Summary
Anthocyanins have crucial biological functions and affect quality of horticultural produce. Anthocyanins accumulate in ripe peach fruit; differential accumulation is observed in deep coloured cultivar ‘Hujingmilu’ and lightly pigmented cultivar ‘Yulu’. The difference was not fully explained by accumulation of total flavonoids and expression of anthocyanin biosynthetic genes. Expression analysis was conducted on a glutathione S‐transferase gene (PpGST1), and it was found that the expression correlated well with anthocyanin accumulation in peach fruit tissues. Functional complementation of the Arabidopsis tt19 mutant indicated that PpGST1 was responsible for transport of anthocyanins but not proanthocyanidins. PpGST1 was localized in nuclei and the tonoplast, including the sites at which anthocyanin vacuolar sequestration occurred. Transient overexpression of PpGST1 together with PpMYB10.1 in tobacco leaves and peach fruit significantly increased anthocyanin accumulation as compared with PpMYB10.1 alone. Furthermore, virus‐induced gene silencing of PpGST1 in a blood‐fleshed peach not only resulted in a reduction in anthocyanin accumulation but also a decline in expression of anthocyanin biosynthetic and regulatory genes. Cis‐element analysis of the PpGST1 promoter revealed the presence of four MYB binding sites (MBSs). Dual‐luciferase assays indicated that PpMYB10.1 bound to the promoter and activated the transcription of PpGST1 by recognizing MBS1, the one closest to the ATG start codon, with this trans‐activation being stronger against the promoter of deep coloured ‘Hujingmilu’ compared with lightly coloured cultivar ‘Yulu’. Altogether, our data provided molecular evidence supporting coordinative regulatory roles of PpGST1 and PpMYB10.1 in anthocyanin accumulation in peach.
Keywords: Anthocyanins, peach, transport, glutathione S‐transferase, MYB10.1, promoter
Introduction
Anthocyanins, a subset of the flavonoids, are a group of water‐soluble pigments widely distributed in the plant kingdom. Anthocyanins are important to plants with multiple biological and physiological functions such as pigmentation, pollinator attraction, reactive oxygen species scavenging, pathogen defence, seed dispersal, abiotic and biotic stress resistance, UV protection, etc., and are also known as potential beneficial compounds for human health due to strong antioxidant activity (Clifford et al., 2015; Tsuda, 2012).
After synthesis, anthocyanins are transported from the cytosol to vacuole for storage. Efforts have been made to understand the molecular mechanisms of anthocyanin transport. At present, three types of mechanism for anthocyanin transport have been proposed: glutathione S‐transferase (GST)‐mediated transport, membrane transporter‐mediated transport and membrane vesicle‐mediated transport (Grotewold and Davis, 2008; Poustka et al., 2007; Zhao, 2015; Zhao and Dixon, 2010). The first mechanism involves GSTs in anthocyanin sequestration from the cytosol into the vacuole (Zhao, 2015). The second relies on transporter proteins located in the tonoplast including ATP‐binding cassette (ABC) proteins and multidrug and toxic extrusion (MATE) transporters (Zhao and Dixon, 2010). The last mechanism is based on the fusion of smaller vesicle‐like structures containing cytoplasmic anthocyanin bodies with the central vacuole (Poustka et al., 2007).
In this study, we focus on GSTs which function in cellular detoxification through the conjugation of glutathione (GSH) to electrophilic compounds (Dixon et al., 2002). In plants, GSTs are encoded by a large gene family and ubiquitously participate in signalling, flavonoid metabolism and response to biotic and abiotic stresses (Loyall et al., 2000; Moons, 2005). So far, numerous GSTs have been identified in plants, including 53 in Arabidopsis thaliana (Sappl et al., 2009), 20 in Dracaena cambodiana (Zhu et al., 2016), 139 in Litchi chinensis (Hu et al., 2016), 61 in Citrus sinensis (Licciardello et al., 2014) and 90 in Solanum lycopersicum (Islam et al., 2017). Among various functions, GSTs are also known to mediate trafficking and accumulation of anthocyanins. Plant GST members can be classified into different subfamilies (TAU group, Phi (F) group, LAMBDA group, THETA group, ZETA group), with anthocyanin‐transporting GSTs belonging to the plant‐specific Phi subfamily (Luo et al., 2018). The absence of anthocyanin‐transporting GST often leads to an anthocyanin‐less phenotype with a reduced amount of pigment, like bz2 (bronze‐2) in maize (Marrs et al., 1995), an9 (anthocyanin 9) in petunia (Alfenito et al., 1998), fl3 (flavonoid 3) in carnation (Larsen et al., 2003) and tt19 (transparent testa 19) in Arabidopsis (Kitamura et al., 2004). In the bz2 knockout mutant, because of the impairment on vacuolar sequestration, the anthocyanins synthesized were retained in the cytosol where they were much more prone to further oxidation and polymerization, and as a result the kernels are brown instead of red (Alfenito et al., 1998). This defect can be complemented by the petunia An9 protein in particle bombardment tests (Mueller et al., 2000). In some plants, a specific GST member can be involved in the vacuolar sequestration of different flavonoid substances. For example, Arabidopsis TT19 is a carrier protein not only for anthocyanin sequestration but also for deposition of proanthocyanidins (PAs) in the seed coat (Sun et al., 2012). However, the functional complementation of Arabidopsis TT19 with petunia An9 only restored the accumulation of anthocyanins but not PAs in seed coat, indicating that An9 is only associated with anthocyanin accumulation (Kitamura et al., 2004).
Recently, anthocyanin‐related GSTs have also been reported in perennial fruit crops. For example, LcGST4 was involved in anthocyanin transport in litchi (Hu et al., 2016). In strawberry, RAP encodes a GST protein critical for anthocyanin accumulation in fruit and foliage (Luo et al., 2018). In grapevine, VviGST3 is related to PA transport while VviGST4 is pivotal for both anthocyanin and PA accumulation (Pérez‐Díaz et al., 2016). MdGSTF6 was shown to be the functional member in anthocyanin sequestration in apple fruit (Jiang et al., 2019).
Peach [Prunus persica L. (Batsch)] is a major deciduous fruit crop worldwide. Red coloration is an essential trait of maturity and quality for peach fruit, which is mainly determined by anthocyanin content. Previous studies have investigated the biosynthesis and transcriptional regulation of anthocyanins in peach (Cheng et al., 2014; Liu et al., 2015; Uematsu et al., 2014; Zhao et al., 2017; Zhou et al., 2015; Zhou et al., 2014), while little is known about the mechanism underlying vacuolar sequestration of anthocyanins in peach fruit.
In our previous study, 54 GSTs were identified in peach according to the transcriptome data set (Zhao et al., 2017). A Phi (F) class GST member, PpGST1, was speculated to participate in vacuolar accumulation of anthocyanins, which was differentially expressed and correlated with anthocyanin content in different peach cultivars under UV irradiation (Zhao et al., 2017). Here, this PpGST1 was shown to be critical for anthocyanin transport and its expression was stimulated by PpMYB10.1 to different extents in a deep coloured cultivar ‘Hujingmilu’ (‘HJ’) and a lightly coloured cultivar ‘Yulu’ (‘YL’). These findings aid in understanding the mechanism of anthocyanin accumulation and also provide potential clues for fruit colour manipulation in peach.
Results
Anthocyanin levels in peach peel during fruit development and ripening
The developmental process of ‘HJ’ and ‘YL’ peach fruits is shown in Figure 1a. During early fruit growth stages, there was no significant difference in peel colour between ‘HJ’ and ‘YL’. The peel of cv. ‘HJ’ was green in colour during the early stages of fruit development, and slightly red pigmentation appeared following natural de‐greening at stage S4, presenting a characteristic red coloration before ripening. The peel of cv. ‘YL’ was green during the early stages and turned to pale green as fruit developed. These observations were in accordance with values of a comprehensive indicator of the colour index of red grapes (CIRG) (Figure 1b). Cyanidin‐3‐glucoside was detected as representative for anthocyanins as previous studies described (Cheng et al., 2014; Zhao et al., 2017). The anthocyanin contents in the peel of cv. ‘HJ’ showed a low concentration only at the early stages and then increased during ripening, with a highest concentration value reached to 1.57 mg/100 gFW at maturity, over 20 times more than ‘YL’ (Figure 1b). Significantly lower content of anthocyanins was observed for ‘YL’ throughout developmental stages, consistent with visual appearances.
Figure 1.
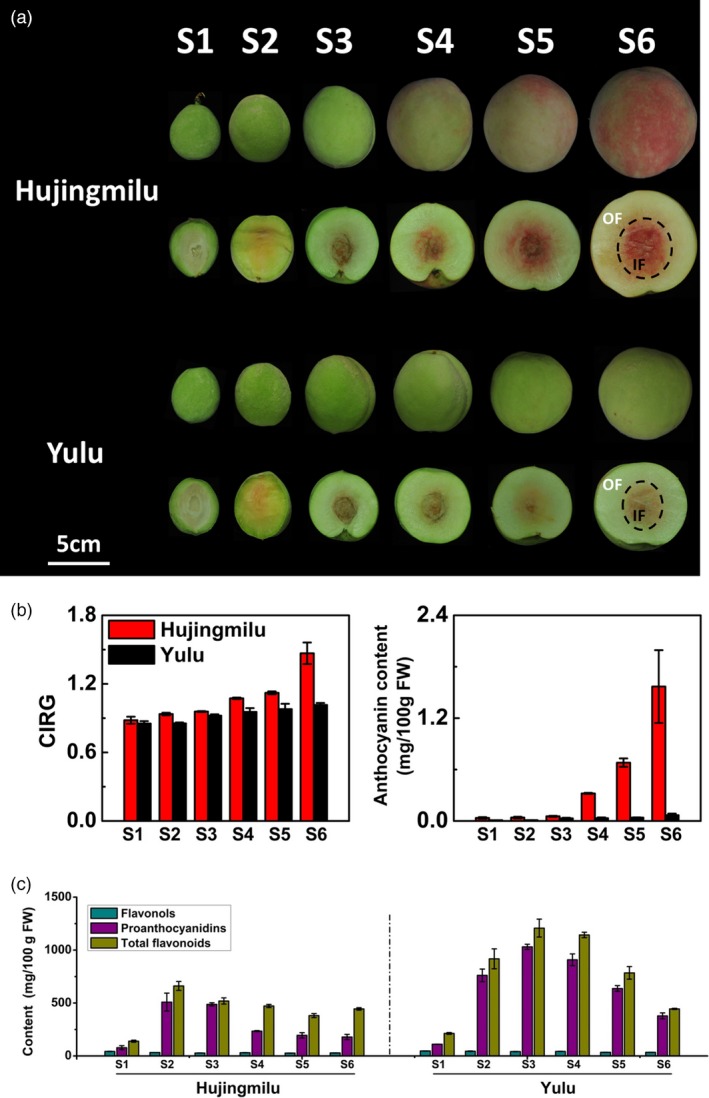
A Photograph of peach fruit (‘Hujingmilu’ and ‘Yulu’) development series (a), the colour index of red grapes (CIRG) values and anthocyanin contents (b) and total flavonoid, proanthocyanidin and flavonol contents (c) in the peel of peach during fruit development and ripening. The developmental stages were indicated as follows: S1, fruitlet; S2, immature green; S3, mature green; S4, breaker; S5, commercial maturity; and S6, fully ripe. OF, outer flesh near the peel; and IF, inner flesh around the stone. Scale bar indicated 5 cm. Data were means (±SE) from three independent biological replicates.
In addition to anthocyanins, other phenolic compounds accumulated in the peel of both ‘HJ’ and ‘YL’ during fruit development as well. The concentrations of total flavonoids, flavonols and PAs had the similar trends in the two cultivars. However, the concentrations were consistently higher in ‘YL’ compared with ‘HJ’. Total flavonoid and PA contents were lower during the early stages of fruit development, and then strongly increased as fruit developed and finally decreased during ripening for both cultivars (Figure 1c). However, flavonols presented a slight reduction in two cultivars with an opposite trend compared with anthocyanins (Figure 1c).
Expression of genes involved in the anthocyanin metabolic pathway
The expression levels of structural genes in the anthocyanin biosynthetic pathway (CHS, CHI, F3H, DFR, ANS and UFGT) and related transcription factors (MYB10.1, MYB10.2, MYB10.3, bHLH3 and WD40‐1) potentially involved were analysed in peel of cv. ‘HJ’ and ‘YL’ during fruit development (Figure 2). Results showed that the expression of early biosynthetic genes (CHS, CHI and F3H) increased and then gradually decreased during fruit growth, which was consistent with the change in total flavonoid content (Figure 1c). Expression of the late biosynthetic genes (DFR, ANS and UFGT) increased in parallel with the accumulation of anthocyanins, and the expression was stronger in peel of cv. ‘HJ’ as compared to cv. ‘YL’, especially UFGT (Figure 2).
Figure 2.

Real‐time quantitative PCR analysis of anthocyanin biosynthetic genes CHS, CHI, F3H, DFR, ANS and UFGT, as well as putative MYB activators MYB10.1, MYB10.2 and MYB10.3, and their partners bHLH3 and WD40‐1 in peel of peach fruits (‘Hujingmilu’ and ‘Yulu’) at different developmental stages. Abbreviations used were as follows: ANS, anthocyanidin synthase/leucoanthocyanidin dioxygenase; bHLH, basic helix–loop–helix; CHS, chalcone synthase; CHI, chalcone isomerase; DFR, dihydroflavonol‐4‐reductase; F3H, flavanone 3‐hydroxylase; and UFGT, UDP‐glucose: flavonoid‐3‐O‐glucosyltransferase. Data were means (±SE) from three independent biological replicates.
The expression levels of anthocyanin regulatory genes were analysed in ‘HJ’ and ‘YL’ (Figure 2). Among three MYB10s (PpMYB10.1, PpMYB10.2 and PpMYB10.3), the expression of PpMYB10.1 was strongest in both cultivars and was consistent with anthocyanin accumulation. The expression of PpMYB10.2 and PpMYB10.3 was low during fruit development in both cultivars. The expression of PpbHLH3 (Prupe.8G242100) and PpWD40‐1 (Prupe.2G319500), implicated in partnering with PpMYB10s, was generally higher than PpMYB10s in both ‘HJ’ and ‘YL’ fruits, increasing in the early stages and then decreasing in the late stages (Figure 2). The expression of PpMYB10.1 was similar to those of anthocyanin biosynthetic genes, supporting the viewpoint that the activation of PpMYB10.1 was pivotal for anthocyanin accumulation.
Transcript levels of PpGST1 corresponded well with anthocyanin accumulation in peach
In our previous research, PpGST1 was implicated in light‐induced anthocyanin accumulation. PpGST1 belongs to the Phi (F) group, which includes anthocyanin‐related GSTs in dicotyledons like AtTT19 (Arabidopsis thaliana), DcGSTF2 (Dianthus caryophyllus), PfGST1 (Perilla frutescens), PhAn9 (Petunia hybrida), CkmGST3 (Cyclamen persicum), CsGST (Citrus sinensis) and VvGST4 (Vitis vinifera) (Zhao et al., 2017). To analyse the possibility of the participation of PpGST1 in anthocyanin accumulation in peach during fruit development, we compared the expression at the different stages of ‘HJ’ and ‘YL’. The expression of PpGST1 in peel increased gradually during fruit ripening which correlated with the anthocyanin contents (Figure 3a). PpGST1, like most of the anthocyanin biosynthesis genes, showed a large difference in expression level between ‘HJ’ and ‘YL’. In flesh, some red spots gradually appeared as fruit matured, indicating that both cultivars could accumulate anthocyanins. We further compared the anthocyanin contents and the PpGST1 expression in the flesh of fully ripe fruit. It was observed that no anthocyanin was accumulated, and the expression level of PpGST1 was undetectable in outer flesh near the peel (OF) of ‘YL’, whereas the expression was detected in ‘HJ’ at fully ripe stage (stage S6). However, expression of PpGST1 was observed in the inner flesh around the stone (IF) of both ‘HJ’ and ‘YL’, with a distinct higher expression in ‘HJ’, which was consistent with anthocyanin contents (Figure 3b).
Figure 3.
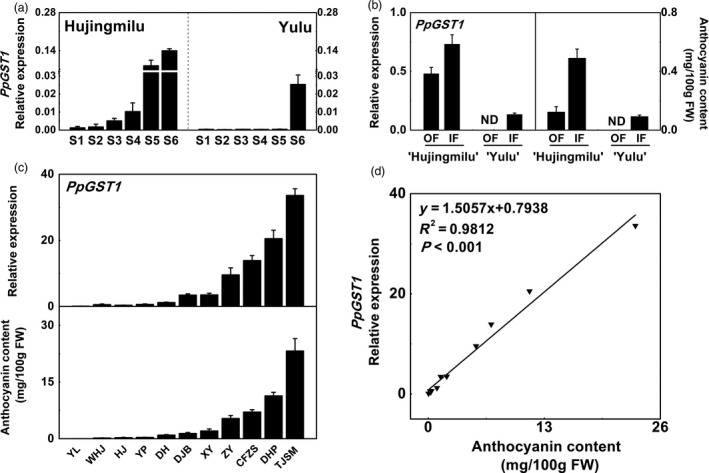
The relationship between PpGST1 (glutathione S‐transferase) expression and anthocyanin accumulation in different parts of fruit and cultivars in peach. (a) Relative expression of PpGST1 in the peel of peach fruit (‘Hujingmilu’ and ‘Yulu’) at different developmental stages. (b) Relative expression of PpGST1 and anthocyanin contents in the outer flesh near the peel (OF) and inner flesh around the stone (IF) of fully ripe fruit (‘Hujingmilu’ and ‘Yulu’). ND, not detectable. (c) Relative expression of PpGST1 and anthocyanin content in the flesh of fully ripe fruit among eleven peach cultivars. Abbreviations used were as follows: CFZS, ‘Cangfangzaosheng’; DH, ‘Dahe’; DHP, ‘Dahongpao’; DJB, ‘Dajiubao’; HJ, ‘Hujingmilu’; TJSM, ‘Tianjingshuimi’; WHJ, ‘Wanhujing’; XY, ‘Xinyu’; YP, ‘Youpan’; YL, ‘Yulu’; and ZY, ‘Zhongyou’. (d) Correlation between the relative expression of PpGST1 and anthocyanin contents in fully ripe fruit of eleven peach cultivars. Data were means (±SE) from three independent biological replicates.
We further analysed the correlation between anthocyanin contents and PpGST1 transcript levels in flesh of mature fruit from eleven peach cultivars (Figure 3c). Anthocyanin contents and PpGST1 expression levels differed in different cultivars, being high in red‐fleshed peach ‘Tianjingshuimi’ (‘TJSM’) and ‘Dahongpao’ (‘DHP’), and were consistent with the coloration of the flesh. Linear regression analysis indicated that the expression of PpGST1 had a strong positive correlation with anthocyanin content among different cultivars (R 2 = 0.98, P < 0.001) (Figure 3d). These results suggested that PpGST1 was the most likely GST member participating in anthocyanin transport in peach.
Functional characterization of PpGST1
To investigate the function of PpGST1 in vivo, a molecular complementation experiment was conducted using Arabidopsis tt19, a knockout mutant of the anthocyanin transporter GST (Sun et al., 2012). Two independent transgenic plants (Line 2 and Line 4) with high expression levels of PpGST1, as revealed by RT‐qPCR, were obtained and used for subsequent characterization (Figure 4a, b). The 35S::PpGST1/tt19 transgenic lines had purple pigmentation in the basal regions of the rosette, leaves and stems as does Arabidopsis wild type, recovering the phenotype in tt19 (Figure 4a). Furthermore, quantification of anthocyanin contents in seedlings of tt19 and transformants confirmed visual inspection, which was about 20 times higher in Line 2 and Line 4 than in tt19 (Figure 4c). However, transgenic lines did not rescue the pale brown seed colour in tt19 (Figure 4a), which differed from the brown seed colour in wild type, indicating that PpGST1 played a different role to TT19 in seed coat pigmentation and did not participate in PA accumulation. Therefore, based on the functional complementation of Arabidopsis tt19 with PpGST1, it was suggested that PpGST1 was responsible for anthocyanin but not PA transport in peach. Furthermore, PpGST1‐GFP was found to be partially co‐localized with a tonoplast marker, as well as in nuclei (Figure S1), supporting the assumption that PpGST1 acted as a carrier in vacuolar sequestration of anthocyanins.
Figure 4.
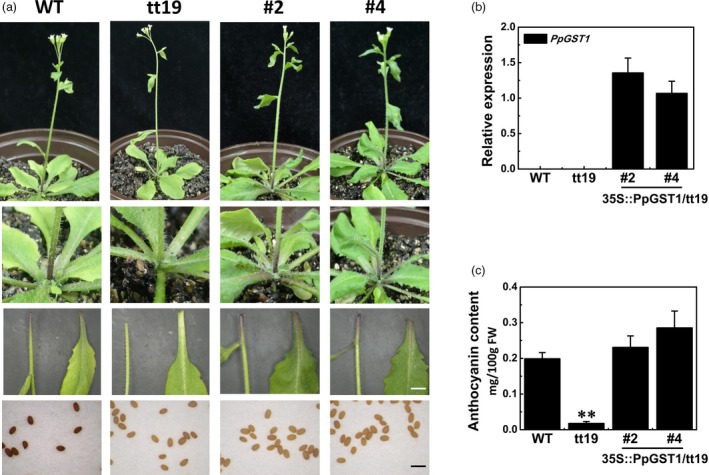
Functional complementation of tt19 mutant with PpGST1 (glutathione S‐transferase). (a) Phenotypic characterization of Arabidopsis wild type (WT), mutant (tt19) and transgenic lines (35S::PpGST1/tt19, Line 2 and Line 4). Images of the adult plants at bolting, the base of the rosette, leaves and stems from the adult plants, fresh seeds were shown. White bar: 0.5 mm; and black bar: 1 mm. (b) Expression analysis of PpGST1 in 35S::PpGST1/tt19 transgenic lines (Line 2 and Line 4) as well as WT and tt19 mutant. (c) Anthocyanin content in 7‐d‐old seedlings (WT, tt19, Line 2 and Line 4) grown on Murashige and Skoog medium supplemented with 5% sucrose. Data were presented as the means (±SE) from three independent biological replicates.
Transient overexpression of PpGST1 in tobacco leaves and peach fruit
Transient overexpression was performed in tobacco leaves and peach fruit to examine the function of PpGST1 in vivo. Tobacco leaves were infiltrated with 35S::PpMYB10.1 (+/− 35S::PpbHLH3) or the combination of 35S::PpMYB10.1 (+/− 35S::PpbHLH3) and 35S::PpGST1, and visible purple patches in the injected areas occurred at one week following certain infiltrations. Only a slight pigmentation was found when leaves were infiltrated with 35S::PpMYB10.1 alone, while an intense pigmentation was observed when co‐infiltrated with 35S::PpbHLH3. Also a stronger purple coloration appeared after the transient overexpression of 35S::PpMYB10.1 together with 35S::PpGST1, both with or without 35S::PpbHLH3 (Figure 5a). However, infiltration with 35S::PpGST1 or 35S::PpbHLH3 or 35S::PpGST1 + 35S::PpbHLH3 in the absence of 35S::PpMYB10.1 did not result in accumulation of anthocyanins, indicating that PpMYB10.1 was essential for regulating anthocyanin biosynthesis while PpbHLH3 and PpGST1 could facilitate increased anthocyanin accumulation.
Figure 5.

Transient overexpression of PpMYB10.1 (+/‐PpbHLH3) or PpGST1 (glutathione S‐transferase) in tobacco (Nicotiana tabacum) leaves (a) and nectarine (cv. ‘Zhongnongjinhui’) fruit at breaker stage (b). G, PpGST1; b, PpbHLH3; M, PpMYB10.1; SK, empty vector. Digital images of infiltration sites were taken at one week after infiltration for tobacco leaves and nectarine fruit, respectively. In each case, one representative example was shown from at least six infiltrated tobacco leaves and ten fruit.
Similar overexpression results were obtained in peach fruit. Clear red colour patches appeared after one week when 35S::PpGST1 and 35S::PpMYB10.1 were simultaneously infiltrated into ‘Zhongnongjinhui’ (‘ZNJH’) nectarine fruit, which was more obvious than the injection site of 35S::PpMYB10.1 alone (Figure 5b). The co‐infiltration of 35S::PpMYB10.1 and 35S::PpGST1 also resulted in red pigmentation in the peel of ‘YL’ (Figure S2), indicating that fruit coloration could be rescued by the overexpression of PpMYB10.1 and PpGST1. However, no obvious pigmentation appeared at infiltration sites after transformation with 35S::PpGST1 alone, possibly due to the low endogenous expression of PpMYB10.1 in these peach fruit. In this study, intense pigmentation could also be induced even without co‐infiltration with 35S::PpbHLH3. This suggested the expression of endogenous PpbHLH3 was strong enough to partner with PpMYB10.1. In summary, deeper colour in tobacco leaves and peach fruit developed following the transient overexpression of PpMYB10.1 together with PpGST1 than PpMYB10.1 alone, supporting that PpGST1 promoted anthocyanin accumulation.
Silencing of PpGST1 reduced anthocyanin accumulation in peach fruit
The virus‐induced gene silencing (VIGS) system was used to further validate the function of PpGST1. Agrobacterium tumefaciens cultures with pTRV1/pTRV2 or pTRV1/pTRV2‐PpGST1 constructs were transiently infiltrated into the blood‐fleshed peach fruit at breaker stage. A decreased pigmentation in the flesh at the injection site was observed at two weeks following the injection with pTRV1/pTRV2‐PpGST1, while no change in flesh colour was found at the site injected with pTRV1/pTRV2. Reduced coloration in the peel around the injection site was also observed (Figure 6a). The anthocyanin content in the flesh around the injection site of pTRV1/pTRV2‐PpGST1 (1.13 mg/100 gFW) was significantly lower compared with corresponding opposite non‐injected site (11.1 mg/100 gFW) in the same fruit and the control (10.82 mg/100 gFW), which correlated well with reduced expression levels of PpGST1 (Figure 6b, c). Interestingly, the genes encoding anthocyanin biosynthetic enzymes and related transcription factors also showed lower expression in treatment of pTRV1/pTRV2‐PpGST1 (Figure 6c). These results showed that PpGST1 had an important role in anthocyanin accumulation in peach.
Figure 6.
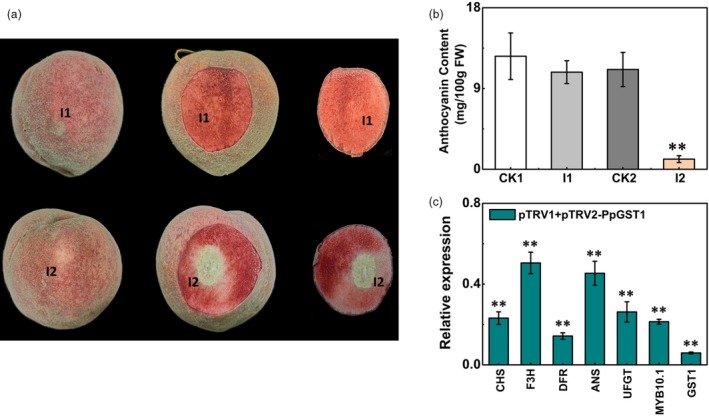
Functional analysis of the PpGST1 (glutathione S‐transferase) gene by TRV (tobacco rattle virus)‐based virus‐induced gene silencing (VIGS). (a) Silencing of the PpGST1 gene in blood‐fleshed peach fruit at breaker stages. I1, infiltration of pTRV1 + pTRV2; I2, infiltration of pTRV1 + pTRV2‐PpGST1. The photograph was taken at two weeks after infiltration. Images of the whole fruit, one side of longitudinal section and the other side of longitudinal section were shown from left to right, respectively. (b) Anthocyanin contents in the flesh around the infiltration sites (CK1, CK2 and I1, I2). CK1, the opposite non‐infiltrated sites of I1; CK2, the opposite non‐infiltrated sites of I2. (c) Relative expression of PpGST1 and anthocyanin structural genes and transcription factors in the flesh around the infiltration sites (I2) at two weeks after infiltration. The expression in the empty vector (pTRV1 + pTRV2) infiltrated tissue (I1) was used as the calibrator (set as 1). Data were means (±SE) from three independent biological replicates and Student’s t‐test was used for statistical analyses compared with corresponding control (**P < 0.01).
Analysis of cis‐elements in the PpGST1 promoter
We cloned the coding region of PpGST1 in ‘HJ’ and ‘YL’, respectively, and found no difference in the amino acid sequence, which was also identical with the reference sequence of PpGST1 (Prupe.3G013600, also previously as ppa011307m). The promoter regions (2000 bp upstream) of PpGST1 were also cloned for both ‘HJ’ and ‘YL’. The promoter sequence of HJ‐PpGST1 was the same as the reference sequence, while there were some differences in the promoter sequence of YL‐PpGST1 as compared with the sequence of HJ‐PpGST1.
To investigate the presence of cis‐acting elements, the promoter sequence of HJ‐PpGST1 was analysed. The presence of putative cis‐regulatory elements in the promoter region of HJ‐PpGST1 was listed in Table S1. TATA‐box and CAAT‐box, which played an important role in transcription initiation, were located, respectively, at −8, +9 and −85 bp upstream of the ATG start codon. Aside from several common cis‐acting elements and core promoter elements, some responsive elements were identified in the PpGST1 promoter as well. These main elements included light‐responsive elements (G‐box, ACE and GATA‐box), hormone‐responsive elements such as ABA‐responsive elements (ABRE), methyl jasmonate (MeJA)‐responsive elements (TGACG‐motif), auxin‐responsive elements (TGA‐element), ethylene‐responsive element (ERE), gibberellin‐responsive element (P‐box) and other stress‐responsive elements (TC‐rich repeats and GC‐motif), indicating that the transcription of PpGST1 might be regulated by various environmental and physiological factors such as light, hormones and abiotic stresses. In addition, four MYB binding sites (MBSs, CAACCA and C/TAACTG) were also observed in PpGST1 promoter region (Table S1). Besides, the promoter sequence of YL‐PpGST1 was also analysed. However, the sequence differences between the promoters isolated from ‘YL’ and ‘HJ’ did not include any of these known motifs.
PpMYB10.1 trans‐activated the PpGST1 promoter
To investigate the effect and the possible role of anthocyanin‐related TFs on transcriptional activity of PpGST1 promoter, dual‐luciferase assays were performed. Compared with the control, PpMYB10.1 showed significant trans‐activation effects on the promoters of both HJ‐PpGST1 (5.3‐fold) and YL‐PpGST1 (3.1‐fold) (Figure 7a). Even stronger activation effects, 8.3‐fold and 5.9‐fold induction on HJ‐PpGST1 and YL‐PpGST1 promoters, respectively, were observed when PpMYB10.1 was co‐infiltrated with PpbHLH3. However, the promoter of neither HJ‐PpGST1 nor YL‐PpGST1 could be activated by PpMYB10.2 or PpMYB10.3 even when co‐infiltrated with PpbHLH3. Infiltration with PpbHLH3 alone also had no activation effect (Figure 7a). The results showed that PpMYB10.1 was responsible for the red coloration in peach by regulating not only anthocyanin biosynthesis but also the transport of anthocyanins as revealed in this study. Noticeably, PpGST1 expression was induced by PpMYB10.1 in ‘HJ’ and ‘YL’ to different degrees, which might be caused by the distinct promoter sequences. As the four MBSs found in PpGST1 promoter (−2000 bp) were identical in ‘HJ’ and ‘YL’, the observed up‐regulation differences in two cultivars were not caused by the coding sequence or MBSs in the promoter.
Figure 7.

PpMYB10.1 activated the PpGST1 (glutathione S‐transferase) promoter in dual‐luciferase transient assays. (a) Effects of PpMYB10.1, PpMYB10.2 or PpMYB10.3 (+/‐PpbHLH3) on the promoter activity of PpGST1 in dual‐luciferase assays. (b) Schematic diagrams of motif mutations for the PpGST1 promoter (cv. ‘Hujingmilu’). (c) Effects of PpMYB10.1 on the activity of original and mutated promoters of PpGST1 in dual‐luciferase assays. The ratio of LUC/REN of the empty vector (SK) plus promoter was used as the calibrator (set as 1). Data were means (± SE) from six independent biological replicates and Student’s t‐test was used for statistical analyses compared with corresponding control (**P < 0.01).
Subsequent experiments were carried out to study the critical MBS elements in the HJ‐PpGST1 promoter. When MBS1 (CAACCA), the one closest to the ATG start codon, was mutated to CTTCCT in HJ‐PpGST1 promoter (PpGST1m1, Figure 7b), trans‐activation activity of PpMYB10.1 was reduced by 72% (Figure 7c). The activity of other mutated PpGST1 promoters (PpGST1m2, PpGST1m3 and PpGST1m4) with the presence of MBS1 but loss of one of the other three MBSs were still significantly enhanced to 4.1‐fold, 5.3‐fold and 4.8‐fold by PpMYB10.1, which showed no significant difference compared to the HJ‐PpGST1 promoter (Figure 7c).
Discussion
GST is a pivotal player in anthocyanin transport and accumulation
Anthocyanins function as UV protectants and bioactive substances in plant life, and contribute to the red‐purple coloration for horticultural plant organs, which is also a key parameter in determining external quality. The biosynthetic and regulatory mechanisms of anthocyanins have been well studied at the genetic and molecular levels.
Anthocyanins are synthesized in the cytosol and accumulate in vacuole. In the past few years, the mechanisms underlying intracellular transport of anthocyanins have become partially unveiled. The delivery of anthocyanins from cytosolic synthesis into vacuolar accumulation needs GST mediation, membrane transport or vesicle trafficking (Zhao, 2015). GSTs comprise a ubiquitous complex gene family in plants with the function to catalyse glutathionylation in a covalent form and serve as non‐enzymatic carrier protein for endogenous compounds that require compartmentalization (Conn et al., 2008; Marrs et al., 1995). The involvement of GSTs in trafficking and accumulation of anthocyanins have been verified in maize (Marrs et al., 1995), petunia (Mueller et al., 2000), Arabidopsis (Sun et al., 2012), cyclamen (Kitamura et al., 2012), perilla (Yamazaki et al., 2008), grape (Conn et al., 2008), apple (Jiang et al., 2019) and litchi (Hu et al., 2016). In this study, we found a GST (PpGST1), with GeneID (Prupe.3G013600, also previously as ppa011307m) later found to be identical to that of an anthocyanin transporter (Riant) related to the variegated coloration of peach flowers (Cheng et al., 2015), was differentially expressed in ‘HJ’ and ‘YL’ throughout developmental stages, which may account for the distinct anthocyanin accumulation in the fruit of two cultivars. Also, the transcription of PpGST1 was consistent with anthocyanin contents of peel and flesh in different cultivars.
The function of GST in anthocyanin transport was verified via application of various mutants and functional complementation. It is well known that bz2, a mutant of a GST in maize, only accumulates anthocyanins in the cytosol (Marrs et al., 1995). Previous research showed that the phenotype of a carnation anthocyanin mutant (fl3) was recovered after heterologous expression of maize BZ2 and petunia An9, and indicated that Fl3 encodes a GST involved in anthocyanin transport (Larsen et al., 2003). In strawberry, a rap mutant caused by a defective GST was found to cause green‐foliage (Luo et al., 2018). Research has also revealed that a GST member can be simultaneously involved in the vacuolar sequestration of different flavonoid substances in some plants. Heterologous overexpression of VviGST4 (Vitis vinifera) and AcGST1 (Actinidia chinensis) in Arabidopsis tt19 mutant can functionally complement both the anthocyanin‐less phenotype in plant and the PA‐deficient phenotype in seed coat (Liu et al., 2019; Pérez‐Díaz et al., 2016). However, in this study, PpGST1 only possessed the capacity to functionally complement the anthocyanin‐less phenotype of Arabidopsis tt19 mutant but not the PA‐deficient phenotype in the seed coat, which was similar to the result of An9 (Petunia hybrida), LcGST4 (Litchi chinensis), RAP (Fragaria ananassa), CsGSTF1 (Camellia sinensis) and MdGSTF6 (Malus domestica) in the tt19 complementation assay (Hu et al., 2016; Jiang et al., 2019; Kitamura et al., 2004; Luo et al., 2018; Wei et al., 2019). These data indicated that PpGST1 plays a pivotal role in anthocyanin transport and differed from AtTT19, VviGST4 and AcGST1 which also participate in seed deposition of PAs.
At present, GSTs specific for transport of other flavonoids, including flavonols and PAs, in peach are unknown. In plants, flavonoid transport mechanisms are diverse and redundant to adapt to the changing environmental conditions (Zhao, 2015). Transport mechanisms such as GST‐mediated, membrane transporter and vesicle trafficking differ in substrate specificity, localization and efficiency for vacuolar sequestration of flavonoids. The major flavonoids in peach are anthocyanins, PAs and flavonols, the content of which varies during fruit development. In conclusion, though the pivotal role of PpGST1 in anthocyanin transport was established, the GSTs responsible for transport of other flavonoids need to be identified and there is a possibility that co‐operation of multiple flavonoid transport mechanisms is present in peach.
It is previously thought that GSTs participate in vacuolar sequestration of anthocyanins through GSH activity (Edwards et al., 2000; Hayes and Pulford, 1995). However, no evidence for the requirement of GSH conjugation in anthocyanin transport has been reported. A bz2 knockout maize mutant could be complemented by the petunia An9 protein, and no anthocyanin‐GSH conjugate was detected (Mueller et al., 2000). In kiwifruit, the recombinant AcGST1 can increase the water solubility of cyanidin‐3‐O‐galactoside and cyanidin‐3‐O‐xylo‐galactoside with no GSH‐conjugated anthocyanins detected (Liu et al., 2019). In this study, localization of PpGST1 revealed its presence in nuclei and the tonoplast, including the sites at which anthocyanin vacuolar sequestration occurs. We hypothesized that PpGST1 bound to anthocyanins covalently to form glutathione cross‐linking complexes to label anthocyanins in order to be recognized by membrane transporter on tonoplast.
Expression of GST is regulated via different means
Due to the importance of GST in anthocyanin accumulation, a number of studies investigated the factors affecting GST expression. In Arabidopsis, TT19 was up‐regulated due to the overexpression of PAP1 (a R2R3 MYB transcription factor) for anthocyanin accumulation (Wangwattana et al., 2008). Earlier work in litchi revealed that the expression of LcGST4 was also activated by LcMYB1 (Hu et al., 2016). Similar results were also reported in strawberry, apple, tea and kiwifruit (Jiang et al., 2019; Liu et al., 2019; Luo et al., 2018; Wei et al., 2019). Data from the dual‐luciferase assays performed in this study suggested that the transcriptional activity of the PpGST1 promoter is directly up‐regulated by PpMYB10.1 rather than PpMYB10.2 and PpMYB10.3, and it was further activated by PpbHLH3 through the interaction between PpMYB10.1 and PpbHLH3. Analysis of cis‐elements indicated that four same MBSs were present in the HJ‐PpGST1 and YL‐PpGST1 promoters, and PpMYB10.1 specifically recognized the MBS1 which is the one closest to the ATG start codon. The differential activation by PpMYB10.1 on HJ‐PpGST1 and YL‐PpGST1 promoters indicated that either a direct interaction or an indirect interaction occurred with other unknown cis‐elements, which needs further investigation. In previous studies, PpMYB10.1 was characterized as a pivotal regulatory transcription factor participating in anthocyanin biosynthesis (Tuan et al., 2015). Here, our results revealed that PpMYB10.1 was also a positive regulator in anthocyanin transport via stimulation of PpGST1 expression.
Some external or internal factors affecting GST expression have also been identified. It was pointed out that the transcription of a litchi GST, LcGST4, was significantly induced by ABA treatment and light (Hu et al., 2016). In our previous research, the transcript level of PpGST1 was stimulated by UV irradiation (Zhao et al., 2017). Here in this study, several hormone‐responsive, light‐responsive and stress‐responsive elements were predicted in the promoter of PpGST1, indicating that the expression of PpGST1 might be regulated by both internal and external factors.
Anthocyanin accumulation is coordinately regulated by both biosynthesis and transport
The accumulation of anthocyanins in peach fruit is related to biosynthetic genes, as well as related transcription factor and transport genes. PpCHS, PpDFR and PpUFGT were considered to be key structural genes in the anthocyanin biosynthetic pathway (Tsuda et al., 2004). In this study, the expression level of PpUFGT showed especially significant difference between ‘HJ’ and ‘YL’ fruits during late developmental stages (Figure 2). PpMYB10.1, PpMYB10.2 and PpMYB10.3 were reported to activate the expression of PpDFR and PpUFGT to regulate anthocyanin biosynthesis (Rahim et al., 2014; Ravaglia et al., 2013), while PpbHLH3 and PpWD40‐1 could promote anthocyanin accumulation (Liu et al., 2015; Tuan et al., 2015). In this study, it was found that both PpMYB10.1 and PpGST1 were indispensable to fruit coloration in peach. The transient overexpression of PpGST1 together with PpMYB10.1 led to much deeper coloration as compared with PpMYB10.1 alone, in accordance with the weaker anthocyanin accumulation in peach fruit after silencing of the PpGST1. This indicates that the expression of PpGST1 is critical for fruit coloration in peach. Anthocyanin accumulation is a complex process which varies in different genetic background of cultivars due to many factors. Our study provides data for further investigation of the molecular mechanisms of anthocyanin transport in peach.
Experimental procedures
Plant materials
The peach [Prunus persica (L.) Batsch] cultivars (cv.) ‘Hujingmilu’ (‘HJ’) and ‘Yulu’ (‘YL’) were maintained in an orchard located at the Fenghua Peach Research Institute (Zhejiang, China). Young leaves were collected in spring. Fruit were collected during development including six stages (S1, fruitlet; S2, immature green; S3, mature green; S4, breaker; S5, commercial maturity; and S6, fully ripe) (Figure 1a). Peel and flesh (including outer flesh near the peel, OF, and inner flesh around the stone, IF, as shown in Figure 1a) samples were separated from 15 fruit, serving as three biological replicates with 5 fruit in each replicate, at each developmental stage, immediately frozen in liquid nitrogen and stored at −80°C for subsequent analysis.
Fully ripe fruit of nine peach cultivars, including ‘Cangfangzaosheng (CFZS)’, ‘Dahe (DH)’, ‘Dahongpao (DHP)’, ‘Dajiubao (DJB)’, ‘Tianjingshuimi (TJSM)’, ‘Wanhujing (WHJ)’, ‘Xinyu (XY)’, ‘Youpan (YP)’ and ‘Zhongyou (ZY)’, were utilized in GST expression analysis. Blood‐fleshed peach and yellow‐fleshed nectarine (‘Zhongnongjinhui, ZNJH’) fruit collected at breaker stage were used in transient expression and virus‐induced gene silencing (VIGS) assays as described below.
Colour measurement
Peel colour was measured with a Hunter Lab Mini Scan XE Plus colorimeter at four evenly distributed equatorial positions around the fruit. A mean value was obtained for each fruit. A comprehensive indicator of the colour index of red grapes (CIRG) was calculated with the formula CIRG = (180‐H)/ (L* + C), while C = (a* 2 + b* 2)0.5 and H = arctan (b*/a*) (Carreño et al., 1995; Usenik et al., 2009; Zhang et al., 2008).
Anthocyanin extraction and HPLC analysis
Extraction and quantification of anthocyanins were carried out as described in our previous study (Zhao et al., 2017). Briefly, 1 g of sample powder was incubated with 5 mL extraction solution (0.05% HCl in methanol) at 4°C for 12 h. After centrifugation, the supernatants were combined and evaporatively dried with a rotary evaporator. The residue was dissolved with 1 mL methanol and filtered with a 0.22 μm Millipore membrane. HPLC analysis was conducted by an Agilent 1260 liquid chromatograph equipped with a ZORBAX SB‐C18 column (4.6 × 250 mm, 5 μm) as described by Cheng et al. (2014).
Proanthocyanidin, total flavonoid and flavonol measurements
Proanthocyanidins (PAs) were extracted and measured following the previously reported method with a minor modification (Wang et al., 2011). Approximately, 0.5 g of sample powder was extracted with 10 mL of anhydrous ethanol overnight in the dark. After centrifugation, 0.5 mL supernatant, 3 mL vanillin (4% w/v) and 1.5 mL concentrated HCl were mixed and put for 15 min at room temperature. Then the absorbance of the reaction mixture was measured at 500 nm by an ultraviolet spectrophotometer. Authentic proanthocyanidin was employed as reference.
Determination of total flavonoids was performed according to the aluminum ion colorimetric assay developed by Chang et al. (2002). Flavonols were measured with a colorimetric protocol of aluminum chloride (Amoussa et al., 2015). The absorbance was measured at 510 nm for flavonoids and at 425 nm for flavonols, respectively. The standard curves were made with quercetin and rutin solutions, for flavonoid and flavonol measurements, respectively. All determinations were performed in triplicate.
DNA, RNA isolation and reverse‐transcription quantitative PCR (RT‐qPCR) analysis
Genomic DNA was isolated from young peach leaves by an improved cetyltrimethylammonium bromide (CTAB) protocol (Chen et al., 2004). Total RNA was extracted according to a modified CTAB method (Shan et al., 2008) for peach fruit tissues and the TRIzol Reagent Kit (Ambion, Hopkinton, MA, USA) for Arabidopsis plants. Total RNA was purified to remove genomic DNA contamination, and first‐strand cDNA synthesis was performed using HiScript® II Q Select RT SuperMix (Vazyme) following the manufacturer’s instructions. RT‐qPCR reactions were carried out with SsoFast EvaGreen Supermix kit (Bio‐Rad, CA, USA) in a CFX96 instrument (Bio‐Rad, CA, USA). The RT‐qPCR programme was initiated with the preliminary step of 3 min at 95°C; followed by 45 cycles of 95°C for 10 s, 60°C for 30 s and then 95°C for 10 s followed by a continuous increase from 65°C to 95°C with a ramp rate of 0.5°C/s for dissociation curve analysis. A peach gene, PpTEF2 (a translation elongation factor gene, GenBank accession No. JQ732180) and an Arabidopsis gene, ATACT2 (actin 2, GenBank accession No. AT3G18780) (Tong et al., 2009; Zhou et al., 2013), were selected as internal controls to normalize expression of all target genes in peach and Arabidopsis, respectively. The primer sequences used for RT‐qPCR are listed in Table S2.
Vector construction
PCR primer sequences involved in vector construction were listed in Table S3. Genomic DNA or cDNA generated from young leaves and fruit were used as templates for sequence amplification. For overexpression, the full‐length coding sequence of PpMYB10.1 (Prupe.3G163100), PpMYB10.2 (Prupe.3G163000), PpMYB10.3 (Prupe.3G163300), PpbHLH3 (Prupe.8G242100) and PpGST1 (Prupe.3G013600) were cloned and recombined into the pGreenII 0029 62‐SK vector (PpMYB10.1‐SK, PpMYB10.2‐SK, PpMYB10.3‐SK, PpbHLH3‐SK and PpGST1‐SK). The promoters of PpGST1 (2000 bp) from both ‘HJ’ and ‘YL’ were amplified and constructed into the pGreen II 0800‐LUC vector, respectively, (HJ‐PpGST1‐LUC and YL‐PpGST1‐LUC). Motifs of PpGST1 promoter were mutated by the Fast Mutagenesis System Kit (Transgene) and then constructed into the pGreen II 0800‐LUC vector, respectively, (PpGST1m1‐LUC, PpGST1m2‐LUC, PpGST1m3‐LUC and PpGST1m4‐LUC). For construction of the vector pTRV2‐PpGST1, a 300‐bp fragment of PpGST1 (1‐300 bp) was amplified and inserted into the vector pTRV2. To observe the subcellular localization, coding sequence of PpGST1 was fused with green fluorescent protein (GFP) and inserted into the pCAMBIA super 1300‐eGFP vector (PpGST1‐GFP). All plant expression constructs were electroporated into Agrobacterium tumefaciens strain GV3101 (MP90) by the GenePulser Xcell™ Electroporation Systems (Bio‐Rad, Hercules, CA, USA).
Stable transformation of Arabidopsis
The Arabidopsis transparent testa19 (tt19) mutant (SALK_105779) in the Columbia genetic background was purchased from The Arabidopsis Information Resource (TAIR). Arabidopsis plants were transformed according to the floral dip method (Zhang et al., 2006). SilwetL‐77, used as a surfactant for Agrobacterium‐based stable transformation in Arabidopsis, was purchased from Real‐Times (Beijing, China). T1 transgenic seeds were screened on half‐strength Murashige and Skoog plates supplemented with 50 mg/mL kanamycin.
Transient overexpression on tobacco leaves and peach fruit
Transient overexpression assays were carried out in tobacco (Nicotiana tabacum) leaves and peach fruits (cv. ‘ZNJH’ and cv. ‘YL’). Briefly, a single colony from Agrobacterium cells was inoculated in 10 mL of liquid LB medium with appropriate antibiotics until A600 reached about 0.8‐1.0. After centrifugation, the cells resuspended in buffer (10 mm MES, 10 mm MgCl2, 150 mm acetosyringone, pH 5.6) to reach an A600 of exactly 0.75 were infiltrated into tobacco leaves of 4‐ to 6‐week‐old plants and peach fruit at breaker stage. The Agrobacterium cultures with empty pGreenII 0029 62‐SK vector (SK) acted as a negative control. Transient expression treatments were performed with three biological replicates and every 6 tobacco plants and at least 10 peach fruit in each replicate.
The infiltrated plants and fruit were placed in a growth chamber (24°C, 60% humidity, 16 h light and 8 h dark) for phenotype identification. Digital photographs of the colour phenotype were taken at one week after infiltration.
The TRV (tobacco rattle virus)‐based virus‐induced gene silencing (VIGS) system on blood‐fleshed peach fruit
The Agrobacterium cultures expressing pTRV1 with pTRV2 or pTRV2‐PpGST1 were mixed in a 1:1 ratio and infiltrated into the flesh of blood‐fleshed peach fruit at breaker stage on the trees. Peach fruit was collected and photographed at two weeks after infiltration. Each treatment was performed with three biological replicates with at least 20 fruit in each replicate.
Dual‐luciferase assay
Dual‐luciferase assays were conducted in Nicotiana benthamiana leaves according to a previous protocol (Zeng et al., 2015). Agrobacterium cultures (strain GV3101) containing the pGreen II 0029 62‐SK vector (SK) with the full‐length coding sequence of PpMYB10.1/2/3, PpbHLH3 and the pGreen II 0800‐LUC (PpGST1‐LUC) vector with the promoter of PpGST1 were transiently expressed in N. benthamiana leaves. Agrobacterium cultures carrying constructs were suspended in infiltration buffer (10 mm MES, 10 mm MgCl2, 150 mm acetosyringone, pH 5.6) to an optimal density (A 600 = 0.75). The mixture of Agrobacterium cultures with transcription factor (1 mL) and the promoter (100 µL) were injected into N. benthamiana leaves together. After three days of infiltration, the activity of transcription factor (PpMYB10.1/2/3, PpbHLH3)‐promoter (PpGST1) interaction was assayed based on the ratio of firefly luciferase (LUC) to renilla luciferase (REN) intensities by using dual‐luciferase reagents (Promega, MI, USA). The LUC/REN value of the empty vector SK on the promoter was set as 1, as a calibrator. The results were calculated from three independent experiments with six replicate reactions in each experiment.
Subcellular localization analysis
To examine the subcellular localization of PpGST1, PpGST1‐GFP was transiently expressed in N. benthamiana leaves by Agrobacterium infiltration (GV3101) with the method described in the dual‐luciferase assay above. Tonoplast marker and transgenic N. benthamiana plant with a red fluorescent nuclear marker (Nucleus‐RFP) were used to identify the intracellular structure. On the third day after infiltration, the GFP florescence of the transiently infected leaves was measured by a Zeiss LSM710NLO confocal laser scanning microscope.
Cis‐acting element analysis
The cis‐acting elements were predicted in PLACE (Plant Cis‐acting Regulatory DNA Elements; ://www.dna.affrc.go.jp/PLACE/signalscan.html) and PlantCARE (://bioinformatics.psb.ugent.be/webtools/plantcare/html/) databases.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
CX and ACA designed the study; CX obtained funding; YZhao, WQ and YZhu carried out the experimental work; YZhao, ACA, KLW and CX contributed to the writing. All authors read and approved the final manuscript.
Supporting information
Figure S1 Subcellular localization of PpGST1 in Nicotiana benthamiana leaves.
Figure S2 Transient overexpression of PpMYB10.1 or PpGST1 in peach (cv. ‘YL’) fruit at breaker stage.
Table S1 Cis‐acting elements found in the upstream region of PpGST1 gene.
Table S2 Primers used for reverse‐transcription quantitative PCR analysis.
Table S3 Primers used for vector construction. The restriction sites are underlined.
Acknowledgements
This study was supported by the National Natural Science Foundation in China (31572102), the Fundamental Research Funds for the Central Universities (2019FZA6010) and the 111 Project (B17039).
Zhao, Y. , Dong, W. , Zhu, Y. , Allan, A. C. , Lin‐Wang, K. and Xu, C. (2020) PpGST1, an anthocyanin‐related glutathione S‐transferase gene, is essential for fruit coloration in peach. Plant Biotechnol J, 10.1111/pbi.13291
References
- Alfenito, M.R. , Souer, E. , Goodman, C.D. , Buell, R. , Mol, J. , Koes, R. and Walbot, V. (1998) Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S‐transferases. Plant Cell, 10, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoussa, M.A. , Sanni, A. and Lagnika, L. (2015) Antioxidant activity and total phenolic, flavonoid and flavonol contents of the bark extracts of Acacia ataxacantha . J. Pharmacogn. Phytochem. 4, 172–178. [Google Scholar]
- Carreño, J. , Martínez, A. , Almela, L. and Fernández‐López, J.A. (1995) Proposal of an index for the objective evaluation of the colour of red table grapes. Food Res. Int. 28, 373–377. [Google Scholar]
- Chang, C.C. , Yang, M.H. , Wen, H.M. and Chern, J.C. (2002) Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug Anal. 10, 178–182. [Google Scholar]
- Chen, K.S. , Li, F. , Xu, C.J. , Zhang, S.L. and Fu, C.X. (2004) An efficient macro‐method of genomic DNA isolation from Actinidia chinensis leaves. Hereditas, 26, 529–531. [PubMed] [Google Scholar]
- Cheng, J. , Wei, G. , Zhou, H. , Gu, C. , Vimolmangkang, S. , Liao, L. and Han, Y.P. (2014) Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 166, 1044–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. , Liao, L. , Zhou, H. , Gu, C. , Wang, L. and Han, Y.P. (2015) A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. J. Exp. Bot. 66, 7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, T. , Howatson, G. , West, D. and Stevenson, E. (2015) The potential benefits of red beetroot supplementation in health and disease. Nutrients, 7, 2801–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn, S. , Curtin, C. , Bézier, A. , Franco, C. and Zhang, W. (2008) Purification, molecular cloning, and characterization of glutathione S‐transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 59, 3621–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, P.D. , Lapthorn, A. and Edwards, R. (2002) Plant glutathione transferases. Genome Biol. 3, 3004.1–3004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R. , Dixon, D.P. and Walbot, V. (2000) Plant glutathione S‐transferases: multifunctional enzymes aiding survival in a hostile world. Trends Plant Sci. 5, 193–198. [DOI] [PubMed] [Google Scholar]
- Grotewold, E. and Davis, K. (2008) Trafficking and sequestration of anthocyanins. Nat. Prod. Commun. 3, 1251–1258. [Google Scholar]
- Hayes, J.D. and Pulford, D.J. (1995) The glutathione S‐transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance part II. CRC Crit. Rev. Biochem. 30, 521–600. [DOI] [PubMed] [Google Scholar]
- Hu, B. , Zhao, J. , Lai, B. , Qin, Y. , Wang, H. and Hu, G. (2016) LcGST4 is an anthocyanin‐related glutathione S‐transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 35, 831–843. [DOI] [PubMed] [Google Scholar]
- Islam, S. , Rahman, I.A. , Islam, T. and Ghosh, A. (2017) Genome‐wide identification and expression analysis of glutathione S‐transferase gene family in tomato: Gaining an insight to their physiological and stress‐specific roles. PLoS ONE, 12, e0187504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Chen, M. , He, N. , Chen, X. , Wang, N. , Sun, Q. , Zhang, T. et al (2019) MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 6, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, S. , Shikazono, N. and Tanaka, A. (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis . Plant J. 37, 104–114. [DOI] [PubMed] [Google Scholar]
- Kitamura, S. , Akita, Y. , Ishizaka, H. , Narumi, I. and Tanaka, A. (2012) Molecular characterization of an anthocyanin‐related glutathione S‐transferase gene in cyclamen. J. Plant Physiol. 169, 636–642. [DOI] [PubMed] [Google Scholar]
- Larsen, E.S. , Alfenito, M.R. , Briggs, W.R. and Walbot, V. (2003) A carnation anthocyanin mutant is complemented by the glutathione S‐transferases encoded by maize Bz2 and petunia An9 . Plant Cell Rep. 21, 900–904. [DOI] [PubMed] [Google Scholar]
- Licciardello, C. , D'Agostino, N. , Traini, A. , Recupero, G.R. , Frusciante, L. and Chiusano, M.L. (2014) Characterization of the glutathione S‐transferase gene family through ESTs and expression analyses within common and pigmented cultivars of Citrus sinensis (L.) Osbeck. BMC Plant Biol. 14, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Song, S. , Yuan, Y.B. , Wu, D.J. , Chen, M.J. , Sun, Q.A. , Zhang, B. et al (2015) Improved peach peel color development by fruit bagging. Enhanced expression of anthocyanin biosynthetic and regulatory genes using white non‐woven polypropylene as replacement for yellow paper. Sci. Hortic. 184, 142–148. [Google Scholar]
- Liu, Y.F. , Qi, Y.W. , Zhang, A.L. , Wu, H.X. , Liu, Z.D. and Ren, X.L. (2019) Molecular cloning and functional characterization of AcGST1, an anthocyanin‐related glutathione S‐transferase gene in kiwifruit (Actinidia chinensis). Plant Mol. Biol. 100, 451–465. [DOI] [PubMed] [Google Scholar]
- Loyall, L. , Uchida, K. , Braun, S. , Furuya, M. and Frohnmeyer, H. (2000) Glutathione and a UV light‐induced glutathione S‐transferase are involved in signaling to chalcone synthase in cell cultures. Plant Cell, 12, 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. , Dai, C. , Li, Y. , Feng, J. , Liu, Z. and Kang, C. (2018) Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 69, 2595–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs, A.K. , Alfenlto, R.M. , Lloyd, M.A. and Walbot, V. (1995) A glutathione S‐transferase involved in vacuolar transfer encoded by the maize gene Bronze‐2 . Nature, 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Moons, A. (2005) Plant hormones regulatory and functional interactions of plant growth regulators and plant glutathione S‐transferases (GSTs). Vitam. Horm. 72, 155–202. [DOI] [PubMed] [Google Scholar]
- Mueller, L.A. , Goodman, C.D. , Silady, R.A. and Walbot, V. (2000) AN9, a petunia glutathione S‐transferase required for anthocyanin sequestration, is a flavonoid‐binding protein. Plant Physiol. 123, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Díaz, R. , Madrid‐Espinoza, J. , Salinas‐Cornejo, J. , González‐Villanueva, E. and Ruiz‐Lara, S. (2016) Differential roles for VviGST1, VviGST3, and VviGST4 in proanthocyanidin and anthocyanin transport in Vitis vinifera . Front. Plant Sci. 7, 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka, F. , Irani, N.G. , Feller, A. , Lu, Y. , Pourcel, L. , Frame, K. and Grotewold, E.A. (2007) Trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum‐to‐vacuole protein‐sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 145, 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim, M.A. , Busatto, N. and Trainotti, L. (2014) Regulation of anthocyanin biosynthesis in peach fruits. Planta, 240, 913–929. [DOI] [PubMed] [Google Scholar]
- Ravaglia, D. , Espley, R.V. , Henry‐Kirk, R.A. , Andreotti, C. and Ziosi, V. (2013) Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappl, P.G. , Carroll, A.J. , Clifton, R. , Lister, R. , Whelan, J. , Harvey, M.A. and Singh, K.B. (2009) The Arabidopsis glutathione transferase gene family displays complex stress regulation and co‐silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 58, 53–68. [DOI] [PubMed] [Google Scholar]
- Shan, L.L. , Li, X. , Wang, P. , Cai, C. , Zhang, B. , Sun, C.D. , Zhang, W.S. et al (2008) Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta, 227, 1243–1254. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Li, H. and Huang, J. (2012) Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant, 5, 387–400. [DOI] [PubMed] [Google Scholar]
- Tong, Z. , Gao, Z. , Wang, F. , Zhou, J. and Zhang, Z. (2009) Selection of reliable reference genes for gene expression studies in peach using real‐time PCR. BMC Mol. Biol. 10, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, T. (2012) Dietary anthocyanin‐rich plants: Biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 56, 159–170. [DOI] [PubMed] [Google Scholar]
- Tsuda, T. , Yamaguchi, M. , Honda, C. and Moriguchi, T. (2004) Expression of anthocyanin biosynthesis genes in the skin of peach and nectarine fruit. J. Am. Soc. Hortic. Sci. 129, 857–862. [Google Scholar]
- Tuan, P.A. , Bai, S.L. , Yaegaki, H. , Tamura, T. , Hihara, S. , Moriguchi, T. and Oda, K. (2015) The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 15, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu, C. , Katayama, H. and Makino, I. (2014) Peace, a MYB‐like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J. Exp. Bot. 65, 1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usenik, V. , Stampar, F. and Veberic, R. (2009) Anthocyanins and fruit colour in plums (Prunus domestica L.) during ripening. Food Chem. 114, 529–534. [Google Scholar]
- Wang, H. , Hu, Z. , Wang, Y. , Chen, H. and Huang, X. (2011) Phenolic compounds and the antioxidant activities in litchi pericarp: Difference among cultivars. Sci. Hortic. 129, 784–789. [Google Scholar]
- Wangwattana, B. , Koyama, Y. , Nishiyama, Y. , Kitayama, M. , Yamazaki, M. and Saito, K. (2008) Characterization of PAP1‐upregulated glutathione S‐transferase genes in Arabidopsis thaliana . Plant Biotechnol. 25, 191–196. [Google Scholar]
- Wei, K. , Wang, L.Y. , Zhang, Y.Z. , Ruan, L. , Li, H.L. , Wu, L.Y. , Xu, L.Y. et al (2019) A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 97, 825–840. [DOI] [PubMed] [Google Scholar]
- Yamazaki, M. , Shibata, M. , Nishiyama, Y. , Springob, K. , Kitayama, M. , Shimada, N. , Aoki, T. et al (2008) Differential gene expression profiles of red and green forms of Perilla frutescens leading to comprehensive identification of anthocyanin biosynthetic genes. FEBS J. 275, 3494–3502. [DOI] [PubMed] [Google Scholar]
- Zeng, J.K. , Li, X. , Xu, Q. , Chen, J.Y. , Yin, X. , Ferguson, I.B. and Chen, K.S. (2015) EjAP2‐1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S. , Niu, Q. and Chua, N. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang, W.S. , Li, X. , Zheng, J.T. , Wang, G.Y. , Sun, C.D. , Ferguson, I.B. and Chen, K.S. (2008) Bioactive components and antioxidant capacity of Chinese bayberry (Myrica rubra Sieb. and Zucc.) fruit in relation to fruit maturity and postharvest storage. Eur. Food Res. Technol. 227, 1091–1097. [Google Scholar]
- Zhao, J. (2015) Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20, 576–585. [DOI] [PubMed] [Google Scholar]
- Zhao, J. and Dixon, R.A. (2010) The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 15, 72–80. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Dong, W.Q. , Wang, K. , Zhang, B. , Allan, A.C. , Lin‐Wang, K. , Chen, K.S. et al (2017) Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Front. Plant Sci. 8, 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Guo, D. , Li, J. , Cheng, J. , Zhou, H. , Gu, C. , Gardiner, S. et al (2013) Coordinated regulation of anthocyanin biosynthesis through photorespiration and temperature in peach (Prunus persica). Tree Genet. Genom. 9, 265–278. [Google Scholar]
- Zhou, Y. , Zhou, H. , Lin‐Wang, K. , Vimolmangkang, S. , Espley, R.V. , Wang, L. , Allan, A.C. et al (2014) Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 14, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Lin‐Wang, K. , Wang, H. and Han, Y.P. (2015) Molecular genetics of blood‐fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Li, H. , Guo, D. , Wang, Y. , Dai, H. , Mei, W. and Peng, S. (2016) Transcriptome‐wide identification and expression analysis of glutathione S‐transferase genes involved in flavonoids accumulation in Dracaena cambodiana . Plant Physiol. Biochem. 104, 304–311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Subcellular localization of PpGST1 in Nicotiana benthamiana leaves.
Figure S2 Transient overexpression of PpMYB10.1 or PpGST1 in peach (cv. ‘YL’) fruit at breaker stage.
Table S1 Cis‐acting elements found in the upstream region of PpGST1 gene.
Table S2 Primers used for reverse‐transcription quantitative PCR analysis.
Table S3 Primers used for vector construction. The restriction sites are underlined.


