Abstract
Friedreich ataxia (FA) is a cardioneurodegenerative disease caused by deficient frataxin expression. This mitochondrial protein has been related to iron homeostasis, energy metabolism, and oxidative stress. Previously, we set up a cardiac cellular model of FA based on neonatal rat cardiac myocytes (NRVM) and lentivirus-mediated frataxin RNA interference. These frataxin-deficient NRVMs presented lipid droplet accumulation, mitochondrial swelling and signs of oxidative stress. Therefore, we decided to explore the presence of protein thiol modifications in this model. With this purpose, reduced glutathione (GSH) levels were measured and the presence of glutathionylated proteins was analyzed. We observed decreased GSH content and increased presence of glutahionylated actin in frataxin-deficient NRVMs. Moreover, the presence of oxidized cysteine residues was investigated using the thiol-reactive fluorescent probe iodoacetamide-Bodipy and 2D-gel electrophoresis. With this approach, we identified two proteins with altered redox status in frataxin-deficient NRVMs: electron transfer flavoprotein-ubiquinone oxidoreductase and dihydrolipoyl dehydrogenase (DLDH). As DLDH is involved in protein-bound lipoic acid redox cycling, we analyzed the redox state of this cofactor and we observed that lipoic acid from pyruvate dehydrogenase was more oxidized in frataxin-deficient cells. Also, by targeted proteomics, we observed a decreased content on the PDH A1 subunit from pyruvate dehydrogenase. Finally, we analyzed the consequences of supplementing frataxin-deficient NRVMs with the PDH cofactors thiamine and lipoic acid, the PDH activator dichloroacetate and the antioxidants N-acetyl cysteine and Tiron. Both dichloroacetate and Tiron were able to partially prevent lipid droplet accumulation in these cells. Overall, these results indicate that frataxin-deficient NRVMs present an altered thiol-redox state which could contribute to the cardiac pathology.
1. Introduction
Friedreich ataxia (FA) is a rare disease caused by decreased expression of frataxin, a nuclear encoded protein with mitochondrial localization. Frataxin is highly expressed in the heart and the nervous system, and consequently these tissues are among the most affected in FA. The first symptoms of the disease are usually neurologic [1], while the primary cause of death in most FA patients is related to cardiomyopathy [2].
Many evidences indicate that at the cellular level frataxin deficiency alters cellular iron homeostasis and causes oxidative stress. Two potential mechanisms have been proposed to explain these phenotypes, the iron-sulfur hypothesis and the iron toxicity hypothesis (reviewed in Refs. [[3], [4], [5], [6]]). The first one suggests that frataxin contributes to iron-sulfur cluster biogenesis, which are cofactors required for the activity of many proteins and also for iron sensing. Therefore, its deficiency would activate an iron deficiency signal that would promote iron uptake. In support of this hypothesis, it has been shown that frataxin localizes to mitochondria where it may interact and regulate the activity of cysteine desulfurase [7]. This enzyme is required for providing sulfide for the biosynthesis of several cofactors, such as iron-sulfur clusters, molybdenum cofactor and lipoic acid [8]. Although this hypothesis is well supported by in vitro data, its major caveat is the absence of iron-sulfur clusters deficiency in many models of frataxin deficiency. The iron toxicity hypothesis is based on the known ability of frataxin to bind iron [9]. According to this hypothesis frataxin deficiency would lead to increased free iron which would catalyze ROS generation.
Oxidative stress in frataxin-deficient cells may also be enhanced by an impaired activation of the Nrf2 signaling pathway [10]. This phenomenon has been observed in several models of the disease and it has been related to actin remodelling, which in turn could be caused by glutahionylation of this protein. This modification is caused by the formation of disulphide bonds between protein thiol groups and glutathione. Glutathione is a cysteine-containing tripeptide present in millimolar concentrations in the cell and involved in a wide range of processes which include: i) serving as electron donor to glutathione peroxidases or peroxiredoxins for scavenging reactive oxygen species [11]; ii) protecting proteins form irreversible thiol modifications or regulating its activity by glutahionylation (which can be reversed by glutaredoxins) [12]; iii) protecting from heavy metals toxicity through its chelation and export [13]; iv) participating in xenobiotic detoxification in collaboration with glutathione S-transfesares [14]. It has also been postulated that glutathione could act as a ligand of the labile iron pool and contribute to cellular iron homestasis [15]. Free glutahione can be found in its reduced form (GSH) or as an oxidized disulphide (GSSG) [16]. In this regard, in several models of FA it has been observed a decreased GSH/GSSG ratio and an increased presence of glutathionylated actin [17,18]. Both events (actin glutathionylation and decreased GSH/GSSG ratio) are indicative of an imbalance in the cellular thiol redox status of frataxin-deficient cells.
Despite cardiomyopathy is the leading cause of death in FA, the specific effects of frataxin deficiency in the heart are poorly understood and very few cellular models have been used to analyse the mechanisms linking frataxin deficiency to impaired cardiac function. In this regard, we have previously investigated the consequences of frataxin deficiency in neonatal cardiac rat myocytes (NRVM) and we have observed that in these cells frataxin deficiency leads to marked mitochondrial disarrangements and impaired lipid metabolism [19]. In this model we have also observed increased protein carbonyl content and increased sensitivity to tert-butyl hydroperoxide, which are clear markers of oxidative stress. However, we had not previously investigated the consequences of frataxin-deficiency on the thiol redox state of NRVM. In the present work we have addressed this issue and we have found that frataxin-deficient NRVM present an altered thiol redox state which impacts on actin, and on the pyruvate dehydrogenase (PDH) complex. We have also observed that an activator of PDH (dichloroacetate, DCA) and an antioxidant (Tiron) can ameliorate the phenotype of frataxin-deficient NRVM.
2. Results
2.1. Analysis of GSH status in frataxin-deficient NRVM
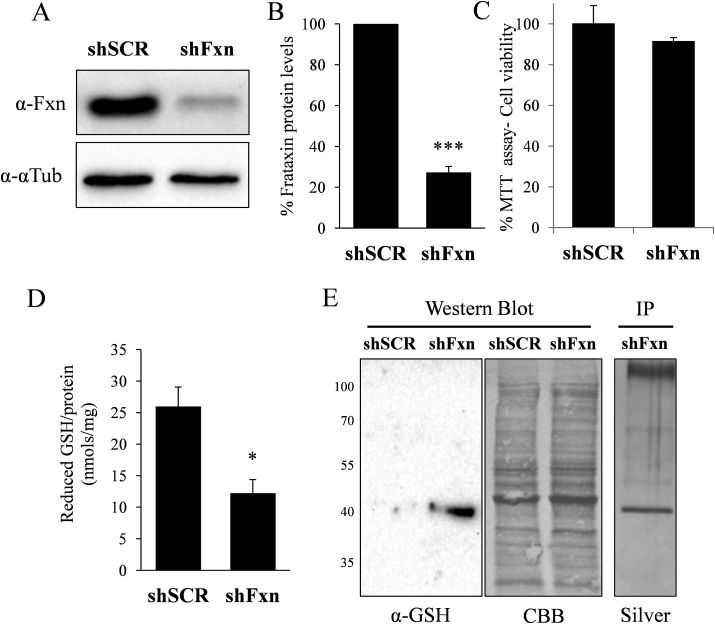
First, we were interested in analyzing the glutathione redox state in the cardiac model of the disease previously developed by our group. This model is based on lentivirus-mediated transduction of shRNAs interfering frataxin into NRVM [19]. As indicated in Fig. 1A, this strategy leads to a marked decrease in frataxin content. In order to analyse the glutathione status in this model, GSH content was measured by HPLC as described under experimental procedures. As shown in Fig. 1D, we observed a marked decrease in GSH in frataxin-deficient NRVM. We next analyzed the potential impact of such decreased GSH content in protein glutathionylation. For this purpose, protein extracts were prepared in non-reducing conditions and analyzed by Western blot using anti-GSH antibodies. Results, shown in Fig. 1E, indicate the appearance of a marked band (40kDa apparent molecular weight) in frataxin-deficient cultures. In order to identify this protein, lysates from frataxin-deficient NRVM were immunoprecipitated with anti-GSH antibodies and the immunoprecipitated proteins separated by SDS-PAGE. Oriole staining revealed the presence of a marked band of 40kDa apparent molecular weight. This band was identified as actin by mass spectrometry (Table 1). Interestingly, actin was previously identified as a glutathionylated protein in FA derived patients fibroblasts [20,21]. Therefore our results confirm that actin is a target of glutathionylation in FA.
Fig. 1.
Frataxin deficiency in rat cardiomyocytes causes GSH depletion and actin glutahionylation. Rat cardiomyocytes were transduced with lentivirus vectors containing shFxn or scrambled shRNA. A, cell lysates were probed with anti-frataxin antibody by Western blot analysis. In each gel lane, 30 ug of protein from whole cell lysates were loaded. B, histogram represents the quantification of the relative expression of frataxin measured by Western blot. Data are represented as means ± SEM from 3 independent cultures. In each experiment, frataxin signal in scrambled control cells were used as a 100% reference value. C, MTT cell viability assay was performed in SCR and shFxn NRVMs. No significant loss in cell viability was observed in fratxin-deficient cultures. D, reduced glutathione content in scrambled and frataxin deficient cells. E, lysates from the indicated NRVM cultures were analyzed by Western blot with anti-GSH antibody under non-reducing conditions. Protein load was verified by post-western Coomassie blue (CBB) staining. Lysates from shFxn cells where also immunoprecipated with anti-GSH antibodies and the immunoprecipitated proteins (IP) separated by SDS-PAGE and silver-stained.
Table 1.
Proteins identified by mass spectrometry. The indicated proteins correspond to the immunoprecipitated band in Fig. 1D (IP) and to the spots from the 2d thiol-proteome analysis shown in Fig. 3. All of them were identified by peptide mass fingerprinting (PMF) using MASCOT as search engine. Table shows the MOWSE (MASCOT) score, number of matched and searched peptides, and sequence coverage of the proteins providing the highest score. MOWSE scores greater than 58 are significant (p<0.01).
| Spot/Band | Uniprot | Protein Name | MOWSE Score | Matched/Searched | Sequence Coverage (%) |
|---|---|---|---|---|---|
| IP | P62738 | Actin, aortic smooth muscle | 89 | 7/14 | 18 |
| P68035 | Actin, alpha cardiac muscle 1 | ||||
| 1 | Q6P6R2 | Dihydrolipoyl dehydrogenase, mitochondrial | 170 | 15/31 | 38 |
| 2 | Q6UPE1 | Electron transfer flavoprotein-ubiquinone oxidoreductase | 374 | 29/34 | 44 |
2.2. Analysis of protein thiol redox state in frataxin-deficient cells
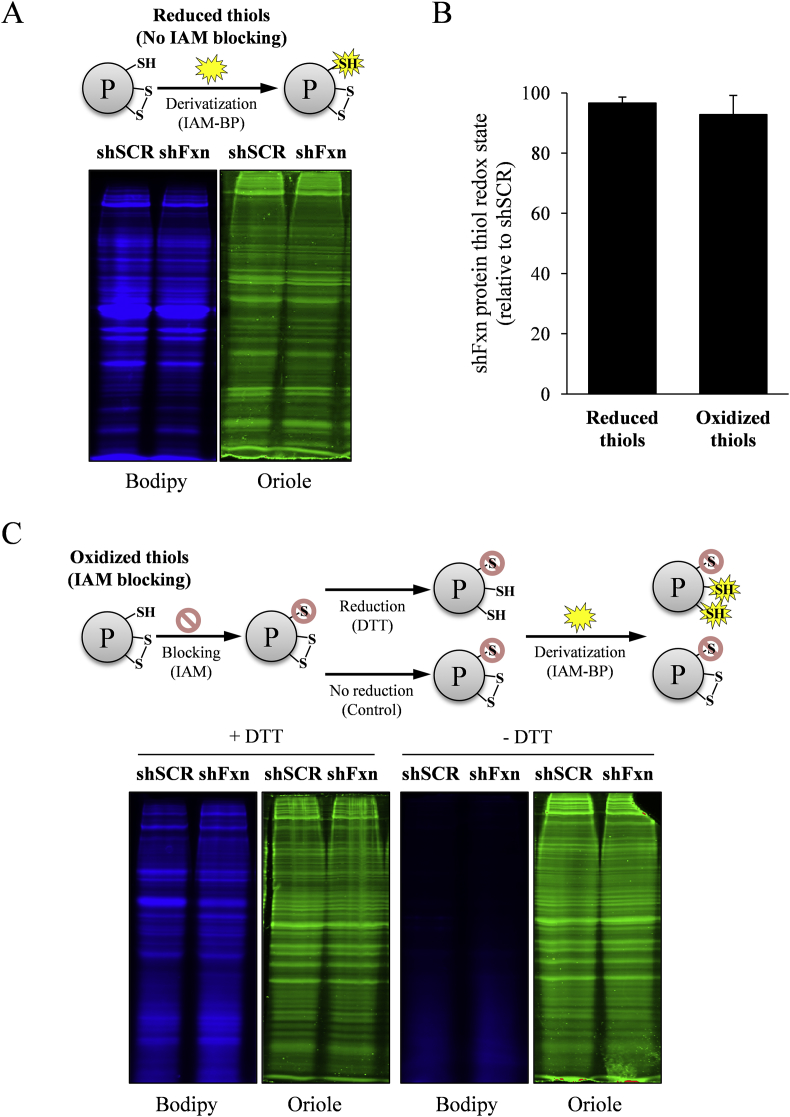
We were also interested in analyzing the presence of alterations in the thiolproteome in frataxin-deficient NRVM. With this aim we developed a strategy to analyse global redox state of thiols in these cells. To analyse the presence of reduced thiols, cells were lysed in the presence of the fluorescent alkylating reagent iodoacetamide-Bodipy (IAM-Bodipy). Protein extracts were subsequently separated by SDS-PAGE and Bodipy fluorescence was detected and quantified in a Chemidoc XRS instrument. Gels were also stained with Oriole total protein stain as a control for protein load. To detect oxidized thiols, cells were lysed in the presence of iodocetamide, a reagent that blocks reduced thiols. Oxidized thiols were then reduced with DTT and further alkylated with IAM-Bodipy. Finally, these extracts were separated by SDS-PAGE and Bodipy fluorescence was detected and quantified. In this case Bodipy fluorescence indicated the presence of oxidized thiols, which had not been blocked with iodocetamide and had been reduced by DTT. As previously, gels were also stained with Oriole. Results from both analyses are shown in Fig. 2. Panel A indicates the workflow for detection of reduced protein thiols and the results obtained. No significant differences were observed in total bodipy fluorescence between SCR and shFXN cells. Panel C shows the workflow for detection of oxidized protein thiols and the results obtained. Samples non-treated with DTT did not show Bodipy staining, confirming the efficiency of the iodoacetamide (IAM) blocking step. In the DTT reduced samples, no significant differences were obtained in total Bodipy fluorescence between both conditions analyzed (quantitative results shown in Fig. 2B). These results indicate that frataxin loss does not lead to a marked alteration of the thiol proteome.
Fig. 2.
Detection of reduced and oxidized protein thiols in lysates from shSCR and shFxn cardiomyocytes. A, detection of reduced protein thiols. Reduced protein thiols where derivatized with Bodipy-IAM, separated by SDS-PAGE and fluorescence signal from Bodipy (blue) acquired in a Chemidoc MP system. Gels where also stained with Oriole protein stain (green). C, detection of oxidized protein thiols. Reduced protein thiols where blocked by IAM and oxidized ones reduced by DTT and derivatized with Bodipy-IAM. B, relative protein thiol redox state of shFxn calculated from the Bodipy signal in A and C. Data are mean +/- SEM from three independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.3. Identification of oxidized proteins by two-dimensional electrophoresis
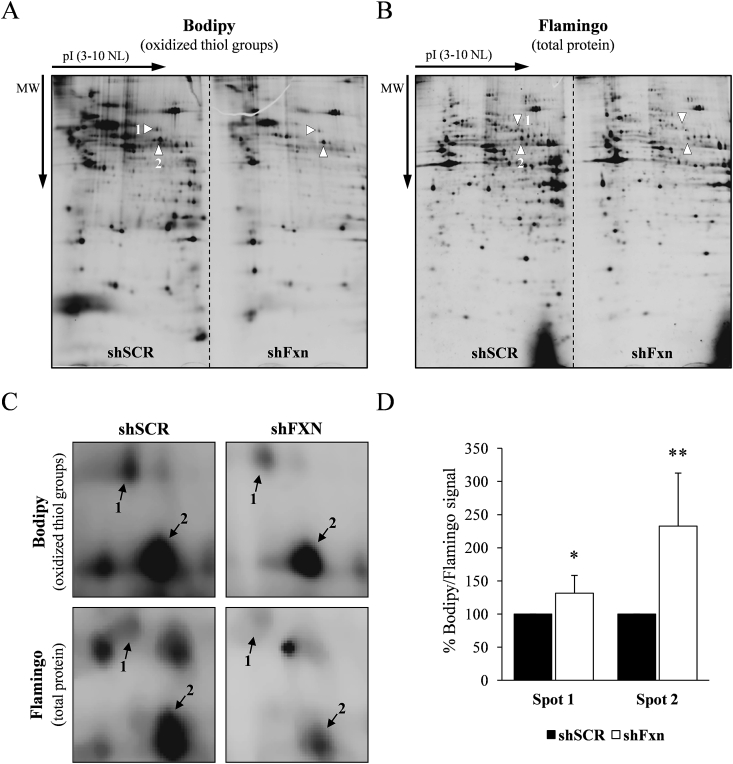
Although in the previous section we did not observe a marked alteration in the redox state of the protein thiol groups, we hypothesized that some specific proteins could be altered. Thus, we subjected these cells to a more detailed analysis using two-dimensional electrophoresis. Whole cell extracts from SCR and frataxin-deficient cardiomyocytes were prepared for detection of oxidized thiols as described previously and precipitated with chloroform/methanol before analysis by 2D electrophoresis. Isoelectric focusing was performed in 11 cm IPG strips, (pH range 3–10, non-linear), while SDS-PAGE was performed in 11% 22 cm x 20,5 cm gels. Two IPG strips were loaded side by side on the same SDS-PAGE gel, one corresponding to SCR samples and the other corresponding to shFxn samples. This approach had been used successfully previously for the detection of carbonylated proteins using fluorescent hydrazides [22]. We analyzed three samples for each condition (each one from an independent experiment). After capturing the Bodipy fluorescence (Fig. 3A), gels where stained with Flamingo total protein stain (Fig. 3B) and images were analyzed with PDQuest software. This analysis allowed us to obtain a Bodipy/flamingo (B/P) ratio of those protein spots which were repetitively detected among the different replicates in both the Bodipy and flamingo channel. We consider this B/P ratio as an estimation of the relative degree of oxidation of the thiol groups of these proteins. Only two spots showed significant differences in the B/P ratio between both experimental conditions. On average, spot 1 presented a 20% higher B/P ratio in frataxin-deficient cultures, while spot 2 presented a 210% increase. A detailed image of the gel region including these spots is shown in Fig. 3C. In order to identify both spots, a preparative 22 x 20,5 cm 2D-gel was prepared in which 200 μg of protein were separated and, after obtaining the Bodipy signal, stained with Oriole total protein stain (Supplemental Fig. 1). Spots were subjected to in gel tryptic digestion and analyzed by mass spectrometry. Spot 1 was identified as ETF dehydrogenase and spot 2 as dihydrolipoyl dehydrogenase (DLDH) (Table 1). We decided to focus on DLDH due to its higher degree of oxidation and also to its potential relationship with frataxin function, as biosynthesis of its substrate (protein-bound lipoic acid) requires an iron-sulfur enzyme and cysteine desulfurase [23].
Fig. 3.
Two-dimensional thiol proteome analysis of NRVM. SCR and frataxin-deficient cultures were processed and analyzed as described in material and methods. Representative analytical gels containing SCR and shFxn samples are shown in A (Bodipy signal, which corresponds to oxidized thiol groups) and B (Flamingo signal, which corresponds to total protein stain). The indicated spots are those showing significant differences in their ratio Bodipy/flamingo; C, amplified image of the gel region containing spots 1 and 2; D, histograms show the relative Bodipy/protein ratio from spots 1 and 2. Data are mean +/- SEM from three independent experiments.
2.4. Analysis of the content of DLDH and protein-bound lipoic acid
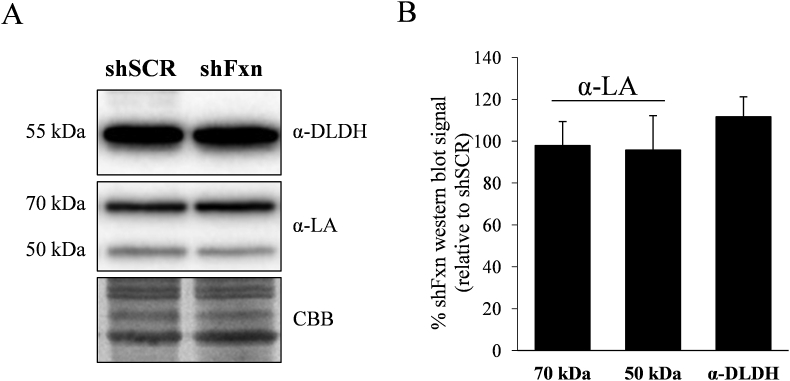
DLDH functions as the E3 component of three enzymatic complexes, pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH) and branched chain α-ketoacid dehydrogenase (BCKDH). These enzymatic complexes are also composed of the dehydrogenase component E1 and the dihydrolipoamide transferase component E2, which contains a covalently-bound lipoic acid. DLDH catalyzes the conversion of this E2-bound dihydrolipoic acid and NAD + into lipoic acid and NADH [24]. We decided to measure the content of DLDH and E2-bound lipoic acid by Western blot. Using antibodies raised against this cofactor, two bands could be appreciated at 70 and 50 kDa apparent molecular weight (Fig. 4A). The 70 kDa band (which is more intense) corresponds to lipoic acid bound to the E2 component from PDH (Dihydrolipoyllysine acetyltransferase, DLAT), while three lipoic acid containing proteins migrate at 50 kDa: the E2 component from KGDH (Dihydrolipoyllysine succinyltransferase, DLST), the E2 component from the branched chain alphaketodehydrogenase complex and the PDH-binding component X (a structural subunit from the PDH complex) [24]. Among these three proteins, DLST is the more abundant in heart according to the PaxDb database. Therefore, we assume that most signal from the 50 kDa band is contributed by DLST. As indicated in Fig. 4B, no significant changes where observed in DLAT-bound lipoic acid nor in the 50 kDa lipoic acid band in shFxn NRVM. This result indicates that protein-bound lipoic acid biosynthesis is not compromised in frataxin-deficient NRVM. Also, no changes were observed in the DLDH protein abundance measured by Western blot.
Fig. 4.
Analysis of lipoic acid containing dehydrogenase complexes by Western blot. A, Western blot analysis of shSCR and shFxn NRVMs using DLDH or lipoic acid (LA) antibodies. B, histogram represents the quantification of the relative Western blot signal (ratio shFxn to shScr). Data are represented as means ± SEM from 3 independent cultures.
2.5. PDH-bound lipoic acid is more oxidized in frataxin-deficient cells
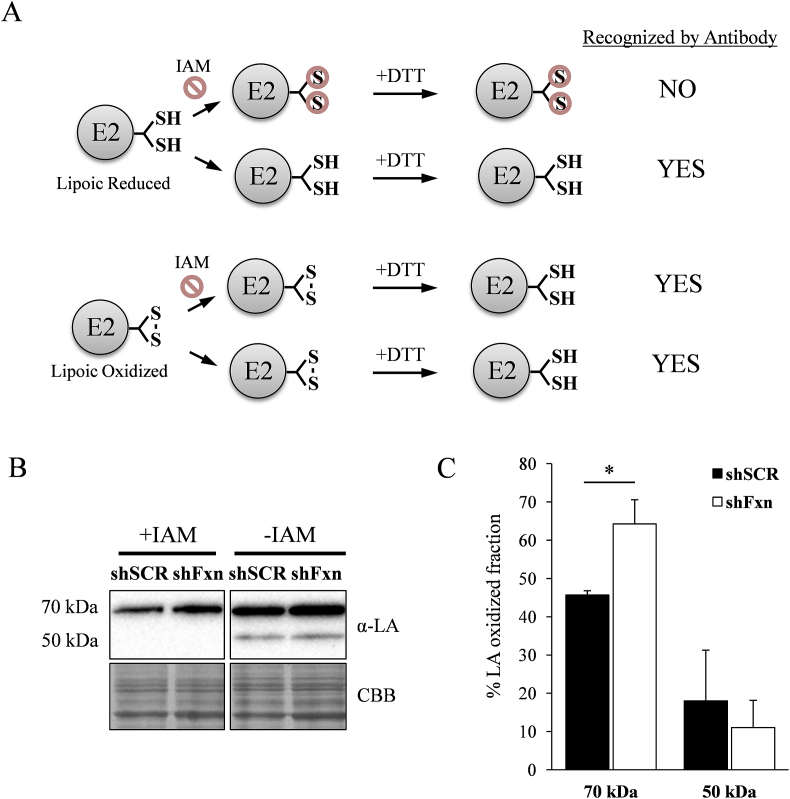
E2-bound lipoic acid contains two thiol groups that are oxidized by DLDH during each catalytic cycle. As we identified DLDH as an oxidized protein, we hypothesized that frataxin-deficient cells could present lipoic acid in a more oxidized form. Therefore we used an approach to analyse the redox state of protein-bound lipoic acid. This approach (summarized in Fig. 5A) was based on the analysis by western-blot of lipoic acid in lysates treated or not with the thiol reactive reagent IAM. Reduced lipoic acid would react with IAM and not be detected by antibodies, while the oxidized one will not react with IAM and, after thiol reduction, could be detected by antibodies. We then calculated the signal ratio between IAM treated and not treated conditions to obtain an estimate of the fraction of protein-bound lipoic acid oxidized under each experimental condition. Results can be observed in figure 5B and 5C. In SCR NRVM the fraction of oxidized DLAT-bound lipoic acid was 45%, while 50kDa lipoic acid was mostly reduced. In shFxn NRVM, the oxidized DLAT fraction increased significantly, while no changes where observed in the 50kDa fraction. These results indicate that DLAT-bound lipoic acid was more oxidized in frataxin-deficient cells.
Fig. 5.
Redox state from protein-bound lipoic acid. NRVMs where lysed in the presence or not of iodoacetamide (IAM), reduced by DTT and analyzed by Western blot using lipoic acid (LA) antibodies. A, schematic representation of the experimental procedure used. Reduced lipoic acid will not be detected in IAM-treated samples. B, representative lipoic acid western from samples treated or not with IAM. C, relative lipoic acid oxidized fraction was calculated from the ratio between the Western blot signal from samples treated or not treated with IAM. Data are represented as means ± SEM from 4 independent experiments. * indicates significant (p<0.05) differences between shSCR and shFxn conditions.
2.6. Analysis of components of pyruvate dehydrogenase complex
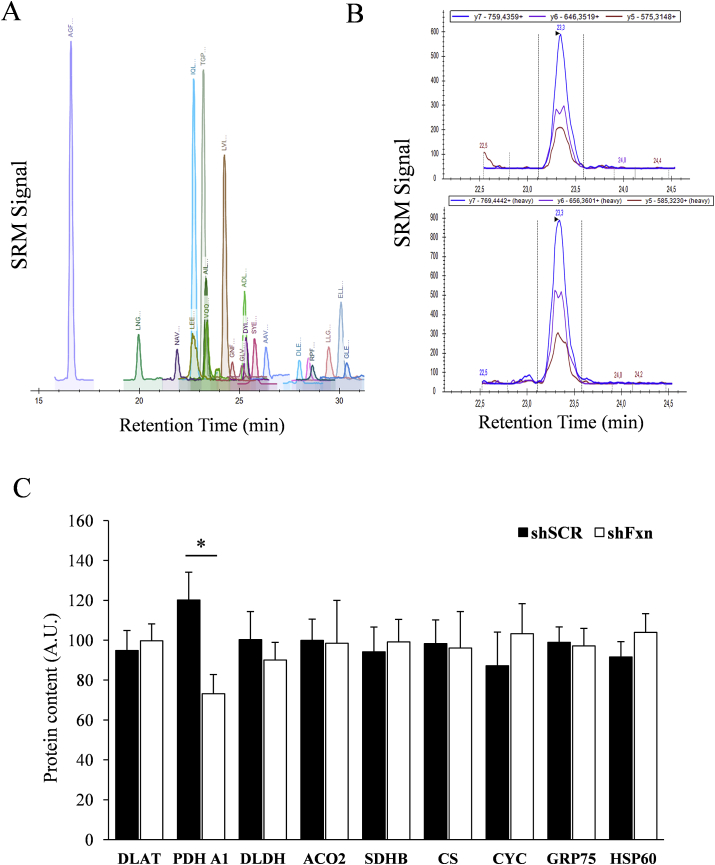
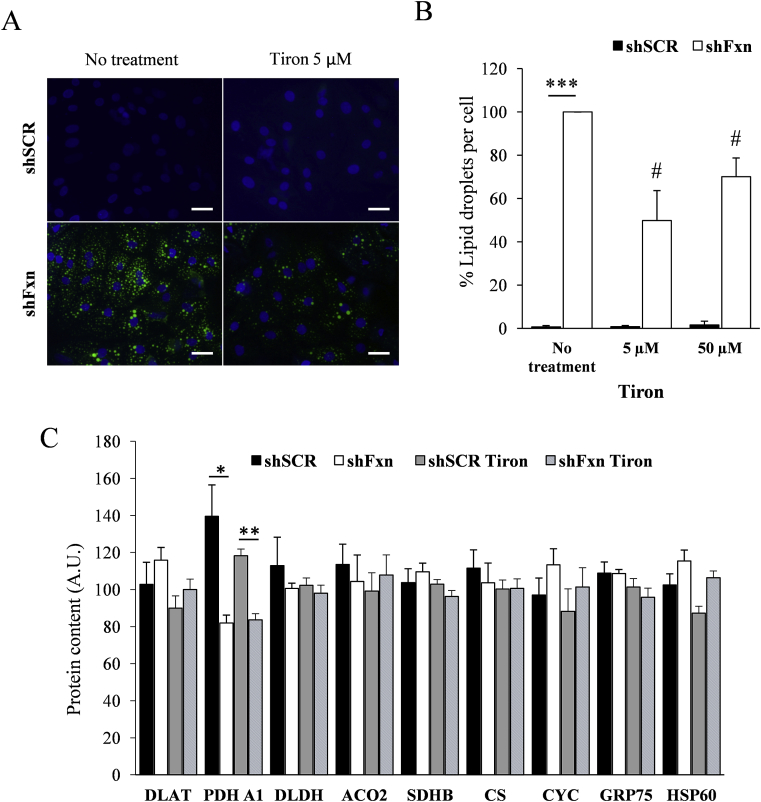
The observation that DLAT-bound lipoic acid was more oxidized in frataxin-deficient cells, prompted us to investigate the content of this protein. We used an SRM-based targeted proteomics approach in which proteotypic peptides from each protein were detected in a LC-triple quadrupole mass spectrometer. For most proteins, this approach allows a higher reproducibility than Western blot and also allows a better comparison between samples and different proteins. We also included in the analysis DLDH, the alpha subunit from the E1 component from PDH (PDH A1), two iron-sulfur containg proteins (Succinate dehydrogenase 2 and Aconitase 2) as well as four additional mitochondrial proteins, HSP60, GRP75, Citrate Synthase (CS) and Cytochrome C (CYC), as a control of mitochondrial content. Actin, tropomiosin, and myosin regulatory light chain were also analyzed and used as housekeeping proteins to normalize protein load among samples. The results from these analyses can be found in Fig. 6. We did not found alterations in the content of DLAT or iron-sulfur containing enzymes. Interestingly, we found that frataxin-deficient NRVM present a significant decrease in the content of PDH A1. This observation confirms that PDH is altered in frataxin-deficient NRVMs.
Fig. 6.
Protein content analysis by targeted proteomics. A, a representative chromatogram showing the SRM traces of the different peptides analyzed in the targeted proteomics experiment. B, representative signals from the three transitions detected for the peptide AILAELTGR, corresponding to PDH A1. Top panel corresponds to the detection of the “light” version of the peptide obtained from the tryptic digestion of whole cells lysates, while low panel corresponds to the detection of the standard, a C13N15 labeled “heavy” version of the peptide. C, Histograms show the relative content (in arbitrary units) of the measured proteins by targeted proteomics. Data are represented as means ± SEM from 4 independent experiments. * indicates significant (p<0.05) differences between shSCR and shFxn conditions.
2.7. Tiron and dichloroacetate supplementation partially prevent lipid droplet accumulation caused by frataxin deficiency
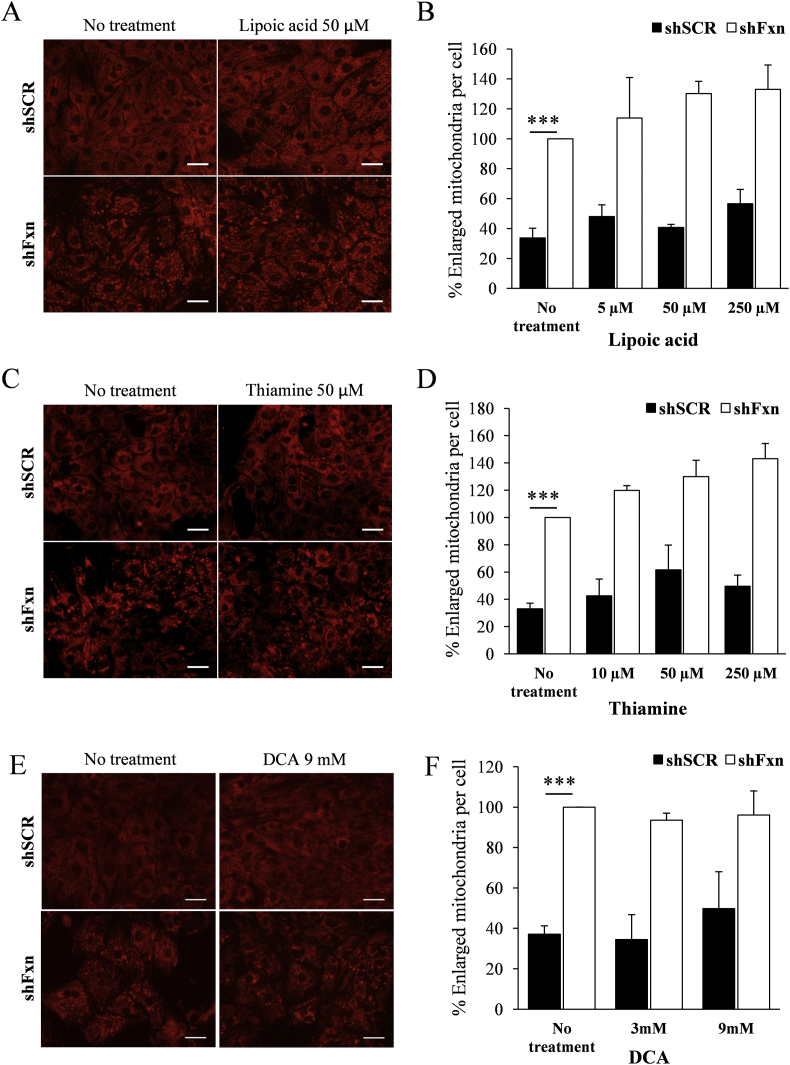
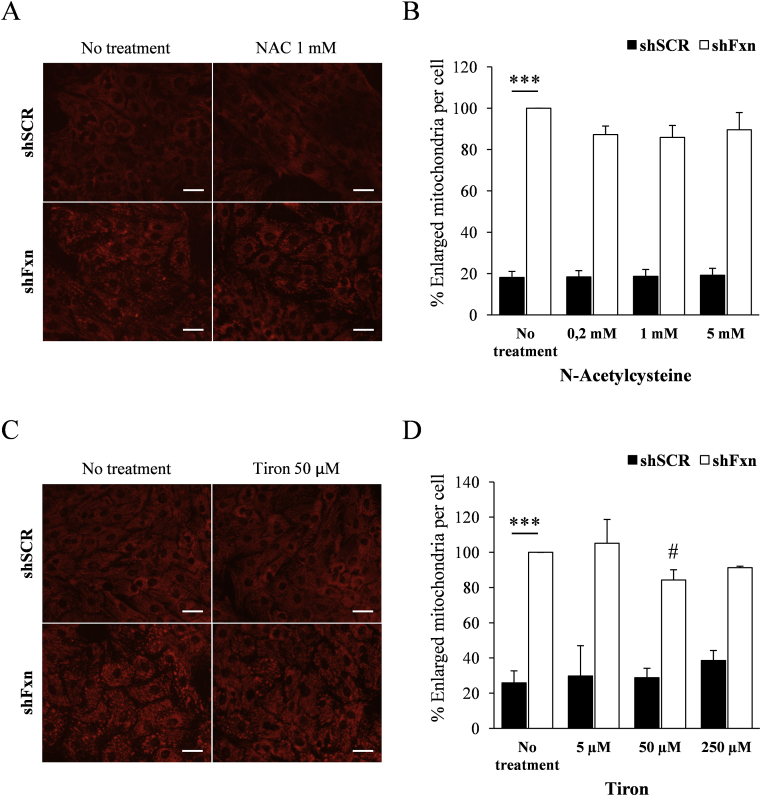
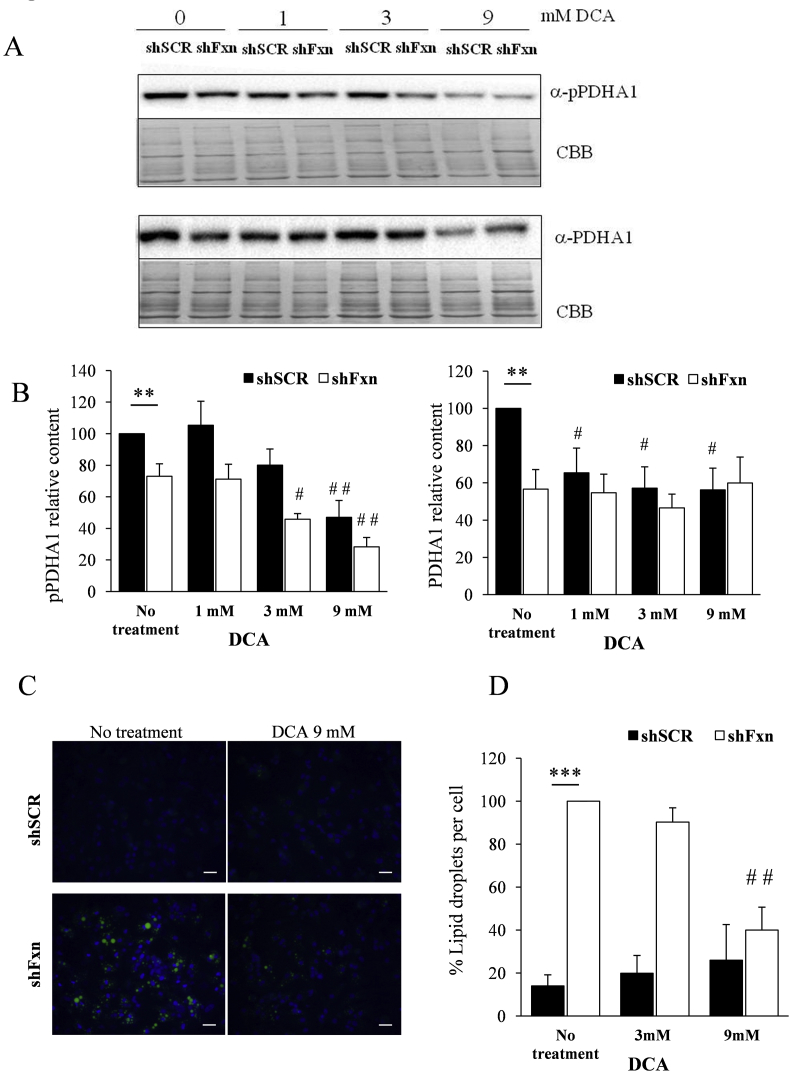
We decided to analyse the potential benefit for frataxin-deficient NRVMs of several compounds impacting on the pathways investigated in the previous sections. The compounds used where expected to work as antioxidants and/or PDH activators, and they were lipoic acid, thiamine (precursor of the PDH cofactor thiamine pyrophosphate), dichloroacetate (DCA, a PDH activator), the glutathione precursor N-acetyl cysteine (NAC), and the superoxide-scavenger mitochondria-targeted Tiron (4,5-dihydroxy-1,3-benzenedisulfonic). First, we tested the effect of supplementation with these compounds on the increased presence of enlarged mitochondria, which can be revealed (and quantified) by TMRM staining and fluorescence microscopy. This phenomenon is caused by mitochondrial permeability transition pore opening and is one of the most marked mitochondrial phenotypes observed in frataxin-deficient NRVMs [25]. Thus, SCR and shFxn NRVM cultures were subjected to different concentrations of the indicated compounds and, after 7 days, stained with TMRM. Images (shown in Fig. 7, Fig. 8) were acquired in an Olympus inverted microscope and the presence of enlarged mitochondria analyzed with ImageJ software. As previously reported, marked differences were observed between SCR and shFxn NRVM in the number of enlarged mitochondria per cell. This phenotype was only slightly reversed by Tiron at 50 μM concentration, while no significant differences in the amount of enlarged mitochondria were observed upon treatment with any of the other compounds tested. Actually, treatment with lipoic acid or thiamine worsened the phenotype. We next decided to analyse the effect of Tiron and DCA on lipid droplet accumulation, which is another consequence of frataxin deficiency in NRVM. The antioxidant tiron and the PDH activator DCA were selected because they presented better results than NAC and thiamine respectively in the enlarged-mitochondria assay. Therefore, presence of lipid droplets was analyzed by Bodipy staining in SCR and shFxn cultures supplemented with these compounds. As shown in Fig. 9, Tiron at 5 and 50 μM concentrations was able to partially rescue the accumulation of lipid droplets, confirming its protective effect on frataxin-deficient NRVM. We also analyzed by targeted proteomics whether Tiron could reverse the content of PDHA1 in frataxin-deficient cells. As shown in Fig. 9, PDHA1 deficiency was not reversed by tiron, indicating that its beneficial effects are not caused by a reversion of PDHA1 deficiency. Regarding DCA, this compound activates PDH by inhibiting pyruvate dehydrogenase kinase, a protein that phosphorylates PDHA1 in order to modulate its activity in response to metabolic requirements [26]. Therefore, we first analyzed by Western blot the effects of DCA supplementation on the content and phosphorylation state of PDHA1. As shown in Fig. 10, these analyses confirmed the decreased content of PDHA1 in frataxin-deficient NRVM (previously observed by targeted proteomics). We could also observe that phosphorylation state of this protein was not markedly altered in frataxin-deficient cells, as signal from the phosphorylated form decreases similarly than total protein signal. The phosphorylated form was significantly decreased in both conditions (SCR and shFxn) in 9 mM DCA-supplemented cultures. This treatment did not change the content of PDHA1 in shFxn, although it decreased its content in SCR. This observation indicated that in shFxn NRVMs, PDHA1 would be activated by DCA at 9 mM concentration. Next, we analyzed the presence of lipid droplets in these cultures by Bodipy staining. As shown in Fig. 10, DCA at 9 mM concentration prevented the accumulation of lipid droplets in frataxin-deficient NRVMs. In summary, these results indicate that neither the antioxidants used nor the activators of PDH are able to prevent markedly the altered mitochondrial phenotype (mitochondria enlargement assay) but may partially reverse the consequences of frataxin deficiency (as indicated by the lipid droplet assay).
Fig. 7.
Supplementation with compounds impacting on PDH. NRVM cultures were supplemented with the indicated concentrations of lipoic acid, thiamine and DCA, and, after 7 days of frataxin depletion, mitochondria were stained with TMRM (A, C and E). The amount of enlarged mitochondria was measured with ImageJ (B, D and F). Graphs represent the relative number of enlarged mitochondria relative to non-treated FXN deficient cells values. Scale bar = 30μm.
Fig. 8.
Supplementation with antioxidants. NRVM cultures were supplemented with the indicated concentrations of N-Acetylcysteine (NAC) and Tiron and, after 7 days of frataxin depletion, mitochondria were stained with TMRM (A and C). The amount of enlarged mitochondria was measured with ImageJ (B and D). Graphs represent the relative number of enlarged mitochondria relative to non-treated FXN deficient cells values. Only 50 μM Tiron treatment shows a significant decrease in the number of enlarged mitochondria in FXN interfered cells. Scale bar = 30 μm. # indicates statistic significance to non-treated shFxn (p<0,05).
Fig. 9.
Tiron supplementation prevents lipid droplet accumulation. NRVM cultures were supplemented with the indicated concentrations of Tiron and, after 7 days of frataxin depletion, lipid droplets were stained with Bodipy 493/503 (A). The number of lipid droplets per cell was measured with ImageJ (B). Graphs represent the relative number of lipid droplets relative to non-treated FXN deficient cells values. C, Histograms show the relative content (in arbitrary units) of the measured proteins by targeted proteomics. Data are represented as means ± SEM from 3 independent experiments. *(p<0.05) and ** (p<0.01) indicate significant differences between shSCR and shFxn conditions. # indicates statistic significance to non-treated shFxn (p<0,05).
Fig. 10.
DCA supplementation decreases PDHA1 phosphorylation and prevents lipid droplet accumulation. NRVM cultures were supplemented with the indicated concentrations of DCA. (A and B), Western blot analysis indicates that DCA at 9 mM inhibits PDHA1 phosphorylation in shFxn without affecting protein levels. (C) Lipid droplets were stained with Bodipy 493/503. (D) The number of lipid droplets per cell was measured with ImageJ. Graphs represent the relative number of lipid droplets relative to non-treated FXN deficient cells values. Data are represented as means ± SEM from 3 independent experiments. *(p<0.05) and ** (p<0.01) indicate significant differences between shSCR and shFxn conditions. # (p<0,05) and ## (p<0,01) indicate statistic significance to non-treated shSCR or shFxn.
3. Discussion
In this work we have performed an analysis of the protein thiol redox state of frataxin-deficient NRVM. The analysis has provided several conclusions. First, frataxin-deficient cardiomyocytes present an altered thiol redox state, revealed by the decreased GSH content and the increased presence of glutahionylated actin. Second, such altered thiol redox state may impact specific proteins (such actin), but there is not a general alteration in the thiol proteome of frataxin-deficient NRVM. Third, alterations can be found in the PDH complex, both in the redox state of DLAT-bound lipoic acid and on the content of its PDH A1 subunit.
Altered GSH/GSSG ratio and increased presence of glutathionylated actin have been previously reported in patient-derived cells (lymphoblasts and fibroblasts) and frataxin-deficient yeast [20,21]. Our results indicate that these alterations can also be observed in a cardiac cell model and therefore confirm that frataxin-deficient cells are experiencing an oxidative stress condition which may cause oxidation of sensitive proteins such actin. However, it is not clear which can be the consequence of such glutahionylation on actin function. This modification is known to impact on actin polymerization [27,28] and could also be related to the impaired NRF2 activation which has been observed in some models of FA [10]. In this regard, the inability to induce NRF2-dependent antioxidant enzymes to counteract oxidative stress could lead frataxin-deficient NRVM to a vicious cycle which would increase its sensitivity towards oxidative stress. Besides actin and DLDH, we have not been able to identify other proteins with an altered thiol redox state. Therefore, we can conclude that there is not a general affectation in the protein thiol redox state. Nevertheless, there could be additional oxidized proteins that have not been detected in this study. This may be due to limitations of the approaches used, which may not be able to detect low abundant proteins or (in the case of 2D electrophoresis) those with transmembrane domains. This limitation could explain why we have only identified two proteins in this analysis. Therefore, the altered thiol redox state observed in this work could have an impact on several cellular processes. In this regard, various types of thiol redox modifications, including S-oxidation (sulfenylation and sulfinylation), S-glutathionylation, and S-nitrosylation have been described to regulate a number of mitochondrial processes ranging from OXPHOS to mitochondrial permeability transition pore opening [29].
The identification of DLDH as an oxidized protein led us to the study of lipoic acid containing proteins. We detected a decreased content of PDHA1 and a higher proportion of oxidized DLAT-bound lipoic acid. Both events could be related, as PDHA1 provides electrons to lipoic acid which are then transferred to DLDH. Therefore, a decreased content of PDHA1 could result in accumulation of oxidized forms of lipoic acid and DLDH. Otherwise, DLAT-bound lipoic acid could be oxidized by oxidized glutathione, as it has been previously described to occur with DLST-bound lipoic acid [30]. This modification, upon rapid disulfide exchange, may produce oxidized lipoic acid and reduced glutathione. In any case, the mechanism causing PDHA1 deficiency in frataxin-deficient NRVMs is not known. A proteomic analysis of frataxin-deficient yeast revealed that mitochondrial Mg-binding proteins (as PDHA1) are prone to experience oxidative modifications due to iron-catalyzed oxidation [31]. Therefore, this mechanism could apply for PDHA1. Nevertheless the PDHA1 yeast orthologue was not identified in that study as an oxidized protein. Also, in the present study Tiron supplementation has not prevented PDHA1 deficiency. This compound has both antioxidant and iron chelating properties and is able to reach mitochondria [32]. Thus, it should be able to prevent the iron-catalyzed oxidation phenomenon observed in yeast. Understanding the mechanism causing PDHA1 deficiency will require further investigation.
Nevertheless, some early investigations reported a dysfunction of lipoamide dehydrogenase [32,33] and of the pyruvate and oxoglutarate dehydrogenase complexes in FA patients [34], although these observations were not confirmed by other studies [35]. Recently, PDH deficiency was found in a proteomic analysis of peripheral blood mononuclear cells from FA patients [36]. Previous studies had also observed low levels of thiamine in the cerebrospinal fluid of patients with FA [37]. Therefore, several evidences point to a potential unbalance in the thiamine and PDH axes in this disease. In this regard, more recently, thiamine administration was tested in a group of FA patients. These patients showed an improvement in neurological symptoms. A subgroup of patients which were subjected to echocardiogram analysis showed also a reduction in interventricular septum thickness. The authors of this study concluded that further studies where mandatory to confirm restorative and neuroprotective action of thiamine in FA [38]. In a similar way, we also conclude that further studies are required to understand the causes of PDHA1 deficiency in frataxin-deficient NRVM and also its potential contribution to pathology. In this regard, the DCA experiments performed in this work indicate that activation of PDH could have a beneficial impact on the metabolic alterations caused by frataxin-deficiency (as DCA prevented the accumulation of lipid droplets). Nevertheless, lipid droplet accumulation is not directly caused by PDHA1 deficiency, as this phenotype is partially prevented by Tiron whithout correcting PDHA1 deficiency. Probably, lipid dropled accumulation is related to several impaired mitochondrial functions caused by frataxin deficiency, and any treatment which partially ameliorates these functions may have a beneficial effect on lipid droplet accumulation. In this regard, we have previously shown that lipid droplet accumulation can be corrected by inhibitors of the mitochondrial permeability transition pore [39].
Besides its relation with the PDH complex, analysis of lipoic acid had an additional interest, because synthesis of this cofactor is dependent on iron-sulfur enzymes. Therefore, it is commonly used as a marker of the presence of these cofactors in the cell. Nevertheless, no differences in the total amount of lipoic acid could be observed between SCR and frataxin-deficient cells. We have also not observed a decreased content of the iron-sulfur enzymes Aconitase 2 and SDH2, a result which is consistent with previous observations reporting normal aconitase and complex II activities in this same model [19]. Overall, we can confirm that iron-sulfur cluster biogenesis is not seriously compromised in frataxin-deficient NRVMs. It is worth mentioning that iron-sulfur cluster deficiency is not always observed in FA models. In this regard, studies in yeast have shown that loss of iron-sulfur enzymes in frataxin-deficient cells would be mostly caused by metabolic remodelling and/or by oxidative stress [40,41].
In summary, the proteomic approach deployed has revealed that frataxin deficiency in NRVM causes an imbalance on the cellular thiol redox state and in PDH enzymatic complex. These results encourage the investigation of these pathways in animal models of the disease. In this regard, FA is a progressive disease, and the cardiac symptoms do not always appear in the early stages. This progressive and chronic condition is not reproduced by our cell culture model. Therefore, it would be interesting to follow the evolution of the alterations found in this work in animal models and correlate their presence with the appearance of cardiac symptoms. It is worth commenting that mutations in PDHA1 cause a variety of clinical signs, which affect metabolic and neurological processes. These symptoms depend on the residual PDH activity presented by the mutated form and, in females, on Chromosome X inactivation (PDHA1 is an X‐chromosomal gene) [42]. Therefore, PDHA1 deficiency could contribute to FA pathology. We have also found that Tiron and DCA have a positive impact on frataxin-deficient cells. This observation should also encourage the testing in animal models of these compounds, which could be in the future included in the FA treatment pipeline.
4. Experimental procedures
4.1. Isolation and culture of NRVM
The investigation with experimental animals conforms to the National Guidelines for the regulation of the use of experimental laboratory animals from the Generalitat de Catalunya and the Government of Spain (article 33.a 214/1997) and was evaluated and approved by the Experimental Animal Ethic Committee of the University of Lleida (CEEA). P3–P4 Sprague–Dawley rat neonates were sacrificed, following the above Guide approved by the CEEA, by decapitation and (NRVM) were obtained from the hearts as described previously [19]. Briefly, ventricles from newborn rats were minced and cells were isolated by 3 subsequent digestion steps with 150U/ml of type-2 collagenase (Worthington, USA) stirring at 37 °C. Non-cardiomyocyte cells were separated from the cardiomyocytes by differential preplating. Cardiomyocytes were seeded on 0.2% gelatin-coated culture dishes (Falcon, Becton & Dickinson, USA) at 7.5·104 cells/cm2, giving a confluent monolayer of spontaneously contracting cells after 24 h. To inhibit the proliferation of cardiac fibroblasts, cells were treated with 10 μg/ml mitomycin C (SIGMA) for 4 h after the seeding time. The culture medium used was DMEM:M199 3:1, 5mM glucose, with 8% horse serum and 4% fetal bovine serum, glutaMAX and HEPES (all from GIBCO). Cardiomyocyte purity was checked by immunofluorescence with a specific cardiac α-actinin antibody (SIGMA) and was found to be higher than 90% after 24 h in vitro. To analyse lipid droplet accumulation fatty acid-albumin solutions were used for FFA supplementation of the culture media. These solutions where prepared as described [19]. These solutions where added to the culture medium to give a final concentration of 30 μM each of linoleic, oleic and stearic acid, and 60 μM palmitic acid.
4.2. Production of lentiviral particles
The shRNA lentiviral plasmids (pLKO.1-puro) for human/mouse/rat frataxin were purchased from SIGMA. The RefSeq used were NM_008044, which correspond to mouse frataxin. The clone TRCN0000197534 (named shFxn in this study) is designed for mouse frataxin interference in a region 100% identical to rat Frataxin. A non-targeted scrambled sequence (the vector SHC002, named shSCR in this study) served as a control. Lentiviral particles were produced by cotransfection of HEK293T cells using the plasmids pMD2.G and psPAX2 (Addgene) and polyethylene as described previously [43]. After 3 days, lentiviruses were collected from the culture medium, centrifuged for 5 min at 1000 rpm, filtered through a 0.45 μm filter (Millipore), concentrated by centrifugation through 100,000 MWCO Vivaspin 20 columns (Sartorius) and stored at −80 °C. Lentiviruses were tittered using the Quicktiter Lentivirus ELISA kit (Cell Biolabs), which detects the amount of p24 protein. For NRVM transduction, 5.5 ng of p24 per 1,000 cells were added to the media 4 h after plating, and 20 h later, culture media was replaced with fresh media. This protocol provided a transduction efficiency greater than 70%.
4.3. Western blot
Cells were first rinsed in ice cold PBS and lysed in 2% SDS, 125mM Tris, pH 6,8 containing a protease inhibitor cocktail (Roche). Protein concentration was determined by the Qubit assay (Invitrogen). After SDS-polyacrylamide gel electrophoresis, proteins were transferred to Immobilon-P or Nitrocellulose membranes. The membranes were probed with the following primary antibodies: Frataxin (sc-25820), DLDH (Sigma SAB2100589), GSH (abCam, ab19534), lipoic acid (Calbiochem, 437695), phosphoPDHA1 (abcam, ab177461), PDHA1 (abcam, ab110334). Detection was performed using peroxidase conjugated secondary antibodies. Image acquisition was performed in a ChemiDoc MP system from Bio-Rad. When required, data was analyzed by Image Lab software (Bio-Rad).
4.4. MTT cell viability assay
For the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay, cells were grown in 96-well plates and 7 days after lentivirus transduction, cells were rinsed and labeled with 0.2 mM MTT for 4 h at 37 °C and 5% CO2 in HBSS with calcium, magnesium and 5 mM glucose. The precipitates were dissolved in DMSO and absorbance was read at 540 nm in a Biotek Power Wave XS.
4.5. Analysis of protein-bound lipoic acid redox state
Cells were first rinsed in ice cold PBS and lysed in 50 μl of lysis buffer (10 mM Tris, 1mM EDTA, 1% SDS) and divided in two parts. To one part, 1/3 volume of loading buffer was added. The other part was incubated for 30 min at 37 °C in the presence of 50 mM IAM before the addition of 1/3 volume of loading buffer. Proteins (10 μg) where separated by SDS-PAGE and after electrophoresis transferred to Immobilon-P membranes. The membranes were probed with antibodies against lipoic acid and Western blot images where acquired in a ChemiDoc MP system from Bio-Rad. Data was analyzed by Image Lab software (Bio-Rad)
4.6. Fluorescent thiol labelling
For labelling reduced protein thiols, cells (750,000) where washed twice with cold PBS and lysed with 200 μl of lysis buffer (10 mM Tris, 1mM EDTA, 1% SDS) plus 0,4 mM IAM-Bodipy (BODIPY™ FL Iodoacetamide, Thermo Fisher ref. D6003) and incubated for 30 min at 37 °C. The IAM moiety of this compound will react with reduced thiol groups, while the Bodipy moiety will provide fluorescence labelling without affecting protein charge. Reaction was stopped by addition of 1/3 volume of loading buffer (20% sacarose, 0,1% sodium azide, 15% β-mercaptoethanol, bromphenol blue). Proteins (10 μg) where separated by SDS-PAGE and after electrophoresis, reduced protein thiols were visualized by epifluorescence in a ChemiDoc MP imager (BioRad) using specific channel settings for Bodipy (Blue LED, 530/28 filter). Once this image was captured, total protein was stained with OrioleTM total protein fluorescent gel stain (Bio-Rad) and the resulting images were superimposed on those obtained from the Bodipy channel. Oriole is a stain for visualization and quantitation of proteins separated by SDS-PAGE. Its fluorescence was recorded in a ChemiDoc MP imager using UV-excitation. Oriole signal does not interfere with Bodipy fluorescence. Images were analyzed with Image Lab software (Bio-Rad). For labelling oxidized protein thiols, cells (750,000) where washed twice with cold PBS and lysed with 200 μl of lysis buffer plus 50 mM IAM. After 30 min of incubation at 37 °C, 1,600 μl of cold acetone where added to the lysate and samples where incubated in ice for 30 min. Proteins where precipitated by centrifugation, washed once with cold acetone, and resuspended in lysis buffer plus 2.5 mM DTT. After 30 min of incubation at 37 °C, proteins where precipitated again with acetone as before. Pellets where resuspended with 10 mM Tris, 1 mM EDTA, 2% SDS, 0.4 mM IAM-Bodipy and incubated for 30 min at 37 °C. For monodimensional SDS-PAGE analysis, reaction was stopped by addition of 1/3 volume of loading buffer and analyzed by SDS-PAGE as indicated for reduced thiols. For two-dimensional electrophoresis, reaction was stopped by addition of 5 mM DTT and analyzed as indicated below.
4.7. GSH quantitation and immunoprecipitation
GSH was analyzed by HPLC chromatography using 5,5′-Ditiobis-(2-nitrobenzoic acid) (DTNB) as derivitizing reagent [44]. Briefly, cells (approximately 200,000) were washed twice with cold PBS and suspended in 75 μl of 0,3% sulfosalicylic acid. This cell suspension was lysed in a Mini-Beadbeater-16 (BioSpec) in the presence of 0.5 mm glass beads. Lysates were centrifuged and 10 μl of the supernatant where mixed with 10 μl PBS and 80 μl DTNB 1 mM (diluted in PBS). This mixture was incubated for 10 min at room temperature and after centrifugation, supernatant analyzed in an Agilent 1260 Infinity HPLC system equiped with an UV–visible detector operating at 330 nm. A Phenomenex Kromasil 5μm 100A (4,6 × 25 mm) column operating at 1 ml/min in isocratic mode was used. Solvent was ammonium acetate 20 mM 10% methanol pH5. For immunoprecipitation, 750,000 cells were lysed with 200 μl of IP solution composed of 50 mM Tris pH 7, 1 mM EDTA, 150 mM NaCl, 0,1% Triton, 0,1% deoxycholate and protease inhibitors. Samples were centrifuged for 3 min at 3000g (4 °C) and supernatant was mixed with a suspension of Protein G DynabeadsTM (ThermoFisher) and anti-GSH antibody (#ab19534). This suspension was incubated for 30 min at 4 °C minutes under agitation. Not-bound proteins were separated from Dynabeads using a magnetic support. After three additional washes with 200 μl of IP solution to minimize non-specific binding to Dynabeads, these were suspended in elution buffer (10 μM DTT in IP solution) and incubated for 10 min to allow reduction of the mixed disufides between glutathione and protein thiols. Eluted protein were separated by SDS-PAGE.
4.8. Two-dimensional electrophoresis
Protein lysates derivatized with IAM-Bodipy where precipitated with methanol/Chloroform. The resulting protein pellet was resuspended in 100 μl 2-D Protein Extraction Buffer -V (PEB-V, GE Healthcare). After 2 h incubation at 25 °C, the solution was centrifuged and the supernatant conserved. Protein concentration was quantified using the Bio-Rad Protein Assay. For analytical gels, a supernatant volume containing 80 μg protein was diluted in 200 μl PEB-V containing 50 mM DTT and 2 μl Bio-lytes. This solution was used to rehydrate 11 cm IPG strips (3–10 NL, Bio-Rad) for 12 h and then focused using a Protean IPG Cell (Bio-Rad) for a total 24000Vhr. After isoelectric focusing, strips were incubated for 15 min in 6 M urea, 2% SDS, 20% glycerol, 375 mM Tris pH 8,8 and 130 mM DTT. Second dimension was performed in an EttanDalt 6-electrophoresis system (GE Healthcare). Two parallel 11-cm strips (shScr and shFxn samples) were loaded side by side on the same 10% polyacrylamide gel (crosslinker ratio 37,5:1). After electrophoresis, oxidized protein thiols were visualized by epifluorescence in a VersaDoc MP4000 imager (BioRad) using specific channel settings for Bodipy. Once this image was captured, the total protein was stained with Flamingo fluorescent gel stain (Bio-Rad) and the resulting images were superimposed on those obtained from the Bodipy channel. Images were analyzed with PDQuest Advanced software (Bio-Rad). Flamingo protein stain was used in 2D-analysis as it provides higher sensitivity than Oriole protein stain.
4.9. Protein identification by peptide mass fingerprinting
Protein spots were excised from gels and subjected to in gel digestion as described [22]. Tryptic peptides were recovered and spotted onto a MALDI plate in the presence of α-cyano-4-hydroxy-cynamic acid. Mass spectra were acquired in a Bruker Ultraflex MALDI-TOF/TOF operating in a reflector positive mode. Proteins were identified by peptide mass fingerprint, searching against UniProt database using MASCOT. Search criteria assumed that peptides were monoisotopic, oxidized at methionine residues (variable modification), and carbamidomethylated at cysteine residues (fixed modification). One maximum missed cleavage was allowed with a maximum peptide mass tolerance of 50 ppm.
4.10. Protein quantitation by targeted proteomics
Cells were resuspended in 125 mM Tris-HCl, 2% SDS plus 1 mM EDTA and a mixture of protease inhibitors, boiled for 3 min and centrifuged (12000 rpm 10 min). Proteins were quantified using the BCA assay (Thermo Scientific) and 30 μg were precipitated with cold acetone (9 vol) and resuspended in 1% sodium deoxycholate, 25 mM ammonium bicarbonate. Then, proteins were subjected to reduction by 12 mM DTT and alquilation by 40 mM IAM. Mass spectrometry grade trypsin (SOLu-Trypsin, Sigma) was added to a final enzyme:substrate ratio of 1:50. After overnight digestion at 37 °C, formic acid was added to precipitate sodium deoxycholate. The resulting peptide mix was purified and enriched using C18 columns (Pierce C-18 Spin Columns, Thermo Scientific). Eluted fraction from the C18 column was evaporated using a Concentrator Plus (Eppendorf) and peptides were resuspended in 3% acetonitrile plus 0,1% formic acid containing a heavy peptide standards mixture. Heavy peptides were obtained from JPT (SpikeTidesTM_L). All peptide samples were analyzed on a triple quadrupole spectrometer (Agilent 6420) equipped with an electrospray ion source. Chromatographic separations of peptides were performed on an Agilent 1200 LC system using a Supelco Bioshell A160 Peptide C18 column (1 mm × 15 cm). Peptides (up to 15 μg of protein digest) were separated with a linear gradient of acetonitrile/water, containing 0.1% formic acid, at a flow rate of 75 μl/min. A gradient from 3 to 60% acetonitrile in 45 min was used. The mass spectrometer was operated in multiple reaction monitoring mode. Transitions were obtained from SRM atlas and imported into Skyline software [45], which was also used to analyse results. In the SRM assays validation phase, the transitions obtained from SRM atlas were analyzed in several runs. Each MRM acquisition was performed with Q1 and Q3 operated at unit resolution. Once validated and optimized, the SRM assays were used to quantify all the analyzed peptides using scheduled SRM mode in a single run (retention time window, 120 s; cycle time, 1 s). For calculating protein content, the light to heavy ratio of each peptide in each sample was first divided by the mean value of that peptide between all samples. Then, this value was normalized to the average value of the housekeeping proteins in each sample. The peptides and transitions analyzed can be found in supplemental table 1.
4.11. Subcellular labelling and image analysis
To analyse mitochondrial swelling, NRVM cells were loaded with 30 nM TMRM for 1 h as described [19]. The number of “balloon-like” swollen mitochondria was assessed using the “analyse particles” tool from ImageJ software. Images were background-subtracted and the following criteria were used to detect particles: size higher than 2 μm2 and circularity 0.5–1. For labelling of lipid droplets, cells were washed with PBS and loaded with 5 μM Bodipy 493/503 (Invitrogen) for 10 min at 37 °C. Nuclei staining with 0.05 μg/ml Hoechst 33258 was done as necessary. An Olympus IX71 microscope equipped with epifluorescence optics and a DP70 CCD camera was used to obtain images with a 40x lens.
4.12. Statistical analysis
All experiments were performed in at least 3 completely independent cardiomyocyte preparations. Values were expressed as mean ± SEM (error bars). The data obtained from the independent experiments were used for statistical analysis. Data obtained was compared with control conditions using Student t-test. The p-values lower than 0.05(*) or 0.01(**) were considered significant.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work has been funded by project SAF2017-83883-R from Ministerio de Economía y Empresa (MINECO, Spain). We thank Isabel Sánchez (Proteomic services, UdL) and Roser Pané for technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101520.
Abbreviations
- FA
Friedreich Ataxia
- NRVM
neonatal rat cardiac myocytes
- DLDH
dihydrolipoyl dehydrogenase
- PDH
pyruvate dehydrogenase
- KGDH
α-ketoglutarate dehydrogenase
- DLAT
dihydrolipoyllysine acetyltransferase
- DLST
dihydrolipoyllysine succinyltransferase
- NAC
N-acetyl cysteine
- IAM
iodoacetamide
- DCA
dichloroacetate
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
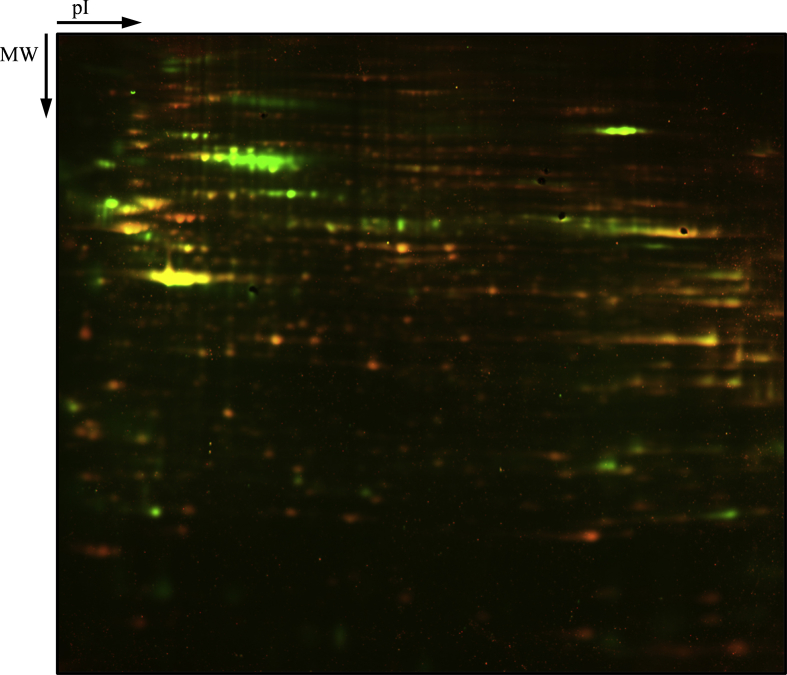
Supplemental Fig. 1.
A preparative gel used to identify protein spots. Green signal corresponds to Bodipy staining (oxidized thiols), while Orange signal to Flamingo stain (total protein stain).
References
- 1.Koeppen A.H. Friedreich’s ataxia: pathology, pathogenesis, and molecular genetics. J. Neurol. Sci. 2011;303:1–12. doi: 10.1016/j.jns.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsou A.Y., Paulsen E.K., Lagedrost S.J., Perlman S.L., Mathews K.D., Wilmot G.R. Mortality in friedreich ataxia. J. Neurol. Sci. 2011;307:46–49. doi: 10.1016/j.jns.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 3.Alsina D., Purroy R., Ros J., Tamarit J. Iron in friedreich ataxia: a central role in the pathophysiology or an epiphenomenon? Pharmaceuticals. 2018;11:89. doi: 10.3390/ph11030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamarit J., Obis È., Ros J. Oxidative stress and altered lipid metabolism in Friedreich ataxia. Free Radic. Biol. Med. 2016;100:138–146. doi: 10.1016/j.freeradbiomed.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Maio N., Jain A., Rouault T.A. Mammalian iron–sulfur cluster biogenesis: recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins. Curr. Opin. Chem. Biol. 2020;55:34–44. doi: 10.1016/j.cbpa.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastore A., Puccio H. Frataxin: a protein in search for a function. J. Neurochem. 2013;126:43–52. doi: 10.1111/jnc.12220. [DOI] [PubMed] [Google Scholar]
- 7.J. Bridwell-Rabb, C. Iannuzzi, A. Pastore, D.P. Barondeau, Effector role reversal during evolution: the case of frataxin in Fe-S cluster biosynthesis, Biochemistry. 51 (n.d.) 92506–92514. doi:10.1021/bi201628j. [DOI] [PMC free article] [PubMed]
- 8.Marelja Z., Stöcklein W., Nimtz M., Leimkühler S. A novel role for human Nfs1 in the cytoplasm: nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J. Biol. Chem. 2008;283:25178–25185. doi: 10.1074/jbc.M804064200. [DOI] [PubMed] [Google Scholar]
- 9.Park S., Gakh O., O'Neill H. a, Mangravita A., Nichol H., Ferreira G.C. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J. Biol. Chem. 2003;278:31340–31351. doi: 10.1074/jbc.M303158200. [DOI] [PubMed] [Google Scholar]
- 10.Paupe V., Dassa E.P., Goncalves S., Auchere F., Lonn M., Holmgren A. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PloS One. 2009;4 doi: 10.1371/journal.pone.0004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deponte M. The incomplete glutathione puzzle: just guessing at numbers and figures? Antioxidants Redox Signal. 2017;27:1130–1161. doi: 10.1089/ars.2017.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero E., Ros J., Belli G., Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Cobbett C., Goldsbrough P. P HYTOCHELATINS and M ETALLOTHIONEINS : roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 14.Board P.G., Menon D. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim. Biophys. Acta Gen. Subj. 2013;1830:3267–3288. doi: 10.1016/j.bbagen.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Hider R.C., Kong X.L. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals. 2011;24:1179–1187. doi: 10.1007/s10534-011-9476-8. [DOI] [PubMed] [Google Scholar]
- 16.Montero D., Tachibana C., Rahr Winther J., Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1:508–513. doi: 10.1016/j.redox.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auchere F., Santos R., Planamente S., Lesuisse E., Camadro J.M. Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich’s ataxia. Hum. Mol. Genet. 2008;17:2790–2802. doi: 10.1093/hmg/ddn178. ddn178 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Piemonte F., Pastore a., Tozzi G., Tagliacozzi D., Santorelli F.M., Carrozzo R. Glutathione in blood of patients with Friedreich’s ataxia. Eur. J. Clin. Invest. 2001;31:1007–1011. doi: 10.1046/j.1365-2362.2001.00922.x. [DOI] [PubMed] [Google Scholar]
- 19.Obis È., Irazusta V., Sanchís D., Ros J., Tamarit J. Frataxin deficiency in neonatal rat ventricular myocytes targets mitochondria and lipid metabolism. Free Radic. Biol. Med. 2014;73:21–33. doi: 10.1016/j.freeradbiomed.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Pastore A., Tozzi G., Gaeta L.M., Bertini E., Serafini V., Di Cesare S. Actin glutathionylation increases in fibroblasts of patients with friedreich’s ataxia. J. Biol. Chem. 2003;278:42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- 21.Bulteau A.L., Planamente S., Jornea L., Dur A., Lesuisse E., Camadro J.M. Changes in mitochondrial glutathione levels and protein thiol oxidation in Δyfh1 yeast cells and the lymphoblasts of patients with Friedreich’s ataxia. Biochim. Biophys. Acta - Mol. Basis Dis. 2012;1822:212–225. doi: 10.1016/j.bbadis.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Tamarit J., de Hoogh A., Obis E., Alsina D., Cabiscol E., Ros J. Analysis of oxidative stress-induced protein carbonylation using fluorescent hydrazides. J. Proteomics. 2012;75:3778–3788. doi: 10.1016/j.jprot.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 23.Ollagnier-de Choudens S., Sanakis Y., Hewitson K.S., Roach P., Baldwin J.E., Münck E. Iron-sulfur center of biotin Synthase and lipoate Synthase. Biochemistry. 2000;39:4165–4173. doi: 10.1021/bi992090u. [DOI] [PubMed] [Google Scholar]
- 24.Mathias R.A., Greco T.M., Oberstein A., Budayeva H.G., Chakrabarti R., Rowland E.A. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purroy R., Britti E., Delaspre F., Tamarit J., Ros J. Mitochondrial pore opening and loss of Ca 2+ exchanger NCLX levels occur after frataxin depletion. Biochim. Biophys. Acta - Mol. Basis Dis. 2018;1864:618–631. doi: 10.1016/j.bbadis.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Tarrado-Castellarnau M., De Atauri P., Cascante M. Oncogenic regulation of tumor metabolic reprogramming. Oncotarget. 2016;7 doi: 10.18632/oncotarget.10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Boja E.S., Tan W., Tekle E., Fales H.M., English S. Reversible glutathionylation regulates actin polymerization in A431 cells. J. Biol. Chem. 2001;276:47763–47766. doi: 10.1074/jbc.C100415200. [DOI] [PubMed] [Google Scholar]
- 28.Dalle-Donne I., Giustarini D., Rossi R., Colombo R., Milzani A. Reversible S-glutathionylation of Cys 374 regulates actin filament formation by inducing structural changes in the actin molecule. Free Radic. Biol. Med. 2003;34:23–32. doi: 10.1016/s0891-5849(02)01182-6. http://www.ncbi.nlm.nih.gov/pubmed/12498976 [DOI] [PubMed] [Google Scholar]
- 29.Mailloux R.J., Jin X., Willmore W.G. Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biol. 2014;2:123–139. doi: 10.1016/j.redox.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLain A.L., Cormier P.J., Kinter M., Szweda L.I. Glutathionylation of α-ketoglutarate dehydrogenase: the chemical nature and relative susceptibility of the cofactor lipoic acid to modification. Free Radic. Biol. Med. 2013;61:161–169. doi: 10.1016/j.freeradbiomed.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irazusta V., Moreno-Cermeño A., Cabiscol E., Ros J., Tamarit J. Major targets of iron-induced protein oxidative damage in frataxin-deficient yeasts are magnesium-binding proteins. Free Radic. Biol. Med. 2008;44:1712–1723. doi: 10.1016/j.freeradbiomed.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Kark R.A., Rodriguez-Budelli M., Blass J.P. Evidence for a primary defect of lipoamide dehydrogenase in Friedreich’s ataxia. Adv. Neurol. 1978;21:163–180. http://www.ncbi.nlm.nih.gov/pubmed/104555 [PubMed] [Google Scholar]
- 33.Mastrogiacomo F., LaMarche J., Dozić S., Lindsay G., Bettendorff L., Robitaille Y. Immunoreactive levels of alpha-ketoglutarate dehydrogenase subunits in Friedreich’s ataxia and spinocerebellar ataxia type 1. Neurodegeneration. 1996;5:27–33. doi: 10.1006/neur.1996.0004. http://www.ncbi.nlm.nih.gov/pubmed/8731379 [DOI] [PubMed] [Google Scholar]
- 34.Blass J.P., Kark R.A.P., Menon N.K., Harris S.E. Low activities of the pyruvate and oxoglutarate dehydrogenase complexes in five patients with friedreich’s ataxia. N. Engl. J. Med. 1976;295:62–67. doi: 10.1056/NEJM197607082950202. [DOI] [PubMed] [Google Scholar]
- 35.Uziel G., Bottacchi E., Moschen G., Giovanardi-Rossi P., Cardace G., Di Donato S. Pyruvate-dehydrogenase complex in ataxic patients: enzyme deficiency in ataxic encephalopathy plus lactic acidosis and normal activity in Friedreich ataxia. Ital. J. Neurol. Sci. 1982;3:317–321. doi: 10.1007/BF02043580. http://www.ncbi.nlm.nih.gov/pubmed/6820014 [DOI] [PubMed] [Google Scholar]
- 36.Pathak D., Srivastava A.K., Padma M.V., Gulati S., Rajeswari M.R. Quantitative proteomic and network analysis of differentially expressed proteins in PBMC of friedreich’s ataxia (FRDA) patients. Front. Neurosci. 2019;13:1054. doi: 10.3389/fnins.2019.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botez M.I., Young S.N. Biogenic amine metabolites and thiamine in cerebrospinal fluid in heredo-degenerative ataxias. Can. J. Neurol. Sci. 2001;28:134–140. doi: 10.1017/s0317167100052811. http://www.ncbi.nlm.nih.gov/pubmed/11383938 [DOI] [PubMed] [Google Scholar]
- 38.Costantini A., Laureti T., Pala M.I., Colangeli M., Cavalieri S., Pozzi E. Long-term treatment with thiamine as possible medical therapy for Friedreich ataxia. J. Neurol. 2016;263:2170–2178. doi: 10.1007/s00415-016-8244-7. [DOI] [PubMed] [Google Scholar]
- 39.Purroy R., Britti E., Delaspre F., Tamarit J., Ros J. Mitochondrial pore opening and loss of Ca2 + exchanger NCLX levels occur after frataxin depletion. Biochim. Biophys. Acta - Mol. Basis Dis. 2018;1864:618–631. doi: 10.1016/j.bbadis.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Moreno-Cermeño A., Alsina D., Cabiscol E., Tamarit J., Ros J. Metabolic remodeling in frataxin-deficient yeast is mediated by Cth2 and Adr1. Biochim. Biophys. Acta. 2013;1833:3326–3337. doi: 10.1016/j.bbamcr.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 41.Alsina D., Ros J., Tamarit J. Nitric oxide prevents Aft1 activation and metabolic remodeling in frataxin-deficient yeast. Redox Biol. 2018;14:131–141. doi: 10.1016/j.redox.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperl W., Fleuren L., Freisinger P., Haack T.B., Ribes A., Feichtinger R.G. The spectrum of pyruvate oxidation defects in the diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis. 2015;38:391–403. doi: 10.1007/s10545-014-9787-3. [DOI] [PubMed] [Google Scholar]
- 43.Mincheva-Tasheva S., Obis E., Tamarit J., Ros J. Apoptotic cell death and altered calcium homeostasis caused by frataxin depletion in dorsal root ganglia neurons can be prevented by BH4 domain of Bcl-xL protein. Hum. Mol. Genet. 2013 doi: 10.1093/hmg/ddt576. [DOI] [PubMed] [Google Scholar]
- 44.Raggi M.A., Mandrioli R., Casamenti G., Musiani D., Marini M. HPLC determination of glutathione and other thiols in human mononuclear blood cells, Biomed. Chromatogr. 1998;12:262–266. doi: 10.1002/(SICI)1099-0801(199809/10)12:5<262::AID-BMC744>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 45.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.