Abstract
Mammalian skeletal muscles comprise different types of muscle fibers, and this muscle fiber heterogeneity is generally characterized by the expression of myosin heavy chain (MyHC) isoforms. A switch in MyHC expression leads to muscle fiber–type transition under various physiological and pathological conditions, but the underlying regulator coordinating the switch of MyHC expression remains largely unknown. Experiments reported in this study revealed the presence of a skeletal muscle–specific antisense transcript generated from the intergenic region between porcine MyHC IIa and IIx and is referred to here as MyHC IIA/X-AS. We found that MyHC IIA/X-AS is identified as a long noncoding RNA (lncRNA) that is strictly expressed in skeletal muscles and is predominantly distributed in the cytoplasm. Genetic analysis disclosed that MyHC IIA/X-AS stimulates cell cycle exit of skeletal satellite cells and their fusion into myotubes. Moreover, we observed that MyHC IIA/X-AS is more enriched in fast-twitch muscle and represses slow-type gene expression and thereby maintains the fast phenotype. Furthermore, we found that MyHC IIA/X-AS acts as a competing endogenous RNA that sponges microRNA-130b (miR-130b) and thereby maintains MyHC IIx expression and the fast fiber type. We also noted that miR-130b was proved to down-regulate MyHC IIx by directly targeting its 3′-UTR. Together, the results of our study uncovered a novel pathway, which revealed that lncRNA derived from the skeletal MyHC cluster could modulate local MyHC expression in trans, highlighting the role of lncRNAs in muscle fiber–type switching.
Keywords: long noncoding RNA (long ncRNA, lncRNA); myogenesis; muscle physiology; microRNA (miRNA); skeletal muscle; lncRNA; muscle fiber type; MyHC; myogenesis
Introduction
Skeletal muscles account for ∼40% of body weight. Apart from the regulation of locomotor activity, these muscles play important roles in energy expenditure, endocrine regulation, and metabolic hemostasis (1). Skeletal muscles are composed of heterogeneous fibers that are broadly classified into two types, oxidative-slow and glycolytic-fast, based upon their oxidative capacity and contractile properties. Adult skeletal muscle fibers are further categorized into I, IIA, IIX, and IIB, primarily expressing skeletal sarcomeric myosin heavy chain (MyHC)2 I, IIa, IIx, and IIb, respectively (2). In general, oxidative-slow myofibers are rich in MyHC I and/or MyHC IIa, whereas glycolytic-fast fibers are characterized by high-MyHC IIb expression. Moreover, IIx myofibers represent the metabolic intermediate type between IIa and IIb myofibers (3).
The compositions of skeletal muscle fiber types undergo highly-dynamic remodeling in response to various functional and metabolic demands in accordance with the expression of MyHC isoforms (4–6). Genes coding mammalian MyHC isoforms are mainly found in two groups, namely cardiac cluster (including MyHC β (or type I) and α) and skeletal cluster (embryonic, IIa, IIx, IIb, and neonatal). Moreover, the switching of adult fast MyHC (IIa, IIx, and IIb) expression during transitions of fiber types occurs in a way similar to their genomic order, and this is normally observed in mammals (3).
In rats, previous studies have suggested that antisense transcripts derived from bidirectional promoters are involved in controlling the coordinated regulation of adjacent MyHC expression, both in heart (8) and skeletal muscles (9, 10). Lnc-mg that partially overlapped with mouse MyHC IIx enhanced myogenesis through the regulation of levels of insulin-like growth factor 2 (IGF2) (11). Another antisense long noncoding RNA (lncRNA) linc-MYH, which is located 50 kb upstream of the mouse MyHC IIa gene, was reported to repress the expression of slow-fiber genes and lock the fast-fiber type in mice (12).
An antisense lncRNA (GenBank accession no. NR_125367) was also observed across human skeletal MyHC cluster. However, information is not yet available about the physiological functions of the antisense transcripts generated from the skeletal MyHC cluster. Therefore, the aim of this study was to identify an antisense lncRNA in the intergenic region of porcine MyHC IIa and IIx that is referred to as MyHC IIA/X-AS. This study has shown that MyHC IIA/X-AS acts as a ceRNA to sponge miR-130b that serves to regulate the MyHC IIx expression and fast myofiber phenotype.
Results
Identification and characterization of MyHC IIA/X-AS in skeletal muscle of pigs
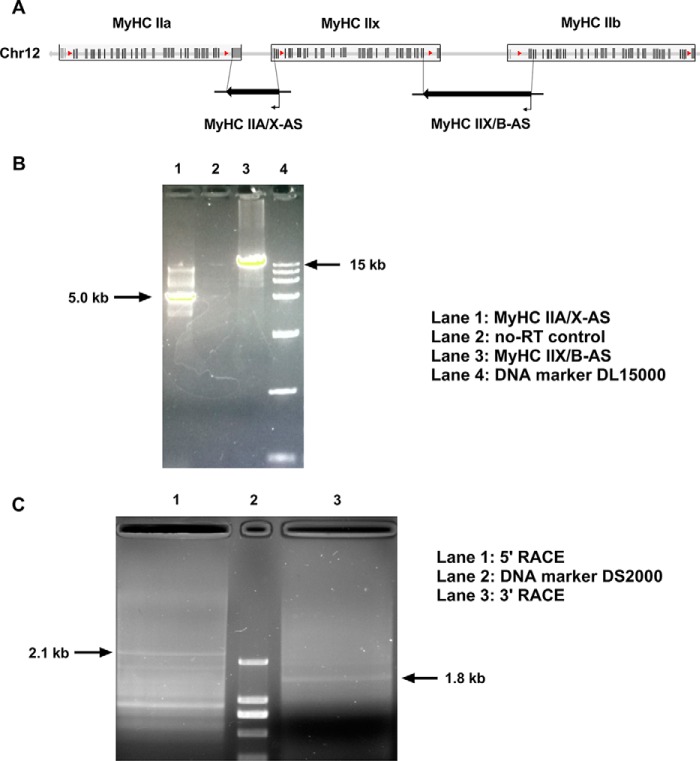
To detect the expression of antisense transcripts within the porcine MyHC II cluster (Fig. 1A), the chain-specific reverse transcription and long fragment PCR amplification were performed using primers overlapping the intergenic regions. The primer sequences are listed in Table S1. In agarose gel electrophoresis, PCR products corresponding to ∼5 and ∼15 kb (Fig. 1B) were consistent with the distance of corresponding primers on the swine genome. This indicated that antisense transcripts were expressed within the intergenic regions of type II MyHCs, hence respectively named as MyHC IIA/X-AS and MyHC IIX/B-AS, on the basis of their positions. To verify whether MyHC IIA/X-AS and MyHC IIX/B-AS were unique transcripts or not, 5′ and 3′ RACE experiments were conducted to identify the 5′ and 3′ ends of MyHC IIA/X-AS. In agarose gel electrophoresis, bands corresponding to the sizes of 2.1 kb 5′ and 1.8 kb 3′ were detected (Fig. 1C), suggesting that MyHC IIA/X-AS and MyHC IIX/B-AS were independent transcripts, and the full size of MyHC IIA/X-AS is about 5 kb, consistent with the results of long fragment PCR (Fig. 1B).
Figure 1.
Identification of the expression of MyHC IIA/X-AS in skeletal muscle of pigs. A, schematic diagram of porcine fast MyHC cluster. B, expression of MyHC IIA/X-AS and MyHC II X/B-AS in Longissimus dorsi of pigs was estimated through chain-specific and long-fragment RT-PCR. C, 5′- and 3′-RACE of MyHC IIA/X-AS were performed using RACE kits.
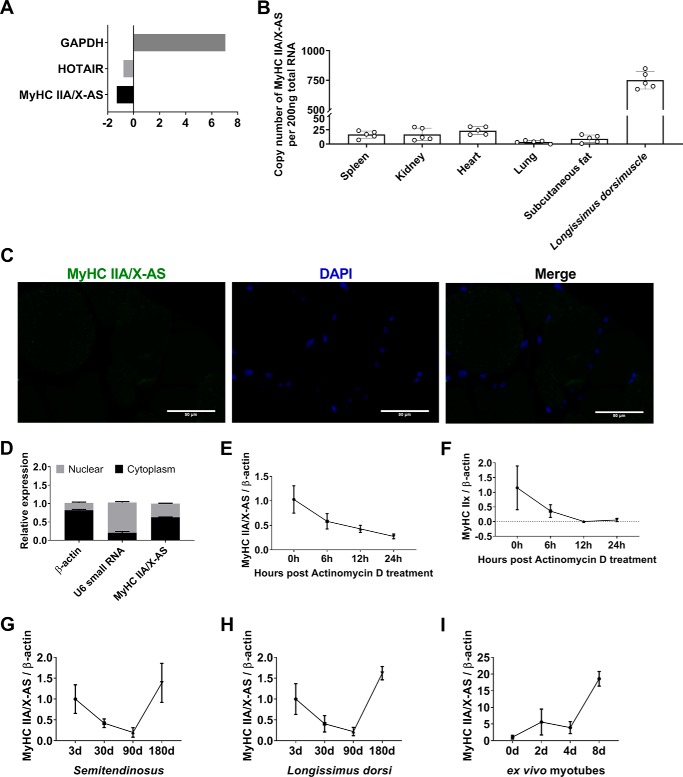
Furthermore, the analysis through an on-line coding potential calculator (CPC) showed no coding potential for MyHC IIA/X-AS using glyceraldehyde-3-phosphate dehydrogenase as the positive control for coding genes and HOTAIR, a well-documented lncRNA (13), as the noncoding control (Fig. 2A). This was also confirmed through the use of another bioinformatics tool, namely the coding potential assessment tool (data not shown). Therefore, these results have shown MyHC IIA/X-AS to be identified as a lncRNA.
Figure 2.
Molecular characteristics of porcine MyHC IIA/X-AS. A, in silico analysis of the coding potential of porcine MyHC IIA/X-AS using CPC. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, expression pattern of MyHC IIA/X-AS in various tissues in 180-day-old pigs was detected through qRT-PCR (n = 5). C, subcellular localization of MyHC IIA/X-AS in longissimus dorsi muscles in 180-day-old pigs was determined through RNA–FISH (n = 3). Scale bar = 50 μm. D, subcellular distribution of MyHC IIA/X-AS in cultured satellite cells was estimated through nucleocytoplasmic separation and qRT-PCR (n = 3). E, half-life of MyHC IIA/X-AS in ex vivo differentiated myotubes was analyzed through qRT-PCR after actinomycin D treatment (n = 3). F, half-life of MyHC IIx in ex vivo differentiated myotubes was estimated through qRT-PCR (n = 3). G, expression pattern of MyHC IIA/X-AS in postnatal semitendinosus was determined through qRT-PCR (n = 5). H, expression pattern of MyHC IIA/X-AS in postnatal longissimus dorsi was detected by qRT-PCR (n = 5). I, expression pattern of MyHC IIA/X-AS during in vitro myogenic differentiation was evaluated through qRT-PCR (n = 3).
RT-qPCR analyses have shown that lncRNA was predominantly expressed in skeletal muscles in 180-day-old pigs (Fig. 2B) and was mainly distributed in the cytoplasm of skeletal myofibers as revealed through RNA–FISH analysis (Fig. 2C). Cellular fractionation also showed high expression of MyHC IIA/X-AS in the cytoplasm of ex vivo cultured myotubes (Fig. 2D). Actinomycin D treatment showed that the half-life of MyHC IIA/X-AS was about 6 h (Fig. 2E), which was longer than the half-life of its neighbor gene, MyHC IIx (Fig. 2F), indicating MyHC IIA/X-AS to be more stable as compared with its neighbor coding genes.
During postnatal skeletal muscle development, the expression patterns of MyHC IIA/X-AS in both semitendinosus (Fig. 2G) and longissimus dorsi (Fig. 2H) were similar and down-regulated in 90-day-old pigs and peaked in 180-day-old pigs. In ex vivo cultured myotubes, the expression of MyHC IIA/X-AS gradually increased during myogenic differentiation (Fig. 2I). Interestingly, it is worth noting that the expression profiles of MyHC IIA/X-AS in postnatal skeletal muscles and cultured myotubes are similar to that of MyHC IIx (Fig. S1), although the correlation was not observed to be significant due to a limited number of samples.
Knockdown of MyHC IIA/X-AS facilitated the proliferation of skeletal satellite cells
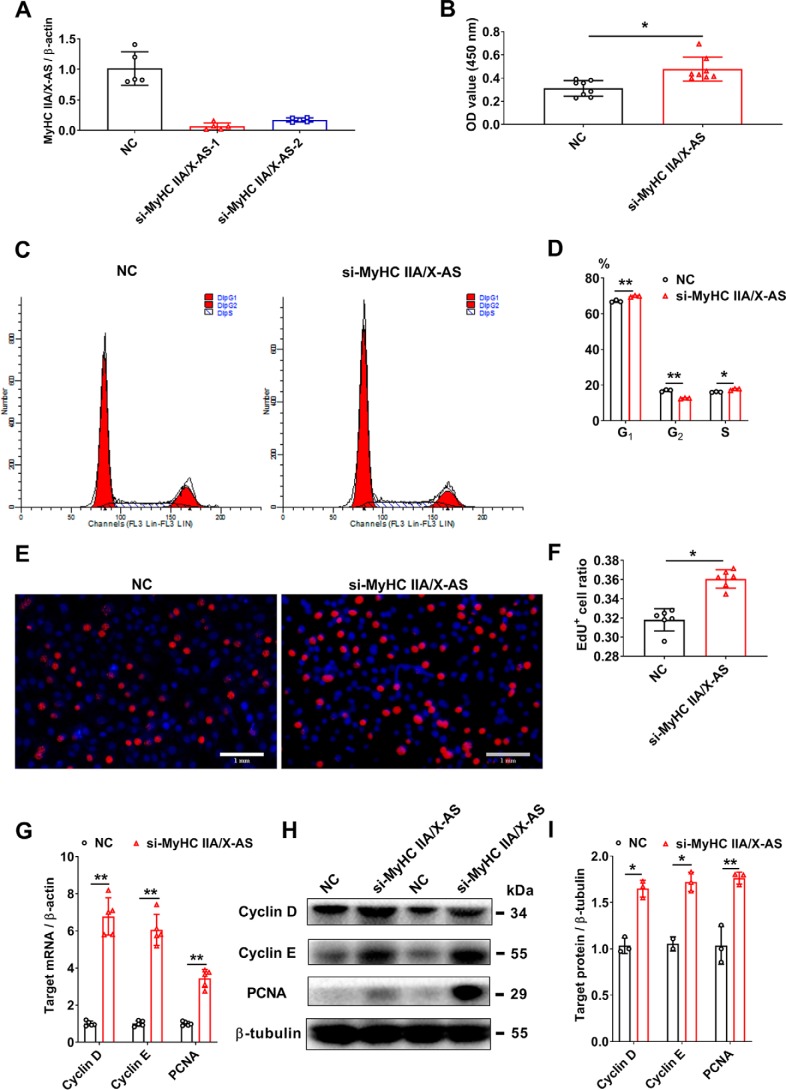
Skeletal satellite cells are resident stem cells that can rapidly enter the cell cycle and fuse with myofibers upon the release of a stimuli. To assess the effect of MyHC IIA/X-AS on satellite cells, two siRNAs were designed to silence the expression of MyHC IIA/X-AS. The si-MyHC IIA/X-AS-1 was used in further experiments due to its higher knockdown efficiency (Fig. 3A). The CCK-8 staining was significantly elevated after 48 h of transfection of si-MyHC IIA/X-AS (Fig. 3B). A higher ratio of G1- and S-phase cells as revealed through flow cytometry analysis indicated that a greater number of cells entered the cell cycle (Fig. 3, C and D). Consistently, more EdU-positive cells were observed 48 h after si-MyHC IIA/X-AS transfection (Fig. 3, E and F). Accordingly, both the mRNA (Fig. 3G) and protein (Fig. 3, H and I) levels of cyclin D, cyclin E, and PCNA were elevated by si-MyHC IIA/X-AS.
Figure 3.
Knockdown of MyHC IIA/X-AS promotes the proliferation of porcine skeletal satellite cells. A, knockdown efficiency of siRNAs targeting MyHC IIA/X-AS was detected through qRT-PCR (n = 5). B, cell numbers were tested by CCK-8 (n = 7). C and D, cell cycles were detected through flow cytometry 48 h post-transfection with MyHC IIA/X-AS siRNA or negative control (NC). n = 3. E and F, DNA duplication was by EdU staining 48 h post-transfection with MyHC IIA/X-AS siRNA or negative control. n = 7. Scale bar = 1 mm. G, mRNA expression levels of cell cycle–related genes were detected through qRT-PCR 48 h post-transfection (n = 5). H and I, protein levels of proliferative proteins were tested by Western blotting 48 h post-transfection (n = 3). *, p < 0.05; **, p < 0.01.
Knockdown of MyHC IIA/X-AS impedes skeletal myogenic differentiation and drives fast–to–slow switch
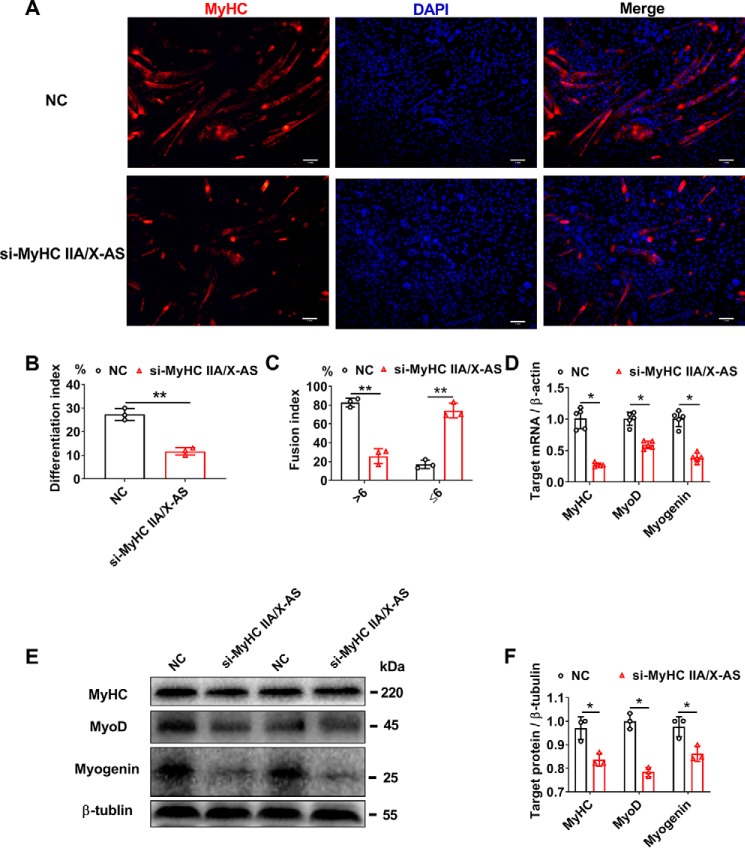
Because of the increasing tendency of MyHC IIA/X-AS during in vitro myogenic differentiation, siRNA was used to evaluate its effects on the myogenic process. The proliferating satellite cells were transfected with si-MyHC IIA/X-AS at 70–80% density, induced into myogenic differentiation until a complete confluence was reached, and then subjected to various assays 6 days post differentiation. Immunofluorescence assay revealed that the ratio of MyHC-positive myotubes was dramatically reduced by si-MyHC IIA/X-AS as indicated by the fusion index and differentiation index (Fig. 4, A–C). Meanwhile, the mRNA (Fig. 4D) and protein (Fig. 4, E and F) expression of MyHC, MyoD, and myogenin, the markers for myogenesis, were also observed to be down-regulated.
Figure 4.
Knockdown of MyHC IIA/X-AS impairs the myogenic differentiation in porcine skeletal satellite cells. A, representative immunofluorescence results for MyHC-positive myotubes 6 days post-myogenic differentiation upon transfection with either MyHC IIA/X-AS siRNA or negative control. Scale bar = 1 mm. n = 3. B, statistical results of differentiation index. The differentiation index was calculated as the percentage of MyHC-positive nuclei among total nuclei (n = 3). C, statistical results of myotube fusion index (n = 3). Myotube fusion index was determined as the distribution of nucleus number in total myotubes. D, mRNA levels of myogenic-related genes were detected through qRT-PCR in myotubes 6 days post-myogenic differentiation (n = 5). E and F, protein levels of myogenic-related genes were detected by Western blotting in myotubes 6 days post-myogenic differentiation (n = 3). NC, negative control. *, p < 0.05; **, p < 0.01.
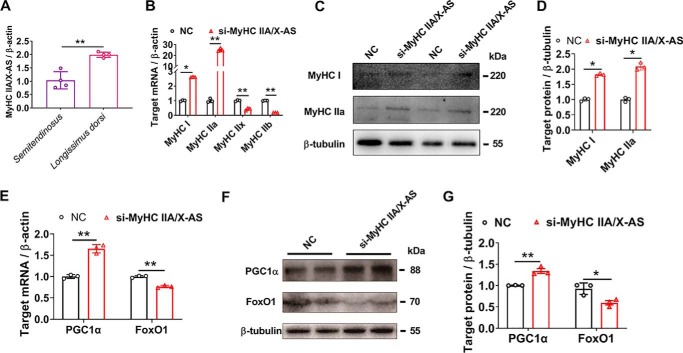
Interestingly, the abundance of MyHC IIA/X-AS was much higher in fast longissimus dorsi as compared with slow semitendinosus (Fig. 5A), indicating a potential function of MyHC IIA/X-AS in maintenance of the fast fiber type. Furthermore, MyHC IIA/X-AS silencing significantly enforced the expression of MyHC I and IIa, and it repressed the expression of MyHC IIx and IIb, indicating a fast–to–slow switch (Fig. 5B). The elevated MyHC I and IIa were further confirmed through Western blot analysis (Fig. 5, C and D). The repressed MyHC IIx proteins are represented in Fig. 9I. However, we failed to detect the changes in MyHC IIb expression at the protein level due to technical difficulties. PGC-1α is a powerful driver for mitochondrial biogenesis in slow-twitch muscle fibers (14), whereas FoxO1 negatively regulates slow-fiber genes (15, 16). In accordance with the expression of MyHC isoforms, PGC-1α was observed to be increased, whereas FoxO1 was observed to be decreased at mRNA (Fig. 5E) and protein (Fig. 5, F and G) levels after si-MyHC IIA/X-AS transfection.
Figure 5.
Reduced MyHC IIA/X-AS induces a fast–to–slow switch. A, expression of MyHC IIA/X-AS in different muscles of 180-day-old pigs was tested through qRT-PCR (n = 5). B, mRNA levels of MyHC isoforms were detected through qRT-PCR in myotubes 6 days post-myogenic differentiation transfected with either MyHC IIA/X-AS siRNA or negative control (NC) (n = 3). C and D, changes of MyHC I and IIa at protein levels were estimated through Western blotting (n = 3). E, mRNA levels of muscle fiber marker genes, PGC-1α and FoxO1, were detected through qRT-PCR in myotubes 6 days post-myogenic differentiation. The cells were transfected with either MyHC IIA/X-AS siRNA or negative control (n = 3). F and G, protein levels of PGC-1α and FoxO1 were determined by Western blotting (n = 3). *, p < 0.05; **, p < 0.01.
Figure 9.
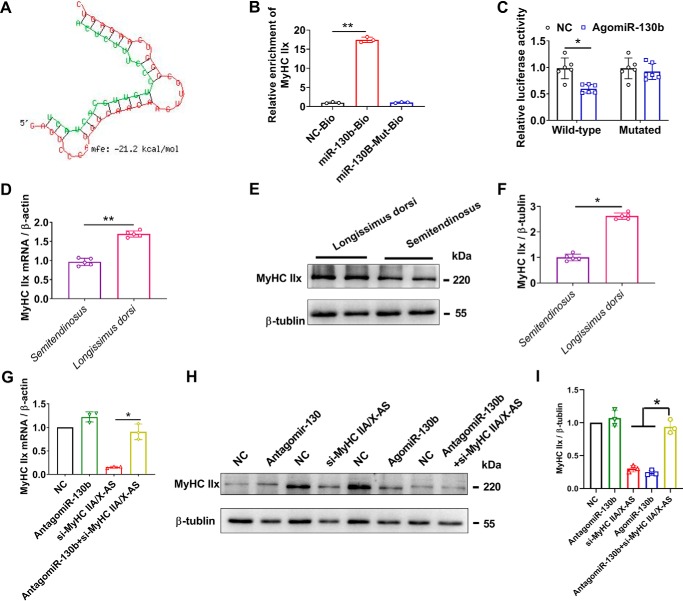
miR-130b targets on the 3′-UTR of MyHC IIx. A, putative binding site of miR-130b on 3′-UTR of MyHC IIx was predicted by RNA-hybrid tool. B, enrichment of MyHC IIx mRNA immunoprecipitated by biotin-labeled miR-130b or miR-130b–Mut probes was determined RNA hybrid qRT-PCR (n = 3). C, statistical results of dual-luciferase reporter assay (n = 5). D, levels of MyHC IIx mRNA in different skeletal muscles of 180-day-old pigs was estimated through qRT-PCR (n = 5). E and F, expression of MyHC IIx at protein levels in different skeletal muscles of 180-day-old pigs was detected by Western blotting (n = 5). G, reduced MyHC IIx was restored by miR-130b inhibitors at mRNA levels (n = 3). H and I, repressed MyHC IIx was rescued by miR-130b inhibitors at the protein level (n = 3). NC, negative control. *, p < 0.05; **, p < 0.01.
MyHC IIA/X-AS acts as a ceRNA sponging miR-130b, a novel regulator of myogenesis
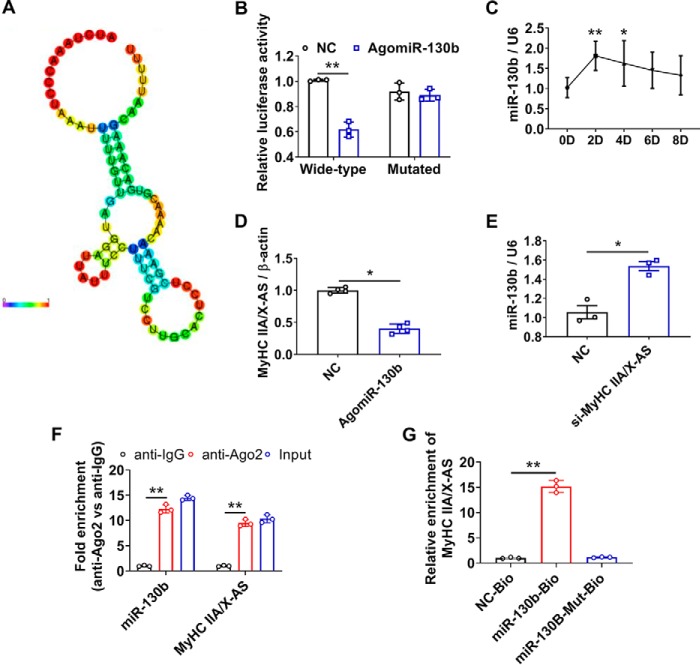
Considering that cytoplasmic lncRNAs always modulate gene expression through sponging miRNAs, we moved planned to identify miRNAs that could bind to MyHC IIA/X-AS. Using the on-line software miRDB (RRID:SCR_010848), a putative binding site of miR-130b in MyHC IIA/X-AS (Fig. 6A) was predicted, and the direct interaction between miR-130b and MyHC IIA/X-AS was further confirmed through dual-luciferase reporter assay (Fig. 6B). The expression of miR-130b rapidly increased upon myogenic stimuli and then gradually decreased during myogenic differentiation (Fig. 6C). Overexpression of miR-130b significantly reduced the levels of MyHC IIA/X-AS (Fig. 6D), and knockdown of MyHC IIA/X-AS slightly but significantly increased the levels of miR-130b (Fig. 6E). Apart from this, significantly enriched miR-130b and MyHC IIA/X-AS were measured through Ago2 RIP assay in combination with qRT-PCR (Fig. 6F). Moreover, MyHC IIA/X-AS was significantly enriched by biotin-labeled miR-130b as compared with the control or mutated miR-130b (Fig. 6G).
Figure 6.
MyHC IIA/X-AS sponges miR-130b as a ceRNA. A, potential miR-130b–binding site in MyHC IIA/X-AS was predicted using RegRNA2.0. B, statistical results of Dual-Luciferase reporter assay (n = 3). C, expression profile of miR-130b during in vitro myogenic differentiation was tested through qRT-PCR (n = 3). D, changes of MyHC IIA/X-AS upon exogenous overexpression of miR-130b were estimated through qRT-PCR in myotubes 6 days post-myogenic differentiation (n = 5). E, expression of miR-130b was significantly altered in myotubes transfected with MyHC IIA//X-AS siRNA (n = 3). F, enrichment of MyHC IIA/X-AS and miR-130b was determined through qRT-PCR in Ago2-RIP assay (n = 3). G, level of MyHC IIA/X-AS mRNA immunoprecipitated by biotin-labeled miR-130b or miR-130b–Mut probes was detected through qRT-PCR (n = 3). NC, negative control. *, p < 0.05; **, p < 0.01.
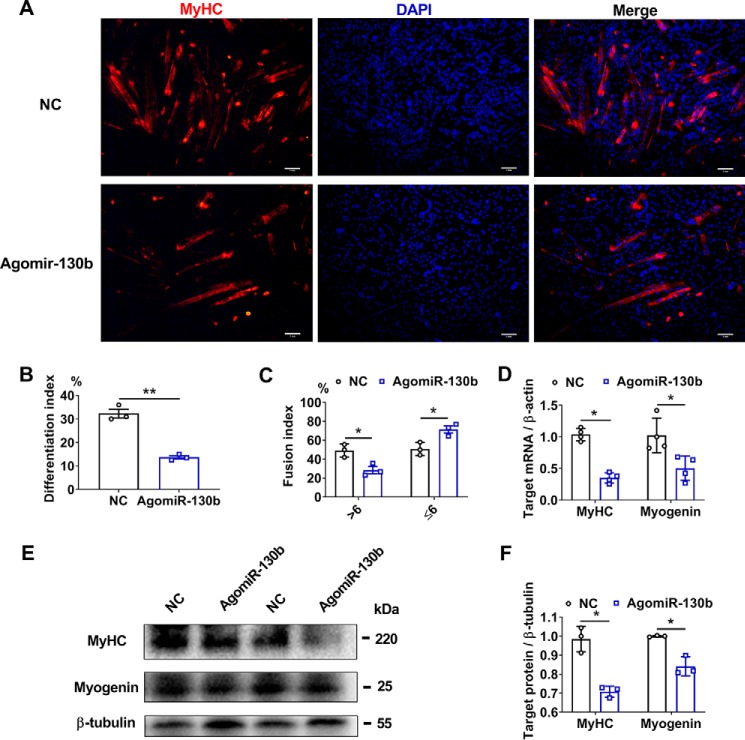
Because of the lack of information about miR-130b in skeletal muscles, we decided to explore the role of miR-130b in skeletal satellite cells. An enhanced miR-130b expression significantly repressed the fusion and formation of MyHC-positive myotubes (Fig. 7, A–C). Meanwhile, the expression levels of myogenic markers, MyHC and myogenin, were also restrained at both mRNA (Fig. 7D) and protein (Fig. 7, E and F) levels.
Figure 7.
Overexpression of miR-130b impedes the myogenic differentiation in porcine skeletal satellite cells. A, representative immunofluorescence staining of MyHC-positive myotubes 6 days post-myogenic differentiation. The cells were transfected with miR-130b agomir or negative control (NC) (n = 3). Scale bar = 1 mm. B, statistical results of differentiation index (n = 3). C, statistical results of fusion index (n = 3). D, changes of myogenic-related genes at mRNA levels after AgomiR-130b transfection were detected through qRT-PCR. Cells were harvested 6 days post-myogenic differentiation (n = 5). E and F, protein levels of myogenic factors were detected by Western blotting 6 days post-myogenic differentiation (n = 3). *, p < 0.05; **, p < 0.01.
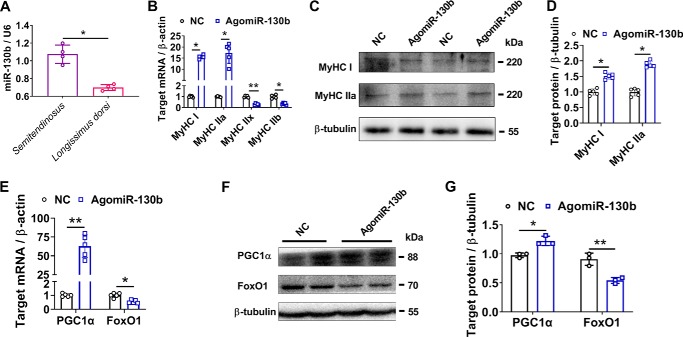
Moreover, in contrast to MyHC IIA/X-AS (Fig. 5A), miR-130b was highly enriched in slow semitendinosus than in fast longissimus dorsi (Fig. 8A). Consistent with the results obtained from MyHC IIA/X-AS siRNA, overexpression of miR-130b also resulted in a fast–to–slow switch that was supported by the expression of MyHC isoforms, PGC1α and FoxO1 (Fig. 8, B–G).
Figure 8.
Enforced overexpression of miR-130b drives a fast–to–slow fiber phenotype. A, expression pattern of miR-130b in skeletal muscles of 180-day-old pigs was analyzed through qRT-PCR (n = 5). B, changes of MyHC isoforms at mRNA levels were detected through qRT-PCR in myotubes upon AgomiR-130b transfection 6 days post-myogenic differentiation (n = 5). C and D, changes of MyHC I and IIa at protein levels upon AgomiR-130b transfection were determined by Western blotting in myotubes 6 days post-myogenic differentiation (n = 3). E, mRNA levels of muscle fiber marker genes, PGC-1α and FoxO1, were detected through qRT-PCR in myotubes 6 days post-myogenic differentiation. The cells were transfected with either miR-130b agomir or negative control (NC) (n = 5). F and G, protein levels of PGC-1α and FoxO1 were evaluated by Western blotting (n = 3). *, p < 0.05; **, p < 0.01.
The on-line tool RNAhybrid was used to verify the target genes of miR-130b, and in silico analysis suggested that MyHC IIx, which is one of the genes reduced by Agomir-130b (Fig. 8B), might be a potential target of miR-130b in skeletal muscles (Fig. 9A). Moreover, MyHC IIx mRNA (primers for 3′-UTR) was significantly enriched by biotin-labeled miR-130b not mutated miR-130b (Fig. 9B). AgomiR-130b could significantly reduce the luciferase of psiCHECKTM-2 constructs containing wild-type 3′-UTR of MyHC IIx instead of the mutated constructs (Fig. 9C), confirming the direct binding of miR-130b and 3′-UTR of MyHC IIx. Moreover, the expression of MyHC IIx was significantly higher in longissimus dorsi as compared with semitendinosus muscles at both mRNA (Fig. 9D) and protein (Fig. 9, E and F) levels, in contrast to with that of miR-130b (Fig. 8A) and consistent with that of MyHC IIA/X-AS (Fig. 5A). Although knockdown of miR-130b by Antagomir-130 had little effect on the MyHC IIx levels. Antagomir-130 could significantly rescue the down-regulated MyHC IIx that was induced by si-MyHC IIA/X-AS at both mRNA (Fig. 9G) and protein levels (Fig. 9, H and I).
Discussion
MyHCs, encoding the molecular motor that impacts contractile velocity and hence muscle performance, are considered as the markers of myofiber types. The organization of skeletal MyHC cluster in pig, human, and mouse is highly conserved as having a similar head–to–tail gene order and intergenic distance (17, 18). Interestingly, the genomic organization of skeletal fast MyHC isoforms (IIA, IIX, and IIB) is similar to the order of increasing ATPase activity (19) during myofiber type switch. This led us to suspect that there might be some regulatory pathways to mediate neighbor MyHCs expression.
Previous studies in rats (9, 10) have suggested that the antisense transcripts might be involved in coordinating the ordinal expression of fast MyHC cluster during fiber type switch. In mice, another antisense lncRNA, adjacent to the fast MyHC cluster, is involved in fast myofiber specialization (12). In this study, the chain-specific and long-fragment RT-PCR in combination with RACE identified an ∼5-kb antisense transcript overlapping the intergenic regions of porcine MyHC IIa and IIx, and hence was named MyHC IIA/X-AS, similar to those observed in rats (9). Furthermore, this study has shown that MyHC IIA/X-AS was an lncRNA.
The specific expression of lncRNA MyHC IIA/X-AS in skeletal muscles indicates its possible role in muscle development. RT-qPCR assay has shown that the expression of MyHC IIA/X-AS was dynamically changed in postnatal longissimus dorsi and semitendinosus muscles. An increasing tendency was observed during the ex vivo myogenic differentiation in porcine satellite cells. These results strongly suggest that MyHC IIA/X-AS might be a critical regulator of skeletal muscle development.
Given the increasing pattern of lncRNA MyHC IIA/X-AS during myogenesis, we decided to explore its function by employing a knockdown strategy. MyHC IIA/X-AS siRNA significantly facilitated the cell cycle of porcine satellite cells and dramatically repressed the myogenic process, indicating that MyHC IIA/X-AS might accelerate satellite cells to exit the cell cycle and thus differentiate into myofibers. The pro-myogenesis role of lncRNA derived from the MyHC cluster was also observed in mice (11). However, MyHC IIA/X-AS was observed to be enriched more in fast longissimus dorsi muscle. Furthermore, the knockdown of MyHC IIA/X-AS significantly up-regulated slow-type genes and down-regulated fast-type gene expression, indicating an attractive role of MyHC IIA/X-AS to maintain myofiber phenotype.
RNA–FISH and cellular fractionation indicated that MyHC IIA/X-AS was predominantly located in the cytoplasm, and cytoplasmic lncRNAs were well-demonstrated to sponge miRNA as ceRNA (20). In silico assay predicted several miRNA-binding sites within MyHC IIA/X-AS, including miR-130b. Dual-luciferase assay of Ago2-RIP combined with miR-130b pulldown confirmed the direct binding of miR-130b to MyHC IIA/X-AS. miR-130b was initially revealed to be correlated with pork quality in a previous microarray assay (21), and the follow-up study revealed that miR-130b was one of cellular ATP regulators by targeting mitochondrial energy metabolism during myogenic differentiation of the C2C12 cell line (22). In this study, the overexpression of miR-130b impaired the myogenic process in porcine satellite cells, consistent with a previous study in chicken primary myoblasts (23). Moreover, in this study, miR-130b was highly expressed in slow semitendinosus, and enforced miR-130b significantly elevated slow-type gene expression, indicating an adverse effect as compared with that of MyHC IIA/X-AS. Therefore, miR-130b might be involved in energy metabolism to maintain the phenotype of myofiber type.
In this study, MyHC IIx (or MYH1) was demonstrated to be the target of miR-130b through miR-130b pulldown and dual-luciferase assay. MyHC IIx is the intermediator between MyHC IIa and IIb, contributing to the fast-fiber phenotype by glycolytic metabolism. In this study, MyHC IIx was highly expressed in fast longissimus dorsi rather than slow semitendinosus. Overexpression of miR-130b and knockdown of MyHC IIA/X-AS both could restrain the expression of MyHC IIx, and miR-130b inhibitor could powerfully restore the repressed MyHC IIx induced by MyHC IIA/X-AS siRNA, supporting a MyHC IIA/X-AS/miR-130b/MyHC IIX axis in skeletal myofibers in pigs.
As described, the genomic organization of skeletal MyHC cluster is highly conserved in mammals (19); however, lncRNAs generated from the skeletal MyHC cluster depend upon various species. In rats, antisense transcripts are derived from the bidirectional promoter, overlapped with the sense MyHC, and involved in fiber type switching possibly by repressing sense MyHC expression (9, 10). In mice, linc-MYH, upstream of the skeletal fast MyHC cluster, coordinates fast MyHC expression sharing a common enhancer (12). Instead, lnc-mg is transcribed from the intergenic region between mouse MyHC IIx and IIb, overlapped with MyHC IIx, whereas lnc-mg appeared to exert no effects on fiber type (11). In this study, porcine MyHC IIA/X-AS, mainly covering the intergenic region between MyHC IIa and IIx, could enhance myogenesis and maintain fast myofiber type partially through sponging miR-130b, thus sustaining, in trans, the expression of MyHC IIx. Therefore, we anticipate that various kinds of antisense lncRNAs produced from the highly-conserved skeletal MyHC cluster might provide flexible control of MyHC expression in a species-dependent way, although the underlying involved mechanism needs further investigation (3).
Experimental procedures
Animal care and procedures
Handling piglets was performed according to the standard procedures approved by the guidelines from the Committee of Animal Care and Usage in Northwest A&F University, Yangling, China. Guanzhong Black pigs used in this study were taken from the experimental pig farm of Northwest A&F University (Yangling, China).
Cell culture and transfection
Primary porcine skeletal muscle satellite cells were isolated from extensor digitorum longus of 3–5-day-old piglets, according to previous reports (7, 24) with some modifications. Briefly, fresh muscles were minced into ∼1-mm3 pieces and digested at 37 °C in 0.1% type I collagenase (Sigma, V900891) in a shaking water bath for 90 min and then switched to 0.25% trypsin for another 30 min. The digest was neutralized using high-glucose DMEM containing 10% FBS. Thereafter, the cell suspension was sequentially filtered through 100-, 70-, and 40-μm strainers to remove tissue debris. The single cells were resuspended in growth medium (DMEM/F-12 supplemented with 20% FBS and 1% penicillin/streptomycin) and plated on noncoated 10-cm dishes. After 3 h, the culture supernatant was gently collected and transferred to new dishes. Again, the culture medium was transferred into Matrigel (BD Biosciences, catalogue no. 356234)-coated plates to obtain the purified satellite cells after 18 h. At ∼90% confluence, the growth medium was replaced with myogenic differentiation media (high-glucose DMEM containing 5% horse serum).
MyHC IIA/X-AS siRNA, miR-130b-5p agomir, and their negative controls were obtained from RiboBio (Guangzhou, China). At ∼70–80% confluence, the oligonucleotides were transfected into primary skeletal satellite cells with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. In proliferation assay, transfection was conducted at ∼60–70% confluence.
RACE
Using SMARTer® RACE 5′/3′ kit (TaKaRa, Japan, catalogue no. 634859), the 5′ and 3′ ends of MyHC IIA/X-AS were determined. Briefly, total RNA was purified from the longissimus dorsi of 180-day-old pigs using TRIzolTM reagent (Invitrogen, 15596018). The RNA integrity number (RIN) was analyzed using Agilent 2200 (Agilent, Palo Alto, CA), and RNA samples with RIN >7.0 were used to perform RACE experiments according to the manufacturer's instructions.
RNA–FISH
The localization of MyHC IIA/X-AS within myofibers was determined through RNA–FISH. Briefly, a special probe targeting MyHC IIA/X-AS was designed and labeled with digoxin (Servicebio, Wuhan, China). Frozen longissimus dorsi muscles were cross-cut into 10-μm sections that were fixed using 100-μl drops of 4% paraformaldehyde with 0.1% polyvinylpyrrolidone for 20 min, and then washed using the washing buffer for 30 min. For hybridization, slices were incubated with 6 ng/μl digoxin-labeled probes in hybridization buffer at 37 °C overnight, and after washing, the slices were blocked using BSA at room temperature. Subsequently, the slices were incubated with anti-digoxin–HRP at 37 °C for 50 min, and then reacted with FITC–TSA for 5 min in the dark followed by washing three times. The nuclei were labeled with DAPI. Images were captured using Nikon microscope.
Subcellular fractionation
The subcellular locations of special genes were identified using the Protein and RNA Isolation System (PARIS)TM kit (Invitrogen, catalog no. AM1921). Briefly, up to 107 cultured satellite cells were homogenized rapidly using the in-cell fraction buffer and incubated for 10 min on ice. Subsequently, the cell lysate was centrifuged at 500 × g for 5 min at 4 °C to carefully separate the cytoplasmic fraction and nuclear pellet. Thereafter, RNA was isolated from both cytoplasm and nuclei. U6 rRNA and protein-coding β-actin were used as positive controls for nuclear and cytoplasmic fractions, respectively.
EdU assay
To assess the cell proliferation, the cell light EdU DNA cell proliferation kit (RiboBio, Guangzhou, China) was used. Primary satellite cells were seeded in 24-well culture plates containing 500 μl of growth media. After 48 h of transfection, the cells were incubated into EdU medium for 2 h, and then observed under a microscope.
Cell counting kit-8 assay
Muscle satellite cells were maintained in six-well plates at a density of 1 × 105 cells/ml. After 48 h, 10 μl of CCK8 reagent was added to each plate, and cells were incubated at 37 °C for 3 h. Subsequently, the absorbance was measured at 450 nm wavelength using a micro-well plate reader.
Immunofluorescence analysis
Differentiated myotubes were fixed using 4% paraformaldehyde, permeabilized using 0.5% Triton X-100, and then blocked in 5% BSA for 30 min. Thereafter, myotubes were sequentially incubated with anti-myosin heavy chain mAb (1:200, catalog no. MAB4470, R&D Systems) overnight and an Alexa Fluor 594–conjugated anti-mouse IgG (1:1000, catalog no SA00006-3, Proteintech, Chicago, IL) for 1 h at 4 °C. Finally, nuclei were stained with DAPI. Images were captured using a fluorescence microscope (Nikon, Tokyo, Japan). Myotubes containing ≤6 and >6 nuclei were counted. The differentiation index was assessed as the percentage of MyHC-positive nuclei among total nuclei, and the myotube fusion index was determined as the distribution of the nuclei number in a total number of myotubes.
Dual-luciferase reporter assay
The putative binding sites (wild and mutated type) of miR-130b within MyHC IIA/X-AS and 3′-UTR of MyHC IIA/X-AS were synthesized using General Biosystems (Chuzhou, Anhui, China) and inserted into psiCHECKTM-2 Vector (Promega, Madison, WI). The psiCHECKTM-2 constructs were miR-130b agomir, and negative control was co-transfected into the human embryonic kidney 293T (HEK293T) cells (ATCC). Cells were harvested and analyzed using the Dual-Luciferase Reporter Assay System (Promega) after 48 h.
Relative quantitative real-time PCR
The total RNA was extracted using TRIzol reagent (Invitrogen), and the concentration and quality of RNA were analyzed using the NanoDrop 2000 (Thermo Fisher Scientific). The cDNA was synthesized using reverse transcription kit (TaKaRa). RT-qPCR was performed using Applied Biosystems qPCR instrument (Thermo Fisher Scientific) with SYBR Green PCR Master Mix (Vazyme, Nanjing, China). The expression levels of the genes of interest were normalized using β-actin (for coding genes) or U6 small RNA (for miRNAs). The sequences of primers for coding genes were synthesized by Invitrogen (Shanghai, China) and are listed in Table 1. The specific primers for miRNAs and U6 were designed and synthesized by RiboBio (Guangzhou, China).
Table 1.
Primers used in RT-qPCR
| Gene symbol | Forward primer | Reverse primer |
|---|---|---|
| β-Actin | GGACTTCGAGCAGGAGATGG | AGGAAGGAGGGCTGGAAGAG |
| MyHC | TTGAAAAGACGAAGCAGCGAC | AGAGAGCGGGACTCCTTCTG |
| MyoD | GCACTACAGTGGCGACTCAGATGC | CACTGTAGTCGGTGTCGTAGCC |
| Myogenin | CATCCAGTACATTGAGCGCCTACAG | GGGAGTTGCATTCACTGGGCAC |
| Cyclin D | TAGGCCCTCAGCCTCACTC | CCACCCCTGGGATAAAGCAC |
| Cyclin E | CAGAGCAGCGAGCAGGAGC | GCAGCTGCTTCCACACCACT |
| PCNA | ATGTTCGAGGCGCGCCTGGTC | CTAAGATCCTTCTTCATCCTC |
| PGC1α | AGCCGTGACCACTGACAACGAG | GCTGCATGGTTCTGAGTGCTAAG |
| FoxO1 | ATGCTCAATCCAGAGGGAGG | ACTCGCAGGCCACTTAGAAAA |
| MyHC I | ACAACCCCTACGATTATGCGT | ACGTCAAAGGCGCTATCCGTG |
| MyHC IIa | AAGTGACTGTGAAAACAGAAGCA | GCAGCCATTTGTAAGGGTTGAC |
| MyHC IIx | CTCCAGGCTGCTTTAGAGGAA | CCTGCTCCTAATCTCAGCATCC |
| MyHC IIb | AAACCACCTCAGAGTTGTGGA | GTTCCGAAGGTTCCTGATTGC |
| MyHC IIA/X-AS for qRT-PCR | TGCTCCTTACCGGAGACTCA | GAGGCGACCGATTGAAGGAA |
| MyHC IIA/X-AS for standard sample in qRT-PCR | TGCTCCTTACCGGAGACTCA | GACAACCCAGCTTGGTGAGA |
| MyHC IIx 3′-UTR for RNA pulldown | TTGCTGAGTCCCAGGTCAAC | AAGTACAAAACAGAGTGACAAAGAT |
| MyHC IIA/X-AS for RNA pulldown | ACTTAGTACATGGTGAAGACCTTT | CGAAACAGTGCAAAACAAAGC |
Absolute quantitative real-time PCR
Absolute RT-PCR was used to analyze the expression levels of MyHC IIA/X-AS in various tissues (spleen, kidney, heart, lung, subcutaneous fat, and longissimus dorsi muscle). Briefly, T-vectors containing a longer fragment of MyHC IIA/X-AS were constructed and diluted in titration to generate the standard curve. Total RNA (2 μg) from each tissue was reverse-transcribed, and cDNA was used as template in real-time PCR. The Ct values were used to calculate the copy numbers of MyHC IIA/X-AS in each sample, according to the standard curve.
RIP assay
Well-differentiated porcine myotubes were subjected to RIP assay after 6 days of induction. Briefly, cells were collected, lysed in complete RIP buffer provided in the EZ-Magna RIP kit (Millipore), and then incubated with RIP buffer containing magnetic beads conjugated to anti-Ago2 antibody (Millipore). Subsequently, the samples were digested with proteinase K, and then the RNA was purified from the precipitates. The concentration of RNA was determined using NanoDrop (Thermo Fisher Scientific). Finally, RT-qPCR was performed to detect the existence of miR-130b and MyHC IIA/X-AS.
miRNA pulldown assay
Porcine satellite cells were transfected with biotinylated mir-130b: WT miR-130b (miR-130b-Bio) or mutated miR-130b (G to C mutation in the MyHC IIA/X-AS-binding sites, miR-130b-MutBio) and biotinylated control (NC-Bio) when 70–80% of confluency was reached and induced into myogenic differentiation at full confluence. Six days after induction, cell lysates were collected and incubated with Dyna M-280 streptavidin magnetic beads (catalog no. 11205D, Thermo Fisher Scientific). The product was then treated with RNase-free DNase I (Roche Applied Science, Mannheim, Germany), and RNA was purified using RNeasy Mini Kit (Qiagen, Frederick, MD). The enrichment of MyHC IIx and MyHC IIA/X-AS were detected through qRT-PCR.
Western blot analysis
Cells were harvested using radioimmunoprecipitation assay lysis buffer (Applygen Technologies Inc., Beijing, China) supplemented with protease and phosphatase inhibitor mixture (Cwbiotech, Jiangsu, China). Protein concentration was determined using the BCA protein assay kit (Cwbiotech), and 25 μg of protein per sample was loaded and separated using a 5% stacking gel and a 10% separating gel. Separated proteins were transferred to polyvinylidene difluoride membrane (Cell Signaling Technology, Boston, MA), which was then blocked in 5% BSA for 2 h at room temperature and incubated with primary antibodies (listed in Table S2) at 4 °C overnight. After washing three times (10 min once) in TBST, membranes were incubated with HRP-conjugated goat anti-mouse IgG (catalog no. BA1050, BosterBio, Wuhan, China) or goat anti-rabbit IgG (catalog no. BA1054, BosterBio) for 1.5 h at 4 °C. Imaging and quantification of the bands were carried out using Gel Doc XR system (Bio-Rad) and Image Lab software (Bio-Rad).
Statistical analysis
Values are presented as the mean ± S.D., and statistical significance of differences was determined by Student's t test or one-way analysis of variance by IBM SPSS Statistics 22.0 (Armonk, NY). A p value of < 0.05 was set as statistically significant.
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Author contributions
M. D. and L. M. data curation; M. D. formal analysis; M. D. writing-original draft; Y. Y. methodology; Y. Y. project administration; X. W. and X. L. writing-review and editing; X. S. and G. Y. resources; X. S. and G. Y. supervision; X. L. conceptualization; X. L. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to Min Du, Washington State University, for the kind suggestion on experiment design and critical commons on manuscript drafting.
This work was supported by National Natural Science Foundation Grant 31501925 and National Key Research and Development Program of China Grant 2018YFD0500402.The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1 and Tables S1 and S2.
- MyHC
- myosin heavy chain
- RACE
- rapid amplification of cDNA ends
- CPC
- coding potential calculator
- qRT
- quantitative RT
- RNA–FISH
- RNA fluorescence in situ hybridization
- EdU
- 5-ethynyl-2′-deoxyuridine
- ceRNA
- competing endogenous RNA
- PCNA
- proliferating cell nuclear antigen
- DAPI
- 4′,6-diamidino-2-phenylindole
- lncRNA
- long noncoding RNA
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- HRP
- horseradish peroxidase
- RIP
- RNA-binding protein immunoprecipitation.
References
- 1. Frontera W. R., and Ochala J. (2015) Skeletal muscle: a brief review of structure and function. Calcif. Tissue Int. 96, 183–195 10.1007/s00223-014-9915-y [DOI] [PubMed] [Google Scholar]
- 2. Schiaffino S. (2018) Muscle fiber type diversity revealed by anti-myosin heavy chain antibodies. FEBS J. 285, 3688–3694 10.1111/febs.14502 [DOI] [PubMed] [Google Scholar]
- 3. Schiaffino S., and Reggiani C. (2011) Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 10.1152/physrev.00031.2010 [DOI] [PubMed] [Google Scholar]
- 4. Carter C. S., Justice J. N., and Thompson L. (2019) Lipotoxicity, aging, and muscle contractility: does fiber type matter? GeroScience 41, 297–308 10.1007/s11357-019-00077-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pattanakuhar S., Pongchaidecha A., Chattipakorn N., and Chattipakorn S. C. (2017) The effect of exercise on skeletal muscle fibre type distribution in obesity: from cellular levels to clinical application. Obes. Res. Clin. Pract. 11, 112–132 10.1016/j.orcp.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 6. Talbot J., and Maves L. (2016) Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 5, 518–534 10.1002/wdev.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li B.-J., Li P.-H., Huang R.-H., Sun W.-X., Wang H., Li Q.-F., Chen J., Wu W.-J., and Liu H.-L. (2015) Isolation, culture and identification of porcine skeletal muscle satellite cells. Asian-Australas. J. Anim. Sci. 28, 1171–1177 10.5713/ajas.14.0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haddad F., Qin A. X., Bodell P. W., Jiang W., Giger J. M., and Baldwin K. M. (2008) Intergenic transcription and developmental regulation of cardiac myosin heavy chain genes. Am. J. Physiol. Heart Circ Physiol. 294, H29–H40 10.1152/ajpheart.01125.2007 [DOI] [PubMed] [Google Scholar]
- 9. Pandorf C. E., Haddad F., Roy R. R., Qin A. X., Edgerton V. R., and Baldwin K. M. (2006) Dynamics of myosin heavy chain gene regulation in slow skeletal muscle: role of natural antisense RNA. J. Biol. Chem. 281, 38330–38342 10.1074/jbc.M607249200 [DOI] [PubMed] [Google Scholar]
- 10. Rinaldi C., Haddad F., Bodell P. W., Qin A. X., Jiang W., and Baldwin K. M. (2008) Intergenic bidirectional promoter and cooperative regulation of the IIx and IIb MHC genes in fast skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R208–R218 10.1152/ajpregu.00134.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu M., Liu J., Xiao J., Yang L., Cai M., Shen H., Chen X., Ma Y., Hu S., Wang Z., Hong A., Li Y., Sun Y., and Wang X. (2017) Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 8, 14718–14718 10.1038/ncomms14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sakakibara I., Santolini M., Ferry A., Hakim V., and Maire P. (2014) Six homeoproteins and a linc-RNA at the fast MYH locus lock fast myofiber terminal phenotype. PLOS Genet. 10, e1004386 10.1371/journal.pgen.1004386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang Q., and Hann S. S. (2018) HOTAIR: an oncogenic long non-coding RNA in human cancer. Cell. Physiol. Biochem. 47, 893–913 10.1159/000490131 [DOI] [PubMed] [Google Scholar]
- 14. Lin J., Wu H., Tarr P. T., Zhang C.-Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., and Spiegelman B. M. (2002) Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- 15. Kamei Y., Miura S., Suzuki M., Kai Y., Mizukami J., Taniguchi T., Mochida K., Hata T., Matsuda J., Aburatani H., Nishino I., and Ezaki O. (2004) Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 279, 41114–41123 10.1074/jbc.M400674200 [DOI] [PubMed] [Google Scholar]
- 16. Xu M., Chen X., Chen D., Yu B., and Huang Z. (2017) FoxO1: a novel insight into its molecular mechanisms in the regulation of skeletal muscle differentiation and fiber type specification. Oncotarget 8, 10662–10674 10.18632/oncotarget.12891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Da Costa N., and Chang K.-C. (2005) Molecular characterisation of the porcine skeletal myosin heavy chain cluster and a major candidate regulatory domain. Arch. Tierzucht. 48, 32–39 [Google Scholar]
- 18. Sun Y. M., Da Costa N., and Chang K. C. (2003) Cluster characterisation and temporal expression of porcine sarcomeric myosin heavy chain genes. J. Muscle Res. Cell Motil. 24, 561–570 10.1023/B:JURE.0000009895.03111.b3 [DOI] [PubMed] [Google Scholar]
- 19. Reiser P. J. (2019) Current understanding of conventional and novel co-expression patterns of mammalian sarcomeric myosin heavy chains and light chains. Arch. Biochem. Biophys. 662, 129–133 10.1016/j.abb.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 20. Kopp F., and Mendell J. T. (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ponsuksili S., Du Y., Hadlich F., Siengdee P., Murani E., Schwerin M., and Wimmers K. (2013) Correlated mRNAs and miRNAs from co-expression and regulatory networks affect porcine muscle and finally meat properties. BMC Genomics 14, 533–533 10.1186/1471-2164-14-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siengdee P., Trakooljul N., Murani E., Schwerin M., Wimmers K., and Ponsuksili S. (2015) MicroRNAs regulate cellular ATP levels by targeting mitochondrial energy metabolism genes during C2C12 myoblast differentiation. PLoS One 10, e0127850 10.1371/journal.pone.0127850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue J., Xue J., Zhang J., Li D., and Jiang L. (2017) miR-130b-3p/301b-3p negatively regulated Rb1cc1 expression on myogenic differentiation of chicken primary myoblasts. Biotechnol. Lett. 39, 1611–1619 10.1007/s10529-017-2400-8 [DOI] [PubMed] [Google Scholar]
- 24. Yang J., Liu H., Wang K., Li L., Yuan H., Liu X., Liu Y., and Guan W. (2017) Isolation, culture and biological characteristics of multipotent porcine skeletal muscle satellite cells. Cell Tissue Bank. 18, 513–525 10.1007/s10561-017-9614-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.