitivity to estrogens on the epigenetic level among women at risk for PPD (Guintivano et al., 2014). Importantly, this effect was strongest in the subpopulation of antenatally depressed women who would remain depressed in the postpartum period, and is consistent with our observations of a significantly lower level in OXTR ERE proximal CpG methylation as well as OXTR gene expression in antenatally depressed as opposed to euthymic PPD cases. A speculative hypothesis is that the ER and SP1 dependent recruitment of epigenetic modifying cofactors may be different in the PPD group and that lower levels of OXTR DNA methylation may be a reflection of a depressed state as opposed to a trait vulnerability to PPD. One such ER binding HP1BP3cofactor involved in chromatin reorganization is the heterochromatin protein 3 binding protein 3 (HP1BP3) gene, which was previously identified by our group to be associated with PPD (Guintivano et al., 2014). While the observed association at OXTR was not affected by differential proportions of cells, our previous work demonstrated that consistent associations across antenatally depressed and euthymic associations with PPD at HP1BP3 were only possible after controlling for cell type levels specific to antenatal depression status (Guintivano et al., 2014; Osborne et al., 2015). Furthermore, recently published observations out of our laboratory demonstrate that epigenetic variation of PPD biomarkers, HP1BP3 and TTC9B, during the first trimester associate with later pregnancy time point hormonal variations (Osborne et al., 2015) and may be linked with the ability of other TTC9 genes to modulate estrogen signaling, while HP1BP3 epigenetic variation was associated with changes in allopregnanolone. As such, early epigenetic variation predisposing to hormonal variation may result in downstream consequences of epigenetic reprogramming such as an estradiol associated decrease in OXTR ERE proximal CpG methylation later in pregnancy and is consistent with the third trimester but not first trimester lower levels of OXTR gene expression observed in antenatally depressed PPD cases in the Prospective Gene Expression cohort. One speculative hypothesis is that depression occurring antenatally and continuing into the postpartum period may be the result of more extreme downstream consequences to depression predisposing factors. These hypotheses warrant further study both to corroborate the differential directional effects of PPD based epigenetic reprogramming at various loci, to determine if these effects are more extreme in cases of depression that manifest antenatally as opposed to in the postpartum period, and to identify the differential involvement of epigenetically modifying cofactors as a function of PPD risk status.
Importantly, many of the previously reported temporal associations of PPD biomarker loci HP1BP3 and TTC9B appeared to be strongest in those antenatally euthymic PPD cases as opposed to antenatally depressed PPD cases (Osborne et al., 2015), which brings up the possibility that OXTR relevant epigenetic variation may be specific only to antenatally depressed PPD cases. This observation is consistent with the observed interaction of estradiol levels and OXTR ERE proximal DNA methylation on the ratio of allopregnanolone to progesterone, as lower levels of allopregnanolone have been associated with depression during pregnancy (Hellgren et al., 2014; Schiller et al., 2014). This assertion is contradicted, however, by Bell et al., who recently identified a PPD specific DNA methylation increase in GG homozygotes of the rs53576 SNP in women who were not antenatally depressed (Bell et al., 2015). As noted in the supplementary material, we did not directly replicate this association; however, did find a moderate and intriguingly similar OXTR DNA methylation PPD specific increase in only antenatally euthymic PPD cases in GA heterozygotes. Notably, limitations of our analysis include a very small sample size as well as the fact that the assayed CpGs were over 1kb away from those assessed by Bell, suggesting that epigenetic associations within the OXTR gene may be dependent on not only the antenatal depression status but also the region of the CpGs analyzed. This reasoning is consistent with recent observations presented by Reinier et al., who identified significantly lower OXTR DNA methylation in depressed women that were specific to a region closer to the first exon, while rs53576 associations appeared to be located closer to exon 2 (Reiner et al., 2015).
The observed reduced OXTR DNA methylation levels were also significantly associated with higher depression rating scores from the HDRS in the FRAMES cohort. Unlike the Johns Hopkins Prospective cohort and the gene expression cohort generated by Mehta et al. (2014), both of which represent high risk cohorts of women with mood disorder, the FRAMES cohort represents women without a previous psychiatric diagnosis. If lower OXTR DNA methylation is the result of an increased sensitivity to estrogen mediated epigenetic reprogramming, the data suggest that such a sensitivity may be a common feature of PPD not confined to women with a pre-existing mood disorder diagnosis. Interestingly, the reduced OXTR DNA methylation observed in the FRAMES cohort are derived from blood taken 6–8 months after parturition, suggesting that such effects are detectable under conditions of normal circulating levels of estradiol. Notably, we would expect some degree of variation due to menstrual cycle fluctuations in the population, which may have decreased statistical power in this cohort. Given that depressive symptoms were assessed at 6–8 months, it remains possible that the observed associations are with cases of major depressive disorder that arose after parturition and may represent a distinct biology to cases of PPD occurring closer to parturition. This interpretation is consistent with the previous reasoning that lower OXTR DNA methylation may mark a depressed state more so than future risk to PPD. Given the possibility that the observed lower DNA methylation is due to estrogen sensitivity, the degree to which this would be generalizable to cases of depression in females arising not during pregnancy or in males requires further study. Unfortunately, in the FRAMES cohort, no metric of early life stress was available.
In the Johns Hopkins Prospective sample, we identified a significant interaction of reported childhood physical or sexual abuse and PPD status on DNA methylation of the OXTR gene. A recently published cross sectional study by Unternaehrer et al., identified an increase in DNA methylation in the OXTR in both men and women with lower scores on the parental Bonding Instrument scale, a measure of maternal care (Unternaehrer et al., 2015). Interestingly, the identified CpGs were located within a region downstream of the CpGs assayed in this study by a mere 80–100 base pairs. Our data indicated a significant increase of OXTR DNA methylation with abuse status in this region specifically in women who did not develop PPD. It is possible this result is due to the above reported PPD specific decreases in OXTR DNA methylation at this locus, which would be expected to compete with any abuse relevant signal and cancel out any observed associations. One caveat to our results is that child sexual abuse was based on retrospective recall in response to a single question and may be subject to recall bias. Furthermore, sexual abuse may represent a very different type of psychosocial stress compared to differences in parental bonding as studied by Unternaehrer et al. Despite this, the region assayed by Unternaehrer et al., and our study resides proximal to an ENCODE implicated ERE. Furthermore, early life stress either derived from childhood sexual abuse (Perroud et al., 2011) or emotional neglect (Provencal and Binder, 2015) is associated with a dysregulated HPA axis and exposure to glucocorticoids (Lightman and George, 2014). Notably, glucocorticoid exposure in vitro can result in increased accessibility of EREs and facilitate estrogen receptor binding (Miranda et al., 2013). In this way, it is possible that early life stress induced glucocorticoid activation may facilitate a differential binding of ER machinery (Lightman and George, 2014) and this in turn may result in differential DNA methylation proximal to the ERE. Importantly, this effect may be expected to occur earlier in life, suggesting further study of trauma induced OXTR methylation and its independence from those etiological factors contributing to PPD associated OXTR methylation is warranted.
Our work provides evidence for the interaction of ERE proximal OXTR DNA methylation with estradiol levels to influence the conversion of progesterone to allopregnanolone. This observation is consistent with work in neonatal rats demonstrating that estradiol exposure at birth results in a decrease in allopregnanolone levels at day 21 and 60 in the hypothalamus, suggesting that estradiol has the capacity to decrease allopregnanolone levels (Berretti et al., 2014). Research out of the nociception field has identified an OXTR mediated increase in the conversion of progesterone to allopregnanolone in response to inflammation induced pain, resulting in prolonged GABAaR induced mIPSPs in downstream lamina II spinal cord neurons (Juif et al., 2013). The authors suggest this effect may be mediated by the recruitment of ERK1/2 based on return to control allopregnanolone levels upon administration of MEK pathway inhibitor PD09859. While ER signaling in the brain can activate the ERK/MEK pathway and lead to downstream epigenetic consequences (Gagnidze et al., 2013), OXTR activation can have a modulatory influence on ERK signaling (Devost et al., 2008). For example, in the myometrium,β (2)AR-mediated ERK1/2 activation requires coactivation of OXTR (Wrzal et al., 2012b) and is blocked by OXTR antagonists (Wrzal et al., 2012a). While OXTR and ER activated intracellular signaling interactions have not been reported in the brain, such a mechanism would be consistent with the observed interactions on anxiolytic neurosteroid allopregnanolone levels and may represent a mechanism by which OXTR affects mood. Prior work has linked mood states with alterations in the ratio of allopregnanolone to its precursors (Girdler et al., 2012; Schiller et al., 2014). Allopregnanolone levels can help to reduce HPA axis activation in response to stress, while depression and anxiety can lead to decreased levels of this neurosteroid (Bali and Jaggi, 2014). In the perinatal period specifically, Deligiannidis et al., in a small sample, found no relationship between pregnancy allopregnanolone and the development of PPD (Deligiannidis et al., 2013), while Hellgren et al., found significantly lower levels of allopregnanolone in depressed pregnant women when compared to healthy controls (Hellgren et al., 2014). It is important to consider that allopregnanolone levels inhibit OXT system activity during pregnancy so as to limit incidence of preterm birth (Brunton et al., 2014). This occurs through allopregnanolone based modulation of GABA signaling on magnocellular OXT neurons (MCNs) in both the paraventricular and super optic nuclei, which in turn project to the pituitary and result in systemic OXT release into the blood (Brunton et al., 2014). Importantly, super optic nucleus derived MCNs also project to OXTR expressing neurons in the central amygdala and represent another pathway by which OXT and the OXT modulating effects of allopregnanolone may regulate mood. (Knobloch et al., 2012). In light of the effects of allopregnanolone on MCN activity, it is clear that any effects of OXTR on allopregnanolone levels would likely feedback and affect the activity of the OXT system activation in the brain, suggesting our data may implicate an epigenetic contribution to such a self regulatory feedback inhibition mechanism. Of note, preterm labor has been associated with elevated PPD rates (Sundaram et al., 2014), suggesting that the investigation of a connection between these systems may be warranted. Importantly, the small sample size from which our observations are derived suggests the interaction of estradiol and OXTR ERE proximal CpG methylation on allopregnanolone levels may be the result of statistical chance. Additionally, the causal direction of the observed associations may be different from our interpretation and should be studied further in systems that allow for a more in depth investigation of the biological interactions of OXTR epigenetic variation and these hormonal systems.
This study has many limitations including variability in the definition of PPD and mood scales used across cohorts as well as a lack of prospective measures of DNA methylation within individuals. Additionally, many of the reported findings derive from small sample sizes and should be interpreted with caution. Despite this, the consistency of the epigenetic associations in numerous diverse cohorts add confidence that a true depression relevant signal exists in the OXTR gene and that further work in larger prospective cohorts is warranted to both replicate and better understand the timing and functional relevance of epigenetic variation in this gene.
Supplementary Material
Acknowledgements
We would like to thank Dr. Summer Rosenstock Ph.D. of the Johns Hopkins Bloomberg School of Public Health, Department of International Health, for guidance on visual presentation of statistical models in this work. We would like to thank The Solomon R. & Rebecca D. Baker Foundation for their generous support of this research. This work was funded in part by a NARSAD 2010 Young Investigator Award to Dr. Kaminsky and by National Institute of Mental Health (NIMH) Grant K23 MH074799-01A2 to Dr. Payne. Prospective human subjects research was conducted under IRB protocol # 00008149 and # 00049309.
Funding
The funding sources had no involvement in the study design, analysis, or decision to publish.
Drs. Kaminsky and Payne are co-inventors listed on a patent for DNA methylation at biomarker loci related to PPD. Under a former option agreement between Physician’s Choice Laboratory Services and the Johns Hopkins University, the University was entitled to fees associated with a PPD biomarker invention mentioned in this article. Dr. Kaminsky was also an unpaid speaker to Physician’s Choice Laboratory Services. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies. Dr. Kaminsky is listed on patents for use of epigenetic information at the SKA2 locus to predict suicidal behavior and PTSD and received consultant fees from Janssen Research and Development, LLC. Dr. Payne received legal consulting fees from Pfizer, Astra Zeneca and Johnson and Johnson and research support from Corcept Therapeutics. Dr. Binder is listed on patent applications related to FKBP5 and ABCB1 as predictors of antidepressant treatment response. Dr. Peter A. Fasching received consulting fees from Roche, Novartis, Teva and Genomic Health, received speaker’s honoraria from Amgen, Roche, Pfizer, Novartis, Genomic Health, Teva, and GSK, and received research grants from Amgen and Novartis.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.psyneuen.2016.04.008.
Conflict of interest
All other authors declare no biomedical financial interests or potential conflicts of interest. All other authors declare no biomedical financial interests or potential conflicts of interest.
References
- Ammerman RT, Putnam FW, Chard KM, Stevens J, Van Ginkel JB, 2012. PTSD in depressed mothers in home visitation. Psychol. Trauma: Theory Res. Pract. Policy 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreen L, Nyberg S, Turkmen S, van Wingen G, Fernandez G, Backstrom T, 2009. Sex steroid induced negative mood may be explained by the paradoxical effect mediated by GABAA modulators. Psychoneuroendocrinology 34, 1121–1132. [DOI] [PubMed] [Google Scholar]
- Aswathi A, Rajendiren S, Nimesh A, Philip RR, Kattimani S, Jayalakshmi D, Ananthanarayanan PH, Dhiman P, 2015. High serum testosterone levels during postpartum period are associated with postpartum depression. Asian J. Psychiatry 17, 85–88. [DOI] [PubMed] [Google Scholar]
- Backstrom T, Haage D, Lofgren M, Johansson IM, Stromberg J, Nyberg S, Andreen L, Ossewaarde L, van Wingen GA, Turkmen S, Bengtsson SK, 2011. Paradoxical effects of GABA-A modulators may explain sex steroid induced negative mood symptoms in some persons. Neuroscience 191, 46–54. [DOI] [PubMed] [Google Scholar]
- Bakermans-Kranenburg MJ, van Ijzendoorn MH, Riem MM, Tops M, Alink LR, 2012. Oxytocin decreases handgrip force in reaction to infant crying in females without harsh parenting experiences. Soc. Cogn. Affective Neurosci 7, 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali A, Jaggi AS, 2014. Multifunctional aspects of allopregnanolone in stress and related disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 48, 64–78. [DOI] [PubMed] [Google Scholar]
- Bartella V, Rizza P, Barone I, Zito D, Giordano F, Giordano C, Catalano S, Mauro L, Sisci D, Panno ML, Fuqua SA, Ando S, 2012. Estrogen receptor beta binds Sp1 and recruits a corepressor complex to the estrogen receptor alpha gene promoter. Breast Cancer Res. Treat. 134, 569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AF, Carter CS, Steer CD, Golding J, Davis JM, Steffen AD, Rubin LH, Lillard TS, Gregory SP, Harris JC, Connelly JJ, 2015. Interaction between oxytocin receptor DNA methylation and genotype is associated with risk of postpartum depression in women without depression in pregnancy. Front. Genet. 6, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretti R, Santoru F, Locci A, Sogliano C, Calza A, Choleris E, Porcu P, Concas A, 2014. Neonatal exposure to estradiol decreases hypothalamic allopregnanolone concentrations and alters agonistic and sexual but not affective behavior in adult female rats. Horm. Behav. 65, 142–153. [DOI] [PubMed] [Google Scholar]
- Bloch M, Schmidt PJ, Danaceau M, Murphy J, Nieman L, Rubinow DR, 2000. Effects of gonadal steroids in women with a history of postpartum depression. Am. J. Psychiatry 157, 924–930. [DOI] [PubMed] [Google Scholar]
- Breese McCoy SJ, 2011. Postpartum depression: an essential overview for the practitioner. South. Med. J. 104, 128–132. [DOI] [PubMed] [Google Scholar]
- Brunton PJ, Russell JA, Hirst JJ, 2014. Allopregnanolone in the brain: protecting pregnancy and birth outcomes. Prog. Neurobiol. 113, 106–136. [DOI] [PubMed] [Google Scholar]
- Bunevicius R, Kusminskas L, Mickuviene N, Bunevicius A, Pedersen CA, Pop VJ, 2009. Depressive disorder and thyroid axis functioning during pregnancy. World J. Biol. Psychiatry 10, 324–329. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE, 2011. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuijpers P, Brannmark JG, van Straten A, 2008. Psychological treatment of postpartum depression: a meta-analysis. J. Clin. Psychol 64, 103–118. [DOI] [PubMed] [Google Scholar]
- Deligiannidis KM, Sikoglu EM, Shaffer SA, Frederick B, Svenson AE, Kopoyan A, Kosma CA, Rothschild AJ, Moore CM, 2013. GABAergic neuroactive steroids and resting-state functional connectivity in postpartum depression: a preliminary study. J. Psychiatr. Res 47, 816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devost D, Wrzal P, Zingg HH, 2008. Oxytocin receptor signalling. Prog. Brain Res. 170, 167–176. [DOI] [PubMed] [Google Scholar]
- Field T, 2011. Prenatal depression effects on early development: a review. Infant Behav. Dev 34, 1–14. [DOI] [PubMed] [Google Scholar]
- Fleming JG, Spencer TE, Safe SH, Bazer FW, 2006. Estrogen regulates transcription of the ovine oxytocin receptor gene through GC-rich SP1 promoter elements. Endocrinology 147, 899–911. [DOI] [PubMed] [Google Scholar]
- Franczak A, Staszkiewicz J, Koziorowski M, Kotwica G, 2002. The influence of estradiol and progesterone on the concentrations of uterine oxytocin receptors and plasma PGFM in response to oxytocin in ovariectomized gilts. Reprod. Nutr. Dev 42, 327–338. [DOI] [PubMed] [Google Scholar]
- Gagnidze K, Weil ZM, Faustino LC, Schaafsma SM, Pfaff DW, 2013. Early histone modifications in the ventromedial hypothalamus and preoptic area following oestradiol administration. J. Neuroendocrinol. 25, 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdler SS, Lindgren M, Porcu P, Rubinow DR, Johnson JL, Morrow AL, 2012. A history of depression in women is associated with an altered GABAergic neuroactive steroid profile. Psychoneuroendocrinology 37, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guintivano J, Arad M, Gould TD, Payne JL, Kaminsky ZA, 2014. Antenatal prediction of postpartum depression with blood DNA methylation biomarkers. Mol. Psychiatry 19, 560–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harony-Nicolas H, Mamrut S, Brodsky L, Shahar-Gold H, Barki-Harrington L, Wagner S, 2014. Brain region-specific methylation in the promoter of the murine oxytocin receptor gene is involved in its expression regulation. Psychoneuroendocrinology 39, 121–131. [DOI] [PubMed] [Google Scholar]
- Hellgren C, Akerud H, Skalkidou A, Backstrom T, Sundstrom-Poromaa I, 2014. Low serum allopregnanolone is associated with symptoms of depression in late pregnancy. Neuropsychobiology 69, 147–153. [DOI] [PubMed] [Google Scholar]
- Hirst KP, Moutier CY, 2010. Postpartum major depression. Am. Fam. Physician 82, 926–933. [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M, 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 (Suppl. 1), S96–104. [DOI] [PubMed] [Google Scholar]
- Huffmeijer R, Tops M, Alink LR, Bakermans-Kranenburg MJ, van Ijzendoorn MH, 2011. Love withdrawal is related to heightened processing of faces with emotional expressions and incongruent emotional feedback: evidence from ERPs. Biol. Psychol. 86, 307–313. [DOI] [PubMed] [Google Scholar]
- Jeong HG, Lim JS, Lee MS, Kim SH, Jung IK, Joe SH, 2013. The association of psychosocial factors and obstetric history with depression in pregnant women: focus on the role of emotional support. Gen. Hosp. Psychiatry 35, 354–358. [DOI] [PubMed] [Google Scholar]
- Josefsson A, Berg G, Nordin C, Sydsjo G, 2001. Prevalence of depressive symptoms in late pregnancy and postpartum. Acta Obstet. Gynecol. Scand. 80, 251–255. [DOI] [PubMed] [Google Scholar]
- Juif PE, Breton JD, Rajalu M, Charlet A, Goumon Y, Poisbeau P, 2013. Long-lasting spinal oxytocin analgesia is ensured by the stimulation of allopregnanolone synthesis which potentiates GABA(A) receptor-mediated synaptic inhibition. J. Neurosci. 33, 16617–16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch HS, Charlet A, Hoffmann LC, Eliava M, Khrulev S, Cetin AH, Osten P, Schwarz MK, Seeburg PH, Stoop R, Grinevich V, 2012. Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73, 553–566. [DOI] [PubMed] [Google Scholar]
- Lightman SL, George CL, 2014. Steroid hormones in 2013: Glucocorticoids: timing, binding and environment. Nat. Rev. Endocrinol 10, 71–72. [DOI] [PubMed] [Google Scholar]
- Magiakou MA, Mastorakos G, Rabin D, Dubbert B, Gold PW, Chrousos GP, 1996. Hypothalamic corticotropin-releasing hormone suppression during the postpartum period: implications for the increase in psychiatric manifestations at this time. J. Clin. Endocrinol. Metab 81, 1912–1917. [DOI] [PubMed] [Google Scholar]
- Mamrut S, Harony H, Sood R, Shahar-Gold H, Gainer H, Shi YJ, Barki-Harrington L, Wagner S, 2013. DNA methylation of specific CpG sites in the promoter region regulates the transcription of the mouse oxytocin receptor. PLoS One 8, e56869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Quast C, Fasching PA, Seifert A, Voigt F, Beckmann MW, Faschingbauer F, Burger P, Ekici AB, Kornhuber J, Binder EB, Goecke TW, 2012. The 5-HTTLPR polymorphism modulates the influence on environmental stressors on peripartum depression symptoms. J. Affect. Disord 136, 1192–1197. [DOI] [PubMed] [Google Scholar]
- Mehta D, Newport DJ, Frishman G, Kraus L, Rex-Haffner M, Ritchie JC, Lori A, Knight BT, Stagnaro E, Ruepp A, Stowe ZN, Binder EB, 2014. Early predictive biomarkers for postpartum depression point to a role for estrogen receptor signaling. Psychol. Med, 1–14. [DOI] [PubMed] [Google Scholar]
- Meltzer-Brody S, Bledsoe-Mansori SE, Johnson N, Killian C, Hamer RM, Jackson C, Wessel J, Thorp J, 2013. A prospective study of perinatal depression and trauma history in pregnant minority adolescents. Am. J. Obstet. Gynecol 208 (21), e211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LJ, 2002. Postpartum depression. JAMA 287, 762–765. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Voss TC, Sung MH, Baek S, John S, Hawkins M, Grontved L, Schiltz RL, Hager GL, 2013. Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 73, 5130–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Williams L, Gatt JM, McAuley-Clark EZ, Dobson-Stone C, Schofield PR, Nemeroff CB, 2014. Variation in the oxytocin receptor gene is associated with increased risk for anxiety, stress and depression in individuals with a history of exposure to early life stress. J. Psychiatr. Res 59, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara MW, 2009. Postpartum depression: what we know. J. Clin. Psychol. 65, 1258–1269. [DOI] [PubMed] [Google Scholar]
- Osborne L, Clive M, Kimmel M, Gispen F, Guintivano J, Brown T, Cox O, Judy J, Meilman S, Brier A, Beckmann MW, Kornhuber J, Fasching PA, Goes F, Payne JL, Binder EB, Kaminsky Z, 2015. Replication of epigenetic postpartum depression biomarkers and variation with hormone levels. Neuropsychopharmacology 41 (6), 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlstein T, Howard M, Salisbury A, Zlotnick C, 2009. Postpartum depression. Am. J. Obstet. Gynecol 200, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CA, Johnson JL, Silva S, Bunevicius R, Meltzer-Brody S, Hamer RM, Leserman J, 2007. Antenatal thyroid correlates of postpartum depression. Psychoneuroendocrinology 32, 235–245. [DOI] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R, Guillaume S, Mouthon D, Stouder C, Dieben K, Huguelet P, Courtet P, Malafosse A, 2011. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl. Psychiatry 1, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Binder EB, 2015. The effects of early life stress on the epigenome: from the womb to adulthood and even before. Exp. Neurol. 268, 10–20. [DOI] [PubMed] [Google Scholar]
- Quinones-Jenab V, Jenab S, Ogawa S, Adan RA, Burbach JP, Pfaff DW, 1997. Effects of estrogen on oxytocin receptor messenger ribonucleic acid expression in the uterus, pituitary, and forebrain of the female rat. Neuroendocrinology 65, 9–17. [DOI] [PubMed] [Google Scholar]
- Reiner I, Van IMH, Bakermans-Kranenburg MJ, Bleich S, Beutel M, Frieling H, 2015. Methylation of the oxytocin receptor gene in clinically depressed patients compared to controls: the role of OXTR rs53576 genotype. J. Psychiatr. Res 65, 9–15. [DOI] [PubMed] [Google Scholar]
- Robertson E, Grace S, Wallington T, Stewart DE, 2004. Antenatal risk factors for postpartum depression: a synthesis of recent literature. Gen. Hosp.Psychiatry 26, 289–295. [DOI] [PubMed] [Google Scholar]
- Schiller CE, Schmidt PJ, Rubinow DR, 2014. Allopregnanolone as a mediator of affective switching in reproductive mood disorders. Psychopharmacology (Berl.) 231, 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrundz M, Bolten M, Nast I, Hellhammer DH, Meinlschmidt G, 2011. Plasma oxytocin concentration during pregnancy is associated with development of postpartum depression. Neuropsychopharmacology 36, 1886–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AS, Tabbaa M, Lei K, Eastham P, Butler MJ, Linton L, Altshuler R, Liu Y, Wang Z, 2015. Local oxytocin tempers anxiety by activating GABA receptors in the hypothalamic paraventricular nucleus. Psychoneuroendocrinology 63, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufia M, Aoun J, Gorsane MA, Krebs MO, 2010. SSRIs and pregnancy: a review of the literature. Encephale 36, 513–516. [DOI] [PubMed] [Google Scholar]
- Studd JW, 2011. A guide to the treatment of depression in women by estrogens. Climacteric. [DOI] [PubMed] [Google Scholar]
- Stuebe AM, Grewen K, Pedersen CA, Propper C, Meltzer-Brody S, 2012. Failed lactation and perinatal depression: common problems with shared neuroendocrine mechanisms? J. Womens Health (Larchmt) 21, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram S, Harman JS, Cook RL, 2014. Maternal morbidities and postpartum depression: an analysis using the 2007 and 2008 Pregnancy Risk Assessment Monitoring System. Women Health Issues 24, e381–388. [DOI] [PubMed] [Google Scholar]
- Thompson RJ, Parker KJ, Hallmayer JF, Waugh CE, Gotlib IH, 2011. Oxytocin receptor gene polymorphism (rs2254298) interacts with familial risk for psychopathology to predict symptoms of depression and anxiety in adolescent girls. Psychoneuroendocrinology 36, 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer E, Meyer AH, Burkhardt SC, Dempster E, Staehli S, Theill N, Lieb R, Meinlschmidt G, 2015. Childhood maternal care is associated with DNA methylation of the genes for brain-derived neurotrophic factor (BDNF) and oxytocin receptor (OXTR) in peripheral blood cells in adult men and women. Stress, 1–11. [DOI] [PubMed] [Google Scholar]
- Viguera AC, Tondo L, Koukopoulos AE, Reginaldi D, Lepri B, Baldessarini RJ, 2011. Episodes of mood disorders in 2,252 pregnancies and postpartum periods. Am. J. Psychiatry 168, 1179–1185. [DOI] [PubMed] [Google Scholar]
- Vivar OI, Zhao X, Saunier EF, Griffin C, Mayba OS, Tagliaferri M, Cohen I, Speed TP, Leitman DC, 2010. Estrogen receptor beta binds to and regulates three distinct classes of target genes. J. Biol. Chem 285, 22059–22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle RJ, Gamble LE, Kershaw YM, Wood SA, Lightman SL, Ingram CD, 2006. Gonadal steroid modulation of stress-induced hypothalamo-pituitary-adrenal activity and anxiety behavior: role of central oxytocin. Endocrinology 147, 2423–2431. [DOI] [PubMed] [Google Scholar]
- Wrzal PK, Devost D, Petrin D, Goupil E, Iorio-Morin C, Laporte SA, Zingg HH, Hebert TE, 2012a. Allosteric interactions between the oxytocin receptor and the beta2-adrenergic receptor in the modulation of ERK1/2 activation are mediated by heterodimerization. Cell. Signal. 24, 342–350. [DOI] [PubMed] [Google Scholar]
- Wrzal PK, Goupil E, Laporte SA, Hebert TE, Zingg HH, 2012b. Functional interactions between the oxytocin receptor and the beta2-adrenergic receptor: implications for ERK1/2 activation in human myometrial cells. Cell. Signal. 24, 333–341. [DOI] [PubMed] [Google Scholar]
- Yim IS, Glynn LM, Schetter CD, Hobel CJ, Chicz-Demet A, Sandman CA, 2010. Prenatal beta-endorphin as an early predictor of postpartum depressive symptoms in euthymic women. J. Affect. Disord 125, 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim IS, Tanner Stapleton LR, Guardino CM, Hahn-Holbrook J, Dunkel Schetter C, 2015. Biological and psychosocial predictors of postpartum depression: systematic review and call for integration. Annu. Rev. Clin. Psychol 11, 99–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.