Abstract
Dynein light chain 8 (LC8) interacts with intrinsically disordered proteins (IDPs) and influences a wide range of biological processes. It is becoming apparent that among the numerous IDPs that interact with LC8, many contain multiple LC8-binding sites. Although it is established that LC8 forms parallel IDP duplexes with some partners, such as nucleoporin Nup159 and dynein intermediate chain, the molecular details of these interactions and LC8's interactions with other diverse partners remain largely uncharacterized. LC8 dimers could bind in either a paired “in-register” or a heterogeneous off-register manner to any of the available sites on a multivalent partner. Here, using NMR chemical shift perturbation, analytical ultracentrifugation, and native electrospray ionization MS, we show that LC8 forms a predominantly in-register complex when bound to an IDP domain of the multivalent regulatory protein ASCIZ. Using saturation transfer difference NMR, we demonstrate that at substoichiometric LC8 concentrations, the IDP domain preferentially binds to one of the three LC8 recognition motifs. Further, the differential dynamic behavior for the three sites and the size of the fully bound complex confirmed an in-register complex. Dynamics measurements also revealed that coupling between sites depends on the linker length separating these sites. These results identify linker length and motif specificity as drivers of in-register binding in the multivalent LC8–IDP complex assembly and the degree of compositional and conformational heterogeneity as a promising emerging mechanism for tuning of binding and regulation.
Keywords: intrinsically disordered protein, nuclear magnetic resonance (NMR), analytical ultracentrifugation, protein dynamic, protein assembly, LC8, multivalency, native mass spectrometry
Introduction
LC84 is a highly conserved protein found across nearly all eukaryotes. First identified as a component of cytoplasmic dynein (1), LC8 is now recognized as a hub protein that interacts with >100 different proteins involved in numerous cellular processes as diverse as synaptic signaling, virus replication, and apoptosis (2–4). LC8 is a homodimer of 10-kDa subunits, with symmetrical binding grooves for binding an eight-amino acid motif in intrinsically disordered protein (IDP) clients (5–7). Thus, LC8 promotes higher-order complex formation by dimerizing client proteins binding on each LC8 protomer.
The short linear recognition sequence includes highly conserved TQT amino acids in the C-terminal half of the motif that are essential for binding (6, 8, 9). High-resolution structures show that this characteristic TQT forms critical interactions that anchor the peptides to LC8 and support high-affinity binding (10). The residues flanking the TQT anchor are less conserved but contribute significantly to the overall binding affinity and correct orientation of the motifs in the binding grooves, supporting the formation of parallel IDP dimers (11). Although the majority of identified LC8-binding motifs have the TQT anchor, some variations in these core residues occur in natural sequences (11, 12).
A number of LC8 client proteins display multiple LC8-binding motifs that are thought to play an important role in their biological functions (13–16). Nup159 from Saccharomyces cerevisiae exhibits five such motifs (17, 18) separated by short linker regions that are generally only two or three amino acids in length. Characterization by negative-stain EM of the LC8–Nup159 complex shows that the protein forms a rigid stacked structure. In contrast, another multivalent LC8-binding partner, ASCIZ (ATMIN-substrate Chk-interacting Zn2+ finger protein), has 11 LC8-binding motifs spaced throughout a ∼460-amino acid intrinsically disordered region (13, 19, 20). The disordered linkers between LC8-binding sites in ASCIZ vary considerably in length, from 3 to 27 amino acids, and could contribute to the heterogeneous population of the LC8–ASCIZ complexes observed by negative-stain EM analysis (13).
To date, ASCIZ is the only known regulator of LC8 transcription (21). ASCIZ has the highest number of LC8-binding sites of any protein identified in either humans or D. melanogaster (19, 22). ASCIZ from D. melanogaster has six predicted binding sites that were confirmed using short synthetic peptides (13). A seventh site (called QT3), which contains a TMT instead of the canonical TQT, does not bind as a synthetic peptide, but larger constructs containing QT3 support binding. Interestingly, ASCIZ binding to LC8 forms heterogenous complexes of varying LC8 occupancy, suggesting that the multiple interaction motifs may act as a sensor for LC8 concentration inside the cell (13, 20).
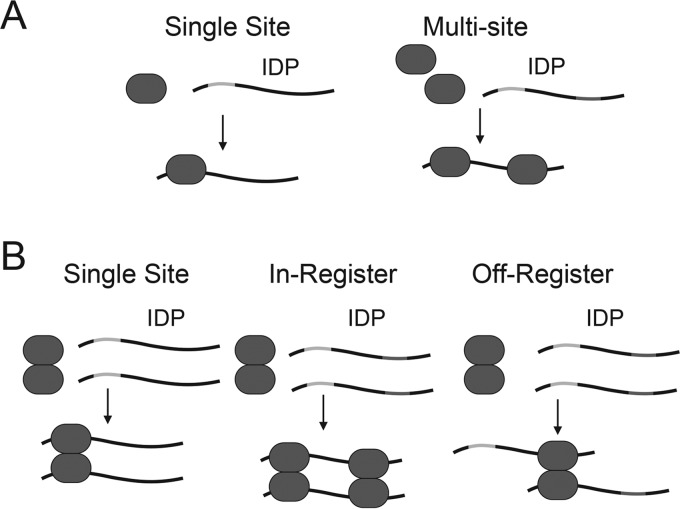
The presence of multiple LC8-binding sites on a given IDP raises important mechanistic questions about LC8 binding. For proteins that present a single IDP-binding site, interaction with a multivalent client results in decoration of the IDP with the binding protein (Fig. 1A). The situation for LC8 is more complicated, because LC8 is a homodimer and presents two parallel binding groves. In IDP clients with only one binding site, LC8 dimer interacts with each of two copies, resulting in formation of an IDP duplex (Fig. 1B). It is tempting to extrapolate this mechanism to IDPs with multiple binding sites, where the LC8 dimers interact with the sites on the client IDPs in a paired or “in-register” fashion. However, this is not the only possible model for LC8 binding to multivalent IDPs. LC8 could bind to any of the available sites, resulting in “off-register” binding. A second factor that could influence the formation of LC8–IDP complexes is the differences in linker length between LC8-binding sites. Indeed, the closely spaced LC8-binding sites in Nup159 appear to yield a relatively in-register and rigid structure, whereas the more varied spacings in ASCIZ yield a more flexible and dynamic structure. These differences in overall flexibility could underlie the functional differences in the respective complexes, where one functions as an assembly scaffold, whereas the other as a molecular sensor.
Figure 1.
Diagram of LC8 interaction models. A, model of a protein with one recognition motif interacting with an IDP client. Interaction with a monovalent IDP results in a 1:1 complex. Interaction with a multivalent IDP results in decoration of the IDP with the binding partner. B, models of homodimeric LC8 interacting with an IDP client. Interaction of LC8 with a monovalent (single site) IDP results in dimerization of the IDP. For the in-register binding model for LC8 interaction with multivalent IDP clients, a given LC8 dimer interacts with the equivalent binding site on each IDP client, resulting in dimerization of the IDP client with multiple bound LC8s. For the off-register binding model for LC8 interaction with multivalent IDP clients, a given LC8 dimer interacts with nonequivalent binding sites, resulting in the formation of dimers and higher-order oligomers. In B, only two LC8-binding sites are shown for clarity.
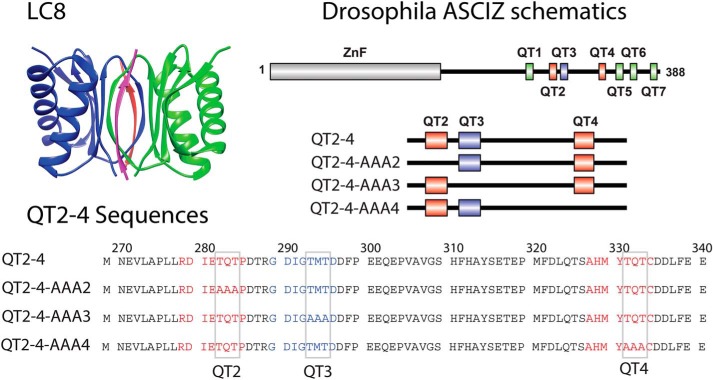
In this study, we address the mechanism of assembly using a multivalent domain of ASCIZ containing the QT2, QT3, and QT4 recognition motifs, referred to as QT2–4 (Fig. 2) (13). This construct was designed to contain 1) multiple LC8-binding sites, 2) varying linker length between motifs, and 3) the QT3 site whose binding to LC8 has not been directly observed. Our study provides the first evidence of in-register binding during complex assembly and suggests a role for linker lengths in modulating flexibility and LC8 occupancy in multivalent LC8–IDP complexes in general.
Figure 2.
Proteins used in this study. Top left panel, ribbon diagram of LC8 showing each monomer in the LC8 homodimer (blue and green). Bound peptides are shown in red and pink. The coordinates were obtained from Protein Data Bank code 2P2T. The LC8 graphic was generated in Chimera (41). Top right panel, schematic diagram of ASCIZ showing the location of the LC8-binding sites and the constructs used in this study, including AAA variants that abolish each recognition motif one at a time. Bottom panel, amino acid sequence of QT2–4. QT2 and QT4 are shown in red. QT3 is shown in blue. Core binding site residues (TQT or TMT) are indicated by gray boxes.
Results
Complex formation monitored by sedimentation velocity analytical ultracentrifugation (SV-AUC) and native ESI-MS
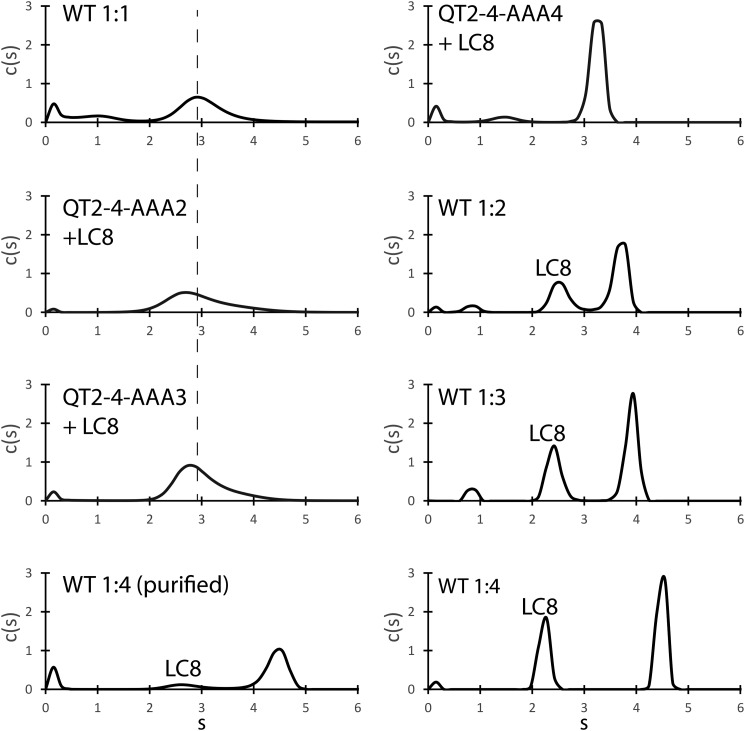
SV-AUC analysis of the complex shows that the proteins are in a dynamic equilibrium at ratios of QT2–4:LC8 up to 1:3 (Fig. 3). At a 1:1 molar ratio of QT2–4 to LC8, the sedimentation coefficient is shifted to ∼2.9 S, larger than the ∼2.5 S observed for LC8 alone and consistent with binding to QT2–4. Higher LC8 concentrations further shift the sedimentation coefficient toward the fully bound state, consistent with incorporation of additional LC8 dimers. We also note that at ratios of 1:2 and greater, we observe a distinct population of unbound LC8. Titrating with a ratio of 1:4 QT2–4:LC8 results in the formation of a fully occupied complex having a sedimentation coefficient of ∼4.5 S. Importantly, we found that the complex is stable, exhibiting the same sedimentation coefficient after purification with size-exclusion chromatography. We did not detect larger order aggregates or oligomers.
Figure 3.
Sedimentation velocity analytical ultracentrifugation of ASCIZ QT2–4 bound to LC8. Populations corresponding to free LC8 are labeled. Experiments were performed at four molar ratios of WT QT2–4 to LC8. For the highest ratio (1:4), the data are shown with and without purification using size-exclusion chromatography to remove excess LC8. The results for mutant QT2–4 are shown after mixing with excess LC8 and purification with size-exclusion chromatography. The dashed line is centered on the main peak for the 1:1 sample.
To isolate the contribution of each binding motif to complex formation, we performed SV-AUC on mutant QT2–4 proteins bound to LC8 (see Fig. 2 for the mutant constructs). QT2–4–AAA4 bound to LC8 exhibited a relatively narrow c(S) distribution with a sedimentation coefficient of ∼3.2 S, lower than the LC8 saturated WT QT2–4 and consistent with a lower-molecular-weight complex. QT2–4–AAA3 and QT2–4–AAA2 bound to LC8 exhibited a broader c(S) profile, with maximum S values at ∼2.8 S and ∼2.7 S, respectively. Increased broadness in the c(S) plot suggests that the complexes formed by QT2–4–AAA3 and QT2–4–AAA2 are more heterogeneous than the complex formed by QT2–4–AAA4. The lower sedimentation coefficients are also consistent with dynamic complexes that average to lower-molecular-weight complexes when compared with WT or QT2–4–AAA4.
Using native electrospray ionization (ESI)–MS measurements of the individual protein subunits along with the mass of the complex(es) allows identification of complex stoichiometries for those formed by QT2–4 bound to LC8. Masses of monomeric QT2–4 (8645.84 ± 0.05 Da) and monomeric LC8 (10,638.5 ± 0.2 Da) were determined first and closely matched the expected masses calculated from each protein sequence (Table 1). Further native mass spectra of both QT2–4 and LC8 acquired at a series of diluted concentrations (Figs. S1 and S2, respectively) confirmed that QT2–4 remains monomeric, whereas LC8 can form dimers in solution.
Table 1.
Expected and measured masses for all proteins and complexes identified by native MS
| Accurate monomer masses | ||
|---|---|---|
| Protein, z+ | Sequence mass (Da) | Measured mass (average ± S.D., Da) |
| QT2–4, 4–7+ | 8592.37 | 8645.84 ± 0.05 |
| LC8, 3–7+ | 10,770.31 | 10,638.52 ± 0.20 |
|
Masses of QT2–4/LC8 species | ||
| Species/complex, z+ | Expected mass (Da) | Measured mass (average ± S.D., Da) |
| LC8 monomer, 3–8+ | 10,638.52 ± 0.20 | 10,753 ± 2 |
| LC8 dimer, 8–10+ | 21,277.0 ± 0.4 | 21,503 ± 1 |
| QT2–4 monomer + 1 LC8 dimer | 29,922.9 ± 0.4 | 30,154.2 ± 0.9 |
| QT2–4 monomer + 2 LC8 dimers | 51,200 ± 1 | 51,765 ± 8 |
| QT2–4 dimer + 3 LC8 dimers | 81,123 ± 1 | 81,925 ± 21 |
| QT2–4 dimer + 4 LC8 dimers | 102,400 ± 2 | 103,497 ± 15 |
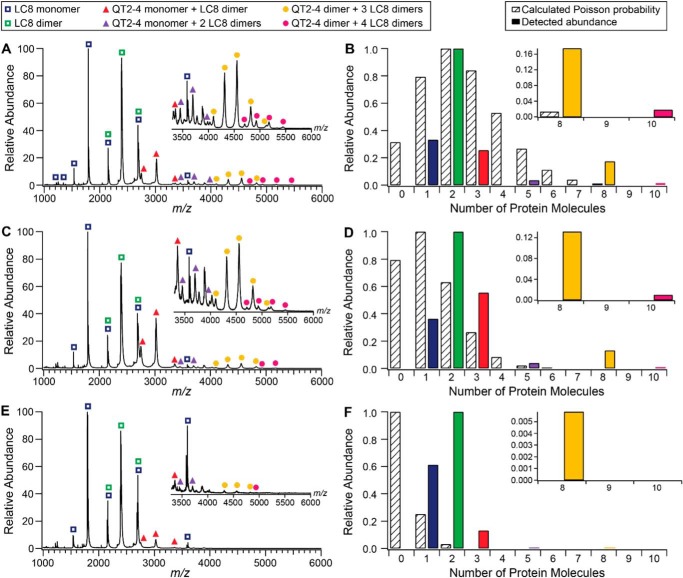
Native ESI-MS of the QT2–4–LC8 complex identified six different species, the associated peaks of which are labeled in Fig. 4. Two of these six correspond to the LC8 monomer and dimer, which are expected to be present upon dilution of the reconstituted complex. The others include QT2–4–LC8 complexes with masses that correspond to QT2–4 monomer bound by one LC8 dimer, QT2–4 monomer bound by two LC8 dimer, QT2–4 dimer bound by three LC8 dimers, and QT2–4 dimer bound by four LC8 dimers (Table 1). Detected complex abundances are shown alongside calculated Poisson probability distributions in Fig. 4. The two smaller complexes of QT2–4 monomer bound by one or two LC8 dimers are detected at higher abundances than predicted by Poisson distributions to the lowest concentration studied and thus may represent complex assembly intermediates. The stoichiometry of the largest complexes identified were confirmed via collision-induced dissociation. Of the two larger complexes, (QT2–4)2(LC8 dimer)3 was consistently detected at higher abundance than (QT2–4)2(LC8 dimer)4. Dilutions of the purified complex (1 μm to 100 nm) show that (QT2–4)2(LC8 dimer)3 is more stable than (QT2–4)2(LC8 dimer)4, with the larger complex not observed at the lowest concentration (Fig. 4). Taken together, we consider the fully formed complex to primarily be the more stable (QT2–4)2(LC8 dimer)3 complex.
Figure 4.
Native mass spectra and abundance distributions of QT2–4–LC8 species at a concentration of 1 μm (A and B), 500 nm (C and D), and 100 nm (E and F). Insets in mass spectra panels (A, C, and E) show the m/z 3300–6000 region of the corresponding mass spectrum within each panel. Peaks associated with each of the six species identified are labeled in the legend. Distribution panels (B, D, and F) display the calculated Poisson probability for 0–10 protein molecules in a droplet of 100-nm radius (striped bars) and the experimentally detected abundances of the six QT2–4–LC8 species (solid bars with colors matching the legend on the left). Probabilities and abundances in each main distribution panel were normalized to the most probable or abundant state. Insets in the distribution panels provide clarity for comparison of the relative calculated Poisson probabilities and abundances of the two largest species detected. For experimentally detected abundances, the six identified species are represented by the number of protein molecules as follows: LC8 monomer (1 molecule), LC8 dimer (2 molecules), QT2–4 monomer + LC8 dimer (3 molecules), QT2–4 monomer + 2 LC8 dimers (5 molecules), QT2–4 dimer + 3 LC8 dimers (8 molecules), and QT2–4 dimer + 4 LC8 dimers (10 molecules).
Structural characterization of QT2–4–LC8 complex
Having determined that the complex composed of two chains of QT2–4 bound by three LC8 dimers (mass of ∼80 kDa) is the most stable complex, we assigned its backbone resonances using triple-labeled (2H, 13C, 15N) protein and a suite of TROSY-based 3D NMR experiments. Structural characterization of LC8 bound to IDP with multiple LC8-binding sites using NMR spectroscopy has been hampered by the disappearance of resonances corresponding to residues associated with LC8 binding (13, 17, 23). Overall peak attenuation is caused by a reduction in global tumbling time, resulting in increased relaxation. To observe these peaks, we used perdeuteration because of its favorable impact on T2 relaxation (24). The spectrum of deuterated QT2–4 bound to LC8 shows that many of the QT2–4 resonances remain observable, and the bound spectrum exhibits a large number of chemical shift perturbations when compared with the spectrum of unbound QT2–4 (Fig. 5 and Fig. S3). Assignments for ∼78% of the backbone amide resonances of QT2–4 bound to LC8 are shown in Fig. 5.
Figure 5.
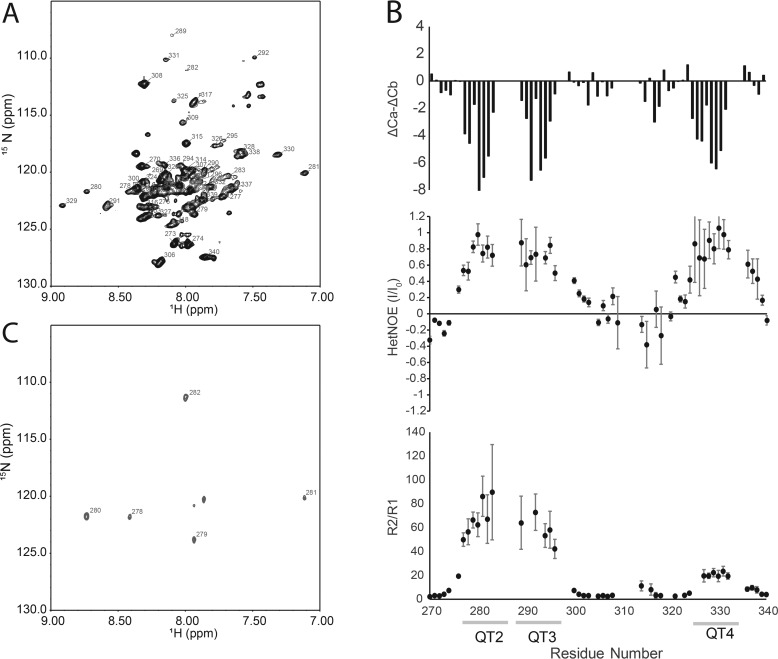
Characterization of LC8 bound to QT2–4. A, assigned TROSY–heteronuclear single quantum coherence spectrum of ASCIZ QT2–4 bound to LC8. B, top panel, ΔCα-ΔCβ chemical shift indexing for ASCIZ QT2–4 bound to LC8. Regions of contiguous negative values indicate β-strand, contiguous positive values indicate α-helix, and values near 0 indicate disorder. Heteronuclear NOE (middle panel) and R2/R1 (bottom panel) dynamics analysis are shown. LC8-binding sites are indicated by the lines below the residue numbers. Gaps are due to unassigned residues or residues that could not be accurately fit because of overlap. C, cross-saturation transfer difference spectrum of ASCIZ QT2–4 bound to LC8 at 1:1 molar ratio of QT2–4 to LC8. Resonances that are saturated by irradiation on the LC8 are labeled and correspond to residues in QT2.
Chemical shift indexing of QT2–4 bound to LC8 reveals that the three LC8-binding sites form a β-strand structure (Fig. 5). This observation is consistent with crystal structures of LC8 bound to short peptides containing an LC8-binding site, where the peptide also adopts β-strand like structure (6, 10, 25). Residues located between QT3 and QT4, such as Gly308 and Thr317, exhibit chemical shifts consistent with a random coil and show modest or no chemical shift perturbations upon binding, indicating that the region between QT3 and QT4 remains unstructured and that their local chemical environment remains largely unchanged by LC8 binding.
It is interesting to note that binding of the QT3 peptide to LC8 was not detected using isothermal titration calorimetry (13), but the QT2–4 construct binds LC8 with a stoichiometry of 3 and a mass consistent with binding at all three sites. These somewhat contradictory results raise the open question: is QT3 an LC8-binding site? Here we show beyond doubt that QT3 binds LC8 in the context of QT2–4 and adopts a β-strand in the bound complex. We identified two glycine residues (Gly289 and Gly292) in QT3 that undergo significant chemical shift perturbations upon LC8 binding when compared with unbound QT2–4 (Fig. S3), whereas Gly308, which is not located in an LC8-binding site, does not undergo any chemical shift perturbation upon addition of LC8, confirming that changes in the spectra are at or near the LC8-binding regions. QT3 residues Thr287, Thr293, and Thr295 shift significantly in the LC8-bound spectrum, further demonstrating that LC8 interacts with QT3 in context of the QT2–4 construct. Although QT3 lacks the canonical TQT, having TMT instead, binding of TMT has been previously reported (12).
Early binding events identified by cross-saturation transfer difference
Cross-saturation transfer difference NMR (26) utilized perdeuterated and 15N-labeled QT2–4 mixed with unlabeled LC8 to probe the interface between LC8 and QT2–4. At a molar ratio of 1:1 QT2–4 to LC8, resonances corresponding to residues Arg278, Asp279, Ile280, and Glu281 showed reduced intensity by saturation on the methyl resonances of LC8, indicating that these residues are in close proximity to LC8. In the absence of LC8, the on-resonance saturation pulse did not result in saturation of any QT2–4 resonances, indicating that any residual protonated QT2–4 that may be present does not significantly influence the spectrum. Interestingly, at this molar ratio, the majority of residues with reduced intensity from the saturation pulse are from QT2, demonstrating that, of the three binding sites in QT2–4, QT2 is most likely to be occupied at substoichiometric LC8 concentrations (Fig. 5). This suggests that the LC8-binding sites in ASCIZ are not equivalent and that LC8 preferentially occupies QT2 over the other sites in QT2–4, even though the affinity for the QT2 linear peptide is only modestly (∼2-fold) tighter than the QT4 linear peptide (13). Bound state peak intensities are generally highest in the QT2 motif at the lower LC8 ratios, consistent with more stable interaction at this site (Fig. S4).
Dynamics analysis of QT2–4 bound to LC8
At a saturating concentration of LC8, where all three sites are fully occupied, R2 increases at each recognition motif, whereas R1 decreases, consistent with increased apparent rotational correlation time and LC8 binding (Fig. S5). A plot of R2/R1 conveys this trend, with a dramatic increase in R2/R1 at each of the LC8-binding regions relative to the linker (Fig. 5). In contrast to the binding regions, the linker exhibits R2/R1 ratios consistent with unstructured protein. Heteronuclear NOE analysis confirms that all three binding regions become structured upon interaction with LC8, with peak intensity ratios approaching 1 (Fig. 5). The linker region remains unstructured with peak intensity ratios near or below 0. Interestingly, the R2/R1 ratio of residues in QT2 and QT3 are similar in magnitude, and both are much larger than the R2/R1 ratio of residues in QT4. This observation indicates that QT2 and QT3, and the associated LC8 dimers, exhibit similar restricted global correlation times and likely tumble as a single unit. In contrast, the lower R2/R1 ratio observed in QT4 suggests that the unstructured linker between QT3 and QT4 decouples motion between these binding sites and allows QT4 to tumble independently of QT2 and QT3.
Discussion
Macromolecular complexes involving IDPs have been implicated in a wide range of biological processes. Key to the function of these complexes is the mechanism by which they form and the overall architecture of the resulting complexes. Our study examined the mechanism and architecture of the complex formed between dimeric LC8 and multivalent QT2–4 with three sites of varying motif specificity and linker lengths separating them. We show the first evidence for preferential binding to one motif that drives in-register complex formation and identify both compositional and conformational heterogeneity as common features of LC8–ASCIZ complexes.
In-register versus off-register binding
As illustrated in Fig. 1, LC8 interacting with a multivalent binding partner could do so either in-register or off-register. To date, there has been little evidence that differentiates between these two models. Our data support in-register binding and lead to an overall binding model that is illustrated in Fig. 6A. In-register binding is consistent with our observation of a single fully bound complex in sedimentation velocity and a single set of resonances in the 15N TROSY NMR spectrum. The saturation transfer difference experiment showed that the majority of saturation transfer is localized to a single binding site at a low ratio of LC8 to QT2–4, an observation that is consistent with in-register binding, because off-register binding would result in saturation transfer at all of the binding sites because of a lack of client site differentiation by LC8. Finally, native ESI-MS shows that the predominant species in the fully bound complex consists of two monomeric QT2–4 chains and three LC8 homodimers, as expected from the in-register binding model. Although other complexes are observed by native ESI-MS, the detected populations indicate that they are minor populations when compared with the in-register complex. Furthermore, we did not observe evidence for these complexes by SV-AUC, consistent with them being lower population states in solution when compared with the in-register complex.
Figure 6.
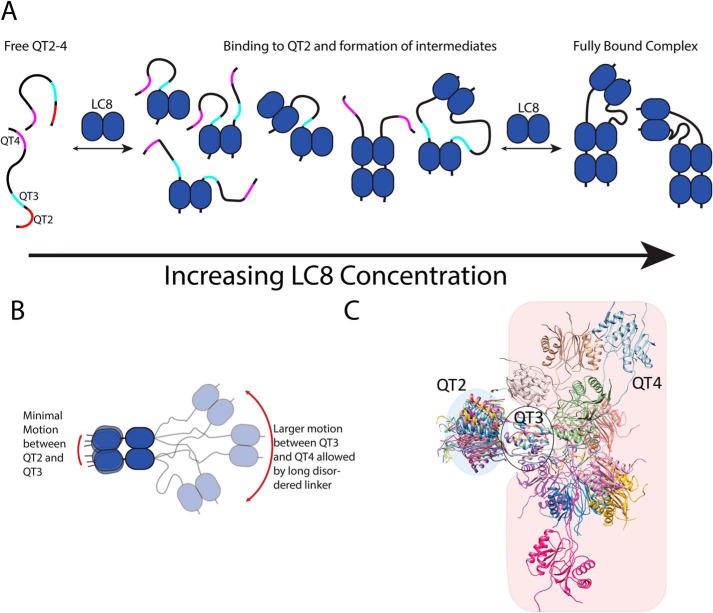
A, model of LC8 binding to QT2–4 showing compositional heterogeneity. The blue ovoids represent LC8, whereas the black lines represent QT2–4. Binding sites on QT2–4 are shown in red (QT2), cyan (QT3), and magenta (QT4). Binding is initiated by interaction of LC8 with QT2, binding to either one chain of QT2–4 or to two chains forming a duplex of QT2–4. Subsequent LC8 homodimers interact with the newly formed duplex. The close proximity of the dimerized QT2–4 chains promote in-register binding. At high concentrations of LC8, a stable complex is formed, with all LC8-binding sites occupied. B, model of QT2–4 bound to LC8 showing conformational heterogeneity. The black lines represent ASCIZ QT2–4. The red arrows indicate the extent of motion imparted by the disordered linkers. Motion between QT2 and QT3 is relatively modest because of the short linker and increases steric constraint between LC8 dimers. Motion between QT3 and QT4 is larger because of the conformational freedom provided by the long disordered linker. C, atomistic model of the relative motional freedom of QT2–4. Models aligned on LC8 bound to QT3. The blue-shaded area emphasizes the freedom of QT2, and the red-shaded area emphasizes the freedom of QT4. The model was generated by fixing the interaction motif and LC8 coordinates to those observed in Protein Data Bank file 2P2T and performing molecular dynamics in XPLOR-NIH.
The R2/R1 data also support in-register binding. If binding were off-register, each LC8 monomer would bind a different LC8 site on QT2–4, resulting in linkers of differing length between each LC8 dimer. In turn, this is expected to give rise to similar R2/R1 values for each binding site, because motion between each binding site would be the result of a mixture of the different possible binding modes. Instead, we see different R2/R1 behavior for QT2 and QT3 versus QT4, as predicted for in-register binding.
In-register binding of stable complexes is observed with EM. Negative-stain EM on the cytoplasmic pore filaments of the nuclear pore complex from yeast shows a rigid, ladder-like assembly of the light chain, Dyn2 (LC8 in yeast), supporting in-register binding (27). Another recent cryo-EM structure of the dynein-2 complex suggests that LC8 forms a complex that is architecturally similar to an in-register complex, having the same ladder-like structure, despite significant sequence differences in the binding motifs (28). With negative-stain EM, it is only possible to image LC8 and not the IDP partner, and therefore in-register binding is inferred and not directly observed. Our work presents direct evidence that LC8 preferentially binds at one site and forms in-register complexes with multivalent ASCIZ QT2–4.
Stepwise dynamic binding
We propose a stepwise model for LC8 interaction with multivalent ASCIZ QT2–4 that is initiated at the QT2 site of highest affinity, as determined from saturation transfer, dimerizing the client protein and orienting the client for in-register binding (Fig. 6A). Subsequent addition of LC8 is supported by the close proximity of the dimerized chains, with weaker binding sites, such as QT3, experiencing enhanced affinity from nearby LC8 interaction. Previous studies on model systems have shown that tethering recognition motifs to binding proteins enhances the interaction by increasing the effective concentration (29, 30). LC8 affinity enhancement caused by bivalency has been fully characterized when other light chains interact with a nearby site (31). Additional intermediate complexes having only a single QT2–4 chain are also possible, as indicated by our native ESI-MS data.
Interestingly, although LC8 initiates binding to QT2–4 by interacting with QT2, subsequent interaction with the other sites appears to be more complicated. We note that the fully bound complex between QT2–4 and LC8 is stable and does not rapidly dissociate, indicating that the overall off rate must be slow. Importantly, if a stable fully bound complex is formed, we would expect to detect the complex in SV-AUC, because the overall off rate would be slow on the sedimentation time scale. However, we do not observe the fully bound complex in the 1:3 or lower ratio complexes, suggesting that the fully bound complex is not formed until high levels of LC8 are present.
The exact nature of the intermediates remains to be fully determined, but our results provide additional insight. SV-AUC analysis of the binding motif mutants show that each site does not contribute equally to overall complex formation. In QT2–4–AAA4, the presence of QT2 and QT3 supports formation of a stable complex indicating that close proximity between QT2 and QT3 enhances binding at QT3. In contrast, mutation of either QT2 or QT3 results in a dynamic mixture of complexes with lower average molecular weights. Taken together, these results provide evidence that linker length plays an important role in multivalent interactions with a longer linker, resulting in complexes with more heterogeneous dynamic equilibrium.
The sedimentation coefficients for the mutant QT2–4 proteins bracket the WT QT2–4 sedimentation coefficient at the 1:1 ratio, suggesting that WT QT2–4 bound to LC8 at the 1:1 ratio is a mixture of complexes that averages between one and two LC8 homodimers bound to QT2–4. The complexes could consist of a mixture of LC8 bound at any of the binding sites, but QT2 is likely more stably occupied based on the saturation transfer results. In the context of the full-length ASCIZ, we also observe a complex dynamic set of intermediates, which tune ASCIZ transcription activity, rather than behaving like a simple on/off switch (13). A set of partially occupied stable complexes also observed in the shorter QT2–4 confirms that a dynamic complex of multiple partially occupied LC8–ASCIZ complexes is a feature of ASCIZ and possibly other multivalent partners.
Complex flexibility
In addition to the compositional heterogeneity that is observed during the titration, the fully occupied LC8/QT2–4 complex has significant conformational heterogeneity, because it is a mixture of flexible and rigid regions, illustrated in Fig. 6 (B and C). The short linker between QT2 and QT3 greatly constricts the overall motion between LC8 homodimers bound at these locations, as inferred from the large R2/R1 values, and forces them to behave as a unit. The long disordered linker between QT3 and QT4 enables QT4 to move more freely with respect to QT3 and QT2. Thus, differences in disordered linker length dictate the relative conformational freedom of the complexes formed by multivalent LC8 binding proteins. Finally, the increased rigidity at QT2 and QT3 may further support in-register binding by limiting interactions between sites flanking the QT2 and QT3 unit. Results with the rabies virus protein, RavP, also show that LC8 binding can alter the conformational space sampled by a protein and limit interactions between sites separated by LC8 (32). With Nup159, LC8 binding forms a rigid structure that is proposed to enhance the cytoplasmic accessibility of Phe-Gly repeats to nuclear transport proteins. Nup159 has short disordered linkers separating the LC8-binding sites, similar to the linker between QT2 and QT3 in QT2–4, and thus would have similar restricted mobility. In contrast to Nup159, QT2–4 retains IDP-like behavior in the region between QT3 and QT4, suggesting that LC8-bound ASCIZ would not adopt a fully rigid structure. Therefore, the mechanism of action for ASCIZ does not rely on formation of a fully rigid structure, as is the case with Nup159, and this added flexibility could offer another layer of regulatory control that contributes to the buffered transcriptional activity observed for ASCIZ.
Conclusions
Here we show that binding of LC8 to a multivalent partner results in formation of an in-register complex. Such in-register binding guides the formation, structure, and function of higher-order complexes. The dynamic behavior of multivalent LC8 complexes is linked to the binding affinity of the individual sites and the length of the intervening disordered linkers. Longer disordered linkers increase flexibility between regions with bound LC8, resulting in both compositional and conformational heterogeneity, whereas shorter linkers can lead to enhanced binding of nearby weaker sites and reduced flexibility in the complex. The combination of motif affinity and specificity, along with variation in disordered linker length in a multivalent partner, is a promising emerging mechanism for an exquisitely tunable system of binding and regulation. We anticipate this mechanism to be general across many biological processes, given the growing number of multivalent LC8 client proteins and the essential role of LC8 in nearly all cellular functions.
Experimental procedures
Cloning, protein expression, and purification
Cloning of Drosophila ASCIZ QT2–4 (ASCIZ residues 271–341) (Fig. 2) with various mutations of recognition motifs was preformed using QuikChange Lightening mutagenesis kit (Agilent). The resulting constructs verified by sequencing are QT2–4–AAA2, QT2–4–AAA3, and QT2–4–AAA4, where the number indicates the LC8 recognition motif whose TQT was replaced with AAA and thus has lost binding at this particular site. Proteins were expressed and purified according to previously published procedures (13). For perdeuteration, Escherichia coli Rosetta DE3 cells, transformed with a pET2Zt2–1a vector with a sequence encoding QT2–4, were grown in Luria broth prepared in 99.9% D2O overnight. Modified M9 minimal medium prepared with 99.9% D2O was inoculated from the overnight culture. 13C and 15N were supplied with uniformly labeled [2H-13C]glucose (0.01 m) and [15N]ammonium chloride (0.02 m), respectively. The cultures were grown at 37 °C to an optical density of 0.6 at 600 nm, induced by adding isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.4 mm, and harvested after 6 h of induction. The proteins were purified under denaturing conditions using TALON His-tag purification protocol (Clontech) and then dialyzed into 20 mm Tris-HCl, 5 mm β-mercaptoethanol, 1 mm sodium azide, pH 8.0 affinity tag cleavage buffer. Complete cleavage of the His tag required incubation with in-house produced His-tagged TEV protease (1:100) for 4 h at room temperature and confirmed on SDS-PAGE. Further purification using anion exchange Macro-Prep High Q support resin (Bio-Rad) was followed by size-exclusion chromatography on a Superdex 75 column (GE Healthcare) in a buffer composed of 20 mm Tris-HCl, 50 mm NaCl, 5 mm β-mercaptoethanol, 1 mm sodium azide, pH 7.5, yielded protein with >95% purity as determined by SDS-PAGE. Protein concentrations were determined by absorbance measurements at 280 nm with extinction coefficients for LC8 of 14,440 m−1 cm−1 and QT2–4 of 2980 m−1 cm−1. When estimating a concentration for SEC-purified complexes, absorbance at 280 nm was still used along with the assumption that the majority of formed complex in solution followed the expected stoichiometry of 1:3 (QT2–4 monomer: LC8 dimer) and that very little excess of either free protein would be present.
Analytical ultracentrifugation
SV-AUC was performed using a Beckman Coulter Optima XL-A analytical ultracentrifuge, equipped with absorbance optics. LC8 was mixed with QT2–4 at ratios of 1:1, 1:2, 1:3, and 1:4 (molar ratio of QT2–4:LC8). An additional sample of QT2–4 was mixed with LC8 at a 1:4 molar ratio and then repurified using gel filtration chromatography on a Superdex 200 column (GE Healthcare). SV-AUC of the QT2–4 AAA complexes was performed on reconstituted complexes purified by gel filtration chromatography. The concentration of protein complex in the final samples was estimated to be 10 μm. Buffer conditions for SV-AUC analysis were 20 mm Tris-HCl, 50 mm NaCl, 5 mm Tris(2-carboxyethyl)phosphine, 1 mm sodium azide, pH 7.5. The complexes were loaded into standard, 12-mm pathlength, two-channel sectored centerpieces and centrifuged at 42,000 rpm and 20 °C. 300 scans were acquired at 280 nm with no interscan delay. The data were fit to a c(S) distribution using the software SEDFIT (33). Buffer density was calculated to be 1.0009 g/ml using Sednterp (34).
Native ESI-MS
Purified samples of the individual proteins (QT2–4 and LC8) and of the QT2–4–LC8 complex were flash-frozen in liquid nitrogen and stored at −80 °C until further use. A 50-μl aliquot of each sample was buffer exchanged into 200 mm ammonium acetate, pH 7.50, using Micro Bio-SpinTM columns (Bio-Rad). All native mass spectra were collected using a Waters Synapt G2-Si TOF mass spectrometer with a nanoelectrospray ionization (nano-ESI) source. A volume of 3–5 μl of sample was loaded into borosilicate capillaries (inner diameter, 0.78 mm) pulled to a fine tip using a Flaming-Brown P-97 micropipette puller (Sutter Instruments). A platinum wire was placed in electrical contact with the solution, and a voltage of +0.5–0.7 kV was applied to the wire to initiate electrospray. The data were acquired with the source at ambient temperature, trap collision energy at 10 V, transfer collision energy at 5 V, and trap gas flow at 10 ml/min. A sampling cone with a small aperture and backing pressure of 1.37 mbar was used in all experiments, and the sampling cone was operated at 25 V (for accurate mass determination), and 50 V (for complex dilution series). Spectra shown were generated by summing data scans collected over 1 min (accurate mass) or 7 min (complex dilution series). A mass calibration profile was generated using cesium iodide clusters prior to acquiring data for accurate mass determination. Complex stoichiometry of the two largest complexes identified was confirmed via collision-induced dissociation and detected abundance distributions of QT2–4–LC8 complexes were assessed in the context of Poisson probability distributions (35). In addition to acquiring data of individual proteins at an initial concentration of 25 μm and of the mixed sample (QT2–4/LC8) at 1 μm, spectra were also acquired for a dilution series of each sample at 10 and 1 μm (individual proteins), 500, 100, and 10 nm (individuals and complex). Poisson probabilities of observing 0–10 protein molecules as nonspecific oligomers were calculated for each of these concentrations using a droplet diameter of 200 nm. Mass spectral peaks were fitted to Gaussian distributions using IGOR Pro, and the resulting areas of each species' charge state peaks were summed to determine abundances for each species observed. Oligomers and complexes detected with relative abundances well above those expected for nonspecific, Poisson-like association occurring during the nanoESI process were determined instead to originate in solution.
NMR experiments
NMR experiments were carried out on an 800-MHz Bruker Avance III HD NMR spectrometer equipped with a 5-mm triple resonance (HCN) cryogenic probe. All NMR data on the complex were collected at 40 °C, because this led to the best overall spectrum of the complex. NMR samples were prepared in 10 mm sodium phosphate, 10 mm NaCl, 1 mm sodium azide, 10 mm Tris(2-carboxyethyl)phosphine, pH 6.5 buffer at a final QT2–4 concentration of 0.5 mm. The samples also contained a protease inhibitor mixture (Roche Applied Science), 10% D2O, and 0.2 mm 2–2 dimethylsilapentane-5-sulfonic acid for 1H chemical shift referencing. Backbone assignments were obtained using a suite of three-dimensional NMR experiments, including HNCA, HNCACB, HNCOCACB, HNCO, and HNCACO. All three-dimensional experiments used TROSY, deuterium decoupling, and nonuniform sampling. Recycle delays were set to 2.5 s for all experiments except HetNOE, which used a recycle delay of 8 s. For the saturation transfer difference experiments (26), saturation was applied during the relaxation delay of a 2D 15N TROSY–heteronuclear single quantum coherence for 4 s using a 50-ms Gaussian pulse with a B1 field strength of 50 Hz. On-resonance and off-resonance saturation was applied at 0.7 and −40 ppm, respectively. Subtraction of the on- and off-resonance spectra is incorporated into the phase cycle. T1 and T2 experiments incorporated temperature compensation. Relaxation delay times for the T1 experiments were 0.02, 0.06, 0.1, 0.2, 0.4, 0.6, 0.8, and 1.2 s. Relaxation delay times for the T2 experiments were 0.0169, 0.0339, 0.0509, 0.0678, 0.0848, 0.1357, 0.1696, and 0.2374 s. Triplicate time points were collected in T1 and T2 experiments for error estimation. NMR data were apodized, zero-filled, Fourier-transformed, phased, and baseline-corrected using nmrPipe (36). The data were apodized with a shifted sine-squared function and zero-filled to twice the original size. Artifacts from nonuniform sampling data collection were removed using SCRUB (37). The data were visualized and analyzed in nmrviewJ or CARA (38). Chemical shift indexing was performed using the ΔCα-ΔCβ method (39, 40).
Author contributions
P. N. R. and E. J. B. conceptualization; P. N. R., J. S. P., and E. J. B. resources; P. N. R., A. D. R., and K. A. J. formal analysis; P. N. R. and A. D. R. visualization; P. N. R., A. D. R., and K. A. J. methodology; P. N. R., K. A. J., and A. D. R. writing-original draft; P. N. R., K. A. J., A. D. R., D. A. S., J. S. P., and E. J. B. writing-review and editing; P. N. R., K. A. J., A. D. R., D. A. S., and H. T. M. H. data curation; A. D. R., J. S. P., and E. J. B. funding acquisition; J. S. P. and E. J. B. supervision; J. S. P. and E. J. B. project administration.
Supplementary Material
Acknowledgments
The authors acknowledge Drs Daniel Zuckerman and Steve Reichow for helpful discussion and Wendy Hare for help with protein preparations.
This work was supported by National Science Foundation Award 1617019. This work was also supported by a National Institutes of Health Grant T32 GM007759 (to A. D. R.). The Oregon State University NMR Facility is supported in part by the National Institutes of Health, HEI Grant 1S10OD018518 (to E. J. B.) and by M. J. Murdock Charitable Trust Grant 2014162 (to E. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
NMR data are available from the Biological Magnetic Resonance Data Bank under BMRB accession number 28033. All remaining data are contained in the article or supporting information.
- LC8
- dynein light chain 8
- IDP
- intrinsically disordered protein
- SV-AUC
- sedimentation velocity analytical ultracentrifugation
- ESI
- electrospray ionization
- TROSY
- tranverse relaxation-optimized spectroscopy.
References
- 1. King S. M., and Patel-King R. S. (1995) The M (r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270, 11445–11452 10.1074/jbc.270.19.11445 [DOI] [PubMed] [Google Scholar]
- 2. Barbar E. (2008) Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47, 503–508 10.1021/bi701995m [DOI] [PubMed] [Google Scholar]
- 3. Rapali P., Szenes Á., Radnai L., Bakos A., Pál G., and Nyitray L. (2011) DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 278, 2980–2996 10.1111/j.1742-4658.2011.08254.x [DOI] [PubMed] [Google Scholar]
- 4. Jespersen N., and Barbar E. (2020) Emerging features of linear motif-binding Hub proteins. Trends Biochem. Sci., in press [DOI] [PubMed] [Google Scholar]
- 5. Fan J., Zhang Q., Tochio H., Li M., and Zhang M. (2001) Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain. J. Mol. Biol. 306, 97–108 10.1006/jmbi.2000.4374 [DOI] [PubMed] [Google Scholar]
- 6. Benison G., Karplus P. A., and Barbar E. (2007) Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J. Mol. Biol. 371, 457–468 10.1016/j.jmb.2007.05.046 [DOI] [PubMed] [Google Scholar]
- 7. Liang J., Jaffrey S. R., Guo W., Snyder S. H., and Clardy J. (1999) Structure of the PIN/LC8 dimer with a bound peptide. Nat. Struct. Biol. 6, 735–740 10.1038/11501 [DOI] [PubMed] [Google Scholar]
- 8. Lo K. W., Naisbitt S., Fan J. S., Sheng M., and Zhang M. (2001) The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J. Biol. Chem. 276, 14059–14066 10.1074/jbc.M010320200 [DOI] [PubMed] [Google Scholar]
- 9. Benison G., and Barbar E. (2009) NMR analysis of dynein light chain dimerization and interactions with diverse ligands. Methods Enzymol. 455, 237–258 10.1016/S0076-6879(08)04209-2 [DOI] [PubMed] [Google Scholar]
- 10. Clark S., Nyarko A., Löhr F., Karplus P. A., and Barbar E. (2016) The anchored flexibility model in LC8 motif recognition: insights from the chica complex. Biochemistry 55, 199–209 10.1021/acs.biochem.5b01099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jespersen N., Estelle A., Waugh N., Davey N. E., Blikstad C., Ammon Y.-C., Akhmanova A., Ivarsson Y., Hendrix D. A., and Barbar E. (2019) Systematic identification of recognition motifs for the hub protein LC8. Life Science Alliance 2, e201900366 10.26508/lsa.201900366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bodor A., Radnai L., Hetényi C., Rapali P., Láng A., Kövér K. E., Perczel A., Wahlgren W. Y., Katona G., and Nyitray L. (2014) DYNLL2 dynein light chain binds to an extended linear motif of myosin 5a tail that has structural plasticity. Biochemistry 53, 7107–7122 10.1021/bi500574z [DOI] [PubMed] [Google Scholar]
- 13. Clark S., Myers J. B., King A., Fiala R., Novacek J., Pearce G., Heierhorst J., Reichow S. L., and Barbar E. J. (2018) Multivalency regulates activity in an intrinsically disordered transcription factor. Elife 7, e40684 10.7554/eLife.40684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark S. A., Jespersen N., Woodward C., and Barbar E. (2015) Multivalent IDP assemblies: Unique properties of LC8-associated, IDP duplex scaffolds. FEBS Lett. 589, 2543–2551 10.1016/j.febslet.2015.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunsch A. K., Hammond D., Lloyd J., Schermelleh L., Gruneberg U., and Barr F. A. (2012) Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J. Cell Biol. 198, 1039–1054 10.1083/jcb.201202112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fejtova A., Davydova D., Bischof F., Lazarevic V., Altrock W. D., Romorini S., Schöne C., Zuschratter W., Kreutz M. R., Garner C. C., Ziv N. E., and Gundelfinger E. D. (2009) Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J. Cell Biol. 185, 341–355 10.1083/jcb.200807155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nyarko A., Song Y., Novácek J., Zídek L., and Barbar E. (2013) Multiple recognition motifs in nucleoporin Nup159 provide a stable and rigid Nup159–Dyn2 assembly. J. Biol. Chem. 288, 2614–2622 10.1074/jbc.M112.432831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stelter P., Kunze R., Flemming D., Höpfner D., Diepholz M., Philippsen P., Böttcher B., and Hurt E. (2007) Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat. Cell Biol. 9, 788–796 10.1038/ncb1604 [DOI] [PubMed] [Google Scholar]
- 19. Rapali P., García-Mayoral M. F., Martínez-Moreno M., Tárnok K., Schlett K., Albar J. P., Bruix M., Nyitray L., and Rodriguez-Crespo I. (2011) LC8 dynein light chain (DYNLL1) binds to the C-terminal domain of ATM-interacting protein (ATMIN/ASCIZ) and regulates its subcellular localization. Biochem. Biophys. Res. Commun. 414, 493–498 10.1016/j.bbrc.2011.09.093 [DOI] [PubMed] [Google Scholar]
- 20. Jurado S., Conlan L. A., Baker E. K., Ng J. L., Tenis N., Hoch N. C., Gleeson K., Smeets M., Izon D., and Heierhorst J. (2012) ATM substrate Chk2-interacting Zn2+ finger (ASCIZ) is a bi-functional transcriptional activator and feedback sensor in the regulation of dynein light chain (DYNLL1) expression. J. Biol. Chem. 287, 3156–3164 10.1074/jbc.M111.306019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jurado S., Gleeson K., O'Donnell K., Izon D. J., Walkley C. R., Strasser A., Tarlinton D. M., and Heierhorst J. (2012) The zinc-finger protein ASCIZ regulates B cell development via DYNLL1 and Bim. J. Exp. Med. 209, 1629–1639 10.1084/jem.20120785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaytseva O., Tenis N., Mitchell N., Kanno S., Yasui A., Heierhorst J., and Quinn L. M. (2014) The novel zinc finger protein dASCIZ regulates mitosis in Drosophila via an essential role in dynein light-chain expression. Genetics 196, 443–453 10.1534/genetics.113.159541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benison G., Nyarko A., and Barbar E. (2006) Heteronuclear NMR identifies a nascent helix in intrinsically disordered dynein intermediate chain: implications for folding and dimerization. J. Mol. Biol. 362, 1082–1093 10.1016/j.jmb.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 24. Venters R. A., Farmer B. T. 2nd, Fierke C. A., and Spicer L. D. (1996) Characterizing the use of perdeuteration in NMR studies of large proteins: 13C, 15N and 1H assignments of human carbonic anhydrase II. J. Mol. Biol. 264, 1101–1116 10.1006/jmbi.1996.0699 [DOI] [PubMed] [Google Scholar]
- 25. Slevin L. K., Romes E. M., Dandulakis M. G., and Slep K. C. (2014) The mechanism of dynein light chain LC8-mediated oligomerization of the Ana2 centriole duplication factor. J. Biol. Chem. 289, 20727–20739 10.1074/jbc.M114.576041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shimada I., Ueda T., Matsumoto M., Sakakura M., Osawa M., Takeuchi K., Nishida N., and Takahashi H. (2009) Cross-saturation and transferred cross-saturation experiments. Prog. Nucl. Magn. Reson. Spectrosc. 54, 123–140 10.1016/j.pnmrs.2008.07.001 [DOI] [Google Scholar]
- 27. Gaik M., Flemming D., von Appen A., Kastritis P., Mücke N., Fischer J., Stelter P., Ori A., Bui K. H., Bassler J., Barbar E., Beck M., and Hurt E. (2015) Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol. 208, 283–297 10.1083/jcb.201411003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toropova K., Zalyte R., Mukhopadhyay A. G., Mladenov M., Carter A. P., and Roberts A. J. (2019) Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat. Struct. Mol. Biol. 26, 823–829 10.1038/s41594-019-0286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krishnamurthy V. M., Semetey V., Bracher P. J., Shen N., and Whitesides G. M. (2007) Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J. Am. Chem. Soc. 129, 1312–1320 10.1021/ja066780e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sørensen C. S., and Kjaergaard M. (2019) Effective concentrations enforced by intrinsically disordered linkers are governed by polymer physics. Proc. Natl. Acad. Sci. U.S.A. 116, 23124–23131 10.1073/pnas.1904813116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hall J., Karplus P. A., and Barbar E. (2009) Multivalency in the assembly of intrinsically disordered Dynein intermediate chain. J. Biol. Chem. 284, 33115–33121 10.1074/jbc.M109.048587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jespersen N. E., Leyrat C., Gérard F. C., Bourhis J. M., Blondel D., Jamin M., and Barbar E. (2019) The LC8-RavP ensemble structure evinces a role for LC8 in regulating lyssavirus polymerase functionality. J. Mol. Biol. 431, 4959–4977 10.1016/j.jmb.2019.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laue T. M. (1992) Computer-aided interpretation of analytical sedimentation data for proteins. Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., and Horton J. C., eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 35. Wheeler L. C., Donor M. T., Prell J. S., and Harms M. J. (2016) Multiple evolutionary origins of ubiquitous Cu2+ and Zn2+ binding in the S100 protein family. PLoS One 11, e0164740 10.1371/journal.pone.0164740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37. Coggins B. E., Werner-Allen J. W., Yan A., and Zhou P. (2012) Rapid protein global fold determination using ultrasparse sampling, high-dynamic range artifact suppression, and time-shared NOESY. J. Am. Chem. Soc. 134, 18619–18630 10.1021/ja307445y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 39. Wishart D. S., and Sykes B. D. (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 40. Wishart D. S., and Case D. A. (2001) Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 338, 3–34 [DOI] [PubMed] [Google Scholar]
- 41. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.