Abstract
Antiviral drugs for managing infections with human coronaviruses are not yet approved, posing a serious challenge to current global efforts aimed at containing the outbreak of severe acute respiratory syndrome–coronavirus 2 (CoV-2). Remdesivir (RDV) is an investigational compound with a broad spectrum of antiviral activities against RNA viruses, including severe acute respiratory syndrome–CoV and Middle East respiratory syndrome (MERS–CoV). RDV is a nucleotide analog inhibitor of RNA-dependent RNA polymerases (RdRps). Here, we co-expressed the MERS–CoV nonstructural proteins nsp5, nsp7, nsp8, and nsp12 (RdRp) in insect cells as a part a polyprotein to study the mechanism of inhibition of MERS–CoV RdRp by RDV. We initially demonstrated that nsp8 and nsp12 form an active complex. The triphosphate form of the inhibitor (RDV-TP) competes with its natural counterpart ATP. Of note, the selectivity value for RDV-TP obtained here with a steady-state approach suggests that it is more efficiently incorporated than ATP and two other nucleotide analogs. Once incorporated at position i, the inhibitor caused RNA synthesis arrest at position i + 3. Hence, the likely mechanism of action is delayed RNA chain termination. The additional three nucleotides may protect the inhibitor from excision by the viral 3′–5′ exonuclease activity. Together, these results help to explain the high potency of RDV against RNA viruses in cell-based assays.
Keywords: plus-stranded RNA virus, viral polymerase, drug development, enzyme inhibitor, nucleoside/nucleotide analog, coronavirus, positive-sense RNA virus, Ebola virus (EBOV), Middle East respiratory syndrome coronavirus (MERS–CoV), SARS–CoV-2, remdesivir, antiviral drug, RNA chain termination, RNA-dependent RNA polymerase (RdRp), viral replicase
Introduction
The emergence of a novel coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS–CoV-2, formerly 2019-nCoV),3 initiated a global effort to identify effective treatments focusing on agents with demonstrated antiviral activity against SARS–CoV, Middle East respiratory syndrome (MERS–CoV), or related positive-sense RNA viruses. Although currently there are no approved antiviral drugs for the treatment of human coronavirus infections available, preclinical data with the nucleotide analog remdesivir (RDV) are promising, and human safety data are available (1). This compound shows a broad spectrum of antiviral activities against several RNA viruses (2–4), including SARS–CoV and MERS–CoV (5). RDV was originally developed for the treatment of Ebola virus disease (2). Cell-culture and animal studies revealed potent antiviral activities against filoviruses, including the Ebola virus (EBOV) (4). Subsequent studies have shown that RDV is also active against coronaviruses with divergent RNA-dependent RNA polymerases (RdRps) (5, 6). However, biochemical data that support these findings and provide a possible mechanism of action are not available.
The triphosphate form of RDV (RDV-TP) was shown to inhibit the RdRp of respiratory syncytial virus (4), Nipah virus, and EBOV (7, 8), which are all nonsegmented negative-sense RNA viruses. Active Ebola RdRp contains the viral L protein in complex with viral protein 35 (9, 10). Viral protein 35 is the functional counterpart of the P protein of respiratory syncytial virus and Nipah (7, 11, 12). Previously, we generated recombinant Ebola RdRp for the study of nucleotide analog inhibitors (8). Enzyme kinetics show that RDV-TP is able to compete with its natural counterpart ATP for incorporation. The selectivity of ATP over the inhibitor is ∼4-fold. Once incorporated at position i, the compound causes inhibition of RNA synthesis predominantly at position i + 5. Delayed chain termination is therefore a plausible mechanism of action.
Progress has also been made in characterizing the SARS–CoV RdRp complex (13–15). Biochemical data suggest that the active complex is composed of at least three viral nonstructural proteins nsp7, nsp8, and nsp12. The RNA polymerase nsp12 alone displays low processivity. Synthesis of longer reaction products require the additional presence of nsp7 and nsp8. Although a heterotrimer was not stable, nsp7 and nsp8 can be linked together to form a complex with nsp12 (15). Here we developed a novel expression system for the MERS–CoV RdRp complex and studied the mechanism of action of remdesivir. Co-expression of the MERS nsp5 protease with nsp7, nsp8, and nsp12 in insect cells yielded a stable complex composed of nsp8 and nsp12. We demonstrate that this complex is active on model primer/template substrates that adequately mimic the elongation state. Most importantly, selectivity measurements determined here under the inherent limitations of the steady-state conditions revealed that incorporation of the inhibitor is more efficient than its natural counterpart, and delayed chain termination is observed at position i + 3.
Results
Expression of MERS–CoV RdRp complex
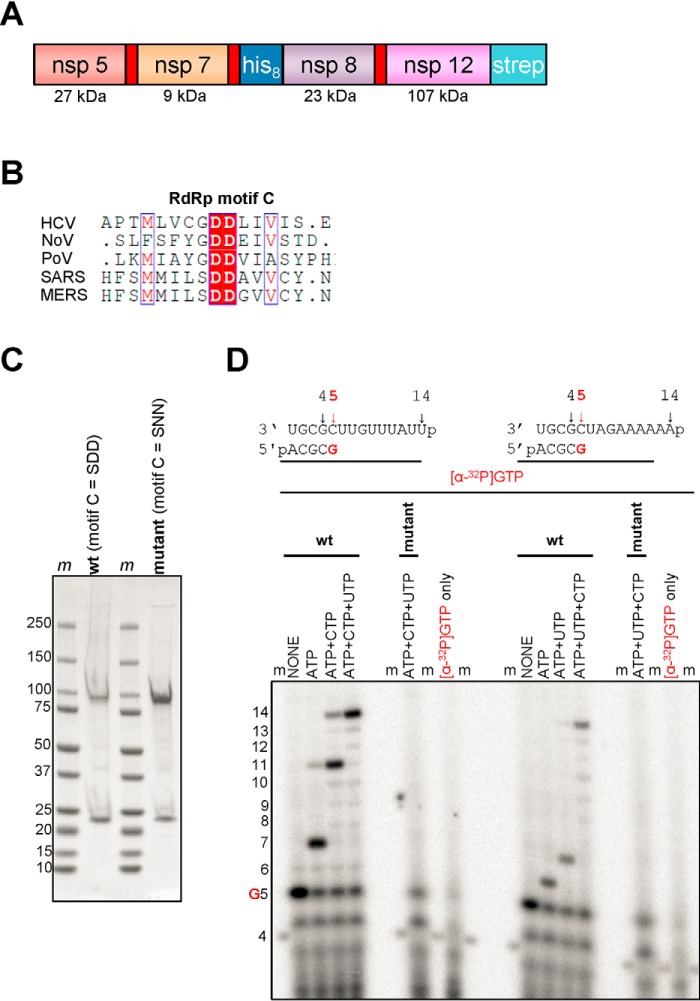
The baculovirus expression system has recently been used to produce recombinant nsp12 from SARS–CoV (13). For SARS–CoV, an active RdRp complex was reconstituted with purified nsp7 and nsp8, with and without a linker, expressed in Escherichia coli (13, 15). Here, we employed an alternative approach whereby MERS nsp5, nsp7, nsp8, and nsp12 were co-expressed in insect cells as a part a polyprotein (NCBI accession no. YP_009047202.1). The polyprotein was post-translationally cleaved by the nsp5 protease at its original cleavage sites (Fig. 1A). We also expressed a MERS–CoV RdRp complex in which the catalytic residues within the conserved motif C (SDD) of nsp12 were mutated (SNN) to generate an inactive RdRp (Fig. 1B) (16).
Figure 1.
Expression, purification, and characterization of the MERS RdRp complex. A, the construct contains nonstructural proteins nsp5, nsp7, nsp8, and nsp12. Red rectangles indicate original nsp5 protease cleavage sites. His8 and Strep indicate the locations of histidine and strep tags, respectively. B, a snapshot of a sequence alignment (T-Coffee) of representative RdRp enzymes from positive-sense RNA genome viruses illustrating sequence conservation within RdRp motif C. C, SDS-PAGE migration pattern of the purified enzyme preparations stained with Coomassie Brilliant Blue G-250 dye. Proteins migrating at ∼100 and ∼25 kDa contain nsp12 and nsp8, respectively. D, RNA synthesis on a short model primer/template substrate. Template and primer were both phosphorylated (p) at their 5′-ends. G indicates incorporation of the radiolabeled nucleotide opposite template position 5. RNA synthesis was monitored with the purified MERS RdRp complex wt (motif C = SDD) and active-site mutant (motif C = SNN) in the presence of NTP combinations designed to generate specific products. Lanes m illustrate the migration pattern of the radiolabeled 4-nucleotide-long primer. HCV, hepatitis C virus; NoV, norovirus; PoV, poliovirus.
Nickel–nitrilotriacetic acid affinity chromatography via the N-terminal eight-histidine tag of the nsp8 protein resulted in a MERS–CoV RdRp complex containing nsp8 (∼23 kDa) and nsp12 (∼110 kDa) (Fig. 1C). Mass spectroscopy confirmed the presence of nsp8 and nsp12, whereas there was no evidence for the presence of nsp7.
The WT MERS–CoV RdRp complex was tested for RNA synthesis on short model primer/template substrates mimicking a random elongation complex during RNA synthesis. We recently used the same model substrates for the study of other RdRp enzymes (Fig. 1D) (8, 10). The 4-nt primer/14-mer templates are designed such that [α-32P]GTP is the first incorporated nucleotide, which labels the RNA products. Reactions containing the MERS–CoV RdRp complex, primer/template, and various combinations of NTPs were initiated by the addition of Mg2+ ions. In the presence [α-32P]GTP alone, the expected 5-mer product is formed. The addition of specific combinations of NTPs generated defined reaction products: [α-32P]GTP and ATP yielded a 7-mer (Fig. 1D, left side of the gel) or a 6-mer product (Fig. 1D, right side of the gel), depending on the template sequence. Similarly, in the presence of [α-32P]GTP, ATP, and CTP (or UTP), the 4-mer primer is extended to yield an 11- or 7-mer depending on the template sequence. The addition of all four NTPs resulted in a 14-mer full-length product. Reactions with the SNN mutant enzyme did not show RNA product formation. The lane marked [α-32P]GTP only illustrates the background signal associated with the [α-32P]GTP preparation in the absence of enzyme. These data confirm that MERS–CoV nsp12 exhibits the observed RdRp activity. It has recently been reported that SARS–CoV nsp8 displays RNA primase activity that yields short (∼6-mer) reaction products (15, 17). However, structural data are inconsistent with the formation of a primase active site in SARS–CoV nsp8 (13), and our data do not provide any evidence for primase activity in MERS–CoV nsp8.
Inhibition of EBOV RdRp and MERS–CoV RdRp with RDV
For EBOV RdRp, it has been challenging to identify a sequence with a single site of incorporation for the RDV. Hence, we devised two different RNA templates that allow multiple and single incorporations, respectively, as shown in Fig. 2A. These sequences were used to compare the inhibitory effects of RDV on EBOV RdRp and MERS RdRp. For EBOV RdRp, we observed delayed chain termination at position i + 5 as previously described (Fig. 2B) (8). However, the template that provides a single site of RDV-TP incorporation also shows reductions in full-length RNA synthesis. In contrast, the MERS RdRp complex yields the full-length RNA product with both sequences. The inhibition patterns with RDV-TP differ markedly from our results with EBOV RdRp. RNA synthesis is arrested at positions i + 3 and i + 4 with a template that provides multiple sites of incorporation of the inhibitor, and the full-length product is only seen as a faint band. The template that allows only a single incorporation event yields RNA synthesis arrest at position i + 3 and an increased amount of the full-length product. Hence, the mechanism of inhibition is likely delayed chain termination for both EBOV RdRp and MERS RdRp, although the specific patterns show subtle differences. In the absence of inhibitor, RNA synthesis and full-length product formation is generally more efficient with MERS–CoV RdRp. This could also help to explain that a small amount of full-length product is still seen with this enzyme in the presence of inhibitor.
Figure 2.
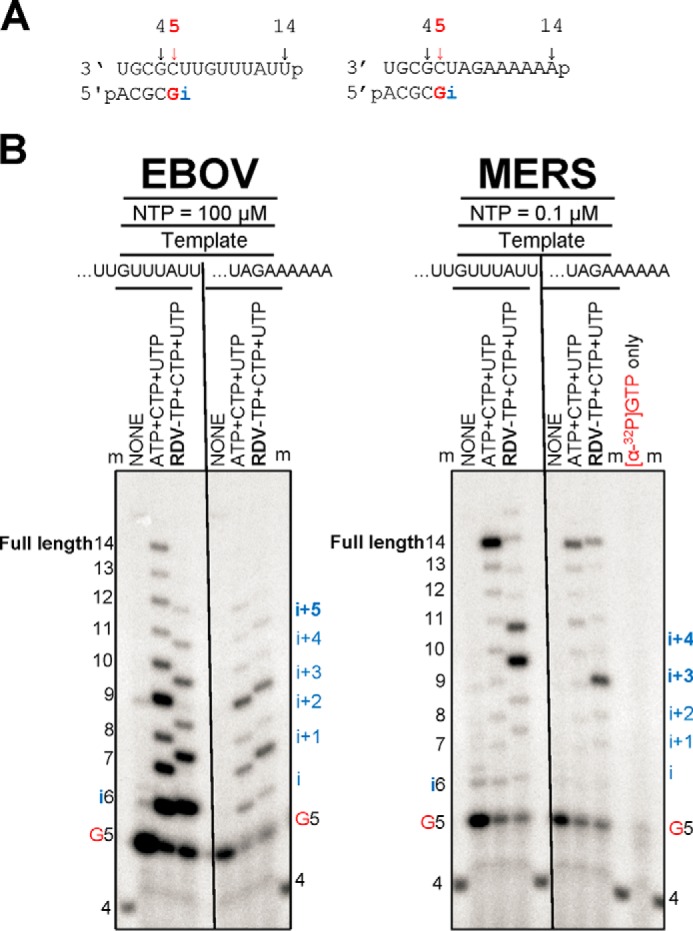
Patterns of inhibition of RNA synthesis with RDV-TP. A, RNA primer/template substrates used to test multiple (left panel) or single (right panel) incorporations of RDV-TP. G indicates incorporation of the radiolabeled nucleotide opposite template position 5. i indicates incorporation site for the first (left panel) or the only (right panel) RDV-TP. Full length indicates the full-template length products of RNA synthesis. B, RDV-TP incorporation was monitored with purified EBOV and MERS RdRp complexes in the presence of the indicated combinations of NTPs and RDV-TP.
Competitive inhibition of RNA synthesis by RDV-TP
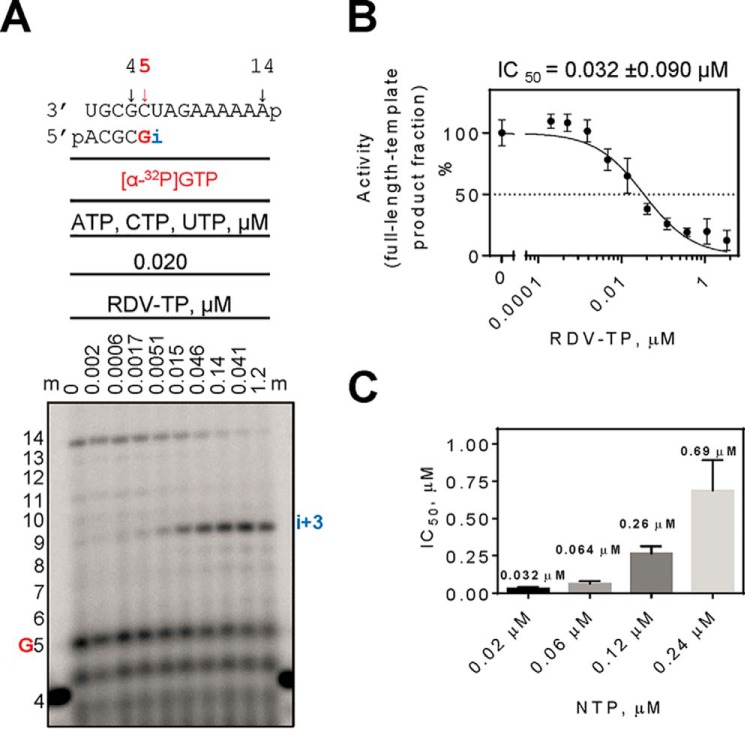
To study whether RDV-TP is able to compete with its natural counterpart ATP, we monitored RNA synthesis at a fixed concentration of NTPs (0.02 μm) and increasing concentrations of RDV-TP (Fig. 3A, top panel). Increasing concentrations of RDV-TP caused a reduction of the 14-mer product caused by increases in RNA synthesis arrest at position i + 3. Formation of the full-length 14-mer product is not evident at RDV-TP concentrations higher than 0.041 μm. Quantification of the data revealed an IC50 for RDV-TP of 0.032 μm under these conditions (Fig. 3B). This value is only 1.5-fold higher than the ATP concentration, which points to an efficient use of the inhibitor. Increasing concentrations of ATP caused a corresponding increase in IC50 values for RDV-TP (Fig. 3C), which shows that RDV-TP is a competitive inhibitor.
Figure 3.
Competition between RDV-TP and ATP. A, the RNA primer/template substrate used in this assay is shown above the gel. G indicates incorporation of the radiolabeled nucleotide opposite template position 5. Position i allows incorporation of ATP or RDV-TP. RNA synthesis was monitored with purified MERS RdRp complex in the presence of 0.02 μm ATP, CTP, and UTP mix and increasing concentrations of RDV-TP as indicated. B, graphic representation and IC50 determination fitting of quantified data from A. The error bars represent standard deviation of the data within four independent experiments. C, graphic representation of the relationship between IC50 values for RDV-TP measured at different NTP concentrations. The average IC50 values for RDV-TP are shown above the corresponding bars. The error bars represent standard deviation of the data within at least three independent experiments.
Selectivity measurements of ATP analogs
To translate our previous findings into quantitative terms, we determined Michaelis–Menten parameters Vmax and Km and calculated the efficiency Vmax/Km of nucleotide incorporation for ATP, RDV-TP, and two other nucleotide analog inhibitors for comparative purpose: ara-ATP and 2′CM-ATP (Fig. 4). ATP, or the ATP analog, was added at increasing concentrations, and the reactions were stopped after 20 min following the addition of Mg2+. This approach allowed us to determine the selectivity for nucleotide incorporation, defined as Vmax/Km (ATP) over Vmax/Km (nucleotide analog) (Table 1). The observed differences in the efficiency of nucleotide analog substrate utilization are driven solely by differences in the respective Km values. The data show an unexpectedly low selectivity values for RDV-TP (0.35-fold), whereas ara-ATP (749-fold), and 2′CM-ATP (165-fold) are both associated with high selectivity values. The selectivity value for RDV-TP below 1 suggests that incorporation of the inhibitor is more efficient as compared with the natural substrate. Commonly, the nucleotide analog is less efficiently incorporated, as seen with ara-ATP and 2′CM-ATP.
Figure 4.
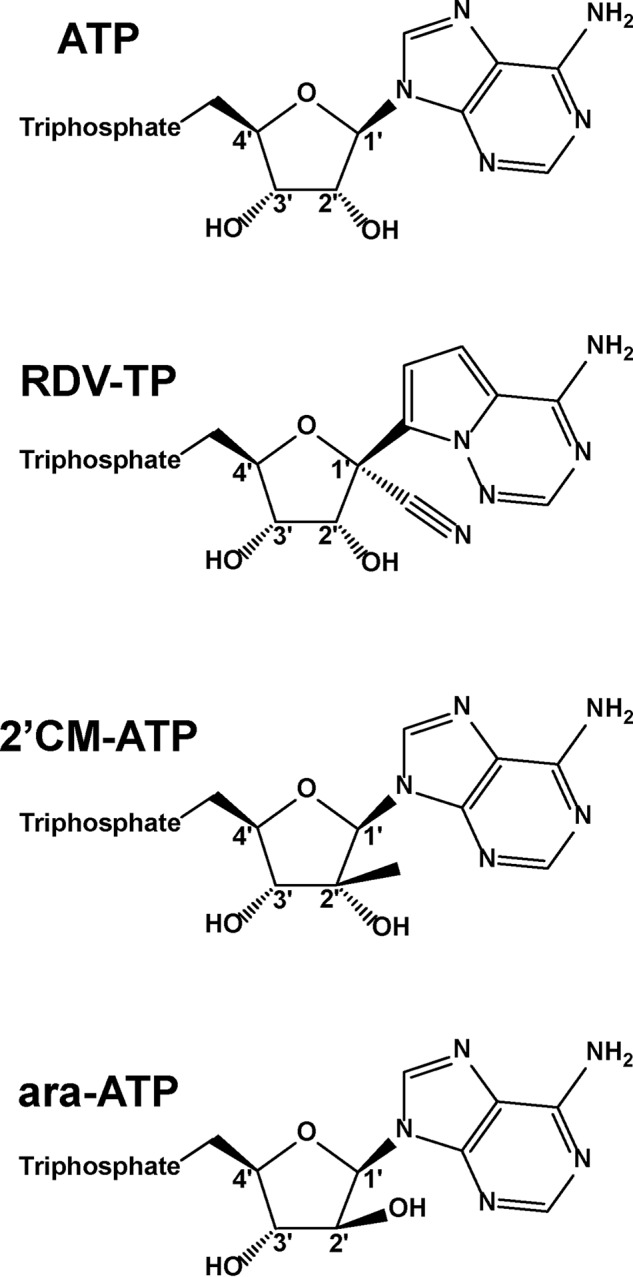
Chemical structures of ATP and ATP analogs.
Table 1.
MERS RdRp complex selectivity values for ATP analogs
| ATP (n = 7) | RDV-TP (n = 6) | Ara-ATP (n = 5) | 2′CM-ATP (n = 5) | |
|---|---|---|---|---|
| Vmax (product fraction)a | 0.47b ± 0.011c | 0.50 ± 0.012 | 0.48 ± 0.019 | 0.47 ± 0.021 |
| % error d | 2 | 2 | 4 | 4 |
| Km (μm)e | 0.017 ± 0.0019 | 0.0063 ± 0.00069 | 13 ± 1.7 | 2.8 ± 0.53 |
| % error | 11 | 11 | 13 | 19 |
| Vmax/Km | 28 | 79 | 0.037 | 0.17 |
| Selectivity (fold)f | 1d | 0.35 | 749 | 165 |
aVmax is a Michaelis–Menten parameter reflecting the maximal velocity of nucleotide incorporation.
bAll reported values have been calculated on the basis of a 9–data point experiment repeated the indicated number of times (n).
c Standard error associated with the fit.
d Reference.
e Km is a Michaelis–Menten parameter reflecting the concentration of the nucleotide substrate at which the velocity of nucleotide incorporation is half of Vmax.
f Selectivity of a viral RNA polymerase for a nucleotide substrate analog is calculated as the ratio of the Vmax/Km values for NTP and NTP analog, respectively.
Discussion
Broad spectrum antivirals or compounds with demonstrated activity against SARS–CoV or MERS–CoV are now considered for the treatment of infection caused by the novel coronavirus SARS–CoV-2. The nucleotide analog RDV has been tested in a randomized, controlled trial for Ebola virus disease (18). Although two other investigational therapies were more efficacious than RDV, an antiviral effect has been demonstrated. This compound also shows a broad spectrum of antiviral activities against coronaviruses in vitro and in animal models (1, 6, 19, 20). Moreover, screening of a compound library of Food and Drug Administration–approved drugs revealed that the HIV type 1 (HIV-1) protease inhibitor lopinavir is active against MERS–CoV (21). This drug is currently tested in a clinical trial for the treatment of MERS–CoV in combination with interferon-β and ritonavir (22), which is used for pharmacological boosting. A recent study compared the efficacy of the combination of lopinavir/ritonavir/interferon-β with RDV in cell culture and showed that RDV was significantly more active against MERS–CoV (1). Here we focused on the biochemical evaluation of RDV to provide a better understanding of its broad spectrum of antiviral activities and the underlying mechanism of action.
In vitro selection experiments with the coronavirus model murine hepatitis virus (5) resulted in two variants with resistance-conferring mutations in nsp12. These data provide strong evidence to show that RdRp is the target for RDV. Our biochemical data corroborate that the RNA polymerase of MERS–CoV is indeed the drug target. We demonstrate that MERS nsp8 and nsp12 form an active, binary complex. RDV-TP is utilized as a substrate and competes with its natural counterpart ATP. Natural nucleotide pools are commonly more efficiently incorporated when compared with nucleotide analogs. However, in this case, we observe that incorporation of the nucleotide analog is significantly more efficient.
Once added into the growing RNA chain, the inhibitor does not cause immediate chain termination. The presence of the 3′-hydroxyl group allows the addition of three more nucleotides until RNA synthesis is arrested at position i + 3. A fraction of the extended primer overcomes this arrest, which can lead to full-length product formation. Full-length product formation is considerably reduced when the inhibitor is added at consecutive sites. The efficient rate of RDV-TP incorporation may translate in multiple incorporation events in vivo, and this could add to the overall potency of the drug. It is important to note that other parameters including the intracellular concentration of the triphosphate form of the inhibitor need to be considered as well.
The different inhibition patterns observed with EBOV RdRp and MERS–CoV RdRp point to subtle differences in the mechanism of action. Delayed chain termination could be based on inhibitor-induced structural changes of the newly synthesized dsRNA that at some point prevent a productive alignment of primer and incoming nucleotide, e.g. through primer/template repositioning (23) or backtracking mechanisms (24). Enzyme-specific interactions with the extended primer may likewise affect the continued extension of the primer, which helps to explain variations in the site of RNA synthesis arrest.
An open question that warrants further investigation is the role of the 3′–5′ exonuclease (nsp14) in susceptibility to RDV. It has previously been reported that nucleotide analog inhibitors can be excised by the viral exonuclease (14). A murine hepatitis virus mutant lacking the 3′–5′ exonuclease activity was shown to be more sensitive to RDV (5). However, given its high potency in cell-based assays, RDV seems to be protected from excision at least to a certain degree. This protection could be provided by the additional three nucleotide following the inhibitor. In contrast, classic chain terminators would be readily accessible for excision. Taken together, this study provides a likely mechanism of action for RDV against coronaviruses. The novel strategy for the expression of MERS–CoV RdRp helps to guide the design of equivalent constructs of related viruses, including SARS–CoV-2.
Experimental procedures
Chemicals
All RNA primers and templates used in this study were 5′-phosphorylated and purchased from Dharmacon (Lafayette, CO). 2′C-methyl-ATP (2′CM-ATP) and RDV-TP were from Gilead Sciences (Foster City, CA). Ara-ATP was purchased from TriLink (San Diego, CA). NTPs were purchased from GE Healthcare. [α-32P]-GTP was purchased from PerkinElmer.
Protein expression and purification
The pFastBac-1 (Invitrogen) plasmid with the codon-optimized synthetic DNA sequences (GenScript, Piscataway, NJ) coding for a portion of MERS–CoV 1ab polyprotein (NCBI accession no. YP_009047202.1) containing only nsp5, nsp7, nsp8, and nsp12 was used as a starting material for protein expression in insect cells (Sf9; Invitrogen). We employed the MultiBac (Geneva Biotech, Indianapolis, IN) system for protein expression in insect cells (Sf9; Invitrogen) according to published protocols (25, 26). MERS–CoV protein complex was purified using nickel–nitrilotriacetic acid affinity chromatography of the nsp8 N-terminal eight-histidine tag according to the manufacturer's specifications (Thermo Scientific). The identities of the purified MERS–CoV proteins were confirmed by MS analysis (Dr. Jack Moore, Alberta Proteomics and Mass Spectrometry, Edmonton, Canada). Expression and purification of EBOV RdRp complex were performed as previously described (10).
RNA synthesis activity
Data acquisition and quantification were done as previously reported by us (8, 10). RNA synthesis assay consisted of mixing (final concentrations) Tris-HCl (pH 8, 25 mm), RNA primer (200 μm), RNA template (1 μm), [α-32P]NTP (0.1 μm), various concentrations and combinations of 100 μm (or as indicated) NTP and NTP analogs, and MERS–CoV RdRp complex (∼0.1 μm) on ice. Reaction mixtures (10 μl) were incubated for 10 min at 30 °C followed by the addition of 5 μl of MgCl2 (2.5 mm). The reactions were stopped after 30 min by the addition of 15 μl of formamide/EDTA (50 mm) mixture and incubated at 95 °C for 10 min. 3-μl reaction samples were subjected to denaturing 8 m urea 20% PAGE to resolve products of RNA synthesis followed by signal quantification (ImageQuant 5.2, GE Healthcare Bio-Sciences) through phosphorimaging (Typhoon TRIO variable mode imager; GE Healthcare Bio-Sciences). For nucleotide incorporation experiments, ATP or the ATP analog inhibitor was added at increasing concentrations, and the reactions were stopped at a fixed time point of 20 min following the addition of Mg2+. The reaction conditions were chosen such that the formation of the G5 product, i.e. the 4-mer primer extended with [α-32P]GTP, was linear with respect to time at the 20-min time point. The product fraction was determined from the total signal in G5 and i6 bands, plotted versus ATP or ATP analog substrate concentrations, and fitted to the Michaelis–Menten equation using GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA) with Vmax and Km parameters. Our assay involves incorporation of [α-32P]GTP into the primer. With this approach it is not possible to quantify the fraction of the extended primer; however, the reaction products are clearly defined by the correct incorporation of [α-32P]GTP. Potentially confounding by-products caused by misaligned primers are invisible. In case of IC50 experiments the full template-length product fractions were plotted versus RDV-TP concentrations and fitted to a log(inhibitor)–versus–normalized response − (variable slope) equation using GraphPad Prism 7.0 to determine the IC50 values for the inhibition of RNA synthesis by RDV-TP. The concentrations of nucleotide substrate (0.02, 0.06, 0.12, and 0.24 μm) in the competition assays were chosen around the Km value for ATP (0.02 μm; Table 1).
Author contributions
C. J. G., E. P. T., and M. G. formal analysis; C. J. G., E. P. T., and M. G. investigation; C. J. G., E. P. T., and M. G. methodology; E. P. T. and M. G. data curation; E. P. T. software; E. P. T. and M. G. validation; E. P. T. and M. G. visualization; E. P. T., J. Y. F., D.P.P., and M. G. writing-review and editing; M. G. conceptualization; M. G. resources; M. G. supervision; M. G. funding acquisition; M. G. writing-original draft; M. G. project administration.
Acknowledgments
We thank Emma Woolner for excellent technical assistance and Dr. Jack Moore at the Alberta Proteomics and Mass Spectrometry facility for MS analysis.
This study was supported by Canadian Institutes of Health Research Grant 159507 (to M. G.) and a grant from the Alberta Ministry of Economic Development, Trade and Tourism by the Major Innovation Fund Program for the AMR–One Health Consortium (to M. G.). M. G. has previously received funding from Gilead Sciences in support for the study of EBOV RdRp inhibition by RDV.
- SARS
- severe acute respiratory syndrome
- CoV
- coronavirus
- RdRp
- RNA-dependent RNA polymerase
- RDV
- remdesivir
- 2′CM
- 2′C-methyl
- MERS
- Middle East respiratory syndrome
- TP
- triphosphate form
- EBOV
- Ebola virus.
References
- 1. Sheahan T. P., Sims A. C., Leist S. R., Schäfer A., Won J., Brown A. J., Montgomery S. A., Hogg A., Babusis D., Clarke M. O., Spahn J. E., Bauer L., Sellers S., Porter D., Feng J. Y., et al. (2020) Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon β against MERS–CoV. Nat. Commun. 11, 222 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel D., Hui H. C., Doerffler E., Clarke M. O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., et al. (2017) Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 60, 1648–1661 10.1021/acs.jmedchem.6b01594 [DOI] [PubMed] [Google Scholar]
- 3. Lo M. K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A. L., Flint M., McMullan L. K., Siegel D., Clarke M. O., Mackman R. L., Hui H. C., Perron M., Ray A. S., Cihlar T., et al. (2017) GS-5734 and its parent nucleoside analog inhibit filo-, pneumo-, and paramyxoviruses. Sci. Rep. 7, 43395 10.1038/srep43395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warren T. K., Jordan R., Lo M. K., Ray A. S., Mackman R. L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H. C., Larson N., Strickley R., Wells J., Stuthman K. S., Van Tongeren S. A., et al. (2016) Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531, 381–385 10.1038/nature17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., et al. (2018) Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9, e00221–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown A. J., Won J. J., Graham R. L., Dinnon K. H. 3rd, Sims A. C., Feng J. Y., Cihlar T., Denison M. R., Baric R. S., and Sheahan T. P. (2019) Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 169, 104541 10.1016/j.antiviral.2019.104541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jordan P. C., Liu C., Raynaud P., Lo M. K., Spiropoulou C. F., Symons J. A., Beigelman L., and Deval J. (2018) Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 14, e1006889 10.1371/journal.ppat.1006889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tchesnokov E. P., Feng J. Y., Porter D. P., and Götte M. (2019) Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses 11, E326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mühlberger E., Weik M., Volchkov V. E., Klenk H. D., and Becker S. (1999) Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73, 2333–2342 10.1128/JVI.73.3.2333-2342.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tchesnokov E. P., Raeisimakiani P., Ngure M., Marchant D., and Götte M. (2018) Recombinant RNA-dependent RNA polymerase complex of Ebola virus. Sci. Rep. 8, 3970 10.1038/s41598-018-22328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deval J., Hong J., Wang G., Taylor J., Smith L. K., Fung A., Stevens S. K., Liu H., Jin Z., Dyatkina N., Prhavc M., Stoycheva A. D., Serebryany V., Liu J., Smith D. B., et al. (2015) Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2′-fluoro-4′-chloromethyl-cytidine triphosphate. PLoS Pathog. 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noton S. L., Deflubé L. R., Tremaglio C. Z., and Fearns R. (2012) The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 8, e1002980 10.1371/journal.ppat.1002980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirchdoerfer R. N., and Ward A. B. (2019) Structure of the SARS–CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferron F., Subissi L., Silveira De Morais A. T., Le N. T. T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., and Imbert I. (2018) Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 115, E162–E171 10.1073/pnas.1718806115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subissi L., Posthuma C. C., Collet A., Zevenhoven-Dobbe J. C., Gorbalenya A. E., Decroly E., Snijder E. J., Canard B., and Imbert I. (2014) One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 111, E3900–E3909 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poch O., Sauvaget I., Delarue M., and Tordo N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874 10.1002/j.1460-2075.1989.tb08565.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imbert I., Guillemot J. C., Bourhis J. M., Bussetta C., Coutard B., Egloff M. P., Ferron F., Gorbalenya A. E., and Canard B. (2006) A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25, 4933–4942 10.1038/sj.emboj.7601368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mulangu S., Dodd L. E., Davey R. T. Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., Ali R., Coulibaly S., Levine A. C., Grais R., Diaz J., et al. (2019) A randomized, controlled trial of Ebola virus disease therapeutics. New Engl. J. Med. 381, 2293–2303 10.1056/NEJMoa1910993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheahan T. P., Sims A. C., Graham R. L., Menachery V. D., Gralinski L. E., Case J. B., Leist S. R., Pyrc K., Feng J. Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M. O., Mackman R. L., et al. (2017) Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, eaal3653 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., and Feldmann H. (2020) Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS–CoV infection. Proc. Natl. Acad. Sci. U.S.A. 201922083 10.1073/pnas.1922083117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Wilde A. H., Jochmans D., Posthuma C. C., Zevenhoven-Dobbe J. C., van Nieuwkoop S., Bestebroer T. M., van den Hoogen B. G., Neyts J., and Snijder E. J. (2014) Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 58, 4875–4884 10.1128/AAC.03011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arabi Y. M., Asiri A. Y., Assiri A. M., Aziz Jokhdar H. A., Alothman A., Balkhy H. H., AlJohani S., Al Harbi S., Kojan S., Al Jeraisy M., Deeb A. M., Memish Z. A., Ghazal S., Al Faraj S., Al-Hameed F., et al. (2020) Treatment of Middle East respiratory syndrome with a combination of lopinavir/ritonavir and interferon-β1b (MIRACLE trial): statistical analysis plan for a recursive two-stage group sequential randomized controlled trial. Trials 21, 8 10.1186/s13063-019-3846-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tchesnokov E. P., Obikhod A., Schinazi R. F., and Götte M. (2008) Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 283, 34218–34228 10.1074/jbc.M806797200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dulin D., Arnold J. J., van Laar T., Oh H. S., Lee C., Perkins A. L., Harki D. A., Depken M., Cameron C. E., and Dekker N. H. (2017) Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Reports 21, 1063–1076 10.1016/j.celrep.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berger I., Fitzgerald D. J., and Richmond T. J. (2004) Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22, 1583–1587 10.1038/nbt1036 [DOI] [PubMed] [Google Scholar]
- 26. Bieniossek C., Richmond T. J., and Berger I. (2008) MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. Chapter 5, Unit 5.20 10.1002/0471140864.ps0520s51 [DOI] [PubMed] [Google Scholar]