Summary
Octoploid strawberry (Fragaria × ananassa Duch.) is a model plant for research and one of the most important non‐climacteric fruit crops throughout the world. The associations between regulatory networks and metabolite composition were explored for one of the most critical agricultural properties in octoploid strawberry, fruit colour. Differences in the levels of flavonoids are due to the differences in the expression of structural and regulatory genes involved in flavonoid biosynthesis. The molecular mechanisms underlying differences in fruit colour were compared between red and white octoploid strawberry varieties. FaMYB genes had combinatorial effects in determining the red colour of fruit through the regulation of flavonoid biosynthesis in response to the increase in endogenous ABA at the final stage of fruit development. Analysis of alleles of FaMYB10 and FaMYB1 in red and white strawberry varieties led to the discovery of a white‐specific variant allele of FaMYB10, FaMYB10‐2. Its coding sequence possessed an ACTTATAC insertion in the genomic region encoding the C‐terminus of the protein. This insertion introduced a predicted premature termination codon, which suggested the loss of intact FaMYB10 protein playing a critical role in the loss of red colour in white octoploid strawberry.
Keywords: FaMYB genes, fruit colour, flavonoids, anthocyanins, Fragaria × ananassa
Introduction
Cultivated strawberry is a non‐climacteric fruit crop, with one of the most complicated genomes among all the crop plants. It carries eight sets of chromosomes (octoploid subspecies in genus Fragaria, 2n = 8x = 56) derived from four different diploid ancestors (Shulaev et al., 2011). Among current cultivated strawberry subspecies, the most popular Fragaria subspecies (Fragaria × ananassa Duch.) was derived from the accidental crossing between the white‐fruited Chilean strawberry subspecies (F. chiloensis (L.) Mill.) and the red‐fruited meadow strawberry subspecies (F. virginiana Mill.) in the Royal Botanical Garden of the French king around 1700 (Finn et al., 2013). This subspecies was subsequently named as Fragaria × ananassa by the great French botanist, Antoine Nicholas Duchesne (Hancock et al., 2008). Due to the superior fruit quality of this hybrid subspecies, it was quickly distributed across the world and gradually displaced different indigenous cultivated subspecies of F. chiloensis in North and South America (Finn et al., 2013). By the middle of the 20th century, Fragaria × ananassa and its derivatives had emerged as the most dominant cultivated strawberry varieties across the world (Hancock et al., 2008). As globalization has progressed, this octoploid cultivated strawberry subspecies has grown to be one of the most important fruit crop plants and its total annual production had reached over 8.11 million tonnes by 2014 (FAO, 2014).
Horticulturists have long been interested in breeding different varieties of plants with fruits having bright colours, saturated hues, sweet taste and high nutritional value, among which attractive fruit colour is one of the most desirable horticultural traits (Kayesh et al., 2013; Pillet et al., 2015). The quantity and variability of flavonoids control strawberry fruit coloration in both wild diploid and cultivated octoploid strawberry varieties (Lin‐Wang et al., 2014; Schaart et al., 2013). Flavonoids have multiple stress‐protecting functions for plants and health‐promoting properties for humans (Allan et al., 2008; Jaakola, 2013; Zhang et al., 2014). Branches of the flavonoid biosynthetic pathway are involved in the production and regulation of anthocyanins, proanthocyanins and flavonols (Figure 1). Pelargonidin and cyanidin are responsible for the bright red colour and dark red colour of ripe fruits, respectively, which are also the major colours in cultivated octoploid strawberry subspecies (Fischer et al., 2014). The key structural genes for flavonoid biosynthesis encode essential biosynthetic enzymes. Expression of early biosynthetic genes (EBG) (Figure 1), including phenylalanine ammonia‐lyase (PAL), cinnamate 4‐hydroxylase (C4H), 4‐coumarate:coenzyme A ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), and late biosynthetic genes (LBG) (Figure 1), including flavonol synthase (FLS) for the formation of flavonols, dihydroflavonol‐4‐reductase/flavanone‐4‐reductase (DFR), anthocyanidin synthase (ANS) and flavonol‐O‐glucosyltransferases (UFGT), is required for the formation of anthocyanins; and expression of leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) is required for the formation of proanthocyanin precursors and proanthocyanins (Figure 1; Fischer et al., 2014; Griesser et al., 2008).
Figure 1.
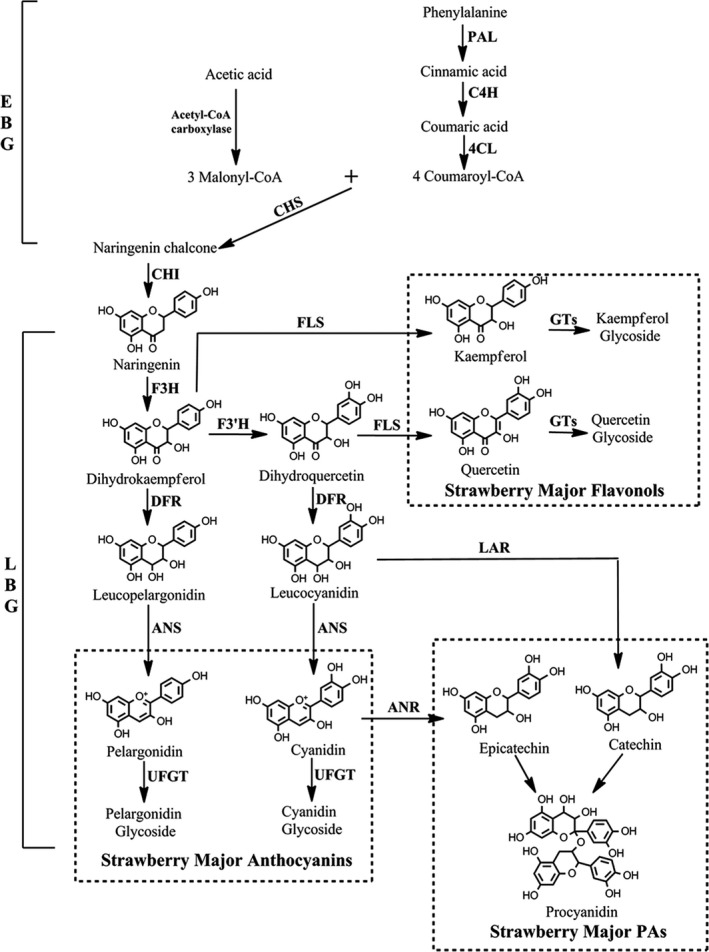
The canonical pathway of flavonoid biosynthesis in strawberry (Fragaria × ananassa) fruits. EBG, early biosynthetic genes; and LBG, late biosynthetic genes.
The differential accumulation of flavonoids is regulated predominantly at the transcriptional level by transcription factors (TFs) (Jin et al., 2016; Lin‐Wang et al., 2010). The R2R3MYB TFs are known to be major players controlling flavonoid biosynthesis during fruit coloration in many species. Another transcription factor family (bHLH) and WD40‐related proteins are also involved in the formation of a conserved ternary MBW complex together with R2R3MYB proteins. The MBW complex plays a critical role in the regulation of anthocyanin and proanthocyanin biosynthesis (Nesi et al., 2000; Walker et al., 1999). Furthermore, the transcript levels of key regulatory genes of flavonoid biosynthesis are tightly regulated during the process of the fruit coloration of different plants, such as Vitis vinifera (grapevine), Malus domestica (apple), Fragaria × ananassa (strawberry) and Prunus avium (sweet cherry; Aharoni et al., 2001; Espley et al., 2007; Jin et al., 2016; Kobayashi et al., 2002). Comparison of the closest diploid species, F. vesca, to octoploid F. × ananassa shows that there are 1616 gene encoding transcription factors in its genome, including 187 MYB and MYB‐related transcription factors (Shulaev et al., 2011). Several recent studies have demonstrated that how several R2R3MYB family members participate in the regulation of branches of flavonoid biosynthesis in the cultivated strawberry variety (F. × ananassa), indicating that FaMYB1 (Paolocci et al., 2011), FaMYB9/FaMYB11 (Schaart et al., 2013), FaMYB5 (Schaart et al., 2013) and FaMYB10 (Kadomura‐Ishikawa et al., 2015; Lin‐Wang et al., 2010) are key TFs involved in regulating flavonoid biosynthesis in response to hormones, abiotic stress, biotic stress, light and circadian rhythms. However, little is known about the regulatory network controlling fruit colour in octoploid strawberry varieties.
To broaden this understanding, ‘Snow Princess’, a white octoploid strawberry variety (WS), and ‘Sweet Charlie’, the red octoploid strawberry (RS) variety, were subjected to qualitative and quantitative LC‐MS/MS analyses of flavonoid composition, co‐expression analysis of regulatory and structural genes related to flavonoid biosynthesis, and sequence analyses of two key TFs for anthocyanin biosynthesis, FaMYB10 and FaMYB1. Based on the homology of R2R3 domains of the MYB transcription factors from different plant species, as well as the red and white strawberry varieties, phylogenetic analysis demonstrated that FaMYB1, FaMYB9, FaMYB10 and FaMYB11 were closely related, structurally to the MYB proteins with known functions in regulating flavonoid metabolism in other species (Dubos et al., 2010; Shulaev et al., 2011). The regulatory network of strawberry fruit coloration was studied using these TFs in combination with the changes in expression of key flavonoid structural genes and the levels of endogenous ABA by principal component analysis. A working model is proposed to explain the combinatorial effects of FaMYB1, FaMYB10; FaMYB9 and FaMYB11 in regulating different branches of flavonoid biosynthesis in ripe fruit.
Allele is one of two or more alternative forms of a gene at the same site in a chromosome, which determine alternative characters that can be passed on from parents to offspring through sexual reproduction. The cultivated strawberry has four subgenomes, each locus can represent up to different alleles in a single individual (Lin‐Wang et al., 2010; Lin‐Wang et al., 2014). Analyses of FaMYB10 and FaMYB1 alleles in WS revealed that FaMYB10‐2, a WS‐specific allele of FaMYB10, contains an insertion (ACTTATAC) resulting in a presumed premature stop codon in the FaMYB10 coding sequence. Interestingly, the genomic sequence of FaMYB10‐2 was intronless, compared to the RS‐specific allele FaMYB10‐1. These structural changes in the FaMYB10‐2 allele are likely to be the primary reasons for the striking colour difference between RS and WS due to the loss of the FaMYB10‐mediated up‐regulation of anthocyanin biosynthesis in ripe fruit. FaMYB1 alleles showed only minor differences between RS and WS varieties.
Results
Analyses of changes in flavonoid composition and endogenous ABA levels during strawberry fruit ripening
Six visual distinguishable developmental stages of strawberry fruits were defined; small green (G1), large green (G2), white (W), turning (T), ripe (R) and over‐ripe (OR), which were minor modifications of stages defined in previous reports (Moyano et al., 1998; Schaart et al., 2013). For WS, these stages were defined by seed colour examined in the field (see Methods; Figure S1) at 13, 17, 24, 30, 46 and 50 days after anthesis (Figure 2a, top), and for RS, these stages were defined by fruit colour at 7, 12, 17, 19, 26 and 30 days after anthesis (Figure 2a, bottom). The coloration of fruit varied significantly between WS and RS (Figure 2a). Fruit coloration is greatly influenced by the differential accumulation of anthocyanins; therefore, anthocyanin contents during fruit development were assessed. In WS, anthocyanins were almost undetectable at any of the six developmental stages; in RS, anthocyanins were undetectable during G1, G2 and W stages, but increased rapidly from 0.13 mg/100 g FW at the T stage to 7.70 mg/100 g FW at the R stage (Figure 2b). Analysis using LC‐MS/MS showed that the major anthocyanin was pelargonidin‐3‐glucoside (Table 1; Figure S2). The concentration of total anthocyanins in WS fruits was 42.43% at the T stage and 0.15% at the R stage (Table S1) of those in RS correspondingly.
Figure 2.
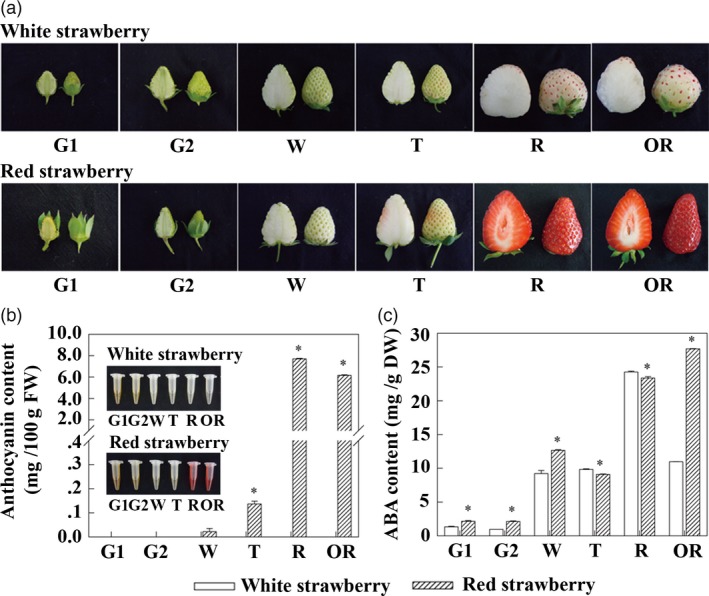
Changes in the anthocyanin content and in endogenous ABA levels during strawberry fruit ripening. (a) The strawberry fruits at the different visual fruit developmental stages used in this study. (b) The anthocyanin content in white and red strawberry varieties. (c) The endogenous ABA levels in white and red strawberry varieties. Asterisks (*) represent that the values of total anthocyanin content (n = 3, ±SE) are significantly different at P < 0.05 as determined using independent t‐test.
Table 1.
Levels of anthocyanidins, flavonols and proanthocyanins in red and white strawberry varieties
| Peak No. | Tentative identification | White strawberry turning phase (μg/g) | Red strawberry turning phase (μg/g) | White strawberry ripe phase (μg/g) | Red strawberry ripe phase (μg/g) | Cat. |
|---|---|---|---|---|---|---|
| 1 | Procyanidin B1 | 819.07 ± 57.61b | 1615.64 ± 103.92c | 329.91 ± 32.55a | 507.02 ± 28.67a | PA |
| 2 | Proanthocyanidins | 765.69 ± 44.69ab | 507.86 ± 42.69a | 592.92 ± 66.43a | 1209.62 ± 189.52b | PA |
| 3 | Cyanidin‐3,5‐diglucoside | 4.52 × 10‐3 ± 0.00a | 5.67 × 10‐3 ± 0.00a | 5.11 × 10‐3 ± 0.00a | 0.01 ± 0.00b | AC |
| 4 | Pelargonidin‐3‐glucoside | 189.89 ± 6.07a | 447.11 ± 7.38a | 188.46 ± 3.35a | 125389.52 ± 8702.50b | AC |
| 5 | Pelargonidin 3‐acetyl‐glucoside | 0.03 ± 0.01a | 0.54 ± 0.04a | N/A | 1029.32 ± 63.37b | AC |
| 6 | Kaempferol‐3‐glucoside | 16.35 ± 1.06a | 228.94 ± 24.77b | 6.14 ± 0.33a | 36.38 ± 2.38a | FL |
| 7 | Kaempfeol‐3‐rutinoside | 1.34 ± 0.22b | 5.96 × 10‐3 ± 0.00a | 5.59 ± 0.36c | N/A | FL |
| 8 | Quercetin 3‐glucoside | 7.78 ± 1.17b | 31.33 ± 1.06d | 1.77 ± 0.30a | 15.91 ± 1.20c | FL |
| 9 | (+/‐)‐Catechin | 1608.66 ± 112.13c | 977.59 ± 32.54b | 161.21 ± 24.11a | 204.30 ± 12.81a | PP |
| 10 | Epicatechin | 14.66 ± 0.48b | 6.12 ± 0.57a | 5.73 ± 0.58a | 5.21 ± 0.43a | PP |
Values represent mean ± SE. a, b, c, d letters indicate statistically significant differences within levels of anthocyanidins, flavonols and proanthocyanins in red and white strawberry varieties (Duncan's honestly significant difference test, p < 0.05).
AC, anthocyanins; FL, flavonols; PA, proanthocyanins; PP, precursors of proanthocyanidins, catechin, and epicatechin.
Proanthocyanins were detected in both RS and WS fruit. The major proanthocyanin was procyanidin B1, and other proanthocyanidins were detected as well (Table 1). In WS, the content of total proanthocyanins was 74.63% at T stage, and 53.76% at R stage, of the values in the red strawberry variety although the major precursors of proanthocyanins accumulated in WS fruit to 165.02% of the levels in RS early during strawberry fruit development (T stage) (Table S1). Flavonols in strawberry fruit contain a major component, Pelargonidin‐3‐glucoside, and a minor component, quercetin‐3‐glucoside (Table 1). The concentration of total flavonols in WS was 9.79% of that in RS fruit at the T stage and 25.82% at the R stage. However, the total amount of flavonols in fruit of both varieties was much lower than either anthocyanins or proanthocyanins, especially at the R stage (Table S1), suggesting that the flavonol biosynthesis was suppressed in ripe fruit. Among the three flavonoids, there were 10 times more anthocyanins than the other flavonoids at the R stage.
Exogenous application of ABA has been reported to affect the accumulation of anthocyanins in strawberry through the action of FaMYB class genes (Ayub et al., 2016; Kadomura‐Ishikawa et al., 2015), so the concentration of endogenous ABA was examined in fruit throughout the fruit developmental stages. There were significant differences in ABA levels at G1, G2 and W stages with slightly higher levels of endogenous ABA in RS compared to WS. Endogenous ABA showed slightly higher levels at the critical stages of fruit coloration, the T and R stages, in WS, but reached its highest level at the OR stage in RS, and at this stage, the endogenous ABA level was significantly higher in RS than WS (Figure 2c). Based on the changes in endogenous ABA levels and flavonoids, the colour difference between WS and RS was most closely associated with the anthocyanin contents at the T and R/OR developmental stages. Therefore, our analyses of molecular mechanisms controlling colour in fruit focused on the T and R stages.
Expression analyses of key genes of flavonoid biosynthesis
Many studies have shown that most structural genes in flavonoid biosynthesis are regulated at the transcriptional level by MYB class transcription factors. RNA‐seq analyses were performed for WS and RS fruit at the T and R developmental stages. Several key structural and regulatory genes involved flavonoid biosynthesis were selected for further analysis using standardized transcript levels, shown as heatmaps (Figure 3a and b). These genes included the structural genes FaPAL, FaCHI, FaCHS, FaC4H, Fa4CL, FaFLS, FaF3' H, FaGT3, FaANR, FaLAR, FaF3H, FaDFR, FaANS and FaUFGT (Figure 3a) and the key regulatory genes FaMYB1, FaMYB10, FaMYB9, FaMYB11, FabHLH3, FabHLH3‐delta and FaWD40 (Figure 3b). A group of EBG genes (FaCHI, FaCHS, FaC4H, and Fa4CL) showed significantly higher expression levels in RS compared to WS at T and R stages, except for the gene, FaPAL, encoding the first committed enzyme of general phenylpropanoid metabolism (Figure 3a). The likely reason that collectively FaPAL transcripts showed little relative difference in transcript levels between WS and RS is that PAL is encoded by multiple genes with differential expression, and while their overall transcript levels may not differ by much, selected isoforms may be differentially expressed in WS and RS fruit. Several structural genes (FaFLS, FaGT3, FaANR, and FaLAR) had low transcript levels in both WS and RS (Figure 3a). Significantly, there was a sharp increase in transcript levels of FaF3H, FaDFR, FaANS, and FaUFGT in RS at T and R stages, which were directly correlated with anthocyanin production. And expression levels of these genes did not increase in WS fruit (Figure 3a).
Figure 3.
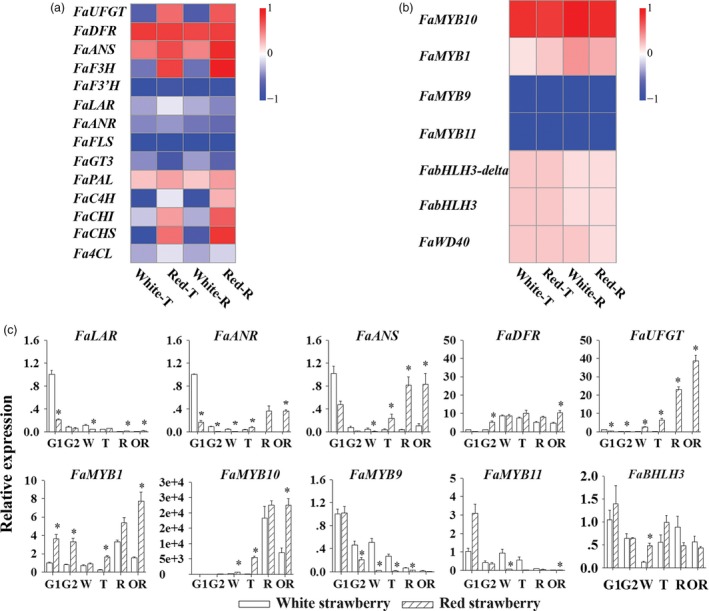
Expression analyses of key genes of flavonoid biosynthesis. (a) RNA‐seq heatmap analysis for transcript levels of structural genes in the white and red strawberry varieties. (b) RNA‐seq heatmap analysis for transcript levels of regulatory genes in the white and red strawberry varieties. (c) The real‐time PCR results for transcript levels of structural genes and regulatory genes in the white and red strawberry varieties. Asterisks (*) represent that the values of the corresponding transcription levels (n = 3, ±SE) are significantly different at p < 0.05 as determined using independent t‐test.
To confirm these differences, the transcript abundance of the structural genes was analysed by qRT‐PCR throughout the six developmental stages of strawberry fruits. Transcript levels were standardized to the Actin gene and shown as bar graphs (Figure 3c, top; Figure S3). The expression levels of key genes FaPAL, FaC4H, Fa4CL, FaCHS and FaCHI, which are responsible for the synthesis of common precursors of flavonoid biosynthesis, were all significantly higher in RS compared to WS, similar to the results of the RNA‐seq analysis (Figure S3). Consistent with the large difference in anthocyanin contents between WS and RS from biochemical assays, the key structural genes, FaANS, FaDFR and FaUFGT (Figure 1) showed the biggest differences in expression levels between the two varieties (Figure 3c, top). In RS, these key anthocyanin biosynthetic genes showed a gradual increase in transcript levels until R and/or OR stages (Figure 3c, top; Table S2). In WS, transcripts of these genes peaked at lower levels during R and/or OR stages (Figure 3c, top). FaUFGT, encoding the last enzyme for anthocyanin biosynthesis, showed the most significant differences between RS and WS, which was 5572.49 times higher in RS relative to WS. As genes involved in proanthocyanin biosynthesis, FaANR and FaLAR showed an early peak at the G1 stage in both strawberry varieties (Figure 3c, top). Both genes exhibited much lower expression levels at G1 stage in RS (Figure 3c; Table S3) compared to WS, in accordance with the significantly higher amounts of both proanthocyanin precursors and proanthocyanins in WS fruit at this stage. FaFLS, which encodes flavonol synthase for flavonol biosynthesis, peaked in transcript levels at the early G2 stage in RS and at W and T stages in WS, and remained at a very low level through fruit maturation stages R and OR in both strawberry varieties, confirming our findings on the low levels of flavonols in mature fruit.
FaANS encodes anthocyanidin synthase and is active as a common biosynthetic step for both anthocyanin and proanthocyanin biosyntheses (Figure 1). FaANS showed high transcript levels at the G1 stage in WS (Figure 3c; Table S2), which was 2 times higher than that in RS at the G1 stage (Table S2), supporting the role of ANS in proanthocyanin biosynthesis. In summary, different branches of flavonoid biosynthesis showed differential transcript levels of key structural genes between RS and WS; the genes responsible for anthocyanin biosynthesis showed the greatest differences between RS and WS. Proanthocyanin biosynthesis is also different in WS compared to RS. The fact that transcript levels of all anthocyanin biosynthetic genes were much lower in ripe fruit of WS than of RS suggested that it was a difference in the regulation of expression of the structural genes encoding enzymes of anthocyanin biosynthesis that gave rise to the differences in colour between WS and RS.
Among the genes encoding FaMYB transcription factors, relative expression levels of FaMYB1, FaMYB10, FaMYB9 and FaMYB11, and related regulatory genes, FabHLH3, FabHLH3‐delta and FaWD40, were compared in WS and RS at T and R stages (Figure 3b). Two distinct patterns of transcript levels between RS and WS were observed for the FaMYB genes. One showed significantly higher transcript levels in RS compared to WS, including FaMYB1 and FaMYB10 (Figure 3b). The other showed significant but much smaller increases in WS compared to RS and included FaMYB9 and FaMYB11 (Figure 3b). Furthermore, most of the genes encoding TFs present in the MBW complex together with FaMYB proteins showed similar transcript levels at T and R stages in RS except for FabHLH3‐delta (Figure 3b). In fact, FabHLH3‐delta has been suggested to be a non‐functional gene lost from the complex regulating the anthocyanin biosynthetic pathway (Schaart et al., 2013).
The expression levels of the FaMYB genes were analysed by qRT‐PCR throughout the development of strawberry fruits and were calculated relative to the Actin gene as bar graphs, including the FaMYB genes and one FaMYB‐related gene, FaWD40 (Figure 3c, bottom; Figure S4). The expression level of FaMYB1 decreased gradually from G1 to T stage in both RS and WS, but started to rise again from the T stage and reached a peak level at the R or the OR stages, respectively (Figure 3c, bottom; Table S4). In RS, there was an early peak of FaMYB1 transcripts at the G1 and G2 stages (Table S4). The transcript levels of FaMYB10 were maintained at a low level from G1 to W but showed a rapid increase from the T stage to a peak level at the R stage in both WS and RS (Table S4). Thereafter, FaMYB10 transcript levels were maintained at peak levels until the OR stage but only in RS (Figure 3c; Table S4). The expression levels of FaMYB9 and FaMYB11 decreased rapidly throughout the development of strawberry fruits (Figure 3c, bottom) from their peak levels at the G1 stage (Table S5), consistent with the expression patterns of their putative target structural genes, LAR and ANR. Analysis of the related genes, FaWD40 and FabHLH3, showed high levels of transcripts at the G2 stage in both RS and WS (Figure 3c; Table S4), and remained at similar levels at the R and OR stages (Table S4), suggesting that the significant differences in anthocyanin contents, between RS and WS, were not due to differences in transcript levels of these genes. The peak level of FaMYB10 expression was much higher than that of FaMYB1 (Figure 3c, bottom), and FaMYB10 also had a higher expression level at G1 than FaMYB9 and FaMYB11 (Table S5), suggesting that FaMYB10 might be involved in both proanthocyanin and flavonol biosyntheses during the early stages of fruit development. Our data suggested that FaMYB10 plays a prominent role in creating the colour difference between RS and WS.
Coding sequences of FaMYB10 are disrupted in WS, potentially contributing to a loss of function allele
Since FaMYB1 and FaMYB10 play critical roles in controlling the coloration of strawberry fruits, the genes encoding these two transcriptional regulators were selected for further analysis in red and white strawberry varieties (Figure 4a). cDNAs of FaMYB1 and FaMYB10 were amplified separately from ripe fruits of red and white strawberry varieties using primers designed on the basis of predicted FaMYB1 and FaMYB10 sequences from F. vesca genomic sequences. Differences in cDNA sizes were examined by gel electrophoresis of the RT‐PCR products (Figure 4b), and the corresponding cDNA bands were excised from the gel and sequenced (Table 2). The FaMYB1 gene produced only two transcript types, a full‐length transcript, FaMYB1‐1 (564 bp), and a partial length transcript, FaMYB1‐2 (561 bp) (Figure 4b). Comparing the FaMYB1‐1 cDNA to the FaMYB1‐2 cDNA, there was a deletion of three nucleotides (at position 441 from the initiating ATG) in FaMYB1‐2, leading to the predicted loss of one amino acid (Asp) in FaMYB1‐2, and a predicted protein of 186 amino acids (Figure 4b). Both FaMYB1‐1 and FaMYB1‐2 transcripts were found in WS and RS fruit with transcript ratios of 12:2 and 7:2, respectively (Table 2), suggesting that FaMYB1‐1 and FaMYB1‐2 alleles probably occur naturally in both octoploid strawberry varieties, in agreement with the similar expression patterns of FaMYB1 in both varieties. Two transcripts for FaMYB10 were also detected, presumably derived from different alleles, namely FaMYB10‐1 (702 bp), encoding a full‐length protein, and FaMYB10‐2 (710 bp), a transcript with an eight‐base insertion (ACTTATAC) relative to FaMYB10‐1. Interestingly, the FaMYB10‐2 transcript was found only in 24 clones from WS, while RS (28 clones) contained only the FaMYB10‐1 transcript (Table 2). Analysis of the coding sequence of FaMYB10‐2 revealed that an 8‐bp (ACTTATAC) insertion occurred at position 491 in FaMYB10‐2, resulting in a premature stop codon and the predicted loss of 54 amino acids from the C‐terminus of the predicted FaMYB10 protein encoded by the FaMYB10‐2 transcript (Figure 4b, 179 amino acid versus 233 amino acid). The predicted truncation of the protein may be the reason for the loss of fruit colour in WS.
Figure 4.
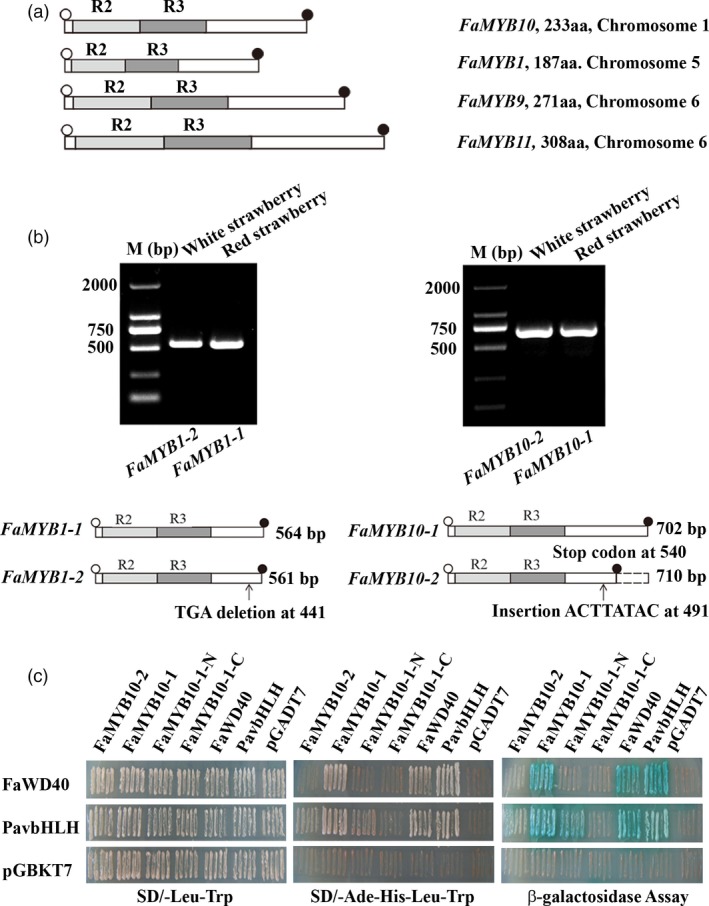
Characterization of FaMYB1 and FaMYB10 coding sequences in the white and red strawberry varieties. (a) Coding domain structures of FaMYB1, FaMYB10, FaMYB9, FaMYB11. (b) The agarose gel of FaMYB1 and FaMYB10 alleles and the missing sequence in MYB10‐2 allele. In the schematic graph, open and filled circles represent start codon and stop codon; light grey and dark grey boxes represent R2 and R3 domains. (c) The yeast two‐hybrid tests for the formation of the ternary complex, MBW by FaMYB10‐1 (red strawberry) and FaMYB10‐2 (white strawberry). Interactions of FaMYB10‐2, FaMYB10‐1, FaMYB10‐1‐N (1‐177 amino acids), FaMYB10‐1‐C (178‐233 amino acids), FaWD40 and PavBHLH with PavbHLH and PavWD40.
Table 2.
Comparison of FaMYB1 and FaMYB10 cDNA sequences in white and red strawberry varieties
| Gene | Allele | White strawberry | Red strawberry | ||
|---|---|---|---|---|---|
| Size (bp) | Ratio | Size (bp) | Ratio | ||
| FaMYB1 | FaMYB1‐1 | 564 | 12/14 | 564 | 7/9 |
| FaMYB1‐2 | 561 | 2/14 | 561 | 2/9 | |
| FaMYB10 | FaMYB10‐1 | – | – | 702 | 28/28 |
| FaMYB10‐2 | 710 | 24/24 | – | – | |
Ratio is the number of sequenced FaMYB1 clones compared to the total number of FaMYB1‐1 and FaMYB1‐2 clones or the number of FaMYB10 clones compared to FaMYB10‐1 and FaMYB10‐2 clones.
In the classic model of the regulation of anthocyanin and proanthocyanin biosynthesis, it has been proposed that there is a MBW complex consisting of MYB proteins, bHLH‐ and WD40‐repeat proteins. The MBW complex provides the core controlling hub for fruit coloration, through the transcriptional regulation of anthocyanin structural genes (D'Amelia et al., 2014; Gonzalez et al., 2008; Jin et al., 2016; Schaart et al., 2013). To test the formation of the MBW complex, FaMYB10‐1 and FaMYB10‐2 alleles from strawberry varieties were cloned into yeast expression vectors and examined in yeast two‐hybrid assays (Y2H assays) together with FaWD40, PavbHLH. Previous studies showed that PavMYB10.1a interacted with PavbHLH and PavWD40, but PavMYB10.1b did not interact with PavbHLH or PavWD40 by Y2H assays (Jin et al., 2016). Hence, PavbHLH was chosen as a partner of FaMYB10‐1 and FaMYB10‐2 alleles. Y2H assays showed that FaMYB10‐1 was able to interact with two regulators, FaWD40 and PavbHLH, while FaMYB10‐2 could not (Figure 4c; Figure S5). To test which region of FaMYB10 protein was required for the formation of the MBW complex, truncated FaMYB10‐1 constructs were designed, FaMYB10‐1‐N, which was shortened by loss of amino acids 178‐233 at the C‐terminus of the protein and FaMYB10‐1‐C, which was shortened by loss of amino acids 1–177 at the N‐terminus of the protein. Y2H assays showed that FaMYB10‐1‐N was able to interact with PavbHLH, while FaMYB10‐1‐C could not (Figure 4c; Figure S5), supporting previous reports that the interaction motif in R2R3MYB proteins lies within R3 of the N‐terminal DNA‐binding domain (Zimmermann et al., 2004). β‐gal staining assays showed positive interactions in yeast containing pGADT7‐FaMYB10‐1 plus pGBKT7‐FaWD40, pGBKT7‐PavbHLH grown on ‐T/‐L/‐H/‐A screening medium, but no growth of yeast containing pGADT7‐FaMYB10‐2 plus pGBKT7‐FaWD40, pGBKT7‐PavBHLH or the empty pGBKT7 vector was observed. The results of β‐galactosidase activity tests were consistent with the idea that FaMYB10‐1 forms a canonical MBW complex with bHLH and WD40, but FaMYB10‐2 cannot form the MBW complex, indicating that the FaMYB10‐2 allele from WS may be unable to induce anthocyanin biosynthesis, which may explain the fruit colour difference between RS and WS.
Transient transformation was used to confirm the function of the FaMYB10 alleles. The HyperTrans expression system was used to deliver gene expression vectors transiently to the abaxial side of leaves of tobacco (Nicotiana benthamiana) and flesh of white strawberry at the W stage. Anthocyanins accumulated in leaves of tobacco infiltrated with the FaMYB10‐1 expression vector plus PavbHLH expression vector and flesh of WS infiltrated with the FaMYB10‐1 expression vector, while anthocyanin was absent with the FaMYB10‐2 expression vector or agroinfiltration solution (CK) alone (Figure 5). After 5 days, anthocyanin accumulation was observed in the tobacco leaves infiltrated with the FaMYB10‐1 expression vector plus PavbHLH expression vector and white fruit infiltrated with the FaMYB10‐1 expression vector, which was not observed with the FaMYB10‐2 expression vector or CK alone (Figure 5a and b). Anthocyanins accumulated to 9.30 mg/100g FW in the tobacco leaves and 13.74mg/100g FW in the white fruit infiltrated with the FaMYB10‐1 expression vector, while anthocyanins were undetectable in leaves infiltrated with the FaMYB10‐2 expression vector or CK (Figure 5c and d). The anthocyanin was identified as cyanidin‐3‐O‐rutinoside in the tobacco leaves and was mainly pelargonidin‐3‐glucoside in the white strawberry fruits infiltrated with the FaMYB10‐1 expression vector (Figure 5e and f), confirming that the HyperTrans experiment had worked. FaMYB10 showed striking differences in transcript levels in tobacco leaves and in the white strawberry fruits infiltrated with the FaMYB10‐2 and FaMYB10‐1 expression vectors, compared to CK (Figure 5g and h). These results confirmed that FaMYB10 genes control anthocyanin and colour production in fruit of octoploid strawberry and that the loss of the functional FaMYB10 protein is critical to the loss of fruit colour in white octoploid strawberry.
Figure 5.
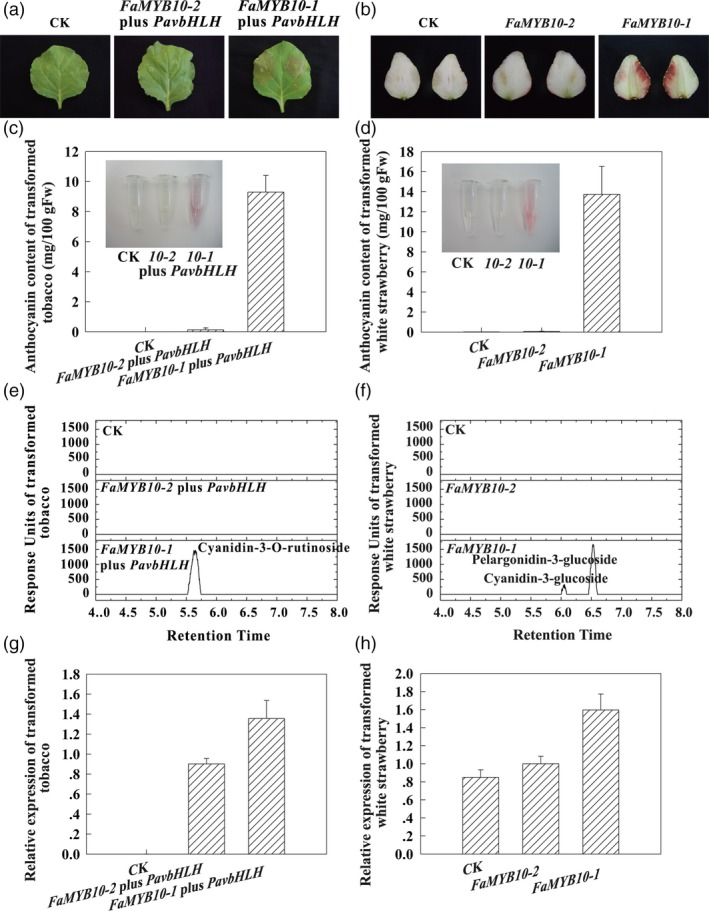
Transient over‐expression of FaMYB10‐1 and FaMYB10‐2. Tobacco (Nicotiana benthamiana) leaves and flesh of white strawberry variety (‘Snow Princess’) were infiltrated with cDNA constructs corresponding to two different FaMYB10 alleles and agroinfiltration solution (CK). (a) The phenotypes of leaves of tobacco after being infiltrated. (b) The phenotypes of fruits of WS after being infiltrated. (c) The anthocyanin contents of extracts of leaves of tobacco after being infiltrated. (d) The anthocyanin contents of extracts of fruits of WS after being infiltrated. (e) The chromatograms of anthocyanins in leaves of tobacco after being infiltrated. (f) The chromatograms of anthocyanins in fruits of WS after being infiltrated. (g) The expression analysis of FaMYB10 in leaves of tobacco after being infiltrated. (h) The expression analysis of FaMYB10 in fruits of WS after being infiltrated.
The genomic sequences of FaMYB10 differ between WS and RS
To investigate whether there were genomic differences between FaMYB1 and FaMYB10 alleles in RS and WS, genomic DNA was amplified by PCR, subcloned and sequenced. Eighty‐seven and 92 independent clones were selected from red and white octoploid strawberry varieties, respectively. Our results demonstrated that the sizes of FaMYB1‐1 and FaMYB1‐2 alleles varied from 1352 bp to 1534 bp (Figure S6; Table 3). The major difference in FaMYB1 genomic sequences appeared in the second intron of FaMYB1 alleles with a variable number of GA repeats (Figure S6), but the distribution of FaMYB1 genomic sequence types was highly similar in the analysed clones between RS and WS (Table 3). However, this was not the case for the analysis of the FaMYB10 genomic sequences. All the analysed genomic sequences of FaMYB10‐1 contained two introns in positions consistent with the intron locations of other R2R3MYB genes (Jin et al., 2016; Kobayashi et al., 2004). The genes encoding FaMYB10‐1 varied in size from 1627 to 1661 bp due to differences in the length of intron 2 (Table 3). FaMYB10‐2 alleles were amplified solely from WS, and strikingly, all clones had no intron sequences (Figure 6a and b; Table 3).
Table 3.
Comparison of FaMYB1 and FaMYB10 genomic sequences in white and red strawberry varieties
| Gene | Type | White strawberry | Red strawberry | ||
|---|---|---|---|---|---|
| Size (bp) | Ratio | Size (bp) | Ratio | ||
| FaMYB1 | FaMYB1‐1 | 1352 | 7/26 | 1352 | 10/61 |
| 1475 | 1/26 | 1475 | 8/61 | ||
| 1490 | 1/26 | 1490 | 2/61 | ||
| 1501 | 5/26 | 1501 | 10/61 | ||
| 1515 | 3/26 | 1515 | 7/61 | ||
| 1534 | 7/26 | 1534 | 17/61 | ||
| FaMYB1‐2 | 1352 | 2/26 | 1352 | 2/61 | |
| 1515 | – | 1515 | 1/61 | ||
| 1534 | – | 1534 | 4/61 | ||
| FaMYB10 | FaMYB10‐1 | – | – | 1627 | 3/72 |
| – | – | 1629 | 3/72 | ||
| – | – | 1631 | 7/72 | ||
| – | – | 1633 | 16/72 | ||
| – | – | 1635 | 27/72 | ||
| – | – | 1637 | 4/72 | ||
| – | – | 1659 | 2/72 | ||
| – | – | 1661 | 10/72 | ||
| FaMYB10‐2 | 710 | 20/20 | – | – | |
After the examination of coding regions in FaMYB1 genomic sequences, if it contained the coding sequence of FaMYB1‐1 or FaMYB1‐2, it was assigned as the genomic sequences for FaMYB1‐1 or FaMYB1‐2; ratio shows the number of FaMYB1‐1 or FaMYB1‐2 clones to total sequenced clones; after examination of coding regions in FaMYB10 genomic sequences, if it contains the coding region of FaMYB10‐1 or FaMYB10‐2, it was assigned to the genomic sequences for FaMYB10‐1 or FaMYB10‐2, respectively; ratio is the number of FaMYB10‐1 or FaMYB10‐2 clones to total sequenced clones.
Figure 6.

Genomic sequence analysis for FaMYB10 alleles in red and white strawberry varieties and phylogenetic analysis of homologous transcriptional regulators. (a) The agarose gel of FaMYB10 alleles. (b) Schematic graph for gene structure of FaMYB10 alleles emphasizing the different regions of FaMYB10 intron examined. (c) Phylogenetic analysis of the flavonoid‐related MYB transcriptional factors based on the amino acid sequences of the MYB proteins. Open and filled black circles represent start codon and stop codon, respectively, in the schematic graph. The grey and dark boxes represent the sequences from online databases and the sequenced FaMYB family gene alleles in the phylogenetic tree.
Phylogenetic analysis of homologous transcriptional regulators
To establish the molecular evolutionary relationship among the colour‐related MYB genes from different plant species, a phylogenetic tree was constructed by the neighbour‐joining method using 40 closely related R2R3‐type MYB genes including strawberry FaMYB1 alleles and FaMYB10 alleles, based on the full‐length deduced amino acid sequences (Figure 6c). The protein products of FaMYB10‐1 and FaMYB10‐2 clustered with all known MYB10‐type proteins that regulate anthocyanin biosynthesis, including MdMYB1, AtMYB75 (production of anthocyanin pigment 1 protein, PAP1), AtMYB90 and VvMYBA1, which grouped in a single clade. In previous studies, MdMYB10, VvMYBA1 and PavMYB10.1 have been demonstrated to control fruit skin colours by controlling anthocyanin biosynthesis (Espley et al., 2007; Jin et al., 2016; Kobayashi et al., 2004), suggesting that FaMYB10 has a very similar, if not identical, function in the regulation of fruit coloration through controlling expression of anthocyanin biosynthetic genes in octoploid strawberry (Figure 6c). Secondly, FaMYB1‐1 and FaMYB1‐2 clustered with other MYB1‐type proteins from different plant species AtMYB6‐like, MdMYB17, MdMYB111, PavMYB111 and VvMYB6 as a single independent clade; FaMYB9 and FaMYB11 cluster in the same clade together with MdMYB9, MdMYB11 and VvMYB308, suggesting that FaMYB9 and FaMYB11 are involved in the control of proanthocyanin biosynthesis similar to MdMYB9 and MdMYB11 in other species (An et al., 2015; Schaart et al., 2013).
Discussion
A regulatory model for the production of flavonoids in octoploid strawberry fruit
In recent years, there has been considerable progress in understanding the molecular mechanisms underpinning the coloration of fruit and flowers through multiple investigations of transcriptional regulation of key genes (Allan et al., 2008; Martin et al., 2010; Schwinn et al., 2014). It has been found that R2R3MYB TFs play critical regulatory roles in determining the accumulation of anthocyanins in several horticultural crops (Jin et al., 2016; Kobayashi et al., 2004; Zhang et al., 2014). In comparison with the known FaMYB genes extracted from the databases, the putative FaMYB1‐1 allele and FaMYB10‐1 allele from RS were included for the analysis of R2R3 domain similarity using conserved regions of homology. The coding sequences of FaMYB1‐1, FaMYB9, FaMYB11 and FaMYB10‐1 all encoded intact R2 and R3 domains (Table 4). Among these FaMYB genes, the shared similarity of the coding sequences in their R2 and R3 domains ranged from 58.3% to 82.6%, but there was very little similarity in their encoded C‐terminal regions ranging from 6.6% to 28.6% (Table 4). These levels of similarity suggest that FaMYB1‐1, FaMYB9, FaMYB11 and FaMYB10‐1 may interact only partially with the same binding partners in the MBW complex (Lin‐Wang et al., 2014; Schaart et al., 2013) and/or may recognize distinct DNA‐binding motifs in their target genes.
Table 4.
Amino acid similarity analysis between the coding sequences of known FaMYB genes and the deduced coding sequence of FaMYB10‐1 from octoploid red strawberry variety
| Percentage Similarity (%) | N‐terminus | R2 domain | R3 domain | C‐terminus |
|---|---|---|---|---|
| FaMYB1‐1 vs FaMYB10‐1 | 14.3 | 58.3 | 82.6 | 6.6 |
| FaMYB9 vs FaMYB10‐1 | 28.6 | 60.4 | 73.9 | 7.1 |
| FaMYB11 vs FaMYB10‐1 | 28.6 | 60.4 | 76.1 | 9.4 |
To dissect further the network of transcriptional regulation in fruit coloration of strawberry varieties, principal component analysis (PCA) was used to decipher the interrelationships between the major FaMYB TFs and the structural genes of flavonoid biosynthesis in response to changes in endogenous ABA levels. PCA is often used to interpret the underlying relationships between multiple variables and reduces the dimensionality of a data set to a few composite variables that retain most of the information in the original variables (Jin et al., 2012; O'Brien et al., 2004; Wang et al., 2012). In the current PCA, the factor loadings mirrored the interrelationships exhibited by anthocyanins, flavonols, proanthocyanidins, transcriptional levels of regulatory and structural genes, and endogenous ABA content in RS and WS at the key developmental stages for fruit coloration, the T and R stages (Figure S7). Two PCA components explained 80.32% of the variance in fruit coloration of red and white strawberry varieties. The first axis explained 55.29% of the variance and was positively correlated with pelargonidin‐3‐glucoside, the transcriptional changes in FaMYB1, FaMYB10, FaCHS, FaCHI, FaF3H, FaANS and FaUFGT and the changes in endogenous ABA levels, indicating that the variation in transcript levels of FaMYB1 and FaMYB10 influenced directly anthocyanin accumulation in response to the high concentrations of endogenous ABA, in accordance with the up‐regulation of expression of key transcriptional factors and structural genes controlling anthocyanin biosynthesis. This axis is referred to hereafter as the ‘anthocyanidin index’ (Figure S7). The second axis explained an additional 25.03% of the variance and was positively correlated with the concentrations of proanthocyanins and proanthocyanin precursors and the transcriptional variation of FaMYB9, FaMYB11, FaLAR and FaFLS, in accordance with key structural and transcription factor genes for the biosynthesis of proanthocyanins and proanthocyanin precursors, indicating that the accumulation of proanthocyanins was closely regulated by FaMYB9 and FaMYB11. This axis is referred to hereafter as the ‘proanthocyanin index’ (Figure S7). Finally, the PCA showed that the accumulation of anthocyanins was, in general, related to the variation of endogenous ABA levels at different maturation stages in strawberry fruit.
Abscisic acid (ABA) is an important hormone associated with fruit maturation processes, such as sugar accumulation, softening, and coloration in sweet cherry (Ayub et al., 2016; Jin et al., 2016; Ren et al., 2010; Shen et al., 2014). In our study, at the early developmental stages, from the G1 to T stages, the level of endogenous ABA was maintained at relatively low levels, compared to R and OR stages in both RS and WS varieties, although the level of endogenous ABA was significantly higher in RS, in accordance with the increase in the levels of transcripts of the key structural genes, FaLAR, and FaANR for proanthocyanin biosynthesis and the higher concentrations of proanthocyanins observed in RS. In contrast, FaMYB10 was expressed at a low level in both strawberry varieties at the early developmental stages, which may explain the low anthocyanin content at these stages. During fruit maturation of strawberry, the endogenous ABA appeared significantly higher from T to OR stages, in accordance with the increase in the transcripts of FaMYB1 and FaMYB10. TFs FaMYB1 and FaMYB10 activated the transcripts of the key EBG genes, including FaPAL, FaCHS, FaCHI and Fa4CL to supply the precursors of flavonoid biosynthesis and the key LBG genes, including FaANS, FaUFGT and FaDFR for anthocyanin biosynthesis. Interestingly, FaMYB1 had been shown to be negatively correlated with anthocyanin biosynthesis, but the results of our PCA and phylogenetic analysis suggested positive functions for FaMYB1, which may include a partial role in promoting the formation of anthocyanin in different tissues, as observed in red strawberry seeds in WS (Figure S1a). Furthermore, FaMYB9/11 are not able to clearly cluster with the key structural genes and the key flavonoid components, suggesting that these R2R3FaMYB genes are probably suppressed in the later developmental stages of strawberry fruit (Schaart et al., 2013).
A regulatory model for major flavonoid components was derived from the above experimental analyses. This model depicts the interrelationships of the key FaMYB genes in regulating the composition of flavonoid components through the differential transcription of key structural genes for anthocyanin, proanthocyanin and flavonol biosynthesis at the late stages of strawberry fruit development (Figure 7). ABA is a signal molecule that promotes FaMYB10 and FaMYB1 expressions, and FaMYB10 or FaMYB1 interacts with bHLH and WD40 to form a putative canonical MBW activation complex, which activates transcription of structural genes and causes anthocyanin accumulation. Moreover, FaMYB9 or FaMYB11 interacts with bHLH and WD40 to form a MBW complex, which activates transcription of structural genes and causes accumulation of proanthocyanins.
Figure 7.
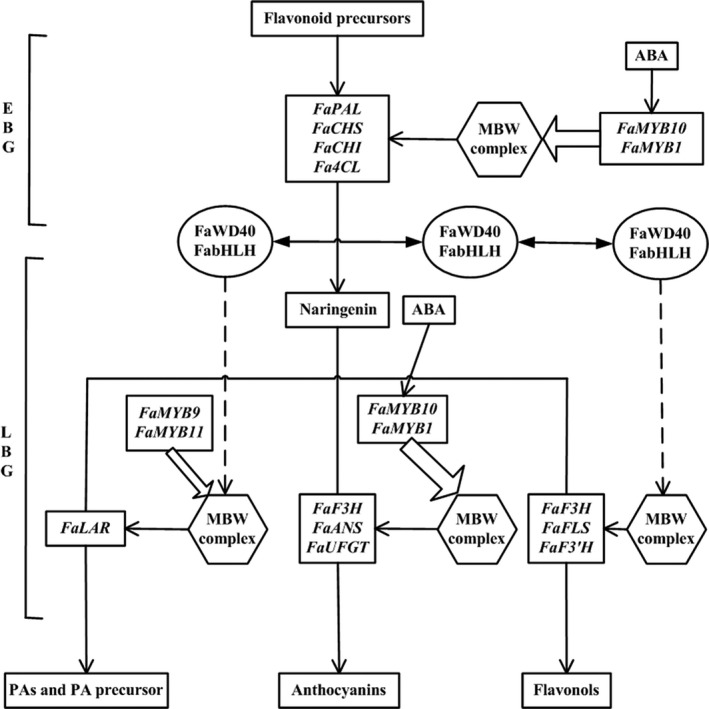
The proposed model for combinatorial effects of FaMYB1, FaMYB10, FaMYB9 and FaMYB11 on the regulation of the flavonoid biosynthetic pathways in response to endogenous ABA maturation signal. ABA is a signal molecule that promotes FaMYB10 and FaMYB1 expressions, and FaMYB10 or FaMYB1 interacts with bHLH and WD40 to form a putative canonical MBW activation complex, which activates transcription of structural genes and causes anthocyanin accumulation. Moreover, FaMYB9 or FaMYB11 interacts with bHLH and WD40 to form a MBW complex, which activates transcription of structural genes and causes accumulation of proanthocyanins.
The R2R3 MYBs control fruit colour, and FaMYB10‐2 leads to coloration loss in the fruits of WS
In many fruits, R2R3 MYB TFs control the expression of structural genes in the anthocyanin biosynthetic pathway. MYB genes play an important role in anthocyanin accumulation in fruit. In sweet cherry, an R2R3MYB PavMYB10.1 controls fruit skin colour. Three functional alleles of the gene, PavMYB10.1a, PavMYB10.1b and PavMYB10.1c, lead to different fruit colours. PavMYB10.1a is dominant to PavMYB10.1b, and PavMYB10.1b is dominant to PavMYB10.1c. The allele PavMYB10.1a is present in sweet cherry with dark red fruit, but not in those with yellow fruit, while the allele PavMYB10.1b is present in sweet cherry with blush fruit, but not in those with yellow fruit. Only the PavMYB10.1c allele produced yellow fruit (Jin et al., 2016). In Citrus, an R2R3 MYB Ruby, a transcriptional activator of anthocyanin production, controls fruit colour. Sicilian blood oranges (Citrus sinensis) arose by insertion of a Copia‐like retrotransposon adjacent to the R2R3MYB gene encoding Ruby. The retrotransposon induces the expression of the Ruby gene in fruit. In different accessions of Citrus species and in domesticated cultivars, the differences in transcript levels of the Ruby caused by point mutations, deletions and insertions of transposable elements lead to natural variation in leaf and flower colours (Butelli et al., 2012, 2017). In apple (Malus domestica), R2R3 MYB TFs responsible for anthocyanin accumulation in fruit are encoded by MdMYB1, MdMYB10 and MdMYBA transcripts, which are likely to be allelic. The promoter of MdMYB10 has an allelic rearrangement that regulates MdMYB10 transcript levels and the subsequent ectopic accumulation of anthocyanins (Allan et al., 2008; Ban et al., 2007; Espley et al., 2007; Lin‐Wang et al., 2010; Takos et al., 2006; Telias et al., 2011).
In strawberry, F. vesca is a diploid species of wild strawberry. A candidate SNP (single nucleotide polymorphism) in FvMYB10 was functionally confirmed to be responsible for yellow‐coloured fruits. The specific SNP (G to C) exhibits a Trp to Ser within the R2 DNA‐binding domain of FaMYB10. All red accessions have G, while all yellow accessions have C (Hawkins et al., 2016). However, we found the conserved G at the R2 DNA‐binding domain in FaMYB10 in octoploid RS and WS. Through the analysis of the sequences of different FaMYB10 alleles, RS carries allele(s) encoding a full‐length transcript, FaMYB10‐1, whereas WS carries only allele(s) with the insertion, which encode a predicted protein with an early termination codon. This truncated FaMYB10 protein was shown to be non‐functional in that it was defective in forming an MBW complex and in activating anthocyanin biosynthesis in N. benthamiana. The insertion, ACTTATAC, in the region of the FaMYB10 gene encoding the C‐terminus of the FaMYB10 protein may explain the loss of anthocyanin biosynthesis in WS due to the loss of transcriptional activation by the MBW complex. The FaMYB10‐2 allele lacks introns, suggesting that it was generated by reverse transcription, in a process that might also have involved transposition. Based on N. benthamiana transient over‐expression of different FaMYB10 alleles, the full‐length FaMYB10‐1 cDNA could induce the accumulation of anthocyanins to visible levels but FaMYB10‐2 could not. Similarly, in transient over‐expression of different FaMYB10 alleles in WS, the full‐length FaMYB10‐1 cDNA developed red pigmentation and complemented the WS phenotype, whereas FaMYB10‐2 did not. These results show that the mutation in FaMYB10‐2 leads to the loss of coloration in the fruits of WS.
Experimental procedures
Plant materials and growth conditions
Red octoploid strawberry variety (Fragaria × ananassa ‘Sweet Charlie’, red strawberry; Chandler et al., 1997) and white octoploid strawberry variety (F. × ananassa ‘Snow Princess’, white strawberry; Zhang et al., 2016) were grown in a greenhouse at the Beijing Academy of Forestry and Pomology Sciences, China. Plant tissues were collected for DNA extraction for gene cloning, for RNA for quantitative PCR and for flavonoid analyses. Based on the seed and fruit colour of ‘Snow Princess’, six visual developmental stages were defined at 13, 17, 24, 30, 46 and 50 day after anthesis. Based on the seed and fruit colour of ‘Sweet Charlie’, six visual developmental stages were defined as: small green fruit stage (G1), big green fruit stage (G2), white fruit stage (W), turning fruit stage (T), ripe fruit stage (R) and over‐ripe fruit stage (OR) at 7, 12, 17, 19, 26 and 30 day after anthesis, as shown in Figure S1 (Moyano et al., 1998; Schaart et al., 2013). For obtaining the experiment material from strawberry fruits, all seeds were removed from the fruit before extraction; fully expanded leaves and fruits of different developmental stages were harvested, immediately frozen in liquid nitrogen and stored at −80 °C until the analysis was performed.
Measurement of total anthocyanin content
Fruits at different developmental stages of both white and red strawberry varieties were harvested, ground and extracted with the extraction solution (1% HCl in methanol) at room temperature and then in the dark for 4 h. The fruit extract was centrifuged at 8500 g for 20 min; the supernatant was collected and transferred to a clean tube and filtered through a 0.2‐μm filter. The total content of anthocyanins was estimated by a pH differential method (Benvenuti et al., 2004; Cheng and Breen, 1991). The total anthocyanin content of individual experimental samples was calculated and estimated as cyanidin‐3‐glucoside equivalents mg per 100 g fresh weight (FW). All analyses were performed with three biological replicates.
Tentative identification and quantification of flavonoid components
The analytical method was modified for the identification and quantification of flavonoid composition, anthocyanins, flavonols and proanthocyanins from strawberry fruit extracts in the fruits of red and white strawberry varieties at T and R stages (Veberic et al., 2015). 10 mg of lyophilized fruit powder was extracted in 1mL of 0.1% acetic acid/methanol solution at 4 °C overnight for different materials at the T and R stage and centrifuged for 10 min at 14 500 g, and then, the supernatant was collected for drying with a vacuum centrifuge concentrator (CV100‐DNA, Baijiu, Beijing, China). The dried experimental materials were stored at −20 °C before analysis. The dried extracts were re‐dissolved in methyl alcohol before analyses and were examined with a ultra‐high‐performance liquid chromatography–mass spectrometry (UPLC‐MS/MS, Waters, Milford, MA) coupled to a triple‐Quadrupole Mass Spectrometry (XEVO®‐TQ) with electrospray ionization (ESI). The separation was carried out with a ZORBAX Eclipse plus C18 (150 mm × 3.0 mm) with particle size of 1.8 µm (Agilent Technologies, Santa Clara, California) at 40°C. The gradient was using 0.1% formic acid (A) and acetonitrile (B) as the mobile phases, 0–1 min (5% B), 1–8 min (5%–30% B), 8‐12 min (40%–95% B), 16‐17 min (95%–100% B), 17–21 min (100% B) and 21–25 min (5% B). The operating conditions were as follows: a flow rate of 0.4 mL/min and positive ion ESI modes, the capillary voltages of 3.0 kV and 16 L/h of nebulization nitrogen flow. The chromatographs were plotted and analysed using OriginPro 2015 (OriginLab Corporation, Northampton, MA).
Compounds were identified by comparison with the retention times of standards, the characteristics of UV‐Vis spectra of peaks and the mass spectrometric information with MassLynx V4.1 software (Waters, Milford, MA). The relative quantification of anthocyanins, flavonoids and proanthocyanins contents was calculated from peak areas of samples based on the intensity of the corresponding standard compounds, including quercetin‐3‐glucoside, kaempferol‐3‐rutinoside, cyanidin‐3,5 ‐diglucoside, pelargonidin‐3‐glucoside, proanthocyanidin B1 and proanthocyanidins, catechin and epicatechin. For compounds lacking corresponding standards, the quantification was carried out using similar compounds. All analysis was performed with three biological replicates.
RNA‐seq analysis
Total RNA was isolated from the fruits of red and white strawberry varieties at T and R stages using an RNA isolation kit (Waryong, Beijing, China). Differential expression of flavonoid biosynthesis genes was analysed using the RPKM method, and the scaled expression levels were displayed as heatmaps (Mortazavi et al., 2008). After the RNA extract had been treated with DNase I, first‐strand cDNA was synthesized with a RevertAid First‐Strand cDNA synthesis kit (Thermo Scientific Inc., MA). A total of 3 μg RNA per sample was used as input material for the RNA sample preparation at T and R stages from the white and red strawberry varieties. Sequencing libraries were generated using NEBNext® Ultra™ RNA LibraryPrep Kit for Illumina® (New England Biolabs, MA) according to the manufacturer's manual. The libraries were sequenced using Illumina HiSeq™ 2000 (Illumina incorporation, San Diego, California). The raw reads were first filtered by removing the adapter sequences and low‐quality sequences and aligned against F. vesca genome sequences. Functional UniGenes were predicted using BLAST program with E‐value threshold of 10−5 to protein databases, including the NCBI non‐redundant (nr) database, the Swiss‐Prot protein database, the Kyoto Encyclopedia of Genes and Genomes database (KEGG) and the Clusters of Orthologous Groups of proteins database (COG/KOG). Differential gene expression analyses were performed using RPKM method, and the scaled expression levels of flavonoid biosynthesis‐related transcription factors and structural genes were displayed as the heatmaps for the samples from the developmental stages, T and R, in both white and red strawberry varieties (Mortazavi et al., 2008). The resulting P values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted P‐value < 0.05 found by DESeq were assigned as differentially expressed. All RNA‐seq analyses were performed using three biological replicates.
Expression analysis of structural genes and transcription factors related to flavonoid biosynthesis
Expression analysis of flavonoid biosynthesis‐related genes was undertaken as described previously (Jin et al., 2016; Livak and Schmittgen, 2001). Total RNA and the first‐strand cDNA were isolated and amplified from different developmental stages of strawberry fruits from G1 to OR. To further investigate various structural genes and transcription factors in the pathway of flavonoid biosynthesis during fruit development, specific primers were designed with Primer 5.0 program (Premier Biosoft International, Palo Alto, CA), as shown in Supplemental Table S6, and acquired from Sangon Inc. (Sangon, Shanghai, China). qRT‐PCR was conducted with on a Bio‐Rad CFX96 Real‐Time PCR Detection System (Bio‐Rad, CA) using 10 μL reaction mixture, which included 5 μL 2 × SYBR Premix (Bio‐Rad, CA), 1 μL forward primer (10 μm), 1 μL reverse primer (10 μm), 1 μL cDNA template (20 ng) and 2 μL ddH2O. PCR conditions were as follows: 1 cycle at 95 °C for 30 s, 40 cycles at 95 °C for 5 s and 1 cycle at 60 °C for 30 s. Finally, the transcription levels of different genes were analysed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). All analyses were performed with three biological replicates.
cDNA and genomic sequence examination of FaMYB1 and FaMYB10 alleles
cDNA and genomic sequence examination of FaMYB1 and FaMYB10 alleles was undertaken as described previously (Jin et al., 2016). Total cDNA was acquired, and total genomic DNA was extracted from 250 mg of fresh leaves with an EZ Spin Column Genomic DNA Isolation Kit (Waryong, Beijing, China). The PCR primers for FaMYB1 and FaMYB10 cDNAs and genomic DNAs were designed with the Primer 5.0 program (Premier Biosoft International, Palo Alto, CA), as shown in Table S6, and acquired from Sangon Inc. (Sangon, Shanghai, China).
For cDNA PCR conditions, 10 μL reaction mixture included 5 μL 2 × SYBR Premix, 1 μL forward primer (10 μm), 1 μL reverse primer (10 μm), 1 μL cDNA template (20 ng) and 2 μL ddH2O; the PCR conditions were as follows: 1 cycle at 95 °C for 30 s, 40 cycles at 95 °C for 5 s and 1 cycle at 60 °C for 30 s.
For genomic DNA PCR conditions, 20 μL PCR mixture included 2 μL 2 × buffer, 1 μL dNTPs (10 mmol), 1 μL primer (10 pmol), 2.5 U Taq polymerase (Sangon, Shanghai, China) and 3 μL genomic DNA (100 ng); the PCR condition was as follows: 1 cycle at 94 °C for 4 min, 35 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min, followed by a final cycle at 72 °C for 10 min. The PCR products were examined on 1% (w/v) agarose gels, stained with ethidium bromide (EtBr) and visualized with a gel imaging system (Gel Doc XR, Bio‐Rad, CA).
The amplified FaMYB1 and FaMYB10 cDNA and genomic DNA were cut from gels according to their predicted cDNA and genomic DNA sizes, and purified using an EZ‐10 Spin Column DNA gel extraction kit (Bio Basic Inc., ON, Canada), and then were cloned into pGEM‐T Easy (Promega, Madison, WI) for amplifying in Escherichia coli strain DH5α. Finally, FaMYB1 and FaMYB10 genes were sequenced.
Yeast two‐hybrid (Y2H) assays
Y2H assays were described previously (Jin et al., 2016). Yeast AH109 cells were co‐transformed with a specific bait and prey constructs according to the manufacturer's instructions (Clontech, CA). For Y2H assays of interactions of FaMYB10 (FaMYB10‐1 and FaMYB10‐2) with two regulators (FaWD40 and PavbHLH), FaMYB10‐1, FaMYB10‐2, FaMYB10‐1‐N (1–531 bp), FaMYB10‐1‐C (532–702 bp), FaWD40 and PavBHLH were amplified and cloned into pGADT7 vector (Clontech, CA); two regulators (FaWD40 and PavBHLH) of the flavonoid biosynthetic pathway were cloned into the pGBKT7 vector (Clontech, CA). All primers are listed in Table S6 and were acquired from Sangon Inc. (Sangon, Shanghai, China). All the constructs were confirmed by sequencing and then transformed into the yeast strain AH109 using the lithium acetate method (Gietz et al., 1995). The subsequent clones were plated onto selective medium lacking Leu and Trp (Leu, leucine; Trp, tryptophan), and finally, putative transformants were transferred onto selective medium lacking Ade, His, Leu and Trp (Ade, adenine; His, histidine). After the selected colonies had been tested, β‐galactosidase assays were undertaken following a yeast protocol (PT3024‐1; Clontech, CA). The selected colonies were transferred to plates containing 40 μg/mL X‐α‐gal (Clontech, CA), and the vector P53 plus SV40 was used to exclude false‐positive activation.
Transient expression in Nicotiana benthamiana leaves and ‘Snow Princess’ fruits
Transient expression of FaMYB10‐1 and FaMYB10‐2 cDNA constructs was carried out using the HyperTrans system (Butelli et al., 2017; Sainsbury et al., 2009). FaMYB10 cDNAs were isolated from the flesh of red octoploid strawberry variety (‘Sweet Charlie’) and the white octoploid strawberry variety (‘Snow Princess’) at the R stage using an RNA isolation kit (Waryong, Beijing, China). PCR amplified (using primers listed in Table S6) was cloned into pEAQ‐HT vectors. The plasmids obtained were introduced into Agrobacterium tumefaciens GV3101. Each fresh colony was inoculated into 10 mL LB with Kan (50 μg/mL) and Rif (50 μg/mL) and grown at 28 °C overnight and then was spun down for 15 min at 4200 g. The supernatant was removed completely, and cells were resuspended in 15 mL agroinfiltration solution (10 mm MgCl2, 10 mm MES pH 5.6). Cultures were spun down for 15 min at 4200 g. The supernatant was removed completely, and cells were resuspended in 10 mL agroinfiltration solution (10 mm MgCl2, 10 mm MES pH 5.6) + 200 μm acetosyringone. Cultures were incubated for 2 h and then diluted to an OD600 of around 0.2. Then, the strains, including agroinfiltration solution (CK), FaMYB10‐1 expression vector plus PavbHLH expression vector, FaMYB10‐2 expression vector plus PavbHLH expression vector, were infiltrated into the abaxial side of leaves of five‐week‐old plants. Leaves were harvested 7 days after infiltration. The strains, including agroinfiltration solution (CK), FaMYB10‐1 expression vector, FaMYB10‐2 expression vector, were infiltrated into the flesh of white octoploid strawberry variety at the W stage. Fruits were harvested 14 days after infiltration. Harvested leaves and fruits were subsequently used for photographs, measurement of total anthocyanin content, HPLC analysis of anthocyanins and expression analysis of the transcription factor FaMYB10.
Phylogenetic analysis of different MYB genes from different plant species
To perform the phylogenetic analysis of flavonoid biosynthesis‐related MYB genes, the MYB gene sequences in octoploid strawberry were first acquired from NCBI database for FaMYB9, FaMYB11, FaMYB1 and FaMYB10 and our consensus sequences of FaMYB1 and FaMYB10 allele sequences were acquired from the alignment of the sequenced clones from red and white strawberry varieties using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and ClustalW (Chenna et al., 2003). Furthermore, to process the previously identified and putative MYB classes of genes in strawberry‐related species, the gene sequences of the identified MYB genes were extracted from NCBI databases using the Blastx program, based on the highest identity of protein sequences to FaMYB9, FaMYB11, FaMYB1 and FaMYB10, respectively. These species included several fruit plant species from the Rosaceae family, such as apple, sweet cherry, wine grape, pears, and Arabidopsis thaliana. There were 40 MYB genes chosen for the phylogenetic analysis in total (Table S7).
To identify the conserved DNA‐binding domains, R2 and R3, and to separate the N and C termini of key strawberry MYB genes, FaMYB9, FaMYB11, FaMYB1 and FaMYB10 genes were predicted and analysed using the Pfam website (http://pfam.xfam.org). The whole amino acid sequences of 40 MYB genes were aligned with ClustalW using MEGA 7.0 software package. Based on the alignment of deduced amino acid sequences, a phylogenetic tree was constructed with 1000 bootstrap replicates by the neighbour‐joining method (NJ) using the MEGA 7.0 software package.
Principal component analysis (PCA) for flavonoid biosynthesis‐related factors
On PCA of flavonoid biosynthesis‐related factors, the analysed data included the scaled measurement of major flavonoids, the scaled transcriptional levels of flavonoid biosynthesis‐related genes and the scaled measurement of endogenous ABA within at least three independent repeats. The data were pre‐treated using a range scaling method (van den Berg et al., 2006). This PCA method was employed to evaluate how explanatory variables and the levels of anthocyanins, flavonols and proanthocyanins were correlated with different structural flavonoid biosynthetic and regulatory genes in response to the changes in endogenous ABA levels at two critical fruit developmental stages, T stage and R stages. Firstly, the PCA factor scores were extracted from the analysed data representing all variables after reducing sixteen variables to two factors. Secondly, the corresponding loading plots were generated with the factor scores from the PCA and displayed as the PCA score plot using SPSS 16.0 (SPSS Inc., Chicago, USA).
Conflict of interest
The authors declare no conflict of interest.
Authors' contributions
W.J. and C.M. designed research; H.W., Y.Y., E.B., M.L., Z.X. and J.D. performed research; H.W., H.Z., J.L., A.W., Y.Z., J.L. and G.W. analysed data; W. J., C.M., H.W. and H.Z., wrote the paper.
Supporting information
Figure S1 Fruit developmental stages of white and red octoploid strawberry varieties.
Figure S2 Chromatographs of anthocyanins, flavonols, and proanthocyanins.
Figure S3 Transcript levels of structural genes and regulatory genes not included in Figure 3 in the white and red strawberry varieties.
Figure S4 Transcript levels of regulatory genes in the white and red strawberry varieties.
Figure S5 The constructs and results from yeast two‐hybrid assay of FaMYB10‐1 or FaMYB10‐2 with two regulators (FaWD40 and PavbHLH) for the formation of MBW ternary complex.
Figure S6 Genomic sequence analysis for FaMYB1 alleles in red and white strawberry varieties.
Figure S7 PCA of key flavonoid components against flavonoid biosynthetic structural genes and transcription‐related genes.
Table S1 Analysis of flavonoid contents.
Table S2 The expression of structural genes, FaANS, FaUFGT, and FaDFR in flavonoid biosynthesis in red and white strawberry varieties.
Table S3 The expression of structural genes, FaLAR, FaANR, and FaFLS in flavonoid biosynthesis in red and white strawberry varieties.
Table S4 The expression of regulatory genes, FaMYB1, FaMYB10, and FaWD40 of flavonoid biosynthesis in red and white strawberry varieties.
Table S5 The expression of regulatory genes, FaMYB9, and FaMYB11 of flavonoid biosynthesis in red and white strawberry varieties.
Table S6 The sequences of the oligonucleotide primers used in this work.
Table S7 Information about the MYB protein sequences used for the construction of the neighbour joining tree.
Acknowledgements
We thank Dr. Yan Zhu from the Institute of Botany, Chinese Academy of Sciences, for experimental technical assistances. We also thank Professor George Lomonossoff from John Innes Centre for providing HyperTrans vector system. This work was supported by the Science and Technology Innovation Ability Construction Projects of Beijing Academy of Agriculture and Forestry Science (KJCX20140202, KJCX20170203 and QNJJ201507), the National Natural Science Foundation for Young Scholars of China (31600418), the National Key R&D Program of China (2016YFE0112400) and the European Union's Horizon 2020 Research and Innovation Program (679303). C.M., E.B. and J.L. were supported by the Institute Strategic Program Understanding and Exploiting Plant and Microbial Secondary Metabolism (BB/J004596/1) from the BBSRC to JIC and by BBSRC Newton Funding to C.M.
We thank Dr. Yan Zhu from the Institute of Botany, Chinese Academy of Sciences, for experimental technical assistances. We also thank Professor George Lomonossoff from John Innes Centre for providing HyperTrans vector system. This work was supported by the Science and Technology Innovation Ability Construction Projects of Beijing Academy of Agriculture and Forestry Science (KJCX20140202, KJCX20170203 and QNJJ201507), the National Natural Science Foundation for Young Scholars of China (31600418), the National Key R&D Program of China (2016YFE0112400) and the European Union's Horizon 2020 Research and Innovation Program (679303). C.M., E.B. and J.L. were supported by the Institute Strategic Program Understanding and Exploiting Plant and Microbial Secondary Metabolism (BB/J004596/1) from the BBSRC to JIC and by BBSRC Newton Funding to C.M.
Wang, H. , Zhang, H. , Yang, Y. , Li, M. , Zhang, Y. , Liu, J. , Dong, J. , Li, J. , Butelli, E. , Xue, Z. , Wang, A. , Wang, G. , Martin, C. and Jin, W. (2020) The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnol J., 10.1111/pbi.13282
Contributor Information
Cathie Martin, Email: cathie.martin@jic.ac.uk.
Wanmei Jin, Email: jwm0809@163.com.
Data availability statement
The FaMYB1, FaMYB10 sequence data for cDNA are available in the GenBank database under the accession numbers MG456857, MG456858, MG456859 and MG456860. RNA‐seq data are available in the CNGB Nucleotide Sequence Archive (https://db.cngb.org/cnsa/) of China National GenBank (CNGB) database with accession number CNP0000406.
References
- Aharoni, A. , De Vos, C.H.R. , Wein, M. , Sun, Z.K. , Greco, R. , Kroon, A. , Mol, J.N.M. et al (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Allan, A.C. , Hellens, R.P. and Laing, W.A. (2008) MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. [DOI] [PubMed] [Google Scholar]
- An, X.H. , Tian, Y. , Chen, K.Q. , Liu, X.J. , Liu, D.D. , Xie, X.B. , Cheng, C.G. et al (2015) MdMYB9 and MdMYB11 are Involved in the regulation of the JA‐induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. [DOI] [PubMed] [Google Scholar]
- Ayub, R.A. , Bosetto, L. , Galvão, C.W. , Etto, R.M. , Inaba, J. and Lopes, P.Z. (2016) Abscisic acid involvement on expression of related gene and phytochemicals during ripening in strawberry fruit Fragaria × ananassa cv. Camino Real. Scientia Horticulturae 203, 178–184. [Google Scholar]
- Ban, Y. , Honda, C. , Hatsuyama, Y. , Igarashi, M. , Bessho, H. and Moriguchi, T. (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. [DOI] [PubMed] [Google Scholar]
- Benvenuti, S. , Pellati, F. , Melegari, M. and Bertelli, D. (2004) Polyphenols, anthocyanins, ascorbic acid, and radical scavenging activity of Rubus, Ribes, and Aronia . J. Food Sci. 69, C164–C169. [Google Scholar]
- van den Berg, R.A. , Hoefsloot, H.C.J. , Westerhuis, J.A. , Smilde, A.K. and van der Werf, M.J. (2006) Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genom. 7, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli, E. , Licciardello, C. , Zhang, Y. , Liu, J. , Mackay, S. , Bailey, P. , Reforgiato‐Recupero, G. et al (2012) Retrotransposons control fruit‐specific, cold‐dependent accumulation of anthocyanins in blood oranges. Plant Cell, 24, 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli, E. , Garcia‐Lor, A. , Licciardello, C. , Las Casas, G. , Hill, L. , Reforgiato Recupero, G. , Keremane, M. et al (2017) Changes in anthocyanin production during domestication of Citrus . Plant Physiol. 173, 2225–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, C.K. , Albregts, E.E. , Howard, C.M. and Brecht, J.K. (1997) 'Sweet Charlie' strawberry. HortScience, 32, 1132–1133. [Google Scholar]
- Cheng, G.W. and Breen, P.J. (1991) Activity of phenylalanine ammonia‐lyase (Pal) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Hortic. Sci. 116, 865–869. [Google Scholar]
- Chenna, R. , Sugawara, H. , Koike, T. , Lopez, R. , Gibson, T.J. , Higgins, D.G. and Thompson, J.D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amelia, V. , Aversano, R. , Batelli, G. , Caruso, I. , Castellano Moreno, M. , Castro‐Sanz, A.B. , Chiaiese, P. et al (2014) High AN1 variability and interaction with basic helix‐loop‐helix co‐factors related to anthocyanin biosynthesis in potato leaves. Plant J. 80, 527–540. [DOI] [PubMed] [Google Scholar]
- Dubos, C. , Stracke, R. , Grotewold, E. , Weisshaar, B. , Martin, C. and Lepiniec, L. (2010) MYB transcription factors in Arabidopsis . Trends Plant Sci. 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Espley, R.V. , Hellens, R.P. , Putterill, J. , Stevenson, D.E. , Kutty‐Amma, S. and Allan, A.C. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10 . Plant J. 49, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2014) FAOstat crops data. Food and Agriculture Organization of the United Nations. http://www.fao.org/faostat/en/#data/QC. [Google Scholar]
- Finn, C.E. , Retamales, J.B. , Lobos, G.A. and Hancock, J.F. (2013) The Chilean Strawberry (Fragaria chiloensis): Over 1000 Years of Domestication. HortScience, 48, 418–421. [Google Scholar]
- Fischer, T.C. , Mirbeth, B. , Rentsch, J. , Sutter, C. , Ring, L. , Flachowsky, H. , Habegger, R. et al (2014) Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria × ananassa). New Phytol. 201, 440–451. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. , Schiestl, R.H. , Willems, A.R. and Woods, R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS‐DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Gonzalez, A. , Zhao, M. , Leavitt, J.M. and Lloyd, A.M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Griesser, M. , Hoffmann, T. , Bellido, M.L. , Rosati, C. , Fink, B. , Kurtzer, R. , Aharoni, A. et al (2008) Redirection of flavonoid biosynthesis through the down‐regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant J. 146, 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.F. , Sjulin, T.M. and Lobos, G.A. (2008) Strawberries In Temperate Fruit Crop Breeding: Germplasm to Genomics (Hancock J.F. ed.), Dordrecht: Springer, Netherlands. [Google Scholar]
- Hawkins, C. , Caruana, J. , Schiksnis, E. and Liu, Z. (2016) Genome‐scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep‐Uk, 6, 29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola, L. (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. [DOI] [PubMed] [Google Scholar]
- Jin, W.M. , Zhang, Q. , Liu, S.Z. , Wei, Q.P. , Cheng, Z.M. , Xue, X.H. and Yang, T.Z. (2012) Genetic diversity of 41 apple rootstocks based on simple sequence repeat markers. J. Am. Soc. Hortic. Sci. 137, 51–56. [Google Scholar]
- Jin, W.M. , Wang, H. , Li, M.F. , Wang, J. , Yang, Y. , Zhang, X.M. , Yan, G.H. et al (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 14, 2120–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadomura‐Ishikawa, Y. , Miyawaki, K. , Takahashi, A. , Masuda, T. and Noji, S. (2015) Light and abscisic acid independently regulated FaMYB10 in Fragaria × ananassa fruit. Planta, 241, 953–965. [DOI] [PubMed] [Google Scholar]
- Kayesh, E. , Shangguan, L.F. , Korir, N.K. , Sun, X. , Bilkish, N. , Zhang, Y.P. , Han, J. et al (2013) Fruit skin color and the role of anthocyanin. Acta Physiol. Plant. 35, 2879–2890. [Google Scholar]
- Kobayashi, S. , Ishimaru, M. , Hiraoka, K. and Honda, C. (2002) Myb‐related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta, 215, 924–933. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S. , Goto‐Yamamoto, N. and Hirochika, H. (2004) Retrotransposon‐induced mutations in grape skin color. Science, 304, 982. [DOI] [PubMed] [Google Scholar]
- Lin‐Wang, K. , Bolitho, K. , Grafton, K. , Kortstee, A. , Karunairetnam, S. , McGhie, T. , Espley, R. et al (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin‐Wang, K. , McGhie, T.K. , Wang, M. , Liu, Y.H. , Warren, B. , Storey, R. , Espley, R.V. et al (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 5, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Martin, C. , Ellis, N. and Rook, F. (2010) Do transcription factors play special roles in adaptive variation? Plant Physiol. 154, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , Mccue, K. , Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Moyano, E. , Portero‐Robles, I. , Medina‐Escobar, N. , Valpuesta, V. , Muñoz‐Blanco, J. and Luis Caballero, J. (1998) A fruit‐specific putative dihydroflavonol 4‐reductase gene is differentially expressed in strawberry during the ripening process. Plant Physiol. 117, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N. , Debeaujon, I. , Jond, C. , Pelletier, G. , Caboche, M. and Lepiniec, L. (2000) The TT8 gene encodes a basic helix‐loop‐helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell, 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, J.J. , Oberbauer, S.F. and Clark, D.B. (2004) Whole tree xylem sap flow responses to multiple environmental variables in a wet tropical forest. Plant Cell Environ. 27, 551–567. [Google Scholar]
- Paolocci, F. , Robbins, M.P. , Passeri, V. , Hauck, B. , Morris, P. , Rubini, A. , Arcioni, S. et al (2011) The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J. Exp. Bot. 62, 1189–1200. [DOI] [PubMed] [Google Scholar]
- Pillet, J. , Yu, H.W. , Chambers, A.H. , Whitaker, V.M. and Folta, K.M. (2015) Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 66, 4455–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, J. , Sun, L. , Wu, J. , Zhao, S. , Wang, C. , Wang, Y. , Ji, K. et al (2010) Cloning and expression analysis of cDNAs for ABA 8′‐hydroxylase during sweet cherry fruit maturation and under stress conditions. J. Plant Physiol. 167, 1486–1493. [DOI] [PubMed] [Google Scholar]
- Sainsbury, F. , Thuenemann, E.C. and Lomonossoff, G.P. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. [DOI] [PubMed] [Google Scholar]
- Schaart, J.G. , Dubos, C. , De La Fuente, I.R. , van Houwelingen, A.M.M.L. , de Vos, R.C.H. , Jonker, H.H. , Xu, W.J. et al (2013) Identification and characterization of MYB‐bHLH‐WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197, 454–467. [DOI] [PubMed] [Google Scholar]
- Schwinn, K.E. , Boase, M.R. , Bradley, J.M. , Lewis, D.H. , Deroles, S.C. , Martin, C.R. and Davies, K.M. (2014) MYB and bHLH transcription factor transgenes increase anthocyanin pigmentation in petunia and lisianthus plants, and the petunia phenotypes are strongly enhanced under field conditions. Front. Plant Sci. 5, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. , Zhao, K. , Liu, L. , Zhang, K. , Yuan, H. , Liao, X. , Wang, Q. et al (2014) A role for PacMYBA in ABA‐regulated anthocyanin biosynthesis in red‐colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 55, 862–880. [DOI] [PubMed] [Google Scholar]
- Shulaev, V. , Sargent, D.J. , Crowhurst, R.N. , Mockler, T.C. , Folkerts, O. , Delcher, A.L. , Jaiswal, P. et al (2011) The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos, A.M. , Jaffe, F.W. , Jacob, S.R. , Bogs, J. , Robinson, S.P. and Walker, A.R. (2006) Light‐induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias, A. , Lin‐Wang, K. , Stevenson, D. , Cooney, J. , Hellens, R. , Allan, A. , Hoover, E. et al (2011) Apple skin patterning is associated with differential expression of MYB10 . BMC Plant Biol. 11, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veberic, R. , Slatnar, A. , Bizjak, J. , Stampar, F. and Mikulic‐Petkovsek, M. (2015) Anthocyanin composition of different wild and cultivated berry species. Lwt‐Food Sci. Technol. 60, 509–517. [Google Scholar]
- Walker, A.R. , Davison, P.A. , Bolognesi‐Winfield, A.C. , James, C.M. , Srinivasan, N. , Blundell, T.L. , Esch, J.J. et al (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell, 11, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wang, X. , Zhao, P. , Zheng, H. , Ren, Y. , Gao, F. and Ouyang, Z. (2012) Transpiration rates of urban trees, Aesculus chinensis . J. Environ. Sci. 24, 1278–1287. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lei J. and Wang G. Eds. (2016) Advances in Strawberry Research V. Beijing: China Agriculture Press. [Google Scholar]
- Zhang, Y. , Butelli, E. and Martin, C. (2014) Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. [DOI] [PubMed] [Google Scholar]
- Zimmermann, I.M. , Heim, M.A. , Weisshaar, B. and Uhrig, J.F. (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B‐like BHLH proteins. Plant J. 40, 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Fruit developmental stages of white and red octoploid strawberry varieties.
Figure S2 Chromatographs of anthocyanins, flavonols, and proanthocyanins.
Figure S3 Transcript levels of structural genes and regulatory genes not included in Figure 3 in the white and red strawberry varieties.
Figure S4 Transcript levels of regulatory genes in the white and red strawberry varieties.
Figure S5 The constructs and results from yeast two‐hybrid assay of FaMYB10‐1 or FaMYB10‐2 with two regulators (FaWD40 and PavbHLH) for the formation of MBW ternary complex.
Figure S6 Genomic sequence analysis for FaMYB1 alleles in red and white strawberry varieties.
Figure S7 PCA of key flavonoid components against flavonoid biosynthetic structural genes and transcription‐related genes.
Table S1 Analysis of flavonoid contents.
Table S2 The expression of structural genes, FaANS, FaUFGT, and FaDFR in flavonoid biosynthesis in red and white strawberry varieties.
Table S3 The expression of structural genes, FaLAR, FaANR, and FaFLS in flavonoid biosynthesis in red and white strawberry varieties.
Table S4 The expression of regulatory genes, FaMYB1, FaMYB10, and FaWD40 of flavonoid biosynthesis in red and white strawberry varieties.
Table S5 The expression of regulatory genes, FaMYB9, and FaMYB11 of flavonoid biosynthesis in red and white strawberry varieties.
Table S6 The sequences of the oligonucleotide primers used in this work.
Table S7 Information about the MYB protein sequences used for the construction of the neighbour joining tree.
Data Availability Statement
The FaMYB1, FaMYB10 sequence data for cDNA are available in the GenBank database under the accession numbers MG456857, MG456858, MG456859 and MG456860. RNA‐seq data are available in the CNGB Nucleotide Sequence Archive (https://db.cngb.org/cnsa/) of China National GenBank (CNGB) database with accession number CNP0000406.


