Abstract
Tumors are characterized by the presence of malignant and non-malignant cells, such as immune cells including macrophages, which are preponderant. Macrophages impact the efficacy of chemotherapy and may lead to drug resistance. In this context and based on our previous work, we investigated the ability to reactivate macrophages by using a proprotein convertases inhibitor. Proprotein convertases process immature proteins into functional proteins, with several of them having a role in immune cell activation and tumorigenesis. Macrophages were treated with a peptidomimetic inhibitor targeting furin, PC1/3, PC4, PACE4, and PC5/6. Their anti-glioma activity was analyzed by mass spectrometry-based proteomics and viability assays in 2D and 3D in vitro cultures. Comparison with temozolomide, the drug used for glioma therapy, established that the inhibitor was more efficient for the reduction of cancer cell density. The inhibitor was also able to reactivate macrophages through the secretion of several immune factors with antitumor properties. Moreover, two proteins considered as good glioma patient survival indicators were also identified in 3D cultures treated with the inhibitor. Finally, we established that the proprotein convertases inhibitor has a dual role as an anti-glioma drug and anti-tumoral macrophage reactivation drug. This strategy could be used together with chemotherapy to increase therapy efficacy in glioma.
Keywords: macrophages, immunotherapy, temozolomide, proprotein convertases, mass spectrometry-based proteomic, glioma
Graphical Abstract
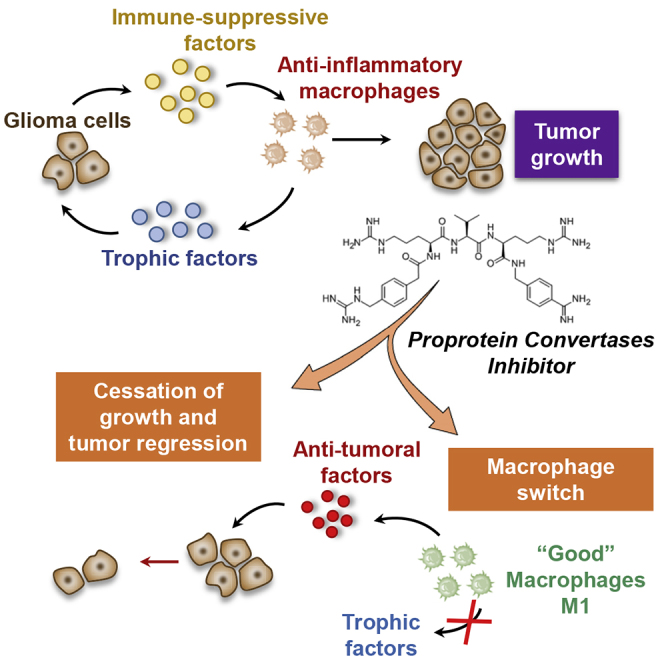
Despite significant advances in cancer treatment, resistance to applied therapy remains a major problem. Salzet and colleagues established that inhibition of proprotein convertases exerts a dual activity at the same time by its anti-tumoral effect on glioma cells and by its ability to switch macrophages to an anti-tumoral phenotype.
Introduction
Proprotein convertases (PCs) are proteases of the subtilisin/kexin family that cleave proproteins through limited proteolysis and convert them into bioactive proteins and peptides.1, 2, 3 Mammalian PCs include PC subtilisin/kexin type 1 (PCSK1), PCSK2, furin, PCSK4, PCSK5, PCSK6, and PCSK7, which are known to cleave proproteins at paired basic residues.4 PCs cleave a variety of precursor proteins within the secretory pathway, including neuropeptides, hormones, growth factors and their respective receptors, adhesion molecules, bacterial toxins, and viral glycoproteins.5 However, deregulation of these enzymes has been associated with pathological conditions, including endocrinopathies,6 Alzheimer’s disease,7 and tumors.8 Among these PCs, furin is one of the major PCs known to be involved in tumor progression. Its expression has been confirmed in a large spectrum of cancers.1 The importance of furin activity for the processing of many cancer-related substrates has been shown, and it has been proposed to promote the malignant phenotype of cancer cells.8,9 Platelet-derived growth factor A (PDGF-A), insulin-like growth factor 1 receptor (IGF-1R), transforming growth factor β (TGF-β), membrane type I-matrix metalloproteinase (MT1-MMP), vascular endothelial growth factor C (VEGF-C), and a disintegrin and metalloproteinase with thrombospondin motifs (ADAM-TS) are the best known cancer-related furin substrates.
Despite significant advances in cancer treatment, resistance to the applied therapy remains a major problem.10,11 The increased efflux of the drugs, the enhancement of repair/increased tolerance to DNA damage, the high anti-apoptotic potential, and the decreased permeability and enzymatic deactivation would enable cancer cells to survive chemotherapy.10 Indeed, multidrug resistance is one of the most significant problems in oncology today.12, 13, 14
Nevertheless, among the emerging therapies that are in development, immunotherapy represents one with the most promise.15 The goal of immunotherapy is to eliminate cancer cells through the transfer of ex vivo expanded and activated immune cells. Immune cells such as dendritic cells (DCs), natural killer (NK) cells, cytotoxic T cells, and cytokine-induced killer (CIK) cells have been investigated as targets for immunotherapy against cancer.15 Additionally, the passive transfer of monoclonal antibodies has been an effective treatment for some cancers.16
Promising trials evaluating the safety and efficacy of immunotherapy for prostate cancer, renal cell carcinoma, and gastric and colorectal cancers have opened the door to research in the area of oncoimmunology, which integrates immunotherapy along with conventional cancer treatments.16 Tumors are heterogeneous and characterized by the presence of many malignant and benign cell types. Among the non-malignant cells, specific immune cells are present, known as tumor-associated macrophages (TAMs), which represent a key element for tumor microenvironment interaction. These TAMs are switched to the protumoral phenotype through cancer cell-tissue microenvironment interaction.15 Thus, one challenge is to reactivate these TAMs and combat cellular immune suppression. PCs have been identified in immune cells and shown to control the Toll-like receptor (TLR)/myeloid differentiating factor 88 (MyD88)/nuclear factor κB (NF-κB)-dependent pathway through PC1/317, 18, 19, 20, 21, 22 and the TLR/MyD88/NF-κB independent pathway through furin in antigen-presenting immune cells such as macrophages or DCs.18,23,24
Interestingly, the inhibition of PC1/3 in knockout (KO) mice or the knock down (KD) of the resident lung macrophage cell line (NR8383) led to a pro-inflammatory response. The NR8383 rat pulmonary macrophage cell line was previously shown as a good model to study the role of PC1/3 in the macrophage innate immune response.18, 19, 20,22 The pro-inflammatory response in NR8383 PC1/3 KO macrophages is characterized by the secretion of chemotactic effectors attracting cytotoxic T cells and anti-tumoral factors against glioma and gynecological cancers.18,19 In this way, finding an inhibitor acting on furin in cancer cells that can also push toward PC1/3 in TAMs can be a novel alternative in cancer immunotherapy. Several inhibitors have been designed against PCs, and several targeting the furin protein25, 26, 27 or PACE428,29 are used for cancer therapy.
In this context, we are using a peptidomimetic PC inhibitor that can act against furin and has been shown to inhibit several other PCs, including PC1/3, PC4, PACE4, and PC5/6, but with no effect on PC2 and PC7. This inhibitor has been developed by Becker et al.30 This is a cell-permeable tripeptide derivative containing an Arg-X-Arg motive that acts as a highly potent and reversible furin inhibitor (Ki of 16 pM) and PC1/3, PC4, PACE4, and PC5/6 inhibitor with almost similar potency. Other peptidomimetic PC inhibitors are used for anti-tumoral assays such as PACE4 inhibitor,31 Multi-Leu peptide PACE4 inhibitor,32 or furin inhibitor.33 These inhibitors are used in tumor cell cultures at a concentration ranging from 25 to 100 μM. For this reason, we investigated, via a concentration/response pharmacodynamic approach (10–600 μM), proteomic platforms, and biochemical anti-tumoral assays, the ability of such a peptidomimetic PC inhibitor to act against glioma cells and macrophages at the same time. We also evaluated its ability to reactivate the macrophages within the tumor. We established that the PC inhibitor is active on glioma cells at a lower dose (2-fold less) than the key drug used in glioma treatment, i.e., temozolomide (TMZ). Moreover, this PC inhibitor is active toward TAMs by reactivating them to be cytotoxic in the intimacy of the tumor environment.
Results
The PC Inhibitor Triggers the Secretion of Anti-tumoral Factors by Macrophages and Decreases C6 Glioma Cell Viability
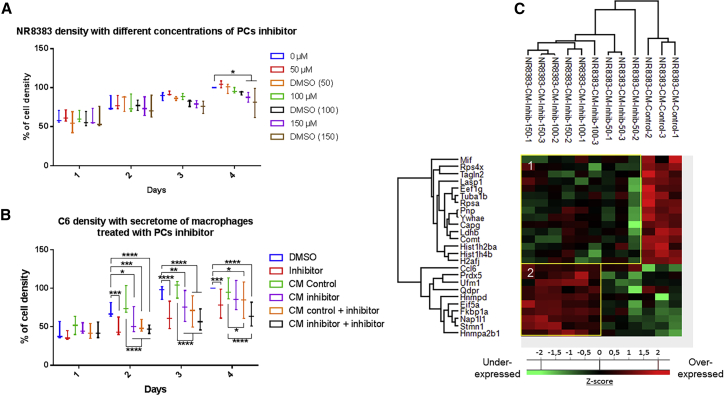
We have previously demonstrated that PC1/3, a PC, is involved in macrophage activation and that its inhibition induces anti-tumoral properties.19 The present study aimed to evaluate the potential use of a peptidomimetic inhibitor of PCs as a potential anti-cancer therapy through tumor inhibition and macrophage reactivation. We first established, using RT-PCR analyses, the expression of PCs in C6 glioma. We previously demonstrated the expression of PC1/3 and furin in NR8383 cells.22 Also, we only observed the expression of PACE4 and furin in C6 glioma. No PC1/3 has been detected (Figure S1). In this context, since the peptidomimetic PC inhibitor (C35H55N15O4) is active toward furin, PACE4, and PC1/3 at the same efficiency (Ki of 16 pM), we selected this PC inhibitor for our study. In order to know whether this PC inhibitor can trigger the secretion of anti-tumoral factors by macrophages, as previously described with the NR8383 PC1/3 KD cell line, we performed a viability assay on the rat C6 glioma cell line (Figure 1B). Based on the literature for the concentration used in cells or tissue cultures for such types of an inhibitor,30, 31, 32, 33 we used this inhibitor at the micromolar range. In the first step, we evaluated the toxicity of the PC inhibitor directly on the macrophages in order to select the right concentration of inhibitor to use. As shown in Figure 1A, the PC inhibitor did not affect the viability of macrophages at 50 and 100 μM. A slight decrease was observed after 4 days of stimulation with 150 μM PC inhibitor due to the toxicity of dimethyl sulfoxide (DMSO), as revealed by the control DMSO (150 μM). At the highest concentration of 300 μM PC inhibitor, the significant impact was shown at 3 days post-treatment (Figure S2A). With a volume of DMSO corresponding to the concentration of 150 μM inhibitor, we noticed a decrease in the proliferation of the macrophages. Therefore, for the following experiment, we treated the macrophages with 100 μM PC inhibitor to avoid the DMSO toxicity effect. We then tested the anti-tumor properties of the conditioned medium of macrophages treated with or without the PC inhibitor at 100 μM during 24 h (Figure 1B). The conditioned medium of the untreated macrophages (CM control) was not toxic for glioma cancer cells since the same viability was identified in the control condition at different time points (control). The conditioned medium of macrophages treated beforehand with the PC inhibitor (100 μM) decreased the viability of the C6 cells (CM inhibitor) by 30% compared to the control and to the CM control as early as 2 days after the incubation of C6 glioma cells with the macrophage-conditioned medium. However, we observed that the inhibitor alone had the same effect as the conditioned medium of macrophages. We cannot exclude that the inhibitor was still present in the medium of macrophages, which may have caused a decrease in C6 cell viability. When we added the inhibitor into the treated macrophage-conditioned medium, an additive effect seemed to be observed after 3 and 4 days, even if it was not statistically significant (CM inhibitor + inhibitor). This cumulative effect was not observed when the inhibitor was added in the conditioned medium of the untreated macrophages (CM control + inhibitor). Therefore, the inhibitor had a direct effect on C6 cancer cells but seemed also to induce the secretion of anti-tumoral factors by macrophages since an effect of the conditioned medium was observed.
Figure 1.
PC Inhibitor Triggers a Molecular Change in Protein Secretion of NR8383 Macrophages
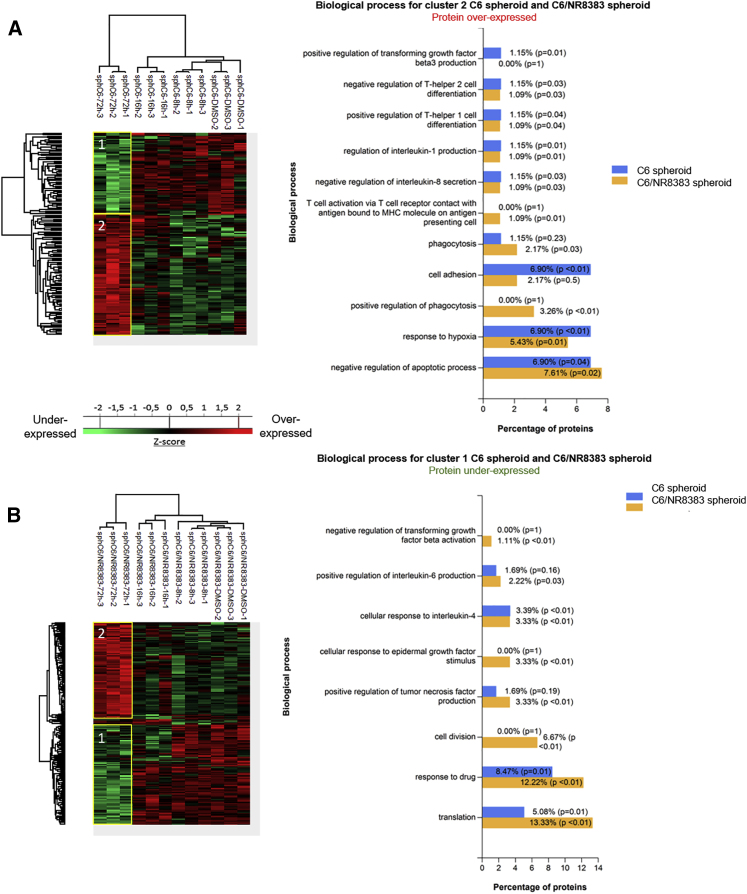
(A and B) The cell density of NR8383 macrophages (A) and C6 rat glioma cells (B) was determined by an MTS assay. The assays were conducted for 1, 2, 3, and 4 days. All results are representative of three independent experiments. Significant differences were identified using a Tukey’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. (A) NR8383 macrophages were incubated with different concentrations of PC inhibitor (50, 100, or 150 μM) or with vehicle (DMSO). (B) The C6 cells were incubated with DMSO (1) or 100 μM PC inhibitor (2) or with NR8383 conditioned media (CM) obtained after 24-h DMSO (3) or 100 μM PC inhibitor (4) treatment. In the condition “CM control + inhibitor” (5) and “CM inhibitor + inhibitor” (6), the cells were incubated with CM from macrophages treated with DMSO (5) or PC inhibitor (6) supplemented with PC inhibitor. (C) NR8383 macrophages were treated or not with PC inhibitor (50, 100, or 150 μM). CM were digested and analyzed with LC-MS/MS. MaxQuant and Perseus software were used for the statistical analysis, and a heatmap was generated to show proteins that were significantly different between treated and untreated NR8383 CM. Two clusters are highlighted (1 and 2).
In order to answer this question, a proteomic study was then conducted to analyze the composition of the conditioned medium of macrophages treated with the PC inhibitor at different concentrations (50, 100, and 150 μM). Shotgun proteomic analysis of secreted proteins was performed 24 h after PC inhibitor treatment of macrophages. A total of 884 secreted proteins were identified. A comparison of the proteins between PC inhibitor-treated macrophages and non-treated macrophages allowed the identification of 25 proteins with significant differential regulation (Figure 1C; Table S1). As a criterion of significance, we applied an ANOVA test with a significance threshold of p < 0.05, and a heatmap was created. Among these proteins, 10 were more secreted by PC inhibitor-treated macrophages at concentrations of 100 and 150 μM compared to control macrophages. Interestingly, some pro-inflammatory phenotype markers were found such as chemokine (C-C motif) ligand 6 (CCL6) and PRDX5 (PRDX5), suggesting that the inhibitor was also able to induce the activation of macrophages as previously described for the NR8383 PC1/3 KD macrophages.19 CCL6 and heterogeneous nuclear ribonucleoprotein D (HNRNPD) proteins, more secreted by treated macrophages, are involved in cytokine signaling. Another interesting protein identified was peptidyl-prolyl cis-trans isomerase (FKBP1A), which is known to have antitumor properties. FKBP1A also prevents TGF-β receptor activation.35 We also noticed that 50 μM PC inhibitor was not enough to induce the secretion of these proteins by macrophages. A concentration of 100 or 150 μM is therefore more suited for our study. Alternatively, 15 proteins were down-secreted by PC inhibitor-treated macrophages at concentrations of 50, 100, and 150 μM compared to control macrophages treated with DMSO (Figure 1C; Table S1). The PC inhibitor seems to impair some macrophage functions as demonstrated by a down-secretion of migration-related proteins (macrophage migration inhibitory factor [MIF] and LIM and SH3 domain protein 1 [LASP1]) and phagocytosis-related proteins (macrophage-capping protein [CAPG] and transgelin-2 [TAGLN2] proteins). Of interest, the macrophage migration inhibitory factor (MIF) is also known to be involved in glioblastoma progression.36 Moreover, several proteins related to the metabolism process such as purine nucleoside phosphorylase (PNP), lactate dehydrogenase B (LDHB), and catechol-O-methyltransferase (COMT) were also less secreted under PC inhibitor treatment of macrophages, indicating a change in macrophage activity. In conclusion, proteins more secreted by macrophages treated with 100 and 150 μM of the inhibitor are more pro-inflammatory, as we previously described for the NR8383 PC1/3 KD cell line. We have also identified some anti-tumoral factors that can explain the decrease of the viability of C6 cells observed in Figure 1B. We have also confirmed these proteomics results on the THP1 human macrophages cell line (Figure S3). Thus, PC inhibition triggers the secretion of pro-inflammatory factors by THP1 as well as NR8383 and decreases the secretion of proteins involved in tumorigenesis processes and the anti-inflammatory microenvironment.
The PC Inhibitor Acts on C6 Glioma Cells by Decreasing Their Viability and Their Expression of Microenvironmental-Related Proteins
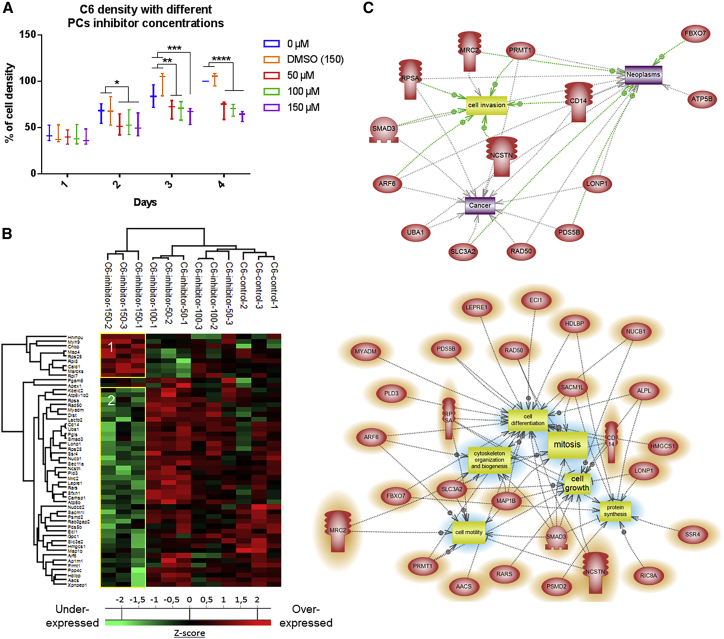
Since the inhibition of PCs in C6 glioma cancer cells triggered a decrease of cell viability, we examined the dose-dependent effects of the PC inhibitor. For that, we have cultured C6 cells with the inhibitor at 50, 100, or 150 μM for 4 days as was performed for macrophages (Figure 2A). We also checked the effect for lower concentrations (1 and 10 μM) and higher concentrations (300 and 400 μM). No effect was observed prior to the 10 μM treatment. At 10 μM, a decrease of C6 cell proliferation was seen, but it was lower than for concentrations above 50 μM (Figure S2B). We also noticed a significant effect at 300 and 400 μM at 2 days post-treatment (Figure S2B). The PC inhibitor decreased the viability of the glioma cells by 40% at any concentration >50 μM used compared to the control without inhibitor at 4 days post-treatment. Interestingly, a volume of DMSO corresponding to the one used at the highest concentration of the inhibitor (400 μM) did not influence the viability of the glioma cells. Therefore, the effect observed was clearly due to the inhibitor.
Figure 2.
PC Inhibitor Toxicity for C6 Rat Glioma Cells
(A) The cell density of C6 rat glioma cells was determined by an MTS assay after stimulation with 50, 100, or 150 μM PC inhibitor or with DMSO. The assays were conducted for 1, 2, 3, and 4 days. All results are representative of three independent experiments. Significant differences were identified using a Tukey’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. (B) C6 cells were treated or not with PC inhibitor (50, 100, or 150 μM) and lysed before FASP and LC-MS/MS analysis. MaxQuant and Perseus software were used for the statistical analysis, and a heatmap was generated to show proteins that were significantly different between treated and untreated C6 in the cell extracts. Two clusters are highlighted (1 and 2). (C) Global pathway analyses of cluster 2.
Next, in order to decipher the molecular impact of the PC inhibitor on C6 glioma cancer cells, we performed a proteomic study of the cellular contents after 24-h treatment with the inhibitor at different concentrations. The control cells (C6-control) were only treated with the volume of DMSO corresponding to the highest concentration of the inhibitor (150 μM). Shotgun proteomic analysis of C6 glioma cells yielded 1,942 protein identifications across all samples. Twelve proteins were exclusive to 50 μM, 33 proteins to 100 μM, 43 proteins to 150 μM, and 26 proteins to the control (Figure S4A; Data S1). Among the 12 proteins identified after the 50 μM PC inhibitor, the immunity-related GTPase family M protein (LRGM) known to activate macrophage migration has been identified as well as the repressor of translation initiation protein that regulates EIF4E activity (Eif4ebp1) and the latexin, which acts as a tumor suppressor.37 For the proteins identified at 100 μM, the TRPM4 transient receptor has been identified as well as semaphorin-3A, known to have a controversial effect on the tumor (promote38 or inhibit39), and netrin receptor UNC5B, which acts toward angiogenesis.40 The proteins detected at 150 μM included the WD repeat-containing protein 26 (Wdr26), an inhibitor of the mitogen-activated protein (MAP) kinase pathway,41 the melanoma-associated antigen D1 (MAGED1), which inhibits cell cycle progression and facilitates NGFR-mediated apoptosis,42 and also SMAD2 and SMAD9, which can switch the macrophage phenotype to M2.43 We then looked at the differentially expressed proteins. Again, as a criterion of significance, we applied an ANOVA significance threshold of p < 0.05, and a heatmap was created, from which 52 proteins showed a significant difference in label-free quantification (LFQ) expression between 150 μM-treated C6 glioma cells and the other conditions (inhibitor at 50 μM, inhibitor at 100 μM, and control). Two main clusters were highlighted (Figure 2B; Data S2). Nine proteins were upregulated in the 150 μM-treated C6 cells when compared to the other conditions, including myristoylated alanine-rich C-kinase substrate (MARCKS), which is involved in the reduction of glioblastoma growth and induction of senescence.36 In contrast, 41 proteins were upregulated in the control cells and the C6 cells treated with lower concentrations of PC inhibitor (50 and 100 μM), corresponding to cluster 2. Such upregulated proteins include PDS5B (PDS5B) and Rab3 GTPase-activating protein non-catalytic subunit (RAB3GAP2), proteins that are implicated in cell proliferation, and C-type mannose receptor 2 (MRC2) protein, which mediates tumor cell invasion. We also identified SMAD3, which is a transcription factor known to be activated after TGF-β binding on its receptor. SMAD3 regulates the expression of target genes involved in angiogenesis, cancer cell migration, and immunosuppression. CD14 was also found to be upregulated in C6 cells untreated or treated with low doses of PC inhibitor. CD14-high cancer cells express higher levels of inflammation mediators and form larger tumors. All of these proteins are downregulated after treatment of the C6 glioma cells with the PC inhibitor at 150 μM. Taken together, 150 μM PC inhibitor seems to prevent the induction of an anti-inflammatory environment by the cancer cells. Subnetwork analyses (Figure 2C) confirmed that proteins involved in neoplasia, cell invasion, and cancer are downregulated for a concentration of 150 μM PC inhibitor.
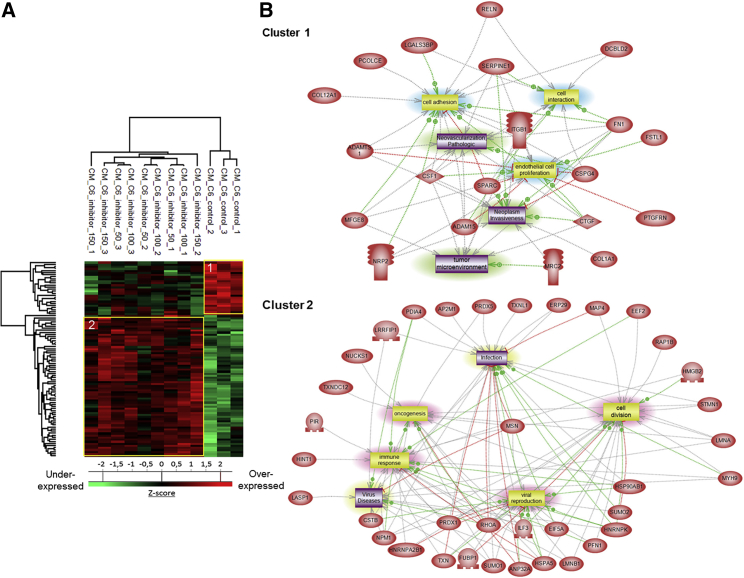
The same approach was then applied to the conditioned medium of C6 glioma cells treated with the PC inhibitor to analyze the secreted factors (Figure 3). The proteomic analysis yielded a total of 1,101 proteins identified across all samples, and none was specific to one concentration of inhibitor (Figure S4B). Ninety-five proteins were statistically significant between the PC inhibitor-treated C6 cells and the untreated C6 cells with a p value of 0.05. Twenty-six proteins were upregulated in the untreated control cells when compared to treated ones, while 69 proteins were significantly upregulated after PC inhibitor stimulation (Figure 3A; Data S3). In the control condition, proteins commonly found in the tumor microenvironment and involved in tumor cell migration, tumor growth, and metastasis were identified, which included the connective tissue growth factor (CTGF), chondroitin sulfate proteoglycan 4 (CSPG4), and a disintegrin and metalloproteinase with thrombospondin motifs 1 (ADAMTS1). Macrophage colony-stimulating factor 1 (CSF1) protein, which has a key role in the recruitment and pro-tumoral activation of macrophages, was shown to be highly secreted by the control C6 glioma cells. Some pro-tumoral proteins were also found such as anamorsin (CIAPIN1), ubiquitin carboxyl-terminal hydrolase 7 (USP7), and cartilage oligomeric matrix protein (COMP). Subnetwork pathway analyses of cluster 1 confirmed pathways involved in neoplasia and cell migration (Figure 3B). All of these proteins and their corresponding pathways (Figure 3B) were less secreted in the presence of the inhibitor at any concentration used. The second cluster groups all proteins that are more secreted in the presence of the PC inhibitor. Among these, we can find some proteins involved in viral reproduction (interleukin enhancer-binding factor 3 [ILF3]) and immune response (heat shock protein 90 alpha family class B member 1 [HSP90AB1], PRDX1). Proteins thioredoxin-like 1 (TXNL1), thioredoxin (TXN), and the peroxiredoxin family members (PRDX1, PRDX2, and PRDX5) reflect an oxidative stress state of the cells caused by the PC inhibitor. Pathways involving immune response activation and oncogenesis decrease have been identified (Figure 3B).
Figure 3.
PC Inhibitor Decreases the Secretion of Tumor Microenvironment-Related Proteins and Increases the Secretion of Immune Proteins
(A) C6 cells were treated or not with PC inhibitor (50, 100, or 150 μM). Conditioned media were digested and analyzed with LC-MS/MS. MaxQuant and Perseus software were used for the statistical analysis, and a heatmap was generated to show proteins that were significantly different between treated and untreated C6 conditioned media. Two clusters are highlighted (1 and 2). (B) Global pathway analyses of clusters 1 and 2.
In conclusion, the inhibitor impacts the C6 glioma cells by reducing their tumorigenesis potential. These results are in line with our previous work showing that the tumorigenesis of ovarian and prostate cancers is inhibited by PACE4 silencing.44 The inhibitor may, therefore, have an effect on several PCs involved in several cancer types.
The PC inhibitor Decreases the Invasion of 3D Mixed Glioma and Macrophage Spheroids
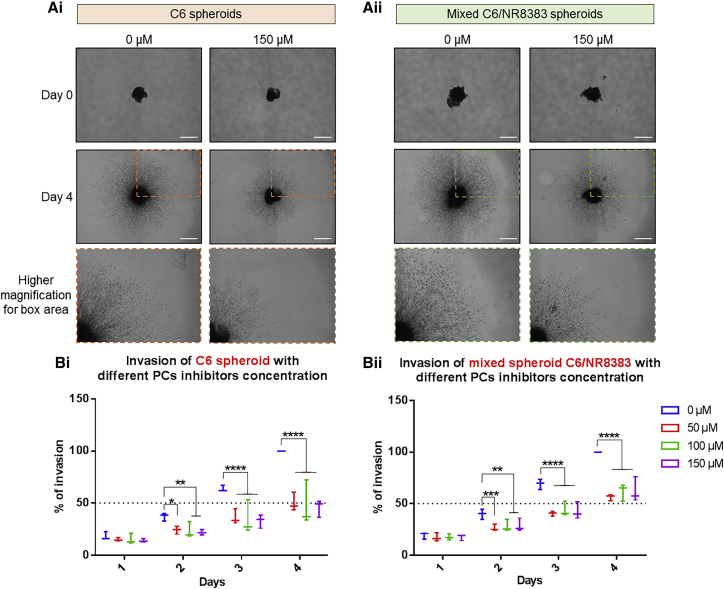
In order to go deeper into the characterization of the effects of the inhibitor on glioma growth, we tested it on 3D micro-tumors. First, we evaluated the distribution of the PC inhibitor in C6 glioma spheroids. Glioma spheroids were treated with 100 μM inhibitor at different time points (8–72 h). Consecutive sections of each spheroid were analyzed to study the distribution of the inhibitor across the entire spheroid. As shown in Figure 4, the PC inhibitor (mass-to-charge ratio [m/z] of 750.5) was detected predominantly into the core region of the spheroids after 16 h of treatment, while no drug signal was observed after 8 h of treatment and in the control, untreated sample. We could observe a slight decrease in the signal at 48 h and a marked decrease at 72 h. Then, we assessed the effects of the inhibitor on the growth and invasion of the cancer cells in 3D. The spheroids were cultured for 4 days in the presence or absence of 50, 100. or 150 μM PC inhibitor. Their growth and the invasion of the matrix by cells migrating out the initial core were monitored during these 4 days.18,45 Spheroids containing only C6 cells (Figure 5Ai) were compared to spheroids containing a mix of C6 and NR8383 cells (Figure 5Aii) to assess the tumor-supportive effects of macrophages as reported in the literature.15,46 For C6 spheroids treated with PC inhibitor, blockage of spheroid growth and invasion started after 2 days of treatment and increased progressively across time (Figure 5Bi). This was observed for all concentrations of PC inhibitor tested. However, this effect was not concentration-dependent since the various concentrations applied gave the same magnitude of inhibition (Table S2). It reached a maximum of 18% on the second day and a maximum of 54.2% on the fourth day. As depicted in Figure 5Bii, in the absence of PC inhibitor treatment, the presence of macrophages did not prevent the spheroids from growing. Conversely, a reduction of spheroid growth and invasion was again observed after the addition of PC inhibitor. Similar to the C6 spheroid, it also started after 2 days of treatment, it intensified in a time-course experiment, it was identical for the various concentrations tested, and it was not concentration-dependent. However, in the presence of macrophages, the magnitude of inhibition exerted by PC inhibitor was slightly less significant. It reached a maximum of 13.4% the second day and a maximum of 43.8% the fourth day. These data suggest that macrophages may exert a slight protective effect on C6 cells against PC inhibitor since the invasion of cancer cells is a bit enhanced in the presence of macrophages. However, the inhibitor still exerts its activity. Altogether, this revealed that PC inhibitor is effective to block spheroid growth and invasion, even in the presence of macrophages.
Figure 4.
PC Inhibitor Was Found at the Core of Mixed C6/NR8383 Spheroids at 16 h of Stimulation until 72 h
Mixed C6/NR8383 spheroids were treated with 100 μM PC inhibitor or with DMSO (control) for different times. Then, they were fixed, frozen, and entirely cut. Lipid images were acquired using a RapiFlex II MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) instrument at 10-μm lateral resolution and spectra were exported in SCiLS Lab version 2015b software. PC inhibitor is represented by an m/z of 750.522. Each row represents the different stimulation time. Each column represents a different slice of the spheroid, from the beginning on the left to the end on the right.
Figure 5.
PC Inhibitor Decreases C6 and Mixed C6/NR8383 Spheroids Invasion
C6 (i) and mixed C6/NR8383 (ii) spheroids were incubated with different concentrations of PC inhibitor (50, 100, 150 μM) or with vehicle (DMSO). Images of spheroids in the collagen matrix were taken every 24 h for 4 days. (A) Representative images of the invasion of C6 (i) spheroids and mixed C6/NR8383 (ii) spheroids in the collagen matrix incubated with DMSO or 150 μM PC inhibitor at day 0 and day 4. All images were acquired with an inverted light microscope at ×5 original magnification. Scale bars, 500 μm. (B) Graphic representation showing the percentage of spheroids invasion with different concentrations of PC inhibitor (0, 50, 100, or 150 μM). Spheroids invasion areas were acquired with in-house software. They are normalized for each day to the relative size of day 0 and transformed into the percentage of invasion. All results are representative of three independent experiments. Significant differences were identified using a Tukey’s multiple comparisons test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
The PC Inhibitor Reactivates Macrophages in Mixed Spheroids
A proteomic study was conducted on C6 and mixed C6/NR8383 spheroids in order to decipher the molecular impact of the PC inhibitor on the C6 glioma cancer cells and macrophages in a tumor microenvironment interaction. Spheroids were treated with 100 μM PC inhibitor for different time points (8, 16, and 72 h). Shotgun proteomic analysis yielded 1,299 protein identifications across all samples. 95 proteins were exclusive to the untreated C6 spheroids and 132 to the untreated mixed spheroids (Figure S5A). These proteins are implicated in different biological processes of tumorigenesis such as positive regulation of cell migration, as well as positive regulation of cell proliferation and the mitotic cell cycle (Figure S5B). Additionally, syntenin-1 positively regulates TGF-β1-mediated SMAD2/3 activation.47 Fourteen proteins were exclusively expressed by mixed spheroids after 72 h of stimulation with PC inhibitor. Among these proteins, TRPV2 is implicated in inflammatory processes and is essential to macrophage functions.48 Adam17 (a substrate of PC1/3) has been described as responsible for cleavage of IL-6R, TNFα, and pro-inflammatory factors.49 In addition, Gpnmb has been found in macrophages upon induction by IFN-γ or lipopolysaccharide (LPS) and was described as a regulator of the pro-inflammatory response.50 We then created two heatmaps (Figures 6A and 6B). Figure 6A represents the heatmap of the 146 proteins that showed a significant difference in LFQ expression between untreated C6 spheroids and C6 spheroids after 72 h of PC inhibitor. Figure 6B represents the heatmap with 187 proteins statistically significant between PC inhibitor-treated mixed spheroids and control mixed spheroids. A difference of protein expression is only observed after 72 h of PC inhibitor treatment for both types of spheroid. Two clusters can be highlighted for both heatmaps. 59 proteins for C6 spheroids (cluster 1, Figure 6A; Data S4) were found in the tumor microenvironment and involved in cell migration and invasion, similar to LAP3, upregulated in glioma or Nampt, known to be involved in M2 orientation and Eef1g, which has been described with a pro-tumorigenic role in brain tumor but also in liver, kidney, and prostate.51, 52, 53 In a same way, 91 proteins for C6/macrophage spheroids were found to be downregulated in cluster 1 at 72 h (Figure 6B; Data S5). Among these proteins, nestin expression is related to poor prognosis in high-grade gliomas, and ACLY (ATP citrate lyase) has been described as a positive regulator of glycolysis in glioblastomas.54,55 Likewise, knocking down nucleolin expression in glioma decreased tumor growth and triggered cell cycle arrest.56 Proteins involved in the inflammatory response and macrophage orientation were also found in this cluster. ROCK2 inhibition seems to decreases M2-like macrophage orientation, and PDLIM1 acts as an inhibitor of NF-κB-mediated inflammation.57,58 Some proteins were found in cluster 1 for both C6 spheroids and mixed C6/macrophage spheroids. Plod1 and Plod2 modulate collagen cross-linking and maturation and thereby contribute to cancer progression.59 Lrrfip1, an inhibitor of NF-κB signaling, has been described as a potential target gene for anti-glioma therapy.60 All of these proteins involved in cancer progression and in the maintenance of tumor microenvironment were under-expressed after 72 h of PC inhibitor treatment (Figure 6C). Alternatively, 87 proteins were found upregulated in cluster 2 for C6 spheroids (Figure 6A). Among these proteins, tenascin-R has been found in low-grade glioma with non-invasive behavior. A decrease in tenascin-R expression is correlated with increasing malignancy. In fact, grade IV glioblastoma has a low level of tenascin-R expression.61 Smad3, described before in proteomic analysis of C6 cells, was also found. In contrast, granulins have been described as an amplifier of acute inflammation and they stimulate the production of neutrophil-attracting chemokines.62 Also, flotillin-1 could have a role in CXCR4-mediated T cell migration, adhesion, and signaling.63 For mixed C6/macrophage spheroids, 92 proteins were found in cluster 2 (Figure 6B). Some markers of activated macrophages and pro-inflammatory response were identified like calectin-3 or Icam1 described as a suppressor of M2 macrophages orientation. Ccdc22, an activator of NF-κB signaling, was also found.64,65 Eps8 protein, involved in phagocytosis by increasing TLR4-Myd88 interaction and Sdha protein, essential for pro-inflammatory polarization of macrophages, were both found in cluster 2 for C6 spheroids and mixed C6/macrophage spheroids,66,67 as well as annexin A1, essential in the innate immune response and inflammatory process and Lta4h, involved in the pro-inflammatory response. All of these immune and inflammatory-related proteins were overexpressed after 72 h of stimulation with PC inhibitor (Figures 6A and 6B). In conclusion, the mixed spheroids treated with the inhibitor are progressing toward a pro-inflammatory response.
Figure 6.
PC Inhibitor Triggers Molecular Changes in Protein Profiles of C6 Spheroid and Mixed C6/NR8383 Spheroids
(A and B) C6 spheroids (A) and mixed C6/NR8383 spheroids (B) were treated with 100 μM PC inhibitor or with DMSO (control) and lysed before FASP and LC-MS/MS analysis. MaxQuant and Perseus software were used for the statistical analysis, and heatmaps were generated to show proteins that were significantly different between treated and untreated spheroids. Different clusters are highlighted. Biological process for clusters of spheroids C6 and C6/NR8383. The analysis was performed with FunRich.
Taken together, based on these proteomics data, signaling pathways related to cancer are modulated under PC inhibition. Moreover, markers of the pro-inflammatory phenotype were also identified in the mixed spheroids condition after inhibitor treatment and can suggest the activation of macrophages.
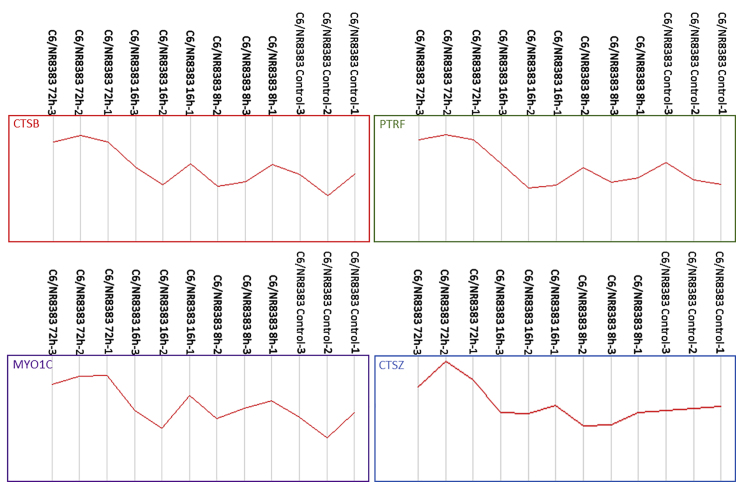
In the same line, cross-analyses were performed between the identified proteins in C6/macrophages at 72 h after treatment and the 104 key genes that were shared between human glioblastoma multiforme (GBM) and lower-grade glioma (LGG), with significant correlation with patients’ survival. This selected list was based on next-generation sequencing data obtained from The Cancer Genome Atlas for gene expression analysis.68 Four proteins have been identified in common (cathepsin B [CTSB], cathepsin Z [CTSZ], myosin 1C [MYO1C], and polymerase I transcript releasing factor [PTRF]) (Figure 7). CTSZ is identified from the list of 10 genes (CTSZ, EFEMP2, ITGA5, KDELR2, MDK, MICALL2, MAP2K3, PLAUR, SERPINE1, and SOCS3) that can potentially serve as indicators to estimate the prognosis of patients with gliomas.68 This gene has been shown to be a good patient survival indicator between GBM and LGG.
Figure 7.
PC Inhibitor Increases the Expression of Biomarkers Related to Glioma Survival in Mixed C6/NR8383 Spheroids
Mixed C6/NR8383 spheroids were treated with 100 μM PC inhibitor for different times (8, 16, and 72 h) or with DMSO (control) and lysed before FASP and LC-MS/MS analysis. MaxQuant and Perseus software were used for the statistical analysis. Graphs represent LFQ intensities showing expression of cathepsin B (CTSB), polymerase I and transcript release factor (PRTF), unconventional myosin-Ic (MYO1C), and cathepsin Z (CTSZ) between mixed C6/NR8383 spheroids treated or not with PC inhibitor are shown. The genes are involved in GBM and LGG patients’ survival. All of these proteins are overexpressed after 72 h of treatment with PC inhibitor.
The PC inhibitor Is More Effective Than TMZ in Reducing the Density of Glioma Cells and Inhibiting C6 Spheroid Growth and Invasion
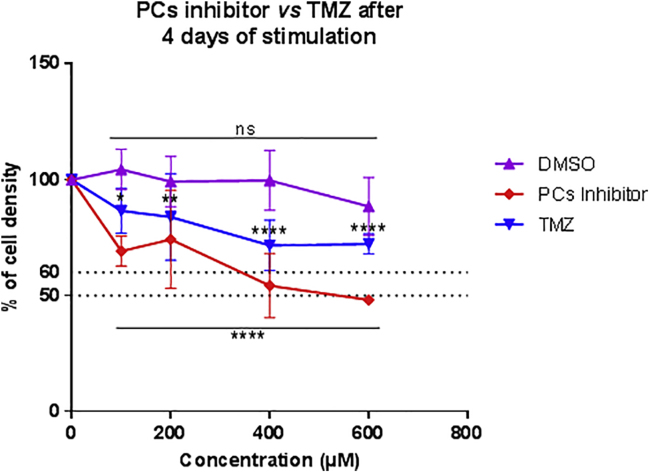
TMZ is one of the main drugs used to treat high-grade gliomas.69 Therefore, in the context of therapy, we wanted to compare the potency of TMZ and PC inhibitor to decrease C6 rat glioma cell density after 4 days of treatment (Figure 8). To determine their respective 50% inhibitory concentrations (IC50s) in vitro, we performed dose-response studies. TMZ and PC inhibitor were dissolved in DMSO. Therefore, for each concentration tested, the same percentage of DMSO was added in control conditions. An inhibitory effect of this compound on C6 rat glioma cell density was only observed for the 600 μM condition. It reduced glioma cell density by 11.5%. This value was subtracted from the values obtained after the treatment of C6 cells with 600 μM TMZ or PC inhibitor (Figure 8). Therefore, only the direct effects exerted by each drug are represented in the graph. These data revealed that TMZ treatment significantly decreased C6 cell density at every concentration tested. The treatment with 100, 200, 400, and 600 μM TMZ induced a diminution of 13.4%, 16.1%, 25.2%, and 27%, respectively (Figure 8). Of note, the IC50 was not attained even after a challenge with a higher concentration, such as 600 μM. The PC inhibitor also significantly reduced C6 cell density at each concentration used. The treatment with 100, 200, 400, and 600 μM PC inhibitor led to a decrease of 30.8%, 25.8%, 47.5%, and 50.2%, respectively (Figure 8), of tumor growth. In comparison with TMZ, the magnitude of decrease of C6 cell density induced by the PC inhibitor was higher for all of the concentrations tested. Moreover, the IC50 was reached after a challenge with 600 μM PC inhibitor. This demonstrated that a lower concentration of PC inhibitor was needed to diminish the density of glioma cells compared to TMZ.
Figure 8.
A Lower Concentration of PC Inhibitor Is Needed to Decrease C6 Cell Density Compared to Temozolomide
The cell density of C6 rat glioma cells was determined by an MTS assay after 4 days of stimulation with different concentrations of PC inhibitor or temozolomide (TMZ) (100, 200, 400, and 600 μM). All results are representative of three independent experiments. Significant differences were identified using Tukey’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. ns, not significant. The cell density of NR8383 cells was determined by an MTS assay for 48, 72, and 96 h with 300 μM PC inhibitor and 600 μM TMZ.
Next, we wanted to compare the effect of TMZ and PC inhibitor on spheroid growth and invasion after 4 days of treatment (Table 1). This experiment was conducted on spheroids containing only C6 cells and on spheroids containing a mix of C6 and NR8383 cells. For the PC inhibitor, we used a concentration of 100 μM since it was efficient to decrease C6 cell density and inhibit spheroid growth and invasion (Figures 2 and 6) without any effect of the DMSO on macrophage survival (Figure 1). The concentration chosen for TMZ was 600 μM since it induced the more important decrease in C6 cell density (Figure 6A). PC inhibitor treatment led to a stronger diminution of C6 spheroid growth and invasion than with TMZ. Indeed, they were reduced by 52.34% with PC inhibitor challenge whereas they were decreased by 25.63% after TMZ treatment. On C6/NR8383 spheroids, the protective effect of macrophages to C6 cells against PC inhibitor was again observed since growth and invasion were only reduced by 38.38%. Conversely, the presence of macrophages did not drop the effect of TMZ since a diminution of 26.16% was found. However, at this concentration, we cannot exclude that DMSO impacted the physiology of macrophages. Altogether, this showed that PC inhibitor inhibited spheroid growth and invasion more efficiently than did TMZ.
Table 1.
PC Inhibitor Is More Efficient than Temozolomide (TMZ) on the Spheroid Invasion Decrease
| Mean of the Percentage of Invasion after 4 Days of Stimulation |
||
|---|---|---|
| 100 μM PC Inhibitor | 600 μM TMZ | |
| C6 spheroids | 47.66% ± 16.41% | 74.37% ± 9.06% |
| p < 0.0001 | p = 0.0037 | |
| Mixed C6/macrophages spheroids | 61.62% ± 6.41% | 73.84% ± 5.49% |
| p < 0.0001 | p < 0.0001 | |
C6 and mixed C6/NR8383 spheroids were incubated with 100 μM PC inhibitor or with 600 μM TMZ. All images were acquired with an inverted light microscope at ×5 original magnification. The percentages of spheroids invasion are shown. All results are representative of three independent experiments. Significant differences were identified using a Tukey’s multiple comparisons test. PC, proprotein convertase; TMZ, temozolomide.
Discussion
In the present study, we have demonstrated that the PC inhibitor acts on macrophage activation, as demonstrated by the proteomic study, without affecting their viability. Some pro-inflammatory phenotype markers were found to be secreted by macrophages after PC inhibitor treatment, such as CCL6 and PRDX5, suggesting that the inhibitor was also able to induce the activation of macrophages as previously described for the NR8383 PC1/3 KD macrophages.19 Moreover, some of the factors secreted by the PC inhibitor-treated macrophages have anti-tumoral effects. For example, FKBP1A, known to have antitumor properties, has been detected.35 The inhibitor seems to orient macrophages toward an anti-tumoral phenotype. Interestingly, the combination of the PC inhibitor and the factors secreted by PC inhibitor-treated macrophages seems to have a cumulative effect by inhibiting C6 glioma cell viability. In addition to influencing macrophages, the inhibitor presents a strong anti-proliferative activity toward the C6 glioma cells directly. It decreases their viability already at 50 μM. The C6 cells are deeply affected by the inhibitor since their intracellular and secreted protein profiles are completely different. In fact, proteins related to cell growth and viability are downregulated. Some anti-tumoral factors are produced by the glioma cells in the presence of PC inhibitors such as latexin, or angiogenesis inhibitors such as UNC95B. Additionally, factors known to be involved in tumor cell migration, tumor growth, and metastasis, such as CTGF, CSPG4, and ADAMTS1, are under-expressed in C6 glioma cells such as some pro-tumoral factors, including CIAPIN1, USP7, and COMP. Proteins involved in immune cell recruitment and activation and extracellular matrix remodeling are also less secreted in the presence of the inhibitor. In fact, CSF1 protein is produced by tumor cells and released in the tumor microenvironment where its main role is the recruitment of macrophages and the promotion of their pro-tumoral phenotype.15 The production of this protein is inhibited in C6 glioma cells under PC inhibitor treatment. Therefore, the inhibitor may modulate the glioma cell microenvironment by regulating the secretion profile of tumor cells. These results suggest that the inhibitor may act both on cancer cells and macrophages in an anti-cancer therapeutic strategy.
Furthermore, a time course proteomic study was realized on C6 and mixed C6/NR8383 spheroids treated or not with the PC inhibitor. This study allowed us to decipher the molecular impact of the PC inhibitor on C6 glioma cancer cells and macrophages in a tumor microenvironment interaction. In this context, the cross-analyses between all proteomic studies revealed a clear differentiation between C6/macrophages treated with the PC inhibitor at 72 h from the other conditions (Figure 6). FunRich analyses have confirmed the presence of specific proteins involved in immune response reactivation such as Th1 cell activation, phagocytosis, and T cell activation via major histocompatibility complex (MHC) presentation. In the same way, compared to C6 alone, C6/macrophage spheroids present a decrease in cell adhesion, response to hypoxia, and no regulation to TGF-β3 production, confirming the reactivation of the immune cells in the tumor microenvironment. CTSZ and PLAUR were also identified in the mixed spheroids and can potentially serve as indicators to estimate the prognosis of patients with gliomas.68 These two genes have been shown to be good patient survival indicators between GBM and LGG.
Collectively, in this work, we have established that the PC inhibitor exerts a dual activity at the same time by its anti-tumoral effect on glioma cells and its ability to switch macrophages to an anti-tumoral phenotype. In this way, we can imagine controlling at a distance macrophage reactivation by using a combination of the PC inhibitor and TLR agonists as previously demonstrated.17,18,20
Materials and Methods
Experimental Design and Statistical Rationale
Shotgun proteomics experiments were conducted in biological triplicates (n = 3). Spheroid studies were conducted in biological triplicates (n = 3), as were all biological assays. For the proteomics statistical analysis, extracted proteins or secreted media proteins presenting as significant by the ANOVA test analysis were used (p < 0.05). Normalization was achieved using a Z score with matrix access by rows. For cell density and invasion tests, results obtained were depicted through a boxplot figure. Significant differences were identified using a Tukey’s multiple comparisons test (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001).
Chemicals and Reagents
Water (H2O), formic acid, acetonitrile (ACN), and trifluoroacetic acid (TFA) were obtained from Biosolve (Dieuze, France). 2,5-dihydroxybenzoic acid (DHB), a ProteoMass MALDI (matrix-assisted laser desorption/ionization) calibration kit, dl-dithiothreitol (DTT), thiourea, TMZ, PC inhibitor (ref. 537076), and iodoacetamide were purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France). Lys-C/trypsin was purchased from Promega (Charbonnieres, France). Polylysine-coated slides, Dulbecco’s modified Eagle’s medium (DMEM), Ham’s F12K medium, heat-inactivated fetal bovine serum (FBS), trypsin, phosphate-buffered saline (PBS), and penicillin and streptomycin were purchased from Thermo Scientific (Braunschweig, Germany). NR8383 is a rat alveolar macrophage cell line (CRL-2192) obtained from the ATCC (USA). The rat C6 glioma cell line was kindly provided by Dr. Bernd Kaina (Institute of Toxicology, University Medical Center, Mainz, Germany).
Cell Culture
Rat alveolar wild-type NR8383 macrophages19 were cultured in Ham’s F12K medium supplemented with 15% FBS and 1% penicillin/streptomycin (100 U/mL). Human THP1 macrophages were cultured in RPMI 1640 medium supplemented with 10% FBS, 1% l-glutamine (2 mM), and 1% penicillin/streptomycin (100 U/mL). Rat glioma C6 cells were cultured in DMEM and supplemented with 10% FBS, 1% l-glutamine (2 mM), and 1% penicillin/streptomycin (100 U/mL). All cell lines were cultured at 37°C in a humidified atmosphere (5% CO2).
RNA Extraction and PCR
The RNA of NR8383 macrophages and C6 glioma cells were extracted with TRIzol reagent. 1 μg of RNA was reverse transcribed and reverse transcriptase-polymerase chain reaction (RT-PCR) was carried out to determine the expression of PCs. The primers used were as follows: (1) actin, forward 5′-TTGTAACCAACTGGGACGATATGG-3′, reverse 5′-GATCTTGATCTTCATGGTGCTAGG-3′, (2) PC1/3, forward 5′-TTGGCTGAAAGGGAAAGG-3′, reverse 5′-ATCTTTGATGATTGCTTTG-3′, (3) furin, forward 5′-CTATGGCTACGGGCTGTTGG-3′, reverse 5′-CCTCGCTGGTATTTTCAATCTC-3′, (4) PACE4, forward 5′-TATGGATTTGGCTTGGTGGATG-3′, reverse 5′-GGCTCCATTCTTTCAACTTTCC-3′, (5) PC4, forward 5′-CTTGTGGCCATCAGACCCTTG-3′, reverse 5′-GAACAGGCAGTGTAGTCGCTG-3′, and (6) PC5/6, forward 5′-AGTGCGCTCCATCTACAAAGC-3′, reverse 5′-GTCAGTGCAGTGATCCGGTC-3′.
Cell Supernatant Collection and Total Protein Extraction
NR8383 and C6 cells were plated on sterile 24-well plates and cultured until they reached confluence. THP1 was plated on sterile six-well plates and differentiated with 10 ng/mL phorbol 12-myristate 13-acetate (PMA) for 24 h. For stimulation, cells were starved overnight in Ham’s F12K medium, DMEM, or RMPI 1640 medium supplemented with 2% FBS. Cells were then stimulated in a serum-free medium with different concentrations of PC inhibitor (50, 100, or 150 μM) with or without DMSO. At 24 h, cell supernatants were centrifuged at 500 × g and passed through a 0.22-μm filter to remove cells and debris. Four hundred microliters of the cell supernatant was collected for each condition. Cells were washed three times with ice-cold PBS and then lysed with radioimmunoprecipitation assay (RIPA) buffer for total protein extraction (150 mM NaCl, 50 mM Tris, 5 mM EGTA, 2 mM EDTA, 100 mM NaF, 10 mM sodium pyrophosphate, 1% Nonidet P-40 (NP-40), 1 mM PMSF, and 1× protease inhibitors). After three 30-s sonications, cell debris was removed by centrifugation (16,000 × g, 10 min, 4°C), supernatants were collected, and protein concentrations were measured using a Bio-Rad protein assay kit, according to the manufacturer’s instructions.
Filter-Aided Sample Preparation
The samples were processed using a shotgun bottom-up proteomic approach. Total protein extract (0.04 mg) was used for filter-aided sample preparation (FASP) digestion as described previously.70, 71, 72 We performed FASP using Microcon YM-10 devices (Millipore) before adding Lys-C trypsin (Promega) for protein digestion (40 μg/mL in 0.05 M Tris-HCl). An equivalent volume of reduction solution (0.1 M DTT) was added to each sample followed by an incubation step at 95°C for 15 min. Then the samples were processed following the FASP protocol using a filter with a nominal molecular mass limit of 10,000 Da (Amicon Ultra-0.5 10K, Millipore). Briefly, each sample was mixed with 200 μL of denaturant buffer (8 M urea, 0.1 M Tris/HCl [pH 8.5]) and transferred to FASP filters. The samples were centrifuged at 14,000 × g, 20°C, for 15 min. For the alkylation step, 100 μL of 0.05 M iodoacetamide in denaturant buffer was added to each sample, followed by incubation in the dark for 20 min at room temperature. Samples were washed twice with 100 μL of denaturant buffer followed by two washes with 100 μL of 0.05 M ammonium bicarbonate (AB) buffer. After each washing step, centrifugation was performed at 14,000 × g, 20°C, for 15 min. The proteins were digested by adding 40 μL of trypsin at 40 μg/mL in AB buffer and then incubated at 37°C overnight. The peptides were eluted by adding 50 μL of saline solution (0.5 M NaCl) and centrifuged at 14,000 × g, 20°C, for 15 min. The digestion was stopped by adding 10 μL of 5% TFA. The samples were desalted using ZipTip C-18 (Millipore) and eluted with a solution of ACN/0.1% TFA (7:3, v/v). The samples were dried with a SpeedVac and resuspended in 20 μL of ACN/0.1% formic acid (0.2:9.8, v/v) just before processing using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Experiments were done in biological triplicates (n = 3).
Proteomics Analysis of Cell Supernatants
Supernatant volumes obtained from the NR8383 and THP1 macrophages and C6 cells treated with PC inhibitor or DMSO were reduced to 100 μL in a SpeedVac. Cell supernatants were denatured with 2 M urea in 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 8.0) by sonication on ice. The proteins were reduced with 10 mM DTT for 40 min at 56°C followed by alkylation with 55 mM iodoacetamide for 40 min in the dark. The iodoacetamide was quenched with 100 mM thiourea. The proteins were digested with 1 μg Lys-C/trypsin mixture (Promega) overnight at 37°C. The digestion was stopped with 0.5% TFA. The samples were desalted using ZipTip C-18 (Millipore) and eluted with a solution of ACN/0.1% TFA (7:3, v/v). The samples were dried with SpeedVac and resuspended in 20 μL of ACN/0.1% formic acid (0.2:9.8, v/v) just before processing using LC-MS/MS. Experiments were done in biological triplicates (n = 3).
LC-MS/MS Analysis
Mass spectrometry proteomics analysis of digested proteins was performed using a nanoAcquity UPLC (ultra-performance LC) system (Waters) coupled with the Q Exactive Orbitrap mass spectrometer (Thermo Scientific) via a nanoelectrospray source. The samples were separated by means of online reversed-phase LC chromatography, using a preconcentration column (nanoAcquity Symmetry C18, 5 μm, 180 μm × 20 mm) and an analytical column (nanoAcquity BEH C18, 1.7 μm, 75 μm × 250 mm). The peptides were separated by applying a linear gradient of ACN in 0.1% formic acid (5%–35%) for 2 h, at a flow rate of 300 nL/min. The Q-Exactive instrument was operated in a data-dependent mode defined to analyze the 10 most intense ions of MS analysis (top 10). The MS analysis was performed with an m/z mass range between 300 and 1,600, a resolution of 70,000 full width at half maximum (FWHM), an automatic gain control (AGC) of 3e6 ions, and a maximum injection time of 120 ms. The MS/MS analysis was performed with an m/z mass range between 200 and 2,000, an AGC of 5e4 ions, a maximum injection time of 60 ms, and a resolution set at 17,500 FWHM.
Protein ID and Data Analysis
Proteins were identified by comparing all MS/MS data with the proteome database of the complete reviewed proteome of Rattus norvegicus (UniProt: R2018_07, release July 2018, 8,054 entries), using MaxQuant software version 1.6.1.0.73,74 Lys-C trypsin specificity was used for the digestion mode with two missed cleavages. The carbamidomethylation of cysteines was set as a fixed modification. N-terminal acetylation and methionine oxidation were selected as the variable modifications. For MS spectra, an initial mass tolerance of 6 ppm was selected, and the MS/MS tolerance was set to 20 ppm for higher energy collisional dissociation (HCD) data.75 For identification, the false discovery rate (FDR) at the peptide spectrum matches (PSMs) and protein level was set to 0.01. Relative, label-free quantification of proteins was performed using the MaxLFQ algorithm integrated into MaxQuant with the default parameters.76 Analysis of the proteins identified was performed using Perseus software (version 1.6.2.1, http://www.perseus-framework.org/).77 The file containing the information from identification was used with hits to the reverse database, and proteins identified with modified peptides and potential contaminants were removed. Then, the LFQ intensity was logarithmized (log2[x]). Categorical annotation of rows was used to define different groups depending on the concentration of the PC inhibitor used (0, 50, 100, or 150 μM). Multiple-sample tests were performed using an ANOVA test with a p value of 0.05 and preserved grouping in randomization. The results were normalized by Z score and represented as hierarchical clustering. Functional annotation and characterization of identified proteins were obtained using PANTHER software (version 14.0, http://www.pantherdb.org) and STRING (version 10.5, http://string-db.org).
Sub-network Enrichment Pathway Analysis
Using Elsevier’s Pathway Studio (version 11.0), all relationships between the differentially expressed proteins among all conditions were depicted based on the Ariadne ResNet.78 For proteins identified in the shotgun analysis after stimulation of cell lines with the PC inhibitor, the subnetwork enrichment analysis (SNEA) algorithm was used to detect the statistically significant altered biological pathways in which the identified proteins are involved. This algorithm uses a Fisher’s statistical test to detect any non-random associations between two categorical variables organized by a specific relationship. Also, this algorithm starts by creating a central “seed” from all of the relevant identities in the database and builds connections with associated entities based on their relationship with the seed. SNEA compares the sub-network distribution to the background distribution using a one-sided Mann-Whitney U test and calculates a p value, thereby representing statistical significance between different distributions. In all analyses that we performed, the GenBank ID was used to form experimental groups based on the different conditions present for analysis. The pathway networks were reconstructed based on biological processes and molecular functions for every single protein, along with its associated targets.
Data and Software Availability
Cancer cell and macrophage proteomics data, including MaxQuant files and annotated MS/MS, have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014679 (user name, reviewer94484@ebi.ac.uk; password, gb85XYvq).
Biological Assays
Generation of NR8383 Supernatants
NR8383 macrophages were treated with or without 100 μM PC inhibitor in complete Ham’s F12K medium for 24 h. At 24 h, cell supernatants were centrifuged at 200 × g and passed through a 0.22-μm filter to remove cells and debris. Supernatants were then used for C6 stimulation.
Cell Density Measured by MTS Assay
C6 or NR8383 cells were seeded into 96-well plates at 30% confluence with different concentrations of the PC inhibitor (0, 0.5, 1, 10, 50, 100, 150, 200, and 400 μM) or with different concentrations of TMZ (0, 100, 200, 400, and 600 μM) or with an equal volume of DMSO. Also, C6 glioma cells were seeded into 96-well plates at 30% confluence with NR8383 supernatants obtained after 24 h of PC inhibitor or DMSO stimulation. These supernatants were supplemented or not with 100 μM PC inhibitor. The assay was observed at 24, 48, 72, or 96 h. CellTiter 96 AQueous One Solution cell proliferation reagent (Promega, Madison, WI, USA) was added to the wells and incubated at 37°C for 1 h protected from light. The absorbance was recorded at 490 nm using a 96-well plate reader. Absorbance values were normalized for each day to the absorbance values measured at day 0 and expressed in percentage of viability. Experiments were performed in biological triplicates (n = 3).
Spheroid Generation and Embedding in a Collagen Matrix
C6 rat glioma cells associated with or without NR8383 cells were resuspended in complete Ham’s F12K medium supplemented with 5% of a collagen mixture at the final concentration of 8,000 cells of each cell line in 20 μl. The collagen mixture was prepared by mixing 2 mL of PureCol bovine collagen type I solution (3 mg/mL; Advanced BioMatrix) with 250 μL of 10× minimal essential medium (MEM) (Sigma- Aldrich) and 500 μL of 0.1 M sodium hydroxide. Cells were cultivated using the hanging drop technique on the lid of a Petri dish with PBS during 72 h at 37°C in a humidified atmosphere (5% CO2). The newly formed C6 spheroids and mixed C6/NR8383 spheroids were then implanted in the center of each well of a 24-well plate coated with a collagen mixture described before (one spheroid per well in 400 μL of collagen mixture per well). After cell spheroid embedding, the plate was incubated for 30 min at standard culture conditions to solidify the collagen. After that, 400 μL of complete Ham’s F12K medium with different concentrations of the PC inhibitor (50, 100, and 150 μM) was overlaid on the collagen matrix in each well. The complete system was incubated for a total of 4 days. Experiments were performed in biological triplicates (n = 3).
Quantification of Spheroid Size and Invaded Area
After the spheroids were embedded, cell invasion of the spheroid was monitored by digital photography using a Leica DM IL LED Fluo inverted light microscope (Leica DFC450C camera) at room temperature, with the Leica Application Suite (LAS, version 4.4). Images were acquired every day (day 0 represents time of embedding in collagen; images were taken immediately after embedding) using a 4×/0.10 objective. Image processing and quantification of spheroids and invasion areas were performed using in-house software. This in-house software takes into account cell density and not the limits of cell migration in the collagen matrix, which is observed independently. The implemented algorithm uses local fluctuations of the image intensity for automated estimation of the invasion magnitude. It is robust enough to handle micrographs of different generation methods and various qualities without the concept of an invasive front of the spheroids.18,45 Data on areas are normalized for each day to the relative size of day 0 and transformed into the percentage of invasion.
MALDI Mass Spectrometry Imaging (MSI) and LC-MS/MS Analysis of Spheroids
Mixed C6/NR8383 spheroids were generated as described before. Spheroids were treated with 100 μM PC inhibitor for 8, 16, 24, 48, and 72 h. After stimulation, they were rinsed with PBS before fixation with 4% paraformaldehyde during 1 h at 4°C and then washed three times in PBS. They were implanted in 175 mg/mL gelatin and frozen at −20°C and −80°C. The entire spheroids were cut into 8-μm sections using a cryostat (Leica Microsystems, Nanterre, France). Sections, obtained after every 40 μm (approximately), were subjected to MSI. These were mounted by finger-thawing on indium tin oxide (ITO)-coated slides. DHB was used as a matrix and was prepared at a concentration of 20 mg/mL in 70:30 methanol/0.1% TFA in H2O. Eight layers of the matrix were deposited using HTX TM-Sprayer (HTX Technologies, Chapel Hill, NC, USA) programmed to spray at a flow rate of 0.125 mL/min. Lipid imaging was performed on a rapifleX MALDI Tissue typer instrument (Bruker Daltonics, Bremen, Germany). The instrument was equipped with a smartbeam 3D laser and was controlled using FlexControl version 4.0 software (Bruker Daltonics). The datasets were recorded in positive reflector mode, and 300 laser shots were accumulated for each raster point at a laser frequency of 10 kHz. Spectra were acquired at a lateral resolution of 10 μm. External calibration was performed using the PepMix standard (Bruker Daltonics). Spectra were acquired between an m/z of 500 and 1,300. For reconstruction of images, the SCiLS Lab software version 2015b (SCiLS, Bremen, Germany) was used.79,80 The data were normalized based on the total ion count (TIC) method.81 For proteomic analysis, C6 and mixed C6/NR8383 spheroids were stimulated with 100 μM PC inhibitor for 8, 16, and 72 h. After stimulation, they were washed three times in PBS. Then, total protein extraction and FASP digestion were performed as described above before the LC-MS/MS analysis.
Author Contributions
Conceptualization, F.R. and M.S.; Methodology, M.D., F.R, M.R., and M.S.; Validation, M.D. and M.R.; Formal Analysis, M.S., F.R., and M.D.; Resources, M.S. and I.F.; Data Curation, M.D., M.S., S.A., F.R., and M.R.; Writing, M.D., M.S., F.R., and M.R.; Writing – Original Draft, M.S. and M.D. Writing – Review & Editing, M.S. M.D., F.R., and I.F.; Supervision, F.R. and M.S.; Project Administration, M.S.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the funding from the Ministère de l’enseignement Supérieur de la Recherche et de l’innovation (MESRI), the Institut National de la Santé et de la Recherche Médicale (INSERM), and the Université de Lille.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.03.005
Contributor Information
Marie Duhamel, Email: marie.duhamel@univ-lille.fr.
Franck Rodet, Email: franck.rodet@univ-lille.fr.
Michel Salzet, Email: michel.salzet@univ-lille.fr.
Supplemental Information
References
- 1.Jaaks P., Bernasconi M. The proprotein convertase furin in tumour progression. Int. J. Cancer. 2017;141:654–663. doi: 10.1002/ijc.30714. [DOI] [PubMed] [Google Scholar]
- 2.Seidah N.G. The proprotein convertases in health and disease. Qatar Found. Annu. Res. Forum Proc. 2012;2012 AESNP11. 10.5339/qfarf.2012.AESNP11#abstract_content. [Google Scholar]
- 3.Day R., Salzet M. The neuroendocrine phenotype, cellular plasticity, and the search for genetic switches: redefining the diffuse neuroendocrine system. Neuroendocrinol. Lett. 2002;23:447–451. [PubMed] [Google Scholar]
- 4.Duckert P., Brunak S., Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 5.Scamuffa N., Calvo F., Chrétien M., Seidah N.G., Khatib A.-M. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 6.Martín M.G., Lindberg I., Solorzano-Vargas R.S., Wang J., Avitzur Y., Bandsma R., Sokollik C., Lawrence S., Pickett L.A., Chen Z. Congenital proprotein convertase 1/3 deficiency causes malabsorptive diarrhea and other endocrinopathies in a pediatric cohort. Gastroenterology. 2013;145:138–148. doi: 10.1053/j.gastro.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcinkiewicz M. βAPP and furin mRNA concentrates in immature senile plaques in the brain of Alzheimer patients. J. Neuropathol. Exp. Neurol. 2002;61:815–829. doi: 10.1093/jnen/61.9.915. [DOI] [PubMed] [Google Scholar]
- 8.Khatib A.-M., Siegfried G., Chrétien M., Metrakos P., Seidah N.G. Proprotein convertases in tumor progression and malignancy: novel targets in cancer therapy. Am. J. Pathol. 2002;160:1921–1935. doi: 10.1016/S0002-9440(10)61140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J., Bassi D.E., Zhang J., Li T., Nicolas E., Klein-Szanto A.J. Transgenic overexpression of the proprotein convertase furin enhances skin tumor growth. Neoplasia. 2012;14:271–282. doi: 10.1593/neo.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 13.Housman G., Byler S., Heerboth S., Lapinska K., Longacre M., Snyder N., Sarkar S. Drug resistance in cancer: an overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 15.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., Cancer Genome Atlas Research Network The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellman I., Coukos G., Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duhamel M., Rodet F., Murgoci A., Wisztorski M., Day R., Fournier I., Salzet M. Proprotein convertase 1/3 inhibited macrophages: a novel therapeutic based on drone macrophages. EuPA Open Proteom. 2016;11:20–22. doi: 10.1016/j.euprot.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duhamel M., Rose M., Rodet F., Murgoci A.N., Zografidou L., Régnier-Vigouroux A., Abeele F.V., Kobeissy F., Nataf S., Pays L. Paclitaxel treatment and proprotein convertase 1/3 (PC1/3) knockdown in macrophages is a promising antiglioma strategy as revealed by proteomics and cytotoxicity studies. Mol. Cell. Proteomics. 2018;17:1126–1143. doi: 10.1074/mcp.RA117.000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duhamel M., Rodet F., Delhem N., Vanden Abeele F., Kobeissy F., Nataf S., Pays L., Desjardins R., Gagnon H., Wisztorski M. Molecular consequences of proprotein convertase 1/3 (PC1/3) inhibition in macrophages for application to cancer immunotherapy: a proteomic study. Mol. Cell. Proteomics. 2015;14:2857–2877. doi: 10.1074/mcp.M115.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duhamel M., Rodet F., Murgoci A.N., Desjardins R., Gagnon H., Wisztorski M., Fournier I., Day R., Salzet M. The proprotein convertase PC1/3 regulates TLR9 trafficking and the associated signaling pathways. Sci. Rep. 2016;6:19360. doi: 10.1038/srep19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Refaie S., Gagnon S., Gagnon H., Desjardins R., D’Anjou F., D’Orléans-Juste P., Zhu X., Steiner D.F., Seidah N.G., Lazure C. Disruption of proprotein convertase 1/3 (PC1/3) expression in mice causes innate immune defects and uncontrolled cytokine secretion. J. Biol. Chem. 2012;287:14703–14717. doi: 10.1074/jbc.M111.323220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagnon H., Refaie S., Gagnon S., Desjardins R., Salzet M., Day R. Proprotein convertase 1/3 (PC1/3) in the rat alveolar macrophage cell line NR8383: localization, trafficking and effects on cytokine secretion. PLoS ONE. 2013;8:e61557. doi: 10.1371/journal.pone.0061557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hipp M.M., Shepherd D., Booth S., Waithe D., Reis e Sousa C., Cerundolo V. The processed amino-terminal fragment of human TLR7 acts as a chaperone to direct human TLR7 into endosomes. J. Immunol. 2015;194:5417–5425. doi: 10.4049/jimmunol.1402703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii N., Funami K., Tatematsu M., Seya T., Matsumoto M. Endosomal localization of TLR8 confers distinctive proteolytic processing on human myeloid cells. J. Immunol. 2014;193:5118–5128. doi: 10.4049/jimmunol.1401375. [DOI] [PubMed] [Google Scholar]
- 25.Coppola J.M., Bhojani M.S., Ross B.D., Rehemtulla A. A small-molecule furin inhibitor inhibits cancer cell motility and invasiveness. Neoplasia. 2008;10:363–370. doi: 10.1593/neo.08166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couture F., D’Anjou F., Desjardins R., Boudreau F., Day R. Role of proprotein convertases in prostate cancer progression. Neoplasia. 2012;14:1032–1042. doi: 10.1593/neo.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longuespée R., Couture F., Levesque C., Kwiatkowska A., Desjardins R., Gagnon S., Vergara D., Maffia M., Fournier I., Salzet M., Day R. Implications of proprotein convertases in ovarian cancer cell proliferation and tumor progression: insights for PACE4 as a therapeutic target. Transl. Oncol. 2014;7:410–419. doi: 10.1016/j.tranon.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couture F., Sabbagh R., Kwiatkowska A., Desjardins R., Guay S.P., Bouchard L., Day R. PACE4 undergoes an oncogenic alternative splicing switch in cancer. Cancer Res. 2017;77:6863–6879. doi: 10.1158/0008-5472.CAN-17-1397. [DOI] [PubMed] [Google Scholar]
- 29.Levesque C., Fugère M., Kwiatkowska A., Couture F., Desjardins R., Routhier S., Moussette P., Prahl A., Lammek B., Appel J.R. The Multi-Leu peptide inhibitor discriminates between PACE4 and furin and exhibits antiproliferative effects on prostate cancer cells. J. Med. Chem. 2012;55:10501–10511. doi: 10.1021/jm3011178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker G.L., Lu Y., Hardes K., Strehlow B., Levesque C., Lindberg I., Sandvig K., Bakowsky U., Day R., Garten W., Steinmetzer T. Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases. J. Biol. Chem. 2012;287:21992–22003. doi: 10.1074/jbc.M111.332643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levesque C., Couture F., Kwiatkowska A., Desjardins R., Guérin B., Neugebauer W.A., Day R. PACE4 inhibitors and their peptidomimetic analogs block prostate cancer tumor progression through quiescence induction, increased apoptosis and impaired neovascularisation. Oncotarget. 2015;6:3680–3693. doi: 10.18632/oncotarget.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwiatkowska A., Couture F., Ait-Mohand S., Desjardins R., Dory Y.L., Guérin B., Day R. Enhanced anti-tumor activity of the Multi-Leu peptide PACE4 inhibitor transformed into an albumin-bound tumor-targeting prodrug. Sci. Rep. 2019;9:2118. doi: 10.1038/s41598-018-37568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassi D.E., Zhang J., Renner C., Klein-Szanto A.J. Targeting proprotein convertases in furin-rich lung cancer cells results in decreased in vitro and in vivo growth. Mol. Carcinog. 2017;56:1182–1188. doi: 10.1002/mc.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fong S., Mounkes L., Liu Y., Maibaum M., Alonzo E., Desprez P.Y., Thor A.D., Kashani-Sabet M., Debs R.J. Functional identification of distinct sets of antitumor activities mediated by the FKBP gene family. Proc. Natl. Acad. Sci. USA. 2003;100:14253–14258. doi: 10.1073/pnas.2332307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Rhun E., Duhamel M., Wisztorski M., Gimeno J.P., Zairi F., Escande F., Reyns N., Kobeissy F., Maurage C.A., Salzet M., Fournier I. Evaluation of non-supervised MALDI mass spectrometry imaging combined with microproteomics for glioma grade III classification. Biochim. Biophys. Acta. Proteins Proteomics. 2017;1865:875–890. doi: 10.1016/j.bbapap.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Basang Z., Ding H., Lu Z., Ning T., Wei H., Cai H., Ke Y. Latexin expression is downregulated in human gastric carcinomas and exhibits tumor suppressor potential. BMC Cancer. 2011;11:121. doi: 10.1186/1471-2407-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Catalano A., Caprari P., Moretti S., Faronato M., Tamagnone L., Procopio A. Semaphorin-3A is expressed by tumor cells and alters T-cell signal transduction and function. Blood. 2006;107:3321–3329. doi: 10.1182/blood-2005-06-2445. [DOI] [PubMed] [Google Scholar]
- 39.Chakraborty G., Kumar S., Mishra R., Patil T.V., Kundu G.C. Semaphorin 3A suppresses tumor growth and metastasis in mice melanoma model. PLoS ONE. 2012;7:e33633. doi: 10.1371/journal.pone.0033633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larrivée B., Freitas C., Trombe M., Lv X., Delafarge B., Yuan L., Bouvrée K., Bréant C., Del Toro R., Bréchot N. Activation of the UNC5B receptor by Netrin-1 inhibits sprouting angiogenesis. Genes Dev. 2007;21:2433–2447. doi: 10.1101/gad.437807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B. A novel long noncoding RNA lncWDR26 suppresses the growth and metastasis of hepatocellular carcinoma cells through interaction with SIX3. Am. J. Cancer Res. 2018;8:688–698. [PMC free article] [PubMed] [Google Scholar]
- 42.Fussbroich B., Wagener N., Macher-Goeppinger S., Benner A., Fälth M., Sültmann H., Holzer A., Hoppe-Seyler K., Hoppe-Seyler F. EZH2 depletion blocks the proliferation of colon cancer cells. PLoS ONE. 2011;6:e21651. doi: 10.1371/journal.pone.0021651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Kuang W., Zhou Q., Zhang Y. TGF-β1 secreted by M2 phenotype macrophages enhances the stemness and migration of glioma cells via the SMAD2/3 signalling pathway. Int. J. Mol. Med. 2018;42:3395–3403. doi: 10.3892/ijmm.2018.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Anjou F., Routhier S., Perreault J.P., Latil A., Bonnel D., Fournier I., Salzet M., Day R. Molecular validation of PACE4 as a target in prostate cancer. Transl. Oncol. 2011;4:157–172. doi: 10.1593/tlo.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cisneros Castillo L.R., Oancea A.-D., Stüllein C., Régnier-Vigouroux A. A novel computer-assisted approach to evaluate multicellular tumor spheroid invasion assay. Sci. Rep. 2016;6:35099. doi: 10.1038/srep35099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantovani A., Schioppa T., Porta C., Allavena P., Sica A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 2006;25:315–322. doi: 10.1007/s10555-006-9001-7. [DOI] [PubMed] [Google Scholar]
- 47.Hwangbo C., Tae N., Lee S., Kim O., Park O.K., Kim J., Kwon S.H., Lee J.H. Syntenin regulates TGF-β1-induced Smad activation and the epithelial-to-mesenchymal transition by inhibiting caveolin-mediated TGF-β type I receptor internalization. Oncogene. 2016;35:389–401. doi: 10.1038/onc.2015.100. [DOI] [PubMed] [Google Scholar]
- 48.Link T.M., Park U., Vonakis B.M., Raben D.M., Soloski M.J., Caterina M.J. TRPV2 has a pivotal role in macrophage particle binding and phagocytosis. Nat. Immunol. 2010;11:232–239. doi: 10.1038/ni.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zunke F., Rose-John S. The shedding protease ADAM17: physiology and pathophysiology. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864(11 Pt B):2059–2070. doi: 10.1016/j.bbamcr.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Ripoll V.M., Irvine K.M., Ravasi T., Sweet M.J., Hume D.A. Gpnmb is induced in macrophages by IFN-γ and lipopolysaccharide and acts as a feedback regulator of proinflammatory responses. J. Immunol. 2007;178:6557–6566. doi: 10.4049/jimmunol.178.10.6557. [DOI] [PubMed] [Google Scholar]
- 51.He X., Huang Q., Qiu X., Liu X., Sun G., Guo J., Ding Z., Yang L., Ban N., Tao T., Wang D. LAP3 promotes glioma progression by regulating proliferation, migration and invasion of glioma cells. Int. J. Biol. Macromol. 2015;72:1081–1089. doi: 10.1016/j.ijbiomac.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 52.Audrito V., Serra S., Brusa D., Mazzola F., Arruga F., Vaisitti T., Coscia M., Maffei R., Rossi D., Wang T. Extracellular nicotinamide phosphoribosyltransferase (NAMPT) promotes M2 macrophage polarization in chronic lymphocytic leukemia. Blood. 2015;125:111–123. doi: 10.1182/blood-2014-07-589069. [DOI] [PubMed] [Google Scholar]
- 53.Hassan M.K., Kumar D., Naik M., Dixit M. The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers. PLoS ONE. 2018;13:e0191377. doi: 10.1371/journal.pone.0191377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arai H., Ikota H., Sugawara K., Nobusawa S., Hirato J., Nakazato Y. Nestin expression in brain tumors: its utility for pathological diagnosis and correlation with the prognosis of high-grade gliomas. Brain Tumor Pathol. 2012;29:160–167. doi: 10.1007/s10014-012-0081-5. [DOI] [PubMed] [Google Scholar]
- 55.Beckner M.E., Fellows-Mayle W., Zhang Z., Agostino N.R., Kant J.A., Day B.W., Pollack I.F. Identification of ATP citrate lyase as a positive regulator of glycolytic function in glioblastomas. Int. J. Cancer. 2010;126:2282–2295. doi: 10.1002/ijc.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Z., Joshi N., Agarwal A., Dahiya S., Bittner P., Smith E., Taylor S., Piwnica-Worms D., Weber J., Leonard J.R. Knocking down nucleolin expression in gliomas inhibits tumor growth and induces cell cycle arrest. J. Neurooncol. 2012;108:59–67. doi: 10.1007/s11060-012-0827-2. [DOI] [PubMed] [Google Scholar]
- 57.Zandi S., Nakao S., Chun K.H., Fiorina P., Sun D., Arita R., Zhao M., Kim E., Schueller O., Campbell S. ROCK-isoform-specific polarization of macrophages associated with age-related macular degeneration. Cell Rep. 2015;10:1173–1186. doi: 10.1016/j.celrep.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ono R., Kaisho T., Tanaka T. PDLIM1 inhibits NF-κB-mediated inflammatory signaling by sequestering the p65 subunit of NF-κB in the cytoplasm. Sci. Rep. 2015;5:18327. doi: 10.1038/srep18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi Y., Xu R. Roles of PLODs in collagen synthesis and cancer progression. Front. Cell Dev. Biol. 2018;6:66. doi: 10.3389/fcell.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Li W., Yang Y., Lu Y., He C., Hu G., Liu H., Chen J., He J., Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 61.Brösicke N., Faissner A. Role of tenascins in the ECM of gliomas. Cell Adhes. Migr. 2015;9:131–140. doi: 10.1080/19336918.2014.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu H., Siegel R.M. Progranulin resolves inflammation. Science. 2011;332:427–428. doi: 10.1126/science.1205992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giri B., Dixit V.D., Ghosh M.C., Collins G.D., Khan I.U., Madara K., Weeraratna A.T., Taub D.D. CXCL12-induced partitioning of flotillin-1 with lipid rafts plays a role in CXCR4 function. Eur. J. Immunol. 2007;37:2104–2116. doi: 10.1002/eji.200636680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang M., Liu J., Piao C., Shao J., Du J. ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis. 2015;6:e1780. doi: 10.1038/cddis.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starokadomskyy P., Gluck N., Li H., Chen B., Wallis M., Maine G.N., Mao X., Zaidi I.W., Hein M.Y., McDonald F.J. CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling. J. Clin. Invest. 2013;123:2244–2256. doi: 10.1172/JCI66466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y.J., Hsieh M.Y., Chang M.Y., Chen H.C., Jan M.S., Maa M.C., Leu T.H. Eps8 protein facilitates phagocytosis by increasing TLR4-MyD88 protein interaction in lipopolysaccharide-stimulated macrophages. J. Biol. Chem. 2012;287:18806–18819. doi: 10.1074/jbc.M112.340935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu J.B.K., Chang T.H., Lee G.A., Lee T.Y., Chen C.Y. Identification of potential biomarkers related to glioma survival by gene expression profile analysis. BMC Med. Genomics. 2019;11(Suppl 7):34. doi: 10.1186/s12920-019-0479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedman H.S., Kerby T., Calvert H. Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 2000;6:2585–2597. [PubMed] [Google Scholar]
- 70.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 71.Wiśniewski J.R. Filter-aided sample preparation: the versatile and efficient method for proteomic analysis. Methods Enzymol. 2017;585:15–27. doi: 10.1016/bs.mie.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 72.Wiśniewski J.R. Quantitative evaluation of filter aided sample preparation (FASP) and multienzyme digestion FASP protocols. Anal. Chem. 2016;88:5438–5443. doi: 10.1021/acs.analchem.6b00859. [DOI] [PubMed] [Google Scholar]
- 73.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 74.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 75.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 76.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 78.Yuryev A., Kotelnikova E., Daraselia N. Ariadne’s ChemEffect and Pathway Studio knowledge base. Expert Opin. Drug Discov. 2009;4:1307–1318. doi: 10.1517/17460440903413488. [DOI] [PubMed] [Google Scholar]
- 79.Quanico J., Hauberg-Lotte L., Devaux S., Laouby Z., Meriaux C., Raffo-Romero A., Rose M., Westerheide L., Vehmeyer J., Rodet F. 3D MALDI mass spectrometry imaging reveals specific localization of long-chain acylcarnitines within a 10-day time window of spinal cord injury. Sci. Rep. 2018;8:16083. doi: 10.1038/s41598-018-34518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mallah K., Quanico J., Trede D., Kobeissy F., Zibara K., Salzet M., Fournier I. Lipid changes associated with traumatic brain injury revealed by 3D MALDI-MSI. Anal. Chem. 2018;90:10568–10576. doi: 10.1021/acs.analchem.8b02682. [DOI] [PubMed] [Google Scholar]
- 81.Jardin-Mathé O., Bonnel D., Franck J., Wisztorski M., Macagno E., Fournier I., Salzet M. MITICS (MALDI Imaging Team Imaging Computing System): a new open source mass spectrometry imaging software. J. Proteomics. 2008;71:332–345. doi: 10.1016/j.jprot.2008.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cancer cell and macrophage proteomics data, including MaxQuant files and annotated MS/MS, have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014679 (user name, reviewer94484@ebi.ac.uk; password, gb85XYvq).