Abstract
Introduction
The pharmacoepigenetics of antipsychotic treatment in severe mental illness is a growing area of research that aims to understand the interface between antipsychotic treatment and genetic regulation. Pharmacoepigenetics may one day assist in identifying treatment response mechanisms or become one of the components in the implementation of precision medicine.
Objectives
To understand the current evidence regarding the effects of antipsychotics on DNA methylation in schizophrenia and bipolar disorder.
Methods
A systematic review with qualitative synthesis was performed through PubMed, Embase, and Psychinfo from earliest date to June 2019. Studies were included if they analyzed DNA methylation in an antipsychotic-treated population of patients with schizophrenia or bipolar disorder. Data extraction occurred via a standardized format and study quality was assessed.
Results
Twenty-nine studies were identified for inclusion. Study design, antipsychotic type, sample source, and methods of DNA methylation measurement varied across all studies. Eighteen studies analyzed methylation in patients with schizophrenia, four studies in patients with bipolar disorder, and seven studies in a combined sample of schizophrenia and bipolar disorder. Twenty-two studies used observational samples whereas the remainder used prospectively-treated samples. Six studies assessed global methylation, five assessed epigenome-wide, and 15 performed a candidate epigenetic study. Two studies analyzed both global and gene-specific methylation, whereas one study performed a simultaneous epigenome-wide and gene-specific study. Only three genes were analyzed in more than one gene-specific study and the findings were discordant.
Conclusions
The state of the pharmacoepigenetic literature on antipsychotic use is still in its early stages and uniform reporting of methylation site information is needed. Future work should concentrate on using prospective sampling with appropriate control groups and begin to replicate many of the novel associations that have been reported.
Keywords: Review, antipsychotic, epigenetic, methylation
Background
Antipsychotic medications are one of the primary pharmacologic modalities for the treatment of severe mental illness, including schizophrenia and bipolar disorder. Despite clear efficacy, antipsychotics do carry a risk of treatment failure, non-adherence, and side effects [1–4]. Following many years of intense research and discovery, pharmacogenetics, the study of the effect of structural gene variation on medication outcomes, is poised to become part of the treatment toolbox for clinicians treating patients with antipsychotic medications [5,6]. In contrast, pharmacoepigenetics, the study of how alterations in gene expression beyond the underlying genetic code influence medication outcomes or how medications influence epigenomics, is still in the beginning stages. Epigenetics encompasses several mechanisms that influence gene expression including DNA methylation, histone modifications, and RNAbased mechanisms such as micro RNA[7]. Although all forms of epigenetic mechanisms have been the subject of study in relation to antipsychotic treatment, most studies have analyzed DNA methylation in one form or another.
The status of genomic methylation (theoretically from 0 to 100%) is measured from broad levels down to individual sites within the genome. Global methylation reflects the methylation status of total genomic content within a given sample. Several methods exist to estimate global methylation status and include the use of restriction enzymes, enzyme-linked immunosorbent assays (ELISA), and surrogate measures that utilize transposable elements that are distributed throughout and make up a considerable portion of the genome (e.g., long interspersed nuclear element; (LINE-1), etc.) [8]. Global estimates of DNA methylation provide an idea of methylation changes at the genome level. However, it does not provide information on changes that may be concentrated in genomic areas of importance that are likely to have effects on gene expression [9].
The next level of measurement, region-specific methylation, can characterize critical regulatory regions, such as CpG islands and promoter areas. There are several techniques that have been developed for the assessment of regional methylation with many being polymerase chain-reaction (PCR) and/or restriction enzyme based [10]. Regional estimates are highly useful for targeting genes of interest, however, as they provide the average methylation for a given region, they may not capture important differences found at the level of the CpG dinucleotide itself. To accomplish measurements at the base pair level, bisulfite sequencing techniques are used. Pyrosequencing is considered the gold standard for smaller genomic regions. Various other strategies, including next generation bisulfite sequencing, are used for greater coverage of the genome [11]. Finally, along with next generation sequencing, array-based analyses are commonly utilized to assess site-specific methylation at hundreds of thousands of sites across the genome [8]. This approach is generally used for discovery or hypothesis-generating studies. The main arrays used in human DNA methylation studies are the Illumina Infinium series (San Diego, CA), which first released an array that assessed 27,000 CpG methylation sites across the genome (HumanMethylation27 BeadChip). Since this original array, updated technologies have been released that have been able to assess approximately 450,000 sites (HumanMethylation450 BeadChip) and now approximately 850,000 sites with the most current version (HumanMethylationEPIC) [12].
There has been a significant amount of interest in the use of epigenetics to explain the interface between genetics and antipsychotic treatment response. With such interest, and the large range of technologies and approaches that can be used in assessing methylation, there are now many studies that have investigated DNA methylation in patients with schizophrenia or bipolar disorder who are on antipsychotic treatment. The purpose of this systematic review was to gather the current evidence aimed at understanding the DNA methylation profiles of patients on antipsychotic medications. The systematic review and summarization of this evidence is intended for researchers and clinicians to understand the current level of evidence that exists and also provide a resource on which future studies can be based.
Methods
This systematic review followed the suggested, structured format for establishing the question, collecting the evidence via a standardized process, evaluating the quality of included studies, creating a summary of evidence, and interpreting the evidence [13–15].
Systematic Review Question
The question that this systematic review aimed to answer was “What is the DNA methylation profile for patients with schizophrenia or bipolar disorder treated with antipsychotics?” The question was posed to capture any description of DNA methylation within the two primary populations treated with antipsychotic medications.
Study Search and Collection
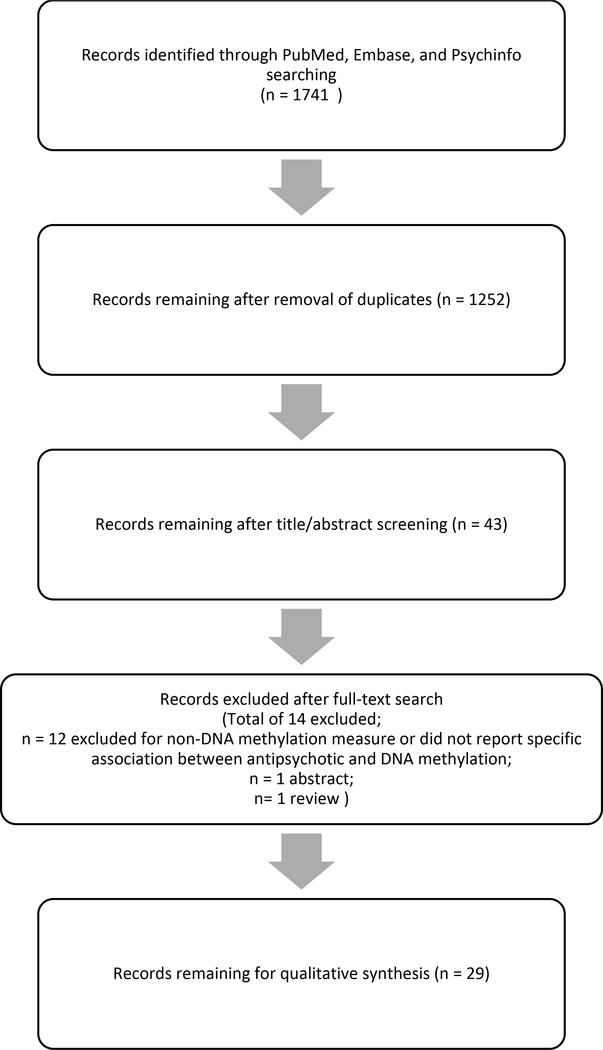
Potential studies for inclusion were searched in PubMed, Embase, and Psychinfo from earliest date to June 2019. Studies were included if they (i) evaluated any measure of DNA methylation in a sample of patients with schizophrenia or bipolar disorder treated with antipsychotics and (ii) presented original, non-duplicated data. Exclusion criteria included (i) non-original research publications such as review papers, abstracts, or commentaries, (ii) non-English studies, or (iii) non-human studies. Studies whose primary objective was to compare DNA methylation based on psychiatric diagnosis were allowed as long as studies explicitly stated patients were on antipsychotics, and the study included a statistical reporting of the association between DNA methylation and antipsychotic treatment. Studies of DNA methylation and psychiatric diagnosis were excluded if (i) antipsychotic medication treatment was not mentioned or (ii) associations between antipsychotic treatment and DNA methylation were not reported in the results section. Studies that used antipsychotic treatment as a covariate in analysis or for the purposes of a sensitivity analysis were excluded as these studies did not report on statistical associations between the measured methylation and antipsychotic use. Search terms entered into each database included a combination of: “antipsychotic”, “neuroleptic”, “[individual FDA or antipsychotic names of FDA or European approved drugs]”, “epigenetic”, “epigenomic”, “methylation”, “schizophrenia,” and “bipolar disorder”. Two authors (KB and AK) independently assessed search results to identify studies that met criteria for inclusion and extracted data. A third author (BS) was utilized for any discordant determinations during search evaluations. Figure 1 shows the PRISMA diagram for the systematic review that yielded 29 studies for inclusion.
Figure 1. PRISMA Flow Diagram of Systematic Review.
The flow diagram shows the results for the initial search and exclusions resulting in 29 studies included in the systematic review.
Data Extraction and Qualitative Review
The final included set of studies were reviewed and data were extracted for the following variables via a standardized collection instrument: study title, authors, study population, study design, subject/patient numbers, antipsychotic(s), comparator group (if applicable), length of treatment (if prospective), tissue assessed, DNA methylation measured, and major findings. Study quality was assessed using the NIH Quality Assessments Tools (https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools). Findings were collected and described in the results section.
RESULTS
Our systematic review yielded 29 studies (Table 1) [16–44]. Eighteen studies analyzed methylation in patients with schizophrenia, four studies in patients with bipolar disorder, and seven studies in a combined sample of patients with schizophrenia and bipolar disorder. The majority (n=22) of studies included observational sampling, such as cross-sectional or case-control studies, and the remaining analyzed changes in a prospective manner. Antipsychotic treatment varied widely, and peripheral blood levels were analyzed in 23 of 29 studies. The sources of DNA consisted of peripheral blood (79%), brain (10%), combined brain and saliva (7%), and skeletal muscle (4%). Of note, only three studies utilizing peripheral blood controlled for the cellular composition in their analyses (all by statistical methods in epigenome-wide studies). Finally, the method of DNA methylation assessment ranged from global to genome-wide and targeted gene analysis. All studies analyzed 5-methylcytosine methylation with the exception of one study who simultaneously analyzed 5-hydroxymethylcytosine gene methylation (an “oxidized derivative” form of 5-methylcytosine) [44]. Unless otherwise stated, all summarized results refer to 5-methylcytosine DNA methylation. Study quality information can be found in the supplementary information.
Table 1.
Studies included in Systematic Review.
| Study | Study Type | Patient population | Antipsychotic type | Sample Type | DNA methylation approach |
|---|---|---|---|---|---|
| Abdolmaleky et. al [16]a | obs | Set 1 (saliva): 30 SCZ;30 HC;15 PR Set 2 (brain): 35 SCZ/BD;35 HC |
NR | saliva and brain | gene-specific |
| Abdolmaleky et al. [17] | obs | 35 SCZ; 35 BD; 35 HC | NR | brain | gene-specific |
| Abdolmaleky et al. [18] | obs | 40 SCZ; 35 BD; 40 HC | NR | brain | gene-specific |
| Abdolmaleky et al. [19] | obs | Set 1 (brain): 35 SCZ/BD;35 HC Set 2 (saliva): 30 SCZ;20 BD;30 HC;20 UPR |
NR | saliva and brain | gene-specific |
| Bönsch et al. [20] | obs | 20 twin pairs discordant for SCZ; 8 pairs concordant for SCZ; 21 HC twin pairs; 21 psychiatric control twin pairs | AAP and TAP | blood | global and gene-specific |
| Bromberg et al. [21] | obs | 28 SCZ; 26 HC | NR | blood | global |
| Burghardt et al. [22]b | obs | 120 BD | AAP | bloodd | EWAS |
| Burghardt et al. [23]c | obs | Discovery Set: 96 SCZ; Validated Set: 166 SCZ | AAP | bloodd | EWAS |
| Burghardt et al.[24] | obs | 30 BD | AAP | skeletal muscle | gene-specific |
| Burghardt et al. [25] | obs | 115 BD | AAP | blood | global |
| Burghardt et al. [26] | obs | 133 SCZ | AAP and TAP | blood | global |
| Cheng et al. [27] | obs and prosp | 60 SCZ; 30 HC | risperidone | blood | gene-specific |
| D’Addario et al.[28] | obs | 24 MDD;24 BD I; 24 BD II; 25 SCZ; 34 HC | AAP and TAP | blood | gene-specific |
| Ellingrod et al. [29] | prosp | 35 SCZ | AAP and TAP | blood | global |
| Houtepen et al. [30] | obs | 172 BD | olanzapine and quetiapine | bloodd | EWAS and gene-specific |
| Kinoshita et al. [31] | prosp | 21 SCZ | olanzapine | blood | EWAS |
| Li et al. [32] | obs | 92 SCZ; 99 BD; 92 HC | AAP and TAP | blood | global |
| Lott et al. [33] | obs | 85 SCZ | NR | blood | gene-specific |
| Melas et al. [34] | obs | 177 SCZ; 171 HC | AAP and TAP | blood | global and gene-specific |
| Mill et al. [35] | obs | 35 SZ; 35 BD; 35 HC | NR | brain | EWAS |
| Miura et al. [36] | prosp | 34 SCZ | risperidone | blood | gene-specific |
| Moons et al. [37] | obs | 438 SCZ | AAP and TAP | blood | gene-specific |
| Nour El Huda et al. [38] | obs | 138 SCZ; 132 HC | AAP and TAP | blood | gene-specific |
| Rukova et al. [39] | prosp | 20 SCZ; 220 HC | NR | blood | EWAS |
| Shi et al. [40] | prosp | 288 SCZ | risperidone | blood | gene-specific |
| Swathy et al. [41] | obs | 184 SCZ; 330 HC | NR | blood | global |
| Tang et al. [42] | prosp | 82 SCZ | AAP and TAP | blood | gene-specific |
| Venugopal et al. [43] | prosp | 47 SCZ; 47 HC | AAP | blood | gene-specific |
| Zong et al. [44] | obs | 279 SCZ; 265 HC | AAP and TAP | blood | gene-specific |
Details of 29 studies included in the systematic review including study design, type of antipsychotic within study, sample type for DNA methylation analysis, and overall DNA methylation strategy. Abbreviations: AAP = atypical antipsychotic; BD = bipolar disorder; EWAS = Epigenome-Wide Association Study; HC = healthy control; MDD = major depressive disorder; NR = not reported; obs = observational; PR = primary relative; pros =prospective; scz = schizophrenia; TAP = typical antipsychotic; UPR = unaffected primary relative
Abdolmakey et al. studies (references 16–19) did not state if patient overlap was present for included samples
Controlled for cell type composition
Global Methylation Studies
Eight studies included a measurement of global methylation (Table 2). The most commonly employed measurement utilized restriction enzymes (five studies) followed by transposable elements (two studies) and ELISA (one study). Six studies included patients with schizophrenia, one study included patients with bipolar disorder, and one study include both patients with bipolar disorder and schizophrenia. All studies reported on associations of typical and/or atypical antipsychotics (AAPs) as a class with DNA methylation, therefore, no study reported associations with individual antipsychotic medications. Five studies included samples on both atypical and typical antipsychotics, one study included only patients on AAPs, and two studies did not specify antipsychotic type. The source for DNA methylation in each study was peripheral blood. Lower global methylation in patients treated with AAPs was observed in three studies [25,26,34] although the comparator groups in each study differed. Increased global methylation was observed in three studies and one study identified increases in responders only [20,32,41]. One study did not identify an effect of antipsychotic dose on global methylation levels [21].
Table 2.
Studies of Global DNA Methylation.
| Study | Global Methylation Method | Objective | Main Finding (s) |
|---|---|---|---|
| Bönsch et al. [20] | Restriction Enzyme-Based | Methylation differences based on diagnosis/groups included and antipsychotic use | Methylation levels were higher in treated patients (similar to HC levels) compared to non-treated patients. |
| Bromberg et al. [21] | Restriction Enzyme-Based | Methylation differences based on homocysteine levels and diagnosis and antipsychotic use | Overall, no associations found between antipsychotic dose or homocysteine and methylation. Interaction noted for increased methylation in female non-smokers compared to female HC non-smokers. |
| Burghardt et al.a [25] | Restriction Enzyme-Based | Methylation levels based on antipsychotic use and antipsychotic-induced insulin resistance | Lower methylation in patients on AAPs versus mood stabilizers. Increased insulin resistance was associated with decreased methylation. |
| Burghardt et al. [26] | LINE-1 transposable elements | Methylation differences related to demographics, clinical variables, antipsychotic use, and MTHFR/COMT genotype | Decreased methylation in MTHFR TT genotype in female patients treated with an AAP. |
| Ellingrod et al. [29] | Restriction Enzyme-Based | Methylation levels following folate treatment in patients with SCZ with metabolic syndrome | Increased methylation after folate treatment. Methylation increases were greatest in patients on olanzapine and clozapine compared to all other types. |
| Li et al. [32] | LINE-1 transposable elements | Methylation levels at three sites based on clinical variables and antipsychotic treatment | Positive correlation of methylation with clozapine treatment and age. Negative correlation with diagnosis and sex. |
| Melas et al. [34] | Restriction Enzyme-Based | Methylation levels based on clinical factors and antipsychotic use | Higher methylation in patients treated with haloperidol compared to all other antipsychotics (2/3rd AAP). Increased methylation in HC versus SCZ (similar level to haloperidol) and in late onset versus early onset SCZ. |
| Swathy et al. [41] | ELISA | Methylation changes post-antipsychotic treatment | Increased methylation in patients with SCZ compared to HCs. Increased methylation in responders compared to HCs and non-responders. |
Details of eight studies that analyzed global methylation in antipsychotic-treated (including typical antipsychotic, atypical antipsychotic, or mixed) patients with schizophrenia or bipolar disorder. Abbreviations: AAP = atypical antipsychotic; COMT = catechol-o-methyl transferase; ELISA = Enzyme-linked immunosorbent assay; HC = healthy control; LINE-1 = Long Interspersed Nuclear Elements; MTHFR = methylene tetrahydrofolate reductase; SCZ = schizophrenia
Genome-wide Methylation Studies
Six studies utilized epigenome-wide technologies to report on DNA methylation profiles in antipsychotic-treated populations within peripheral blood (five studies) or brain (one study) (Table 3). Three studies included in patients with schizophrenia, two included patients with bipolar disorder, and one included a combined schizophrenia/bipolar sample. Patients within these studies were on AAPs (two studies) or did not state the type of antipsychotic (two studies). One study only included patients on olanzapine or quetiapine and the remaining study only included patients on olanzapine. Two studies analyzed epigenome-wide changes prospectively, one compared to mood stabilizers and two evaluated antipsychotic side effects. All studies utilized array approaches to interrogate genome-wide methylation with four studies utilizing the Illumina HumanMethylation technology. Within these genome-wide studies, methylation changes within a wide number of genes and pathways was observed. For example, pathway changes were identified in lipid metabolism, the Wnt/β-catenin, neurogenesis, immune function, cell substrate/matrix adhesion, and mitochondrial.
Table 3.
Studies of Epigenome-wide Association.
| Study | EWAS Method | Objective | Main Finding (s) |
|---|---|---|---|
| Burghardt et al.a [22] | Illumina 450K | Identify methylation associated with antipsychotic-induced insulin resistance | Two sites associated with AAP-induced insulin resistance after correction for multiple testing. The top site was in an intergenic region and the second site was in the FAR2 gene. The FAR2 site was replicated in an additional sample. |
| Burghardt et al. [23] | Illumina 450K | Determine the methylation profile related to metabolic syndrome in SCZ patients on antipsychotics | Sites within five genes found to be associated with metabolic syndrome and the top pathway from a bioinformatic analysis was the Wnt/β-catenin pathway. Additionally, sex-specific sites associated with metabolic syndrome were also identified. Validation occurred in additional sample set for two genes (CDH22 and MAP3K13 genes). |
| Houtepen et al. [30] | Illumina 27K (n=122) and 450K (n=50) | Identify changes based on AAP and mood stabilizer use | No individual methylation sites were significant after multiple testing correction. Methylation changes in pathways related to neurogenesis, embryonic function, regulatory, and immune function were associated with quetiapine treatment. |
| Kinoshita et al. [31] | Illumina 450K | Identify changes after 1 year of clozapine treatment | 29,134 CpG sites showed significant changes in DNA methylation. A higher number of sites had decreased DNA methylation compared to increased methylation. These sites were associated with the cell substrate adhesion and cell matrix adhesion pathways. A site within the CREBBP gene was the sole site associated with psychotic symptom changes after correction for multiple testing. |
| Mill et al. [35] | In-house CpG island microarray | Determine DNA methylation profiles in major psychosis and with lifetime antipsychotic treatment | Male patients with SCZ had lower methylation in the MAP2K1 gene which correlated with lifetime antipsychotic use. A similar trend was observed in female patients with SCZ but this did not remain significant with a correction for multiple testing. Overall, psychiatric patients had differential methylation in “Mitochondrial” pathways. |
| Rukova et al. [39] | Agilent Human DNA Methylation Microarray | Identify the DNA methylation profiles after prospective treatment with antipsychotics | Differentially methylated regions differed from controls before and after treatment and were also dependent on sex and remission status. |
Details of six studies that analyzed epigenome-wide methylation arrays in antipsychotic-treated patients with schizophrenia or bipolar disorder. AAP = atypical antipsychotic; CDH22 = Cadherin 22; CREBBP = CREB Binding Protein; EWAS = Epigenome-Wide Association Study; FAR2 = Fatty acyl CoA reductase; MAP3K13 = Mitogen-Activated Protein Kinase Kinase Kinase 13; MAP2K1 = Mitogen-Activated Protein Kinase Kinase 1; SCZ = schizophrenia
Gene-Specific Studies
Targeted gene methylation analyses were performed in 18 studies with patients with schizophrenia or bipolar disorder who were treated with antipsychotics (Table 4). Eleven studies included samples from patients with schizophrenia, five studies included both patients with schizophrenia and bipolar disorder, and two studies only included patients with bipolar disorder. Seven studies included samples treated with either AAP or typical antipsychotics, six studies included only AAP-treated samples, and five studies did not specify antipsychotic type. Most studies included any type of antipsychotic in the specified class, whereas three studies only included patients on risperidone and one study included patients on olanzapine or quetiapine. The methodology of gene-specific analyses varied across studies, with pyrosequencing and methylation-specific PCR as the most common methods (five studies each). Similarly, the genes of interest varied across studies. Thirty-three different genes were analyzed and only the COMT, HTR2a, and 5-HTT genes were analyzed in more than one study. Studies were mixed in identifying increased (eight studies), decreased (five studies), or no change (five studies) in gene-specific methylation within antipsychotic-treated patients.
Table 4.
Studies of Gene-Specific Methylation.
| Study | Gene-specific Methoda | Gene(s) analyzed | Objective | Main Finding (s) |
|---|---|---|---|---|
| Abdolmaleky et. al [16]b | Illumina 27K, qMSP | DTNBP1 | Identify changes with diagnosis and antipsychotic treatment | In saliva and brain, antipsychotic-naïve patients had higher methylation compared to patients on antipsychotic and controls. There was a trend of decreased methylation with increased lifetime use of antipsychotics. |
| Abdolmaleky et al. [17] | qMSP and bisulfite sequencing | HTR2A | Role of HTR2A methylation in SCZ/BD pathogenesis and treatment | CpG-specific methylation higher in SCZ/BD compared to HCs. Antipsychotic use was associated with higher methylation compared to drug-free patients however there was no difference between AAP and typical antipsychotic use. |
| Abdolmaleky et al. [18] | qMSP and bisulfite sequencing | COMT | Evaluate gene-environment interactions and their effect on correlations with diagnosis and treatment | Lower methylation in SCZ and BD compared to HCs. There was no observed effect of antipsychotic or mood stabilizer use on these observed differences. |
| Abdolmaleky et al. [19] | Illumina 27K/450K, qMSP | 5-HTT | Identify changes with diagnosis and treatment | In saliva and brain, increased methylation in drug-naïve compared to HCs and patients on antipsychotics. Longer use of antipsychotics associated with decreased methylation. |
| Bönsch et al. [20] | qMSP | Reelin, SOX10 | Methylation differences based on diagnosis/groups included and antipsychotic use | Lower Reelin methylation in SCZ twins on antipsychotics compared to drug-free SCZ twins. |
| Burghardt et al.[24] | MS-HRM PCR | AKT1, AKT2, AKT3 | Identify effect of antipsychotic treatment and insulin resistance | Increased DNA methylation observed for AKT1, AKT2 but not AKT3 in AAP versus patients on mood stabilizer. Trends observed between AKT2methylation and insulin resistance. |
| Cheng et al. [27] | PSQ | DRD4 | Understand the role of methylation in SCZ susceptibility and treatment | Increased methylation in SCZ versus HC. No significant difference between AAPs used (quetiapine, risperidone, clozapine) but quetiapine had increased methylation in both males and females compared to HCs (other AAPs did not). |
| D’Addario et al.[28] | PSQ | CNR1 | Identify changes with diagnosis and antipsychotic type | Increased methylation in patients with SCZ but no effect of antipsychotic type was found. |
| Houtepen et al. [30] | Illumina 27K/450K | RELN, SLC1A2, MTNR1A, IGF2, H19, BDNF, SLC6A4, GAD1 | Examine DNA methylation signatures of psychiatric medication in BD subjects | No associations detected for any included medication after correction for multiple testing. |
| Lott et al. [33] | PSQ | COMT | Assess relationship between COMT methylation and metabolic syndrome in patients on AAPs | Patients with the COMT val/val genotype and metabolic syndrome had higher methylation of COMT. This relationship was further influenced by the amount of physical activity (increased activity associated with decreased methylation). |
| Melas et al. [34] | PSQ | COMT and 5-HTT | Methylation levels compared to HCs | Higher COMT methylation in SCZ (all antipsychotic treated) versus HC. No differences in 5-HTT detected. |
| Miura et al. [36] | Bisulfite sequencing | ANKK1 | Analyze the effect of risperidone treatment on methylation | Of the 35 CpG sites analyzed, one site showed increased methylation in responders versus non-responders but did not remain significant after correction for multiple testing. |
| Moons et al. [37] | EpiTYPER | IGF2 | Determine associations between methylation and antipsychotic-induced metabolic side effects | Nominal significant association between a single CpG site in IGF2 and hip circumference. No associations with methylation found. |
| Nour El Huda et. al [38] | MethyLight | COMT | Determine the effect of diagnosis and clinical variables on methylation | Decreased methylation observed in patients with SCZ versus HC. Lower methylation observed in AAP (and in risperidone only) group compared to typical antipsychotic group. |
| Shi et al. [40] | MassARRAY | CYP3A4, CYP2D6, HTR2A, ABCB1, DRD2 | Analyze methylation based on risperidone treatment and response | Decreased methylation in CYP3A4 and CYP2D6 genes associated with response to risperidone. No significant associations observed for HTR2A, ABCB1, or DRD2 gene methylation. |
| Tang et al. [42] | PSQ | HTR1A | Understand the effect of methylation on response to antipsychotics | One site correlated with change in symptom scores however, it was not dependent on antipsychotic type. |
| Venugopal et al. [43] | Bisulfite sequencing | IL-6 | Determine the effect of AAP treatment on methylation | Reduced methylation in SCZ prior to treatment compared to HC. AAP treatment increased methylation to levels similar to HC. |
| Zong et al. [44] | EpiMark Restriction Enzyme qPCR | GABRB2 5-mC and 5-hmC | Effect of diagnosis and clinical variables on 5-mC and 5-hmC of GABRB2 gene | Both GABRB2 5-mC and 5-hmC were increased in SCZ compared to HCs. Olanzapine and ziprasidone treatment associated with increased 5-mC levels (no effect on 5-hmC levels). |
Details of 18 studies that analyzed gene-specific methylation in antipsychotic-treated patients with schizophrenia or bipolar disorder. Common gene nomenclature provided in table. Abbreviations: 5-mc = 5-methylcytosine; 5-hmC = 5-hydroxymethylcytosine; AAP = atypical antipsychotic; BD = bipolar disorder; CpG = cytosine-phosphate-guanine dinucleotide; HC = healthy controls; MS-HRM = Methylation Sensitive – High Resolution Melting PCR; PSQ = pyrosequencing; qMSP = quantitative methylation specific PCR; SCZ = schizophrenia
Studies that utilized arrays only analyzed a specified set of candidate genes and did not perform an epigenome-wide associate study
Discussion
The goal of this study was to perform a systematic review to better understand the current evidence regarding DNA methylation profiles of antipsychotic-treated patients with bipolar disorder and schizophrenia. Within the included studies, the majority (62%) analyzed associations in patients with a schizophrenia diagnosis, whereas a smaller proportion reported associations in patients with bipolar disorder (14%) or a combined sample (24%). Across the 29 included studies, the type of antipsychotic was not uniform and included typical, atypical, or multiple concurrent antipsychotics. Furthermore, class effects of antipsychotics (rather than a single or few antipsychotics) were more commonly studied and often effects of antipsychotics represented secondary analyses in studies of methylation changes based on psychiatric diagnosis. The studies included in this review used various approaches to assess DNA methylation including non-specific, global methylation, genome-wide discovery with epigenome arrays, and targeted analyses of candidate genes/pathways. The findings among these studies were mixed in that there was no uniform identification of increased or decreased DNA methylation in antipsychotic-treated patients, nor were any hits replicated in a rigorous manner. It appears that the effects of antipsychotics are dependent on the study design, population under study, and the gene(s) being studied. Given the heterogeneity identified here, it is critical that future clinical epigenetic studies of antipsychotics carefully choose sample populations (e.g., targeting a specific diagnosis with a well-defined antipsychotic exposure), methylation analysis methodology, sample sources, and gene locations that will allow for the development of an evidence base with sufficient rigor and quality for clinical application.
Methylation assessments of the COMT, HTR2a, and 5-HTT genes were included in more than one study. Two studies identified increased COMT methylation; the first in patients with schizophrenia on antipsychotics compared to healthy controls and the second in patients with schizophrenia with metabolic syndrome [33,34]. In contrast, two studies found decreased COMT methylation in patients with schizophrenia versus controls and furthermore, in one study, lower methylation was also specifically associated with atypical antipsychotic treatment [18,38]. 5-HTT findings were also mixed with one study finding decreased methylation with long-term antipsychotic use whereas another study found no differences [19,34]. In one study, HTR2A methylation was found to be higher with antipsychotic use compared to non-treated or healthy controls, however, another study found no difference [17,40].
Although several reasons could lead to the discordant findings among studies analyzing the same genes (e.g., study design, patient population, etc), one primary issue among gene-specific studies is a lack of uniformity in assessing a gene’s methylation status. This may include the sample source and technologies used to determine methylation, which varied among the studies described in this review. Additionally, the target area of a given gene can vary widely especially since most technologies described here only analyze a very small portion or section of a gene. Although technologies like bisulfite sequencing exist to analyze an entire gene, this is currently limited by its expense and need for advanced bioinformatic processing approaches. For example, in the two studies that analyzed 5HT2RA, one stated that they “screened [a] CpG-rich spot in the upstream of promoter region or within candidate genes” [40] whereas the other stated that they analyzed “HTR2A promoter” methylation [17]. The latter study provides some more details including primers sets, base pair distances from the assumed transcription start site, and an external reference [45] that provides their methods for identifying CpG islands near promoter regions. Despite the base pair distances for the HTR2A analysis provided in the latter study, neither study provided exact genomic coordinates for their analyzed region of interest or were fully transparent regarding the number of CpG methylation sites included in their analysis (although the [40] provides more information for their statistically significant associations). This demonstrates that the descriptions, either within the manuscript or available supplementary material, of the exact genomic locations analyzed may not be known, thus making comparisons difficult and potentially unreliable. Future publications should consider utilizing supplementary material to report specific genomic locations assessed including number of CpG sites, location of CpGs, and primer(s) used to analyze methylation (if applicable). Such information should be provided regardless of the significance of the statistical associations described within the study. Creating a more transparent and uniform reporting of gene-specific methylation assessment will allow for future strategies to replicate previous studies and potentially combine results in using meta-analytic strategies.
The epigenome-wide studies identified numerous sites, genes, and pathways associated with antipsychotic treatment. Many of the reported associations are novel in nature and occurred in relatively small sample sizes given the large number of comparisons performed within each study. Therefore, although promising, findings from these exploratory studies should be considered preliminary until further replication is pursued. A few studies did include a replication cohort to validate their epigenome-wide candidates, however, independent replication did not occur in any study; thus the rigor of replication was low. Pathway analyses were included in three studies that identified several potential pathways impacted by methylation changes including mitochondrial, Wnt/β-catenin, adipogenesis, immune-related, neurogenesis, and regulatory [30,35,46]. Some of these pathways have been investigated for molecular changes (e.g., protein, gene variation, etc.) in response to antipsychotic medications in both human and non-human studies [47–50]. Overall, these epigenome-wide studies have identified both novel and known pathways that should be furthered pursued for their roles in antipsychotic treatment response and side effects.
Most studies included in this review utilized clinically-accessible samples (i.e., blood or saliva) for DNA methylation measurements. This does increase the potential clinical utility of methylation associations identified in these tissues, however, it may not reflect the brain, which is the main site of interest in psychiatric disease treatment. A few studies utilized saliva and post-mortem brain samples and found similar DNA methylation changes in both tissues suggesting some changes in the brain may be reflected in the periphery. This is further supported in studies whose primary purpose is to identify DNA methylation correlations across various tissues within the brain [51]. Of note, peripheral blood DNA methylation has been found to have lower correlation with brain DNA methylation whereas saliva appears to have a higher correlation. Such nuances should be considered when interpreting the utility of such studies (e.g., biomarker versus mechanism) [52]. The only other tissue utilized in the included studies was skeletal muscle, which was specifically chosen as a candidate tissue for the study of antipsychotic-associated insulin resistance [53]. Future work will need to take into consideration the source of DNA methylation assessment and what role this plays in the interpretation of findings.
The associations described within these studies evaluated several relationships to antipsychotic use. These relationships included: (i) a basic effect of antipsychotics compared to healthy controls or another medication class, (ii) an effect of length of treatment on DNA methylation, (iii) associations of antipsychotic response (based on psychiatric scale measurements) to DNA methylation changes, and (iv) associations between antipsychotic side effects and DNA methylation changes. This wide range of assessment may partly explain the observed contradictory changes in DNA methylation (hyper versus hypomethylation). Nevertheless, this range of work is also useful in delineating the epigenetic underpinnings of antipsychotic efficacy versus side effects, which may or may not be distinct. Given the low response rates, high rates of nonadherence, and high mortality rates, this work may well create an evidence base on which future studies can pursue multi-gene approaches to understand treatment mechanisms or improve treatment outcomes.
Some limitations to the current body of research regarding DNA methylation and antipsychotics should be considered. First, the populations, treatment, methodology, and targets of DNA methylation analysis varied widely across the studies. The most powerful study design to identify causal DNA methylation changes secondary to antipsychotic treatment would be through prospective studies. Within this systematic review, 8 of 29 studies included a prospectively treated population, however, none included placebo comparators. This is further reflected in the quality assessments of each study (supplementary information) included in this review, which did not identify a study that met every quality criteria. As discussed, the source of DNA methylation measurement varied among studies, with the most common source being peripheral blood. The use of this tissue in understanding AAP effects should be taken with caution, especially if the study utilized no method to correct for potential cell type bias inherent to peripheral blood. It is evident from the included studies that more uniform approaches are required in assessing associations between DNA methylation and antipsychotics as the current heterogeneity among the studies makes it impossible to critically evaluate and combine findings. Finally, there is a current lack of replication and/or validation of most of the findings described within the studies included in this review, suggesting that the findings to date are not yet suitable for further clinical application at this time.
Conclusions and Considerations for Future Research
It remains to be seen if pharmacoepigenetics will prove useful in the same manner as pharmacogenetics, or if it will have a unique role in clinical applications or used in the evidence base for treatment mechanisms and future treatment development. Such future use of DNA methylation in antipsychotic treatment is not out of the realm of possibility given that DNA methylation has been used to support clinical treatment decisions in cancer [54], prenatal testing [55], forensic testing [56], and in the development of drugs [57]. Until such time, it is imperative that studies of DNA methylation in antipsychotic-treated populations utilize high quality methodologies (from primer design to methylation assessment) that allow for a robust evidence base that can be used in future research that would inform clinical decisions. Upcoming research should focus on assessing previously identified pathways from either pre-clinical or clinical studies in prospectively-treated populations with adequate control groups in order to identify the most relevant and powerful associations between antipsychotics and DNA methylation. Strategies, such as Mendelian Randomization, may be a useful method for estimating the causal associations between DNA methylation and antipsychotic treatment in observational studies while accounting for possible confounding effects. This approach has been successfully utilized in other disease states, such as dyslipidemia, and its application in epigenetics is growing [58,59]. Future work should simultaneously assess critical tissues of interest related to antipsychotic effects (e.g., brain for efficacy, metabolic tissues for side effects, etc.) alongside clinically accessible tissues (e.g., saliva, blood, etc.) to identify robust associations that would have potential for clinical utility. Additionally, studies should utilize technical or statistical approaches to control for the heterogeneous cellular composition of peripheral blood. Studies should also include clearly defined populations with inclusion and exclusion criteria that target specific diagnoses and/or diagnostic sub-types (e.g., bipolar disorder I, etc.) with clear exposures of specific individual antipsychotic medications. Future studies and reviews should attempt to clarify associations of DNA methylation with antipsychotic treatment versus disease development itself as it is possible that the two are linked. This work will be critical to defining epigenetic mechanisms that are relevant to both disease and treatment outcome. Employing these approaches, among others, will be vital to the improvement of future application of epigenetics in antipsychotic treatment including replication and new discovery.
Supplementary Material
Acknowledgements
This work was supported in part by grants from NIH/NIDDK R01DK081750 (ZY), R01DK107666 (ZY), K23DK118199 (KB), and L30DK110823 (KB).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
REFERENCES
- 1.Leucht S, Tardy M, Komossa K, Heres S, Kissling W, Salanti G, et al. Antipsychotic drugs versus placebo for relapse prevention in schizophrenia: a systematic review and meta-analysis. Lancet 2012, 379, 2063–2071. [DOI] [PubMed] [Google Scholar]
- 2.Tiihonen J, Mittendorfer-Rutz E, Majak M, Mehtälä J, Hoti F, Jedenius E, Enkusson D, et al. Real-World Effectiveness of Antipsychotic Treatments in a Nationwide Cohort of 29 823 Patients With Schizophrenia. JAMA psychiatry 2017, 74, 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García S, Martínez-Cengotitabengoa M, López-Zurbano S, Zorrilla I, López P, Vieta E, et al. Adherence to Antipsychotic Medication in Bipolar Disorder and Schizophrenic Patients: A Systematic Review. Journal of clinical psychopharmacology 2016, 36, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascade E, Kalali AH, Mehra S, Meyer JM Real-world Data on Atypical Antipsychotic Medication Side Effects. Psychiatry (Edgmont (Pa. : Township)) 2010, 7, 9–12. [PMC free article] [PubMed] [Google Scholar]
- 5.Eum S, Lee AM, Bishop JR Pharmacogenetic tests for antipsychotic medications: clinical implications and considerations. Dialogues Clin Neurosci 2016, 18, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousman CA CYP2D6 testing to guide risperidone and aripiprazole therapy. The lancet. Psychiatry 2019, 6, 362–364. [DOI] [PubMed] [Google Scholar]
- 7.Skvortsova K, Iovino N, Bogdanovic O Functions and mechanisms of epigenetic inheritance in animals. Nature reviews. Molecular cell biology 2018, 19, 774–790. [DOI] [PubMed] [Google Scholar]
- 8.Kurdyukov S, Bullock M DNA Methylation Analysis: Choosing the Right Method. Biology (Basel) 2016, 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du X, Han L, Guo AY, Zhao Z Features of methylation and gene expression in the promoter-associated CpG islands using human methylome data. Comparative and functional genomics 2012, 2012, 598987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer BD A Practical Guide to the Measurement and Analysis of DNA Methylation. American journal of respiratory cell and molecular biology 2019, 61, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barros-Silva D, Marques CJ, Henrique R, Jeronimo C Profiling DNA Methylation Based on Next-Generation Sequencing Approaches: New Insights and Clinical Applications. Genes (Basel) 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran S, Arribas C, Esteller M Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 2016, 8, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan KS, Kunz R, Kleijnen J, Antes G Five steps to conducting a systematic review. Journal of the Royal Society of Medicine 2003, 96, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan K, Kunz R, Kleijnen J, Antes G Systematic reviews to support evidence-based medicine, Crc Press: 2011. [Google Scholar]
- 15.Wright RW, Brand RA, Dunn W, Spindler KP How to write a systematic review. Clinical Orthopaedics and Related Research (1976–2007) 2007, 455, 23–29. [DOI] [PubMed] [Google Scholar]
- 16.Abdolmaleky HM, Pajouhanfar S, Faghankhani M, Joghataei MT, Mostafavi A, Thiagalingam S Antipsychotic drugs attenuate aberrant DNA methylation of DTNBP1 (dysbindin) promoter in saliva and post-mortem brain of patients with schizophrenia and psychotic bipolar disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2015, 168, 687–696. [DOI] [PubMed] [Google Scholar]
- 17.Abdolmaleky HM, Yaqubi S, Papageorgis P, Lambert AW, Ozturk S, Sivaraman V, et al. Epigenetic dysregulation of HTR2A in the brain of patients with schizophrenia and bipolar disorder. Schizophrenia research 2011, 129, 183–190. [DOI] [PubMed] [Google Scholar]
- 18.Abdolmaleky HM, Cheng K. h., Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Human molecular genetics 2006, 15, 3132–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdolmaleky HM, Nohesara S, Ghadirivasfi M, Lambert AW, Ahmadkhaniha H, Ozturk S, et al. DNA hypermethylation of serotonin transporter gene promoter in drug naive patients with schizophrenia. Schizophrenia research 2014, 152, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bönsch D, Wunschel M, Lenz B, Janssen G, Weisbrod M, Sauer H Methylation matters? Decreased methylation status of genomic DNA in the blood of schizophrenic twins. Psychiatry research 2012, 198, 533–537. [DOI] [PubMed] [Google Scholar]
- 21.Bromberg A, Levine J, Nemetz B, Belmaker R, Agam G No association between global leukocyte DNA methylation and homocysteine levels in schizophrenia patients. Schizophrenia research 2008, 101, 50–57. [DOI] [PubMed] [Google Scholar]
- 22.Burghardt KJ, Goodrich JM, Dolinoy DC, Ellingrod VL Gene-specific DNA methylation may mediate atypical antipsychotic-induced insulin resistance. Bipolar disorders 2016, 18, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burghardt KJ, Goodrich JM, Lines BN, Ellingrod VL The Influence of Metabolic Syndrome and Sex on the DNA Methylome in Schizophrenia. International journal of genomics 2018, 10.1155/2018/8076397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burghardt KJ, Seyoum B, Dass S, Sanders E, Mallisho A, Yi Z Association of Protein Kinase B (AKT) DNA Hypermethylation with Maintenance Atypical Antipsychotic Treatment in Patients with Bipolar Disorder. Pharmacotherapy 2018, April;38(4):428–435.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burghardt KJ, Goodrich JM, Dolinoy DC, Ellingrod VL DNA methylation, insulin resistance and second-generation antipsychotics in bipolar disorder. Epigenomics 2015, 7, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burghardt KJ, Pilsner JR, Bly MJ, Ellingrod VL DNA methylation in schizophrenia subjects: gender and MTHFR 677C/T genotype differences. Epigenomics 2012, 4, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng J, Wang Y, Zhou K, Wang L, Li J, Zhuang Q, Xu X, et al. Male-specific association between dopamine receptor D4 gene methylation and schizophrenia. PloS one 2014, 9, e89128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Addario C, Micale V, Di Bartolomeo M, Stark T, Pucci M, Sulcova A, et al. A preliminary study of endocannabinoid system regulation in psychosis: Distinct alterations of CNR1 promoter DNA methylation in patients with schizophrenia. Schizophrenia research 2017, 188, 132–140. [DOI] [PubMed] [Google Scholar]
- 29.Ellingrod VL, Grove TB, Burghardt KJ, Taylor SF, Dalack G The effect of folate supplementation and genotype on cardiovascular and epigenetic measures in schizophrenia subjects. Npj Schizophrenia 2015, 1, 15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houtepen LC, van Bergen AH, Vinkers CH, Boks MP DNA methylation signatures of mood stabilizers and antipsychotics in bipolar disorder. Epigenomics 2016, 8, 197–208. [DOI] [PubMed] [Google Scholar]
- 31.Kinoshita M, Numata S, Tajima A, Yamamori H, Yasuda Y, Fujimoto M, et al. Effect of clozapine on DNA methylation in peripheral leukocytes from patients with treatment-resistant schizophrenia. International journal of molecular sciences 2017, 18, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Yang Q, Hou Y, Jiang T, Zong L, Wang Z, et al. Hypomethylation of LINE-1 elements in schizophrenia and bipolar disorder. Journal of psychiatric research 2018, 107, 68–72. [DOI] [PubMed] [Google Scholar]
- 33.Lott SA, Burghardt PR, Burghardt KJ, Bly MJ, Grove TB, Ellingrod VL The influence of metabolic syndrome, physical activity and genotype on catechol-O-methyl transferase promoter-region methylation in schizophrenia. The pharmacogenomics journal 2013, 13, 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melas PA, Rogdaki M, Osby U, Schalling M, Lavebratt C, Ekstrom TJ Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2012, 26, 2712–2718. [DOI] [PubMed] [Google Scholar]
- 35.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. American journal of human genetics 2008, 82, 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miura I, Kunii Y, Hino M, Hoshino H, Matsumoto J, Kanno-Nozaki K, et al. DNA methylation of ANKK1 and response to aripiprazole in patients with acute schizophrenia: A preliminary study. Journal of psychiatric research 2018, 100, 84–87. [DOI] [PubMed] [Google Scholar]
- 37.Moons T, De Hert M, Kenis G, Viechtbauer W, van Os J, Gohlke H, et al. No association between genetic or epigenetic variation in insulin growth factors and antipsychotic-induced metabolic disturbances in a cross-sectional sample. Pharmacogenomics 2014, 15, 951–962. [DOI] [PubMed] [Google Scholar]
- 38.Nour El Huda AR, Norsidah KZ, Nabil Fikri MR, Hanisah MN, Kartini A, Norlelawati AT DNA methylation of membrane-bound catechol-O-methyltransferase in Malaysian schizophrenia patients. Psychiatry and clinical neurosciences 2018, 72, 266–279. [DOI] [PubMed] [Google Scholar]
- 39.Rukova B, Staneva R, Hadjidekova S, Stamenov G, Milanova V, Toncheva D Whole genome methylation analyses of schizophrenia patients before and after treatment. Biotechnology & Biotechnological Equipment 2014, 28, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Li M, Song C, Xu Q, Huo R, Shen L, et al. Combined study of genetic and epigenetic biomarker risperidone treatment efficacy in Chinese Han schizophrenia patients. Translational psychiatry 2017, 7, e1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swathy B, Saradalekshmi KR, Nair IV, Nair C, Banerjee M Understanding the influence of antipsychotic drugs on global methylation events and its relevance in treatment response. Epigenomics 2018, 10, 233–247. [DOI] [PubMed] [Google Scholar]
- 42.Tang H, Dalton CF, Srisawat U, Zhang ZJ, Reynolds GP Methylation at a transcription factor-binding site on the 5-HT1A receptor gene correlates with negative symptom treatment response in first episode schizophrenia. International Journal of Neuropsychopharmacology 2014, 17, 645–649. [DOI] [PubMed] [Google Scholar]
- 43.Venugopal D, Shivakumar V, Subbanna M, Kalmady SV, Amaresha AC, Agarwal SM, et al. Impact of antipsychotic treatment on methylation status of Interleukin-6 [IL-6] gene in Schizophrenia. Journal of psychiatric research 2018, 104, 88–95. [DOI] [PubMed] [Google Scholar]
- 44.Zong L, Zhou L, Hou Y, Zhang L, Jiang W, Zhang W, et al. Genetic and epigenetic regulation on the transcription of GABRB2: Genotype-dependent hydroxymethylation and methylation alterations in schizophrenia. Journal of psychiatric research 2017, 88, 9–17. [DOI] [PubMed] [Google Scholar]
- 45.Abdolmaleky HM, Smith CL, Zhou JR, Thiagalingam S Epigenetic alterations of the dopaminergic system in major psychiatric disorders. Methods in molecular biology (Clifton, N.J.) 2008, 448, 187–212. [DOI] [PubMed] [Google Scholar]
- 46.Burghardt KJ, Goodrich JM, Lines BN, Ellingrod VL The Influence of Metabolic Syndrome and Sex on the DNA Methylome in Schizophrenia. International journal of genomics 2018, 2018, 8076397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton LP, Honardoust D, Mouyal J, Rajakumar N, Rushlow WJ Activation of the canonical Wnt pathway by the antipsychotics haloperidol and clozapine involves dishevelled-3. Journal of neurochemistry 2007, 102, 153–169. [DOI] [PubMed] [Google Scholar]
- 48.Best L, Yates AP, Reynolds GP Actions of antipsychotic drugs on pancreatic beta-cell function: contrasting effects of clozapine and haloperidol. Journal of psychopharmacology (Oxford, England) 2005, 19, 597–601. [DOI] [PubMed] [Google Scholar]
- 49.Kristóf E, Doan-Xuan QM, Sárvári AK, Klusóczki Á, Fischer-Posovszky P, Wabitsch M, et al. Clozapine modifies the differentiation program of human adipocytes inducing browning. Translational psychiatry 2016, 6, e963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Amin MM, Nasir Uddin MM, Mahmud Reza H Effects of antipsychotics on the inflammatory response system of patients with schizophrenia in peripheral blood mononuclear cell cultures. Clinical psychopharmacology and neuroscience : the official scientific journal of the Korean College of Neuropsychopharmacology 2013, 11, 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT,, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Translational psychiatry 2019, 9, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD Epigenetic Research in Neuropsychiatric Disorders: the “Tissue Issue”. Current behavioral neuroscience reports 2016, 3, 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdul-Ghani MA, DeFronzo RA Pathogenesis of insulin resistance in skeletal muscle. Journal of biomedicine & biotechnology 2010, 2010, 476279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Werner RJ, Kelly AD, Issa JJ Epigenetics and Precision Oncology. Cancer journal (Sudbury, Mass.) 2017, 23, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee DE, Kim SY, Lim JH, Park SY, Ryu HM Non-Invasive Prenatal Testing of Trisomy 18 by an Epigenetic Marker in First Trimester Maternal Plasma. PloS one 2013, 8, e78136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidaki A, Kayser M From forensic epigenetics to forensic epigenomics: broadening DNA investigative intelligence. Genome biology 2017, 18, 238–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganesan A Epigenetic drug discovery: a success story for cofactor interference. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 2018, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sayols-Baixeras S, Tiwari HK, Aslibekyan SW Disentangling associations between DNA methylation and blood lipids: a Mendelian randomization approach. BMC proceedings 2018, 12, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Relton CL, Davey Smith G Mendelian randomization: applications and limitations in epigenetic studies. Epigenomics 2015, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.