Summary
Kernel size is an important trait determining cereal yields. In this study, we cloned and characterized TaDA1, a conserved negative regulator of kernel size in wheat (Triticum aestivum). The overexpression of TaDA1 decreased the size and weight of wheat kernels, while its down‐regulation using RNA interference (RNAi) had the opposite effect. Three TaDA1‐A haplotypes were identified in Chinese wheat core collections, and a haplotype association analysis showed that TaDA1‐A‐HapI was significantly correlated with the production of larger kernels and higher kernel weights in modern Chinese cultivars. The haplotype effect resulted from a difference in TaDA1‐A expression levels between genotypes, with TaDA1‐A‐HapI resulting in lower TaDA1‐A expression levels. This favourable haplotype was found having been positively selected during wheat breeding over the last century. Furthermore, we demonstrated that TaDA1‐A physically interacts with TaGW2‐B. The additive effects of TaDA1‐A and TaGW2‐B on kernel weight were confirmed not only by the phenotypic enhancement arising from the simultaneous down‐regulation of TaDA1 and TaGW2 expression, but also by the combinational haplotype effects estimated from multi‐environment field data from 348 wheat cultivars. A comparative proteome analysis of developing transgenic and wild‐type grains indicated that TaDA1 and TaGW2 are involved in partially overlapping but relatively independent protein regulatory networks. Thus, we have identified an important gene controlling kernel size in wheat and determined its interaction with other genes regulating kernel weight, which could have beneficial applications in wheat breeding.
Keywords: TaDA1, kernel size, kernel weight, additive effect, TaGW2, Triticum aestivum
Introduction
Common wheat (Triticum aestivum L., AABBDD) is a major staple crop in the world. With growing global population and increasing demand for wheat, wheat yields must be further improved. Grain weight and size are major components of wheat yield and are therefore key targets for the further improvement of this crop (Li and Yang, 2017; Mohler et al., 2016; Xie et al., 2015). Many genes regulating wheat grain weights have been identified using comparative genomics approaches, including TaGW2 (Qin et al., 2017; Simmonds et al., 2016; Su et al., 2011), Ta CKX2 (Zhang et al., 2012), TaGS5 (Ma et al., 2016; Wang et al., 2015), TaCYP78A3 (Ma et al., 2015), TaTGW6 (Hanif et al., 2016; Hu et al., 2016), TaFlo2‐A1 (Sajjad et al., 2017), and TaAGP‐S1 and TaAGP‐L (Hou et al., 2017). Based on the knowledge of their counterparts in rice (Oryza sativa), these genes are predicted to function in diverse signalling pathways, including proteasomal degradation, phytohormone signalling, and starch biosynthesis (Zuo and Li, 2014). Although they are known to regulate grain size or grain filling, the interactions between these different regulators and the overall regulatory network underlying grain development remain to be elucidated.
The ubiquitin–proteasome pathway plays a key role in controlling organ size in dicot and monocot plants (Brinton et al., 2018; Li and Li, 2016; Shi et al., 2019). Recently, increasing evidence has indicated that the ubiquitin receptor DA1, a conserved component of the ubiquitin–proteasome pathway, has important functions in regulating seed size. In Arabidopsis thaliana, DA1 functions partially redundantly with one of its homologs, DA1‐RELATED 1 (DAR1), to control seed size by restricting the period of cell proliferation in maternal integuments (Dong et al., 2017; Li et al., 2008). The dominant‐negative mutant da1‐1 carries a G‐to‐A mutation in DA1, causing an R‐to‐K change in the conserved amino acid at position 358 (DA1R358K). This da1‐1 allele dramatically increases the seed and organ size of Arabidopsis plants because the mutant DA1R358K protein negatively impacts the function of DA1 and DAR1 (Li et al., 2008). In rapeseed (Brassica napus) and maize (Zea mays), the overexpression of a similar mutant form of DA1 or DAR1 homologs also improved their seed weights and organ sizes, thereby increasing the overall grain yield and biomass (Wang et al., 2017a; Xie et al., 2018). Furthermore, a loss‐of‐function T‐DNA DA1 mutant (da1‐ko1) in Arabidopsis also caused an increase in seed size, indicating that DA1 can be studied relatively independently of DAR1 in regulating seed size (Dong et al., 2017).
The RING finger‐type E3 ubiquitin ligases BIG BROTHER (BB) and DA2 restrict organ growth (Disch et al., 2006; Xia et al., 2013) and function synergistically with DA1 in organ size determination (Li et al., 2008; Xia et al., 2013). A recent study showed that BB and DA2 ubiquitylate DA1 and activate its peptidase activity, and then activated DA1 cleaves and destabilizes BB and DA2 in a feedback mechanism (Dong et al., 2017). In addition, DA2 physically interacts with DA1, which may enable DA1 to recognize specific substrates for degradation (Xia et al., 2013). DA2 shares significant sequence similarity with GW2, which is known to determine grain weight (Song et al., 2007), and the overexpression of DA2 and OsGW2 in Arabidopsis both restricted seed growth, similar to the phenotype change in rice (Xia et al., 2013). GW2 and its homologs have been confirmed to be conserved negative regulators of grain size and weight in various crops including rice (Song et al., 2007), maize (Li et al., 2010), and wheat (Qin et al., 2017; Su et al., 2011). Both DA1 and GW2 (or DA2) therefore have conserved functions in kernel size control which implies that their interaction in the ubiquitin–proteasome pathway may also be conserved in plants.
In this study, we isolated DA1 in wheat using a bioinformatics approach and mapped the TaDA1 homoeologs on chromosomes 2A, 2B, and 2D. Transgenic analysis showed that the overexpression of TaDA1 in wheat causes a decline in kernel size and weight, while the down‐regulation of TaDA1 using RNA interference (RNAi) had the opposite effect, indicating that TaDA1 has a conserved function in the negative regulation of kernel size. Our haplotype association analysis also demonstrated that, in modern Chinese cultivars, the favourable haplotype of TaDA1‐A was significantly associated with higher kernel weights. Moreover, we found that TaDA1‐A physically interacted with TaGW2‐B and verified their additive effects not only through the enhanced kernel size phenotype generated by the simultaneous down‐regulation of TaDA1 and TaGW2 in wheat, but also using a combinational analysis of TaDA1‐A and TaGW2‐B haplotypes in the association population. Our findings provide evidence that TaDA1 is an important gene controlling kernel size in wheat, and it can be potentially applied in combination with other yield‐related genes in wheat high‐yield breeding.
Results
Cloning and expression analysis of TaDA1 homoeologs in wheat
Based on the reported Arabidopsis and maize DA1 sequences (Li et al., 2008; Xie et al., 2018), we identified DA1 homologs in wheat through a bioinformatics search on Ensembl Plants (IWGSC, 2018). TaDA1 was identified as the closest homolog of AtDA1 and ZmDA1, and its homoeologs were located on chromosomes 2A, 2B, and 2D (Table S1; Figure S1). The coding regions of TaDA1‐A, ‐B, and ‐D are 6363, 6660, and 6180 bp in length, respectively, and each consists of 11 exons and 10 introns (Figure S2). TaDA1‐A, ‐B, and ‐D all comprise 504 amino acids and share high sequence similarities with AtDA1 (~60%) and ZmDA1 (~86%), especially in the conserved UIM and LIM domains (Li et al., 2008; Figure S3). This high level of evolutionary conservation suggests that TaDA1 may have similar biological functions to AtDA1 and ZmDA1.
We first investigated the subcellular localization of TaDA1 in wheat protoplasts. The coding sequences (CDSs) of the TaDA1 homoeologs were fused in‐frame with the gene encoding green fluorescent protein (GFP), and the resulting constructs were introduced into wheat protoplasts using the PEG method (Zheng et al., 2014). As shown in Figure 1a, the GFP fluorescence patterns of the fused TaDA1‐A, ‐B, or ‐D proteins were distributed throughout the cytoplasm and nucleus, indicating that these TaDA1 homoeologs have similar subcellular localizations. Next, we analysed the tissue‐specific expression patterns of the TaDA1 homoeologs using quantitative real‐time PCR (qRT‐PCR) with genome‐specific primers (Table S2). The TaDA1 homoeologs were all constitutively expressed in various wheat tissues but showed higher expression levels in the leaves (seedling and elongation stages) and young spikes (Figure 1b). Among the three homoeologs, TaDA1‐B and TaDA1‐D were predominantly expressed in vegetative organs such as the leaves and roots, while TaDA1‐A was predominantly expressed in reproductive organs such as the young spikes, implying that they might play partially redundant but somewhat divergent roles in regulating the development of different organs.
Figure 1.
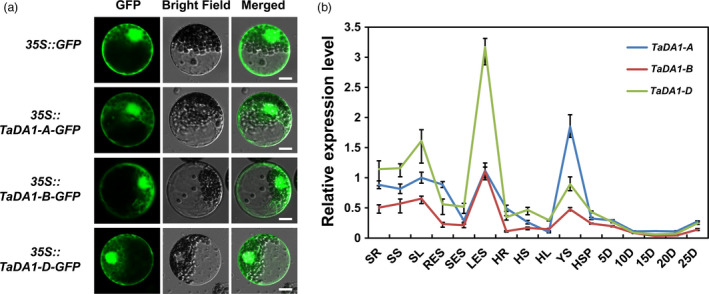
Characterization of TaDA1 homoeologs. (a) Subcellular localization of TaDA1 homoeologs in wheat protoplasts. GFP and TaDA1s‐GFP fusions under the control of the CaMV 35S promoter were transiently expressed in wheat protoplasts. Bar = 10 µm. (b) Tissue expression patterns of the three homoeologs in various Chinese Spring tissues. SR, seedling roots; SS, seedling stems; SL, seedling leaves; RES, roots at elongation stage; SES, stems at elongation stage; LES, leaves at elongation stage; HR, roots at heading stage; HS, stems at heading stage; HL, leaves at heading stage; YS, 1–2‐cm young spikes; HSP, spikes at heading stage; 5D–25D, grains at 5–25 days post‐anthesis, respectively. Normalized values of TaDA1 expression relative to Actin were given as mean ± SD from three replicates.
TaDA1 negatively regulates kernel weight and size in wheat
To study the physiological function of TaDA1 in wheat, we generated TaDA1‐A overexpression lines and RNAi transgenic plants in the hexaploid wheat cultivar Fielder using Agrobacterium‐mediated transformation (Wang et al., 2017b). The total expression levels of TaDA1 were examined in the transgenic plants using qRT‐PCR. As shown in Figure 2a, the mRNA levels of TaDA1 were significantly increased in all three representative overexpression lines (OE‐1–3; P < 0.01) and were reduced in the three independent RNAi lines (Ri‐1–3; P < 0.01). Compared with the WT control, the overexpression lines had smaller kernels, with 1.7–5.6% shorter kernel lengths (KLs) and 3.4–5.1% narrower kernel widths (KWs; Figure 2b–e). The RNAi lines produced significantly larger kernels than the WT, with approximately 2.5–3.5% longer KLs and 4.0–6.8% wider KWs when grown under field conditions (Figure 2b–e). Consistent with these results, the thousand‐kernel weight (TKW) of the overexpression lines and RNAi lines was 2.9–4.5 g lower and 3.6–5.9 g higher than that of the WT, respectively (Figure 2c). The phenotypic effects of the transgenic lines were negatively related to the expression levels of TaDA1; for example, the OE‐1 line had the highest TaDA1 expression level among the three overexpression lines and demonstrated the strongest reductions in kernel size and weight (Figure 2a–e). No significant differences were observed in other yield‐related traits between the transgenic lines and the WT, including the tiller number, grain number per spike, spike length, and spikelet number per spike (Figure S4). Our results show that the down‐regulation of TaDA1 might be effective for improving wheat yields, and confirm that TaDA1 is a conserved negative regulator of kernel size and weight.
Figure 2.
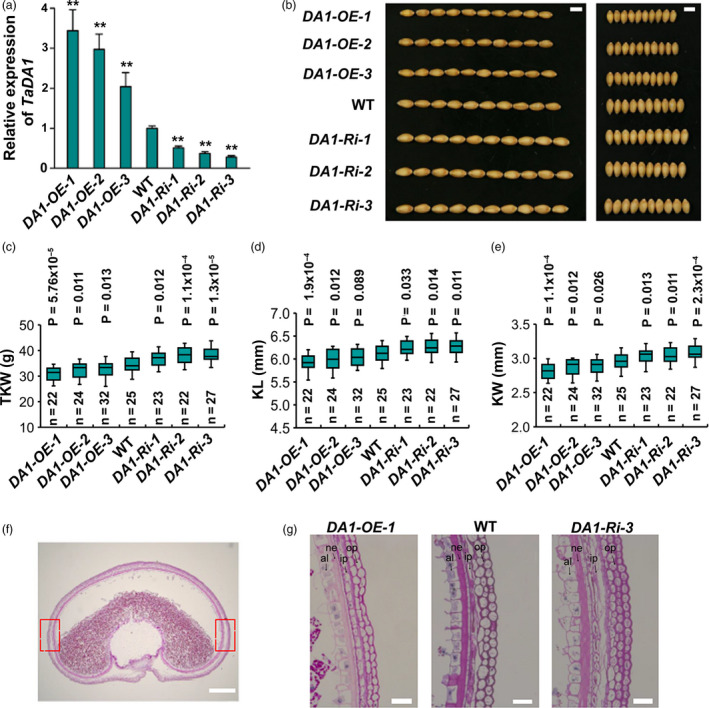
Phenotypic comparison of kernel traits among TaDA1 overexpression lines, RNAi lines, and the WT under field conditions. (a) Expression levels of TaDA1 in three representative T2 generation overexpression lines (OE‐1–3), RNAi lines (Ri‐1–3), and the WT. The values are presented as mean ± SD. **P < 0.01 (ANOVA) indicates a significant difference to the WT. (b) Comparison of the kernel phenotypes among these genotypes. Bar = 5 mm. (c) Thousand‐kernel weights (TKW), (d) kernel lengths (KL), and (e) kernel widths (KW) of these genotypes. Box plots showing the components of descriptive statistics including sample size (n), medians (black lines), upper and lower quartiles (box edges), minimums and maximums (whiskers), and P values (ANOVA). (f) A representative section of the middle portion of a developing grain of Fielder (WT) wheat at 15 DPA. Bar = 500 µm. (g) Comparison of the outer pericarp cells in the TaDA1 overexpression line (DA1‐OE‐1), RNAi line (DA1‐Ri‐3), and WT. Each image was taken at the boxed areas indicated in (f). Op, outer pericarp; ip, inner pericarp; ne, nucellar epidermis; al, aleurone layer. Bars = 50 µm.
TaDA1 restricts the proliferation of maternal pericarp cells
To test whether TaDA1 affects maternal pericarp cell growth, we observed the outer layer cells in grains harvested at 15 days post‐anthesis (DPA) from DA1‐overexpressing plants (DA1‐OE‐1), DA1 RNAi plants (DA1‐Ri‐3), and the WT (Figure 2f, g). Compared with the WT, the DA1‐Ri‐3 plants produced more outer pericarp cells and consequently formed a wider pericarp cell layer (Figure 2g, Figure S5a). By contrast, the outer pericarps of the DA1‐OE‐1 plants contained approximately 8% fewer cells than those of the WT (Figure 2g, Figure S5a). Furthermore, the cell length was slightly reduced in the outer pericarp of the DA1‐Ri‐3 plants (Figure S5b). We also observed similar alteration of thickness in other cell layers including inner pericarp and nucellar epidermis among DA1‐Ri‐3, DA1‐OE‐1, and the WT (Figure 2g). These results confirmed that TaDA1 has a conserved function in restricting maternal cell proliferation.
TaDA1‐A haplotypes were associated with kernel weight and size in modern Chinese cultivars
Since TaDA1‐A was the most highly expressed homoeolog in the reproductive organs, we concluded that it might play the largest role in affecting kernel weight and size. Specific primers were designed to obtain the genomic sequence of TaDA1‐A, including the 2.1‐kb promoter region and the 6.4‐kb coding region (Table S2). By sequencing 36 highly diverse Chinese wheat accessions (Table S3), we detected 19 natural variations present in the TaDA1‐A genomic regions, most of which were located in the introns or promoter regions (Figure 3a). Only three SNPs were located in exons, all of which caused amino acid substitutions (Figure 3a). These variations resulted in three distinct haplotypes (TaDA1‐A‐HapI/II/III) in the common wheat cultivars surveyed. Based on two of the exonic SNPs (T/A at 2,590 bp and G/A at 4,437 bp), we developed two dCAPS (derived cleaved amplified polymorphic sequence) markers to distinguish the three haplotypes of TaDA1‐A (Figure S6).
Figure 3.
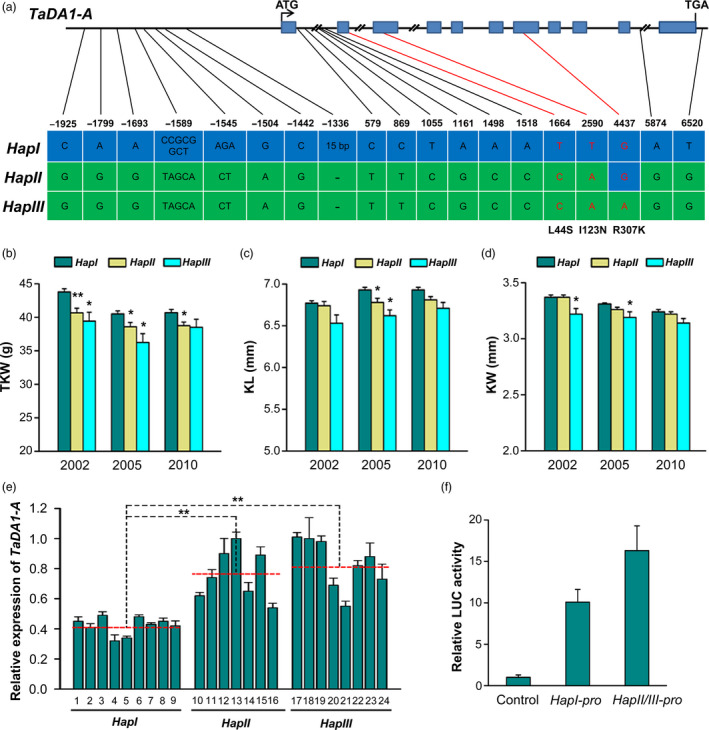
TaDA1‐A haplotype identification and haplotype association analysis with kernel traits in modern Chinese cultivars. (a) Variations in the TaDA1‐A promoter and genomic regions could be divided into three haplotypes (TaDA1‐A‐HapI/II/III). The blue boxes indicate exons. The SNPs in red cause an amino acid substitution (L44S, I123N, and R307K). (b–d) Association of TaDA1‐A haplotypes with the thousand‐kernel weights (TKW; b), kernel lengths (KL; c), and kernel widths (KW, d) of 348 modern Chinese cultivars in three environments (Luoyang, 2002; Luoyang, 2005; Shunyi, 2010). The number of HapI, HapII, and HapIII cultivars is 209, 116, and 23, respectively. The values are presented as mean ± SE. *P < 0.05 and **P < 0.01 (ANOVA) indicate significant differences from HapI. (e) Differential expression levels of TaDA1‐A in the 10‐DPA grains of 24 wheat accessions with different haplotypes. The red‐dashed lines indicate the average expression levels of each haplotype accession. 1, Shanxibaimai; 2, Baiqimai; 3, Xiaobaimang; 4, Dahongmai; 5, Huangguaxian; 6, Chushanbao; 7, Dabaimai; 8, Yuqiumai; 9, Chinese Spring; 10, Hongjinmai; 11, Xiaokouhong; 12, Daimanghongmai; 13, Zhuoludongmai; 14, Hongmai; 15, Youmangbaifu; 16, Shuilizhan; 17, Jiahongmai; 18, Lanhuamai; 19, Baiqiumai; 20, Chejianzi; 21, Baimangmai; 22, Banjiemang; 23, Bianbachunmai; and 24, Dianxihongkeyangmai. **P < 0.01 (ANOVA). (f) Relative LUC activity (LUC/REN) from the transient expression analysis of TaDA1‐A promoters with HapI and HapII/III. The blank pGreenII vector was used as control.
After using these dCAPS markers to genotype 348 modern Chinese cultivars released since the 1940s (Table S4), we performed an association analysis between the TaDA1‐A haplotypes and 11 agronomic traits collected in multiple environments. Consistent with its function in controlling kernel size and weight, the TaDA1‐A haplotypes were significantly correlated with TKW, KL, and KW (Figure 3b–d). The mean TKW of TaDA1‐A‐HapI was significantly higher than those of the TaDA1‐A‐HapII/III plants (3.1–4.3 g higher in 2002, 1.9–4.2 g higher in 2005, and 2.0–2.2 g higher in 2010; Figure 3b, Table S5). Similar results were found in KL and KW, except that no significant differences were observed in the KLs of HapI and HapII plants (Figure 3c, d). To further verify the results in the genetic populations, we detected an F16 segregating population derived from the cross Chuan 35050 × Shannong 483 (Xu et al., 2017). The mean TKW of TaDA1‐A‐HapI over four environments was significantly higher than that of TaDA1‐A‐HapII (Figure S7). In summary, these results indicate that TaDA1‐A‐HapI is the favourable haplotype because it is associated with larger kernels and higher TKWs in modern Chinese cultivars.
Variations in the promoter region of TaDA1‐A lead to significant differences in expression among haplotypes
To explore the possible reasons for these distinct haplotype effects, we first focused on the three exonic SNPs, all of which caused amino acid changes (L44S, I123N, and R307K, respectively; Figure 3a, Figure S8a). Because the L44S and I123N mutations were near the UIM domains responsible for the ubiquitin‐binding activity of DA1 (Li et al., 2008), we expressed maltose‐binding protein (MBP)‐fused N‐terminal TaDA1‐A‐HapI/II/III proteins (amino acids 1–140) containing the UIM domains in Escherichia coli. After incubation with ubiquitin‐agarose beads, no significant differences in the ubiquitin‐binding activity were detected among the HapI/II/III proteins, indicating that the L44S and I123N mutations did not affect TaDA1‐A ubiquitin binding (Figure S8b). We further expressed the full‐length HapI/II/III proteins in Schizosaccharomyces pombe using the pREP1 vector, as described previously (Lin et al., 2012; Maundrell, 1990). The HapI/II/III proteins were all similarly able to inhibit cell division in these assays (Figure S8c); therefore, our results reduce the possibilities that the exonic SNPs are responsible for the phenotypic differences among the TaDA1‐A haplotypes.
As reported previously (Hou et al., 2017; Su et al., 2011), many of the natural variations present in promoter regions affect gene expression. It is therefore also possible that large variations in the TaDA1‐A promoter region cause the observed haplotype effects by modifying the expression level of this gene. To test this hypothesis, we examined TaDA1‐A expression in the 10‐DPA grains of wheat accessions with different TaDA1‐A haplotypes. As shown in Figure 3e, TaDA1‐A expression was significantly lower in the HapI accessions than in the HapII and HapIII accessions, consistent with the observation that TaDA1 is a negative regulator of kernel size and HapI is associated with a higher TKW and larger kernels. We further compared HapI and HapII/III promoter driven luciferase (LUC) activities through transient expression assay in tobacco (N. benthamiana) leaves as described (Hellens et al., 2005). As expected, the HapI promoter had significantly lower relative LUC activity (LUC/REN) compared with the HapII/III promoter (Figure 3f). These results suggest that variations in the TaDA1‐A promoter region cause the main haplotype effects in modern Chinese cultivars.
TaDA1‐A‐HapI underwent positive selection during modern wheat breeding
The geographic distribution of the TaDA1‐A haplotypes was evaluated in both landraces and modern cultivars from China’s ten ecological zones (Hou et al., 2014; Ma et al., 2016). The frequency of the favourable haplotype HapI was increased in the modern cultivars relative to the landraces in the major Chinese production zones (Figure S9a, b). By contrast, the frequency of HapIII declined during the transition from landraces to modern cultivars. We next investigated the frequency change of the TaDA1‐A haplotypes during Chinese wheat breeding since the 1940s. The frequency of HapI showed a dramatic increase in the 1950s and has remained almost constant (~60%) since the 1960s, whereas the frequency of HapIII continuously decreased over the same period and almost disappeared after the 1980s (Figure S9c). In addition, HapI has become the dominant haplotype in the modern cultivars around the world, especially those developed in Europe or by CIMMYT (the International Maize and Wheat Improvement Center; Figure S9d). The presence of the favourable TaDA1‐A‐HapI haplotype is consistent with the yields of these lines (FAO Data, http://www.fao.org/docrep/006/y4011e/y4011e04.htm#TopOfPage). These results provide strong evidence that TaDA1‐A‐HapI underwent positive selection in global wheat breeding for higher yields.
TaDA1‐A physically interacts with TaGW2‐B
In Arabidopsis, DA1 physically interacts with DA2, a RING finger‐type E3 ubiquitin ligase (Xia et al., 2013). Because DA2 shares significant sequence similarity with GW2 in rice and wheat (Figure S10), we inferred that TaDA1 might also interact with TaGW2 in wheat. Using a yeast two‐hybrid system, we confirmed that TaDA1‐A could interact with TaGW2‐B (Figure 4a). To further verify that TaDA1‐A interacts with TaGW2‐B, we performed a firefly luciferase complementation imaging (LCI) assay in tobacco (N. benthamiana) cells. The co‐infiltration of TaDA1‐A‐nLUC and cLUC‐TaGW2‐B resulted in luminescence generated by the complemented luciferase, providing further evidence that TaDA1 and TaGW2 interact in vivo (Figure 4b). We also performed an in vitro pull‐down assay, which revealed that MBP‐TaDA1‐A but not the MBP control interacted with His‐TaGW2‐B (Figure 4c). These results confirmed that TaDA1‐A physically interacts with TaGW2‐B.
Figure 4.
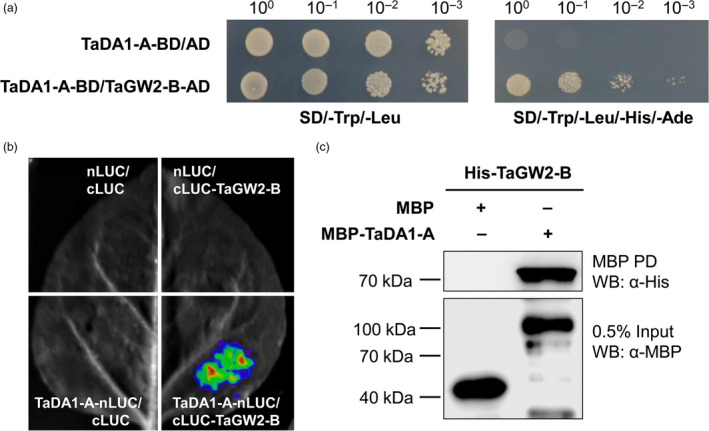
Protein–protein interactions between TaDA1‐A and TaGW2‐B. (a) Yeast two‐hybrid assay showing the interaction between TaDA1‐A and TaGW2‐B. BD, GAL4 DNA binding domain; AD, GAL4 activation domain. The yeast strains were serially diluted (100–10‐3) before spotting on selection medium. SD/‐Trp/‐Leu, synthetic dextrose medium lacking Trp and Leu; SD/‐Trp/‐Leu/‐His/‐Ade, synthetic dextrose medium lacking Trp, Leu, His and Ade. (b) LCI assay showing that TaDA1‐A physically interacts with TaGW2‐B in tobacco cells. nLUC, N‐terminal part of LUC; cLUC, C‐terminal part of LUC. The nLUC and cLUC derivative constructs were transformed into A. tumefaciens and then co‐infiltrated into tobacco (N. benthamiana). The LUC signals were collected 48–72 h after infiltration. (c) In vitro pull‐down assay showing that MBP‐TaDA1‐A, but not MBP itself, can pull down the His‐TaGW2‐B protein.
The simultaneous silencing of TaDA1 and TaGW2 has an additive effect on kernel size and weight
In wheat, there is increasing evidence to suggest that TaGW2 is a dominant‐negative regulator of kernel size and weight (Hong et al., 2014; Qin et al., 2017; Su et al., 2011; Zhang et al., 2018a). To confirm this effect, we generated TaGW2 RNAi lines in the hexaploid wheat cultivar Fielder, which produced much larger kernels than the WT (5.7–7.8% larger in KL and 16.1–20.8% larger in KW; Figure S11a–e). Accordingly, the TKW of the TaGW2 RNAi lines was 9.5–12.7 g higher than the WT (Figure S11c), showing that TaGW2 silencing had a stronger effect than TaDA1 silencing on increasing TKW (Figure 2c).
To study the genetic interactions of TaDA1 and TaGW2, we simultaneously silenced TaDA1 and TaGW2 in wheat by crossing the GW2‐Ri‐3 and DA1‐Ri‐3 transgenic lines (Figure 5a). The phenotypic results showed that both the GW2‐Ri‐3/DA1‐Ri‐3 and GW2‐Ri‐3 plants had larger kernels and higher TKWs than the DA1‐Ri‐3 plants, but GW2‐Ri‐3/DA1‐Ri‐3 showed a more significant increase in TKW and KW (Figure 5b–e). Compared with DA1‐Ri‐3, GW2‐Ri‐3/DA1‐Ri‐3 showed a 20% increase in TKW and an 11% increase in KW (Figure 5c–e). Therefore, GW2‐Ri‐3/DA1‐Ri‐3 had a greater increase in kernel size and TKW than both DA1‐Ri‐3 and GW2‐Ri‐3, suggesting that TaDA1 and TaGW2 have genetically additive effects on these traits.
Figure 5.
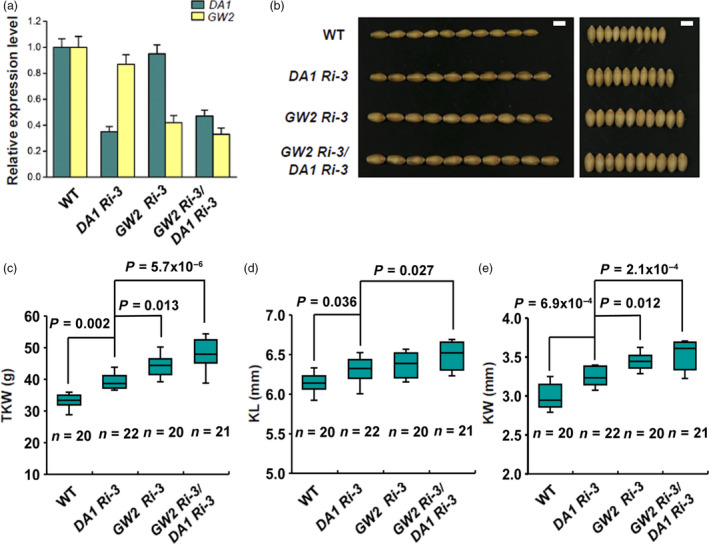
Genetically additive effect of TaDA1 and TaGW2 on kernel size and weight in wheat. (a) Expression levels of TaDA1 and TaGW2 in WT, the TaDA1 RNAi line (DA1‐Ri‐3), TaGW2 RNAi line (GW2‐Ri‐3), and double RNAi line (GW2‐Ri‐3/DA1‐Ri‐3). (b) Comparison of kernel phenotypes between these genotypes. Bar = 5 mm. (c) Thousand‐kernel weights (TKW), (d) kernel lengths (KL), and (e) kernel widths (KW) of these genotypes. Box plots showing the sample size (n), medians (black lines), upper and lower quartiles (box edges), minimums and maximums (whiskers), and P values (ANOVA).
The TaDA1‐A and TaGW2‐B haplotypes have additive effects on TKW
Because both TaDA1 and TaGW2 are negative regulators of kernel size and weight and can physically interact with each other, we speculated that the TaDA1‐A and TaGW2‐B haplotypes may have a combinational effect on TKW. The results of our present and previous association studies (Qin et al., 2017) revealed that the TaDA1‐A and TaGW2‐B haplotypes can both be divided into high‐TKW haplotypes (TaDA1‐A‐HapI and TaGW2‐B‐HapI/II) and low‐TKW haplotypes (TaDA1‐A‐HapII/III and TaGW2‐B‐HapIII/IV). Wheat accessions displaying these four combinations were divided into groups of high‐/high‐ (TaDA1‐A/TaGW2‐B), high‐/low‐, low‐/high‐, and low‐/low‐TKW haplotypes, designated HapC1–C4, respectively (Figure 6a, Table S4). Among the four combinations, HapC1 (high‐TaDA1‐A/high‐TaGW2‐B) was associated with the highest TKW in the modern Chinese cultivars (44.4 g, 41.1 g, and 41.1 g in 2002, 2005, and 2010, respectively), while the TKW of HapC4 (low‐TaDA1‐A/low‐TaGW2‐B) was the lowest among four haplotypes (Figure 6b), demonstrating an additive effect for the low‐ and high‐TaDA1‐A and TaGW2‐B haplotypes. Although the difference was not significant, the TKW of HapC3 (low‐TaDA1‐A/high‐TaGW2‐B) was higher than that of HapC2 (high‐TaDA1‐A/low‐TaGW2‐B), implying that the TaGW2‐B haplotypes had a stronger effect on TKW than the TaDA1‐A haplotypes (Figure 6b). Our data demonstrate the presence of an additive effect of the TaDA1‐A and TaGW2‐B haplotypes on the regulation of TKW.
Figure 6.
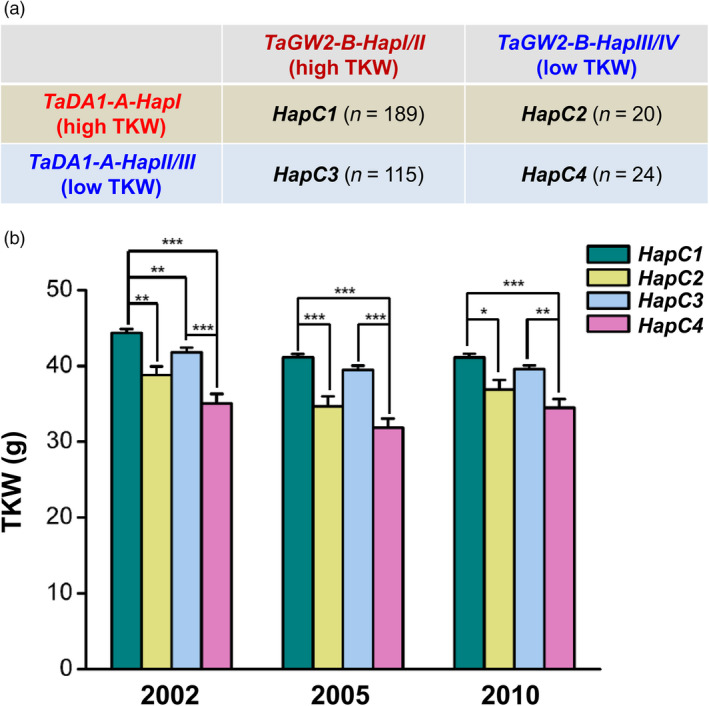
Combination of the TaDA1‐A and TaGW2‐B haplotypes with different effects on thousand‐kernel weights (TKW) in wheat. (a) Combinations of TaDA1‐A and TaGW2‐B high‐TKW and low‐TKW haplotypes (HapC1–C4). The number of HapC1–C4 is 189, 20, 115, and 24, respectively, in 348 modern Chinese cultivars. (b) Association of combinational haplotypes (HapC1–C4) with TKW in 348 modern Chinese cultivars grown in three environments (Luoyang, 2002; Luoyang, 2005; Shunyi, 2010). The values are presented as mean ± SE. *P < 0.05, **P < 0.01, and ***P < 0.001 (ANOVA) indicate significant differences between these combinations.
TaDA1 and TaGW2 function in partially overlapping but relatively independent protein networks
TaDA1 and TaGW2 are both involved in the ubiquitin–proteasome pathway; therefore, their functions are related to substrate degradation. We performed a comparative proteome assay to identify proteins affected by TaDA1 and TaGW2. In grains at 10 DPA, a total of 95 differently abundant proteins were identified between WT and DA1‐Ri‐3, 77 of which were up‐regulated and 18 down‐regulated in DA1‐Ri‐3 (Figure 7a, b; Table S6). By contrast, 249 differently abundant proteins were identified between the WT and GW2‐Ri‐3, including 157 up‐regulated proteins and 92 down‐regulated proteins in GW2‐Ri‐3. This indicated that TaGW2 silencing evoked a more significant alteration of the abundance of downstream proteins than TaDA1 silencing, which correlated with the finding that TaGW2 had a more dominant effect on TKW than TaDA1.
Figure 7.
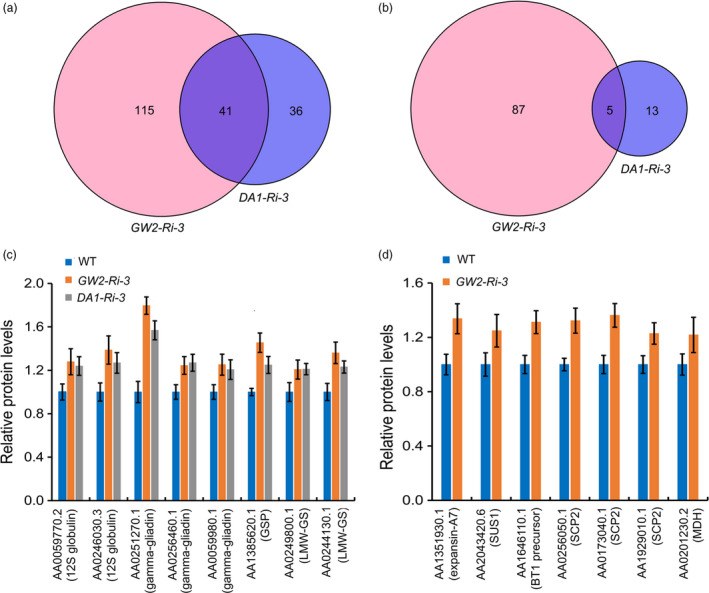
Comparative proteome analysis identified differently abundant proteins in 10‐DPA grains of DA1‐Ri‐3 and GW2‐Ri‐3. (a, b) Number of up‐regulated (a) and down‐regulated (b) proteins in DA1‐Ri‐3 and GW2‐Ri‐3 compared with the WT control. (c) Representative up‐regulated proteins identified both in DA1‐Ri‐3 and GW2‐Ri‐3. 12S globulin, 12S seed storage globulin 1; GSP, grain softness protein; LMW‐GS, low‐molecular‐weight glutenin subunit. (d) Representative up‐regulated proteins identified only in GW2‐Ri‐3. SUS1, sucrose synthase 1; BT1, ADP‐glucose brittle‐1 transporter precursor; SCP2, serine carboxypeptidase 2; MDH, malate dehydrogenase. The values are presented as mean ± SD.
DA1‐Ri‐3 and GW2‐Ri‐3 were found to share 41 up‐regulated proteins and five down‐regulated proteins relative to the WT (Figure 7a, b), which indicated that TaDA1 and TaGW2 function in partially overlapping regulatory pathways. The up‐regulated proteins were mainly involved in the biosynthesis of seed storage proteins, including multiple 12S seed storage globulins, low‐molecular‐weight glutenin subunits, gamma‐gliadins, and grain softness protein (Figure 7c, Table S6). Moreover, many of the differently abundant proteins were unique to DA1‐Ri‐3 or GW2‐Ri‐3, indicating that TaDA1 and TaGW2 act relatively independently of each other; for example, several proteins related to cell proliferation and cell expansion were significantly up‐regulated in GW2‐Ri‐3 (Figure 7d, Table S6). Three serine carboxypeptidases were up‐regulated in GW2‐Ri‐3. This group of proteins was previously reported to promote cell division and act as positive regulators of kernel size (Ma et al., 2016). Expansin A7 was also found to be up‐regulated in GW2‐Ri‐3 (Figure 7d). The expansin proteins are highly related to cell expansion, and expansin‐like A1 was the first verified substrate of GW2 in rice (Choi et al., 2018). Furthermore, several crucial proteins involved in starch synthesis are also up‐regulated in GW2‐Ri‐3, including ADP‐glucose brittle‐1 transporter precursor and sucrose synthase 1 (Figure 7d).
Discussion
In plants, seed size is a critical trait controlling seedling vigour and early growth, but it is also a key factor in determining grain yields in crops. Recently, the ubiquitin–proteasome pathway was identified as a conserved regulatory pathway of seed size in model plants (Li and Li, 2016). DA1, a core component of this regulatory pathway, acts partially redundantly with DAR1 to control seed size (Dong et al., 2017; Li et al., 2008). In this study, we cloned the DA1 homoeologs in wheat and revealed their high protein sequence similarities to AtDA1, especially in the functional domains such as the UIMs and LIM (Figure S3), implying that they have similar protein structures and functions. Disruption of the DA1 genes causes similar phenotypes in Arabidopsis and wheat, demonstrating their functional similarity; da1‐ko1, a loss‐of‐function T‐DNA mutant in Arabidopsis, produced larger seeds than the WT (Dong et al., 2017), while silencing of TaDA1 using RNAi also led to larger kernels in transgenic wheat (Figure 2b). AtDA1 was previously shown to physically interact with AtDA2 (Xia et al., 2013), while TaDA1 was shown to interact with TaGW2 (Figure 4a–c). AtDA2 and TaGW2 are similar RING‐type E3 ubiquitin ligases (Figure S10); therefore, TaDA1 interacts with similar proteins to AtDA1. These findings suggest that DA1 is a conserved negative regulator of seed size in dicots and monocots, with a high level of conservation that facilitates the elucidation of its role and its application in crop yield research. Many other components of the ubiquitin–proteasome pathway share conserved sequences and functions (Li and Li, 2016); therefore, future research should build on these characterized proteins to elucidate the function of other conserved proteins in the ubiquitin–proteasome pathway.
Many variations identified in mutant studies result in dramatic changes of protein functions, including missense mutations, premature stop codon mutations, and frame‐shift mutations. Unlike artificial mutations, most natural variations with a phenotypic effect are located in regulatory regions such as the promoters and introns. Previous studies of DA1 focused on the AtDA1R358K mutation in the da1‐1 dominant‐negative mutant. This mutation inhibited the activities of DA1 and DAR1, significantly increasing seed sizes in Arabidopsis, as well as transgenic rapeseed and maize (Li et al., 2008; Wang et al., 2017a; Xie et al., 2018); however, this mutation has not been identified in natural variations so far. In this study, we sequenced many natural variations in TaDA1‐A, most of which were located in the promoter and intronic regions (Figure 3a). Although we identified three exonic SNPs that introduced missense mutations, they did not cause obvious changes in the functions of the mutant TaDA1‐A proteins (Figure S8). By contrast, the variations in the promoter regions altered the expression level of TaDA1‐A, providing a convincing explanation for the phenotypic differences among the three TaDA1‐A haplotypes (Figure 3a, b). Similar regulatory patterns have also been observed in many other yield‐related genes, including TaGW2‐A (Su et al., 2011), TaGW2‐B (Qin et al., 2017), and TaAGP‐L‐B (Hou et al., 2017). Variations in regulatory regions are increasingly believed to be advantageous for the rapid adaptation of crops by enabling gene expression levels to be fine‐tuned to their environments; therefore, these variations are potential target sites in future wheat breeding and selection.
Marker‐assisted selection (MAS) is a highly efficient method for the genetic improvement of crops. The process of wheat breeding largely involves the accumulation of favourable alleles or haplotypes and can therefore be dramatically sped up using MAS (Gupta et al., 2010). In this study, we developed two dCAPS markers to distinguish three haplotypes of TaDA1‐A (Figure S6). Our association analysis indicated that the TaDA1‐A haplotypes were significantly correlated with differences in the kernel size and weights of modern Chinese cultivars (Figure 3b–d). TaDA1‐A‐HapI, the favourable haplotype producing larger kernel sizes and higher TKWs, was gradually accumulated over the course of the wheat breeding process (Figure S9a–c); however, further increases in the frequency of this haplotype are still possible as it is not yet saturated in modern cultivars (Figure S9c). Our TaDA1‐A molecular markers therefore have the potential to be applied in the MAS breeding for high TKWs in wheat. Previous studies have confirmed that combinations of favourable alleles or haplotypes associated with higher TKWs show strong addictive effects, including TaGW2‐A‐HapA and TaGW2‐B‐HapI (Qin et al., 2017), TaSus2‐A‐HapA and TaSus1‐B‐HapT (Hou et al., 2014), and TaAGP‐S1‐A‐HapI and TaAGP‐L‐B‐HapI (Hou et al., 2017). TaDA1‐A physically interacts with TaGW2‐B, providing a strong basis for the study of their genetically additive effects. We revealed that the combinational effect of the favourable TaDA1‐A and TaGW2‐B haplotypes (TaDA1‐A‐HapI and TaGW2‐B‐HapI/II) was significantly higher than the phenotypic effect of any single favourable haplotype (Figure 6a, b). In conclusion, pyramiding favourable haplotypes of TaDA1‐A, TaGW2‐B, and other major genes controlling yield‐related traits, will be more effective in future wheat yield improvement.
TaGW2 showed a stronger effect than TaDA1 on kernel size and weight in wheat, both in the haplotype association analysis and in a transgenic phenotypic assay (Figure 5b–e). The likely explanation is that DA1 acts partially redundantly with its homologs such as DAR1 (Li et al., 2008), and that the down‐regulation of DA1 alone has only a limited impact on the activities of its related proteins. Despite this, manipulating TaDA1 expression still alters kernel traits in wheat (Figure 2a–e), indicating that DA1 and its homologs are potential target genes for the future breeding of high‐yielding wheat cultivars. Recently, gene editing has been widely applied to improve the agronomic traits of crops (Wang et al., 2014; Zhang et al., 2018a). It is possible that this technique can be used to knock out DA1 and its close homologs (Zhang et al., 2018b), potentially generating a much greater impact on wheat kernel size. Furthermore, our proteomic data showed that TaDA1 and TaGW2 affected partially overlapping but relatively independent protein networks involved in diverse pathways (Figure 7a, b), including protein and starch biosynthesis, cell proliferation, and cell expansion. These proteins might promote rapid grain filling and enhance the final kernel size and TKW of the transgenic plants. Further work will be needed to identify the specific substrates of DA1 and GW2 and clarify their functional mechanisms in regulating kernel size in wheat.
Experimental procedures
Plant materials and phenotype assessment
A total of 505 Chinese wheat accessions including 157 landraces and 348 modern cultivars were used for the marker screening and association analyses, as previously described (Hou et al., 2017). Agronomic trait data for these wheat accessions were collected from plants grown in Luoyang, Henan Province, China in 2002 and 2005, and in Shunyi, Beijing, China in 2010 (Table S4). The field planting design and agronomic trait evaluation were described previously (Su et al., 2011). In addition, 384 European, 429 North American, 53 CIMMYT (the International Maize and Wheat Improvement Center), 82 former USSR, and 51 Australian modern wheat cultivars were used to investigate the global distribution of TaDA1‐A haplotypes in cultivars released during the last century (Hou et al., 2014). For tissue‐specific expression analysis, different tissue samples were collected from wheat plants grown in the field at seedling stage, elongation stage, and heading stage, respectively. The TaDA1 and TaGW2 transgenic wheat and wild type were grown in the field at the CAAS experimental station in Beijing under natural conditions. For each T2 generation transgenic line, the phenotypes of at least 20 plants were analysed. Kernel traits of transgenic lines were measured after harvesting according to Su et al. (2011).
RNA extraction and qRT‐PCR analysis
Total RNA was extracted from various Chinese Spring tissues using an RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). The cDNA was synthesized using the FastQuant RT Kit (Tiangen Biotech) and subjected to qRT‐PCR analysis on a LightCycler 96 Real‐Time PCR system (Roche Applied Science, Penzberg, Germany) using the SYBR Premix Ex Taq (Takara Bio, Beijing, China), as previously described (Ma et al., 2016). Genome‐specific primers of TaDA1 homoeologs were validated by sequencing. The wheat Actin gene was used as the internal reference (Li et al., 2015). The relative expression of each gene was calculated as a fold change using the comparative C T method (Livak and Schmittgen, 2001). All assays were performed three times in independent experiments. Primers used in this study are listed in Table S2.
Subcellular localization
The full‐length CDSs of the TaDA1 homoeologs were amplified from Chinese Spring seedling cDNA sample and sub‐cloned into the pJIT163‐GFP vector (Zheng et al., 2014). The verified fusion constructs and the control pJIT163‐GFP vector were introduced into wheat mesophyll protoplasts using the PEG‐mediated method as previously described (Zheng et al., 2014). After a 16‐h incubation, GFP fluorescence was monitored using a LSM880 laser‐scanning confocal microscope (Carl Zeiss, Jena, Germany).
Generation of overexpression and RNAi lines
For the TaDA1 overexpression construct, the full‐length TaDA1‐A CDS was amplified from Chinese Spring seedling cDNA sample and cloned into the BamHI and SpeI sites of the pWMB006 vector under the control of the maize ubiquitin promoter Ubi (Wang et al., 2017b). The resulting construct was double‐digested using HindIII and EcoRI, and then cloned into the corresponding sites of the pCAMBIA3301 vector to generate the TaDA1 overexpression construct. To generate the TaDA1 RNAi construct, a 403‐bp sequence derived from the TaDA1‐A CDS (nucleotides 418–820) was amplified and sequentially cloned into the BamHI/KpnI and SacI/SpeI sites of the pWMB006 vector. The resulting construct was double‐digested using HindIII and EcoRI, and then cloned into the corresponding sites of the pCAMBIA3301 vector to generate the TaDA1 RNAi construct. Similarly, a 402‐bp sequence from the TaGW2‐A CDS (nucleotides 302–703) was used to generate the TaGW2 RNAi construct.
All the constructs were introduced into Agrobacterium tumefaciens strain EHA105 and transformed into immature embryos of wheat (variety Fielder) following a modified Agrobacterium‐mediated transformation method (Wang et al., 2017b). The phenotypic traits of the T2 generation transgenic lines were measured at the Chinese Academy of Agricultural Sciences experimental field in Beijing, China. To assess phenotypic differences between transgenic lines and wild type, we performed a one‐way analysis of variance (ANOVA) using SPSS 16.0 (IBM Corporation, Armonk, NY) and a Tukey test at a significance level of P < 0.05.
Microscopy and measurement of cell length
The 15‐DPA developing grains were collected from DA1‐OE‐1, DA1‐Ri‐3, and WT plants, fixed for 24 h in FAA solution (70% alcohol, 5% acetic acid, and 0.02% formaldehyde; v/v), dehydrated in a series of ethanol solutions (75%, 85%, 90%, 95%, and 100%; v/v), and infiltrated with Paraplast Regular (Sigma‐Aldrich, St. Louis, MO). Tissue sections (4‐μm thick) were cut on a microtome (Leica Microsystems, Nussloch, Germany), stained using periodic acid–Schiff reagent for 30 min, and then counter‐stained with Luxol Fast Blue stain, as described previously (Wu et al., 2016). The sections were imaged under a SteREO Discovery V20 microscope (Carl Zeiss). To measure the cell length, approximately 100 outer pericarp cells were analysed in each of at least six different grain sections using the ZEN 2012 Image Processing software (Carl Zeiss).
TaDA1‐A haplotype discovery and association analysis
A total of 36 wheat accessions (24 modern cultivars and 12 landraces) were initially analysed for any TaDA1‐A sequence variations (Table S3). Specific primers were designed to amplify the 2.1‐kb promoter region and the 6.4‐kb coding region of TaDA1‐A (Table S2). The resulting DNA fragments were purified and sequenced on an Applied Biosystems 3730XI DNA Analyzer (Thermo Fisher Scientific, Carlsbad, CA). Sequence variations (SNPs and InDels) were identified using DNASTAR (http://www.dnastar.com/). Molecular markers of the TaDA1‐A haplotypes were developed based on two SNPs (T/A at 2590 bp and G/A at 4437 bp). A previously described method (Hou et al., 2014; Ma et al., 2016) was used to perform the haplotype association analysis. Briefly, the variance analyses were performed using SPSS 16.0 (IBM Corporation). Phenotypic differences between haplotypes were determined using a one‐way ANOVA and a Tukey test at a significance level of P < 0.05.
Transient promoter activity assay
The transient promoter activity assay was performed using a dual‐luciferase system in N. benthamiana leaves as described (Hellens et al., 2005). Briefly, the 2.1‐kb promoter sequences from different TaDA1‐A haplotypes were cloned into the pGreenII 0800‐Luc reporter plasmid. The resulting derivative constructs and pSoup‐p19 were transformed into Agrobacterium tumefaciens strain GV3101, and then co‐infiltrated into N. benthamiana leaves. After 48 h incubation, the assay was performed using the Dual‐Luciferase Reporter Assay System (Promega, Madison, WI), and the relative LUC activity (LUC/REN) was measured by a GloMax‐Multi luminescence reader (Promega).
Yeast two‐hybrid assay and LCI assay
The yeast two‐hybrid assay was carried out as described previously (Wu and Li, 2017). The TaDA1‐A and TaGW2‐B CDSs were amplified and sub‐cloned into the pGBKT7 and pGADT7 vectors, respectively, to generate the TaDA1‐A‐BD and TaGW2‐B‐AD plasmids. After the co‐transformation of both plasmids into yeast AH109 cells, the interaction between the expressed proteins was determined by the growth of the co‐transformants on a selection medium (SD/‐Trp/‐Leu/‐His/‐Ade), following the Yeast Protocols Handbook (Takara Bio).
The LCI assay for the interaction between TaDA1‐A and TaGW2‐B was performed in N. benthamiana leaves as described previously (Liu et al., 2017). The full‐length TaDA1‐A and TaGW2‐B CDSs were fused with the N‐terminal and C‐terminal regions of the LUC reporter gene, respectively (TaDA1‐A‐nLUC and cLUC‐TaGW2‐B), and transformed into Agrobacterium tumefaciens strain GV3101. Agrobacteria harbouring the nLUC and cLUC derivative constructs were co‐infiltrated into N. benthamiana, and the LUC activity was imaged and analysed 48–72 h after infiltration using the NightSHADE LB 985 Plant Imaging System (Berthold Technologies, Bad Wildbad, Germany).
Protein expression and in vitro pull‐down assay
The full‐length TaDA1‐A and TaGW2‐B CDSs were amplified and inserted into the pMAL‐c2X (New England Biolabs, Beverly, MA) and pET32a (Merck Millipore, Darmstadt, Germany) vectors, respectively. The resulting plasmids were transformed into Escherichia coli strain Rosetta (DE3) and induced using 0.4 mm isopropyl β‐D‐thiogalactopyranoside (IPTG) for 6 h at 20 °C. For the protein purification, the cultured cells were centrifuged at 6000 g at 4 °C for 5 min and then lysed using sonication. MBP‐TaDA1‐A and MBP proteins were purified using amylose resin (New England Biolabs) and the His‐TaGW2‐B protein was purified using Ni‐NTA resin (Qiagen, Hilden, Germany), according to the manufacturer’s instructions.
The in vitro pull‐down assay was performed as described previously (Dong et al., 2018). Briefly, 2 μg His‐TaGW2‐B protein was mixed with 2 μg MBP‐TaDA1‐A or MBP protein in 500 μl binding buffer [20 mm Tris (pH 7.4), 200 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 mm DTT, and 1 × protease inhibitor (Roche, Indianapolis, IN)], and then pulled down overnight using amylose resin (New England Biolabs) at 4 °C. The amylose resin was washed six times with binding buffer, and the proteins that bound to the resin were eluted by boiling the samples in 50 μl SDS‐PAGE loading buffer for 5 min. The eluted proteins were separated using a 12% SDS‐PAGE and detected by immunoblotting with anti‐MBP and anti‐His antibodies (CWbiotech, Beijing, China).
Proteome analysis
Approximately 2 g of developing 10‐DPA grains were collected from DA1‐Ri‐3, GW2‐Ri‐3, and WT plants and used for protein extraction. The total proteins were extracted from three biological replicates using extraction buffer (8 m urea, 30 mm HEPES, 1 mm PMSF, 2 mm EDTA, 10 mm DTT, and 55 mm IAM). Protein digestion was performed following the filter‐aided sample preparation (FASP) method, as described previously (Wiśniewski et al., 2009). Peptides from each sample were labelled with isobaric mass tags for relative and absolute quantitation (iTRAQ) using an iTRAQ Reagent‐8Plex Multiplex Kit (Sciex, Framingham, MA).
The liquid chromatography–mass spectrometry (LC‐MS)/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled with a Dionex Ultimate 3000 Nano LC system (Thermo Fisher Scientific). The mass spectrometer was operated in a data‐dependent mode with a full MS scan from 350–2000 m/z, a full scan resolution at 70 000, and MS/MS scan resolution at 17 500. The MS/MS spectra were searched against the IWGSC Triticum aestivum protein database (http://plants.ensembl.org/index.html) using the SEQUEST engine and quantified using Proteome Discoverer (version 1.4; Thermo Fischer Scientific). Differently abundant proteins between the WT and DA1‐Ri‐3 or GW2‐Ri‐3 (in each of the three biological replicates) were defined as having a ratio of ≥1.2 or ≤0.8 between samples, determined using an ANOVA at a significance level of P < 0.05.
Conflict of interest
The authors declare that they have no conflict of interests.
Author contributions
X.Z, T.L., and C.H. designed the research and analysed the data; H.L., T.L., H.F.L., K.W., Y.W., and L.Q. performed the experiments; T.L. and H.L. wrote the article; X.Z. supervised and revised the writing of the article; D.A. contributed during revision, providing an F16 segregating population and phenotype data used for association analysis.
Supporting information
Figure S1 PCR‐based chromosome mapping of the three TaDA1 homoeologs in Chinese Spring (CS) nullisomic‐tetrasomic lines.
Figure S2 Gene structures of the TaDA1 homoeologs.
Figure S3 Sequence alignment of DA1 proteins from wheat, Arabidopsis, and maize.
Figure S4 Comparison of other yield‐related traits between the WT and TaDA1 transgenic lines under field conditions.
Figure S5 Comparison of cell number and cell length in the outer pericarp tissues of DA1‐OE‐1, WT, and DA1‐Ri‐3.
Figure S6 Development of molecular markers for the TaDA1‐A haplotypes based on the T/A SNP at position 2590 and the G/A SNP at position 4437.
Figure S7 The effect of TaDA1‐A haplotypes on TKW in an F16 segregation population derived from Chuan 35050 × Shannong 483.
Figure S8 Biochemical functional analysis of proteins encoded by the three TaDA1‐A haplotypes.
Figure S9 Global selection and frequency change of the TaDA1‐A haplotypes in modern cultivars.
Figure S10 Sequence alignment of GW2 proteins from wheat and rice, and the DA2 protein from Arabidopsis.
Figure S11 The down‐regulation of TaGW2 led to significantly increased kernel sizes and weights in wheat.
Table S1 Gene IDs or accession numbers
Table S2 Primers used in this study
Table S3 The 36 wheat accessions used for polymorphism discovery
Table S4 Genotype and phenotype of the 348 Chinese modern cultivars
Table S5 Association results of the phenotypes and TaDA1‐A haplotypes of 348 modern cultivars grown in three different environments
Table S6 Differently abundant proteins in the 10‐DPA grains of DA1‐Ri‐3, GW2‐Ri‐3, and the WT
Acknowledgements
The authors would like to thank Dr. Jiaqiang Sun and Dr. Jie Liu (Institute of Crop Science, CAAS) for their assistance with the transient LUC activity assay. This research was financially supported by grants from the National Key Research and Development Program of China (2016YFD0100302), the National Animal and Plant Transgenic Project (2016ZX08009001), Natural Science Foundation of China (31671687), and CAAS‐Innovation Project.
Liu, H. , Li, H. , Hao, C. , Wang, K. , Wang, Y. , Qin, L. , An, D. , Li, T. and Zhang, X. (2020) TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol. J., 10.1111/pbi.13298
Contributor Information
Tian Li, Email: litian@caas.cn.
Xueyong Zhang, Email: zhangxueyong@caas.cn.
References
- Brinton, J. , Simmonds, J. and Uauy, C. (2018) Ubiquitin‐related genes are differentially expressed in isogenic lines contrasting for pericarp cell size and grain weight in hexaploid wheat. BMC Plant Biol. 18, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, B.S. , Kim, Y.J. , Markkandan, K. , Koo, Y.J. , Song, J.T. and Seo, H.S. (2018) GW2 functions as an E3 ubiquitin ligase for rice expansin‐like 1. Int. J. Mol. Sci. 19, 1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch, S. , Anastasiou, E. , Sharma, V.K. , Laux, T. , Fletcher, J.C. and Lenhard, M. (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage‐dependent manner. Curr. Biol. 16, 272–279. [DOI] [PubMed] [Google Scholar]
- Dong, H. , Dumenil, J. , Lu, F.H. , Na, L. , Vanhaeren, H. , Naumann, C. , Klecker, M. et al. (2017) Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis . Genes Dev. 31, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Yan, S. , Liu, J. , Liu, P. and Sun, J. (2018) TaCOLD1 defines a new regulator of plant height in bread wheat. Plant Biotechnol. J. 17, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P.K. , Langridge, P. and Mir, R.R. (2010) Marker‐assisted wheat breeding: present status and future possibilities. Mol. Breeding, 26, 145–161. [Google Scholar]
- Hanif, M. , Gao, F. , Liu, J. , Wen, W. , Zhang, Y. , Rasheed, A. , Xia, X. et al. (2016) TaTGW6‐A1, an ortholog of rice TGW6, is associated with grain weight and yield in bread wheat. Mol. Breeding, 36, 1. [Google Scholar]
- Hellens, R.P. , Allan, A.C. , Friel, E.N. , Bolitho, K. , Grafton, K. , Templeton, M.D. , Karunairetnam, S. et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y. , Chen, L. , Du, L.P. , Su, Z. , Wang, J. , Ye, X. and Qi, L. (2014) Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct. Integr. Genomics, 14, 341–349. [DOI] [PubMed] [Google Scholar]
- Hou, J. , Jiang, Q. , Hao, C. , Wang, Y. , Zhang, H. and Zhang, X. (2014) Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 164, 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, J. , Li, T. , Wang, Y. , Hao, C. , Liu, H. and Zhang, X. (2017) ADP‐glucose pyrophosphorylase genes, associated with kernel weight, underwent selection during wheat domestication and breeding. Plant Biotechnol. J. 15, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M.J. , Zhang, H.P. , Cao, J.J. , Zhu, X.F. , Wang, S.X. , Jiang, H. , Wu, Z.Y. et al. (2016) Characterization of an IAA‐glucose hydrolase gene TaTGW6 associated with grain weight in common wheat (Triticum aestivum L.). Mol. Breeding, 36, 25. [Google Scholar]
- IWGSC . (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- Li, N. and Li, Y. (2016) Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. [DOI] [PubMed] [Google Scholar]
- Li, W. and Yang, B. (2017) Translational genomics of grain size regulation in wheat. Theor. Appl. Genet. 130, 1765–1771. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zheng, L. , Corke, F. , Smith, C. and Bevan, M.W. (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana . Genes Dev. 15, 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Li, L. , Yang, X. , Warburton, M.L. , Bai, G. , Dai, J. , Li, J ., et al. (2010) Relationship, evolutionary fate and function of two maize co‐orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Ma, L. , Geng, Y. , Hao, C. , Chen, X. and Zhang, X. (2015) Small RNA and degradome sequencing reveal complex roles of miRNAs and their targets in developing wheat grains. PLoS ONE 10, e0139658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q. , Wang, D. , Dong, H. , Gu, S. , Cheng, Z. , Gong, J. , Qin, R. et al. (2012) Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1. Nat. Commun. 3, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Cheng, X. , Liu, P. and Sun, J. (2017) miR156‐targeted SBP‐box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 174, 1931–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, M. , Wang, Q. , Li, Z. , Cheng, H. , Li, Z. , Liu, X. , Song, W. et al. (2015) Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. Plant J. 83, 312–325. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Li, T. , Hao, C. , Wang, Y. , Chen, X. and Zhang, X. (2016) TaGS5‐3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 14, 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K. (1990) nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265, 10857–10864. [PubMed] [Google Scholar]
- Mohler, V. , Albrecht, T. , Castell, A. , Diethelm, M. , Schweizer, G. and Hartl, L. (2016) Considering causal genes in the genetic dissection of kernel traits in common wheat. J. Appl. Genet. 57, 467–476. [DOI] [PubMed] [Google Scholar]
- Qin, L. , Hao, C. , Hou, J. , Wang, Y. , Li, T. , Wang, L. , Ma, Z. et al. (2017) Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2 . BMC Plant Biol. 14, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjad, M. , Ma, X. , Khan, S.H. , Shoaib, M. , Song, Y. , Yang, W. , Zhang, A. et al. (2017) TaFlo2‐A1, an ortholog of rice Flo2, is associated with thousand grain weight in bread wheat (Triticum aestivum L.). BMC Plant Biol. 17, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, C. , Ren, Y. , Liu, L. , Wang, F. , Zhang, H. , Tian, P. , Pan, T. et al. (2019) Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 180, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds, J. , Scott, P. , Brinton, J. , Mestre, T.C. , Bush, M. , Del Blanco, A. , Dubcovsky, J. et al. (2016) A splice acceptor site mutation in TaGW2‐A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 129, 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.J. , Huang, W. , Shi, M. , Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Su, Z. , Hao, C. , Wang, L. , Dong, Y. and Zhang, X. (2011) Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeolleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Zhang, X. , Chen, F. and Cui, D. (2015) A single‐nucleotide polymorphism of TaGS5 gene revealed its association with kernel weight in Chinese bread wheat. Front. Plant Sci. 6, 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.L. , Tang, M.Q. , Chen, S. , Zheng, X.F. , Mo, H.X. , Li, S.J. , Wang, Z. et al. (2017a) Down‐regulation of BnDA1, whose gene locus is associated with the seeds weight, improves the seeds weight and organ size in Brassica napus . Plant Biotechnol. J. 15, 1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Liu, H. , Du, L. and Ye, X. (2017b) Generation of marker‐free transgenic hexaploid wheat via an Agrobacterium‐mediated co‐transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 15, 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski, J.R. , Zougman, A. , Nagaraj, N. and Mann, M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- Wu, X.Y. and Li, T. (2017) A Casein Kinase II phosphorylation site in AtYY1 affects its activity, stability, and function in the ABA response. Front. Plant Sci. 8, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Liu, J. , Li, D. and Liu, C.M. (2016) Rice caryopsis development II: dynamic changes in the endosperm. J. Integr. Plant Biol. 58, 786–798. [DOI] [PubMed] [Google Scholar]
- Xia, T. , Li, N. , Dumenil, J. , Li, J. , Kamenski, A. , Bevan, M.W. , Gao, F. et al. (2013) The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis . Plant Cell, 25, 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q. , Mayes, S. and Sparkes, D.L. (2015) Carpel size, grain filling, and morphology determine individual grain weight in wheat. J. Exp. Bot. 66, 6715–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, G. , Li, Z. , Ran, Q. , Wang, H. and Zhang, J. (2018) Over‐expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnol. J. 16, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.F. , Li, S.S. , Li, L.H. , Ma, F.F. , Fu, X.Y. , Shi, Z.L. , Xu, H.X. et al. (2017) QTL mapping for yield and photosynthetic related traits under different water regimes in wheat. Mol Breeding, 37, 34. [Google Scholar]
- Zhang, L. , Zhao, Y.L. , Gao, L.F. , Zhao, G.Y. , Zhou, R.H. , Zhang, B.S. and Jia, J.Z. (2012) TaCKX6‐D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 195, 574–584. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Li, D. , Zhang, D. , Zhao, X. , Cao, X. , Dong, L. , Liu, J. et al. (2018a) Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Zhang, R. , Song, G. , Gao, J. , Li, W. , Han, X. , Chen, M. et al. (2018b) Targeted mutagenesis using the Agrobacterium tumefaciens‐mediated CRISPR‐Cas9 system in common wheat. BMC Plant Biol. 18, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Liu, H. , Wang, Y. , Wang, L. , Chang, X. , Jing, R. , Hao, C. et al. (2014) TEF‐7A, a transcript elongation factor gene, influences yield‐related traits in bread wheat (Triticum aestivum L.). J. Exp. Bot. 65, 5351–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J. and Li, J. (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 PCR‐based chromosome mapping of the three TaDA1 homoeologs in Chinese Spring (CS) nullisomic‐tetrasomic lines.
Figure S2 Gene structures of the TaDA1 homoeologs.
Figure S3 Sequence alignment of DA1 proteins from wheat, Arabidopsis, and maize.
Figure S4 Comparison of other yield‐related traits between the WT and TaDA1 transgenic lines under field conditions.
Figure S5 Comparison of cell number and cell length in the outer pericarp tissues of DA1‐OE‐1, WT, and DA1‐Ri‐3.
Figure S6 Development of molecular markers for the TaDA1‐A haplotypes based on the T/A SNP at position 2590 and the G/A SNP at position 4437.
Figure S7 The effect of TaDA1‐A haplotypes on TKW in an F16 segregation population derived from Chuan 35050 × Shannong 483.
Figure S8 Biochemical functional analysis of proteins encoded by the three TaDA1‐A haplotypes.
Figure S9 Global selection and frequency change of the TaDA1‐A haplotypes in modern cultivars.
Figure S10 Sequence alignment of GW2 proteins from wheat and rice, and the DA2 protein from Arabidopsis.
Figure S11 The down‐regulation of TaGW2 led to significantly increased kernel sizes and weights in wheat.
Table S1 Gene IDs or accession numbers
Table S2 Primers used in this study
Table S3 The 36 wheat accessions used for polymorphism discovery
Table S4 Genotype and phenotype of the 348 Chinese modern cultivars
Table S5 Association results of the phenotypes and TaDA1‐A haplotypes of 348 modern cultivars grown in three different environments
Table S6 Differently abundant proteins in the 10‐DPA grains of DA1‐Ri‐3, GW2‐Ri‐3, and the WT


