Summary
Recent advances in genome engineering technologies based on designed endonucleases (DE) allow specific and predictable alterations in plant genomes to generate value‐added traits in crops of choice. The EXZACT Precision technology, based on zinc finger nucleases (ZFN), has been successfully used in the past for introduction of precise mutations and transgenes to generate novel and desired phenotypes in several crop species. Current methods for delivering ZFNs into plant cells are based on traditional genetic transformation methods that result in stable integration of the nuclease in the genome. Here, we describe for the first time, an alternative ZFN delivery method where plant cells are transfected with ZFN protein that eliminates the need for stable nuclease genomic integration and allows generation of edited, but not transgenic cells or tissues. For this study, we designed ZFNs targeting the wheat IPK1 locus, purified active ZFN protein from bacterial cultures, complexed with cell‐penetrating peptides (CPP) and directly transfected the complex into either wheat microspores or embryos. NGS analysis of ZFN‐treated material showed targeted edits at the IPK1 locus in independent experiments. This is the first description of plant microspore genome editing by a ZFN when delivered as a protein complexed with CPP.
Keywords: wheat, genome editing, ZFN, microspores
Introduction
Bread wheat (Triticum aestivum L.) is a source of ~ 20% of the world’s food and is considered one of the most important staple crops. It is believed that allohexaploid structure of its genome (AABBDD, 2n = 6x = 42) made wheat adaptable to a wide range of climatic conditions (International Wheat Genome Sequencing, 2014; Matsuoka, 2011; Yang et al., 2018; Yang et al., 2014). At the same time, wheat belongs to the cool season crops; therefore, increase in the global temperature can significantly hamper its cultivation in the world. In fact, it has been predicted that increase in global temperature by 1 ºC can lead to decline in global wheat yield by up to 6.4% (Liu et al., 2016). This is especially disturbing since in order to meet global food demand of the growing population, we need to increase wheat production by about 70% by 2050 (Foley et al., 2011; Tilman et al., 2002). Therefore, significant efforts must be placed to develop new tools to speed up generation of the improved cultivars which can consistently deliver high yield under progressively changing climate conditions.
Conventional breeding practice of the small grain cereals relies on efficient production of fully homozygous plants with the fixed beneficial traits. Some of the methods available to breeders include single seed descent (SSD) (Allard, 1999), pollination with alien species followed by embryo rescue and immature microspore culture (IMC) (Daniel et al., 2005; Zheng, 2003). The SSD method as well as double‐haploid (DH) methods have their benefits for generation of breeding populations for phenotypic evaluation of allelic effects(Knott and Kumar, 1975; Tee and Qualset, 1975)(Santra et al., 2012).In case of DH methods, haploid plant material is the choice to regenerate a population of mature green plants that eventually are treated with the chromosome doubling agent to produce double‐haploid lines. The ability to work with haploid cells/tissues is desirable for biotechnology applications since it significantly reduces time and cost required for generation of complete homozygous lines with the altered genome. Microspores are predecessors of male gametes that under certain conditions can undergo embryogenesis in vitro to produce green plants. Being single cell and haploid in nature, the cells provide an opportunity to produce homozygous plants with edited or disrupted locus without germ‐line chimerism in a single generation. In comparison to the conventional breeding practices, the approach significantly reduces time and resources required for bringing the plant with novel trait and can also be used for ‘variation breeding’, a process to generate crop mutant lines with desirable characteristics to be bred with other cultivars to bring new useful germplasms to the market place (Santra et al., 2012).
Recent advances in genome editing (GE) technology resulted in wide adoption of the method for rapid and efficient creation of crops with novel traits (Wang et al., 2014). There is still some uncertainty of how genome‐edited crops will be regulated depending on geographies (Jones, 2015). Some of the concerns are stemming from the type of molecules used to deliver DE (e.g. DNA, RNA or protein) inside the plant cells. If the enzymes (ZFN, TALEN or Cas9/gRNA) are delivered in the form of DNA, there is a risk of unintentional extraneous integration of the complete or fragmented plasmid in the genome, therefore triggering regulation statutes pertaining to transgenic plants. This issue was recently addressed in the elegant studies on lettuce, corn and wheat where purified Cas9 protein complexed with gRNA was delivered into plant cells/tissues with subsequent regeneration of edited plants (Liang et al., 2017; Svitashev et al., 2016; Woo et al., 2015). In all of these studies, diploid somatic cells/tissues (protoplasts and callus) were used for the introduction of edits at the target site using either PEG‐mediated transformation or particle bombardment. During preparation of this manuscript, a report was published describing optimization of GE in wheat microspores using CRISPR/Cas9 DNA constructs delivered by electroporation (Bhowmik et al., 2018).
We provide evidence for the first time of genome editing through direct delivery of purified ZFN proteins into unmodified microspores with intact walls. We used purified ZFN monomers from bacterial culture complexed with cell‐penetrating peptides (CPP) and internalized into the plant cells (Bilichak et al., 2015). As a target, we chose the INOSITOL PENTAKISPHOSPHATE KINASE 1 (IPK1) gene, the product of which is involved in catalysing the final steps of phytic acid production, which is an anti‐nutritional component of feed grain (Shukla et al., 2009). We recovered edits at the target site for both – microspores and haploid embryo‐like structures (ELS). Curiously, although ZFN‐IPK1 was designed to target conservative region in both subgenomes A and B, we observed clear dominance of indels, as a result of ZFN activity, only at the subgenome A.
Results
Design, purification and testing cleavage activity of ZFN protein in vitro
Wheat IPK1 homologs were mapped to chromosomes 2AL, 2BL and 2DL. Sequencing of homoeologs from cultivar AC Andrew confirmed their identity to those identified using IWGSC database. The A and B copies share 91% identity at the DNA level and the D copy contains a low confidence protein‐coding gene. Therefore, zinc finger domains (Figure S1) were designed to target a 41‐nt conserved region at either exon 2 or 3 of the IPK1 gene for either subgenome A or B, respectively (gene model IDs TraesCS2A02G497700 and TraesCS2B02G525900, Figure 1a, Figure S2). The monomers were separated by 6 nt from each other to facilitate dimerization of the FokI restriction domains over the target site. OP2 nuclear localization sequence from Z. mays was fused in frame at the 5´ end of the ZFN monomer sequence (Figure S1) (Elango et al., 2012).
Figure 1.

Recognition sites, purification and cleavage activity of ZFN‐IPK1 protein in vitro. (a – schematic map of the IPK1 homoeologs for subgenomes A and B showing binding sites for ZFN‐L and ZFN‐R. b – left and right ZFN monomers purified from bacterial culture and analysed on 10% PAGE, showing ZFN‐L and ZFN‐R proteins (42 and 46 kDa, respectively), L – BLUelf prestained protein ladder (GeneDireX, Inc.); c – cleavage activity of purified ZFN‐IPK1 in vitro; 300 ng of each ZFN monomer was combined together and incubated for different period of time with 100 ng of pUC18::IPK1 plasmid that carries ZFN recognition sequences and linearized with KpnI. Products were analysed on a 1% agarose gel. Expected cleavage products of 527 bp and 3,325 bp are seen in ZFN‐treated samples and indicated by arrowheads (ZFN+), L – GeneRuler DNA Ladder mix (Thermo Fisher).
ZFN monomers were cloned in frame at the 5´ region with 6xHis tag sequence into pET45b(+) bacterial expression vector and proteins were subsequently purified from bacterial culture using AKTA Purifier FPLC system and 1 mL His‐Trap HP column (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The elution fractions were analysed using a PAGE gel and those with more than 90% purity were combined and concentrated using Amicon Ultra 10K spin filtration units (Figure 1b). During this process, protein instability was observed, with protein precipitation when concentrations reached 1 µg/µL. To alleviate stability issues, concentration of the monomers was kept at around 500 ng/µL. The two monomers mixed together demonstrated specific and reproducible cleavage activity in vitro when combined with the linearized plasmid containing IPK1 gene fragment (Figure 1c, Figures S3 and S7).
CPP‐mediated transfection of wheat microspores with Alexa Fluor‐labelled ZFN proteins and assessment of cleavage activity in the cells
ZFNs belong to the supercharged proteins with a high positive charge in the range of 32 – 36 at pH 7 (isoelectric point – 9.9) (Gaj et al., 2012). Previously, we have reported that positively charged proteins tend to be trapped at the negatively charged surface of the exine – the outer layer of microspores that hampers CPP‐mediated delivery of the cargos (Bilichak et al., 2015). Therefore, it was essential for us to visually assess CPP‐mediated transfection of the cells by ZFN protein. For this, ZFN‐R monomer was labelled at the N‐terminus with Alexa Fluor 647 (AlF647) and was complexed with cys(Npys)‐(D‐R)9 (Figure S4) to facilitate formation of (D‐R)9‐ZFN‐R‐AlF647 through asymmetrical disulphide bond that can be dissociated under reducing conditions inside a cell (Liu et al., 2014). For each transfection, 7 µg of the labelled protein was used and three different weight ratios of ZFN‐R‐AlF647 to cys(Npys)‐(D‐R)9 were tested (20:1, 10:1 and 1:2). Although all combinations resulted in the visual confirmation of ZFN protein delivery inside the microspore cells (Figure 2b), the 1:2 weight ratio (7 µg of ZFN‐R‐AlF647 to 14 µg of cys(Npys)‐(D‐R)9) turned out to be cytotoxic to the cells as was evident by plasmolysis of the cytoplasm. The protein was localized to the cytoplasm and putatively to nucleus. Additionally, it was also trapped at the outer surface of the microspores. Therefore, 5 ng/µL of final CPP concentration was used for the following steps of ZFN protein delivery into microspore cells.
Figure 2.

Transfection of wheat microspores with ZFN monomers using CPPs. a – experimental design for transfection of either wheat microspores or haploid embryos with ZFN monomers combined with either cys(Npys)‐(D‐R)9 or cys(Npys)‐(BP100)2K8 CPP. b – transfection of wheat microspores with ZFN‐R monomer labelled with Alexa Fluor 647 using cys(Npys)‐(D‐R)9 CPP and visualization using direct or fluorescence microscopy. 7 µg of ZFN‐R protein was used for every transfection, treatment was carried out for 2 consecutive days. Examination of transfected microspores was done using confocal laser scanning microscope Olympus, FV1000 with 594/633 laser line. Negative control is a no treatment sample for visualisation of microspores’ autofluorescence. DIC – differential interference contrast image.
Two different CPPs, cys(Npys)‐(D‐R)9 (Anaspec) and cys(Npys)‐(BP100)2K8 (CanPeptide Inc), were tested for their ability to deliver ZFN monomers into wheat microspores to induce indels at the target site (Figure S4), whereas the cys(Npys)‐(D‐R)9 CPP was shown to promote internalization of the TALEN proteins inside the animal cells with subsequent introduction of edits at the target site (Liu et al., 2014), the (BP100)2K8 CPP was able to deliver proteins into Arabidopsis mesophyll cells (Ng et al., 2016). Both CPPs belong to positively charged peptides with the former one having charge at pH 7 of 8.76 and the latter 17.74. To test the activity of ZFN proteins at the target site in planta, final concentration of either of the CPPs during the transfection process was kept at 5 ng/µL (3.51 and 1.3 µm for cys(Npys)‐(D‐R)9 and cys(Npys)‐(BP100)2K8, respectively). Following transfection, microspores were harvested, genomic DNA (gDNA) was isolated and the target region was amplified with primers specific to either subgenome A or B. We detected up to 10‐fold enrichment of indels at the IPK1 locus in subgenome A for microspores treated with ZFN‐(BP100)2K8 as compared to control (Figure 3a). Similarly, samples transfected with ZFN‐(D‐R)9 also demonstrated up to 6 times increase in the level of indels at the target site as compared to control. Both insertions and deletions were detected at the target site (Figure 3b). Interestingly, no cleavage activity above background was detected at the subgenome B for either of the treatments. In an attempt to elevate ZFN cleavage activity at subgenome B, we tested different chemicals that can either increase stability of ZFN proteins inside the cells (cOmplete™, Mini, EDTA‐free protease inhibitor cocktail and MG132 proteasome inhibitor) or promote opening of the chromatin through higher level of histone acetylation (trichostatin A), thus making DNA more accessible for the cleavage (Chakrabarti et al., 2019; Liu et al., 2014; Ramakrishna et al., 2013). However, neither of the treatments resulted in significant increase in the level of indels over the control (Figures S5 and S6), suggesting that some other factors, like DNA methylation, may be involved in accessibility/repair of the locus at the subgenome B (Voytas, 2013).
Figure 3.

Analysis of ZFN‐IPK1 activity at the target site following transfection of wheat microspores with ZFN‐CPP complexes. a – the bar graphs represent the percentage of reads with targeted indels after background correction (BC) at either subgenome A or B (sgA and sgB, respectively) analysed using NGS. Treatments were done with 1 µg of ZFN‐IPK1 protein (total for both monomers) that was combined with 1 µg of either of the CPPs, bar marked with asterisk indicates statistically significant difference as compared to corresponding control (Student’s t‐test, P < 0.1); b – representative indels at the subgenome A target site for both CPPs used. Multiple sequence alignment of the processed reads was done using MAFFT aligner tool in Unipro UGENE program (Okonechnikov et al., 2012). Ct – no transfection control. All treatments were done in two biological replicates.
Evaluation of ZFN cleavage activity in ELS either regenerated from transfected microspores or directly treated with CPP‐ZFN complexes
To follow transmission of indels into the following stages of microspore androgenesis, we analysed edits at the target site in the pooled samples of embryo‐like structures (ELSes) at 3 weeks of culture regenerated from transfected cells. Although we detected up to 10‐fold enrichment of indels for some of the treatments (ZFN‐R9‐sgA) as compared to control, we also observed difference among three biological replicates which could be due to variation in transfection efficiency (Figure 4 a). Overall, deletions were detected for most of the samples treated with ZFN/CPP complexes.
Figure 4.

Analysis of edits at ZFN‐IPK1 target site in ELS regenerated from transfected microspores. a – the bar graphs represent the percentage of reads with targeted indels after background correction (BC) at subgenome A (sgA) analysed using NGS; b – representative indels at the IPK1‐sgA site in ELS regenerated from microspores transfected with ZFN‐cys‐cys‐CPP. Multiple sequence alignment of the processed reads was done using MAFFT aligner tool in Unipro UGENE program (Okonechnikov et al., 2012). Ct – no transfection control. All treatments were done in three biological replicates.
Previously, it has been shown that CPPs can deliver protein cargos into plant somatic cells (Ng et al., 2016). To demonstrate versatility of the developed method for GE in somatic cells, we tested the ability of CPP‐ZFN complexes to introduce edits at the IPK1 locus in haploid ELSes regenerated from untreated microspores. Complexes were formulated in vitro as described previously and pooled ELSes at 3 weeks of culture were collected and incubated with different amounts of CPP‐ZFN conjugates for 2 consecutive days. Since the ELS has higher tolerance to the CPPs and can continue embryogenic development following transfection (P. Maheswhari, A. Bilichak, & F. Eudes, unpublished data), the transfection attempts included treatments starting from 0 to up to 20 µg of CPP‐ZFN complexes (Figure 5).
Figure 5.
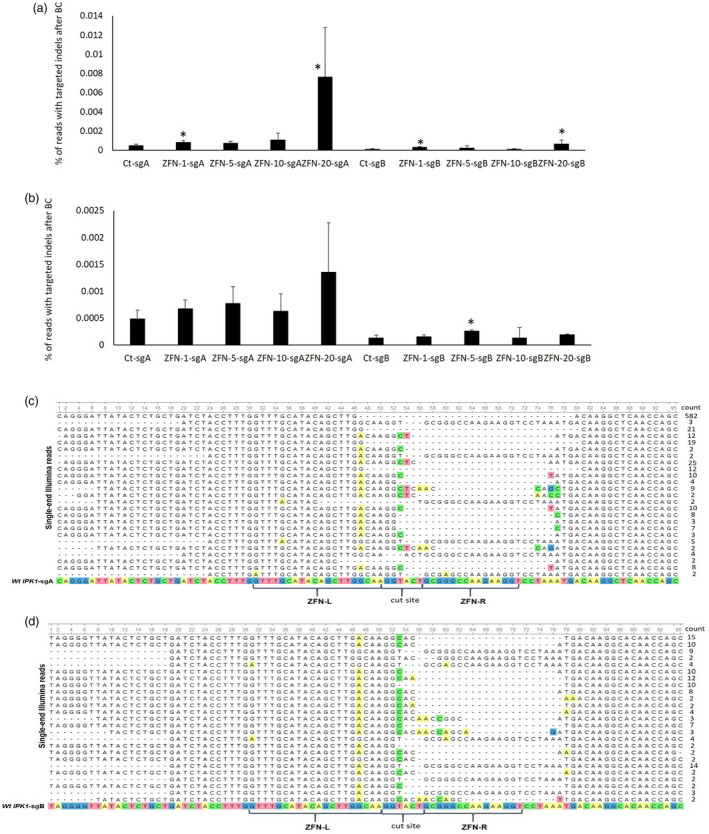
Cleavage activity of ZFN‐IPK1 protein at either subgenome A (sgA) or B (sgB) following transfection of wheat haploid ELS (3 weeks old) with different CPP‐ZFN complexes. A and B – the bar graphs represent the percentage of reads with targeted indels after background correction (BC) at subgenomes A and B as analysed using NGS for either cys(Npys)‐(D‐R)9 (A) or cys(Npys)‐(BP100)2K8 (B) CPP, respectively. Ct – no transfection control, ZFN‐1, ZFN‐5, ZFN‐10 and ZFN20 – ELS transfected with either 1, 5, 10 or 20 µg of ZFN‐IPK1 (total amount for both monomers) protein combined with the corresponding weight of either cys(Npys)‐(D‐R)9 or cys(Npys)‐(BP100)2K8. Experiment was done in 2 biological and 3 technical reps, the bars marked with asterisks indicate statistically significant difference as compared to corresponding controls (Student’s t‐test, P < 0.1). C and D ‐ representative types of edits detected using NGS at either subgenome A or B for both CPPs, respectively. Multiple sequence alignment of the processed reads was done using MAFFT aligner tool in Unipro UGENE program (Okonechnikov et al., 2012).
ELS transfected with the highest amount of CPP‐ZFN complex (ZFN‐20‐sgA for both CPPs) demonstrated the highest level of indels at the target site. Treatment of haploid embryos with 20 µg of ZFN‐cys‐cys‐(D‐R)9 complex resulted in more than 15‐fold enrichment of indels at the target region as compared to control for the subgenome A. Overall, the level of indels was higher for the subgenome A as compared to the B subgenome, which is consistent with the previous experiment where microspores were used as the target cells (Figure 3). At the same time, delivery of ZFN protein with cys(Npys)‐(D‐R)9 resulted in a higher level of indels recorded as compared to the cys(Npys)‐(BP100)2K8 CPP which contrasts with the previous experiment. In general, deletions and substitutions were observed for both subgenomes (Figure 5c and 5d).
Discussion
GE technology mediated by DE has added flexibility to the breeders’ toolbox for the development of varieties with beneficial traits in a timely and cost‐efficient manner. Robust combination of the tissue culture and cargo delivery technologies still remains a bottleneck for the wide adoption of the GE methods in the number of crops (Liang et al., 2017). Here, for the first time, we present an approach of nucleic acid‐free genome editing directly in isolated predecessors of plant gametes by delivery of purified ZFN proteins using CPPs. The developed method eliminates many concerns associated with similar nucleic acid‐driven technologies. Unlike the protein‐mediated disruption of the target loci, the DNA‐mediated method possesses a risk of unintentional integration of the construct coding for the designed nuclease (Gorbunova and Levy, 1997; Zhang et al., 2010). Long‐term presence and expression of the designed nuclease, in turn, could lead to mutation or alteration of the expression level of the unrelated loci, potentially causing collateral issues. Brief introduction of the nuclease in the form of protein also potentially decreases the chance of off‐target effects associated with the DNA‐based methods, since it provides a narrow window for cleavage activity following which the protein is degraded (Liang et al., 2017; Woo et al., 2015). The technology can potentially have broad applications in plant biotechnology and CPPs can be complexed with a variety of designed endonucleases (Custers, 2003; Jahne‐Gartner and Lorz, 1999; Jain et al., 1996; Pauk et al., 2000). Overall, positively charged CPPs would be preferred peptides to use for delivery of cargos into plant cells.
There are a scarce number of reports indicating the preferred DNA repair pathway (non‐homologous end joining versus homologous recombination) in uninucleate microspores. It has been shown that the cells are sensitive to mutation of OsGEN1 gene involved in homologous recombination DNA repair in rice and initiate programmed cell death during the first asymmetric mitotic division (Wang et al., 2017). This can potentially suggest that the HR pathway is essential for male gametogenesis and microspores possess an active DNA repair mechanism. At the same time, it is hard to speculate whether the role of the pathway remains the same after microspores undergo reprograming during IMC. NGS analysis of microspores transfected with CPP‐ZFN complex demonstrated enrichment of edits at the target site as compared to non‐treated control reads (Figure 3). At the same time, it is worth noting that analysis of indels in the microspore population was challenging since the number of reads had inversion segments following ZFN cutting site at the 3' region that were not observed for the sequences generated from ELSes (data not shown). This, apparently, could be caused by programmed cell death and gDNA fragmentation in the microspore population that is usually observed during cell culturing (Sinha et al., 2016). Nevertheless, the indels that were introduced during microspore transfection were also propagated into the following stages of androgenesis as evident by the NGS analysis of regenerated ELSes (Figure 4). Together, these data suggest that green plants with potential edits at the target site could also be recovered.
The ability to induce edits in the ELSes regenerated from untreated microspores indicates that, apparently, other somatic tissues of the variety of plants where tissue culture was established can also be used as material for generation of indels using appropriate optimized CPP combined with DE protein. Additionally, CPP delivery technology eliminates the requirement of using resource intense and cumbersome operation process of equipment such as biolistic chamber for protein delivery. It still has to be shown whether method presented in this study can induce mutations at all three copies of the gene in haploid tissues of wheat. Surprisingly, our results demonstrated that ZFN‐IPK1 protein designed to target conserved sequences in both subgenomes A and B was able to introduce only insignificant level of indels at the B copy as compared to A as a result of its activity (Figures 3 and 5). Further optimization of the technology includes testing CPPs with different properties for their ability to deliver DE in the form of proteins into plant cells and including DNA demethylation agents (e.g. zebularine) to potentially decrease inhibitory effect of DNA methylation at the endogenous target loci. The technology presented herein is not limited only to DE and potentially can be applied to deliver other chimeric enzymes for targeted genome editing or gene expression modulation in a non‐transgenic way (Chaikind et al., 2016; Konermann et al., 2015; Shimatani et al., 2017). Further effort to demonstrate if the developed method can result in regeneration of double‐haploid plants with the edits at the target site is in progress.
Methods
Cloning of IPK1 homolog from wheat cultivar AC Andrew
Wheat IPK1 homolog for subgenome A was identified using BLASTN search with Zea mays IPK1 against Triticum aestivum IWGSC genomic sequence using Ensembl Plants database (Shukla et al., 2009). The homolog was mapped to chromosome 2AL (TraesCS2A02G497700). The B and D copies were identified by BLASTN search against the 2A copy and were mapped to 2BL (TraesCS2B02G525900) and 2DL (TraesCS2D02G612600LC.1) chromosomes. All three copies were amplified from AC Andrew gDNA using subgenome specific primers (Table S1), cloned and sequenced to confirm sequence identity with the published database of IWGSC. ZFN constructs were designed to target specifically IPK1‐sgA and IPK1‐sgB copies.
Cloning of recombinant plasmids
Several ZFNs targeting endogenous IPK1 gene were designed by Sangamo Biotherapeutics and tested in a yeast proxy assay (Ainley et al., 2013). Active yeast candidates were cloned into monocot expression vectors for plant cell testing. All ZFN constructs carried a Zea mays OP2 nuclear localization sequence at the 5´ end (Maddaloni et al., 1989). For the current study, ZFN monomers were cloned from their expression plasmids into bacterial protein expression vector pET45b(+)(Novagen). The left (L) and right (R) ZFN monomers were released from vectors by double digestion with NcoI/AvrII followed by gel purification. Eventually, ZFN monomers were PCR‐amplified from the previously digested products using Phusion high‐fidelity DNA polymerase (Thermo Fisher Scientific, Winnipeg, MB, Canada) with the primers AB001 and AB002 (Table S1). PCR products were digested with AvrII, gel purified and ligated in‐frame with 6xHis tag at the 5´ end into previously dephosphorylated pET45b(+)/PmlI/AvrII expression vector to generate the plasmids pET45b(+)::ZFN‐L and pET45b(+)::ZFN‐R. Correct construction of ZFN expression cassettes was verified by sequencing.
To assess ZFN cleavage activity in vitro, an IPK1 fragment containing ZFN recognition site was amplified form the subgenome B using primers AB016 and AB030 and cloned into linearized pUC18 plasmid using blunt‐end cloning. Resulting pUC18::IPK1 vector was linearized with KpnI and was used as a substrate to test ZFN cleavage activity in vitro.
Of the ZFNs tested, a candidate demonstrating consistent in vitro cleavage activity was selected for further analysis. This ZFN targets a conserved 41 nucleotide sequence in the IPK1 locus with the 5´ and 3´ zinc finger monomers recognizing 20 (TTGCCAaGCTGTAtGCAAAC) and 15 nt (GCGGGCCAAGAAGGT) sequences, respectively. ZFN target site in subgenome A is located in exon 2 and exon 3 is the target for the subgenome B.
Overexpression and purification of ZFN monomers from bacterial culture
Purification of ZFN proteins was done according to protocols described in (Gaj et al., 2012; Tovkach et al., 2011). pET45b(+)::ZFN‐L and pET45b(+)::ZFN‐R plasmids were transformed into chemically competent cells – E. coli BL21 (DE3) (Novagene). A single colony was added to 20 mL of LB medium in the presence of 100 μg/mL of carbenicillin. Bacteria were grown overnight at 37°C with shaking at 250 rpm. The following day, 500 mL of LB medium supplemented with 100 μm ZnCl2 and 100 μg/ml of carbenicillin was inoculated with 20 mL of the overnight culture and incubated at 37°C with shaking at 250 rpm. The culture was grown to an optical density of 0.5 at 600 nm (OD600) and the protein expression was induced with 1 mm isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) and incubated at 22°C with shaking for 4 h. Bacteria were harvested by centrifugation at 3,000 g for 10 min at 4°C, and the pellet was resuspended in native lysis buffer (20 mm HEPES, 300 mm NaCl, 100 μm ZnCl2 and 5% glycerol, pH 7.5) supplemented with EDTA‐free protease inhibitor cocktail (Roche). Cells were disrupted by sonication (50% intensity, 30 s on, 1 min off time, four cycles, model Q55, Qsonica), and the insoluble debris were removed by centrifugation (18,500 g, 1 h at 4°C). The clarified supernatant was applied directly onto 1 ml His‐Trap column (GE Healthcare Life Sciences) equilibrated with the native lysis buffer using an FPLC system (AKTA purifier, GE Healthcare Life Sciences). The elution of bound protein was done using a linear gradient of His‐Tag buffer B (20 mm HEPES, 300 mm NaCl, 100 μm ZnCl2, 5% glycerol and 500 mm imidazole, pH 7.5) from 0 to 100%. The fractions, containing more than 90% pure protein were combined and used for further experiments.
In vitro ZFN cleavage activity
Activity of purified ZFN proteins was assessed using an in vitro cleavage assay (Tovkach et al., 2011). For this, 300 ng of each ZFN monomer was combined together and incubated with 100 ng of pUC18::IPK1 plasmid (Figure S3) linearized with KpnI that carries ZFN recognition sequences in 1xNEB4 buffer in a total volume of 20 μL at 37°C for up to 60 min. Following incubation, the digestion mix was loaded directly on 1% agarose gel and analysed for the presence of the cleavage products.
Labelling of ZFN monomers with the fluorophore and in vitro conjugation to CPPs
To assess CPP‐mediated delivery of ZFN proteins, ZFN‐R monomer was labelled with Alexa Fluor 647 (AlF‐647) protein labelling kit (ThermoFisher Scientific, cat.no. A20173) according to the manufacture’s protocol. The labelled protein was purified using gel filtration column, followed by concentration using Amicon Ultra‐15 10 K centrifugal filter units. The last step served as a quality control to verify the absence of a free dye following purification. The labelled protein was analysed by 10% PAGE followed by scanning the gel using fluorescent scanner to verify labelling of ZFN‐R protein (data not shown). ZFN proteins possess a single free Cys residue near the N‐terminus that allows for their stable conjugation to CPP molecule using 3‐nitro‐2‐pyridylsulphenyl (Npys) group (Chen et al., 2013). Conjugation reaction was done with 7 µg of ZFN‐R‐AlF647 (1.59 µm in His‐Tag buffer B, pH 7.5) by adding different amounts of cys(Npys)‐(D‐R)9 (Anaspec) and incubating the mixture for 2 h at room temperature. 100,000 cells in CIMC7 wash medium (Bilichak et al., 2015; Bilichak et al., 2018) were directly mixed with (D‐R)9‐ZFN‐R‐AlF647 conjugate and the volume was adjusted to 100 µL. Transfection was done for 2 consecutive days followed by washing of the microspores for 3 times with CIMC7 medium. The visual examination of transfected microspores was done using confocal laser scanning microscope Olympus, FV1000 with 594/633 laser line.
Transfection of microspores and haploid embryos with CPP‐ZFN conjugate
Isolation of wheat microspores from immature spikes of cultivar AC Andrew was done essentially as described previously (Bilichak et al., 2015; Bilichak et al., 2018). Transfection of microspores for NGS analysis of edits at the target site was done by mixing together 500 ng each of left and right ZFN monomers together with 1 µg of either cys(Npys)‐(D‐R)9 or cys(Npys)‐(BP100)2K8 CPPs in 100 µL of CIMC7 medium, incubating the mixture for at least 1 h at room temperature and adding the solution to 100,000 cells. The total volume was brought to 200 µL, and transfection was carried either for one or two days (Figure 2a). Following the incubation step, the microspores were collected and gDNA was isolated using DNeasy Plant Mini kit (Qiagen).
To regenerate haploid embryo‐like structures (ELS), either treated or non‐treated microspores were transferred to 35 mm Petri dishes containing 3.3 mL of the CIMC7 medium (CIMC7 wash medium supplemented with 0.25 mm of Ficoll PM400; Sigma, Oakville, ON, Canada) and 5 wheat ovaries (Asif et al., 2014). The culture was incubated for 3 weeks at 28 ºC in the dark and ELS either were harvested for gDNA isolation (ELS regenerated from treated microspores) or were used for transfection with CPP‐ZFN complexes. The transfection of ELS was done essentially as with microspores followed by incubation for two consecutive days and isolation of gDNA.
Treatment of microspores with protease or histone deacetylase inhibitors.
To improve genome‐editing efficiency at the subgenome B locus, microspores were transfected with CPP‐ZFN complexes as described above, but in the presence of either MG132 (5 µm, cat. no. M7449, Sigma), 1x cOmplete™, Mini, EDTA‐free protease inhibitor cocktail (PIC, Roche) or trichostatin A (0.01 µm TSA, Sigma). Following incubation, microspores were collected and gDNA isolated for NGS analysis. Either untreated or CPP‐ZFN transfected microspores in the absence of inhibitors were used as controls.
Generation of NGS library and bioinformatic analysis of indels at the target site
The NGS library was generated by amplification of IPK1 target regions using primers specific for SNPs in subgenomes at both 5' and 3' regions in regard to ZFN cut site (AB352 + AB353 and AB094 + AB095, respectively). 100 ng of gDNA was used for amplification of the target regions using either Q5 or Phusion DNA polymerase (NEB). Amplification was done for 35 cycles in total volume of 30 µl each. This was followed by gel purification of PCR fragments. Every primer combination had a unique barcode for discrimination among samples during bioinformatic processing of the NGS data. Sequencing of the NGS libraries was conducted using Illumina NextSeq platform with single‐end sequencing runs and 150 cycles. Each sample generated from 2.5 to 10 million HQ reads with the overall average of 7 million HQ reads per sample. Reads were trimmed to 80 bp for the analysis and processing of the data was done by Dow AgroSciences LLC bioinformatic pipeline (Elango et al., 2012). Briefly, trimmed reads were first binned based on their unique barcodes and further processed to identify high‐quality trimmed reads using fastq‐mcf (‐q option set to 33) (https://github.com/ExpressionAnalysis/ea-utils). Next, a dictionary of unique sequences, and their frequencies, was extracted from the total set of high‐quality trimmed reads and each unique sequence was aligned against both forward and reverse strands of the endogenous IPK1 target sequence using the EMBOSS package (v6.3.1) with the custom parameters (Rice et al., 2000). The resulting alignments were filtered using several sequence similarity criteria to identify the subset of high‐quality alignments with insertions or deletions that span the ZFN cutting site. Background correction was done by removing sequences with point mutations or less than 2 indels from both sets and activity is determined relative to the control sample. The method has been used to show editing activity of ZFNs in multiple crops, in multiple tissue types (embryos/protoplasts/suspensions) (Sastry‐dent et al., 2018a, 2018b). The frequencies of the identified high‐quality alignments were represented as percentage of high‐quality trimmed reads sequenced. Multiple sequence alignment of the reads showing indels at the ZFN cutting site was done and visualized using MAFFT aligner tool in Unipro UGENE program (Okonechnikov et al., 2012). Summary of the NGS analysis for all figures is available in the Data S1. FASTQ NGS files of processed reads were deposited to Sequence Read Archive, submission: SUB3678301.
Author contribution
B.A. – designed and executed experiments, analysed data and wrote the manuscript, S.D.L. – initiated the project, provided ZFN constructs, input on study design, analysed data, S.S. – analysed NGS data, S.M. – generated libraries and provided NGS support, S.P. – initiated the project, provided guidance on ZFN construct design and commented on manuscript, W.S. – initiated the project, provided guidance on ZFN construct design, J.F. – provided support with microspore cell culture, E.F. – initiated and designed the project. All authors reviewed the manuscript.
Conflict of interests
S.D.L., S.S., S.M., S.P. and W.S. are employees of Corteva Agriscience, The Agriculture Division of DowDuPont, Dow AgroSciences LLC, USA. We declare no non‐financial competing interests.
Data Availability Statement
Summary of the NGS analysis for all figures is available in the supplementary dataset. FASTQ NGS files of processed reads were deposited to Sequence Read Archive, submission: SUB3678301.
Supporting information
Figure S1 Amino acid sequences of ZFN‐IPK1‐L and ZFN‐IPK1‐R.
Figure S2 gDNA sequences of the 5' and 3' regions for IPK1‐subgenome A and B, respectively.
Figure S3 pUC18::IPK1 plasmid used for assessment of ZFN cleavage activity in vitro.
Figure S4 Amino acid sequence of peptides used in this study.
Figure S5 Cleavage activity of ZFN‐IPK1 at subgenome B following transfection of wheat microspores with different amounts of ZFN‐cys‐cys‐(D‐R)9 complex either in the presence or absence of protease inhibitor MG132.
Figure S6 Cleavage activity of ZFN‐IPK1 protein at either subgenome A (sgA) or B (sgB) following transfection of wheat microspores with two different CPPs in the presence of cOmplete™, Mini, EDTA‐free protease inhibitor cocktail (PIC, Roche) and trichostatin A (TSA, Sigma).
Figure S7 Full size pictures of gels for Figures 1B and C.
Table S1 List of primers used in this study.
Data S1 NGS data of indels analysis.
Acknowledgments
We would like to thank to Dr. John Laurie for comments on the manuscript. We also would like to express our gratitude to Justin Luu for the technical support. Financial support from A‐Base funding, Growing Forward 2 – AgriInnovation program (Agri‐science Project No. P343) and Dow AgroSciences LLC are recognized.
Bilichak, A. , Sastry‐Dent, L. , Sriram, S. , Simpson, M. , Samuel, P. , Webb, S. , Jiang, F. and Eudes, F. (2020) Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell‐penetrating peptide complexes. Plant Biotechnol. J., 10.1111/pbi.13296
References
- Ainley, W.M. , Sastry‐Dent, L. , Welter, M.E. , Murray, M.G. , Zeitler, B. , Amora, R. , Corbin, D.R. et al. (2013) Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. [DOI] [PubMed] [Google Scholar]
- Allard, R.W. (1999) Principles of Plant Breeding. Hoboken, NJ: Wiley. [Google Scholar]
- Asif, M. , Eudes, F. , Randhawa, H. , Amundsen, E. and Spaner, D. (2014) Phytosulfokine alpha enhances microspore embryogenesis in both triticale and wheat. Plant Cell . Tissue Organ Cult. (PCTOC) 116, 125–130. [Google Scholar]
- Bhowmik, P. , Ellison, E. , Polley, B. , Bollina, V. , Kulkarni, M. , Ghanbarnia, K. , Song, H. et al. (2018) Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 8, 6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak, A. , Luu, J. and Eudes, F. (2015) Intracellular delivery of fluorescent protein into viable wheat microspores using cationic peptides. Front. Plant Sci. 6, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak, A. , Luu, J. , Jiang, F. and Eudes, F. (2018) Identification of BABY BOOM homolog in bread wheat. Agri. Gene 7, 43–51. [Google Scholar]
- Chaikind, B. , Bessen, J.L. , Thompson, D.B. , Hu, J.H. and Liu, D.R. (2016) A programmable Cas9‐serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nuc. Acids Res. 44, 9758–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, A.M. , Henser‐Brownhill, T. , Monserrat, J. , Poetsch, A.R. , Luscombe, N.M. and Scaffidi, P. (2019) Target‐Specific Precision of CRISPR‐Mediated Genome Editing. Mol. Cell 73, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Jaafar, L. , Agyekum, D.G. , Xiao, H. , Wade, M.F. , Kumaran, R.I. , Spector, D.L. et al. (2013) Receptor‐mediated delivery of engineered nucleases for genome modification. Nucleic Acids Res 41, e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custers, J.B.M. (2003). Microspore culture in rapeseed (Brassica napus L.). In Doubled Haploid Production in Crop Plants: A Manual ( Maluszynski, M. , Kasha, K.J. , Forster, B.P. and Szarejko, I. eds). Dordrecht, The Netherlands: Springer, Netherlands. [Google Scholar]
- Daniel, G. , Baumann, A. and Schmucker, S. (2005) Production of wheat doubled haploids (Triticum aestivum L.) by wheat x maize crosses using colchicine enriched medium for embryo regeneration. Cereal Res. Comm. 33, 461–468. [Google Scholar]
- Elango, N.I. , Petolino, J.F. , Sriram, S. and Sastry‐Dent, L. (2012). Data Analysis of DNA Sequences. United States patent application 20120173153.
- Foley, J.A. , Ramankutty, N. , Brauman, K.A. , Cassidy, E.S. , Gerber, J.S. , Johnston, M. , Mueller, N.D. et al. (2011) Solutions for a cultivated planet. Nature 478, 337–342. [DOI] [PubMed] [Google Scholar]
- Gaj, T. , Guo, J. , Kato, Y. , Sirk, S.J. and Barbas, C.F. (2012) Targeted gene knockout by direct delivery of zinc‐finger nuclease proteins. Nat. Meth. 9, 805–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova, V. and Levy, A.A. (1997) Non‐homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25, 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing, C. (2014) A chromosome‐based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. [DOI] [PubMed] [Google Scholar]
- Jahne‐Gartner, A. and Lorz, H. (1999) Protocols for anther and microspore culture of barley. Methods Mol. Biol. 111, 269–279. [DOI] [PubMed] [Google Scholar]
- Jain, S.M. , Sopory, S.K. and Veilleux, R.E. (1996) In vitro haploid production in higher plants. Dordrecht; Boston: Kluwer Academic Publishers. [Google Scholar]
- Jones, H.D. (2015) Regulatory uncertainty over genome editing. Nat. Plants 1, 14011. [DOI] [PubMed] [Google Scholar]
- Knott, D.R. and Kumar, J. (1975) Comparison of early generation yield testing and a single seed descent procedure in wheat breeding. Crop Sci. 15, 295–299. [Google Scholar]
- Konermann, S. , Brigham, M.D. , Trevino, A.E. , Joung, J. , Abudayyeh, O.O. , Barcena, C. , Hsu, P.D. et al. (2015) Genome‐scale transcriptional activation by an engineered CRISPR‐Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Chen, K. , Li, T.D. , Zhang, Y. , Wang, Y. , Zhao, Q. , Liu, J. et al. (2017) Efficient DNA‐free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Comm. 8, 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Gaj, T. , Patterson, J.T. , Sirk, S.J. and Barbas III, C.F. (2014) Cell‐penetrating peptide‐mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS ONE 9, e85755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Asseng, S. , Muller, C. , Ewert, F. , Elliott, J. , Lobell, D.B. , Martre, P. et al. (2016) Similar estimates of temperature impacts on global wheat yield by three independent methods. Nat. Climate Change 6, 1130–1136. [Google Scholar]
- Maddaloni, M. , di Fonzo, N. , Hartings, H. , Lazzaroni, N. , Salamini, F. , Thompson, R. and Motto, M. (1989) The sequence of the zein regulatory gene opaque‐2 (O2) of Zea Mays. Nucleic Acids Res 17, 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, Y. (2011) Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52, 750–764. [DOI] [PubMed] [Google Scholar]
- Ng, K.K. , Motoda, Y. , Watanabe, S. , Sofiman Othman, A. , Kigawa, T. , Kodama, Y. and Numata, K. (2016) Intracellular Delivery of Proteins via Fusion Peptides in Intact Plants. PLoS ONE 11, e0154081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov, K. , Golosova, O. , Fursov, M. and Team, U. (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Pauk, J. , Puolimatka, M. , Lökös Tóth, K. and Monostori, T. (2000) In vitro androgenesis of triticale in isolated microspore culture. Plant Cell, Tissue Organ Cult. 61, 221. [Google Scholar]
- Ramakrishna, S. , Kim, Y.‐H. and Kim, H. (2013) Stability of zinc finger nuclease protein is enhanced by the proteasome inhibitor MG132. PLoS ONE 8, e54282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, P. , Longden, I. and Bleasby, A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–283. [DOI] [PubMed] [Google Scholar]
- Santra, M. , Ankrah, N. , Santra, D.K. and Kidwell, K.K. (2012) An Improved Wheat Microspore Culture Technique for the Production of Doubled Haploid Plants. Crop Sci. 52, 2314–2320. [Google Scholar]
- Sastry‐Dent, L.A. , Cao, Z. , Sriram, S. , Webb, S.R. and Camper, D.L. (2018a) Optimal maize loci. United States patent application, 10093940.
- Sastry‐Dent, L.A. , Cao, Z. , Sriram, S. , Webb, S.R. , Camper, D.L. and Ainley, M.W. (2018b) Optimal soybean loci. United States patent application 9909131.
- Shimatani, Z. , Kashojiya, S. , Takayama, M. , Terada, R. , Arazoe, T. , Ishii, H. , Teramura, H. et al. (2017) Targeted base editing in rice and tomato using a CRISPR‐Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. [DOI] [PubMed] [Google Scholar]
- Shukla, V.K. , Doyon, Y. , Miller, J.C. , Dekelver, R.C. , Moehle, E.A. , Worden, S.E. , Mitchell, J.C. et al. (2009) Precise genome modification in the crop species Zea mays using zinc‐finger nucleases. Nature 459, 437–441. [DOI] [PubMed] [Google Scholar]
- Sinha, R.K. , Pospisil, P. , Maheshwari, P. and Eudes, F. (2016) Bcl‐2 big up tri, open21 and Ac‐DEVD‐CHO Inhibit Death of Wheat Microspores. Front. Plant Sci. 7, 1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev, S. , Schwartz, C. , Lenderts, B. , Young, J.K. and Cigan, A.M. (2016) Genome editing in maize directed by CRISPR‐Cas9 ribonucleoprotein complexes. Nat. Comm. 7, 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee, T.S. and Qualset, C.O. (1975) Bulk populations in wheat breeding: comparison of single‐seed descent and random bulk methods. Euphytica 24, 393–405. [Google Scholar]
- Tilman, D. , Cassman, K.G. , Matson, P.A. , Naylor, R. and Polasky, S. (2002) Agricultural sustainability and intensive production practices. Nature 418, 671–677. [DOI] [PubMed] [Google Scholar]
- Tovkach, A. , Zeevi, V. and Tzfira, T. (2011) Expression, purification and characterization of cloning‐grade zinc finger nuclease. J. Biotechnol. 151, 1–8. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F. (2013) Plant genome engineering with sequence‐specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. [DOI] [PubMed] [Google Scholar]
- Wang, Y.P. , Cheng, X. , Shan, Q.W. , Zhang, Y. , Liu, J.X. , Gao, C.X. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Higgins, J.D. , He, Y. , Lu, P. , Zhang, D. and Liang, W. (2017) Resolvase OsGEN1 mediates DNA repair by homologous recombination. Plant Physiol. 173, 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, J.W. , Kim, J. , IL Kwon, S. , Corvalan, C. , Cho, S.W. , Kim, H. , Kim, S.G. et al.(2015) DNA‐free genome editing in plants with preassembled CRISPR‐Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–U156. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Zhao, L. , Zhang, H. , Yang, Z. , Wang, H. , Wen, S. , Zhang, C. et al. (2014) Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl Acad. Sci. USA 111, 11882–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Yang, Z. , Zhao, L. , Sun, F. and Liu, B. (2018) A newly formed hexaploid wheat exhibits immediate higher tolerance to nitrogen‐deficiency than its parental lines. BMC Plant Biol. 18, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Maeder, M.L. , Unger‐Wallace, E. , Hoshaw, J.P. , Reyon, D. , Christian, M. , Li, X.H. et al. (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl Acad. Sci. USA 107, 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M.Y. (2003) Microspore culture in wheat (Triticum aestivum) – doubled haploid production via induced embryogenesis. Plant Cell, Tissue Organ Cult. 73, 213–230. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Amino acid sequences of ZFN‐IPK1‐L and ZFN‐IPK1‐R.
Figure S2 gDNA sequences of the 5' and 3' regions for IPK1‐subgenome A and B, respectively.
Figure S3 pUC18::IPK1 plasmid used for assessment of ZFN cleavage activity in vitro.
Figure S4 Amino acid sequence of peptides used in this study.
Figure S5 Cleavage activity of ZFN‐IPK1 at subgenome B following transfection of wheat microspores with different amounts of ZFN‐cys‐cys‐(D‐R)9 complex either in the presence or absence of protease inhibitor MG132.
Figure S6 Cleavage activity of ZFN‐IPK1 protein at either subgenome A (sgA) or B (sgB) following transfection of wheat microspores with two different CPPs in the presence of cOmplete™, Mini, EDTA‐free protease inhibitor cocktail (PIC, Roche) and trichostatin A (TSA, Sigma).
Figure S7 Full size pictures of gels for Figures 1B and C.
Table S1 List of primers used in this study.
Data S1 NGS data of indels analysis.
Data Availability Statement
Summary of the NGS analysis for all figures is available in the supplementary dataset. FASTQ NGS files of processed reads were deposited to Sequence Read Archive, submission: SUB3678301.


