Abstract
2′,5′/3′,5′-cGMP-AMP (cGAMP) is a second messenger produced in response to cytosolic dsDNA that activates the stimulator of interferon genes (STING) pathway. We recently discovered that cGAMP is exported by cancer cells and that this extracellular signal is an immunotransmitter key to tumor detection and elimination by the innate immune system. The enhancement of extracellular cGAMP levels therefore holds great promise for managing cancer. However, there is still much more to understand about the basic biology of cGAMP before its full therapeutic potential can be realized. To answer these questions, we must be able to detect and quantitate cGAMP with an assay that is high-throughput, sensitive, and precise. Existing assays fall short of these needs. Here, we describe the development of cGAMP-Luc, a coupled enzyme assay that relies on the degradation of cGAMP to AMP by ectonucleotide pyrophosphatase phosphodiesterase 1 (ENPP1) and an optimized assay for the detection of AMP by luciferase. We also developed STING-CAP, a STING-mediated method to concentrate and purify cGAMP from any type of biological sample. We conclude that cGAMP-Luc is an economical high-throughput assay that matches the accuracy of and surpasses the detection limit of MS, the current gold standard of cGAMP quantitation. We propose that cGAMP-Luc is a powerful tool that may enable discoveries that advance insights into extracellular cGAMP levels in healthy and diseased tissues, such as cancer.
Keywords: cyclic nucleotide, enzyme, mass spectrometry (MS), analytical chemistry, drug discovery, ATP, assay development, cancer, coupled-enzyme assay, extracellular cGAMP, STING, cyclic nucleotide
Introduction
Innate immunity is our first line of defense against foreign pathogens and cancers. It is activated when pattern or damage recognition receptors detect pathogen or damage-associated molecular patterns. One such molecular pattern is cytosolic dsDNA, which signals the presence of viral infection, cell damage, and cancer. In cancer cells, cytosolic dsDNA originates from chromosomal instability and the formation of micronuclei (1–3). Once in the cytosol, this dsDNA activates the enzyme cGMP/AMP synthase (cGAS)2 to synthesize 2′,5′/3′,5′-cAMP-GMP (cGAMP) from ATP and GTP (4–9). cGAMP binds to and activates the protein stimulator of interfering genes (STING), leading to the production of Type I interferons (10–13). These potent cytokines drive tumor regression both directly (14) and through the activation of the adaptive immune system (15, 16). Since its discovery in 2008, the cytosolic dsDNA sensing STING pathway has taken center stage in cancer drug development.
We recently discovered that extracellular cGAMP is an immunotransmitter produced and exported by cancer cells (17) and imported by host cells (18, 19). Extracellular cGAMP plays a major role in tumor detection and elimination; we demonstrated through in vivo experiments that depleting extracellular cGAMP in tumors represses innate immune activation (17). We also found that enhancing extracellular cGAMP through the inhibition of its degradation or the catalysis of its synthesis with ionizing radiation diminishes tumors (17). The enhancement of extracellular cGAMP therefore contains great potential for the treatment of cancer. Nonhydrolyzable analogs of cGAMP are currently in Phase I clinical trials for the treatment of metastatic solid tumors with promising results (SCR_002309: NCT03172936 and NCT03010176).
To realize the full therapeutic potential of extracellular cGAMP, there is still much more to be understood about the basic biology of cGAMP. Among the immediately apparent questions are how ENPP1 inhibitors can be screened in a high-throughput manner, how cGAMP exporters can be identified in cancer cells, how cancer cells can be stimulated to export more cGAMP, and which cell types import and respond to cGAMP to mediate the immune response. Outside of cancer immunology, the roles that extracellular cGAMP plays in homeostasis and other diseases are largely unexplored.
To begin to address these questions, we must first be able to measure cGAMP using an assay accessible to academic laboratories, pharmaceutical companies, and hospitals. Such an assay must have a wide dynamic range, as physiological cGAMP concentrations range from low nanomolar to high micromolar (17). In addition, this assay must be sufficiently precise to study significant biological processes that may rely only on subtle changes in cGAMP concentrations. Ideally, such an assay is also suitable for high-throughput genetic and drug screens.
Current methods to quantitate cGAMP fall short of these standards. We previously developed a quantitative LC tandem MS (LC-MS/MS) technique with a wide dynamic range and detection limit of 4 nm (17), but it is low-throughput and requires equipment and expertise beyond the range of most laboratories. cGAMP ELISAs are commercially available, but like all ELISAs, they are expensive and have a narrow detection range. A riboswitch for cGAMP has also been developed, but its Kd of 13 μm limits its utility in detecting and quantitating physiologically relevant concentrations of cGAMP (20). Here, we describe the development of cGAMP-Luc, a sensitive, precise, and high-throughput coupled enzyme assay based on luciferase that can quantitate cGAMP in a range of complex biological samples.
Results
cGAMP can be detected but not quantitated by pairing ENPP1 with commercially available AMP quantitation kits
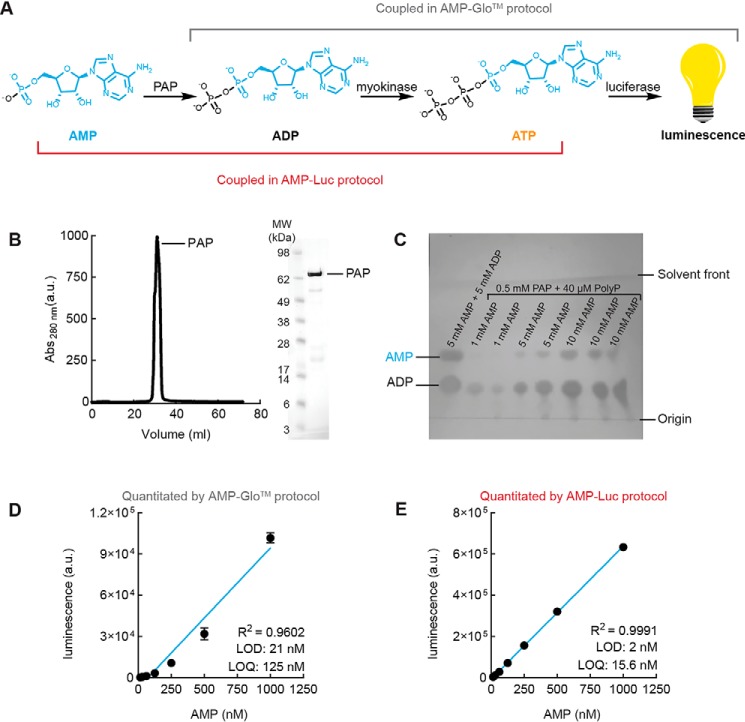
The general scheme of the cGAMP-Luc assay is described in Fig. 1A. Our overall strategy takes advantage of our discovery of ENPP1, the dominant hydrolase of cGAMP (21). In this assay, cGAMP is first cleaved into AMP and GMP by ENPP1. We considered alternative cGAMP hydrolases, but they are ultimately incompatible with this assay; poxvirus immune nucleases (poxins) do not cleave cGAMP to AMP (22), and snake venom phosphodiesterase (6) cannot be purified in large quantities as it is harvested from snake venom. Following ENPP1 digestion of cGAMP, the resulting AMP is subsequently converted to ADP by the enzyme polyphosphate:AMP phosphotransferase (PAP) at the expense of polyphosphate (23, 24). ADP is then phosphorylated to ATP by myokinase. ATP is used by luciferase to oxidize luciferin in a concentration-dependent manner to generate a luminescent species. The final ATP concentration is thus quantitated on a luminescence plate reader and related back to the original cGAMP concentration.
Figure 1.
cGAMP can be detected but not quantitated by pairing ENPP1 with commercially available AMP quantitation kits. A, scheme of cGAMP-Luc assay. B, purification of ENPP1 by size-exclusion chromatography. C, 2′3′-cGAMP hydrolysis reaction monitored by TLC assay and visualized by autoradiography. Data are representative of three independent experiments. D, cGAMP standard curve generated by ENPP1 and AMP-GloTM. Data are mean ± S.E. (error bars) (n = 2). E, AMP standard curve generated by AMP-GloTM. Data are mean ± S.E. (n = 4) with error bars too small to visualize. F, ATP standard curve generated by CellTiter-Glo®. Data are mean ± S.E. (n = 3) with error bars too small to visualize.
To accurately quantitate low nanomolar concentrations of cGAMP, all enzymatic steps of cGAMP-Luc should operate at nanomolar concentrations and couple with each other in a linear fashion. We purified ENPP1 (Fig. 1B) and first tested whether it could convert nanomolar amounts of cGAMP to AMP despite a Km of 15 μm for cGAMP. We incubated a trace amount of [32P]cGAMP with 350 nm cGAMP and 10 nm ENPP1 for 16 h and analyzed the reaction mixture using TLC (Fig. 1C). We observed complete cleavage of cGAMP to Pi with a trace amount of the desired AMP intermediate. This is in agreement with our previous observation (21) that trace amounts of phosphatases from the purified cGAS protein are sufficient to cleave most of the AMP intermediate to Pi. To avoid phosphatase contaminants and the subsequent loss of AMP as the key analyte, all of the cold cGAMP used in the rest of this study was subjected to HPLC purification. Regardless, the demonstrated complete cleavage of crude hot [32P]cGAMP indicates that the quantitation of cGAMP via its cleavage product AMP is a viable strategy.
We then attempted to generate a standard curve by coupling our ENPP1 cleavage of cGAMP to AMP with a commercially available AMP quantitation assay, AMP-GloTM. Biological samples likely contain a significant amount of AMP that does not originate from cGAMP. To ensure specificity for cGAMP, we plotted standard curves after taking the difference in luminescence signals of samples that were and were not subjected to ENPP1 cleavage. The generated cGAMP standard curve has a quantitation limit of nearly 200 nm (Fig. 1D), which is 50 times worse than our LC-MS/MS method (17). Similarly, the standard curve of AMP generated by AMP-GloTM has a quantitation limit of 125 nm (Fig. 1E). In contrast, the standard curve of ATP alone is linear and quantitative until 16 nm (Fig. 1F). The limit of quantitation of ATP is the theoretical limit of quantitation of cGAMP. To improve the quantitation of cGAMP to approach this theoretical limit, we began by optimizing the quantitation of AMP.
Optimization of AMP quantitation assay (AMP-Luc) with new quantitation limit of 16 nm
In the commercially available AMP-GloTM assay kit, the detection of ATP by luciferase is coupled to its synthesis from ADP. We hypothesized that the quantitation limit of AMP would improve if we allowed ATP to accumulate to greater concentrations before its detection by luciferase. To test this hypothesis, we assembled the components of this assay ourselves and uncoupled these two steps. As shown in the scheme in Fig. 2A, we instead coupled the generation of ADP and ATP from AMP by the enzymes PAP and myokinase.
Figure 2.
Optimization of AMP detection method (AMP-Luc) with new quantitation limit of 16 nm. A, scheme of coupled enzymatic steps in AMP-GloTM versus AMP-Luc protocol. B, Purification of PAP by anion exchange. C, PAP activity assay monitored by TLC and visualized under short-wavelength (254-nm) UV light source. D, AMP standard curve generated using the AMP-GloTM coupling scheme, with all enzymes either purified (ENPP1, PAP) or purchased (myokinase, luciferase) and assembled in-house. Data are mean ± S.E. (error bars) (n = 3). E, AMP standard curve generated using the AMP-Luc coupling scheme, with all enzymes either purified or purchased and assembled in-house. Data are mean ± S.E. (n = 3) with error bars too small to visualize.
As the first step to assembling our own AMP quantitation assay, we expressed and purified PAP from Escherichia coli (Fig. 2B) and verified its activity by TLC. In 2 h, the conversion of 1 mm AMP to ADP by 0.5 mm PAP was nearly complete (Fig. 2C), suggesting that our purified PAP enzyme is active.
To test our hypothesis that rearranging the coupled steps in the AMP assay would lead to an improved quantitation limit, we generated AMP standard curves with both the old and new coupling schemes. We combined PAP with a commercially available myokinase enzyme to convert AMP to ATP and used CellTiter-Glo® to quantitate the resulting ATP. The quantitation limit with the old coupling scheme was 125 nm, similar to that generated by the AMP-GloTM assay (Figs. 1E and 2D). Under the new coupling scheme, the standard curve of AMP was much more linear and had a quantitation limit of 15 nm (Fig. 2E), confirming our hypothesis that we could improve the quantitation limit of AMP by rearranging the coupled steps in the assay. We named this new AMP quantitation assay AMP-Luc.
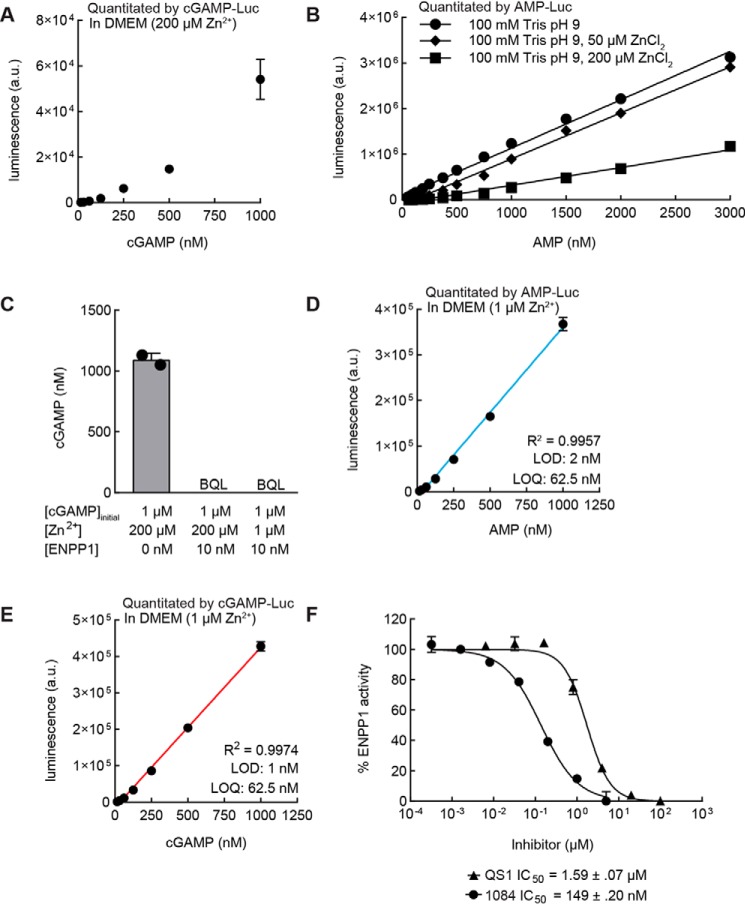
Optimization of ENPP1 cleavage of cGAMP to yield quantitation limit of 62.5 nm
We combined our new AMP-Luc assay with ENPP1 to generate a cGAMP standard curve but found that it was still nonlinear (Fig. 3A). After confirming that any GMP from the cGAMP digestion would not affect any components of AMP-Luc (Fig. S1), we hypothesized that this nonlinearity was due to added components from the digestion of cGAMP by ENPP1. Zn2+ is a known inhibitor of myokinase, and there is 200 μm ZnCl2 present in our previously reported ENPP1 buffer (21, 25). Indeed, standard curves of AMP generated with buffers containing 50 or 200 μm ZnCl2 show a concentration-dependent decrease in luminescence signal (Fig. 3B). Reducing the amount of ZnCl2 present in the buffer from 200 to 1 μm did not affect ENPP1 activity (Fig. 3C) and had minimal effect on the detection limit of AMP (Fig. 3D). With this minimal ENPP1 buffer, we obtained a linear cGAMP standard curve with a detection limit of 1 nm and a quantitation limit of 62.5 nm (Fig. 3E). With this optimized protocol, we achieved a smaller S.E. in the cGAMP standard curve than in the unoptimized protocol used to generate Fig. 1D. We named this fully optimized series of enzymatic reactions the cGAMP-Luc assay.
Figure 3.
Optimization of ENPP1 cleavage of cGAMP to yield quantitation limit of 62.5 nm. A, cGAMP standard curve with high-zinc ENPP1 buffer quantitated by cGAMP-Luc assay. Data are mean ± S.E. (error bars) (n = 3). B, AMP standard curve quantitated by an AMP-Luc assay (n = 1; data representative of two independent experiments). R2 is 0.9945 for 0 μm Zn2+, 0.9955 for 50 μm Zn2+, and 0.9648 for 100 μm Zn2+. C, 2′,3′-cGAMP hydrolysis reactions. Products of reactions were submitted for analysis by LC-MS/MS. Data are mean ± S.D. (error bars) (n = 2). D, AMP standard curve with low-zinc ENPP1 buffer quantitated by an AMP-Luc assay. Data are mean ± S.E. (n = 2). E, cGAMP standard curve with low-zinc ENPP1 buffer quantitated by cGAMP-Luc assay. Data are mean ± S.E. (n = 4). F, a serial dilution of ENPP1 inhibitors QS1 and 1084 was incubated with 10 nm ENPP1 and 5 μm cGAMP for 2 h at room temperature. The resulting AMP was quantitated via the AMP-Luc assay (n = 3 for 1084, n = 2 for QS1).
Use of cGAMP-Luc to accurately measure IC50 of ENPP1 inhibitors in buffer
With the linearity of the cGAMP standard curve established, we hypothesized that we could utilize the cGAMP-Luc assay to assess the potency of ENPP1 inhibitors. We incubated 5 μm cGAMP and 10 nm ENPP1 with a serial dilution of known ENPP1 inhibitors, STF-1084 (17) and QS1 (26). After a 2-h incubation, we heat-inactivated ENPP1 to terminate the reaction and quantitated the resulting AMP. The cGAMP-Luc assay determined IC50 values of 149 ± 20 nm for STF-1084 and 1.59 ± .07 μm for QS1 (Fig. 3F). These values agree well with our previously reported values of 110 ± 10 nm for STF-1084 and 6.4 ± 3.2 μm for QS1, determined by measuring the degradation of 32P-labeled cGAMP by ENPP1 in the presence of ENPP1 inhibitors (17). This experiment further validates the ability of the cGAMP-Luc assay to be quantitative across a wide range of cGAMP concentrations.
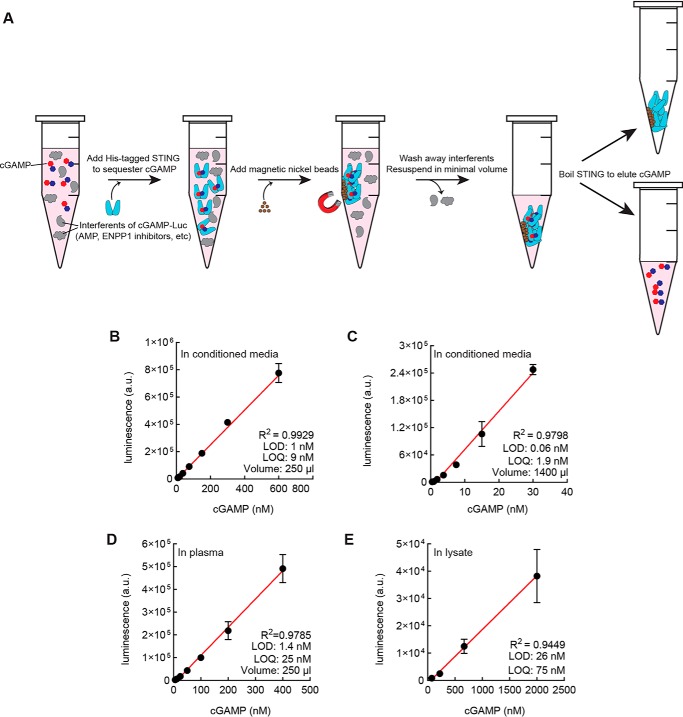
Development of STING-CAP for the quantitation of cGAMP in complex biological samples
Our ultimate objective is to develop an assay to quantitate cGAMP in a wide array of biological samples. Ideally, such an assay would be able to measure intracellular and extracellular cGAMP levels during homeostasis and in cancer. To preserve cGAMP in these biological samples before measurement, ENPP1 inhibitors are required; ENPP1 potently degrades extracellular cGAMP as a result of its ubiquitous expression on the plasma membrane and in serum (21). To avoid the interference of ENPP1 inhibitors in the ENPP1-dependent portion of our cGAMP-Luc assay, we sought to develop a method that would allow us to purify cGAMP away from these inhibitors. Such a method would also preferentially be able to concentrate cGAMP and remove any other analytes, such as AMP, from complex culture medium that could lead to false positives in the assay.
To this end, we developed STING-CAP (STING-mediated concentration and purification of cGAMP). In this method, biological samples containing cGAMP are treated with His-tagged STING. The resulting cGAMP:STING complexes are collected by magnetic nickel beads, which are subsequently washed and boiled in a minimal volume of buffer to elute cGAMP (Fig. 4A). To boost the robustness of the assay, we added Tween 20 and imidazole to prevent the nonspecific binding of unwanted proteins and molecules to the nickel beads, as well as protease inhibitors to prevent STING degradation. We optimized each step of this procedure to ensure maximal recovery of cGAMP (Fig. S2, A–D). To increase the ease of use of cGAMP-Luc, we also developed an alternate method of STING-CAP that replaces the magnetic nickel beads with standard nonmagnetic nickel beads and filter columns with no loss of detection limit (Fig. S2E).
Figure 4.
STING-CAP and cGAMP-Luc enable the quantitation of cGAMP in complex biological samples. A, scheme of STING column purification and concentration of cGAMP (1). Complex biological mixtures containing cGAMP are incubated with His-tagged STING (2). Magnetic nickel beads are added to collect the STING-cGAMP complexes (3). A series of washes reduce complexity of medium while the STING-cGAMP complexes are retained on the magnetic nickel beads. On the final wash, cGAMP-STING complexes are resuspended in a minimal volume (4). cGAMP is eluted by heat inactivation of STING. B, cGAMP standard curve in 250 μl of conditioned medium from the WT 293T cell line was subjected to STING-CAP with 1 μm STING dimer and quantitated by a cGAMP-Luc assay. Data are mean ± S.E. (error bars) (n = 3). C, cGAMP standard curve in 1400 μl of conditioned medium from the WT 293T cell line was subjected to STING-CAP with 700 nm STING dimer and quantitated by a cGAMP-Luc assay. Data are mean ± S.E. (n = 3). D, cGAMP standard curve in 250 μl of human plasma was subjected to STING-CAP with 1 μm STING dimer and quantitated by a cGAMP-Luc assay. Data are mean ± S.E. (n = 3). E, cGAMP standard curve in lysate from the WT 293T cell line was subjected to STING-CAP with 1 μm STING and quantitated by a cGAMP-Luc assay. The x axis represents intracellular concentration of cGAMP. Data are mean ± S.E. (n = 2).
We paired STING-CAP and cGAMP-Luc to generate cGAMP standard curves in conditioned medium and human plasma. We collected conditioned medium from WT 293T cells, which do not produce any appreciable levels of cGAMP (17). Two standard curves were generated by spiking cGAMP into 250 μl or 1400 μl of medium. From a volume of 250 μl, we obtained a quantitation limit of 9 nm (Fig. 4B), and from a volume of 1400 μl, we obtained a quantitation limit of 1.9 nm (Fig. 4C). In human plasma, we obtained a standard curve with a quantitation limit of 25 nm from 250 μl of plasma (Fig. 4D).
We next tested whether cGAMP-Luc could be used to quantitate intracellular cGAMP from cell lysate. We lysed WT 293T cells in hypotonic buffer and added known concentrations of cGAMP into the lysate. Following complete lysis, we corrected the osmolarity of the buffer and performed the same STING-CAP and cGAMP-Luc assay steps as in the quantitation of extracellular cGAMP. The standard curve of intracellular cGAMP was generated with a detection limit of 26 nm from 20 million cells (Fig. 4E).
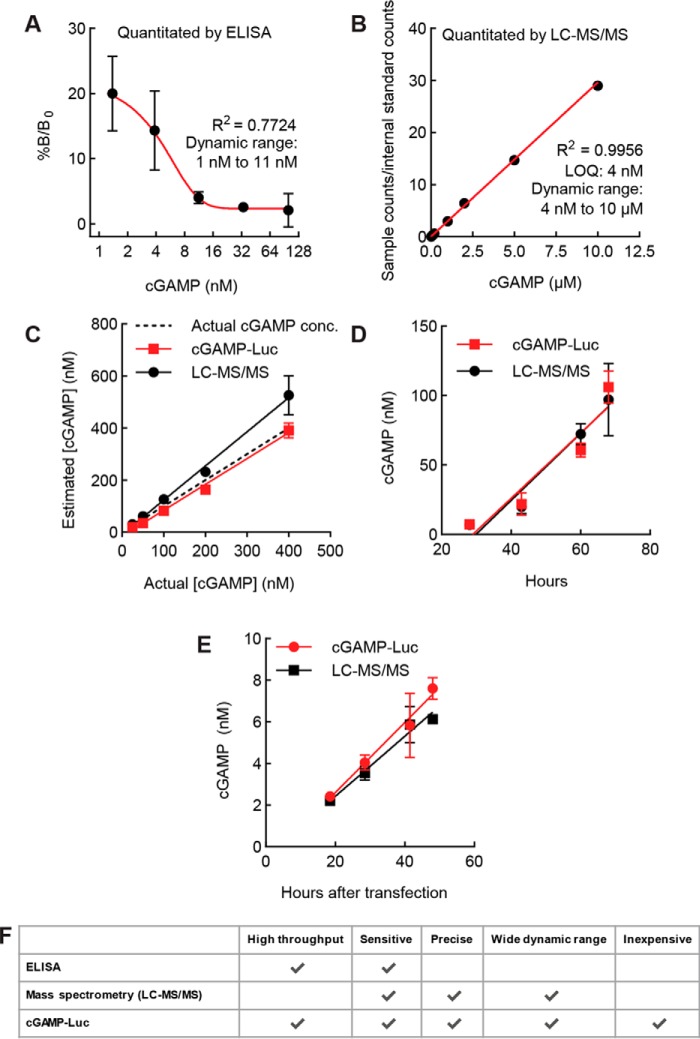
Comparison of cGAMP-Luc with existing cGAMP quantitation methods
With the cGAMP-Luc assay fully optimized, we sought to compare it with other methods of cGAMP quantitation. We first compared cGAMP-Luc with the commercially available cGAMP ELISA. Although expensive, the cGAMP ELISA is high-throughput in principle. However, the ELISA yielded a cGAMP standard curve with a dynamic range that spans barely an order of magnitude (Fig. 5A). This limited dynamic range makes the ELISA impractical for most applications, as several dilutions of a sample are required in order to find one that falls within the quantitation range of the standard curve.
Figure 5.
Comparison of cGAMP-Luc with existing cGAMP quantitation methods. A, cGAMP standard curve in 50 μl of conditioned medium from the WT 293T cell line was quantitated by cGAMP ELISA. Data are mean ± S.E. (error bars) (n = 2). B, LC-MS/MS cGAMP standard curve. A series of cGAMP dilutions with an internal standard of 500 nm isotopically labeled cGAMP was submitted for LC-MS/MS. The ratio of counts of unlabeled cGAMP to isotopically labeled internal standard counts is represented on the y axis. The standard curve was weighted by 1/x2 (n = 1; data representative of 10 independent experiments). C, samples of cGAMP in conditioned medium from WT 293T cells were subjected to STING-CAP with 700 nm STING dimer and quantitated via cGAMP-Luc assay and LC-MS/MS. R2 is 0.8770 for cGAMP-Luc and 0.8576 for LC-MS/MS. Data are mean ± S.E. (n = 2). D, conditioned serum-free medium was collected at time points (28, 43, 60, and 68 h) from a cGAS+/+ ENPP1−/− 293T cell line, subjected to STING-CAP with 900 nm STING dimer, and quantitated via both cGAMP-Luc and LC-MS/MS. R2 is 0.9884 for cGAMP-Luc and 0.9775 for LC-MS/MS. Data are mean ± S.E. (n = 3). E, conditioned medium supplemented with 50 μm STF-1084 was collected from HeLa cells transfected with dsDNA at the indicated time points, subjected to STING-CAP with 700 nm STING dimer, and quantitated via both cGAMP-Luc and LC-MS/MS. R2 is 0.8771 for cGAMP-Luc and 0.9216 for LC-MS/MS. Untransfected controls assayed via cGAMP-Luc and LC-MS/MS had signals below quantitation limits (data not shown). Data are mean ± S.E. (n = 3). F, comparison of attributes of all cGAMP quantitation methods.
We next compared cGAMP-Luc with our previously developed MS method, the current gold standard of cGAMP quantitation. LC-MS/MS generates a linear standard curve with a quantitation limit of 4 nm, and cGAMP-Luc is capable of generating a standard curve spanning the same range (Fig. 5B and Fig. S3). To compare the quantitative abilities of the cGAMP-Luc assay and the LC-MS/MS assay, we prepared samples with known amounts of cGAMP and quantitated them utilizing the previously generated standard curves. At a wide range of concentrations of cGAMP, both assays were able to quantitate cGAMP with an average of ∼80% accuracy (Fig. 5C). In addition, we took time points of conditioned medium from a cGAS-overexpressing 293T line and used both assays to quantitate the cGAMP concentrations in these samples. The concentrations of cGAMP determined by the two assays agreed well with an average difference of less than 5% (Fig. 5D). We also measured the export of cGAMP from HeLa cells stimulated by dsDNA (Fig. 5E). The cGAMP-Luc assay is therefore as precise and sensitive an assay as LC-MS/MS, with additional high-throughput capacity. A summary of the advantages of each assay for cGAMP quantitation is listed in Fig. 5F.
Discussion
Here, we developed the cGAMP-Luc assay as the first high-throughput and quantitative detection assay for cGAMP at physiologically relevant concentrations. With the assistance of STING-CAP, the STING-mediated concentration, and purification of cGAMP, virtually any amount of cGAMP can be assayed from any sample type. cGAMP-Luc surpasses the detection limit and matches the accuracy of MS, the current gold standard for cGAMP quantitation, with the added benefit of accessibility to most biology laboratories. cGAMP-Luc can therefore replace MS as the standard technique for quantitating cGAMP.
cGAMP-Luc is a powerful tool for answering questions that rely on accurate quantitation of cGAMP. For example, conventional cancer treatments, including radiation and chemotherapy, have the potential to stimulate cancer cells to produce and export cGAMP through the activation of cGAS by DNA damage. cGAMP-Luc can be used to understand which of these existing treatments can stimulate cGAMP synthesis and export. In addition, these existing treatments can be combined to investigate how they might synergize with each other and inform strategies for improving the efficacy of these treatments.
Finally, although an importer of cGAMP has been identified, exporters of cGAMP have not yet been reported. Knowing the identities of the possible exporters of cGAMP is critical to fully understanding the transport mechanisms of cGAMP and its extracellular biology, which may lead to novel drug targets. Because of its precision, sensitivity, and high throughput capability, cGAMP-Luc can be used as the readout in an RNAi screen to identify potential exporters of cGAMP.
Materials and methods
Expression and purification of recombinant proteins
PAP
The DNA sequence encoding polyphosphate:AMP phosphotransferase (GenBankTM accession number AB092983) was synthesized by Integrated DNA Technologies. The DNA fragment was inserted into the SapI and XhoI sites of the pTB146 His-SUMO vector (a generous gift from T. Bernhard, Harvard Medical School) using isothermal assembly, and the resulting plasmid was transformed into DH5α cells.
PAP was expressed in BL21-DES cells grown in 2XYT with ampicillin (100 ng/ml). Cells were induced at A600 = 1.0 with 0.7 mm isopropyl 1-thio-β-d-galactopyranoside and grown overnight at 16 °C. Induced cells were pelleted and resuspended in buffer with 50 mm Tris, pH 7.5, 400 mm NaCl, and 1× cOmplete protease inhibitor mixture (Roche Applied Science). Cells were snap-frozen in liquid nitrogen and lysed by two freeze-thaw cycles. Following lysis, cells were sonicated (4 × 30 s at 35% power), and cell debris was removed by ultracentrifugation for 45 min at 40,000 rcf. Supernatant was supplemented with His-Pur cobalt resin (Thermo Fisher Scientific) and incubated for 2 h at 4 °C. Resin was washed with 50 column volumes of buffer made of 50 mm Tris, pH 7.5, 150 mm NaCl (1× TBS), and 10 mm imidazole. PAP was eluted with 600 mm imidazole in 1× TBS. PAP was dialyzed against 50 mm Tris, pH 7.5, overnight, loaded onto a 1-ml HiTrap Q anion-exchange column (GE Healthcare) using an Akta FPLC (GE Healthcare), and eluted with a NaCl gradient. All fractions containing protein from anion exchange were collected and dialyzed overnight into 1× TBS supplemented with 40 μm polyphosphate to prevent aggregation of PAP. Following dialysis, PAP was snap-frozen at concentrations of 1 mg/ml. An average of 10 mg of PAP was obtained from 1 liter of bacterial culture. Proteins were analyzed by SDS-PAGE and stained with InstantBlue.
To test PAP activity, AMP was incubated with 0.5 mm PAP and 30 mm polyphosphate for 2 h at room temperature. A 2-μl volume from the reaction was spotted on a silica TLC plate (Millipore Sigma) and allowed to dry for a minimum of 15 min. The TLC was developed in a solvent of 11:7:2 n-propanol/ammonium hydroxide/water (27). The plate was dried and imaged with a camera while exposed to UV light with wavelength 254 nm.
STING
The DNA sequence encoding the cytosolic domain of human STING (amino acids 137–379) was PCR-amplified from HEK 293 cell cDNA using Phusion High-Fidelity DNA polymerase (Thermo). The PCR product was inserted into the SapI and XhoI sites of pTB146 using isothermal assembly. The H232R variant was introduced via site-directed mutagenesis. Mutated plasmids were cloned using pfuTurbo DNA polymerase (Agilent), and parent plasmids were degraded by DpnI (New England Biolabs). All plasmids were transformed into DH5α cells.
His-tagged STING was expressed in Rosetta component cells grown in 2XYT with ampicillin (100 ng/ml). Cells were induced at A600 = 0.8 with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and grown overnight at 16 °C. Cells were pelleted, resuspended in buffer with 50 mm phosphate, pH 7.5, 400 mm NaCl, and 1× cOmplete protease inhibitor mixture (Roche Applied Science). Cells were snap-frozen in liquid nitrogen and lysed by two freeze-thaw cycles. Following lysis, cells were sonicated (4 × 30 s at 35% power), and cell debris was removed by ultracentrifugation for 45 min at 40,000 rcf. His-Pur cobalt resin (Thermo Fisher Scientific) was added to the supernatant and incubated for 2 h at 4 °C. Resin was washed with 50 column volumes of buffer made of 50 mm phosphate, pH 7.5, 150 mm NaCl (1× PBS), and 10 mm imidazole. STING was eluted with 600 mm imidazole in 1× PBS. All fractions from elution were collected and dialyzed overnight into 1× PBS. Proteins were analyzed by SDS-PAGE and stained with InstantBlue. STING concentration was determined by UV-visible spectroscopy on a Nanodrop 2000 spectrophotometer according to the relationship, 1 Abs = 1 mg/ml at λmax 280 nm.
ENPP1
Recombinant mouse ENPP1 was produced as described previously (28, 29). Proteins were analyzed by SDS-PAGE and stained with InstantBlue.
Cell culture
The HEK 293T and HeLa cell lines were procured from ATCC. The HEK 293T cGAS ENPP1−/− cell line originates from Ref. 17. All cell lines were maintained in a 5% CO2 incubator at 37 °C. 293T and Hela cell lines were maintained in DMEM (Corning Cellgro) supplemented with 10% (v/v) fetal bovine serum (Atlanta Biologics) and 100 units/ml penicillin-streptomycin (Thermo Fisher Scientific).
cGAMP export assay in cancer cell lines
In 293T cGAS ENPP1−/− cells
As previously described (17), 293T cGAS ENPP1−/− cells were plated in tissue culture–treated plates coated with 2% PurCol (Advanced BioMatrix). At the start of the experiment, the medium was gently removed and replaced with serum-free DMEM supplemented with 1% insulin-transferrin-selenium-sodium pyruvate (Thermo Fisher Scientific) and 100 units/ml penicillin-streptomycin. Medium was collected at the indicated times and centrifuged at 1000 rcf for 5 min at room temperature. Supernatant was removed and analyzed for cGAMP content.
In HeLa cells
Cells were plated in 6-well tissue culture–treated plates. At the start of the experiment, the medium was removed and replaced with 1.5 ml of DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin-streptomycin, and 50 μm STF-1084. Immediately after the medium change, cells were transfected with Fugene 6 (Promega) according to the manufacturer's instructions, with 3.4 μl of Fugene reagent and 1.125 μg of empty pcDNA6 plasmid used per well. Untransfected controls were treated with Fugene only. Medium was collected at the indicated times and centrifuged at 1000 rcf for 5 min at room temperature. Supernatant was removed, subjected to STING-CAP, and analyzed for cGAMP content.
[32P]cGAMP degradation TLC assay
[32P]cGAMP was synthesized as described previously (17). In brief, [32P]cGAMP was synthesized by incubating unlabeled ATP (1 mm) and GTP (1 mm) doped with [32P]ATP. Nucleotides were combined with 2 μm purified recombinant porcine cGAS in 20 mm Tris, pH 7.5, 2 mm MgCl2, and 100 μg/ml herring testes DNA overnight at room temperature. The remaining nucleotide starting materials were degraded with alkaline phosphatase for 4 h at 37 °C.
To test the activity of ENPP1, 1 nm [32P]cGAMP and 350 nm cGAMP were incubated in the presence or absence of 10 nm ENPP1 overnight in DMEM supplemented with ENPP1 buffer (100 mm Tris, pH 8, 150 mm NaCl, 2 mm CaCl2, 200 μm ZnCl2). As described previously (30), a 2-μl volume from the reaction was spotted on a silica TLC plate (Millipore Sigma) and allowed to dry for a minimum of 15 min. The TLC was developed in a solvent containing 85% ethanol and 5 mm NH4HCO3. Plates were dried and exposed to a Storage Phosphor Screen (Molecular Dynamics) overnight before being imaged with a Typhoon 9400 Imager (Molecular Dynamics).
Quantitation of ATP
ATP (Sigma–Aldrich) was dissolved in water to a concentration of 10 mm. Concentration was verified by UV-visible spectroscopy on a Nanodrop 2000 spectrophotometer according to extinction coefficient ϵ = 15.4 × 103 m−1 cm−1 at λmax 260 nm. A serial dilution of ATP was performed in water. To generate the standard curve, 20 μl of CellTiter-Glo® solution (Promega) was added to 20 μl of each ATP standard in a 384-well flat white plate (Corning). After a 10-min benchtop incubation at room temperature, luminescence was quantitated on a Spark® multimode microplate reader (Tecan) with an integration time of 1 s. Default settings on GraphPad Prism 8.0.0 were used to plot standard curves. For limits of detection and quantitation calculations, see “cGAMP-Luc.”
Quantitation of AMP
For standard curves, AMP (Sigma–Aldrich) was dissolved in 50 mm Tris, pH 7.5, to a concentration of 20 mm. Concentration was verified by UV-visible spectroscopy on a Nanodrop 2000 spectrophotometer according to extinction coefficient ϵ = 15.4 × 103 m−1 cm−1 at λmax 260 nm.
AMP-GloTM
The AMP-GloTM assay (Promega) was performed as advised by the manufacturer.
AMP-Luc
A solution of 145 μm PAP, 0.2 units of myokinase (Sigma), 80 mm Tris, pH 7.5, 100 μg/ml Prionex, 10 mm MgCl2, and 40 μm polyphosphate was made immediately before performing the assay. 10 μl of this solution was added to 10 μl of each sample of AMP and incubated for 3 h at room temperature in a 384-well flat white plate. Following this incubation, 20 μl of CellTiter-Glo® solution was added. After a 10-min incubation at room temperature with no shaking, luminescence was quantitated on a Spark® Multimode Microplate Reader with an integration time of 1 s. Prior to plotting the standard curves, luminescence signals from controls containing no AMP were averaged and subtracted from luminescence signals of all other samples. Default settings on GraphPad Prism 8.0.0 were used to plot standard curves.
Quantitation of cGAMP
For standard curves, 2′,3′-cGAMP was synthesized according to protocols described previously (18, 30). Concentration was verified by UV-visible spectroscopy on a Nanodrop 2000 spectrophotometer according to extinction coefficient ϵ = 25.1 × 103 m−1 cm−1 at λmax 256 nm.
cGAMP ELISA
cGAMP ELISA was purchased from Cayman and performed as recommended by the manufacturer. Default settings on Prism 8.0.0 were used to fit data to four-parameter logistic curves.
LC-MS/MS
LC-MS/MS was performed as described (17) to quantitate cGAMP.
cGAMP-Luc
20× solutions of ENPP1 buffer (2 m Tris, pH 8.0, 3 m NaCl, 40 mm CaCl2, 4 m ZnCl2) or minimal ENPP1 buffer (1 m Tris, pH 8.0, 2.75 m NaCl, 20 μm ZnCl2) were formulated. In a 384-well PCR plate (USA Scientific), 12 μl of each cGAMP sample was supplemented with 0.7 μl of ENPP1 buffer and 0.12 μl of 1 μm ENPP1 or 0.12 μl of ENPP1 dilution buffer (0.1% Nonidet P-40 and 1× TBS, pH 7.5) to a final volume of 13.25 μl. After overnight incubation at room temperature, the reaction was terminated at 90 °C for 10 min in an Applied Biosystems ViiA 7 real-time PCR system. 10 μl of this solution was quantitated according to the AMP-Luc protocol described above. Prior to plotting the standard curves, luminescence signals from samples not subjected to ENPP1 digestion were subtracted from luminescence signals from samples subjected to ENPP1 digestion prior to plotting standard curves. The resulting luminescence values for control samples containing no cGAMP were then averaged and subtracted from all other samples. Default settings in GraphPad Prism 8.0.0 were used to plot standard curves unless otherwise specified. By literature precedent, standard curves spanning 3 or more orders of magnitude were weighted by 1/x2 in the determination of the best-fit line (31). The error in cGAMP measurement increases proportionally as the concentration of cGAMP increases. The best fit is therefore obtained by using a linear regression that weights the points by the inverse-squared values (1/x2) so that the increased error of the assay at the highest concentrations of cGAMP does not dominate the determination of the best-fit line.
Calculations of limits of detection and quantitation
By literature precedent, the limit of detection (LOD) is calculated as LOD = 3.3 × (Sy/S), where Sy is the S.D. of the blank samples and S is the slope of the line of best fit (32–35). The limit of detection calculated by this formula is typically low but is reasonable as the S.D. of blank samples is small (e.g. compare luminescence signals between 15 nm AMP and 0 nm AMP in Fig. S4A).
By literature precedent, the limit of quantification (LOQ) can be calculated similarly, as LOQ = 10 × (Sy/S). However, this method of calculating the LOQ yields a limit that is artificially low. For example, in Fig. S4A, this method of calculating the LOQ yields a limit of 2.8 nm. However, at 15 nm AMP, the concentration of AMP back-calculated from the line of best fit differs from the actual concentration by nearly 300%. The high error is likely a result of the standard curve not being entirely linear at its lowest concentrations. The LOQ is thus defined as the lowest concentration in the standard curve that can be quantified by the line of best fit with an error of less than 25%. For the example in Fig. S4, this method yields an LOQ of 62.5 nm. The cutoffs recommended by the FDA are 20% for chromatographic assays and 25% for ligand-binding assays (36). cGAMP-Luc is closer in nature to a ligand-binding assay, so the cutoff of 25% was chosen.
The use of two orthogonal methods to calculate the LOD and LOQ allows for the most information to be gleaned from the assay. The LOD is significantly lower than the LOQ, as the low S.D. of standards in the assay enables detection of low concentrations of cGAMP. The more stringent method of calculating the LOQ simultaneously ensures accurate quantitation of cGAMP.
Assay for inhibition of ENPP1 by small molecules
QS1, STF-1084, and STF-1623 were synthesized as described previously (17, 26). For inhibition experiments, 10 nm ENPP1 was incubated with a serial dilution of ENPP1 inhibitor and 5 μm cGAMP in a buffer containing 50 mm Tris, pH 7.5, 250 mm NaCl, 0.5 mm CaCl2, and 1 μm ZnCl2. After 3.5 h of incubation at room temperature, the reaction was terminated at 90 °C for 10 min in an Applied Biosystems ViiA 7 real-time PCR system. 10 μl of this solution was quantitated according to the AMP-Luc protocol described above. Inhibition curves were fit to obtain IC50 values with GraphPad Prism 8.0.0. IC50 values were converted to Ki,app values using the Cheng–Prusoff equation, Ki,app = (IC50)/(1 + [S]/Km).
STING-CAP
Synthesis of [32P]cGAMP
For all experiments involving STING-CAP, radiolabeled [32P]cGAMP was synthesized and purified as described previously (30).
Optimization of STING-CAP
To test the binding capacity of STING, 1 nm [32P]cGAMP was incubated with either 10 or 700 nm STING in 100 μl of DMEM for 2 min. The mixture was briefly centrifuged (10,000 rcf for 10 s) through a filter with a 3-kDa molecular mass cutoff. 2 μl of the flow-through was added to 10 ml of Bio-Safe II scintillation mixture (Research Products International). Radioactivity of the flow-through was measured on a LS 6500 scintillation counter (Beckman Coulter). Samples were normalized to 0 nm STING condition.
To test the binding capacity of magnetic nickel beads, cGAMP (200 nm cold and 0.5 nm [32P]cGAMP as a tracer) was incubated with 700 nm STING and magnetic nickel beads for 1 h in a volume of 250 μl of DMEM. After being washed in 1 ml of PBS, pH 8.0, with 0.1% Tween 20 (PBST) and 10 mm imidazole and 1 ml of PBST, cGAMP:STING:bead complexes were resuspended in a volume of 100 μl of PBS and added to 10 ml of Bio-Safe II scintillation mixture. Radioactivity of the flow-through was measured on an LS 6500 scintillation counter. Samples were normalized to the total radioactivity in 250 μl of 1 nm [32P]cGAMP.
To test the loss of cGAMP during STING-CAP, 700 nm STING, 160 nm cGAMP, and 0.33 nm [32P]cGAMP as a tracer in a 250-μl reaction were incubated with 2.5 μl of magnetic nickel beads for 2 h. Samples were washed with 1 ml each of TBST, pH 7.5 with 10 mm imidazole, PBST, pH 8.0, and TBST, pH 7.5. 100 μl of each step in the procedure (flow-through, washes, final sample of beads) was added to 10 ml of Bio-Safe II scintillation mixture. Radioactivity of the flow-through was measured on a LS 6500 scintillation counter.
To optimize cGAMP elution from STING, 750 nm STING, 12.5 nm cGAMP, and 2.5 μl of magnetic nickel beads were incubated in a volume of 1000 μl of DMEM for 2 h at room temperature. Samples were washed with 2 × 1 ml of PBST, pH 8.0, resuspended in a volume of 30 μl DMEM, and boiled at either 75 or 95 °C in increments of 0, 5, 15, or 30 min. Beads were collected by magnet, and supernatant was subjected to a cGAMP-Luc assay.
Use of STING-CAP for extracellular cGAMP (in conditioned medium and plasma)
Samples were supplemented to a final concentration of 5 μm STF-1084, 10 mm imidazole, pH 7.5, 75 mm phosphate, pH 8.0, 0.05% Tween 20, 1× cOmplete protease inhibitor mixture, and STING. The necessary STING concentration was determined by Equation 1, assuming a Kd of 5 nm of STING for cGAMP (9).
| (Eq. 1) |
Pierce nickel-nitrilotriacetic acid magnetic agarose beads (Thermo Scientific) were washed with 2 × 1 ml of PBS, pH 8.0, with 0.1% Tween 20. 1 μl of settled bead volume was used for every 6 μg of STING. Magnetic nickel beads were incubated with STING between 4 and 16 h at 4 °C. Beads were collected a DynaMag-2 Magnet (Thermo Fisher Scientific) and washed with 1 ml each of 1× PBS, 10 mm imidazole, and 1× PBS before transfer to a 96-well PCR plate (Thermo Scientific). Beads were collected on a Magnetic Stand-96 (Thermo Fisher Scientific), and the supernatant was removed. 30 μl of DMEM was added, and beads were heated at 80 °C for 10 min to elute cGAMP. Beads were collected by magnet, and supernatant was subjected to cGAMP-Luc assay.
Use of STING-CAP with nonmagnetic nickel beads
Samples were supplemented to a final concentration of 5 μm STF-1084, 10 mm imidazole, pH 7.5, 75 mm phosphate, pH 8.0, 0.05% Tween 20, 1× cOmplete protease inhibitor mixture, and STING. The necessary STING concentration was determined by Equation 1, assuming a Kd of 5 nm of STING for cGAMP (9).
HisPur nickel-nitrilotriacetic acid resin (Thermo Scientific) was washed with 2 × 1 ml of PBS, pH 8.0, with 0.1% Tween 20. 1 μl of settled bead volume was used for every 0.5 μg of STING. Beads were transferred to a Micro Bio-Spin Chromatography Column (Bio-Rad) and centrifuged for 15 s at 200 rcf. Beads were washed with 500 μl of PBST, pH 8.0, with 10 mm imidazole and 2 × 500 μl of PBST, pH 8.0, and centrifuged for 30 s at 200 rcf between each wash. To elute, beads were incubated with 35 μl of TBS, pH 7.5, with 300 mm imidazole for 15 min and spun for 1 min at 20,000 rcf. Eluate was heated at 80 °C for 10 min to release cGAMP from STING and subjected to a cGAMP-Luc assay.
Use of STING-CAP for intracellular cGAMP
The standard curve for intracellular cGAMP was generated in the WT 293T cell line. Cells were harvested from plates with trypsin-EDTA (Thermo Fisher Scientific), washed with PBS, and counted. 20 million cells were used per point in the standard curve. Each aliquot of 20 million cells was lysed in 200 μl of hypotonic buffer containing 20 mm Tris, pH 7.5, 0.05% Tween 20, 5 mm MgCl2, 0.5 mm CaCl2, 5 μm STF-1084, 3 units of Dnase I (Millipore Sigma), and cGAMP. To calculate the necessary cGAMP concentrations, a volume of 60 μl was assumed for 20 million WT 293T cells. Cells were shaken at room temperature for 30 min and boiled at 95 °C for 10 min to ensure complete lysis. Lysed cells were spun at 20,000 rcf at room temperature for 10 min. Supernatant was removed, supplemented with 20× TBS to correct the hypotonicity of the solution, and subjected to STING concentration and purification as described previously.
Data availability
All data are contained within this paper.
Author contributions
R. E. M., J. A. C., and L. L. conceptualization; R. E. M. data curation; R. E. M. formal analysis; R. E. M. and L. L. funding acquisition; R. E. M. investigation; R. E. M. and J. A. C. methodology; R. E. M. writing-original draft; L. L. supervision; L. L. project administration; L. L. writing-review and editing.
Supplementary Material
Acknowledgments
We thank the following people: J. Brown for ENPP1 purification, C. Ritchie for [32P]cGAMP synthesis, S. Ergun for STING cloning, K. Nguyen and the Stanford BioADD core facility for cGAMP LC-MS/MS analysis, Dr. D. Herschlag and Dr. A. Straight for assay conception, and all members of the Li laboratory for valuable discussions.
This work was supported by the National Science Foundation Graduate Research Fellowship Program Grant 1656518 (to R. E. M.) and National Institutes of Health Grant DP2CA228044 (to L. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- cGAS
- cGMP/AMP synthase
- STING
- stimulator of interferon genes
- STING-CAP
- STING-mediated concentration and purification of cGAMP
- PAP
- polyphosphate:AMP phosphotransferase
- cGAMP
- 2′,5′/3′,5′-cGMP-AMP
- rcf
- relative centrifugal force
- DMEM
- Dulbecco's modified Eagle's medium
- LOD
- limit of detection
- LOQ
- limit of quantification
- a.u.
- arbitrary units.
References
- 1. Mackenzie K. J., Carroll P., Martin C.-A., Murina O., Fluteau A., Simpson D., Olova N., Sutcliffe H., Rainger J. K., Leitch A., Osborn R. T., Wheller A. P., Nowotny M., Gilbert N., Chandra T., et al. (2017) cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harding S. M., Benci J. L., Irianto J., Discher D. E., Minn A. J., and Greenberg R. A. (2017) Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakhoum S. F., Ngo B., Laughney A. M., Cavallo J. A., Murphy C. J., Ly P., Shah P., Sriram R. K., Watkins T. B. K., Taunk N. K., Duran M., Pauli C., Shaw C., Chadalavada K., Rajasekhar V. K., et al. (2018) Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 10.1038/nature25432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., Hopfner K. P., Ludwig J., and Hornung V. (2013) CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 10.1038/nature12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., and Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diner E. J., Burdette D. L., Wilson S. C., Monroe K. M., Kellenberger C. A., Hyodo M., Hayakawa Y., Hammond M. C., and Vance R. E. (2013) The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 3, 1355–1361 10.1016/j.celrep.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun L., Wu J., Du F., Chen X., and Chen Z. J. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., and Chen Z. J. (2013) Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 51, 226–235 10.1016/j.molcel.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishikawa H., and Barber G. N. (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 13. Jin L., Waterman P. M., Jonscher K. R., Short C. M., Reisdorph N. A., and Cambier J. C. (2008) MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 10.1128/MCB.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Apelbaum A., Yarden G., Warszawski S., Harari D., and Schreiber G. (2013) Type I interferons induce apoptosis by balancing cFLIP and caspase-8 independent of death ligands. Mol. Cell. Biol. 33, 800–814 10.1128/MCB.01430-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corrales L., Woo S.-R., and Gajewski T. F. (2013) Extremely potent immunotherapeutic activity of a STING agonist in the B16 melanoma model in vivo. J. Immunother. Cancer 1, O15 10.1186/2051-1426-1-S1-O15 [DOI] [Google Scholar]
- 16. Woo S. R., Fuertes M. B., Corrales L., Spranger S., Furdyna M. J., Leung M. Y. K., Duggan R., Wang Y., Barber G. N., Fitzgerald K. A., Alegre M. L., and Gajewski T. F. (2014) STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 10.1016/j.immuni.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carozza J. A., Böhnert V., Nguyen K. C., Skariah G., Shaw K. E., Brown J. A., Rafat M., von Eyben R., Graves E. E., Glenn J. S., Smith M., and Li L. (2020) Extracellular cGAMP is a cancer-cell-produced immunotransmitter involved in radiation-induced anticancer immunity. Nat. Cancer. 1, 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ritchie C., Cordova A. F., Hess G. T., Bassik M. C., and Li L. (2019) SLC19A1 is an importer of the immunotransmitter cGAMP. Mol. Cell. 75, 372–381.e5 10.1016/j.molcel.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luteijn R. D., Zaver S. A., Gowen B. G., Wyman S. K., Garelis N. E., Onia L., McWhirter S. M., Katibah G. E., Corn J. E., Woodward J. J., and Raulet D. H. (2019) SLC19A1 transports immunoreactive cyclic dinucleotides. Nature 573, 434–438 10.1038/s41586-019-1553-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bose D., Su Y., Marcus A., Raulet D. H., and Hammond M. C. (2016) An RNA-based fluorescent biosensor for high-throughput analysis of the cGAS-cGAMP-STING pathway. Cell Chem. Biol. 23, 1539–1549 10.1016/j.chembiol.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L., Yin Q., Kuss P., Maliga Z., Millán J. L., Wu H., and Mitchison T. J. (2014) Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10, 1043–1048 10.1038/nchembio.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eaglesham J. B., Pan Y., Kupper T. S., and Kranzusch P. J. (2019) Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS–STING signalling. Nature 566, 259–263 10.1038/s41586-019-0928-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Resnick S. M., and Zehnder A. J. B. (2000) In vitro ATP regeneration from polyphosphate and AMP by polyphosphate:AMP phosphotransferase and adenylate kinase from Acinetobacter johnsonii 210A. Appl. Environ. Microbiol. 66, 2045–2051 10.1128/AEM.66.5.2045-2051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kameda A., Shiba T., Kawazoe Y., Satoh Y., Ihara Y., Munekata M., Ishige K., and Noguchi T. (2001) A novel ATP regeneration system using polyphosphate-AMP phosohotransferase and polyphosphate kinase. J. Biosci. Bioeng. 91, 557–563 10.1016/S1389-1723(01)80173-0 [DOI] [PubMed] [Google Scholar]
- 25. Noda L. (1957) Adenosine Triphosphate-Adenosine Monophosphate Transphosphorylase. J. Biol. Chem. 226, 541–549 [PubMed] [Google Scholar]
- 26. Shayhidin E. E., Forcellini E., Boulanger M. C., Mahmut A., Dautrey S., Barbeau X., Lagüe P., Sévigny J., Paquin J. F., and Mathieu P. (2015) Quinazoline-4-piperidine sulfamides are specific inhibitors of human NPP1 and prevent pathological mineralization of valve interstitial cells. Br. J. Pharmacol. 172, 4189–4199 10.1111/bph.13204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ablasser A., Schmid-Burgk J. L., Hemmerling I., Horvath G. L., Schmidt T., Latz E., and Hornung V. (2013) Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 10.1038/nature12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kato K., Nishimasu H., Mihara E., Ishitani R., Takagi J., Aoki J., and Nureki O. (2012) Expression, purification, crystallization and preliminary X-ray crystallographic analysis of Enpp1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 778–782 10.1107/S1744309112019306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato K., Nishimasu H., Okudaira S., Mihara E., Ishitani R., Takagi J., Aoki J., and Nureki O. (2012) Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 16876–16881 10.1073/pnas.1208017109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ritchie C., Cordova A. F., and Li L. (2019) In response to Luteijn et al.: concerns regarding cGAMP uptake assay and evidence that SLC19A1 is not the major cGAMP importer in human PBMCs. bioXriv 10.1101/798397 [DOI] [Google Scholar]
- 31. Gu H., Liu G., Wang J., Aubry A. F., and Arnold M. E. (2014) Selecting the correct weighting factors for linear and quadratic calibration curves with least-squares regression algorithm in bioanalytical LC-MS/MS assays and impacts of using incorrect weighting factors on curve stability, data quality, and assay performance. Anal. Chem. 86, 8959–8966 10.1021/ac5018265 [DOI] [PubMed] [Google Scholar]
- 32. Muranaka L. S., Giorgiano T. E., Takita M. A., Forim M. R., Silva L. F. C., Coletta-Filho H. D., Machado M. A., and de Souza A. A. (2013) N-Acetylcysteine in agriculture, a novel use for an old molecule: focus on controlling the plant–pathogen Xylella fastidiosa. PLoS ONE 8, e72937 10.1371/journal.pone.0072937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shrivastava A., and Gupta V. B. (2011) Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chronicles Young Sci. 2, 21–25 10.4103/2229-5186.79345 [DOI] [Google Scholar]
- 34. Expert Working Group (Quality) of the International Conference on 1 Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) (1996) Guidance for Industry: Q2B Validation of Analytical Procedures: Methodology, United States Food and Drug Administration, Silver Spring, MD [Google Scholar]
- 35. Diana F. J. (2008) Method validation and transfer. in Handbook of Stability Testing in Pharmaceutical Development: Regulations, Methodologies, and Best Practices (Huynh-Ba K., ed) p. 171, Springer, New York [Google Scholar]
- 36. United States Food and Drug Administration (2018) Bioanalytical Method Validation Guidance for Industry. United States Food and Drug Administration, Silver Spring, MD [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this paper.