Abstract
Oligomers of β-amyloid 42 (Aβ42), rather than fibrils, drive the pathogenesis of Alzheimer's disease (AD). In particular, toxic oligomeric species called protofibrils (PFs) have attracted significant attention. Herein, we report RNA aptamers with higher affinity toward PFs derived from a toxic Aβ42 dimer than toward fibrils produced from WT Aβ42 or from a toxic, conformationally constrained Aβ42 variant, E22P–Aβ42. We obtained these RNA aptamers by using the preincubated dimer model of E22P–Aβ42, which dimerized via a linker located at Val-40, as the target of in vitro selection. This dimer formed PFs during incubation. Several physicochemical characteristics of an identified aptamer, E22P–AbD43, suggested that preferential affinity of this aptamer toward PFs is due to its higher affinity for the toxic dimer unit (KD = 20 ± 6.0 nm) of Aβ42 than for less-toxic Aβ40 aggregates. Comparison of CD data from the full-length and random regions of E22P–AbD43 suggested that the preferential binding of E22P–AbD43 toward the dimer might be related to the formation of a G-quadruplex structure. E22P–AbD43 significantly inhibited the nucleation phase of the dimer and its associated neurotoxicity in SH-SY5Y human neuroblastoma cells. Of note, E22P–AbD43 also significantly protected against the neurotoxicity of WT Aβ42 and E22P–Aβ42. Furthermore, in an AD mouse model, E22P–AbD43 preferentially recognized diffuse aggregates, which likely originated from PFs or higher-order oligomers with curvilinear structures, compared with senile plaques formed from fibrils. We conclude that the E22P–AbD43 aptamer is a promising research and diagnostic tool for further studies of AD etiology.
Keywords: Alzheimer disease, amyloid-beta (AB), aggregation, aptamer, circular dichroism (CD), immunohistochemistry, G-quadruplex, oligomer, transgenic mice, RNA
Introduction
Amyloid fibrils of senile plaques in Alzheimer's disease (AD)5 consist mainly of the 40- or 42-mer β-amyloid proteins (Aβ40 and Aβ42), which are produced from the Aβ precursor protein (1, 2). The ability of Aβ42 to aggregate and its neurotoxicity is higher than that of Aβ40 (3). Cumulative evidence shows that metastable oligomers of Aβ42 can induce neuronal death and cognitive dysfunction, which is in contrast to end-stage mature fibrils whose contribution to AD pathology is lower (4). Notably, these toxic oligomers include protofibrils (PFs) or higher-order oligomers with a curvilinear structure (5). Although some less toxic higher-order oligomers exist, most of the higher-order oligomers such as PFs (6, 7), the Aβ-derived diffusible ligands (8), and amylospheroid (9) are potently neurotoxic. “Toxic oligomers” are defined in this work as the constituents of these higher-order oligomers (24–700-mer) that are neurotoxic. The development of oligomer-specific inhibitors is imperative for making meaningful progress toward developing AD therapies without adverse effects.
Aptamers are ssDNA or RNA oligonucleotides used for molecular recognition of diverse targets for diagnostic and therapeutic applications (10). However, current application of aptamers to the field of Aβ research remains limited. There are several published aptamers targeting Aβ including RNA aptamers targeting Aβ40 fibrils (11, 12), RNA aptamers targeting other amyloid fibrils and Aβ40 fibrils (13), DNA aptamers developed against α-synuclein but recognizing Aβ40 oligomers (14) and DNA aptamers targeting Aβ40 oligomers (15). Because more aggregative Aβ42 exists in a more dynamic conformational equilibrium of monomeric Aβ42 (16), it is difficult to isolate and characterize aptamers targeting the short-lived, metastable structures of toxic Aβ42 oligomers, unlike less-toxic Aβ40 oligomers, for in vitro selection (known as the systematic evolution of ligands by exponential enrichment (SELEX)).
Based on systematic proline replacement and solid-state NMR Irie and co-workers (17, 18) determined the toxic conformer of Aβ42 to have a turn at Glu-22 and Asp-23, whereas the nontoxic conformer of Aβ42 has a turn at Gly-25 and Ser-26 (Fig. 1A, toxic conformation theory). The E22P mutation induces the toxic conformation of Aβ42 and Aβ40, which tends to stabilize oligomer formation and promotes the neurotoxicity (Fig. 1A) (18, 19). We recently developed a covalently linked dimer model of E22P–Aβ42 using an l,l-2,6-diaminopimelic acid (DAP) linker at Val-40 in the C-terminal hydrophobic core (Fig. 1A, E22P–V40DAP–Aβ42 dimer), which plays an important role in the formation of toxic oligomers. This dimer model produced quasistable PFs, which were toxic oligomers, and had a neurotoxic effect on SH-SY5Y human neuroblastoma cells following incubation (20). The current report describes a comprehensive study on the development of RNA aptamers with potent affinity for the toxic dimer model of Aβ42 (E22P–V40DAP–Aβ42 dimer), and we discuss their application to therapeutics and diagnostics based on experiments using an AD mouse brain.
Figure 1.
Selection of RNA aptamers targeting PFs based on the toxic conformation theory of Aβ42. A, toxic conformation theory of Aβ42. The toxic conformer of Aβ42 forms a turn at Glu-22 and Asp-23, and the E22P mutation enhances the ratio of the toxic Aβ42 conformer. The covalently linked dimer of E22P–Aβ42 has a DAP linker attached at Val-40 in the C-terminal hydrophobic core (E22P–V40DAP–Aβ42 dimer). This dimer produces quasistable PFs (50–100 kDa) after incubation for 48 h at 37 °C (20). B, progression of SELEX monitored by the filter membrane method using real-time PCR. The ratio of bound RNA for each round of selection was plotted as a percentage of total RNA used for the corresponding cycle. The ratios of Aβ to RNA are Aβ:RNA = 400:100 pmol (rounds 1–5), Aβ:RNA = 400:400 pmol (round 6), and Aβ:RNA = 400:800 pmol (round 7). R7, enriched RNA pool after round seven. C, screening of the 36 binding monoclones toward Aβ42 PFs by the filter membrane method using the RiboGreen fluorescent dye. R0, initial RNA pool before selection. D, comparison of the affinity of each clone (E22P–AbD4, –AbD31, and –AbD43) toward PFs and comparison with fibrils produced from WT Aβ42 or E22P–Aβ42. The data are expressed as the means ± S.D. (n = 3). ***, p < 0.001. n.s., not significant.
Results
Isolation of RNA aptamers targeting the preincubated E22P–V40DAP–Aβ42 dimer and their recognition toward PFs
As a model target molecule for the development of an aptamer toward PFs, the E22P–V40DAP–Aβ42 dimer (9 kDa) was initially used as a model target for selection of candidate aptamers. The selection was performed using a column where an E22P–V40DAP–Aβ42 dimer biotinylated at the N terminus (termed the biotin–E22P–V40DAP–Aβ42 dimer) was immobilized, and the binding rate of RNA nucleotides to Aβ was calculated by real-time PCR. However, RNA pools failed to be enriched even after several rounds of SELEX (data not shown). Preincubation (37 °C, 24 h) with the E22P–V40DAP–Aβ42 dimer that generates a lower-molecular-mass oligomer of Aβ42 than PFs was also unsuccessful (data not shown).
To further enlarge the molecular mass of the target for potential selection, an extended preincubation period with the E22P–V40DAP–Aβ42 dimer (37 °C, 48 h) was next used for the selection process in combination with the method of membrane filtering (molecular mass cutoff, 50 kDa). As reported previously (20), the biotin–E22P–V40DAP–Aβ42 dimer formed PFs after incubation for 48 h, as determined by transmission EM (TEM) analysis (Fig. S1A). The RNA-binding ratio gradually increased after seven rounds of selection, meaning that the RNA pool (∼26 kDa) was effectively enriched (Fig. 1B). Given unsuccessful enrichment in the two trials stated above using the Aβ-immobilized column, it should be noted that the use of membrane filtering (Fig. S2) may be more effective for the selection of aptamers targeting oligomeric assemblies including ordered structures such as PFs. The column matrix may have prevented association of dimer model, which could be required for the enrichment of aptamers. Alternatively, steric hindrance of PFs at the N-terminal region may have affected the selection process.
After cloning of the enriched RNA pools after round 7, 36 identical monoclones were obtained. This was followed by binding tests based on the membrane filter method. As shown in Fig. 1C, only three monoclones (E22P–AbD4, –AbD31, and –AbD43) showed higher affinity toward the PFs than the enriched RNA pool after round 7 (R7). The binding of the initial RNA pool to the PFs (i.e. before selection) was confirmed to be negligible.
Because the Aβ42 monomer can partially form the toxic conformation (18), there was the possibility that the obtained monoclones would also bind to fibrils produced from WT Aβ42 or E22P–Aβ42. Using a membrane filter, all three aptamers (E22P–AbD4, –AbD31, and –AbD43) were shown to exhibit significant preferential binding toward PFs when compared with these two types of fibrils (Fig. 1D).
G-quadruplex structure of RNA aptamers is involved in their binding to PFs
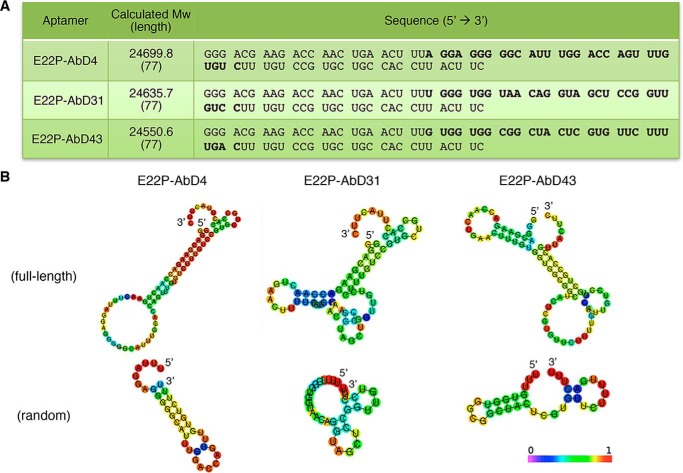
CentroidFold was used to predict the folding of the RNA aptamers (21). As shown in Fig. 2B, the 77-nt RNA sequences (Fig. 2A) of E22P–AbD4, –AbD31, and –AbD43 folded to primarily form stem–loop intramolecular base pairing as a hairpin structure. Alignment search, classification, and local supermotif analysis did not find a specific common domain among these aptamers based on analysis with Clustal Omega (22) (Fig. S3A) and Multiple EM for Motif Elucidation (23) (Fig. S3B).
Figure 2.
Analysis of the sequence of RNA aptamers targeting PFs. A, sequences of E22P–AbD4, –AbD31, and –AbD43 and their calculated molecular mass with lengths. Bold letters indicate random regions. B, the secondary structure of each aptamer (both full-length and random) predicted by CentroidFold analysis. Each predicted base pair is colored by heat color gradation from purple to red, which corresponds to base pairing probability scores of from 0 to 1, respectively.
Aptamers can fold into various secondary structures that exhibit high binding capacity, such as stem–loop, G-quadruplex, and three-way junction structures (10). Given the slightly higher content of guanine in the obtained aptamers (Fig. 2A: E22P–AbD4, 29%; E22P–AbD31, 27%; and E22P–AbD43, 26%), parallel, anti-parallel, and hybrid G-quadruplex structures are predicted for these aptamers (24, 25). Based on CD spectra used to analyze their secondary structure, E22P–AbD4, –AbD31, and –AbD43 exhibited a negative peak at ∼240 nm and a positive peak at ∼265 nm, suggesting that the RNA aptamers formed G-quadruplexes (Fig. 3A), where four guanines are held in a square planar G-tetrad arrangement through Hoogsteen hydrogen bonding (25).
Figure 3.
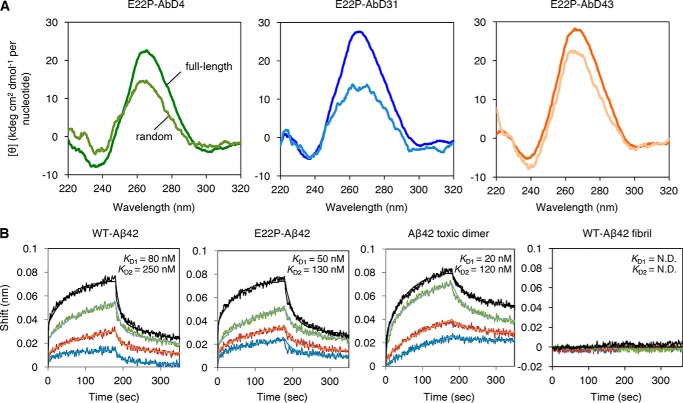
Analysis of the secondary structure and binding constants of RNA aptamers targeting PFs. A, CD spectra of the full-length (dark colors) and random region sequence as E22P–AbD4(Ran29), –AbD31(Ran29), and –AbD43(Ran29) (light colors) of each aptamer (10 μm; E22P–AbD4, –AbD31, and –AbD43). B, BLI sensorgram and curve fitting (2:1 heterogeneous ligand-binding model) of E22P–AbD43 as a ligand to WT Aβ42, E22P–Aβ42, Aβ42 toxic dimer (the E22P–V40DAP–Aβ42 dimer), and WT Aβ42 fibril as the analyte with the concentration of Aβ shown in blue for 200 nm, orange for 400 nm, green for 800 nm, and black for 1600 nm. The KD1 and KD2 values are indicated. N.D., not determined.
The random region sequence of each aptamer was prepared to elucidate the structure of these RNA aptamers. Although the ellipticity of each random region sequence at the maximum absorption wavelength (∼265 nm) decreased, the difference in the ellipticity of E22P–AbD43 between the full-length sequence and the random region sequence was relatively small when compared with the results for the other two aptamers (Fig. 3A).
Next, biolayer interferometry (BLI) measurements were performed to determine the kinetics and affinity of the interaction between the RNA aptamer and Aβ. For this purpose, the 5′-biotinylated RNA clones were immobilized onto a streptavidin biosensor, and Aβ was used as the analyte. We observed highly stable complexes for all three aptamers with PFs, characterized by dissociation constants (KD) in the nanomolar range using the appropriate fitting of the 2:1 heterogeneous ligand-binding model of the full-length sequence (77 nt) contingent on the lower χ2 coefficient (Fig. S4A). Table S1 provides a summary of the binding kinetics between the three aptamers and PFs.
The dissociation constant (KD) of the random region sequence (29 nt) of E22P–AbD4, –AbD31, and –AbD43 for Aβ as E22P–AbD4(Ran29), -AbD31(Ran29), and -AbD43(Ran29) was derived from curve fitting of the data with a 1:1 interaction contingent on the lower χ2 coefficient. The truncation of E22P–AbD4 and -AbD31 moderately enhanced the affinity, whereas the affinity of E22P–AbD43(Ran29) was lower than that of full-length E22P–AbD43 (Figs. S4A and S5 and Table S1). Notably, the constant regions may assist the binding of the E22P–AbD43 aptamer. An alternative explanation might be that the constant regions are occluding the random fragment region of E22P–AbD4 and E22P–AbD31 but not that of E22P–AbD43. Comparing the CD absorption data for the full-length and random region of E22P–AbD43 (Fig. 3A), the preferential binding of E22P–AbD43 toward PFs might be partly related to the formation of a G-quadruplex structure. Attenuated total reflection spectroscopy (ATR)–FTIR (Fig. S6) of E22P–AbD43 showed absorptions at 1649 (guanine carbonyl group) cm−1, which is suggestive of the possible presence of a G-quadruplex structure according to previous combination studies with vibrational CD (26) using anti-Aβ aptamer forming G-quadruplex as a reference (14, 27).
Characterization of E22P–AbD43 for its preferential recognition unit of PFs
To obtain information on how E22P–AbD43 can preferentially recognize PFs, BLI tests were conducted using WT Aβ42, E22P–Aβ42, and the E22P–V40DAP–Aβ42 dimer as analytes. As shown in Fig. 3B, E22P–AbD43 demonstrated 2–8-fold stronger binding affinity toward the dimer model (KD1 = 20 ± 6.0 nm) than toward both monomers of WT Aβ42 (KD1 = 80 ± 8.3 nm) or E22P–Aβ42 (KD1 = 50 ± 5.0 nm), indicating that a minimum unit of recognition of PFs by E22P–AbD43 is the dimer. Table S1 provides a summary of the binding kinetics between the E22P–AbD43 and each Aβ42.
Furthermore, the binding affinity of E22P–AbD43 to WT Aβ40 and PFs prepared from WT Aβ40 basically according to the developed protocol (28) was weaker than WT Aβ42 and PFs model produced from the E22P–V40DAP–Aβ42 dimer, respectively (Fig. S4B). In addition, E22P–AbD43 did not react with WT Aβ40 fibril (Fig. S4B) similar to WT Aβ42 fibril (Fig. 3B). These results suggest the preferable binding of E22P–AbD43 to toxic Aβ42 aggregates rather than less-toxic Aβ40 aggregates.
An additional binding assay using fluorescence polarization for E22P–AbD43 showed a similar tendency (Fig. S7; KD = 54 ± 1.1 nm for Aβ42 toxic dimer, KD = 160 ± 1.4 nm for WT Aβ42, KD = 120 ± 1.2 nm for E22P–Aβ42). E22P–AbD43 did not bind to WT Aβ42 fibril. The slight difference of KD values between BLI and fluorescence polarization experiments might be ascribed to the presence or absence of immobilization of one side to the support. The immobilization of analytes/ligands might help stabilize their conformation to modulate the binding. The advantages and limitations of these methods were compared by Rossi and Taylor (29).
E22P–AbD43 prevents the formation of Aβ42 aggregates and their neurotoxicity
Because the binding affinity of E22P–AbD43 toward PFs (KD = 150 ± 11 nm) was the most potent among the three aptamers examined (Fig. S4A and Table S1), we selected full-length E22P–AbD43 for the following studies on anti-Aβ properties. The full-length sequence, which should be more resistant to degradation, is also more appropriate for cell culture tests than the corresponding random region. We next determined whether E22P–AbD43 could prevent the aggregation of WT Aβ42. Using TEM, WT Aβ42 was shown to form typical long amyloid fibrils, whereas in the presence of E22P–AbD43 WT Aβ42 formed short or globular (curvilinear) aggregates (Fig. 4A).
Figure 4.
Effects of E22P–AbD43 on the aggregation of Aβ42 and its neurotoxicity. A, TEM analysis to evaluate the aggregates produced from Aβ42 (25 μm) after incubation with E22P–AbD43 (50 μm) for 24 h at room temperature. Arrows and arrowheads indicate fibrils and PFs, respectively. Scale bar, 200 nm. B and C, Th-S assay to evaluate aggregation of B WT Aβ42 or E22P–Aβ42 (10 μm). C, the E22P–V40DAP–Aβ42 dimer (5 μm) after incubation with E22P–AbD43 (40 μm) for the indicated period at 25 °C. The data are expressed as the means ± S.D. (n = 3). In B, the inset figure indicates the magnified graph of E22P–Aβ42 (4–12 h). D and E, MTT assay to evaluate the neurotoxicity of WT Aβ42 or E22P–Aβ42 (1 μm) (D) and the E22P–V40DAP–Aβ42 dimer (0.5 μm) (E) using SH-SY5Y cells after a 16-h incubation period with E22P–AbD43 (1, 2, or 4 μm) at 37 °C. The data are expressed as the means ± S.D. (n = 3): ###, p < 0.001 versus vehicle alone; *, p < 0.05; **, p < 0.01 and ***, p < 0.001. n.s., not significant. Veh, vehicle; Apt, aptamer.
Naiki and Gejyo (31) have proposed a nucleation-dependent polymerization model for Aβ42 that is composed of nucleation and elongation phases. During the nucleation phase, the Aβ42 monomer gradually forms low molecular mass oligomers (called “nuclei”), followed by the elongation phase where each nucleus acts as a template and associates with monomers for polymerization and the subsequent formation of higher-order oligomers and fibrils (30). The thioflavin-T (Th-T) assay (31) is often used to quantify the inhibition of Aβ42 aggregation by test compounds, but RNA nucleotides themselves enhance the fluorescence of the Th-T reagent (32). The thioflavin-S (Th-S) can be used instead of Th-T because Th-S exhibits almost no nonspecific interaction with RNA.
In the Th-S fluorescence assay performed in the present study, the peptides were used at 10 μm effective concentration of the monomer (i.e. the monomer was used at 10 μm, and the dimer was used at 5 μm). Both WT Aβ42 (0–2 h and 2–6 h) and E22P–Aβ42 (0–4 h and 4–12 h) exhibited two sigmoid-like curve points in the process of Th-S fluorescence. These curves may reflect a conformational transition or equilibrium during the nucleation of Aβ. E22P–AbD43 moderately retarded the nucleation phase (4–12 h) of E22P–Aβ42, but it did not significantly slow the nucleation of WT Aβ42 (Fig. 4B). Incidentally, a known drawback of the Th-T or Th-S method is the gradual decrease in Th-T or Th-S signal during the later stage of the elongation phase of Aβ, possibly because of the limited surface area of mature fibrils with a high core density to which Th-T or Th-S can bind. This may explain why the signals from the elongation phase of both WT Aβ42 and E22P–Aβ42 in the presence of E22P–AbD43 did not decrease (Fig. 4B). Furthermore, to evaluate the effect of E22P–AbD43 on the nucleation phase of Aβ42, the E22P–V40DAP–Aβ42 dimer was used. Notably, E22P–AbD43 delayed the nucleation of the E22P–V40DAP–Aβ42 dimer by almost 2 days (Fig. 4C). These findings suggest that E22P–AbD43 may prevent the formation of PFs by interacting with the E22P–V40DAP–Aβ42 dimer.
Because E22P–AbD43 may interact with PFs during the nucleation phase, we investigated the effect of E22P–AbD43 on the neurotoxicity of the E22P–V40DAP–Aβ42 dimer, as well as WT Aβ42 and E22P–Aβ42. For this purpose, we used cultures of SH-SY5Y human neuroblastoma cells that are a typical model of neuronal cells (Fig. 4, D and E). The peptides were added to the culture medium at 1 μm effective concentration of the monomer (i.e. the monomer was used at 1 μm, and the dimer was used at 0.5 μm). After incubation for 16 h, which is appropriate for the evaluation of Aβ oligomers using E22P–AbD43 based on the previous study (20), the neurotoxicity caused by the E22P–V40DAP–Aβ42 dimer was significantly reduced by E22P–AbD43 (1, 2, and 4 μm) in a dose-dependent manner (Fig. 4E). The aptamer itself at the maximum concentration (4 μm) was inactive. E22P–AbD43 was also significantly protective against the neurotoxicity of WT Aβ42 and E22P–Aβ42 after a 16-h incubation (Fig. 4D). The R7 pool of RNA exhibited almost no alternation on the cytotoxicity of all Aβ samples tested (WT Aβ42, E22P–Aβ42, and E22P–V40DAP–Aβ42 dimer) (Fig. 4, D and E). These findings indicate that E22P–AbD43 can preferentially delay the onset of nuclei-driven toxic oligomerization in the pathogenesis of AD.
E22P–AbD43 recognizes diffuse aggregates rather than senile plaques in an AD mouse
We next performed histochemical analysis of E22P–AbD43 using an AD mouse brain (Tg2576/PS2) model (33) at the age of 6 months. E22P–AbD43 recognized diffuse aggregates of Aβ mainly in the cerebral cortex and hippocampus regions (Fig. 5A), whereas an anti-Aβ-N terminus antibody (82E1) (34) mainly stained senile plaques with dense cores (Fig. 5B). Given that the diffuse halo surrounding the plaque cores contains immature or oligomeric aggregates (35), the diffuse aggregates detected by E22P–AbD43 might have originated from PFs or higher-order oligomers with curvilinear structures derived from Aβ, as likely observed in a previous report (36). Interestingly, 82E1 reacted with diffuse aggregates and senile plaques, possibly because the N termini of the PFs were exposed. It is noteworthy that E22P–AbD43 exclusively bound to diffuse aggregates in both the cerebral cortex and hippocampus based on counting the numbers of diffuse aggregates and senile plaques (Fig. S8). Using multiphoton microscopy, Farrar et al. (37) demonstrated that the β55 aptamer developed by Ylera et al. (11) simultaneously stained the diffuse halos of the amyloid of angiopathic lesions as well as dense-core Aβ plaques in Tg2576/PS1 mice.
Figure 5.

Histochemical analysis of AD mouse brains using E22P–AbD43. A and B, histochemical staining of AD mouse brain sections from Tg2576/PS2 was performed with E22P–AbD43 (400 nm) (A) together with the anti-N terminus of the Aβ (82E1) (B) antibody (1 μg/ml). The slices were exposed to formic acid for antigen activation by autoclaving. High magnification images (scale bar, 50 μm) of the area (scale bar, 500 μm) inside the rectangles (dashed line: cerebral cortex, solid line: hippocampus) are shown within each picture in A and B, respectively. Arrowheads represent diffuse aggregates, and arrows represent senile plaques, respectively.
Discussion
In the field of AD research, antibodies against Aβ have entered clinical trials as potential therapeutics and diagnostics. Irie and colleagues recently developed 11A1 (38) and 24B3 (20) antibodies, which target the toxic conformation and toxic oligomers of Aβ42, respectively. Both of these toxic moieties of Aβ42 include a turn structure at Glu-22 and Asp-23. Aβ40 can also form toxic oligomers derived from a toxic conformation, as exemplified by the work of Irie et al. (39), and 11A1 also binds to Aβ40, albeit at a slightly lower affinity toward Aβ42 (38). Despite various advances in antibody research, a weakness of antibodies and protein-based drugs is their potential inherent immunogenicity because of their chronic usage in animals and humans. Aptamers are a suitable alternative with comparable or potentially even superior affinity and selectivity when compared with that of antibodies, and RNA and DNA aptamers can be synthesized on a larger scale at a lower cost.
Recently, Kageyama et al. (40) demonstrated the presence of an Aβ toxic conformer in the human inferior parietal cortex several decades before the onset of AD using the 11A1 antibody. Because an RNA aptamer (β55) recognizing Aβ40 fibrils (11) stained the diffuse halos of an amyloid in addition to dense-core Aβ plaques in Tg2576/PS1 mice (37), E22P–AbD43 targeting more pathogenic Aβ42 may represent an ultra-early diagnostic tool of AD. Moreover, the strategy of stabilizing unstable oligomers of Aβ42 by covalent bonding within the C-terminal region followed by preincubation was an effective approach to enrich RNA pools, and such a strategy may be helpful for the discovery of “unstable” target-based drugs in other fields of amyloid research. The changes of the SELEX target from a dimer-immobilized bead to nonimmobilized PFs may have contributed to the more efficient aptamer enrichment program used herein and the selection of E22P–AbD43.
Unlike RNA pools, DNA pools failed to be enriched by using preincubated PFs despite several trials (data not shown). Although it remains controversial whether RNA or DNA aptamers are more useful for development as anti-Aβ aptamers, there are reports based on histochemical analysis that indicate that senile plaques composed of amyloid fibrils inherently contain mRNA (41, 42). To conclude, the present findings demonstrate the development of RNA aptamers against PFs prepared from the E22P–V40DAP–Aβ42 dimer with a toxic conformation as the smallest unit of PFs, and their possible application as therapeutic and diagnostic tools for AD was evaluated. The relatively stronger recognition by E22P–AbD43 of the Aβ42 dimer model rather than PFs (Fig. 3B and Fig. S4A and Table S1) indicates that Aβ42 dimers dissociate at equilibrium in PFs, because the formation of soluble PFs is a reversible reaction (5). This is the first report of RNA aptamers that target the toxic dimer of Aβ42. The affinity (KD = 20 ± 6.0 nm) of E22P–AbD43 toward Aβ42 toxic dimer may not still be sufficient for the therapeutic application; however, this KD value is extensively lower than any previously reported anti-Aβ aptamers (e.g. KD values of anti-Aβ40 aptamers (12) are 10–20 μm). Further trials to enhance the stability of toxic Aβ oligomers are currently underway.
Tsukakoshi et al. (27) reported that a telomestatin derivative increased the binding capacity of G-quadruplex–forming aptamers by enhancing π–π stacking on the G-tetrad plane. Because the planarity derives from α,β-unsaturated carbonyl groups in not only curcumin but also noncatechol-type flavonoids (e.g. morin and datiscetin), targeting His-13,14 and Phe-19,20 through π–π stacking for intercalation into β-sheet regions could contribute to the suppression of Aβ42 oligomerization (43, 44). We expect that the guanine bases in E22P–AbD43 could generate a scaffold like a flat G-quadruplex structure (Fig. 3A) that may target similar residues to these phytochemicals. BLI studies including the analysis for the percentage contributions from KD1 and KD2 implied a 2:1 heterogeneous ligand-binding model as the binding mode of full-length E22P–AbD43 to Aβ42 dimers (Tables S1 and S2), which possibly suggests the existence of two independent binding sites within E22P–AbD43 and the generation of complexes comprising an Aβ42 dimer:E22P–AbD43 ratio of 2:1 or 1:1. Structural analysis of these complexes by crystallographic analysis is now underway. Additionally, the formation of trimer-induced oligomers, which E22P–AbD43 may not readily recognize, may also have an impact on Aβ42-induced neurotoxicity.
This report also describes the unprecedented application of RNA aptamers to histochemical staining of AD mouse brains. Considering the preferential recognition by E22P–AbD43 of prematured aggregates of Aβ in AD mice (Fig. 5A), the application of this aptamer in early diagnosis or positron emission tomography imaging of amyloidogenesis for determining the progression of AD has potential. Further experiments are required to shorten the length of the RNA aptamers to facilitate their passage across the blood–brain barrier and their applications in AD therapy.
Experimental procedures
Preparation of PFs
The E22P–V40DAP–Aβ42 dimer or biotin–E22P–V40DAP–Aβ42 dimer, in which Val-40 of Aβ42 was replaced with DAP as a cross-linker, was synthesized using a previously described procedure (20) with a slight modification to the HPLC condition required for purification. Briefly, the E22P–V40DAP–Aβ42 dimer was purified using a YMC-Pack ODS-A column (20-mm inner diameter × 150 mm; YMC, Kyoto, Japan) attached to an HPLC instrument (Waters model 600E with a 2487 UV detector) with elution at 8.0 ml/min and a 80-min exponential gradient (curve 7 in Waters model 600E program) of 30–60% CH3CN containing 0.1% TFA. Subsequent serial purification was performed using a YMC-Pack Protein RP column (20-mm inner diameter × 150 mm; YMC) with elution at 8.0 ml/min using an 80-min linear gradient (curve 6 in Waters model 600E program) of 20–60% CH3CN containing 0.1% TFA and an x-Bridge Peptide BEH C18 OBDTM prep column (300Å, 5 μm, 19-mm inner diameter × 150 mm; Waters, Milford, MA) with elution at 8.0 ml/min using a 60-min linear gradient (curve 6 in Waters model 600E program) of 15–40% CH3CN containing 0.1% NH4OH. Preincubation of the dimer model at 37 °C for 48 h led to the production of quasistable PFs, as observed previously (20).
Purification of the biotin–E22P–V40DAP–Aβ42 dimer was carried out using a YMC-Pack ODS-A column (20-mm inner diameter × 150 mm; YMC) with elution at 8.0 ml/min and a 80-min exponential gradient (curve 7 in Waters model 600E program) of 30–60% CH3CN containing 0.1% TFA. Subsequent purification was performed using an x-Bridge Peptide BEH C18 OBDTM prep column (300Å, 5 μm, 19-mm inner diameter × 150 mm; Waters, Milford, MA) with elution at 8.0 ml/min and a 80-min exponential gradient (curve 7 in Waters model 600E program) of 10–50% CH3CN containing 0.1% NH4OH. The preincubation of the biotinylated dimer model at 37 °C for 48 h led to the production of quasistable PFs, and the presence of PFs was confirmed by TEM (Fig. S1A). PFs were also prepared from WT Aβ40 by incubation at 37 °C for 48 h basically according to the developed protocol (28) using a Millipore Amicon Ultra-0.5 centrifugal filter unit (molecular mass cutoff, 50 kDa; Millipore). The presence of PFs from WT Aβ40 was also confirmed by TEM (Fig. S1B), in which the morphology was similar to that of the work of Teplow and co-workers (28). The slight difference between A and B in Fig. S1 might be due to the difference of Aβ used for preparation (i.e. Fig. S1A: biotin–E22P–V40DAP–Aβ42 dimer, Fig. S1B: WT Aβ40). For preparation of fibrils of WT Aβ40 and WT Aβ42, the pellets were obtained by centrifugation at 20,130 × g at 4 °C for 1 h after each Aβ solution was incubated for 2 weeks at 37 °C.
HFIP treatment of Aβ
For treatment with 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP; Wako, Osaka, Japan), each Aβ was dissolved in HFIP at 1 mg/ml. After incubation at room temperature for 30 min, the solution was sonicated for 5 min and dried in vacuo. The resultant Aβ film was stored at −80 °C until use.
Preparation of the initial RNA pool
A DNA library consisting of a randomized region of 29 nucleotides (A:T:G:C = 25:25:25:25%), 5′- and 3′-primer regions and thymine linkers of three nucleotides between the randomized region and two primer regions (library: 5′-TAG AGA TAA TAC GAC TCA CTA TAG GGA CGA AGA CCA ACT GAA C-ttt-N29-ttt-GTC CGT GCT GCC ACC TTA CTT C-3′) was designed. DNA library and both primers were purchased from either ThermoFisher or Eurofins (Tokyo, Japan). PCR was performed to amplify with the 5′-primer (5′-TAG AGA TAA TAC GAC TCA CTA TAG GGA CGA AGA CCA ACT GAA C-3′) and 3′-primer (5′-GAA GTA AGG TGG CAG CAC GGA C-3′) under the following conditions: 20 cycles at 98 °C for 10 s (denaturing), 53 °C for 30 s (annealing), and 72 °C for 5 s (extension) using the TaKaRa Ex Taq hot start version kit on a TaKaRa PCR thermal cycler dice mini (TaKaRa). Following purification of the double-stranded, PCR-amplified DNA template with the QIAquick PCR purification kit (Qiagen), RiboMAX large-scale RNA production system T7 (Promega, Madison, WI) was used to generate the RNA pool for selection. After phenol-chloroform extraction and desalting using Illustra MicroSpin G-25 columns (GE Healthcare), the integrity of RNA was confirmed by electrophoresis using 6% EDTA–urea acrylamide gels (Invitrogen) and stained by SYBR Green (TaKaRa). RNA quantification was performed using a UV BioPhotometer (Eppendorf). In the SELEX or binding assays (Fig. 1, B–D), the ratio of bound RNA was calculated by real-time PCR (real-time PCR CFX96; Bio-Rad) using the PrimeScriptTM RT reagent kit (Perfect Real Time, TaKaRa) and TB GreenTM premix Ex TaqTM II (Tli RNaseH Plus, TaKaRa). The fibrils induced from WT Aβ42 and E22P–Aβ42 were synthesized as reported previously (45) and were prepared so that each Aβ dissolved in 0.1% NH4OH at 250 μm, followed by 10-fold dilution with PBS to a final concentration of 25 μm and then subjected to incubation at 37 °C for 48 h. The RNA pool or the aptamer was denatured at 90 °C for 10 min and renatured rapidly on ice for 10 min for refolding before use in the following studies.
SELEX for RNA aptamers using PFs as targets
Selection using Aβ42 toxic dimer as target
The biotin dimer (biotin–E22P–V40DAP–Aβ42 dimer) treated with HFIP was dissolved in 0.1% NH4OH at 250 μm, followed by 10-fold dilution with PBS (50 mm sodium phosphate, and 100 mm NaCl, pH 7.4) to a final concentration of 25 μm. To avoid excessive aggregation of Aβ42, dimers without preincubation were incubated with Dynabeads 280-M streptavidin (Veritas, Tokyo, Japan) in PBS at 4 °C for 1 h. The renatured RNA pool was incubated with the biotin dimer–immobilized streptavidin beads (RNA pool:E22P–Aβ42 dimer = 100:400 pmol) in PBS at 4 °C for 2 h. After removing unbound RNA by washing with PBS, Aβ was denatured by a SDS buffer (SDS buffer: 3.5 mm SDS, 0.5 m ammonium acetate, 10 mm magnesium acetate, 1 mm EDTA) at 70 °C for 10 min. Bound RNA was purified as described above. Purified RNA was reverse-transcribed and amplified by PCR. The negative selection of the RNA pool against streptavidin beads was performed before round 8 to remove nonspecific RNAs that bind the beads. Although nine rounds were used, significant enrichment was not observed (data not shown).
Selection using the preincubated Aβ42 toxic dimer as target
The biotin dimer treated with HFIP was dissolved in 0.1% NH4OH at 250 μm, followed by 10-fold dilution with PBS to a final concentration of 25 μm. Initially, the renatured RNA pool was incubated with the preincubated dimer model (biotin–E22P–V40DAP–Aβ42 dimer) (RNA:E22P–Aβ42 dimer = 250:1000 pmol) in PBS at 37 °C for 24 h. The resulting mixture was incubated with Dynabeads M-280 streptavidin beads (Veritas) in PBS at 25 °C for 1 h. After removing unbound RNA by washing with PBS, Aβ was denatured by the SDS buffer at 70 °C for 10 min. Bound RNA was purified by extraction with phenol-chloroform–isoamyl alcohol and chloroform–isoamyl alcohol, followed by ethanol precipitation. Purified RNA was reverse-transcribed into cDNA by the ImProm-II reverse transcription system (Promega) and amplified by PCR. The negative selection of RNA pool against streptavidin beads was performed before round 3 to remove nonspecific RNAs that bind the beads. Although five rounds were used, the significant enrichment was not observed (data not shown).
Selection using PFs as target
The dimer (E22P–V40DAP–Aβ42 dimer) treated with HFIP was dissolved in 0.1% NH4OH at 250 μm, followed by 10-fold dilution with PBS to a final concentration of 25 μm. To enlarge the molecular mass of the oligomer, the dimers were further preincubated at 37 °C for 48 h. The formation of PFs was confirmed by TEM analysis (Fig. S1A). The renatured RNA pool was incubated with mixture of PFs (RNA pool:E22P–Aβ42 dimer = 100:400 pmol) in PBS at 25 °C for 1 h. By using a Millipore Amicon Ultra-0.5 centrifugal filter unit (molecular mass cutoff, 50 kDa; Millipore), bound RNA with PFs were collected. After washing with PBS, Aβ was denatured by the SDS buffer at 70 °C for 10 min. Bound RNA was purified as described above. Purified RNA was reverse-transcribed and was amplified by PCR. Over the course of SELEX, the ratio of RNA:Aβ was increased gradually from 1:4 to 1:1 to 2:1 in the first five rounds, next round, and final round, respectively. After seven rounds, a significant enrichment was observed (Fig. 1B).
BLI measurements
BLI experiments were conducted at 30 °C with an OctetRED96 (ForteBio, Menlo Park, CA). The biotinylation of RNA aptamers were conducted by the 5′ EndTag nucleic acid labeling system (Vector) using biotin–PEG6–maleimide (TCI, Tokyo, Japan) and according to the manufacture's protocol. The RNA oligonucleotide of the random region sequence of E22P–AbD43 was purchased from FASMAC (Kanagawa, Japan). Biotinylated RNA aptamers were immobilized on a streptavidin biosensor that was subjected to 10 min of rehydration in PBS buffer before carrying out the binding experiments. The immobilization of the biotinylated aptamers to the sensor was performed with 200 μl of 1 μm biotinylated aptamers in 96-well black plate for 840 s followed by a 180-s incubation of the sensor in PBS buffer. The binding reaction occurred in 200 μl of PBS buffer containing 0.05% Tween 20 (IBL, Gunma, Japan) including various concentrations of HFIP-treated Aβ (200–1,600 nm) with agitation under the condition of association for 180 s and dissociation for 180 s.
The sensorgrams were better fitted globally to a 2:1 heterogeneous ligand-binding model or a 1:1 binding model contingent on the lower χ2 coefficient, and the association rate kon (kon1 and kon2), the dissociation rate koff (koff1 and koff2), and KD (KD1 and KD2) were obtained by fitting the data through data analysis software from ForteBio (Table S1). For a reference, the kinetic parameters for kon, koff, and KD in the corresponding binding model contingent on the higher χ2 coefficient are shown in Table S2. The percent contributions from KD1 and KD2 were shown in the case of the 2:1 heterogeneous ligand model. The contribution of KD1 was larger than that of KD2 (Tables S1 and S2). Because the association signal shifts of aptamer for WT Aβ42 fibril and WT Aβ40 fibril were below the signal to noise, the curve fitting was not performed, and kinetic parameters were not able to be calculated (Tables S1 and S2). In general, the 2:1 heterogeneous ligand-binding model simply assumes binding with analyte at two independent ligand sites of the 1:1 binding model. Each ligand site binds the analyte independently with a different rate constant with an additional parameter to account for percentage of binding contributed by each interaction. χ2 coefficient is simply the square of the sum of deviations between the data and the fit curve. Other plausible and equally feasible synergistic 2:1 heterogeneous ligand-binding models were not considered to analyze the BLI-binding data.
Fluorescence polarization
The fluorescence polarization binding assays between FITC-labeled aptamer (20 nm) and HFIP-treated Aβ (10−9.2–10−6.5 M) were performed with a Multilabel plate reader ARVO X5 (PerkinElmer). FITC-labeled aptamer was prepared by the 5′ EndTag nucleic acid labeling system (Vector) using fluorescein-5-maleimide (TCI) according to the manufacturer's protocol. The samples in 200 μl of PBS plus potassium (PBS-K; 10 mm sodium phosphate, 140 mm NaCl, pH 7.4) were incubated at room temperature for 1 h in a 96-well black plate. The excitation and emission wavelengths were 485 and 538 nm, respectively, and then fluorescence polarization was calculated. The resulting millipolarization (mP) values were plotted against concentration using GraphPad Prism version 6 (GraphPad Software, San Diego, CA).
Sequencing and prediction of secondary structure of the RNAs
After enrichment of bound RNA, the individual aptamer clones were obtained by ligation of the PCR products using the TOPO TA cloning kit (Invitrogen). Cloned DNA was sequenced at Eurofins. Alignment search, classification, and local super-motif analysis were performed based on sequences of 36 unique aptamers by Clustal Omega (22) (Fig. S3A) and Multiple EM for motif elucidation (23) (Fig. S3B). To identify common secondary structural motifs, RNA sequences were folded using the energy minimization algorithm of CentroidFold (21) (Fig. 2B).
CD spectropolarimetry
CD spectra were measured on a JASCO J-805 spectropolarimeter using a 0.1-mm path length quartz cell, as described previously (20). After dilution of the RNA solution (final concentration, 10 μm) with PBS containing 1 mm EDTA, an aliquot (200 μl) was loaded into the quartz cell, and the CD spectrum was recorded over the wavelength range of 220–320 nm. The spectra are shown after subtraction of the spectrum of the vehicle alone.
ATR-FTIR
FTIR was measured using the ATR method as described previously (19). A droplet (5∼10 μg) of aptamer solution with PBS containing 1 mm EDTA was loaded and then air-dried on an ATR unit in the FTIR 470plus spectrometer (Jasco, Tokyo, Japan). To improve the signal-to-noise ratio, 128 scans were accumulated, and the spectra were recorded at a resolution of 4.0 cm−1. The spectra are shown after suppression of water vapor signal.
TEM
The aggregates of each Aβ solution (25 μm) in PBS after 24 or 48 h incubation in the absence or presence of E22P–AbD43 (50 μm) were examined under a TEM (JEM-1400; JEOL, Tokyo, Japan). After each Aβ aggregate was centrifuged (4 °C, 17,900 × g, 10 min), the supernatant was removed from the pellet. The resultant pellet was gently resuspended in water (100 μl) using a vortex for 1 min just before TEM analysis. The sample suspension (15 μl) was applied to a 400-mesh carbon-coated copper grid (thickness, 20–25 nm; Veco, Eerbeek, The Netherlands) and incubated for 5 min before being negatively stained twice with 2% uranyl acetate. Stained samples were subjected to TEM.
Th-S aggregation assay
The aggregation of each Aβ in the presence of RNA aptamers was evaluated by a Th-S (Sigma–Aldrich) fluorescence assay based on the Th-T assay developed by Naiki and Gejyo (31) with modifications, because the inherent nonspecific binding of Th-S to nucleic acids is much lower than that of Th-T (32). The procedure has been described previously (43). In brief, WT Aβ42 and E22P–Aβ42, in addition to the E22P–V40DAP–Aβ42 dimer, were dissolved in 0.1% NH4OH at 100 or 50 μm, and E22P–AbD43 was dissolved in RNase-free water (Invitrogen) at 200 μm, followed by dilution with PBS to the desired concentration (WT Aβ42 and E22P–Aβ42, 10 μm; E22P–V40DAP–Aβ42 dimer, 5 μm; E22P–AbD43, 40 μm). The fluorescence intensity was measured by excitation at 430 nm and emission at 485 nm using a microplate reader (Fluoroskan Ascent; ThermoFisher, Rockford, IL).
MTT assay
SH-SY5Y cells (ATCC, Manassas, VA) maintained in a mixed medium containing equal amounts of Eagle's minimal essential medium (Wako) and Ham's F12 medium (Wako) containing 10% fetal bovine serum (Biological Industries) were used as a typical neuronal cell model to estimate the neurotoxicity of each Aβ (WT Aβ42, E22P–Aβ42, and the E22P–V40DAP–Aβ42 dimer) with slight modifications to the described method (20). Briefly, Aβ, E22P–AbD43, or R7 were dissolved in 0.1% NH4OH or in RNase-free water to make a 12× stock before being diluted with culture medium to the desired final concentration. After preincubating 120 μl of Aβ with E22P–AbD43 or R7 for 30 min at room temperature, the culture medium used on nearly confluent cells (1 × 104 cells/well) for overnight adaptation was exchanged with the preincubated solution (120 μl). After incubation for 16 h at 37 °C, 15 μl/well of Dye solution from the CellTiter 96 nonradioactive cell proliferation assay kit (Promega) was added to the cells, followed by incubation for 4 h at 37 °C. The solubilization/stop solution (100 μl/well) was subsequently added to the cells. The cell lysate was incubated overnight in the dark at room temperature before performing measurements at 570 nm with a microplate reader (Multiskan FC; ThermoFisher). The absorbance obtained by adding the vehicle (0.1% NH4OH + RNase-free water) was taken as 100%.
Histochemical staining
All experimental procedures were performed in accordance with specified guidelines for the care and use of laboratory animals and were approved by the Animal Care and Use Committee of Chiba University. Five-micrometer-thick coronal paraffin-embedded sections were prepared from 4% paraformaldehyde-fixed brain hemispheres of an AD mouse brain (Tg2576/PS2) (33) at the age of 6 months. After deparaffinization and hydration, the slices were autoclaved at 120 °C for 20 min for antigen activation. To inactivate the endogenous peroxidase, the brain sections were soaked in methanol with 0.1% H2O2 at room temperature for 30 min. After washing with ice-cold PBS-K containing 0.02% Tween 20 (PBS-T), blocking was performed in a blocking buffer, PBS-T with 10 μg/ml BSA (Nacalai) and 10 μg/ml yeast tRNA (Nacalai, Kyoto, Japan) at room temperature for 60 min. Biotinylated E22P–AbD43 (400 nm) diluted by RNase-free water (Invitrogen) with 1 mm EDTA was applied at room temperature for 60 min, followed by reaction with horseradish peroxidase (HRP)–conjugated avidin by the VECTORSTAIN ABC HRP kit (Vector) for 30 min at room temperature. Alternatively, the sections were treated with 82E1 (1 μg/ml) diluted with PBS-T at room temperature for 60 min, followed by reaction with the biotinylated secondary antibody for 30 min at room temperature before incubation with HRP-conjugated avidin by the VECTORSTAIN ABC HRP kit (Vector) for 30 min at room temperature. To visualize the signals, brain sections were treated with 3,3′-diaminobenzidine (Dojindo, Kumamoto, Japan) at 37 °C for 12 h (E22P–AbD43) or at room temperature for 12 min (82E1). The nuclei were stained with hematoxylin reagent (Wako). After dehydration and soaking in xylene, the brain sections were mounted with Permount (FALMA).
Statistical analysis
Statistical analysis was performed using the scientific data analysis software GraphPad Prism version 6 (GraphPad Software) with one-way analysis of variance followed by Tukey's test. Statistical significance is indicated in figure legends as ###, p < 0.001 versus vehicle alone; *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Author contributions
K. M. and K. I. conceptualization; K. M., Y. O., and N. I. data curation; K. M., Y. O., N. I., T. S., and K. I. formal analysis; K. M. and K. I. supervision; K. M. and K. I. funding acquisition; K. M., Y. O., A. S., H. U., and N. I. investigation; K. M. writing-original draft; T. A., K. T., T. S., and K. I. resources; K. M. and K. I. validation; K. I. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Prof. Hirohide Saito (CiRA, Kyoto University) for helpful discussions of aptamer selection, Prof. Masaya Nagao and Dr. Taiho Kambe (Graduate School of Biostudies, Kyoto University) for accessibility to real-time PCR, and Mr. Takeshi Seguchi (Primetech, Tokyo, Japan) for technical assistance of BLI measurement. We thank the Edanz Group for editing a draft of this manuscript.
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grants 26221202 (to K. I. and K. M.) 16H06194 (to K. M.), and 22603006 (to K. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S8.
- AD
- Alzheimer's disease
- Aβ
- amyloid β-protein
- ATR
- attenuated total reflection
- BLI
- biolayer interferometry
- DAP
- l,l-2,6-diaminopimelic acid
- HFIP
- 1,1,1,3,3,3-hexafluoro-2-propanol
- HRP
- horseradish peroxidase
- PF
- protofibril
- SELEX
- systematic evolution of ligands by exponential enrichment
- TEM
- transmission electron microscopy
- Th-S
- thioflavin-S
- Th-T
- thioflavin-T
- R7
- enriched RNA pool after round seven
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Glenner G. G., and Wong C. W. (1984) Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 10.1016/S0006-291X(84)80190-4 [DOI] [PubMed] [Google Scholar]
- 2. Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., and Beyreuther K. (1985) Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. U.S.A. 82, 4245–4249 10.1073/pnas.82.12.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haass C., and Selkoe D. J. (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 10.1038/nrm2101 [DOI] [PubMed] [Google Scholar]
- 4. Roychaudhuri R., Yang M., Hoshi M. M., and Teplow D. B. (2009) Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 284, 4749–4753 10.1074/jbc.R800036200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watanabe-Nakayama T., Ono K., Itami M., Takahashi R., Teplow D. B., and Yamada M. (2016) High-speed atomic force microscopy reveals structural dynamics of amyloid β1–42 aggregates. Proc. Natl. Acad. Sci. U.S.A. 113, 5835–5840 10.1073/pnas.1524807113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harper J. D., Wong S. S., Lieber C. M., and Lansbury P. T. (1997) Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem. Biol. 4, 119–125 10.1016/S1074-5521(97)90255-6 [DOI] [PubMed] [Google Scholar]
- 7. Walsh D. M., Lomakin A., Benedek G. B., Condron M. M., and Teplow D. B. (1997) Amyloid β-protein fibrillogenesis: detection of a protofibrillar intermediate. J. Biol. Chem. 272, 22364–22372 10.1074/jbc.272.35.22364 [DOI] [PubMed] [Google Scholar]
- 8. Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., and Klein W. L. (1998) Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 10.1073/pnas.95.11.6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoshi M., Sato M., Matsumoto S., Noguchi A., Yasutake K., Yoshida N., and Sato K. (2003) Spherical aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate Tau protein kinase I/glycogen synthase kinase-3β. Proc. Natl. Acad. Sci. U.S.A. 100, 6370–6375 10.1073/pnas.1237107100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou J., and Rossi J. (2017) Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 10.1038/nrd.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ylera F., Lurz R., Erdmann V. A., and Fürste J. P. (2002) Selection of RNA aptamers to the Alzheimer's disease amyloid peptide. Biochem. Biophys. Res. Commun. 290, 1583–1588 10.1006/bbrc.2002.6354 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi T., Tada K., and Mihara H. (2009) RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol. Biosyst. 5, 986–991 10.1039/b903391b [DOI] [PubMed] [Google Scholar]
- 13. Rahimi F., Murakami K., Summers J. L., Chen C. H., and Bitan G. (2009) RNA aptamers generated against oligomeric Aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS One 4, e7694 10.1371/journal.pone.0007694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsukakoshi K., Abe K., Sode K., and Ikebukuro K. (2012) Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal. Chem. 84, 5542–5547 10.1021/ac300330g [DOI] [PubMed] [Google Scholar]
- 15. Chakravarthy M., AlShamaileh H., Huang H., Tannenberg R. K., Chen S., Worrall S., Dodd P. R., and Veedu R. N. (2018) Development of DNA aptamers targeting low-molecular-weight amyloid-β peptide aggregates in vitro. Chem. Commun. (Camb.) 54, 4593–4596 10.1039/C8CC02256A [DOI] [PubMed] [Google Scholar]
- 16. Irie K., Murakami K., Masuda Y., Morimoto A., Ohigashi H., Hara H., Ohashi R., Takegoshi K., Fukuda H., Nagao M., Shimizu T., and Shirasawa T. (2007) The toxic conformation of the 42-residue amyloid β peptide and its relevance to oxidative stress in Alzheimer's disease. Mini-Rev. Med. Chem. 7, 1001–1008 10.2174/138955707782110187 [DOI] [PubMed] [Google Scholar]
- 17. Morimoto A., Irie K., Murakami K., Masuda Y., Ohigashi H., Nagao M., Fukuda H., Shimizu T., and Shirasawa T. (2004) Analysis of the secondary structure of β-amyloid (Aβ42) fibrils by systematic proline replacement. J. Biol. Chem. 279, 52781–52788 10.1074/jbc.M406262200 [DOI] [PubMed] [Google Scholar]
- 18. Masuda Y., Uemura S., Ohashi R., Nakanishi A., Takegoshi K., Shimizu T., Shirasawa T., and Irie K. (2009) Identification of physiological and toxic conformations in Aβ42 aggregates. ChemBioChem. 10, 287–295 10.1002/cbic.200800411 [DOI] [PubMed] [Google Scholar]
- 19. Murakami K., Irie K., Morimoto A., Ohigashi H., Shindo M., Nagao M., Shimizu T., and Shirasawa T. (2003) Neurotoxicity and physicochemical properties of Aβ mutant peptides from cerebral amyloid angiopathy: implication for the pathogenesis of cerebral amyloid angiopathy and Alzheimer's disease. J. Biol. Chem. 278, 46179–46187 10.1074/jbc.M301874200 [DOI] [PubMed] [Google Scholar]
- 20. Murakami K., Tokuda M., Suzuki T., Irie Y., Hanaki M., Izuo N., Monobe Y., Akagi K., Ishii R., Tatebe H., Tokuda T., Maeda M., Kume T., Shimizu T., and Irie K. (2016) Monoclonal antibody with conformational specificity for a toxic conformer of amyloid β42 and its application toward the Alzheimer's disease diagnosis. Sci. Rep. 6, 29038 10.1038/srep29038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato K., Hamada M., Asai K., and Mituyama T. (2009) CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37, W277–W280 10.1093/nar/gkp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 23. Bailey T. L., and Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 24. Ambrus A., Chen D., Dai J., Bialis T., Jones R. A., and Yang D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 10.1093/nar/gkl348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burge S., Parkinson G. N., Hazel P., Todd A. K., and Neidle S. (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 10.1093/nar/gkl655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andrushchenko V., Tsankov D., Krasteva M., Wieser H., and Bour P. (2011) Spectroscopic detection of DNA quadruplexes by vibrational circular dichroism. J. Am. Chem. Soc. 133, 15055–15064 10.1021/ja204630k [DOI] [PubMed] [Google Scholar]
- 27. Tsukakoshi K., Ikuta Y., Abe K., Yoshida W., Iida K., Ma Y., Nagasawa K., Sode K., and Ikebukuro K. (2016) Structural regulation by a G-quadruplex ligand increases binding abilities of G-quadruplex-forming aptamers. Chem. Commun. (Camb.) 52, 12646–12649 10.1039/C6CC07552E [DOI] [PubMed] [Google Scholar]
- 28. Ono K., Condron M. M., Ho L., Wang J., Zhao W., Pasinetti G. M., and Teplow D. B. (2008) Effects of grape seed-derived polyphenols on amyloid β-protein self-assembly and cytotoxicity. J. Biol. Chem. 283, 32176–32187 10.1074/jbc.M806154200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rossi A. M., and Taylor C. W. (2011) Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 6, 365–387 10.1038/nprot.2011.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Serio T. R., Cashikar A. G., Kowal A. S., Sawicki G. J., Moslehi J. J., Serpell L., Arnsdorf M. F., and Lindquist S. L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 10.1126/science.289.5483.1317 [DOI] [PubMed] [Google Scholar]
- 31. Naiki H., and Gejyo F. (1999) Kinetic analysis of amyloid fibril formation. Methods Enzymol. 309, 305–318 10.1016/S0076-6879(99)09022-9 [DOI] [PubMed] [Google Scholar]
- 32. Sugimoto S., Arita-Morioka K., Mizunoe Y., Yamanaka K., and Ogura T. (2015) Thioflavin T as a fluorescence probe for monitoring RNA metabolism at molecular and cellular levels. Nucleic Acids Res. 43, e92 10.1093/nar/gkv338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toda T., Noda Y., Ito G., Maeda M., and Shimizu T. (2011) Presenilin-2 mutation causes early amyloid accumulation and memory impairment in a transgenic mouse model of Alzheimer's disease. J. Biomed. Biotechnol. 2011, 617974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horikoshi Y., Mori T., Maeda M., Kinoshita N., Sato K., and Yamaguchi H. (2004) Aβ N-terminal-end specific antibody reduced β-amyloid in Alzheimer-model mice. Biochem. Biophys. Res. Commun. 325, 384–387 10.1016/j.bbrc.2004.10.039 [DOI] [PubMed] [Google Scholar]
- 35. Koffie R. M., Meyer-Luehmann M., Hashimoto T., Adams K. W., Mielke M. L., Garcia-Alloza M., Micheva K. D., Smith S. J., Kim M. L., Lee V. M., Hyman B. T., and Spires-Jones T. L. (2009) Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. U.S.A. 106, 4012–4017 10.1073/pnas.0811698106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., and Glabe C. G. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 10.1126/science.1079469 [DOI] [PubMed] [Google Scholar]
- 37. Farrar C. T., William C. M., Hudry E., Hashimoto T., and Hyman B. T. (2014) RNA aptamer probes as optical imaging agents for the detection of amyloid plaques. PLoS One 9, e89901 10.1371/journal.pone.0089901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murakami K., Horikoshi-Sakuraba Y., Murata N., Noda Y., Masuda Y., Kinoshita N., Hatsuta H., Murayama S., Shirasawa T., Shimizu T., and Irie K. (2010) Monoclonal antibody against the turn of the 42-residue amyloid β protein at positions 22 and 23. ACS Chem. Neurosci. 1, 747–756 10.1021/cn100072e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Irie Y., Murakami K., Hanaki M., Hanaki Y., Suzuki T., Monobe Y., Takai T., Akagi K. I., Kawase T., Hirose K., and Irie K. (2017) Synthetic models of quasi-stable amyloid β40 oligomers with significant neurotoxicity. ACS Chem. Neurosci. 8, 807–816 10.1021/acschemneuro.6b00390 [DOI] [PubMed] [Google Scholar]
- 40. Kageyama Y., Saito A., Pletnikova O., Rudow G. L., Irie Y., An Y., Murakami K., Irie K., Resnick S. M., Fowler D. R., Martin L. J., and Troncoso J. C. (2018) Amyloid β toxic conformer has dynamic localization in the human inferior parietal cortex in absence of amyloid plaques. Sci. Rep. 8, 16895 10.1038/s41598-018-35004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ginsberg S. D., Galvin J. E., Chiu T. S., Lee V. M., Masliah E., and Trojanowski J. Q. (1998) RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 96, 487–494 10.1007/s004010050923 [DOI] [PubMed] [Google Scholar]
- 42. Ginsberg S. D., Crino P. B., Hemby S. E., Weingarten J. A., Lee V. M., Eberwine J. H., and Trojanowski J. Q. (1999) Predominance of neuronal mRNAs in individual Alzheimer's disease senile plaques. Ann. Neurol. 45, 174–181 [DOI] [PubMed] [Google Scholar]
- 43. Sato M., Murakami K., Uno M., Nakagawa Y., Katayama S., Akagi K., Masuda Y., Takegoshi K., and Irie K. (2013) Site-specific inhibitory mechanism for amyloid β42 aggregation by catechol-type flavonoids targeting the Lys residues. J. Biol. Chem. 288, 23212–23224 10.1074/jbc.M113.464222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanaki M., Murakami K., Akagi K., and Irie K. (2016) Structural insights into mechanisms for inhibiting amyloid β42 aggregation by non-catechol-type flavonoids. Bioorg. Med. Chem. 24, 304–313 10.1016/j.bmc.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 45. Murakami K., Irie K., Ohigashi H., Hara H., Nagao M., Shimizu T., and Shirasawa T. (2005) Formation and stabilization model of the 42-mer Aβ radical: implications for the long-lasting oxidative stress in Alzheimer's disease. J. Am. Chem. Soc. 127, 15168–15174 10.1021/ja054041c [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.