Abstract
Background & Aims
Human studies examining associations between circulating levels of insulin-like growth factor 1 (IGF1) and insulin-like growth factor binding protein 3 (IGFBP3) and colorectal cancer risk have reported inconsistent results. We conducted complementary serologic and Mendelian randomization (MR) analyses to determine whether alterations in circulating levels of IGF1 or IGFBP3 are associated with colorectal cancer development.
Methods
Serum levels of IGF1 were measured in blood samples collected from 397,380 participants from the UK Biobank, from 2006 through 2010. Incident cancer cases and cancer cases recorded first in death certificates were identified through linkage to national cancer and death registries. Complete follow-up was available through March 31, 2016. For the MR analyses, we identified genetic variants associated with circulating levels of IGF1 and IGFBP3. The association of these genetic variants with colorectal cancer was examined with 2-sample MR methods using genome-wide association study consortia data (52,865 cases with colorectal cancer and 46,287 individuals without [controls])
Results
After a median follow-up period of 7.1 years, 2665 cases of colorectal cancer were recorded. In a multivariable-adjusted model, circulating level of IGF1 associated with colorectal cancer risk (hazard ratio per 1 standard deviation increment of IGF1, 1.11; 95% confidence interval [CI] 1.05–1.17). Similar associations were found by sex, follow-up time, and tumor subsite. In the MR analyses, a 1 standard deviation increment in IGF1 level, predicted based on genetic factors, was associated with a higher risk of colorectal cancer risk (odds ratio 1.08; 95% CI 1.03–1.12; P = 3.3 × 10–4). Level of IGFBP3, predicted based on genetic factors, was associated with colorectal cancer risk (odds ratio per 1 standard deviation increment, 1.12; 95% CI 1.06–1.18; P = 4.2 × 10–5). Colorectal cancer risk was associated with only 1 variant in the IGFBP3 gene region (rs11977526), which also associated with anthropometric traits and circulating level of IGF2.
Conclusions
In an analysis of blood samples from almost 400,000 participants in the UK Biobank, we found an association between circulating level of IGF1 and colorectal cancer. Using genetic data from 52,865 cases with colorectal cancer and 46,287 controls, a higher level of IGF1, determined by genetic factors, was associated with colorectal cancer. Further studies are needed to determine how this signaling pathway might contribute to colorectal carcinogenesis.
Keywords: CRC, Risk Factors, Signal Transduction, GWAS
Abbreviations used in this paper: BMI, body mass index; CI, confidence interval; CRP, C-reactive protein; GWAS, genome-wide association studies; HbA1c, glycolated hemoglobin; HR, hazard ratio; ICC, intraclass correlation coefficient; IGF1, insulin-like growth factor 1; IGFBP3, insulin-like growth factor binding protein 3; MR, Mendelian randomization; MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier; OR, odds ratio; SD, standard deviation; SHBG, sex hormone binding globulin; SNP, single nucleotide polymorphism
Graphical abstract
What You Need to Know.
Background and Context
Human studies examining associations between circulating levels of insulin-like growth factor 1 (IGF1) and insulin-like growth factor binding protein 3 (IGFBP3) and colorectal cancer risk have reported inconsistent results
New Findings
In an analysis of blood samples from almost 400,000 participants in the UK Biobank, we found an association between circulating level of IGF1 and colorectal cancer. Using genetic data on 52,865 persons with colorectal cancer and 46,287 persons without, a higher level of IGF1, determined by genetic factors, was also associated with colorectal cancer. Further studies are needed to determine how this signaling pathway might contribute to colorectal carcinogenesis.
Limitations
This is an association study between measured and genetically determined blood levels of proteins and cancer incidence.
Impact
Changes in IGF signaling might contribute to colorectal carcinogenesis, but further studies are needed to determine the mechanisms of this process.
Insulin-like growth factor-1 (IGF1) has mitogenic and anti-apoptotic effects and has been implicated in the development and progression of several cancers.1,2 The bioactivity of IGF1 is partially regulated through insulin-like growth factor binding proteins (IGFBPs-), with approximately 80% bound to IGFBP3.3 In addition to its IGF-binding properties, IGFBP3 also has been shown to exhibit direct antiproliferative and pro-apoptotic effects.4,5 Multiple epidemiological studies have investigated the associations of circulating IGF1 and IGFBP3 levels with colorectal cancer risk.6, 7, 8, 9, 10, 11, 12, 13, 14 However, most of these studies were of relatively small size (<500 colorectal cancer cases) and reported inconsistent results. As such, there is currently a lack of consensus as to whether higher circulating levels of IGF1 and lower levels of IGFBP3 are risk factors for colorectal cancer.
To address this, we conducted complementary serologic and Mendelian randomization (MR) analyses to examine the role of circulating IGF1 and IGFBP3 in colorectal cancer development. We investigated how prediagnostic circulating levels of IGF1 are related to colorectal cancer risk in the UK Biobank study, a large prospective cohort. We then used a 2-sample MR approach to obtain causal estimates of the associations by combining genetic variants associated with circulating IGF1 and IGFBP3 levels in genome-wide association studies (GWAS) and then assessing the association of these variants with colorectal cancer risk in a large consortium of 52,865 colorectal cancer cases and 46,287 controls.15
Methods
UK Biobank: Serologic Analysis
Study participants
The UK Biobank is a prospective cohort study that aims to investigate the genetic, lifestyle, and environmental causes of a range of diseases.16,17 This research has been conducted using the UK Biobank Resource under application number 25897. Between 2006 and 2010, 502,656 adults aged between 40 and 69 years (229,182 men and 273,474 women) were recruited. All participants were registered with the UK National Health Service and lived within approximately 25 miles (40 km) of 1 of the 22 study assessment centers. The UK Biobank invited approximately 9.2 million people to participate through postal invitation with a telephone follow-up, with a response rate of 5.7%. The UK Biobank has approval from the North West Multi-centre Research Ethics Committee, the National Information Governance Board for Health and Social Care in England and Wales, and the Community Health Index Advisory Group in Scotland. In addition, an independent Ethics and Governance Council was formed in 2004 to oversee the UK Biobank’s continuous adherence to the Ethics and Governance Framework that was developed for the study (http://www.ukbiobank.ac.uk/ethics/). All participants provided written informed consent. During the baseline recruitment visit, participants were asked to complete a self-administered touchscreen questionnaire, which included questions on sociodemographics (including age, sex, education, and Townsend deprivation score), health and medical history, lifestyle exposures (including smoking habits, dietary intakes, and alcohol consumption), early life exposures, and medication use. At the baseline visit, participants also underwent physical measurements, including body weight, height, and waist circumference. Blood samples were collected from all participants at recruitment and from a subset of approximately 20,000 participants who re-attended the assessment center between 2012 and 2013 for a repeat assessment visit. Blood samples were labeled, centrifuged, and stored at −80°C.
Exclusions before the onset of analyses were participants with prevalent cancer at recruitment (n = 27,264); missing data on body size measurements (n = 3032); prevalent type-2 diabetes or unknown diabetes status at recruitment (as diabetes medications can affect circulating levels of IGF proteins18; n = 26,698); those who reported oral contraceptive and menopausal hormone use at recruitment (as oral estrogens exert a strong first-pass effect on the liver that alters hepatic protein production and changes circulating levels of multiple hormones, including IGF-system markers18; n = 19,802); and participants without an IGF1 measurement (n = 28,480). Our analysis therefore included 397,380 participants.
Blood collection and laboratory methods
As part of the UK Biobank Biomarker Project,19 serum levels of IGF1 (DiaSorin Liaison XL), testosterone, and sex hormone binding globulin (SHBG) (all Beckman Coulter DXI 800) were determined by a chemiluminescent immunoassay. The Immuno-turbidimetric method (Beckman Coulter DXI 800) was used to assay serum high sensitivity C-reactive protein (CRP) levels. Glycated hemoglobin (HbA1c) levels were determined using the high-performance liquid chromatography Variant II Turbo 2.0 system (Bio-Rad, Hercules, CA). Full details on assay performance have been published.19 In summary, average within-laboratory (total) coefficient of variation for low, medium, and high internal quality control level samples for each biomarker ranged from 1.7% to 15.3% (for IGF1, the coefficients of variation ranged from 5.3% to 6.2%).19 A total of 16,357 participants had IGF1 levels measured in blood samples collected at both the recruitment and repeat assessment visit (median of 4 years apart).
Assessment of outcome
Incident cancer cases and cancer cases recorded first in death certificates within the UK Biobank cohort were identified through linkage to national cancer and death registries. Complete follow-up was available through March 31, 2016, for England and Wales and October 31, 2015, for Scotland. Cancer incidence data were coded using the 10th Revision of the International Classification of Diseases (ICD-10). Proximal colon cancers included those found within the cecum, appendix, ascending colon, hepatic flexure, transverse colon, and splenic flexure (C18.0–18.5). Distal colon cancers included those found within the descending (C18.6) and sigmoid (C18.7) colon. Overlapping (C18.8) and unspecified (C18.9) lesions of the colon were included in colon cancers only. Cancer of the rectum included cancers occurring at the recto-sigmoid junction (C19) and rectum (C20).
Statistical analysis
To assess reproducibility between the 2 measurements of IGF1 available in a subsample of participants, we calculated intraclass correlation coefficients (ICC) by dividing the between-person variance by the sum of the between-person and within-person variances.
Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated using Cox proportional hazards models. Age was the primary time variable in all models. Time at entry was age at recruitment. Exit time was age at whichever of the following came first: colorectal cancer diagnosis, death, or the last date at which follow-up was considered complete. Models were stratified by age at recruitment in 5-year categories, Townsend deprivation index quintiles, and region of the recruitment assessment centre. Deviations from proportionality were assessed using an analysis of Schoenfeld residuals,20 with no evidence of nonproportionality detected. IGF1 was modeled on the continuous scale (per 1-standard deviation [SD] increment of log-IGF1) and with participants grouped into sex-specific quintiles of circulating levels. Continuous scale models were additionally corrected for regression dilution using regression dilution ratios obtained from participants with repeated IGF1 measurement.21,22 To obtain corrected HRs, the log HRs and their standard errors were divided by the regression dilution ratio for IGF1 (0.76) and then exponentiated.23
The multivariable model (model 1) was adjusted for a set of a priori–determined colorectal cancer risk factors, namely waist circumference, total physical activity, height, alcohol consumption frequency, smoking status and intensity, frequency of red and processed meat consumption, family history of colorectal cancer, educational level, regular aspirin/ibuprofen use, and ever use of hormone replacement therapy. We also additionally adjusted the multivariable models (model 2) for markers of inflammation, sex hormones, and glycemic pathways that are known to interrelate/cross talk with IGF-system markers,18 and have been linked to colorectal cancer risk in some studies,24, 25, 26, 27 namely CRP, testosterone, SHBG, and HbA1c. Statistical tests for trend were calculated using the ordinal quintile IGF1 entered into the model as a continuous variable. Analyses were also conducted by sex and anatomic subsite (colon, proximal colon, distal colon, and rectal cancer). Heterogeneity of associations by sex and across subsite was assessed by calculating χ2 statistics. Possible nonlinear effects were modeled using restricted cubic spline models with 5 knots placed at Harrell’s default percentiles of circulating IGF1 levels.28
The associations between circulating IGF1 and colorectal cancer were further assessed across subgroups of body mass index (BMI; <25, ≥25 kg/m2), height (below or above median), age at recruitment (<60 years, ≥60 years), follow-up time (<5 years, ≥5 years), smoking status (never, former, current), and circulating levels (below or above median) of CRP, HbA1c, testosterone, and SHBG. Interaction terms (multiplicative scale) between these variables and circulating IGF1 levels were included in separate models and the statistical significance of the cross-product terms were evaluated using the likelihood ratio test. In a sensitivity analysis, we excluded those participants with circulating levels of HbA1c ≥48 mmol/mol (or 6.5%, the cutoff for type-2 diabetes).
Statistical tests were all 2-sided and P < 0.05 was considered statistically significant.
Mendelian Randomization
We conducted 2-sample MR analyses, in which 2 different independent study samples (GWAS) were used to estimate the single nucleotide polymorphism (SNP)-risk factor (circulating IGF1 and IGFBP3 levels) and SNP-outcome (colorectal cancer) associations to allow causal effect estimates of risk factor-outcome association to be obtained.
Genetic determinants of IGF1 and IGFBP3
Genetic markers for circulating IGF1 and IGFBP3 levels comprised SNPs that were identified (P <5 × 10–8) from the largest GWAS to date.29,30 For IGF1, this GWAS was of 358,072 participants from the UK Biobank.29 The GWAS analyses of IGFBP3 combined data on 18,995 individuals from 13 studies.30 All participants were of European ancestry. From the genome-wide significant variants identified in these GWAS, we excluded correlated SNPs based on a linkage disequilibrium level of R2 < 0.01. Consequently, the instruments for IGF1 (413 SNPs spanning 22 autosomes) and IGFBP3 (4 SNPs spanning 3 autosomes) explained 9.4% (F-statistic 89.9) and 6.1% (F-statistic 308.4) of variability in circulating levels, respectively. Summary information on the genetic instruments and the effect estimates for each individual SNP regarding their association with IGF1 and IGFBP3 levels are presented in Supplementary Tables 1 and 2.
Data on colorectal cancer
Summary data for the associations of the IGF1- and IGFBP3-related genetic variants with colorectal cancer were obtained from a GWAS of 99,152 participants (52,865 colorectal cancer cases and 46,287 controls). The GWAS data were from a meta-analysis within the ColoRectal Transdisciplinary Study, the Colon Cancer Family Registry, and the Genetics and Epidemiology of Colorectal Cancer consortium.15 Imputation was performed using the Haplotype Reference Consortium r1.0 reference panel and the regression models were further adjusted for age, sex, genotyping platform (whenever appropriate), and genomic principal components as detailed here.31 Strength-of-association estimates for each individual SNP with colorectal cancer are presented in Supplementary Table 2.
Statistical power
The a priori statistical power was calculated using an online tool at http://cnsgenomics.com/shiny/mRnd/.32 Given a type 1 error of 5%, we had sufficient power (>80%) to detect an odds ratio (OR) per 1 SD for colorectal cancer risk of ≤0.94/≥1.06 for IGF1 and ≤0.93/≥1.08 for IGFBP3.
Statistical analysis
A 2-sample MR approach using summary data and a likelihood-based approach was implemented. Likelihood-based MR analyses are considered the most accurate method to estimate causal effects when there is a continuous log-linear association between risk factor and disease risk. The causal estimate of on (), assumed to be the same for all genetic variants (), is obtained from the likelihood function of the following model33,34:
MR results correspond to an OR per 1-SD increment in genetically predicted circulating levels of IGF1 and IGFBP3. Heterogeneity of associations by sex and across colorectal anatomic subsites was assessed by calculating χ2 statistics. Cochran’s Q statistics quantified heterogeneity across the individual SNPs. Sensitivity analyses were used to check and correct for the presence of pleiotropy in the causal estimates. To evaluate the extent to which directional pleiotropy (nonbalanced horizontal pleiotropy in the MR risk estimates) may have affected the causal estimates for the IGF1 and colorectal cancer association, we used an MR-Egger regression approach.35 We also computed OR estimates using the complementary weighted-median method that can give valid MR estimates under the presence of horizontal pleiotropy when up to 50% of the included instruments are invalid.36 The MR-PRESSO (Mendelian Randomization Pleiotropy RESidual Sum and Outlier) distortion test was used to estimate if horizontal pleiotropy caused by any identified outlier SNPs biased the effect estimates (P < .05).37 As a visual evaluation of pleiotropy, we also provided funnel plots depicting the weight exerted on the effect estimate along the y-axis and estimates of the effect on colorectal cancer along the x-axis for each SNP used in the corresponding instrumental variable. For the IGFBP3 instrument that had 4 SNPs, we conducted leave-one-out analyses to assess the influence of individual variants on the observed associations.
Results
UK Biobank: Serologic Analysis
After a median follow-up time of 7.1 years, 2665 cases of colorectal cancer (1539 in men and 1126 in women) were recorded. Compared with those in the lowest quintile, participants in the highest circulating IGF1 quintile were younger, taller, had lower BMI and waist circumference, were more likely to be never smokers, and less likely to be daily or almost daily consumers of alcohol and to be regular aspirin/ibuprofen users (Table 1). In addition, participants in the highest circulating IGF1 quintile had lower circulating CRP, testosterone, SHBG, and HbA1c levels.
Table 1.
Characteristics of UK Biobank Study Participants by Category of Circulating IGF1 levels (n = 397,380 participants
| Baseline characteristic | IGF1 levels |
||||
|---|---|---|---|---|---|
| Q1 | Q2 | Q3 | Q4 | Q5 | |
| IGF1, nmol/La | 14.3 (2.2) | 18.6 (1.1) | 21.4 (0.9) | 24.1 (1.0) | 29.5 (3.9) |
| Colorectal cancer (n cases) | 595 | 586 | 528 | 475 | 481 |
| Age at recruitment, ya | 59.1 (7.2) | 57.6 (7.6) | 56.3 (7.9) | 55.1 (8.2) | 53.2 (8.3) |
| Women (%) | 55.5 | 52.5 | 52.5 | 52.5 | 52.5 |
| Body mass index (kg/m2)a | 28.3 (5.4) | 27.4 (4.7) | 27.1 (4.4) | 26.8 (4.2) | 26.6 (4.0) |
| Waist circumference (cm)a | 92.7 (14.2) | 90.4 (13.1) | 89.5 (12.7) | 88.9 (12.4) | 88.2 (12.2) |
| Height (cm)a | 167.7 (9.2) | 168.4 (9.3) | 168.9 (9.3) | 169.2 (9.3) | 169.8 (9.3) |
| Total physical activity (MET hours per wk) (%) | |||||
| <10 | 23.8 | 21.8 | 21.1 | 21.2 | 21.0 |
| ≥60 | 22.8 | 23.1 | 22.7 | 22.1 | 21.3 |
| Smoking status (%) | |||||
| Never | 50.3 | 53.3 | 55.5 | 57.2 | 59.5 |
| Current | 11.8 | 10.9 | 10.2 | 10 | 9.8 |
| Alcohol consumption (%) | |||||
| Never | 9.7 | 7.4 | 6.9 | 6.6 | 6.7 |
| Daily or almost daily | 24.2 | 22.7 | 21.4 | 19.4 | 16.2 |
| Socioeconomic status (Townsend deprivation index) (%) | |||||
| Highest quintile | 22.2 | 19.2 | 18 | 18 | 17.8 |
| Family history (first degree relative) of colorectal cancer (%) | |||||
| Yes | 11.5 | 11.2 | 10.7 | 10.5 | 10.1 |
| Regular aspirin/ibuprofen use (%) | |||||
| Yes | 26.8 | 25.6 | 24.8 | 24.8 | 24.8 |
| Red and processed meat (%) | |||||
| <1 occasion per week | 10.3 | 10 | 9.9 | 9.9 | 10.2 |
| ≥3 occasions per week | 23.4 | 22.3 | 21.8 | 21.2 | 20.7 |
| Ever menopausal hormone use (%)b | |||||
| Yes | 43.8 | 38.1 | 33.8 | 30 | 24.9 |
| CRP (mg/L)a | 3.4 (5.1) | 2.6 (4.1) | 2.3 (3.8) | 2.1 (3.7) | 1.8 (3.8) |
| Testosterone (nmol/L)a | 7.0 (6.3) | 6.9 (6.2) | 6.8 (6.1) | 6.7 (6.1) | 6.6 (6.0) |
| SHBG (nmol/L)a | 55.4 (28.6) | 52.2 (25.8) | 50.5 (24.9) | 49.1 (24.2) | 46.5 (23.3) |
| HbA1c (mmol/mol)a | 35.9 (5.3) | 35.4 (4.5) | 35.1 (4.4) | 34.9 (4.2) | 34.7 (4.5) |
MET, metabolic equivalents.
Mean and SD.
Among women only.
The reproducibility (ICC) of IGF1 levels measured at both the recruitment and repeat assessment visit (n = 16,357 participants; median of 4 years apart) was 0.78 (95% CI 0.77–0.79).
Association between circulating IGF1 levels and colorectal cancer risk
For men and women combined, higher circulating IGF1 levels were associated with elevated colorectal cancer risk in the multivariable models (model 1, HR comparing quintile 5 vs 1 [q5 – q1] = 1.24, 95% CI 1.10–1.40; P-trend = .006) (Table 2). This positive association strengthened after additional adjustment for circulating levels of CRP, HbA1c, testosterone, and SHBG (model 2, HR[q5 – q1] = 1.34, 95% CI 1.18–1.52; P-trend < 0.0001). In the restricted cubic spline model, no deviation from linearity for the relationship between IGF1 and colorectal cancer was observed (P-nonlinear = 0.91). In the continuous multivariable model 2, adjusted for circulating levels of CRP, HbA1c, testosterone, and SHBG, a 1-SD increment of IGF1 was associated with an 8% higher colorectal cancer risk (HR 1.08; 95% CI 1.04–1.13). Correction for regression dilution resulted in a stronger positive association (HR per 1-SD increment of IGF1, HR 1.11; 95% CI 1.05–1.17) (Table 2; Figure 1). Similar magnitude positive associations were found for men and women (P-heterogeneity = .1), and across anatomic subsites (colon vs rectal, P-heterogeneity = 1; proximal colon vs distal colon, P-heterogeneity = 0.8) (Table 2).
Table 2.
Risk (HRs) of Colorectal Cancer Associated With Circulating IGF1 Levels in the UK Biobank
| Q1 | Q2 | Q3 | Q4 | Q5 | P-trend | HR per 1-SD increment | HR per 1-SD increment (adjusted)a | |
|---|---|---|---|---|---|---|---|---|
| Quintile cutpoints (nmol/L) | ||||||||
| Men | <17.6 | 17.6-<20.6 | 20.6-<23.1 | 23.1-<26.1 | ≥26.1 | |||
| Women | <16.3 | 16.3-<19.5 | 19.5-<22.3 | 22.3-<25.5 | ≥25.5 | |||
| Colorectal cancer | ||||||||
| Both sexes | ||||||||
| n cases | 595 | 586 | 528 | 475 | 481 | |||
| Model 1 | 1 | 1.11 (0.99–1.24) | 1.09 (0.97–1.23) | 1.07 (0.95–1.21) | 1.24 (1.10–1.40) | 0.006 | 1.05 (1.01–1.10) | 1.07 (1.02–1.13) |
| Model 2 | 1 | 1.14 (1.01–1.28) | 1.14 (1.01–1.28) | 1.14 (1.00–1.29) | 1.34 (1.18–1.52) | <0.0001 | 1.08 (1.04–1.13) | 1.11 (1.05–1.17) |
| Men | ||||||||
| Model 2 | 1 | 1.09 (0.94–1.27) | 1.09 (0.93–1.27) | 1.09 (0.93–1.28) | 1.29 (1.09–1.51) | 0.009 | 1.06 (1.01–1.12) | 1.09 (1.01–1.17) |
| Women | ||||||||
| Model 2 | 1 | 1.20 (1.01–1.44) | 1.21 (1.00–1.45) | 1.20 (0.99–1.46) | 1.41 (1.15–1.72) | 0.003 | 1.11 (1.04–1.18) | 1.14 (1.05–1.24) |
| Colon cancer | ||||||||
| Both sexes | ||||||||
| n cases | 411 | 378 | 342 | 306 | 322 | |||
| Model 1 | 1 | 1.05 (0.91–1.21) | 1.04 (0.90–1.20) | 1.02 (0.88–1.19) | 1.24 (1.06–1.44) | 0.03 | 1.05 (1.00–1.10) | 1.07 (1.00–1.14) |
| Model 2 | 1 | 1.08 (0.94–1.25) | 1.09 (0.94–1.26) | 1.09 (0.94–1.27) | 1.35 (1.16–1.57) | 0.001 | 1.09 (1.03–1.14) | 1.11 (1.04–1.19) |
| Men | ||||||||
| Model 2 | 1 | 1.07 (0.88–1.29) | 1.05 (0.86–1.28) | 1.09 (0.88–1.34) | 1.28 (1.04–1.57) | 0.04 | 1.07 (1.00–1.14) | 1.09 (0.99–1.19) |
| Women | ||||||||
| Model 2 | 1 | 1.10 (0.89–1.35) | 1.13 (0.91–1.40) | 1.09 (0.87–1.37) | 1.42 (1.13–1.78) | 0.01 | 1.10 (1.02–1.18) | 1.13 (1.03–1.24) |
| Proximal colon cancer | ||||||||
| Both sexes | ||||||||
| n cases | 209 | 210 | 189 | 140 | 159 | |||
| Model 1 | 1 | 1.15 (0.95–1.39) | 1.13 (0.93–1.38) | 0.93 (0.75–1.16) | 1.23 (1.00–1.52) | 0.37 | 1.04 (0.97–1.11) | 1.05 (0.96–1.15) |
| Model 2 | 1 | 1.19 (0.98–1.45) | 1.20 (0.98–1.47) | 1.01 (0.81–1.26) | 1.36 (1.10–1.69) | 0.07 | 1.08 (1.00–1.15) | 1.10 (1.01–1.21) |
| Men | ||||||||
| Model 2 | 1 | 1.18 (0.90–1.55) | 1.05 (0.79–1.40) | 0.99 (0.73–1.34) | 1.27 (0.94–1.71) | 0.4 | 1.06 (0.96–1.17) | 1.07 (0.94–1.22) |
| Women | ||||||||
| Model 2 | 1 | 1.19 (0.90–1.58) | 1.35 (1.02–1.79) | 1.00 (0.73–1.39) | 1.45 (1.05–1.99) | 0.11 | 1.09 (0.99–1.20) | 1.12 (0.98–1.27) |
| Distal colon cancer | ||||||||
| Both sexes | ||||||||
| n cases | 181 | 151 | 141 | 152 | 145 | |||
| Model 1 | 1 | 0.95 (0.77–1.18) | 0.98 (0.78–1.22) | 1.15 (0.92–1.44) | 1.24 (0.99–1.55) | 0.03 | 1.06 (0.99–1.15) | 1.09 (0.99–1.20) |
| Model 2 | 1 | 0.97 (0.78–1.21) | 1.01 (0.81–1.27) | 1.21 (0.97–1.51) | 1.32 (1.05–1.67) | 0.005 | 1.09 (1.01–1.18) | 1.12 (1.01–1.24) |
| Men | ||||||||
| Model 2 | 1 | 0.99 (0.74–1.32) | 1.06 (0.79–1.42) | 1.21 (0.90–1.62) | 1.21 (0.89–1.65) | 0.1 | 1.05 (0.95–1.17) | 1.07 (0.94–1.23) |
| Women | ||||||||
| Model 2 | 1 | 0.95 (0.68–1.34) | 0.97 (0.68–1.38) | 1.21 (0.85–1.71) | 1.47 (1.03–2.09) | 0.02 | 1.13 (1.01–1.27) | 1.18 (1.01–1.37) |
| Rectal cancer | ||||||||
| Both sexes | ||||||||
| n cases | 184 | 208 | 186 | 169 | 159 | |||
| Model 1 | 1 | 1.23 (1.01–1.51) | 1.20 (0.98–1.48) | 1.18 (0.95–1.46) | 1.26 (1.01–1.56) | 0.09 | 1.06 (0.99–1.14) | 1.08 (0.99–1.18) |
| Model 2 | 1 | 1.26 (1.03–1.54) | 1.24 (1.01–1.53) | 1.23 (0.99–1.53) | 1.33 (1.06–1.66) | 0.03 | 1.08 (1.01–1.16) | 1.11 (1.01–1.22) |
| Men | ||||||||
| Model 2 | 1 | 1.13 (0.89–1.44) | 1.15 (0.90–1.48) | 1.09 (0.84–1.42) | 1.30 (1.00–1.69) | 0.11 | 1.06 (0.97–1.16) | 1.08 (0.96–1.21) |
| Women | ||||||||
| Model 2 | 1 | 1.58 (1.11–2.26) | 1.48 (1.02–2.15) | 1.58 (1.08–2.32) | 1.43 (0.94–2.16) | 0.12 | 1.13 (1.00–1.28) | 1.18 (1.00–1.38) |
NOTE. Model 1: Multivariable Cox regression model using age as the underlying time variable and stratified by sex, Townsend deprivation index (quintiles), region of the recruitment assessment center, and age at recruitment. Models adjusted for waist circumference (per 5 cm), total physical activity (<10, 10 to <20, 20 to <40, 40 to <60, ≥60 metabolic equivalent hours per week, unknown), height (per 10 cm), alcohol consumption frequency (never, special occasions only, 1–3 times per month, 1–2 times per week, 3–4 times per week, daily/almost daily, unknown), smoking status and intensity (never, former, current- <15 per day, current- ≥15 per day, current- intensity unknown, unknown), frequency of red and processed meat consumption (<2, 2 to <3, 3 to <4, ≥4 occasions per week, unknown), family history of colorectal cancer (no, yes, unknown), educational level (Certificates of secondary education [CSEs]/Ordinary [O]-levels/General Certificates of Secondary Education [GCSEs] or equivalent, National Vocational Qualification [NVQ]/Higher National Diploma [HND]/Higher National Certificate [HNC]/Advanced [A]-levels/Advanced Subsidiary [AS]-levels or equivalent, other professional qualifications, college/university degree, none of the above, unknown), regular aspirin/ibuprofen use (no, yes, unknown), ever use of hormone replacement therapy (no, yes, unknown).
Model 2: Model 1 plus additional adjustment for circulating levels (sex-specific quintiles, missing/unknown) of CRP (mg/L), HbA1c (mmol/mol), testosterone (nmol/L), and SHBG (nmol/L).
HRs per SD increment were additionally corrected for regression dilution using a regression dilution ratio (0.76) obtained from the subsample of participants with repeat IGF1 measurements.
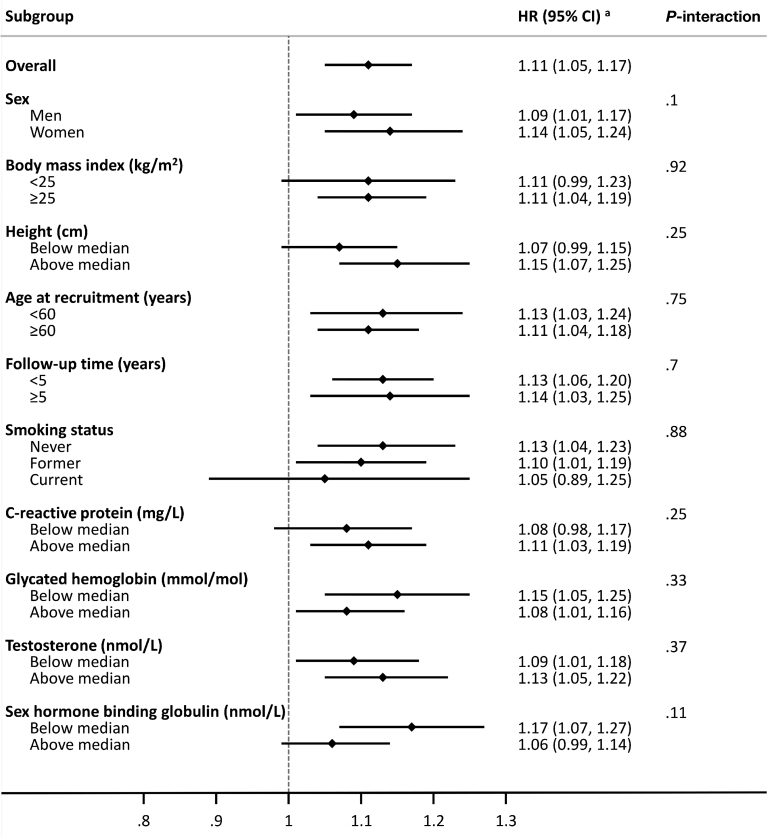
Figure 1.
Subgroup analyses of the association between circulating IGF1 levels and colorectal cancer risk in the UK Biobank. HR per 1-SD increment in circulating IGF1 levels. Multivariable Cox regression model using age as the underlying time variable and stratified by sex, Townsend deprivation index (quintiles), region of the recruitment assessment center, and age at recruitment. Models adjusted for waist circumference (per 5 cm), total physical activity (<10, 10 to <20, 20 to <40, 40 to <60, ≥60 metabolic equivalent hours per week, unknown), height (per 10 cm), alcohol consumption frequency (never, special occasions only, 1–3 times per month, 1–2 times per week, 3–4 times per week, daily/almost daily, unknown), smoking status and intensity (never, former, current- <15 per day, current- ≥15 per day, current- intensity unknown, unknown), frequency of red and processed meat consumption (<2, 2 to <3, 3 to <4, ≥4 occasions per week, unknown), family history of colorectal cancer (no, yes, unknown), educational level (Certificates of secondary education [CSEs]/Ordinary [O]-levels/General Certificates of Secondary Education [GCSEs] or equivalent, National Vocational Qualification [NVQ]/Higher National Diploma [HND]/Higher National Certificate [HNC]/Advanced [A]-levels/Advanced Subsidiary [AS]-levels or equivalent, other professional qualifications, college/university degree, none of the above, unknown), regular aspirin/ibuprofen use (no, yes, unknown), ever use of hormone replacement therapy (no, yes, unknown), and circulating levels (sex-specific quintiles, missing/unknown) of CRP (mg/L), HbA1c (mmol/mol), testosterone (nmol/L), and SHBG (nmol/L). aHRs per SD increment were corrected for regression dilution using a regression dilution ratio (0.76) obtained from the subsample of participants with repeat IGF1 measurements. Median values: height = 176 cm for men and 162 cm for women; CRP = 1.3 mg/L; HbA1c = 35 mmol/mol; testosterone = 1 nmol/L for women and 11.8 nmol/L for men; SHBG = 56.3 nmol/L for men and 37.2 nmol/L for women.
The positive association observed between circulating IGF1 levels and colorectal cancer was evident across subgroups of BMI, height, age at recruitment/blood collection, follow-up time, smoking status, and circulating levels of CRP, HbA1c, testosterone, and SHBG (all P-interactions ≥ .1) (Figure 1). In a sensitivity analysis, similar relationships for circulating IGF1 levels and colorectal cancer were found when participants with HbA1c levels ≥48 mmol/mol (or 6.5%, cutoff for type-2 diabetes), were excluded from the analyses (Supplementary Table 3).
Mendelian Randomization
Association between genetically determined circulating IGF1 levels and colorectal cancer risk
We estimated that a 1-SD increment in genetically determined IGF1 levels was associated with an 8% higher colorectal cancer risk (OR 1.08; 95% CI 1.03–1.12; P value = 3.3 × 10–4) (Table 3). Positive associations of similar magnitude were found for men and women when analyzed separately (P-heterogeneity = .24), and by anatomic subsite (colon vs rectal, P-heterogeneity = 1; proximal colon versus distal colon, P-heterogeneity = .55) (Table 3). Evidence of effect heterogeneity Cochran’s Q P values < .001) and horizontal pleiotropy (MR-PRESSO P values <1 × 10−4) were observed for each model (Supplementary Table 4), but there was little evidence of directional pleiotropy (MR-Egger intercept P values >.20) for all models, except for distal colon cancer (MR-Egger intercept P value = .02). However, the corrected estimates for distal colon cancer and all other models from the MR-Egger and the weighted-median approach analyses replicated the initial positive effect estimates (Table 3; Supplementary Table 4), and the MR-PRESSO test identified a few outlier SNPs that did not distort the effect estimates (MR-PRESSO P values >.8). Finally, the funnel plot for the IGF1 instruments indicated a symmetric distribution of effect estimates (Supplementary Figure 1).
Table 3.
MR Estimates Between Circulating Levels of IGF1 and IGFBP3 and Risk of Colorectal Cancer (n = 52,865 Colorectal Cancer Cases and n = 46,287 Controls)
| MR |
|||
|---|---|---|---|
| Likelihood-based approach |
Weighted median approach |
MR-Egger test |
|
| OR (95% CI) per 1-SD increment | OR (95% CI) per 1-SD increment | OR (95% CI) per 1-SD increment | |
| IGF1 | |||
| Colorectal cancer | |||
| Both sexes | 1.08 (1.03–1.12) | 1.12 (1.04–1.20) | 1.11 (0.98–1.27) |
| Men | 1.11 (1.05–1.17) | 1.07 (0.97–1.18) | 1.18 (1.00–1.38) |
| Women | 1.05 (0.99–1.11) | 1.07 (0.97–1.18) | 1.06 (0.89–1.25) |
| Colon cancer | |||
| Both sexes | 1.06 (1.01–1.11) | 1.12 (1.03–1.23) | 1.14 (0.98–1.32) |
| Proximal colon cancer | |||
| Both sexes | 1.04 (0.97–1.11) | 1.08 (0.97–1.21) | 1.02 (0.84–1.23) |
| Distal colon cancer | |||
| Both sexes | 1.07 (1.00–1.14) | 1.10 (0.98–1.23) | 1.26 (1.05–1.50) |
| Rectal cancer | |||
| Both sexes | 1.06 (1.00–1.14) | 1.01 (0.90–1.13) | 1.09 (0.91–1.30) |
| IGFBP3 | |||
| Colorectal cancer | |||
| Both sexes | 1.12 (1.06–1.18) | 1.14 (1.07–1.20) | 1.18 (1.07–1.31) |
| Men | 1.12 (1.04–1.21) | 1.13 (1.04–1.22) | 1.14 (0.98–1.34) |
| Women | 1.12 (1.04–1.21) | 1.13 (1.04–1.22) | 1.25 (1.18–1.32) |
| Colon cancer | |||
| Both sexes | 1.11 (1.04–1.19) | 1.13 (1.05–1.21) | 1.27 (1.16–1.38) |
| Proximal colon cancer | |||
| Both sexes | 1.12 (1.03–1.22) | 1.12 (1.02–1.23) | 1.25 (1.21–1.29) |
| Distal colon cancer | |||
| Both sexes | 1.10 (1.01–1.20) | 1.13 (1.03–1.24) | 1.29 (1.01–1.65) |
| Rectal cancer | |||
| Both sexes | 1.12 (1.04–1.22) | 1.14 (1.05–1.25) | 1.18 (1.01–1.39) |
Supplementary Figure 1.
Funnel plots of risk estimates of (A) IGF1 and (B) IGFBP3 with colorectal cancer against instrumental strength. Instrumental strength is SNP to colorectal cancer effect corrected by SNP to IGF1 or IGFBP3 standard error of the effect. X-axis is in logarithmic scale. P values are 2-sided, MR test.
Association between genetically determined circulating IGFBP3 levels and colorectal cancer risk
A 1-SD increment in genetically determined IGFBP3 levels was associated with a 12% higher colorectal cancer risk (OR 1.12; 95% CI 1.06–1.18; P = 4.2 × 10–5) (Table 3). Similar positive associations were found for men and women when analyzed separately (P-heterogeneity = 1), and by anatomic subsite (colon vs rectal, P-heterogeneity = .87; proximal colon vs distal colon, P-heterogeneity = .77) (Table 3). Evidence of effect heterogeneity was weak (P > .10) for all models except colon cancer and distal colon cancer models (P values .04 and .02, respectively). The MR-PRESSO test identified possible horizontal pleiotropy for colon cancer and distal colon cancer (P < .05). However, little evidence of directional pleiotropy was found for all models (MR-Egger intercept P > .15), and the estimates from the weighted-median approach and MR-Egger were consistent with those of likelihood-based approach (Table 3; Supplementary Table 4). The leave-one-out analysis revealed that the positive association between IGFBP3 and colorectal cancer was driven by 1 variant, rs11977526, located in the IGFBP3 gene region (OR per SD increment in IGFBP3 with rs11977526 excluded = 1.02; 95% CI 0.92–1.13; P = .7) (Supplementary Table 5).
Discussion
In serologic analyses of UK Biobank data, we found that higher prediagnostic levels of circulating IGF1 were associated with greater colorectal cancer risk, with similar-strength positive associations found by sex and colorectal anatomical subsite. Concordant with this, the MR analyses revealed a positive effect of IGF1 on colorectal cancer risk, suggesting that higher circulating IGF1 levels may have a causal role in the development of this malignancy. Collectively, these findings provide strong support for a role of the IGF-pathway in colorectal tumorigenesis.
Despite extensive experimental evidence indicating a role for IGF1 in colorectal tumorigenesis, results from previous epidemiological studies (most including <500 colorectal cancer cases) have been inconsistent, with null or weak positive associations found that did not meet the statistical significance threshold.6, 7, 8, 9, 10, 11 Our study, using UK Biobank data, is unique in that rather than following a nested study design including a subset of the cohort, IGF1 levels were measured in all study participants. Further, we were able to control statistically for other serologic risk factors that are related to both circulating IGF1 levels and colorectal cancer risk (CRP, testosterone, SHBG, and HbA1c). Our study was the largest to date to examine the IGF1 and colorectal cancer relationship (including >2600 incident cases) which meant we were sufficiently powered to study the IGF1 and colorectal cancer relationship by sex, anatomic subsite of tumor, and across subgroups of other risk factors. We found no heterogeneity for the positive association by sex, anatomic subsite, follow-up time, and other colorectal cancer risk factors.
Despite the robustness of this positive IGF1 and colorectal cancer association, as with all observational studies, these analyses are vulnerable to the inherent biases of this study design, namely residual confounding and reverse causality. MR analyses are less susceptible to such biases because alleles are randomly assigned during meiosis and germline genetic variants are unaffected by disease process.38 Our MR analyses yielded strikingly similar estimates of effect for IGF1 and colorectal cancer to our UK Biobank serologic analyses. Similar to the serologic analyses we found no heterogeneity by sex or anatomic subsite in our MR analyses. An important assumption of MR is that the genetic variants do not influence the outcome through a pathway independent of the main exposure of interest (horizontal pleiotropy). We conducted multiple sensitivity analyses to test for the influence of pleiotropy on our causal estimates, and our results for IGF1 were robust according to these various tests.
Experimental data support a role for IGF1 and its downstream signaling pathways in colorectal tumorigenesis. IGF1 can promote cellular proliferation through the activation of the mitogen activated protein kinase and the phosphoinositide 3-kinase pathways.39 Colonocytes express IGF1-receptors and these are frequently overexpressed in neoplastic cells.40 IGF1 has been shown to promote cellular proliferation in colorectal tissue, and the blockade of the IGF1-receptor by a monoclonal antibody inhibits cell proliferation.41
Potentially modifiable determinants of circulating IGF1 levels include dietary protein intake,42, 43, 44 BMI,43 and physical activity.45 In addition, a number of pharmacologic agents targeting the IGF system have been developed, although, as of yet, they have not been successful in treating colorectal cancer patients in clinical trials.46 Although circulating IGF1 levels are modifiable, it is currently unknown how long an intervention aimed at altering IGF1 concentrations would have to be applied to have measurable effects. Our result provides possible evidence for a causal role of circulating IGF1 levels in colorectal cancer development and will hopefully reinvigorate efforts to develop and introduce interventions targeting the IGF system for colorectal cancer prevention in susceptible individuals.
The bioactivity of IGF1 is partially regulated through IGFBPs, with most bound to IGFBP3. Higher levels of IGFBP3 both increase the serologic binding capacity for IGF1, but also lower circulating IGF1 bioavailability.47 As well as IGF-binding properties, IGFBP3 has been shown to induce apoptosis and reduce proliferation in colon cancer cell lines.4,5 The positive association we found in our MR analyses for IGFBP3 and colorectal cancer is inconsistent with its anticipated anti-tumorigenic effects. Epidemiologic studies examining the IGFBP3 and colorectal cancer association have reported mixed findings, with inverse, positive, and null results all previously reported.6,7,10, 11, 12, 13, 14 The positive effect estimate in our MR analysis was driven solely by 1 variant, rs11977526, with a null result found when this variant was excluded. Although rs11977526 is located in close proximity to the IGFBP3 gene, this variant has also been associated with several anthropometric traits48 and circulating levels of IGF2,49 for which there is evidence of a role in colorectal cancer development. The IGF2 gene is imprinted and loss of imprinting has been detected in colorectal cancer tumor tissue.50 Stroma-induced IGF2 has been shown to promote colon cancer progression in a paracrine and autocrine manner.51 A meta-analysis of 3 prospective studies reported an almost 2-fold higher colorectal cancer risk when the highest and lowest groups of circulating IGF2 levels were compared.52 MR analyses for IGF2 are currently limited as a GWAS for circulating IGF2 has not been conducted. Similarly, for IGFBP3 and other IGF system components (including bioavailable IGF1 levels), more precise genetic instruments are now required to help disentangle the possible biological effects of specific ligands and binding proteins on colorectal cancer development.
This was the largest and most comprehensive study that used 2 complementary study designs to examine the role of IGF1 on colorectal cancer development. A limitation of our analysis is that IGF1 levels were measured only once at baseline in all participants, and it is possible that these measurements may not reflect exposure levels across time. However, our reproducibility analysis in a subset of the cohort found an ICC value of 0.78 between IGF1 measurements collected 4 years apart, indicating that a single measurement provides a good estimate of medium to longer-term exposures. The availability of repeat IGF1 measurements also allowed us to correct for regression dilution bias, and thus limiting the effects of measurement error and within-person variability on our risk estimates21; the positive associations we found were stronger after regression dilution correction. A limitation of our MR analyses is that our use of summary-level data precluded the investigation of nonlinear effects and subgroup analyses by other risk factors (eg, age, BMI, smoking). However, our serologic analysis, which yielded a similar overall risk estimate to our MR result, found no evidence of a nonlinear association for IGF1 and colorectal cancer and little heterogeneity across subgroups of other risk factors.
In conclusion, our complementary serologic and MR analyses provide strong support for a positive association of circulating IGF1 levels on colorectal cancer risk. This result suggests that diet/lifestyle42, 43, 44,53 or pharmacologic1 interventions targeting the IGF system may offer a promising strategy in reducing the risk of colorectal cancer.
Acknowledgments
This work was conducted using the UK Biobank Resource under Application Number 25897 and we express our gratitude to the participants and those involved in building the resource. UK Biobank is an open access resource. Bona fide researchers can apply to use the UK Biobank dataset by registering and applying at http://ukbiobank.ac.uk/register-apply/. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Author contributions: Conceived and designed the experiments: NM, MJG. Analyzed the data: RCT, NM, MJG. This article was written by NM and MJG taking into account the comments and suggestions of the coauthors. All coauthors had the opportunity to comment on the analysis and interpretation of the findings and approved the final version for publication.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding RMM was supported by the NIHR Biomedical Research Centre at University Hospitals Bristol NHS Foundation Trust and the University of Bristol. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the Department of Health.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2019.12.020.
Funding (continued)
Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO): National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088, R01 CA059045, R01201407). Genotyping/Sequencing services were provided by the Center for Inherited Disease Research (CIDR) (X01-HG008596 and X-01-HG007585). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268201200008I. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA015704.
ASTERISK: a Hospital Clinical Research Program (PHRC-BRD09/C) from the University Hospital Center of Nantes (CHU de Nantes) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC).
The ATBC Study is supported by the Intramural Research Program of the U.S. National Cancer Institute, National Institutes of Health, and by U.S. Public Health Service contract HHSN261201500005C from the National Cancer Institute, Department of Health and Human Services.
CLUE II: Funding to support CLUE II, for studies on colorectal cancer in the cohort, and for the investigators were from grants from the National Cancer Institute (U01 CA86308, Early Detection Research Network; P30 CA006973), National Institute on Aging (U01 AG18033), and the American Institute for Cancer Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
COLO2&3: National Institutes of Health (R01 CA60987).
ColoCare: This work was supported by the National Institutes of Health (grant numbers R01 CA189184 (Li/Ulrich), U01 CA206110 (Ulrich/Li/Siegel/Figueireido/Colditz, 2P30CA015704- 40 (Gilliland), R01 CA207371 (Ulrich/Li)), the Matthias Lackas-Foundation, the German Consortium for Translational Cancer Research, and the EU TRANSCAN initiative.
The Colon Cancer Family Registry (CCFR) Illumina GWAS was supported by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (grant numbers U01 CA122839, R01 CA143247). The CCFR participant recruitment and collection of data and biospecimens used in this study were supported by the NCI/NIH (grant number U01 CA167551) and through NCI/NIH cooperative agreements with the following Colon CFR centers: Australasian Colorectal Cancer Family Registry (grant numbers U01 CA074778 and U01/U24 CA097735), USC Consortium Colorectal Cancer Family Registry (grant numbers U01/U24 CA074799), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (grant number U01/U24 CA074800), Ontario Familial Colorectal Cancer Registry (grant number U01/U24 CA074783), Seattle Colorectal Cancer Family Registry (grant number U01/U24 CA074794), and University of Hawaii Colorectal Cancer Family Registry (grant number U01/U24 CA074806)’ The content of this manuscript does not necessarily reflect the views or policies of the NCI, NIH or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
COLON: The COLON study is sponsored by Wereld Kanker Onderzoek Fonds, including funds from grant 2014/1179 as part of the World Cancer Research Fund International Regular Grant Programme, by Alpe d’Huzes and the Dutch Cancer Society (UM 2012–5653, UW 2013-5927, UW2015-7946), and by TRANSCAN (JTC2012-MetaboCCC, JTC2013-FOCUS). The Nqplus study is sponsored by a ZonMW investment grant (98-10030); by PREVIEW, the project PREVention of diabetes through lifestyle intervention and population studies in Europe and around the World (PREVIEW) project which received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant no. 312057; by funds from TI Food and Nutrition (cardiovascular health theme), a public–private partnership on precompetitive research in food and nutrition; and by FOODBALL, the Food Biomarker Alliance, a project from JPI Healthy Diet for a Healthy Life.
Colorectal Cancer Transdisciplinary (CORECT) Study: The CORECT Study was supported by the National Cancer Institute, National Institutes of Health (NCI/NIH), U.S. Department of Health and Human Services (grant numbers U19 CA148107, R01 CA81488, P30 CA014089, R01 CA197350,; P01 CA196569; R01 CA201407) and National Institutes of Environmental Health Sciences, National Institutes of Health (grant number T32 ES013678).
CORSA: “Österreichische Nationalbank Jubiläumsfondsprojekt” (12511) and Austrian Research Funding Agency (FFG) grant 829675.
CPS-II: The American Cancer Society funds the creation, maintenance, and updating of the Cancer Prevention Study-II (CPS-II) cohort. This study was conducted with Institutional Review Board approval.
CRCGEN: Colorectal Cancer Genetics & Genomics, Spanish study was supported by Instituto de Salud Carlos III, co-funded by FEDER funds –a way to build Europe– (grants PI14-613 and PI09-1286), Agency for Management of University and Research Grants (AGAUR) of the Catalan Government (grant 2017SGR723), and Junta de Castilla y León (grant LE22A10-2). Sample collection of this work was supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncología de Catalunya (XBTC), Plataforma Biobancos PT13/0010/0013 and ICOBIOBANC, sponsored by the Catalan Institute of Oncology.
Czech Republic CCS: This work was supported by the Grant Agency of the Czech Republic (grants CZ GA CR: GAP304/10/1286 and 1585) and by the Grant Agency of the Ministry of Health of the Czech Republic (grants AZV 15-27580A and AZV 17-30920A).
DACHS: This work was supported by the German Research Council (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, CH 117/1-1, HO 5117/2-1, HE 5998/2-1, KL 2354/3-1, RO 2270/8-1 and BR 1704/17-1), the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany, and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A and 01ER1505B).
DALS: National Institutes of Health (R01 CA48998 to M. L. Slattery).
EDRN: This work is funded and supported by the NCI, EDRN Grant (U01 CA 84968-06).
EPIC: The coordination of EPIC is financially supported by the European Commission (DGSANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRCItaly and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPICOxford) (United Kingdom).
Funding: (EPICOLON) This work was supported by grants from Fondo de Investigación Sanitaria/FEDER (PI08/0024, PI08/1276, PS09/02368, P111/00219, PI11/00681, PI14/00173, PI14/00230, PI17/00509, 17/00878, Acción Transversal de Cáncer), Xunta de Galicia (PGIDIT07PXIB9101209PR), Ministerio de Economia y Competitividad (SAF07-64873, SAF 2010-19273, SAF2014-54453R), Fundación Científica de la Asociación Española contra el Cáncer (GCB13131592CAST), Beca Grupo de Trabajo “Oncología” AEG (Asociación Española de Gastroenterología), Fundación Privada Olga Torres, FP7 CHIBCHA Consortium, Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR, Generalitat de Catalunya, 2014SGR135, 2014SGR255, 2017SGR21, 2017SGR653), Catalan Tumour Bank Network (Pla Director d’Oncologia, Generalitat de Catalunya), PERIS (SLT002/16/00398, Generalitat de Catalunya), CERCA Programme (Generalitat de Catalunya) and COST Action CA17118. CIBERehd is funded by the Instituto de Salud Carlos III.
ESTHER/VERDI. This work was supported by grants from the Baden-Württemberg Ministry of Science, Research and Arts and the German Cancer Aid.
Harvard cohorts (HPFS, NHS, PHS): HPFS is supported by the National Institutes of Health (P01 CA055075, UM1 CA167552, U01 CA167552, R01 CA137178, R01 CA151993, R35 CA197735, K07 CA190673, and P50 CA127003), NHS by the National Institutes of Health (R01 CA137178, P01 CA087969, UM1 CA186107, R01 CA151993, R35 CA197735, K07CA190673, and P50 CA127003) and PHS by the National Institutes of Health (R01 CA042182).
Hawaii Adenoma Study: NCI grants R01 CA72520.
HCES-CRC: the Hwasun Cancer Epidemiology Study–Colon and Rectum Cancer (HCES-CRC; grants from Chonnam National University Hwasun Hospital, HCRI18007-1).
Kentucky: This work was supported by the following grant support: Clinical Investigator Award from Damon Runyon Cancer Research Foundation (CI-8); NCI R01CA136726.
LCCS: The Leeds Colorectal Cancer Study was funded by the Food Standards Agency and Cancer Research UK Programme Award (C588/A19167).
MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 509348, 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database.
MEC: National Institutes of Health (R37 CA54281, P01 CA033619, and R01 CA063464).
MECC: This work was supported by the National Institutes of Health, U.S. Department of Health and Human Services (R01 CA81488 to SBG and GR).
MSKCC: The work at Sloan Kettering in New York was supported by the Robert and Kate Niehaus Center for Inherited Cancer Genomics and the Romeo Milio Foundation. Moffitt: This work was supported by funding from the National Institutes of Health (grant numbers R01 CA189184, P30 CA076292), Florida Department of Health Bankhead-Coley Grant 09BN-13, and the University of South Florida Oehler Foundation. Moffitt contributions were supported in part by the Total Cancer Care Initiative, Collaborative Data Services Core, and Tissue Core at the H. Lee Moffitt Cancer Center & Research Institute, a National Cancer Institute-designated Comprehensive Cancer Center (grant number P30 CA076292).
NCCCS I & II: We acknowledge funding support for this project from the National Institutes of Health, R01 CA66635 and P30 DK034987.
NFCCR: This work was supported by an Interdisciplinary Health Research Team award from the Canadian Institutes of Health Research (CRT 43821); the National Institutes of Health, U.S. Department of Health and Human Serivces (U01 CA74783); and National Cancer Institute of Canada grants (18223 and 18226). The authors wish to acknowledge the contribution of Alexandre Belisle and the genotyping team of the McGill University and Génome Québec Innovation Centre, Montréal, Canada, for genotyping the Sequenom panel in the NFCCR samples. Funding was provided to Michael O. Woods by the Canadian Cancer Society Research Institute.
NSHDS: Swedish Cancer Society; Cancer Research Foundation in Northern Sweden; Swedish Research Council; J C Kempe Memorial Fund; Faculty of Medicine, Umeå University, Umeå, Sweden; and Cutting-Edge Research Grant from the County Council of Västerbotten, Sweden.
OFCCR: National Institutes of Health, through funding allocated to the Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); see CCFR section above. Additional funding toward genetic analyses of OFCCR includes the Ontario Research Fund, the Canadian Institutes of Health Research, and the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation.
OSUMC: OCCPI funding was provided by Pelotonia and HNPCC funding was provided by the NCI (CA16058 and CA67941).
PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH,
PMH-SCCFR: National Institutes of Health (R01 CA076366 to P. Newcomb) and U01CA074794 (to J. Potter).
SEARCH: The University of Cambridge has received salary support in respect of PDPP from the NHS in the East of England through the Clinical Academic Reserve. Cancer Research UK (C490/A16561); the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge.
SELECT: Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award Numbers U10 CA37429 (CD Blanke), and UM1 CA182883 (CM Tangen/IM Thompson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SMS and REACH: This work was supported by the National Cancer Institute (grant P01 CA074184 to J.D.P. and P.A.N., grants R01 CA097325, R03 CA153323, and K05 CA152715 to P.A.N., and the National Center for Advancing Translational Sciences at the National Institutes of Health (grant KL2 TR000421 to A.N.B.-H.)
The Swedish Low-risk Colorectal Cancer Study: The study was supported by grants from the Swedish research council; K2015-55X-22674-01-4, K2008-55X-20157-03-3, K2006-72X-20157-01-2 and the Stockholm County Council (ALF project).
Swedish Mammography Cohort and Cohort of Swedish Men: This work is supported by the Swedish Research Council /Infrastructure grant, the Swedish Cancer Foundation, and the Karolinska Institutés Distinguished Professor Award to Alicja Wolk.
UK Biobank: This research has been conducted using the UK Biobank Resource under Application Number 8614
VITAL: National Institutes of Health (K05 CA154337).
WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
Acknowledgements (continued)
ASTERISK: We are very grateful to Dr. Bruno Buecher without whom this project would not have existed. We also thank all those who agreed to participate in this study, including the patients and the healthy control persons, as well as all the physicians, technicians and students.
CLUE II: We appreciate the continued efforts of the staff members at the Johns Hopkins George W. Comstock Center for Public Health Research and Prevention in the conduct of the CLUE II study. Cancer incidence data for CLUE were provided by the Maryland Cancer Registry, Center for Cancer Surveillance and Control, Maryland Department of Health, 201 W. Preston Street, Room 400, Baltimore, MD 21201, http://phpa.dhmh.maryland.gov/cancer, 410-767-4055. We acknowledge the State of Maryland, the Maryland Cigarette Restitution Fund, and the National Program of Cancer Registries of the Centers for Disease Control and Prevention for the funds that support the collection and availability of the cancer registry data.
COLON and NQplus: the authors would like to thank the COLON and NQplus investigators at Wageningen University & Research and the involved clinicians in the participating hospitals.
CORSA: We kindly thank all those who contributed to the screening project Burgenland against CRC. Furthermore, we are grateful to Doris Mejri and Monika Hunjadi for laboratory assistance.
CPS-II: The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
Czech Republic CCS: We are thankful to all clinicians in major hospitals in the Czech Republic, without whom the study would not be practicable. We are also sincerely grateful to all patients participating in this study.
DACHS: We thank all participants and cooperating clinicians, and Ute Handte-Daub, Utz Benscheid, Muhabbet Celik and Ursula Eilber for excellent technical assistance.
EDRN: We acknowledge all the following contributors to the development of the resource: University of Pittsburgh School of Medicine, Department of Gastroenterology, Hepatology and Nutrition: Lynda Dzubinski; University of Pittsburgh School of Medicine, Department of Pathology: Michelle Bisceglia; and University of Pittsburgh School of Medicine, Department of Biomedical Informatics.
EPIC: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
EPICOLON: We are sincerely grateful to all patients participating in this study who were recruited as part of the EPICOLON project. We acknowledge the Spanish National DNA Bank and the Biobank of Hospital Clínic–IDIBAPS for the availability of the samples. The work was carried out (in part) at the Esther Koplowitz Centre, Barcelona.
Harvard cohorts (HPFS, NHS, PHS): The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the HPFS, NHS and PHS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Kentucky: We would like to acknowledge the staff at the Kentucky Cancer Registry.
LCCS: We acknowledge the contributions of Jennifer Barrett, Robin Waxman, Gillian Smith and Emma Northwood in conducting this study.
NCCCS I & II: We would like to thank the study participants, and the NC Colorectal Cancer Study staff.
NSHDS: We thank all participants in the NSHDS cohorts and the staff at the Department of Biobank Research, Umeå University, as well as Kerstin Näslund, formerly of the Department of Medical Biosciences, and Inger Cullman, Department of Chemistry, both at Umeå University for excellent technical assistance.
PLCO: The authors thank the PLCO Cancer Screening Trial screening center investigators and the staff from Information Management Services Inc and Westat Inc. Most importantly, we thank the study participants for their contributions that made this study possible.
PMH-SCCFR: The authors would like to thank the study participants and staff of the Hormones and Colon Cancer study.
SEARCH: We thank the SEARCH team.
SELECT: We thank the research and clinical staff at the sites that participated on SELECT study, without whom the trial would not have been successful. We are also grateful to the 35,533 dedicated men who participated in SELECT.
WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf
Supplementary Table 1.
Summary Information on the IGF1 and IGFBP3 Genetic Instruments Used in the Mendelian Randomization Analyses
Supplementary Table 2.
Association Parameters of Instrumental SNPs Used in the IGF1 and IGFBP3 Genetic Instruments
| SNP | Chr | Position | Genea | Effect allele | Other allele | Association parameters for the exposure |
Association parameters for colorectal cancer |
Association parameters for colorectal cancer men |
Association parameters for colorectal cancer women |
Association parameters for colon cancer |
Association parameters for proximal colon cancer |
Association parameters for distal colon cancer |
Association parameters for rectal cancer |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | SE | Effect | SE | Effect | SE | Effect | SE | Effect | SE | Effect | SE | Effect | SE | Effect | SE | ||||||
| IGF1 - n SNPs = 413 | |||||||||||||||||||||
| rs903908 | 1 | 2,202,967 | SKI | T | C | −0.016 | 0.003 | −0.006 | 0.009 | −0.014 | 0.013 | 0.003 | 0.013 | −0.016 | 0.011 | −0.006 | 0.014 | −0.023 | 0.014 | 0.016 | 0.014 |
| rs17393144 | 1 | 9,210,262 | MIR34A | G | A | −0.016 | 0.003 | 0.006 | 0.010 | 0.019 | 0.013 | −0.007 | 0.014 | 0.005 | 0.011 | 0.012 | 0.015 | −0.002 | 0.015 | −0.005 | 0.015 |
| rs112436634 | 1 | 10,637,709 | PEX14 | C | T | −0.016 | 0.003 | 0.001 | 0.009 | −0.014 | 0.013 | 0.014 | 0.013 | −0.002 | 0.011 | −0.014 | 0.015 | 0.018 | 0.015 | 0.017 | 0.015 |
| rs17037452 | 1 | 11,895,675 | CLCN6 | A | G | 0.023 | 0.003 | 0.000 | 0.012 | −0.011 | 0.017 | 0.011 | 0.018 | 0.000 | 0.015 | 0.029 | 0.019 | −0.029 | 0.019 | −0.003 | 0.019 |
| rs36086195 | 1 | 16,510,894 | ARHGEF19−AS1 | T | C | −0.019 | 0.003 | 0.018 | 0.009 | 0.012 | 0.013 | 0.028 | 0.013 | 0.020 | 0.011 | 0.003 | 0.014 | 0.043 | 0.014 | 0.010 | 0.014 |
| rs12723255 | 1 | 21,233,570 | EIF4G3 | T | C | −0.017 | 0.003 | 0.002 | 0.009 | 0.004 | 0.013 | 0.001 | 0.013 | 0.003 | 0.011 | −0.002 | 0.014 | 0.008 | 0.014 | 0.010 | 0.014 |
| rs6701954 | 1 | 22,022,176 | USP48 | T | G | 0.014 | 0.003 | 0.005 | 0.009 | 0.007 | 0.012 | −0.001 | 0.013 | −0.008 | 0.011 | 0.007 | 0.014 | −0.022 | 0.014 | 0.014 | 0.014 |
| rs76914895 | 1 | 23,292,603 | LACTBL1 | T | C | −0.027 | 0.005 | 0.046 | 0.019 | 0.054 | 0.026 | 0.038 | 0.027 | 0.030 | 0.022 | 0.026 | 0.029 | 0.054 | 0.030 | 0.093 | 0.029 |
| rs2075995 | 1 | 23,847,464 | E2F2 | A | C | −0.014 | 0.003 | 0.003 | 0.009 | 0.011 | 0.012 | −0.003 | 0.013 | 0.008 | 0.011 | −0.004 | 0.014 | 0.024 | 0.014 | 0.014 | 0.014 |
| rs2802330 | 1 | 26,466,831 | PDIK1L | A | G | −0.031 | 0.003 | 0.003 | 0.012 | −0.001 | 0.016 | 0.004 | 0.016 | 0.011 | 0.014 | 0.025 | 0.018 | −0.009 | 0.018 | −0.004 | 0.018 |
| rs17360994 | 1 | 27,278,573 | KDF1 | C | T | −0.042 | 0.005 | −0.012 | 0.017 | −0.003 | 0.023 | −0.023 | 0.024 | −0.006 | 0.020 | −0.012 | 0.026 | 0.022 | 0.026 | −0.005 | 0.026 |
| rs569356 | 1 | 29,136,686 | OPRD1 | A | G | −0.027 | 0.004 | 0.007 | 0.013 | −0.016 | 0.018 | 0.035 | 0.019 | 0.005 | 0.016 | 0.019 | 0.020 | −0.001 | 0.021 | 0.012 | 0.020 |
| rs3131646 | 1 | 40,383,552 | MYCL | G | T | 0.016 | 0.003 | −0.006 | 0.010 | −0.006 | 0.013 | −0.008 | 0.014 | −0.013 | 0.012 | −0.029 | 0.015 | 0.002 | 0.015 | 0.002 | 0.015 |
| rs61780439 | 1 | 41,490,177 | SLFNL1−AS1 | G | A | 0.021 | 0.003 | 0.004 | 0.011 | 0.012 | 0.015 | −0.001 | 0.015 | −0.011 | 0.013 | −0.013 | 0.017 | −0.007 | 0.017 | 0.013 | 0.017 |
| rs2819336 | 1 | 44,015,809 | PTPRF | C | T | −0.027 | 0.003 | −0.004 | 0.009 | −0.003 | 0.013 | −0.006 | 0.013 | −0.007 | 0.011 | −0.019 | 0.014 | 0.001 | 0.015 | 0.008 | 0.014 |
| rs7539178 | 1 | 65,383,002 | JAK1 | A | C | −0.026 | 0.004 | −0.024 | 0.013 | −0.026 | 0.018 | −0.023 | 0.018 | −0.036 | 0.016 | −0.047 | 0.020 | −0.008 | 0.021 | −0.015 | 0.020 |
| rs1046011 | 1 | 65,898,996 | LEPR / LEPROT | C | T | −0.021 | 0.003 | 0.011 | 0.010 | 0.006 | 0.014 | 0.017 | 0.014 | 0.005 | 0.012 | −0.001 | 0.015 | 0.014 | 0.015 | 0.015 | 0.015 |
| rs1430753 | 1 | 68,692,642 | WLS | G | A | −0.021 | 0.003 | −0.011 | 0.011 | −0.012 | 0.016 | −0.010 | 0.016 | −0.007 | 0.013 | 0.000 | 0.017 | −0.020 | 0.018 | −0.041 | 0.017 |
| rs165316 | 1 | 91,533,297 | RPL5P6 | A | G | −0.073 | 0.003 | −0.018 | 0.011 | −0.045 | 0.015 | 0.009 | 0.016 | −0.037 | 0.013 | −0.035 | 0.017 | −0.033 | 0.017 | 0.006 | 0.017 |
| rs1825813 | 1 | 92,708,973 | C1orf146 | G | A | −0.023 | 0.003 | −0.027 | 0.011 | −0.044 | 0.016 | −0.009 | 0.016 | −0.037 | 0.013 | −0.034 | 0.017 | −0.040 | 0.018 | −0.019 | 0.017 |
| rs599839 | 1 | 109,822,166 | PSRC1/CELSR2 | A | G | −0.031 | 0.003 | 0.006 | 0.011 | 0.013 | 0.015 | −0.002 | 0.015 | 0.009 | 0.013 | −0.004 | 0.016 | 0.022 | 0.017 | 0.029 | 0.016 |
| rs45505697 | 1 | 153,651,058 | NPR1 | C | A | 0.030 | 0.005 | 0.011 | 0.020 | −0.009 | 0.028 | 0.037 | 0.029 | 0.021 | 0.024 | 0.015 | 0.031 | 0.023 | 0.032 | 0.022 | 0.031 |
| rs1127313 | 1 | 154,556,425 | ADAR | G | A | 0.024 | 0.003 | 0.009 | 0.009 | 0.007 | 0.012 | 0.013 | 0.013 | 0.007 | 0.011 | −0.001 | 0.014 | 0.017 | 0.014 | 0.019 | 0.014 |
| rs77369503 | 1 | 163,027,266 | RGS4 | G | A | 0.045 | 0.007 | −0.004 | 0.028 | −0.002 | 0.038 | −0.013 | 0.041 | −0.027 | 0.033 | −0.048 | 0.042 | −0.051 | 0.043 | 0.012 | 0.042 |
| rs75681856 | 1 | 174,916,323 | RABGAP1L | C | T | −0.023 | 0.004 | 0.006 | 0.013 | 0.016 | 0.019 | −0.007 | 0.019 | −0.004 | 0.016 | −0.010 | 0.021 | −0.007 | 0.021 | 0.015 | 0.021 |
| rs12749024 | 1 | 176,522,365 | PAPPA2 | C | T | −0.075 | 0.004 | −0.008 | 0.013 | −0.019 | 0.018 | 0.000 | 0.018 | −0.011 | 0.015 | −0.008 | 0.020 | −0.009 | 0.020 | −0.013 | 0.020 |
| rs10913351 | 1 | 177,447,742 | AL122019.1 | G | A | −0.032 | 0.005 | 0.021 | 0.018 | 0.028 | 0.026 | 0.017 | 0.026 | 0.020 | 0.022 | 0.022 | 0.028 | 0.023 | 0.029 | 0.004 | 0.028 |
| rs11577063 | 1 | 179,341,999 | AXDND1 | G | T | −0.020 | 0.003 | −0.011 | 0.011 | 0.007 | 0.015 | −0.031 | 0.015 | −0.011 | 0.013 | −0.025 | 0.016 | −0.002 | 0.017 | −0.020 | 0.016 |
| rs143885630 | 1 | 183,482,785 | SMG7 | G | A | 0.030 | 0.004 | −0.013 | 0.014 | −0.013 | 0.019 | −0.008 | 0.020 | −0.014 | 0.017 | −0.045 | 0.022 | 0.016 | 0.022 | 0.019 | 0.022 |
| rs940400 | 1 | 200,269,134 | LINC00862 | C | A | 0.025 | 0.004 | 0.012 | 0.014 | 0.016 | 0.019 | 0.007 | 0.020 | 0.023 | 0.017 | 0.001 | 0.021 | 0.014 | 0.022 | −0.016 | 0.021 |
| rs7545345 | 1 | 205,690,941 | NUCKS1 | T | C | −0.026 | 0.004 | 0.000 | 0.013 | 0.007 | 0.019 | −0.007 | 0.019 | 0.002 | 0.016 | −0.017 | 0.021 | 0.016 | 0.021 | −0.013 | 0.021 |
| rs2724373 | 1 | 207,999,200 | C1orf132 | C | T | 0.019 | 0.003 | −0.014 | 0.009 | −0.003 | 0.013 | −0.030 | 0.013 | −0.024 | 0.011 | −0.024 | 0.014 | −0.022 | 0.015 | −0.011 | 0.014 |
| rs10779509 | 1 | 209,728,370 | AL023754.1 | T | C | −0.014 | 0.003 | −0.012 | 0.009 | −0.024 | 0.013 | 0.000 | 0.013 | −0.016 | 0.011 | −0.013 | 0.014 | −0.021 | 0.014 | −0.017 | 0.014 |
| rs340837 | 1 | 214,162,734 | PROX1 | T | G | −0.021 | 0.003 | −0.025 | 0.009 | −0.034 | 0.012 | −0.011 | 0.013 | −0.032 | 0.011 | −0.029 | 0.014 | −0.029 | 0.014 | −0.019 | 0.014 |
| rs12141189 | 1 | 221,053,545 | HLX | C | T | −0.045 | 0.003 | 0.023 | 0.010 | 0.015 | 0.015 | 0.033 | 0.015 | 0.031 | 0.013 | 0.033 | 0.016 | 0.027 | 0.017 | 0.000 | 0.016 |
| rs4306136 | 1 | 221,608,720 | AL360013.2 | A | G | 0.017 | 0.003 | 0.014 | 0.009 | 0.001 | 0.013 | 0.029 | 0.013 | 0.010 | 0.011 | 0.026 | 0.014 | 0.001 | 0.014 | 0.022 | 0.014 |
| rs708108 | 1 | 228,189,855 | WNT3A | C | T | −0.015 | 0.003 | −0.007 | 0.009 | −0.009 | 0.013 | −0.009 | 0.013 | −0.007 | 0.011 | −0.013 | 0.014 | −0.001 | 0.014 | −0.011 | 0.014 |
| rs684818 | 1 | 234,854,779 | AL160408.6 | T | C | 0.024 | 0.003 | 0.030 | 0.009 | 0.028 | 0.012 | 0.029 | 0.013 | 0.021 | 0.011 | 0.019 | 0.014 | 0.027 | 0.014 | 0.038 | 0.014 |
| rs7517340 | 1 | 243,710,190 | AKT3 | C | T | 0.035 | 0.003 | −0.001 | 0.011 | 0.018 | 0.016 | −0.020 | 0.016 | −0.009 | 0.014 | −0.011 | 0.018 | 0.003 | 0.018 | 0.013 | 0.018 |
| rs35135518 | 2 | 16,120,506 | RN7SL104P | T | C | 0.029 | 0.004 | −0.006 | 0.015 | −0.006 | 0.020 | −0.006 | 0.021 | 0.004 | 0.018 | −0.002 | 0.024 | 0.001 | 0.024 | −0.004 | 0.024 |
| rs12710648 | 2 | 17,989,500 | SMC6 | A | G | 0.017 | 0.003 | −0.003 | 0.009 | −0.003 | 0.013 | 0.000 | 0.013 | −0.002 | 0.011 | 0.015 | 0.014 | −0.015 | 0.014 | 0.004 | 0.014 |
| rs6760135 | 2 | 26,088,769 | ASXL2 | C | T | −0.050 | 0.003 | −0.002 | 0.011 | 0.001 | 0.016 | −0.005 | 0.016 | −0.010 | 0.013 | −0.006 | 0.017 | −0.015 | 0.017 | 0.004 | 0.017 |
| rs1260326 | 2 | 27,730,940 | GCKR | C | T | 0.063 | 0.003 | 0.036 | 0.009 | 0.043 | 0.013 | 0.029 | 0.013 | 0.035 | 0.011 | 0.019 | 0.014 | 0.052 | 0.014 | 0.034 | 0.014 |
| rs11677980 | 2 | 30,522,137 | LBH | A | G | −0.015 | 0.003 | −0.014 | 0.010 | 0.002 | 0.014 | −0.031 | 0.014 | −0.011 | 0.012 | −0.006 | 0.015 | −0.014 | 0.015 | −0.017 | 0.015 |
| rs7574340 | 2 | 40,621,239 | SLC8A1 | C | T | −0.017 | 0.003 | 0.012 | 0.009 | 0.025 | 0.013 | 0.000 | 0.014 | 0.019 | 0.012 | 0.021 | 0.015 | 0.016 | 0.015 | 0.005 | 0.015 |
| rs6544549 | 2 | 42,693,056 | KCNG3 | T | C | 0.024 | 0.004 | 0.026 | 0.013 | 0.022 | 0.018 | 0.027 | 0.018 | 0.025 | 0.016 | 0.041 | 0.020 | 0.007 | 0.020 | 0.039 | 0.020 |
| rs62136965 | 2 | 44,347,953 | snRNA | T | C | −0.037 | 0.006 | 0.022 | 0.022 | 0.001 | 0.030 | 0.051 | 0.032 | 0.031 | 0.026 | 0.025 | 0.034 | 0.035 | 0.034 | −0.012 | 0.033 |
| rs3791679 | 2 | 56,096,892 | EFEMP1 | A | G | −0.018 | 0.003 | 0.020 | 0.010 | 0.031 | 0.014 | 0.011 | 0.015 | 0.021 | 0.012 | 0.013 | 0.016 | 0.032 | 0.016 | 0.023 | 0.016 |
| rs12471768 | 2 | 64,928,603 | SERTAD2 | C | T | 0.022 | 0.003 | −0.012 | 0.010 | −0.004 | 0.014 | −0.020 | 0.014 | −0.021 | 0.012 | −0.038 | 0.015 | 0.000 | 0.015 | 0.000 | 0.015 |
| rs702878 | 2 | 65,702,609 | AC007389.1 | A | G | 0.014 | 0.003 | 0.011 | 0.009 | 0.020 | 0.012 | 0.003 | 0.013 | 0.014 | 0.011 | 0.009 | 0.014 | 0.009 | 0.014 | 0.015 | 0.014 |
| rs35641591 | 2 | 70,323,994 | PCBP1−AS1 | C | T | 0.050 | 0.006 | 0.070 | 0.027 | 0.031 | 0.037 | 0.103 | 0.038 | 0.040 | 0.032 | 0.032 | 0.041 | 0.039 | 0.043 | 0.071 | 0.042 |
| rs6749680 | 2 | 73,685,852 | ALMS1 | A | G | 0.015 | 0.003 | −0.001 | 0.009 | −0.001 | 0.013 | −0.003 | 0.013 | −0.004 | 0.011 | 0.009 | 0.014 | −0.016 | 0.014 | −0.004 | 0.014 |
| rs73954943 | 2 | 111,890,432 | BCL2L11 | G | A | −0.031 | 0.005 | −0.032 | 0.018 | −0.069 | 0.025 | −0.003 | 0.025 | −0.036 | 0.022 | −0.044 | 0.028 | −0.047 | 0.029 | −0.041 | 0.029 |
| rs7578633 | 2 | 113,978,650 | PAX8 | C | T | −0.018 | 0.003 | −0.017 | 0.009 | −0.013 | 0.013 | −0.021 | 0.013 | −0.014 | 0.011 | −0.035 | 0.014 | −0.003 | 0.014 | −0.021 | 0.014 |
| rs17050272 | 2 | 121,306,440 | AC073257.2 | G | A | 0.024 | 0.003 | 0.010 | 0.009 | 0.012 | 0.013 | 0.007 | 0.013 | 0.003 | 0.011 | −0.010 | 0.014 | 0.018 | 0.014 | 0.011 | 0.014 |
| rs58387407 | 2 | 152,924,773 | CACNB4 | A | G | −0.018 | 0.003 | −0.002 | 0.011 | 0.013 | 0.015 | −0.017 | 0.016 | −0.018 | 0.013 | −0.022 | 0.017 | −0.023 | 0.017 | 0.017 | 0.017 |
| rs2674492 | 2 | 172,422,338 | CYBRD1 | G | A | −0.014 | 0.003 | 0.027 | 0.009 | 0.029 | 0.013 | 0.022 | 0.013 | 0.028 | 0.011 | 0.033 | 0.014 | 0.029 | 0.015 | 0.028 | 0.014 |
| rs17400325 | 2 | 178,565,913 | PDE11A | C | T | 0.054 | 0.006 | 0.004 | 0.023 | −0.004 | 0.031 | 0.011 | 0.033 | 0.016 | 0.027 | 0.003 | 0.035 | 0.052 | 0.035 | 0.011 | 0.034 |
| rs6435156 | 2 | 203,425,475 | BMPR2 | C | T | 0.024 | 0.003 | 0.015 | 0.010 | 0.013 | 0.014 | 0.017 | 0.015 | 0.013 | 0.012 | 0.015 | 0.016 | 0.020 | 0.016 | 0.025 | 0.016 |
| rs1427676 | 2 | 204,741,166 | CTLA4 | T | C | 0.015 | 0.003 | 0.025 | 0.009 | 0.026 | 0.013 | 0.022 | 0.013 | 0.025 | 0.011 | 0.008 | 0.015 | 0.035 | 0.015 | 0.030 | 0.015 |
| rs13418037 | 2 | 218,314,141 | DIRC3 | C | T | −0.020 | 0.003 | −0.006 | 0.011 | −0.017 | 0.016 | 0.003 | 0.016 | −0.002 | 0.014 | 0.002 | 0.018 | −0.010 | 0.018 | −0.036 | 0.018 |
| rs62182127 | 2 | 219,279,588 | VIL1 | A | G | 0.019 | 0.003 | 0.017 | 0.009 | 0.005 | 0.013 | 0.029 | 0.013 | 0.019 | 0.011 | 0.012 | 0.014 | 0.024 | 0.014 | 0.027 | 0.014 |
| rs11678946 | 2 | 222,302,730 | EPHA4 | C | A | −0.014 | 0.003 | 0.003 | 0.009 | 0.003 | 0.012 | 0.003 | 0.013 | −0.007 | 0.011 | 0.005 | 0.014 | −0.024 | 0.014 | 0.012 | 0.014 |
| rs4402747 | 2 | 225,457,173 | CUL3 | G | A | −0.016 | 0.003 | 0.003 | 0.009 | 0.002 | 0.012 | 0.002 | 0.013 | −0.004 | 0.011 | −0.006 | 0.014 | 0.007 | 0.014 | 0.013 | 0.014 |
| rs17323117 | 2 | 230,162,971 | PID1 | A | G | −0.029 | 0.005 | −0.014 | 0.017 | −0.022 | 0.024 | 0.000 | 0.025 | −0.029 | 0.020 | −0.064 | 0.026 | 0.001 | 0.027 | 0.035 | 0.027 |
| rs1465529 | 2 | 231,039,037 | SP110 | T | C | 0.019 | 0.003 | 0.009 | 0.010 | −0.003 | 0.014 | 0.018 | 0.014 | 0.017 | 0.012 | 0.017 | 0.015 | 0.020 | 0.015 | 0.007 | 0.015 |
| rs6437249 | 2 | 242,175,331 | HDLBP | C | T | 0.019 | 0.003 | −0.018 | 0.010 | −0.014 | 0.014 | −0.021 | 0.014 | −0.010 | 0.012 | −0.008 | 0.015 | −0.016 | 0.015 | −0.030 | 0.015 |
| rs7625680 | 3 | 11,378,069 | ATG7 | A | G | 0.015 | 0.003 | 0.005 | 0.009 | 0.004 | 0.013 | 0.007 | 0.013 | 0.007 | 0.011 | 0.012 | 0.014 | −0.004 | 0.014 | −0.002 | 0.014 |
| rs1822825 | 3 | 12,449,963 | PPARG | A | G | −0.014 | 0.003 | 0.005 | 0.009 | 0.014 | 0.012 | −0.005 | 0.013 | 0.013 | 0.011 | 0.001 | 0.014 | 0.026 | 0.014 | 0.002 | 0.014 |
| rs2607748 | 3 | 14,158,725 | CHCHD4 | T | C | −0.017 | 0.003 | −0.007 | 0.009 | 0.002 | 0.012 | −0.017 | 0.013 | −0.005 | 0.011 | 0.000 | 0.014 | −0.005 | 0.014 | −0.020 | 0.014 |
| rs11717397 | 3 | 23,368,583 | UBE2E2 | G | A | 0.015 | 0.003 | −0.001 | 0.009 | −0.027 | 0.012 | 0.027 | 0.013 | −0.008 | 0.011 | −0.017 | 0.014 | −0.002 | 0.014 | 0.018 | 0.014 |
| rs2362755 | 3 | 24,716,668 | THRB−AS1 | G | T | −0.016 | 0.003 | 0.006 | 0.009 | −0.001 | 0.012 | 0.012 | 0.013 | 0.004 | 0.011 | −0.016 | 0.014 | 0.026 | 0.014 | −0.009 | 0.014 |
| rs11928797 | 3 | 33,457,493 | UBP1 | A | C | 0.030 | 0.004 | −0.003 | 0.015 | 0.003 | 0.021 | −0.005 | 0.022 | −0.017 | 0.018 | −0.023 | 0.023 | 0.004 | 0.024 | 0.008 | 0.023 |
| rs33969824 | 3 | 42,679,777 | NKTR | G | T | 0.020 | 0.004 | 0.002 | 0.013 | 0.014 | 0.018 | −0.014 | 0.019 | 0.030 | 0.016 | 0.036 | 0.021 | 0.020 | 0.021 | −0.011 | 0.021 |
| rs12491473 | 3 | 46,989,904 | CCDC12 | G | A | 0.020 | 0.003 | 0.019 | 0.009 | 0.025 | 0.012 | 0.009 | 0.013 | 0.019 | 0.011 | 0.024 | 0.014 | 0.011 | 0.014 | 0.021 | 0.014 |
| rs2228561 | 3 | 48,628,014 | COL7A1 | A | G | 0.020 | 0.004 | 0.005 | 0.014 | 0.007 | 0.019 | 0.004 | 0.020 | 0.007 | 0.017 | −0.008 | 0.022 | 0.017 | 0.022 | −0.003 | 0.022 |
| rs4768 | 3 | 49,758,764 | RNF123 | A | G | −0.015 | 0.003 | 0.008 | 0.010 | 0.005 | 0.013 | 0.007 | 0.014 | 0.010 | 0.012 | 0.021 | 0.015 | 0.000 | 0.015 | −0.006 | 0.015 |
| rs9809209 | 3 | 51,281,664 | DOCK3 | G | A | −0.017 | 0.003 | 0.007 | 0.009 | 0.009 | 0.013 | 0.005 | 0.013 | 0.014 | 0.011 | 0.007 | 0.014 | 0.030 | 0.015 | −0.026 | 0.014 |
| rs112893170 | 3 | 57,211,863 | IL17RD | T | C | 0.020 | 0.003 | 0.012 | 0.012 | 0.013 | 0.017 | 0.012 | 0.017 | 0.019 | 0.014 | −0.003 | 0.019 | 0.043 | 0.019 | 0.008 | 0.019 |
| rs7628689 | 3 | 88,216,647 | C3orf38 | G | A | 0.029 | 0.003 | 0.014 | 0.013 | 0.016 | 0.017 | 0.010 | 0.018 | 0.009 | 0.015 | 0.000 | 0.020 | 0.026 | 0.020 | 0.023 | 0.020 |
| rs3772102 | 3 | 98,502,628 | ST3GAL6 | T | G | −0.020 | 0.003 | 0.006 | 0.009 | 0.007 | 0.013 | 0.004 | 0.013 | 0.008 | 0.011 | 0.004 | 0.014 | 0.023 | 0.014 | 0.018 | 0.014 |
| rs62280667 | 3 | 101,084,604 | SENP7 | T | C | −0.028 | 0.003 | −0.006 | 0.009 | −0.005 | 0.013 | −0.008 | 0.013 | −0.006 | 0.011 | 0.001 | 0.015 | 0.000 | 0.015 | 0.011 | 0.015 |
| rs62263345 | 3 | 107,252,190 | BBX | A | G | 0.028 | 0.004 | −0.002 | 0.014 | −0.012 | 0.019 | 0.006 | 0.019 | 0.008 | 0.016 | 0.007 | 0.021 | 0.013 | 0.021 | −0.011 | 0.021 |
| rs13069961 | 3 | 124,358,715 | KALRN | A | G | −0.018 | 0.003 | 0.001 | 0.011 | 0.004 | 0.015 | −0.002 | 0.015 | −0.003 | 0.013 | 0.002 | 0.017 | −0.008 | 0.017 | 0.002 | 0.017 |
| rs687339 | 3 | 135,932,359 | AC092991.1 | T | C | 0.040 | 0.003 | 0.012 | 0.010 | −0.003 | 0.015 | 0.027 | 0.015 | 0.014 | 0.013 | −0.001 | 0.016 | 0.011 | 0.017 | 0.004 | 0.016 |
| rs811332 | 3 | 138,078,348 | MRAS | C | T | 0.019 | 0.003 | −0.021 | 0.011 | −0.009 | 0.016 | −0.035 | 0.016 | −0.035 | 0.013 | −0.039 | 0.017 | −0.026 | 0.018 | 0.015 | 0.018 |
| rs55717031 | 3 | 138,848,505 | MRPS22 | G | T | 0.032 | 0.003 | −0.001 | 0.010 | 0.006 | 0.014 | −0.003 | 0.014 | 0.012 | 0.012 | 0.019 | 0.015 | 0.005 | 0.015 | −0.007 | 0.015 |
| rs6440008 | 3 | 141,154,542 | ZBTB38 | T | C | 0.035 | 0.003 | −0.007 | 0.009 | −0.015 | 0.013 | −0.002 | 0.013 | −0.017 | 0.011 | −0.026 | 0.014 | −0.004 | 0.015 | 0.000 | 0.014 |
| rs73238159 | 3 | 142,078,759 | XRN1 | T | C | −0.025 | 0.004 | −0.010 | 0.013 | −0.019 | 0.018 | −0.003 | 0.019 | −0.003 | 0.016 | 0.010 | 0.021 | −0.003 | 0.021 | −0.002 | 0.021 |
| rs13073970 | 3 | 170,630,520 | EIF5A2 | G | T | −0.025 | 0.003 | 0.009 | 0.011 | 0.001 | 0.015 | 0.017 | 0.015 | 0.008 | 0.013 | 0.000 | 0.017 | 0.013 | 0.017 | 0.004 | 0.017 |
| rs56062334 | 3 | 172,299,226 | LINC02068 | T | C | 0.017 | 0.003 | −0.007 | 0.009 | −0.017 | 0.013 | 0.004 | 0.013 | −0.006 | 0.011 | −0.007 | 0.015 | −0.004 | 0.015 | −0.021 | 0.015 |
| rs9819762 | 3 | 178,914,879 | PIK3CA | T | C | 0.019 | 0.003 | −0.030 | 0.011 | −0.023 | 0.016 | −0.038 | 0.016 | −0.040 | 0.013 | −0.053 | 0.017 | −0.011 | 0.018 | −0.031 | 0.017 |
| rs66707192 | 3 | 186,382,065 | HRG | G | A | 0.018 | 0.003 | 0.012 | 0.010 | 0.025 | 0.014 | 0.000 | 0.014 | 0.007 | 0.012 | 0.003 | 0.016 | 0.005 | 0.016 | 0.001 | 0.016 |
| rs13108218 | 4 | 3,443,931 | HGFAC | G | A | 0.017 | 0.003 | 0.002 | 0.009 | −0.005 | 0.013 | 0.008 | 0.013 | −0.009 | 0.011 | −0.002 | 0.014 | −0.002 | 0.015 | 0.011 | 0.014 |
| rs1055582 | 4 | 39,700,173 | UBE2K | C | T | 0.027 | 0.003 | −0.021 | 0.009 | −0.016 | 0.012 | −0.027 | 0.013 | −0.027 | 0.011 | −0.027 | 0.014 | −0.020 | 0.014 | −0.010 | 0.014 |
| rs62302688 | 4 | 46,448,465 | GABRA2 | G | A | 0.039 | 0.004 | −0.006 | 0.017 | 0.022 | 0.023 | −0.033 | 0.024 | 0.017 | 0.020 | −0.007 | 0.026 | 0.025 | 0.026 | −0.035 | 0.025 |
| rs115805235 | 4 | 69,764,890 | AC021146.3 | C | T | 0.039 | 0.006 | −0.032 | 0.023 | −0.080 | 0.032 | 0.022 | 0.033 | −0.025 | 0.027 | −0.032 | 0.035 | −0.008 | 0.036 | −0.037 | 0.035 |
| rs16845929 | 4 | 72,017,058 | SLC4A4 | C | T | 0.032 | 0.005 | 0.003 | 0.019 | 0.018 | 0.027 | −0.016 | 0.028 | 0.004 | 0.023 | 0.027 | 0.030 | −0.019 | 0.030 | 0.012 | 0.030 |
| rs2280099 | 4 | 90,035,549 | TIGD2 | G | A | 0.025 | 0.003 | −0.006 | 0.011 | −0.011 | 0.016 | −0.004 | 0.016 | 0.012 | 0.014 | 0.037 | 0.018 | −0.003 | 0.018 | −0.017 | 0.018 |
| rs35036084 | 4 | 97,552,791 | LINC02267 | T | C | 0.017 | 0.003 | 0.007 | 0.009 | 0.009 | 0.013 | 0.006 | 0.013 | 0.003 | 0.011 | 0.002 | 0.014 | −0.004 | 0.014 | 0.017 | 0.014 |
| rs6532798 | 4 | 100,054,827 | ADH4 | T | C | 0.037 | 0.003 | 0.003 | 0.010 | −0.009 | 0.013 | 0.018 | 0.014 | 0.009 | 0.011 | 0.005 | 0.015 | 0.005 | 0.015 | 0.006 | 0.015 |
| rs62342064 | 4 | 104,665,972 | TACR3 | C | T | 0.022 | 0.004 | −0.011 | 0.014 | −0.002 | 0.020 | −0.021 | 0.021 | 0.002 | 0.017 | 0.017 | 0.022 | −0.017 | 0.023 | −0.009 | 0.022 |
| rs17429745 | 4 | 106,038,169 | AC096577.1 | G | T | 0.026 | 0.003 | 0.022 | 0.010 | 0.043 | 0.013 | −0.002 | 0.014 | 0.019 | 0.011 | 0.005 | 0.015 | 0.032 | 0.015 | 0.027 | 0.015 |
| rs3804173 | 4 | 121,719,923 | PRDM5 | A | G | −0.020 | 0.003 | −0.004 | 0.009 | 0.007 | 0.013 | −0.020 | 0.013 | −0.005 | 0.011 | −0.023 | 0.015 | 0.007 | 0.015 | −0.019 | 0.015 |
| rs111443396 | 4 | 124,773,202 | LINC01091 | T | C | −0.026 | 0.004 | 0.014 | 0.014 | 0.020 | 0.019 | 0.008 | 0.020 | −0.006 | 0.016 | −0.011 | 0.021 | 0.004 | 0.021 | 0.048 | 0.021 |
| rs7667562 | 4 | 129,133,826 | LARP1B | C | A | 0.016 | 0.003 | 0.017 | 0.010 | 0.007 | 0.014 | 0.032 | 0.014 | 0.029 | 0.012 | 0.036 | 0.016 | 0.020 | 0.016 | 0.006 | 0.016 |
| rs6827641 | 4 | 145,653,694 | GYPA/HHIP | C | T | −0.014 | 0.003 | −0.020 | 0.009 | −0.032 | 0.012 | −0.008 | 0.013 | −0.023 | 0.011 | −0.007 | 0.014 | −0.037 | 0.014 | −0.017 | 0.014 |
| rs6853741 | 4 | 148,982,559 | ARHGAP10 | A | G | 0.024 | 0.003 | −0.004 | 0.010 | −0.014 | 0.014 | 0.008 | 0.015 | −0.019 | 0.012 | −0.023 | 0.016 | −0.016 | 0.016 | 0.013 | 0.016 |
| rs62334147 | 4 | 169,345,005 | DDX60L | T | C | −0.019 | 0.003 | −0.006 | 0.012 | 0.003 | 0.016 | −0.012 | 0.017 | 0.003 | 0.014 | 0.012 | 0.018 | −0.009 | 0.018 | −0.004 | 0.018 |
| rs4394044 | 4 | 186,607,420 | SORBS2 | T | C | 0.014 | 0.003 | −0.008 | 0.009 | 0.007 | 0.012 | −0.024 | 0.013 | −0.007 | 0.011 | −0.010 | 0.014 | −0.007 | 0.014 | −0.007 | 0.014 |
| rs9292578 | 5 | 35,230,075 | PRLR | C | A | 0.040 | 0.006 | 0.024 | 0.022 | 0.049 | 0.031 | −0.002 | 0.032 | 0.022 | 0.027 | −0.016 | 0.035 | 0.050 | 0.036 | 0.019 | 0.035 |
| rs6895953 | 5 | 39,084,471 | RICTOR | G | A | 0.024 | 0.003 | −0.026 | 0.009 | −0.012 | 0.012 | −0.041 | 0.013 | −0.029 | 0.011 | −0.042 | 0.014 | −0.017 | 0.014 | −0.009 | 0.014 |
| rs72758321 | 5 | 41,464,841 | PLCXD3 | A | G | −0.047 | 0.006 | 0.003 | 0.025 | 0.002 | 0.035 | −0.006 | 0.035 | −0.037 | 0.029 | −0.059 | 0.038 | −0.004 | 0.039 | 0.037 | 0.039 |
| rs6180 | 5 | 42,719,239 | GHR | C | A | −0.035 | 0.003 | −0.005 | 0.009 | −0.001 | 0.012 | −0.007 | 0.013 | −0.005 | 0.011 | -0.019 | 0.014 | 0.010 | 0.014 | 0.003 | 0.014 |
| rs12520263 | 5 | 44,122,508 | RNU6−381P | G | T | −0.017 | 0.003 | 0.010 | 0.010 | 0.004 | 0.014 | 0.015 | 0.014 | 0.016 | 0.012 | 0.035 | 0.016 | −0.001 | 0.016 | 0.018 | 0.016 |
| rs7719168 | 5 | 53,292,390 | ARL15 | A | C | −0.030 | 0.004 | −0.024 | 0.014 | −0.038 | 0.020 | −0.008 | 0.021 | −0.025 | 0.017 | −0.023 | 0.022 | −0.027 | 0.022 | −0.023 | 0.022 |
| rs28650790 | 5 | 55,861,464 | C5orf67 | C | T | −0.018 | 0.003 | −0.008 | 0.011 | 0.016 | 0.016 | −0.032 | 0.016 | −0.019 | 0.014 | −0.031 | 0.017 | −0.009 | 0.018 | 0.014 | 0.018 |
| rs1498603 | 5 | 58,333,125 | PDE4D | T | G | 0.031 | 0.005 | 0.032 | 0.019 | 0.055 | 0.026 | 0.009 | 0.026 | 0.035 | 0.023 | 0.056 | 0.030 | 0.009 | 0.030 | 0.010 | 0.029 |
| rs11954036 | 5 | 59,028,853 | PDE4D | T | C | 0.037 | 0.003 | 0.006 | 0.009 | −0.001 | 0.013 | 0.011 | 0.013 | 0.001 | 0.011 | −0.004 | 0.015 | 0.011 | 0.015 | −0.001 | 0.014 |
| rs80170948 | 5 | 64,020,316 | SREK1IP1 | T | G | −0.039 | 0.006 | 0.007 | 0.025 | 0.034 | 0.034 | −0.023 | 0.035 | 0.018 | 0.029 | 0.002 | 0.038 | 0.009 | 0.039 | 0.011 | 0.038 |
| rs2227819 | 5 | 76,012,745 | F2R | C | T | −0.022 | 0.004 | 0.022 | 0.015 | 0.008 | 0.021 | 0.036 | 0.021 | 0.020 | 0.018 | 0.037 | 0.023 | 0.004 | 0.023 | 0.039 | 0.023 |
| rs12108803 | 5 | 77,158,507 | TBCA | T | G | −0.033 | 0.006 | −0.008 | 0.022 | −0.021 | 0.031 | 0.008 | 0.031 | −0.006 | 0.026 | −0.020 | 0.034 | 0.009 | 0.035 | −0.011 | 0.034 |
| rs840809 | 5 | 87,173,927 | TMEM161B | A | C | 0.016 | 0.003 | −0.022 | 0.010 | −0.003 | 0.014 | −0.044 | 0.014 | −0.034 | 0.012 | −0.029 | 0.015 | −0.040 | 0.015 | −0.002 | 0.015 |
| rs13178887 | 5 | 88,355,993 | MEF2C−AS1 | T | C | 0.023 | 0.003 | 0.013 | 0.009 | 0.002 | 0.013 | 0.023 | 0.013 | 0.016 | 0.011 | 0.015 | 0.014 | 0.015 | 0.014 | −0.002 | 0.014 |
| rs2366398 | 5 | 89,437,963 | LINC01339 | G | T | −0.018 | 0.003 | 0.012 | 0.011 | 0.010 | 0.015 | 0.015 | 0.015 | 0.009 | 0.013 | −0.006 | 0.016 | 0.032 | 0.017 | 0.013 | 0.016 |
| rs26822 | 5 | 102,518,795 | PPIP5K2 | A | G | −0.017 | 0.003 | −0.009 | 0.010 | −0.007 | 0.013 | −0.012 | 0.014 | −0.014 | 0.012 | −0.024 | 0.015 | −0.002 | 0.015 | −0.017 | 0.015 |
| rs73271090 | 5 | 132,313,550 | AC010240.1 | G | A | 0.044 | 0.003 | 0.010 | 0.012 | 0.019 | 0.017 | 0.002 | 0.018 | 0.003 | 0.015 | −0.008 | 0.019 | 0.003 | 0.020 | −0.005 | 0.019 |
| rs329122 | 5 | 133,864,599 | JADE2 | G | A | −0.018 | 0.003 | 0.003 | 0.009 | 0.003 | 0.012 | 0.002 | 0.013 | 0.006 | 0.011 | 0.018 | 0.014 | −0.003 | 0.014 | 0.018 | 0.014 |
| rs11242236 | 5 | 134,586,980 | C5orf66 | G | A | 0.025 | 0.003 | 0.013 | 0.009 | 0.021 | 0.012 | 0.006 | 0.013 | 0.018 | 0.011 | 0.025 | 0.014 | 0.018 | 0.014 | 0.005 | 0.014 |
| rs2348604 | 5 | 136,809,831 | SPOCK1 | T | C | −0.016 | 0.003 | −0.004 | 0.010 | −0.008 | 0.014 | −0.001 | 0.014 | −0.011 | 0.012 | −0.005 | 0.015 | −0.020 | 0.016 | 0.004 | 0.015 |
| rs3734166 | 5 | 137,665,323 | CDC25C | A | G | 0.028 | 0.003 | 0.004 | 0.010 | 0.014 | 0.014 | −0.005 | 0.014 | 0.008 | 0.012 | 0.005 | 0.015 | 0.011 | 0.016 | −0.008 | 0.015 |
| rs2042253 | 5 | 143,059,433 | MIR5197 | T | C | 0.023 | 0.003 | −0.023 | 0.010 | −0.045 | 0.014 | −0.001 | 0.014 | −0.017 | 0.012 | −0.027 | 0.016 | −0.004 | 0.016 | −0.021 | 0.016 |
| rs35668185 | 5 | 168,256,455 | SLIT3 | T | C | 0.056 | 0.003 | −0.002 | 0.011 | −0.001 | 0.015 | −0.001 | 0.015 | 0.003 | 0.013 | 0.012 | 0.017 | −0.009 | 0.017 | −0.008 | 0.017 |
| rs13168379 | 5 | 173,382,761 | CPEB4 | G | A | −0.031 | 0.005 | 0.003 | 0.017 | −0.003 | 0.024 | 0.011 | 0.024 | 0.022 | 0.021 | 0.024 | 0.028 | 0.018 | 0.028 | −0.006 | 0.027 |
| rs17714046 | 5 | 180,661,980 | TRIM41 | C | T | 0.042 | 0.006 | 0.026 | 0.024 | −0.002 | 0.033 | 0.051 | 0.034 | 0.032 | 0.028 | 0.022 | 0.037 | 0.045 | 0.037 | −0.009 | 0.037 |
| rs584955 | 6 | 7,097,141 | RREB1 | A | G | 0.036 | 0.006 | 0.037 | 0.021 | 0.018 | 0.030 | 0.053 | 0.031 | 0.035 | 0.026 | 0.034 | 0.034 | 0.039 | 0.034 | 0.022 | 0.034 |
| rs2296198 | 6 | 18,399,750 | RNF144B | C | T | 0.016 | 0.003 | −0.001 | 0.010 | −0.004 | 0.014 | 0.002 | 0.015 | −0.021 | 0.012 | −0.024 | 0.016 | −0.017 | 0.016 | −0.005 | 0.016 |
| rs72828596 | 6 | 19,183,591 | AL589647.1 | G | A | −0.019 | 0.004 | −0.012 | 0.014 | −0.019 | 0.019 | −0.006 | 0.019 | −0.040 | 0.016 | −0.038 | 0.021 | −0.059 | 0.021 | 0.028 | 0.021 |
| rs73382439 | 6 | 20,404,420 | E2F3 | C | T | 0.019 | 0.003 | 0.005 | 0.011 | 0.000 | 0.016 | 0.009 | 0.016 | 0.010 | 0.014 | 0.015 | 0.018 | 0.000 | 0.018 | −0.003 | 0.018 |
| rs17258904 | 6 | 21,928,131 | CASC15 | A | G | −0.017 | 0.003 | −0.005 | 0.010 | −0.011 | 0.014 | 0.004 | 0.014 | 0.002 | 0.012 | −0.007 | 0.015 | 0.011 | 0.016 | −0.008 | 0.015 |
| rs1165196 | 6 | 25,813,150 | SLC17A1 | A | G | −0.029 | 0.003 | −0.007 | 0.009 | −0.022 | 0.012 | 0.010 | 0.013 | 0.002 | 0.011 | 0.010 | 0.014 | 0.005 | 0.014 | −0.014 | 0.014 |
| rs13195402 | 6 | 26,463,575 | BTN2A1 | T | G | −0.038 | 0.004 | 0.042 | 0.019 | 0.026 | 0.026 | 0.058 | 0.027 | 0.037 | 0.022 | 0.066 | 0.028 | 0.014 | 0.029 | 0.058 | 0.028 |
| rs16897515 | 6 | 27,278,020 | POM121L2 | A | C | −0.023 | 0.003 | 0.036 | 0.012 | 0.022 | 0.017 | 0.051 | 0.017 | 0.038 | 0.014 | 0.051 | 0.018 | 0.036 | 0.019 | 0.058 | 0.018 |
| rs33932084 | 6 | 28,268,824 | PGBD1 | G | A | −0.034 | 0.004 | 0.054 | 0.016 | 0.041 | 0.022 | 0.066 | 0.023 | 0.051 | 0.019 | 0.096 | 0.024 | 0.020 | 0.025 | 0.087 | 0.024 |
| rs9267488 | 6 | 31,514,247 | ATP6V1G2−DDX39B/ATP6V1G2 | G | A | −0.031 | 0.004 | 0.051 | 0.014 | 0.059 | 0.020 | 0.044 | 0.020 | 0.048 | 0.017 | 0.057 | 0.022 | 0.037 | 0.022 | 0.092 | 0.022 |
| rs1150752 | 6 | 32,064,726 | TNXB | C | T | −0.031 | 0.004 | 0.058 | 0.015 | 0.066 | 0.021 | 0.047 | 0.021 | 0.054 | 0.018 | 0.066 | 0.023 | 0.041 | 0.023 | 0.096 | 0.022 |
| rs12194618 | 6 | 38,091,030 | ZFAND3 | G | A | −0.017 | 0.003 | 0.007 | 0.009 | 0.011 | 0.013 | 0.002 | 0.013 | 0.009 | 0.011 | −0.003 | 0.014 | 0.016 | 0.015 | −0.002 | 0.014 |
| rs7740433 | 6 | 42,908,013 | CNPY3 | A | G | 0.017 | 0.003 | 0.003 | 0.011 | −0.001 | 0.015 | 0.004 | 0.015 | 0.018 | 0.013 | 0.012 | 0.016 | 0.018 | 0.017 | −0.021 | 0.016 |
| rs998584 | 6 | 43,757,896 | VEGFA | A | C | 0.020 | 0.003 | 0.005 | 0.009 | −0.020 | 0.012 | 0.033 | 0.013 | 0.009 | 0.011 | 0.012 | 0.014 | 0.008 | 0.014 | 0.007 | 0.014 |
| rs6924225 | 6 | 45,584,732 | RUNX2 | G | A | 0.019 | 0.003 | 0.005 | 0.012 | 0.020 | 0.017 | −0.011 | 0.017 | −0.006 | 0.014 | −0.019 | 0.019 | −0.004 | 0.019 | 0.006 | 0.019 |
| rs2397112 | 6 | 52,684,333 | GSTA6P | A | G | 0.019 | 0.003 | 0.002 | 0.009 | 0.008 | 0.013 | −0.004 | 0.013 | −0.003 | 0.011 | −0.001 | 0.014 | 0.005 | 0.014 | 0.005 | 0.014 |
| rs9361489 | 6 | 79,816,785 | PHIP | T | C | 0.022 | 0.003 | 0.008 | 0.009 | −0.004 | 0.012 | 0.021 | 0.013 | 0.018 | 0.011 | 0.036 | 0.014 | 0.009 | 0.014 | −0.003 | 0.014 |
| rs6916994 | 6 | 87,991,236 | GJB7 | C | T | 0.029 | 0.003 | −0.005 | 0.009 | −0.001 | 0.012 | −0.006 | 0.013 | −0.008 | 0.011 | −0.030 | 0.014 | 0.015 | 0.014 | 0.009 | 0.014 |
| rs670049 | 6 | 100,087,024 | PRDM13 | A | C | 0.019 | 0.003 | −0.005 | 0.009 | 0.002 | 0.013 | −0.012 | 0.013 | −0.003 | 0.011 | −0.011 | 0.014 | 0.001 | 0.015 | −0.004 | 0.014 |
| rs9322822 | 6 | 105,369,598 | LIN28B−AS1 | C | T | 0.015 | 0.003 | 0.003 | 0.009 | 0.016 | 0.013 | −0.011 | 0.014 | 0.007 | 0.011 | 0.006 | 0.015 | 0.006 | 0.015 | 0.001 | 0.015 |
| rs4946810 | 6 | 107,420,270 | BEND3 | A | C | −0.016 | 0.003 | 0.006 | 0.009 | 0.004 | 0.013 | 0.011 | 0.013 | 0.016 | 0.011 | 0.012 | 0.015 | 0.020 | 0.015 | −0.004 | 0.014 |
| rs218291 | 6 | 108,467,024 | OSTM1/OSTM1−AS1 | G | A | 0.016 | 0.003 | 0.011 | 0.009 | 0.014 | 0.013 | 0.007 | 0.013 | 0.007 | 0.011 | 0.002 | 0.014 | 0.009 | 0.015 | 0.021 | 0.014 |
| rs9398171 | 6 | 108,983,527 | FOXO3 | T | C | 0.050 | 0.003 | 0.012 | 0.010 | 0.023 | 0.013 | −0.002 | 0.014 | 0.022 | 0.012 | 0.027 | 0.015 | 0.024 | 0.015 | −0.001 | 0.015 |
| rs41285260 | 6 | 126,661,502 | CENPW | T | G | 0.039 | 0.004 | -0.007 | 0.017 | -0.012 | 0.024 | -0.005 | 0.024 | -0.012 | 0.020 | 0.004 | 0.026 | -0.039 | 0.027 | 0.008 | 0.026 |
| rs9321106 | 6 | 128,355,316 | PTPRK | A | G | 0.018 | 0.003 | 0.016 | 0.012 | 0.018 | 0.017 | 0.014 | 0.018 | 0.004 | 0.015 | 0.008 | 0.019 | 0.001 | 0.019 | 0.030 | 0.019 |
| rs9398891 | 6 | 129,314,749 | LAMA2 | C | T | −0.017 | 0.003 | −0.001 | 0.010 | −0.002 | 0.013 | 0.001 | 0.014 | 0.000 | 0.011 | −0.002 | 0.015 | 0.005 | 0.015 | 0.007 | 0.015 |
| rs3890746 | 6 | 130,371,055 | L3MBTL3 | T | C | −0.020 | 0.003 | 0.004 | 0.009 | 0.014 | 0.012 | −0.004 | 0.013 | −0.006 | 0.011 | −0.033 | 0.014 | 0.020 | 0.014 | 0.017 | 0.014 |
| rs2786185 | 6 | 147,595,554 | STXBP5 | G | A | 0.019 | 0.003 | 0.005 | 0.009 | 0.019 | 0.012 | −0.008 | 0.013 | 0.004 | 0.011 | −0.009 | 0.014 | 0.012 | 0.014 | −0.001 | 0.014 |
| rs7774230 | 6 | 152,164,239 | ESR1 | C | T | −0.026 | 0.003 | 0.011 | 0.009 | 0.007 | 0.012 | 0.016 | 0.013 | 0.008 | 0.011 | 0.013 | 0.014 | 0.004 | 0.014 | 0.010 | 0.014 |
| rs790513 | 6 | 154,420,368 | OPRM1 | C | A | 0.025 | 0.003 | 0.006 | 0.010 | −0.015 | 0.014 | 0.027 | 0.015 | −0.001 | 0.012 | −0.027 | 0.016 | 0.012 | 0.016 | 0.011 | 0.016 |
| rs7758644 | 6 | 156,583,467 | snoRNA | C | A | −0.019 | 0.003 | 0.008 | 0.012 | 0.014 | 0.017 | 0.000 | 0.018 | 0.004 | 0.015 | 0.004 | 0.019 | 0.008 | 0.019 | 0.019 | 0.019 |
| rs3127579 | 6 | 160,674,632 | SLC22A2 | G | A | −0.033 | 0.004 | 0.012 | 0.013 | 0.028 | 0.019 | −0.008 | 0.019 | −0.004 | 0.016 | −0.016 | 0.021 | 0.005 | 0.021 | 0.028 | 0.021 |
| rs12211977 | 6 | 161,252,770 | lincRNA | G | A | −0.023 | 0.004 | 0.010 | 0.016 | 0.040 | 0.022 | −0.022 | 0.022 | 0.024 | 0.018 | 0.061 | 0.024 | −0.004 | 0.024 | −0.007 | 0.024 |
| rs504371 | 6 | 165,724,052 | C6orf118 | C | A | −0.015 | 0.003 | −0.015 | 0.009 | −0.006 | 0.013 | −0.023 | 0.013 | −0.018 | 0.011 | −0.027 | 0.014 | −0.006 | 0.015 | −0.015 | 0.014 |
| rs4709995 | 6 | 166,313,447 | PDE10A | C | T | −0.042 | 0.003 | −0.022 | 0.009 | −0.023 | 0.013 | −0.020 | 0.013 | −0.026 | 0.011 | −0.020 | 0.014 | −0.041 | 0.014 | −0.049 | 0.014 |
| rs7802508 | 7 | 1,191,689 | ZFAND2A | G | A | −0.021 | 0.003 | −0.006 | 0.009 | −0.012 | 0.013 | 0.001 | 0.013 | −0.007 | 0.011 | −0.012 | 0.014 | −0.001 | 0.014 | −0.023 | 0.014 |
| rs12699547 | 7 | 2,015,970 | MAD1L1 | C | T | 0.021 | 0.003 | 0.005 | 0.009 | −0.009 | 0.013 | 0.022 | 0.013 | 0.008 | 0.011 | 0.004 | 0.014 | 0.010 | 0.015 | 0.016 | 0.014 |
| rs1182174 | 7 | 2,875,420 | GNA12 | G | A | −0.021 | 0.003 | 0.028 | 0.010 | 0.029 | 0.014 | 0.026 | 0.014 | 0.037 | 0.012 | 0.036 | 0.015 | 0.031 | 0.015 | 0.003 | 0.015 |
| rs2250243 | 7 | 6,690,240 | ZNF316 | T | C | −0.024 | 0.003 | −0.011 | 0.010 | −0.009 | 0.014 | −0.015 | 0.015 | −0.010 | 0.012 | −0.008 | 0.016 | −0.014 | 0.016 | −0.016 | 0.016 |
| rs4719393 | 7 | 14,219,213 | DGKB | T | G | 0.027 | 0.003 | −0.008 | 0.010 | −0.010 | 0.013 | −0.005 | 0.014 | −0.008 | 0.012 | −0.013 | 0.015 | −0.004 | 0.015 | −0.009 | 0.015 |
| rs10252510 | 7 | 31,023,108 | GHRHR | G | A | 0.020 | 0.003 | 0.001 | 0.010 | −0.009 | 0.014 | 0.012 | 0.015 | −0.001 | 0.012 | −0.018 | 0.016 | 0.015 | 0.016 | 0.007 | 0.016 |
| rs7790246 | 7 | 32,976,416 | AVL9/RP9P | C | T | −0.017 | 0.003 | −0.007 | 0.010 | 0.000 | 0.014 | −0.013 | 0.014 | 0.001 | 0.012 | 0.004 | 0.015 | 0.005 | 0.015 | 0.006 | 0.015 |
| rs1050327 | 7 | 44,808,017 | ZMIZ2 | G | A | −0.017 | 0.003 | −0.010 | 0.009 | −0.008 | 0.012 | −0.013 | 0.013 | −0.005 | 0.011 | −0.003 | 0.014 | −0.007 | 0.014 | −0.010 | 0.014 |
| rs870796 | 7 | 45,426,435 | ELK1P1 | G | A | 0.017 | 0.003 | −0.007 | 0.009 | −0.006 | 0.013 | −0.010 | 0.013 | −0.004 | 0.011 | 0.002 | 0.014 | −0.014 | 0.014 | −0.014 | 0.014 |
| rs2270628 | 7 | 45,949,570 | IGFBP3 | C | T | −0.033 | 0.003 | 0.023 | 0.011 | 0.030 | 0.015 | 0.019 | 0.016 | 0.024 | 0.013 | 0.017 | 0.017 | 0.043 | 0.018 | 0.048 | 0.017 |
| rs74657816 | 7 | 46,670,682 | HMGN1P19 | T | G | 0.047 | 0.005 | 0.012 | 0.019 | 0.002 | 0.026 | 0.021 | 0.027 | −0.004 | 0.022 | 0.004 | 0.029 | −0.022 | 0.029 | −0.006 | 0.029 |
| rs12538762 | 7 | 47,264,328 | TNS3 | C | T | 0.026 | 0.004 | 0.001 | 0.014 | 0.002 | 0.020 | −0.003 | 0.020 | −0.004 | 0.017 | −0.002 | 0.022 | −0.006 | 0.023 | −0.012 | 0.022 |
| rs6974707 | 7 | 55,982,894 | ZNF713 | G | A | −0.019 | 0.003 | 0.009 | 0.011 | 0.024 | 0.015 | −0.008 | 0.015 | 0.004 | 0.013 | −0.008 | 0.016 | 0.023 | 0.017 | 0.012 | 0.017 |
| rs35862187 | 7 | 69,625,029 | AUTS2 | A | G | 0.031 | 0.006 | 0.002 | 0.020 | −0.030 | 0.028 | 0.038 | 0.029 | −0.015 | 0.024 | −0.036 | 0.031 | −0.003 | 0.031 | −0.012 | 0.031 |
| rs17145738 | 7 | 72,982,874 | TBL2 | C | T | −0.034 | 0.004 | −0.016 | 0.014 | −0.018 | 0.019 | −0.016 | 0.019 | −0.021 | 0.016 | −0.005 | 0.021 | −0.030 | 0.022 | −0.034 | 0.021 |
| rs411717 | 7 | 94,033,031 | COL1A2 | C | T | −0.015 | 0.003 | 0.009 | 0.009 | 0.017 | 0.012 | −0.001 | 0.013 | 0.001 | 0.011 | −0.007 | 0.014 | 0.010 | 0.014 | 0.022 | 0.014 |
| rs34670419 | 7 | 99,130,834 | ZKSCAN5 | G | T | −0.036 | 0.006 | 0.001 | 0.024 | −0.020 | 0.033 | 0.022 | 0.034 | −0.024 | 0.028 | −0.028 | 0.036 | −0.020 | 0.037 | 0.045 | 0.037 |
| rs34312198 | 7 | 99,674,870 | ZNF3 | C | A | −0.024 | 0.004 | −0.020 | 0.014 | −0.021 | 0.020 | −0.020 | 0.021 | −0.026 | 0.017 | −0.020 | 0.022 | −0.021 | 0.023 | −0.010 | 0.022 |
| rs7783012 | 7 | 114,116,881 | FOXP2 | A | G | −0.016 | 0.003 | 0.010 | 0.009 | 0.017 | 0.012 | 0.004 | 0.013 | 0.011 | 0.011 | 0.018 | 0.014 | 0.005 | 0.014 | 0.015 | 0.014 |
| rs12666306 | 7 | 115,082,406 | AC073901.1 | G | A | 0.017 | 0.003 | −0.001 | 0.009 | 0.017 | 0.012 | −0.020 | 0.013 | −0.007 | 0.011 | −0.003 | 0.014 | −0.010 | 0.014 | 0.017 | 0.014 |
| rs2896395 | 7 | 127,511,705 | SND1 | C | T | 0.015 | 0.003 | −0.010 | 0.010 | −0.016 | 0.014 | −0.003 | 0.014 | −0.018 | 0.012 | −0.015 | 0.015 | −0.021 | 0.015 | −0.007 | 0.015 |
| rs11556924 | 7 | 129,663,496 | ZC3HC1 | T | C | −0.016 | 0.003 | 0.037 | 0.009 | 0.032 | 0.013 | 0.043 | 0.013 | 0.043 | 0.011 | 0.030 | 0.015 | 0.047 | 0.015 | 0.033 | 0.015 |
| rs207212 | 7 | 130,547,217 | LINC00513 | C | T | 0.028 | 0.004 | 0.058 | 0.016 | 0.075 | 0.022 | 0.045 | 0.022 | 0.068 | 0.019 | 0.057 | 0.024 | 0.063 | 0.025 | 0.049 | 0.024 |
| rs1986692 | 7 | 133,743,393 | EXOC4 | A | G | −0.015 | 0.003 | 0.004 | 0.009 | −0.003 | 0.013 | 0.011 | 0.013 | 0.008 | 0.011 | 0.012 | 0.014 | 0.003 | 0.014 | −0.004 | 0.014 |
| rs273956 | 7 | 137,603,188 | CREB3L2 | G | A | −0.021 | 0.003 | 0.011 | 0.009 | 0.007 | 0.013 | 0.016 | 0.013 | 0.012 | 0.011 | 0.016 | 0.014 | 0.023 | 0.015 | 0.021 | 0.014 |
| rs114949263 | 7 | 150,498,245 | TMEM176B/TMEM176A | T | C | 0.027 | 0.004 | −0.013 | 0.014 | −0.004 | 0.020 | −0.020 | 0.021 | −0.027 | 0.017 | −0.019 | 0.022 | −0.044 | 0.022 | −0.023 | 0.022 |
| rs10246481 | 7 | 156,184,748 | lincRNA | A | G | −0.015 | 0.003 | 0.004 | 0.009 | 0.004 | 0.013 | 0.006 | 0.013 | 0.006 | 0.011 | 0.018 | 0.014 | −0.020 | 0.014 | 0.002 | 0.014 |
| rs9657541 | 8 | 10,643,164 | SOX7/PINX1/PINX1 | C | T | 0.020 | 0.003 | −0.002 | 0.011 | −0.010 | 0.016 | 0.007 | 0.016 | −0.006 | 0.013 | 0.004 | 0.017 | −0.015 | 0.017 | −0.002 | 0.017 |
| rs76393968 | 8 | 16,282,937 | MSR1 | G | A | 0.060 | 0.010 | 0.035 | 0.034 | 0.018 | 0.048 | 0.060 | 0.051 | 0.029 | 0.044 | 0.036 | 0.058 | 0.025 | 0.059 | 0.015 | 0.057 |
| rs1495741 | 8 | 18,272,881 | NAT2 | A | G | −0.026 | 0.003 | 0.010 | 0.010 | 0.010 | 0.015 | 0.009 | 0.015 | 0.011 | 0.013 | 0.012 | 0.016 | 0.013 | 0.017 | 0.012 | 0.016 |
| rs11782452 | 8 | 26,361,601 | BNIP3L | G | A | 0.015 | 0.003 | −0.002 | 0.009 | 0.000 | 0.013 | −0.005 | 0.013 | −0.002 | 0.011 | 0.009 | 0.014 | −0.009 | 0.014 | −0.001 | 0.014 |
| rs56352849 | 8 | 73,769,173 | KCNB2 | G | A | −0.016 | 0.003 | −0.011 | 0.010 | −0.012 | 0.014 | −0.005 | 0.014 | −0.017 | 0.012 | −0.038 | 0.015 | −0.002 | 0.016 | −0.010 | 0.015 |
| rs1431015 | 8 | 77,131,580 | HNF4G | C | T | 0.020 | 0.003 | 0.008 | 0.009 | 0.014 | 0.012 | −0.001 | 0.013 | 0.003 | 0.011 | 0.008 | 0.014 | −0.001 | 0.014 | 0.016 | 0.014 |
| rs6473015 | 8 | 78,178,485 | lincRNA | A | C | −0.019 | 0.003 | −0.028 | 0.010 | −0.025 | 0.014 | −0.034 | 0.014 | −0.024 | 0.012 | −0.031 | 0.015 | −0.019 | 0.015 | −0.035 | 0.015 |
| rs445036 | 8 | 81,408,409 | ZBTB10 | T | C | 0.019 | 0.003 | −0.007 | 0.010 | −0.008 | 0.013 | −0.007 | 0.014 | 0.004 | 0.011 | 0.015 | 0.015 | −0.012 | 0.015 | −0.029 | 0.015 |
| rs1786342 | 8 | 101,676,363 | SNX31 | T | C | 0.017 | 0.003 | 0.022 | 0.009 | 0.030 | 0.013 | 0.011 | 0.013 | 0.023 | 0.011 | 0.014 | 0.014 | 0.032 | 0.014 | 0.016 | 0.014 |
| rs60862542 | 8 | 109,275,071 | EIF3E | G | A | 0.017 | 0.003 | −0.005 | 0.011 | −0.018 | 0.015 | 0.008 | 0.015 | −0.010 | 0.013 | −0.007 | 0.016 | −0.015 | 0.017 | −0.010 | 0.016 |
| rs2737205 | 8 | 116,610,180 | TRPS1 | C | T | −0.023 | 0.003 | −0.013 | 0.009 | −0.005 | 0.013 | −0.021 | 0.013 | −0.028 | 0.011 | −0.023 | 0.014 | −0.034 | 0.014 | 0.010 | 0.014 |
| rs2978062 | 8 | 134,571,618 | ST3GAL1 | T | G | −0.019 | 0.003 | −0.007 | 0.013 | 0.009 | 0.018 | −0.023 | 0.018 | −0.004 | 0.015 | 0.018 | 0.020 | −0.035 | 0.020 | 0.019 | 0.020 |
| rs716100 | 8 | 135,661,278 | ZFAT | G | A | −0.019 | 0.003 | −0.017 | 0.009 | −0.018 | 0.013 | −0.013 | 0.013 | 0.002 | 0.011 | −0.001 | 0.015 | 0.005 | 0.015 | −0.039 | 0.015 |
| rs12549853 | 8 | 145,020,636 | PLEC | G | A | −0.016 | 0.003 | 0.011 | 0.009 | 0.009 | 0.013 | 0.015 | 0.013 | 0.006 | 0.011 | 0.014 | 0.014 | −0.004 | 0.014 | 0.009 | 0.014 |
| rs10114121 | 9 | 19,440,136 | ACER2 | G | A | −0.020 | 0.004 | −0.021 | 0.012 | −0.025 | 0.017 | −0.018 | 0.018 | −0.019 | 0.015 | −0.013 | 0.019 | −0.023 | 0.019 | −0.019 | 0.019 |
| rs10757291 | 9 | 22,161,884 | CDKN2B−AS1 | A | G | −0.019 | 0.003 | 0.007 | 0.009 | 0.010 | 0.012 | 0.006 | 0.013 | 0.008 | 0.011 | 0.018 | 0.014 | −0.008 | 0.014 | 0.011 | 0.014 |
| rs10811787 | 9 | 22,871,816 | AL391117.1 | T | C | −0.015 | 0.003 | −0.009 | 0.009 | −0.011 | 0.012 | −0.009 | 0.013 | −0.010 | 0.011 | −0.020 | 0.014 | −0.008 | 0.014 | −0.002 | 0.014 |
| rs11557154 | 9 | 34,107,505 | DCAF12 | T | C | 0.024 | 0.004 | 0.036 | 0.013 | 0.038 | 0.018 | 0.029 | 0.018 | 0.045 | 0.016 | 0.051 | 0.020 | 0.041 | 0.021 | 0.050 | 0.020 |
| rs10869022 | 9 | 74,057,313 | TRPM3 | C | T | 0.021 | 0.003 | 0.005 | 0.011 | 0.027 | 0.015 | −0.016 | 0.015 | 0.020 | 0.013 | 0.019 | 0.017 | 0.024 | 0.017 | −0.020 | 0.017 |
| rs2378662 | 9 | 86,707,289 | AL390838.1 | A | G | −0.017 | 0.003 | 0.004 | 0.009 | −0.007 | 0.012 | 0.019 | 0.013 | 0.008 | 0.011 | 0.020 | 0.014 | −0.003 | 0.014 | 0.018 | 0.014 |
| rs10908903 | 9 | 92,228,559 | GADD45G | T | G | 0.015 | 0.003 | 0.015 | 0.009 | 0.012 | 0.012 | 0.019 | 0.013 | 0.013 | 0.011 | 0.019 | 0.014 | 0.007 | 0.014 | 0.014 | 0.014 |
| rs1055710 | 9 | 96,214,928 | FAM120AOS | A | G | −0.018 | 0.003 | −0.013 | 0.009 | −0.014 | 0.013 | −0.011 | 0.013 | −0.011 | 0.011 | −0.039 | 0.015 | 0.017 | 0.015 | 0.000 | 0.015 |
| rs75660441 | 9 | 97,662,448 | C9orf3 | A | G | 0.039 | 0.005 | −0.029 | 0.018 | −0.024 | 0.026 | −0.030 | 0.026 | −0.032 | 0.022 | −0.049 | 0.028 | −0.016 | 0.029 | −0.038 | 0.028 |
| rs28831479 | 9 | 98,254,526 | PTCH1 | C | A | 0.022 | 0.003 | 0.001 | 0.010 | 0.007 | 0.014 | −0.002 | 0.014 | 0.002 | 0.012 | −0.016 | 0.016 | 0.015 | 0.016 | 0.001 | 0.016 |
| rs7034716 | 9 | 101,858,382 | TGFBR1 | C | T | 0.015 | 0.003 | 0.013 | 0.010 | −0.030 | 0.014 | 0.006 | 0.014 | −0.008 | 0.012 | 0.004 | 0.016 | −0.024 | 0.016 | −0.016 | 0.016 |
| rs6479003 | 9 | 102,948,685 | INVS | G | A | 0.024 | 0.004 | 0.022 | 0.016 | 0.035 | 0.022 | 0.006 | 0.023 | 0.029 | 0.019 | 0.025 | 0.025 | 0.015 | 0.025 | 0.021 | 0.024 |
| rs41277821 | 9 | 109,689,972 | ZNF462 | T | C | 0.062 | 0.009 | 0.005 | 0.038 | 0.013 | 0.054 | −0.005 | 0.055 | −0.018 | 0.046 | −0.007 | 0.060 | −0.021 | 0.062 | 0.021 | 0.058 |
| rs7872812 | 9 | 119,341,544 | ASTN2 | C | T | −0.026 | 0.004 | −0.007 | 0.013 | −0.023 | 0.017 | 0.004 | 0.018 | −0.003 | 0.015 | 0.019 | 0.019 | −0.027 | 0.019 | −0.017 | 0.019 |
| rs13301073 | 9 | 128,284,378 | MAPKAP1 | G | A | 0.022 | 0.003 | 0.005 | 0.009 | 0.000 | 0.013 | 0.012 | 0.013 | 0.008 | 0.011 | 0.017 | 0.014 | −0.005 | 0.015 | −0.005 | 0.014 |
| rs1832007 | 10 | 5,254,847 | AKR1C4 | A | G | −0.057 | 0.003 | −0.034 | 0.012 | −0.045 | 0.017 | −0.023 | 0.018 | −0.043 | 0.015 | −0.043 | 0.019 | −0.055 | 0.020 | −0.009 | 0.019 |
| rs2801482 | 10 | 12,459,773 | CAMK1D | A | G | −0.050 | 0.008 | −0.026 | 0.026 | −0.016 | 0.036 | −0.037 | 0.037 | 0.003 | 0.032 | −0.010 | 0.041 | 0.033 | 0.041 | −0.021 | 0.040 |
| rs7921105 | 10 | 13,535,398 | BEND7 | T | C | −0.016 | 0.003 | −0.001 | 0.009 | −0.003 | 0.012 | 0.002 | 0.013 | 0.008 | 0.011 | −0.001 | 0.014 | 0.010 | 0.014 | −0.003 | 0.014 |
| rs11012712 | 10 | 21,760,015 | − | C | T | 0.022 | 0.003 | −0.026 | 0.011 | −0.021 | 0.016 | −0.032 | 0.016 | −0.027 | 0.014 | −0.039 | 0.017 | −0.012 | 0.018 | −0.035 | 0.017 |
| rs10047326 | 10 | 22,839,463 | PIP4K2A | A | C | 0.017 | 0.003 | −0.011 | 0.009 | 0.013 | 0.013 | −0.034 | 0.013 | −0.010 | 0.011 | −0.019 | 0.014 | −0.005 | 0.014 | −0.026 | 0.014 |
| rs293275 | 10 | 53,215,020 | PRKG1 | T | C | −0.014 | 0.003 | 0.003 | 0.009 | 0.009 | 0.012 | −0.005 | 0.013 | 0.013 | 0.011 | 0.023 | 0.014 | 0.005 | 0.014 | −0.008 | 0.014 |
| rs10821713 | 10 | 62,055,781 | ANK3 | C | T | −0.017 | 0.003 | −0.004 | 0.009 | −0.004 | 0.012 | −0.004 | 0.013 | 0.006 | 0.011 | −0.007 | 0.014 | 0.018 | 0.014 | −0.009 | 0.014 |
| rs7910087 | 10 | 77,209,145 | LRMDA | C | T | −0.017 | 0.003 | −0.009 | 0.009 | −0.020 | 0.012 | 0.001 | 0.013 | −0.002 | 0.011 | −0.009 | 0.014 | 0.006 | 0.014 | −0.020 | 0.014 |
| rs4418728 | 10 | 94,839,724 | CYP26A1 | G | T | 0.024 | 0.003 | −0.027 | 0.009 | −0.029 | 0.012 | −0.025 | 0.013 | −0.029 | 0.011 | −0.030 | 0.014 | −0.028 | 0.014 | −0.022 | 0.014 |
| rs116454156 | 10 | 95,347,041 | FFAR4 | A | G | 0.078 | 0.010 | −0.013 | 0.033 | −0.007 | 0.047 | −0.024 | 0.049 | −0.006 | 0.040 | −0.003 | 0.051 | −0.015 | 0.053 | −0.006 | 0.051 |
| rs11187838 | 10 | 96,038,686 | PLCE1 | G | A | 0.024 | 0.003 | −0.006 | 0.009 | −0.002 | 0.012 | −0.010 | 0.013 | 0.000 | 0.011 | 0.005 | 0.014 | −0.004 | 0.014 | −0.006 | 0.014 |
| rs10509746 | 10 | 102,656,897 | PAX2 | C | T | 0.027 | 0.003 | 0.006 | 0.009 | 0.031 | 0.013 | −0.021 | 0.013 | 0.000 | 0.011 | −0.002 | 0.014 | 0.001 | 0.014 | 0.011 | 0.014 |
| rs4917962 | 10 | 103,931,931 | NOLC1 | G | T | −0.024 | 0.004 | −0.024 | 0.013 | −0.008 | 0.018 | −0.042 | 0.019 | −0.017 | 0.016 | 0.004 | 0.021 | −0.032 | 0.021 | −0.021 | 0.021 |
| rs12244851 | 10 | 114,773,926 | TCF7L2 | C | T | −0.015 | 0.003 | −0.010 | 0.010 | −0.004 | 0.013 | −0.014 | 0.014 | −0.009 | 0.011 | 0.008 | 0.015 | −0.021 | 0.015 | −0.005 | 0.015 |
| rs3858325 | 10 | 117,988,795 | GFRA1 | C | T | −0.019 | 0.003 | 0.006 | 0.009 | 0.000 | 0.012 | 0.011 | 0.013 | 0.004 | 0.011 | 0.015 | 0.014 | −0.007 | 0.014 | 0.011 | 0.014 |
| rs61867536 | 11 | 1,513,700 | MOB2 | C | T | −0.018 | 0.003 | −0.030 | 0.009 | −0.019 | 0.012 | −0.044 | 0.013 | −0.023 | 0.011 | −0.016 | 0.014 | −0.025 | 0.014 | −0.054 | 0.014 |
| rs3213223 | 11 | 2,156,930 | IGF2/INS−IGF−2 | G | A | −0.076 | 0.003 | 0.007 | 0.011 | 0.011 | 0.015 | 0.004 | 0.016 | 0.021 | 0.013 | 0.028 | 0.017 | 0.012 | 0.017 | −0.023 | 0.017 |
| rs11029620 | 11 | 3,771,924 | NUP98 | C | T | 0.022 | 0.003 | −0.004 | 0.011 | −0.009 | 0.015 | 0.003 | 0.015 | 0.004 | 0.013 | −0.012 | 0.017 | 0.010 | 0.017 | −0.011 | 0.017 |
| rs67257872 | 11 | 8,530,218 | STK33 | A | G | 0.014 | 0.003 | −0.003 | 0.009 | −0.003 | 0.012 | 0.000 | 0.013 | 0.000 | 0.011 | 0.000 | 0.014 | 0.010 | 0.014 | 0.000 | 0.014 |
| rs7947951 | 11 | 13,356,030 | ARNTL | G | A | 0.020 | 0.003 | −0.007 | 0.009 | −0.008 | 0.013 | −0.006 | 0.013 | −0.016 | 0.011 | −0.016 | 0.015 | −0.015 | 0.015 | −0.001 | 0.015 |
| rs72858776 | 11 | 15,772,953 | AC087379.1 | G | T | 0.030 | 0.005 | −0.010 | 0.016 | −0.006 | 0.022 | −0.010 | 0.023 | −0.008 | 0.019 | −0.014 | 0.025 | −0.002 | 0.026 | −0.010 | 0.025 |
| rs11024614 | 11 | 18,326,758 | HPS5 | T | C | −0.023 | 0.003 | −0.008 | 0.009 | −0.013 | 0.013 | −0.001 | 0.013 | −0.004 | 0.011 | −0.007 | 0.014 | −0.001 | 0.014 | −0.021 | 0.014 |
| rs34452566 | 11 | 27,793,470 | AC103796.1 | G | T | −0.018 | 0.003 | −0.010 | 0.012 | 0.004 | 0.016 | −0.026 | 0.017 | −0.004 | 0.014 | −0.002 | 0.018 | −0.010 | 0.018 | −0.021 | 0.018 |
| rs11031058 | 11 | 30,375,889 | ARL14EP | C | T | −0.022 | 0.003 | 0.015 | 0.012 | 0.019 | 0.017 | 0.008 | 0.017 | 0.014 | 0.014 | 0.013 | 0.018 | 0.018 | 0.019 | 0.007 | 0.018 |
| rs117529631 | 11 | 46,159,633 | AC024475.4 | C | T | −0.041 | 0.006 | −0.035 | 0.023 | −0.058 | 0.031 | −0.014 | 0.032 | −0.024 | 0.027 | −0.015 | 0.035 | −0.037 | 0.036 | −0.013 | 0.035 |
| rs5896 | 11 | 46,745,003 | F2 | T | C | 0.036 | 0.004 | −0.023 | 0.013 | −0.022 | 0.018 | −0.021 | 0.019 | −0.023 | 0.016 | −0.016 | 0.021 | −0.028 | 0.021 | −0.026 | 0.021 |
| rs1051006 | 11 | 47,306,585 | MADD | A | G | 0.040 | 0.003 | −0.009 | 0.012 | −0.013 | 0.016 | −0.004 | 0.017 | −0.012 | 0.014 | −0.011 | 0.018 | −0.019 | 0.019 | −0.017 | 0.018 |
| rs1039481 | 11 | 48,182,237 | PTPRJ | A | G | −0.042 | 0.003 | −0.013 | 0.010 | 0.000 | 0.014 | −0.027 | 0.015 | −0.010 | 0.012 | −0.005 | 0.016 | −0.013 | 0.016 | −0.011 | 0.016 |
| rs202676 | 11 | 49,227,620 | FOLH1 | G | A | 0.021 | 0.003 | 0.002 | 0.010 | −0.005 | 0.015 | 0.008 | 0.015 | 0.003 | 0.013 | 0.008 | 0.016 | 0.001 | 0.017 | −0.017 | 0.017 |
| rs10769621 | 11 | 49,860,463 | TRIM51FP | T | C | 0.020 | 0.003 | 0.012 | 0.010 | 0.002 | 0.013 | 0.025 | 0.014 | 0.011 | 0.012 | 0.017 | 0.015 | 0.004 | 0.015 | 0.009 | 0.015 |
| rs11230983 | 11 | 55,541,284 | OR5D13 | A | G | 0.035 | 0.004 | 0.009 | 0.014 | −0.002 | 0.019 | 0.023 | 0.019 | 0.015 | 0.016 | 0.010 | 0.021 | 0.013 | 0.022 | 0.001 | 0.021 |
| rs78460947 | 11 | 56,143,715 | OR8U1 | G | A | 0.042 | 0.006 | −0.006 | 0.027 | 0.023 | 0.037 | −0.035 | 0.038 | −0.013 | 0.032 | 0.019 | 0.041 | −0.020 | 0.043 | −0.029 | 0.041 |
| rs146345029 | 11 | 59,596,007 | GIF | G | A | −0.034 | 0.006 | 0.052 | 0.022 | 0.051 | 0.032 | 0.053 | 0.032 | 0.036 | 0.027 | 0.003 | 0.034 | 0.067 | 0.036 | 0.051 | 0.035 |
| rs174554 | 11 | 61,579,463 | FADS1/FADS2 | A | G | 0.022 | 0.003 | 0.061 | 0.009 | 0.062 | 0.013 | 0.062 | 0.013 | 0.060 | 0.011 | 0.045 | 0.015 | 0.081 | 0.015 | 0.051 | 0.015 |
| rs117104648 | 11 | 65,543,736 | AP5B1 | T | C | −0.036 | 0.005 | 0.001 | 0.019 | 0.008 | 0.027 | 0.000 | 0.027 | −0.010 | 0.023 | −0.012 | 0.029 | −0.016 | 0.030 | 0.018 | 0.029 |
| rs12790261 | 11 | 66,988,048 | KDM2A | C | A | −0.031 | 0.005 | −0.013 | 0.019 | −0.003 | 0.027 | −0.016 | 0.028 | −0.015 | 0.023 | 0.006 | 0.030 | −0.024 | 0.031 | −0.030 | 0.030 |
| rs4980661 | 11 | 69,306,579 | CCND1 | A | G | 0.014 | 0.003 | −0.012 | 0.009 | −0.001 | 0.012 | −0.024 | 0.013 | −0.014 | 0.011 | −0.010 | 0.014 | −0.020 | 0.014 | −0.002 | 0.014 |
| rs2512525 | 11 | 77,923,019 | USP35 | T | C | 0.024 | 0.003 | −0.012 | 0.012 | −0.018 | 0.016 | −0.005 | 0.017 | −0.007 | 0.014 | −0.017 | 0.018 | −0.005 | 0.019 | −0.032 | 0.018 |
| rs61904289 | 11 | 85,994,731 | AP003084.1 | C | T | −0.016 | 0.003 | −0.002 | 0.009 | 0.001 | 0.013 | −0.004 | 0.014 | −0.005 | 0.011 | 0.000 | 0.015 | −0.010 | 0.015 | 0.006 | 0.015 |
| rs625245 | 11 | 94,192,103 | MRE11 | T | G | −0.016 | 0.003 | 0.009 | 0.009 | −0.005 | 0.013 | 0.025 | 0.013 | 0.016 | 0.011 | 0.022 | 0.015 | 0.005 | 0.015 | 0.006 | 0.014 |
| rs35023999 | 11 | 113,266,411 | ANKK1 | C | A | 0.015 | 0.003 | 0.006 | 0.009 | 0.003 | 0.012 | 0.009 | 0.013 | 0.011 | 0.011 | 0.026 | 0.014 | −0.006 | 0.014 | −0.007 | 0.014 |
| rs10892564 | 11 | 120,224,650 | ARHGEF12 | A | G | −0.017 | 0.003 | 0.012 | 0.009 | −0.001 | 0.013 | 0.028 | 0.013 | 0.008 | 0.011 | 0.011 | 0.014 | 0.013 | 0.014 | 0.005 | 0.014 |
| rs4936759 | 11 | 122,763,516 | C11orf63 | C | T | 0.016 | 0.003 | 0.005 | 0.009 | 0.015 | 0.013 | −0.001 | 0.013 | 0.006 | 0.011 | 0.008 | 0.014 | 0.009 | 0.014 | 0.014 | 0.014 |
| rs10893499 | 11 | 126,241,979 | ST3GAL4 | G | A | 0.022 | 0.004 | 0.025 | 0.013 | 0.001 | 0.018 | 0.050 | 0.018 | 0.022 | 0.016 | 0.024 | 0.020 | 0.018 | 0.020 | 0.046 | 0.020 |
| rs11064536 | 12 | 905,582 | WNK1 | T | C | 0.020 | 0.003 | −0.016 | 0.012 | 0.016 | 0.017 | −0.051 | 0.017 | −0.021 | 0.014 | −0.022 | 0.019 | −0.025 | 0.019 | −0.024 | 0.019 |
| rs2856321 | 12 | 11,855,773 | ETV6 | A | G | −0.026 | 0.003 | 0.006 | 0.009 | 0.022 | 0.013 | −0.009 | 0.013 | −0.003 | 0.011 | 0.001 | 0.015 | −0.006 | 0.015 | 0.018 | 0.015 |
| rs10841649 | 12 | 20,954,879 | SLCO1B3 | C | T | 0.021 | 0.004 | −0.002 | 0.013 | 0.010 | 0.019 | −0.011 | 0.019 | −0.010 | 0.016 | 0.014 | 0.021 | −0.017 | 0.021 | 0.016 | 0.021 |
| rs9738365 | 12 | 31,997,635 | − | C | A | −0.058 | 0.003 | 0.000 | 0.010 | 0.007 | 0.014 | −0.009 | 0.014 | −0.007 | 0.012 | −0.001 | 0.016 | −0.005 | 0.016 | 0.015 | 0.016 |
| rs12231073 | 12 | 38,526,901 | − | G | T | −0.017 | 0.003 | 0.011 | 0.009 | 0.022 | 0.012 | 0.001 | 0.013 | 0.010 | 0.011 | 0.013 | 0.014 | 0.012 | 0.014 | 0.008 | 0.014 |
| rs11175935 | 12 | 40,693,806 | LRRK2 | G | T | 0.020 | 0.003 | 0.000 | 0.011 | 0.019 | 0.015 | −0.020 | 0.016 | −0.006 | 0.013 | −0.016 | 0.017 | 0.006 | 0.018 | −0.002 | 0.017 |
| rs247917 | 12 | 46,265,916 | ARID2 | C | T | −0.015 | 0.003 | 0.000 | 0.009 | 0.010 | 0.012 | −0.009 | 0.013 | −0.001 | 0.011 | −0.006 | 0.014 | 0.006 | 0.014 | −0.007 | 0.014 |
| rs117564283 | 12 | 52,300,110 | ACVRL1 | C | T | −0.029 | 0.005 | 0.037 | 0.018 | 0.026 | 0.025 | 0.046 | 0.026 | 0.045 | 0.022 | 0.053 | 0.028 | 0.024 | 0.028 | −0.006 | 0.028 |
| rs773116 | 12 | 56,486,159 | ERBB3 | G | A | 0.016 | 0.003 | 0.008 | 0.009 | 0.008 | 0.013 | 0.010 | 0.013 | 0.017 | 0.011 | 0.010 | 0.014 | 0.037 | 0.014 | 0.000 | 0.014 |
| rs78607331 | 12 | 57,648,644 | R3HDM2 | T | C | −0.037 | 0.006 | 0.025 | 0.025 | 0.015 | 0.034 | 0.040 | 0.036 | −0.001 | 0.030 | −0.019 | 0.039 | 0.037 | 0.039 | 0.048 | 0.037 |
| rs4547160 | 12 | 63,503,650 | AVPR1A | G | T | −0.018 | 0.003 | −0.018 | 0.009 | −0.014 | 0.013 | −0.020 | 0.013 | −0.015 | 0.011 | −0.008 | 0.015 | −0.031 | 0.015 | −0.013 | 0.015 |
| rs1351394 | 12 | 66,351,826 | HMGA2 | C | T | 0.024 | 0.003 | 0.018 | 0.009 | 0.018 | 0.012 | 0.015 | 0.013 | 0.012 | 0.011 | 0.018 | 0.014 | 0.001 | 0.014 | 0.023 | 0.014 |
| rs2230281 | 12 | 89,917,518 | GALNT4/POC1B−GALNT4 | G | A | −0.016 | 0.003 | −0.010 | 0.010 | 0.009 | 0.014 | −0.033 | 0.014 | −0.013 | 0.012 | −0.021 | 0.015 | −0.002 | 0.016 | 0.002 | 0.015 |
| rs10777540 | 12 | 94,150,321 | CRADD | T | G | −0.018 | 0.003 | −0.014 | 0.009 | −0.023 | 0.012 | −0.005 | 0.013 | −0.016 | 0.011 | −0.023 | 0.014 | −0.012 | 0.014 | −0.013 | 0.014 |
| rs10860237 | 12 | 98,157,010 | AC007424.1 | G | A | −0.030 | 0.003 | −0.003 | 0.009 | −0.013 | 0.013 | 0.007 | 0.014 | 0.001 | 0.012 | 0.000 | 0.015 | 0.006 | 0.015 | −0.007 | 0.015 |
| rs11111274 | 12 | 102,838,128 | IGF1 | A | G | −0.080 | 0.003 | −0.013 | 0.010 | −0.019 | 0.014 | −0.007 | 0.014 | −0.015 | 0.012 | 0.002 | 0.016 | −0.041 | 0.016 | 0.002 | 0.016 |
| rs10745954 | 12 | 103,483,094 | AC068643.1 | G | A | 0.015 | 0.003 | 0.003 | 0.009 | −0.003 | 0.012 | 0.008 | 0.013 | 0.005 | 0.011 | 0.010 | 0.014 | 0.008 | 0.014 | 0.000 | 0.014 |
| rs7314285 | 12 | 111,522,026 | CUX2 | T | G | −0.052 | 0.005 | 0.011 | 0.017 | −0.006 | 0.024 | 0.033 | 0.024 | 0.005 | 0.021 | −0.019 | 0.027 | 0.029 | 0.028 | 0.002 | 0.027 |
| rs1061657 | 12 | 115,108,136 | TBX3 | T | C | −0.022 | 0.003 | −0.032 | 0.010 | −0.018 | 0.014 | −0.047 | 0.014 | −0.029 | 0.012 | −0.023 | 0.016 | −0.029 | 0.016 | −0.044 | 0.015 |
| rs2460488 | 12 | 116,187,660 | − | G | A | 0.026 | 0.003 | 0.008 | 0.012 | 0.008 | 0.016 | 0.008 | 0.017 | 0.009 | 0.014 | 0.005 | 0.018 | 0.009 | 0.019 | 0.004 | 0.018 |
| rs1800574 | 12 | 121,416,864 | HNF1A | T | C | 0.145 | 0.007 | 0.003 | 0.027 | −0.009 | 0.038 | 0.019 | 0.039 | 0.019 | 0.032 | −0.013 | 0.042 | 0.071 | 0.041 | 0.050 | 0.041 |
| rs11057265 | 12 | 123,805,950 | SBNO1 | G | A | 0.044 | 0.007 | 0.011 | 0.027 | −0.013 | 0.037 | 0.035 | 0.038 | 0.005 | 0.032 | −0.017 | 0.041 | 0.028 | 0.043 | 0.040 | 0.042 |
| rs9532512 | 13 | 40,769,897 | LINC00598 | G | A | −0.043 | 0.003 | −0.010 | 0.011 | −0.004 | 0.016 | −0.012 | 0.016 | −0.015 | 0.013 | −0.014 | 0.017 | −0.027 | 0.018 | −0.010 | 0.017 |
| rs1170158 | 13 | 42,701,941 | DGKH | T | G | 0.021 | 0.003 | −0.021 | 0.011 | −0.036 | 0.015 | −0.007 | 0.016 | −0.034 | 0.013 | −0.032 | 0.017 | −0.038 | 0.017 | −0.013 | 0.017 |
| rs1535793 | 13 | 47,154,966 | LRCH1 | A | G | 0.024 | 0.003 | 0.033 | 0.010 | 0.024 | 0.014 | 0.042 | 0.015 | 0.036 | 0.012 | 0.031 | 0.016 | 0.045 | 0.016 | 0.028 | 0.016 |
| rs118081390 | 13 | 49,671,053 | FNDC3A | G | A | 0.028 | 0.005 | −0.018 | 0.018 | −0.049 | 0.025 | 0.014 | 0.025 | −0.020 | 0.021 | −0.012 | 0.027 | −0.041 | 0.027 | −0.019 | 0.027 |
| rs9573360 | 13 | 74,771,429 | KLF12 | A | C | 0.014 | 0.003 | −0.010 | 0.009 | 0.003 | 0.012 | −0.022 | 0.013 | −0.011 | 0.011 | −0.019 | 0.014 | −0.004 | 0.014 | −0.017 | 0.014 |
| rs71432868 | 13 | 106,559,402 | SNORA25 | T | C | −0.028 | 0.005 | 0.016 | 0.022 | 0.023 | 0.030 | 0.009 | 0.031 | 0.032 | 0.026 | 0.038 | 0.034 | 0.020 | 0.034 | 0.011 | 0.034 |
| rs9583151 | 13 | 107,666,257 | − | C | T | 0.014 | 0.003 | 0.001 | 0.009 | 0.000 | 0.012 | 0.001 | 0.013 | 0.001 | 0.011 | 0.011 | 0.014 | −0.012 | 0.014 | −0.001 | 0.014 |
| rs7323205 | 13 | 110,365,525 | LINC00676 | C | T | 0.015 | 0.003 | −0.004 | 0.009 | −0.014 | 0.013 | 0.004 | 0.013 | −0.008 | 0.011 | 0.005 | 0.014 | −0.022 | 0.015 | 0.002 | 0.014 |
| rs6602909 | 13 | 114,551,993 | GAS6 | T | C | −0.020 | 0.003 | −0.009 | 0.010 | −0.001 | 0.014 | −0.016 | 0.014 | −0.009 | 0.012 | −0.007 | 0.016 | −0.004 | 0.016 | 0.006 | 0.015 |
| rs8017377 | 14 | 24,883,887 | NYNRIN | A | G | −0.017 | 0.003 | −0.010 | 0.009 | −0.012 | 0.012 | −0.007 | 0.013 | −0.007 | 0.011 | −0.011 | 0.014 | −0.007 | 0.014 | −0.028 | 0.014 |
| rs28396553 | 14 | 36,673,392 | lincRNA | T | C | 0.015 | 0.003 | 0.021 | 0.009 | 0.018 | 0.013 | 0.025 | 0.013 | 0.015 | 0.011 | 0.036 | 0.014 | −0.004 | 0.014 | 0.010 | 0.014 |
| rs33912345 | 14 | 60,976,537 | SIX6 | A | C | −0.023 | 0.003 | 0.008 | 0.009 | 0.023 | 0.013 | −0.009 | 0.013 | 0.007 | 0.011 | 0.013 | 0.014 | 0.001 | 0.014 | 0.028 | 0.014 |
| rs79936318 | 14 | 64,315,556 | SYNE2 | G | A | −0.017 | 0.003 | −0.007 | 0.012 | −0.017 | 0.017 | 0.009 | 0.018 | −0.010 | 0.015 | −0.045 | 0.019 | 0.027 | 0.020 | 0.001 | 0.019 |
| rs168961 | 14 | 69,282,930 | ZFP36L1 | A | G | −0.018 | 0.003 | −0.004 | 0.009 | −0.013 | 0.012 | 0.006 | 0.013 | −0.004 | 0.011 | −0.004 | 0.014 | 0.001 | 0.014 | −0.016 | 0.014 |
| rs75088740 | 14 | 69,819,101 | GALNT16 | G | A | −0.019 | 0.003 | −0.021 | 0.012 | −0.018 | 0.017 | −0.020 | 0.017 | −0.025 | 0.014 | −0.030 | 0.018 | −0.019 | 0.019 | −0.010 | 0.019 |
| rs13379043 | 14 | 74,250,126 | ELMSAN1 | T | C | 0.025 | 0.003 | 0.016 | 0.010 | 0.008 | 0.014 | 0.024 | 0.014 | 0.011 | 0.012 | 0.024 | 0.016 | 0.002 | 0.016 | 0.002 | 0.016 |
| rs175043 | 14 | 75,471,803 | EIF2B2 | G | A | 0.018 | 0.003 | −0.021 | 0.009 | −0.026 | 0.012 | −0.017 | 0.013 | −0.023 | 0.011 | −0.037 | 0.014 | −0.005 | 0.014 | −0.024 | 0.014 |
| rs1061638 | 14 | 77,928,525 | AHSA1 | G | A | 0.018 | 0.003 | 0.008 | 0.010 | 0.013 | 0.013 | 0.007 | 0.014 | 0.008 | 0.011 | 0.013 | 0.015 | −0.001 | 0.015 | 0.005 | 0.015 |
| rs10145154 | 14 | 79,939,525 | NRXN3 | C | T | −0.018 | 0.003 | −0.006 | 0.011 | −0.002 | 0.016 | −0.012 | 0.016 | 0.000 | 0.013 | 0.011 | 0.017 | −0.009 | 0.017 | −0.009 | 0.017 |
| rs78598185 | 14 | 92,791,479 | SLC24A4 | A | G | −0.029 | 0.004 | 0.003 | 0.015 | 0.004 | 0.021 | 0.004 | 0.022 | 0.036 | 0.018 | 0.028 | 0.024 | 0.041 | 0.024 | −0.023 | 0.023 |
| rs1115897 | 14 | 93,910,816 | UNC79 | A | C | 0.021 | 0.003 | 0.012 | 0.010 | 0.015 | 0.013 | 0.008 | 0.014 | 0.002 | 0.012 | 0.017 | 0.015 | −0.018 | 0.015 | 0.003 | 0.015 |
| rs28929474 | 14 | 94,844,947 | SERPINA1 | T | C | −0.063 | 0.009 | −0.097 | 0.035 | −0.139 | 0.051 | −0.033 | 0.050 | −0.117 | 0.043 | −0.116 | 0.055 | −0.087 | 0.057 | −0.069 | 0.055 |
| rs10136874 | 14 | 101,202,022 | DLK1 | G | T | 0.023 | 0.003 | −0.006 | 0.009 | −0.017 | 0.013 | 0.007 | 0.013 | 0.002 | 0.011 | 0.011 | 0.014 | −0.008 | 0.014 | −0.021 | 0.014 |
| rs17747633 | 15 | 40,916,237 | KNL1 | G | A | 0.015 | 0.003 | 0.026 | 0.009 | 0.029 | 0.013 | 0.022 | 0.013 | 0.026 | 0.011 | 0.037 | 0.014 | 0.019 | 0.014 | 0.028 | 0.014 |
| rs62020698 | 15 | 43,237,414 | UBR1 | C | T | 0.047 | 0.004 | 0.008 | 0.016 | 0.011 | 0.023 | −0.001 | 0.024 | 0.026 | 0.020 | 0.052 | 0.026 | −0.005 | 0.026 | −0.023 | 0.025 |
| rs55707100 | 15 | 43,820,717 | MAP1A | T | C | −0.151 | 0.008 | −0.007 | 0.026 | −0.011 | 0.036 | −0.006 | 0.037 | −0.002 | 0.030 | −0.067 | 0.040 | 0.071 | 0.039 | −0.001 | 0.040 |
| rs4273010 | 15 | 44,947,434 | SPG11 | T | C | 0.128 | 0.008 | 0.014 | 0.029 | 0.008 | 0.040 | 0.023 | 0.043 | −0.016 | 0.035 | 0.024 | 0.046 | −0.066 | 0.045 | 0.027 | 0.045 |
| rs4545755 | 15 | 51,549,044 | MIR4713HG/CYP19A1 | G | A | 0.016 | 0.003 | 0.018 | 0.009 | 0.019 | 0.012 | 0.015 | 0.013 | 0.014 | 0.011 | 0.025 | 0.014 | 0.013 | 0.014 | 0.023 | 0.014 |
| rs12442867 | 15 | 62,489,128 | AC126323.2 | C | A | −0.017 | 0.003 | 0.003 | 0.009 | −0.020 | 0.013 | 0.027 | 0.013 | 0.004 | 0.011 | 0.010 | 0.014 | −0.005 | 0.014 | 0.008 | 0.014 |
| rs79076440 | 15 | 63,803,863 | USP3 | A | G | 0.019 | 0.003 | −0.024 | 0.012 | −0.023 | 0.017 | −0.024 | 0.017 | −0.023 | 0.014 | −0.040 | 0.018 | −0.017 | 0.019 | −0.026 | 0.018 |
| rs10851736 | 15 | 64,940,718 | ZNF609 | C | T | −0.027 | 0.005 | 0.013 | 0.015 | −0.003 | 0.021 | 0.029 | 0.022 | 0.001 | 0.018 | −0.010 | 0.024 | 0.000 | 0.024 | 0.011 | 0.024 |
| rs8024330 | 15 | 67,443,926 | SMAD3 | C | T | 0.018 | 0.003 | −0.015 | 0.010 | −0.018 | 0.013 | −0.013 | 0.014 | −0.021 | 0.012 | −0.014 | 0.015 | −0.033 | 0.015 | −0.019 | 0.015 |
| rs8033075 | 15 | 68,353,652 | PIAS1 | A | G | 0.045 | 0.005 | 0.032 | 0.021 | 0.013 | 0.030 | 0.053 | 0.030 | 0.018 | 0.025 | 0.018 | 0.033 | 0.005 | 0.033 | 0.034 | 0.033 |
| rs5742915 | 15 | 74,336,633 | PML | C | T | 0.025 | 0.003 | −0.003 | 0.009 | 0.001 | 0.013 | −0.007 | 0.013 | −0.005 | 0.011 | 0.000 | 0.014 | −0.002 | 0.014 | 0.002 | 0.014 |
| rs12593755 | 15 | 89,111,712 | AC013489.2 | G | T | −0.016 | 0.003 | −0.002 | 0.009 | 0.013 | 0.013 | −0.019 | 0.013 | −0.005 | 0.011 | −0.018 | 0.014 | 0.017 | 0.014 | −0.005 | 0.014 |
| rs11856160 | 15 | 93,452,846 | CHD2 | A | G | 0.021 | 0.003 | 0.003 | 0.012 | 0.006 | 0.017 | 0.002 | 0.018 | 0.002 | 0.015 | 0.018 | 0.019 | −0.011 | 0.020 | 0.013 | 0.019 |
| rs12912439 | 15 | 95,828,705 | LINC01197 | C | T | −0.022 | 0.003 | −0.005 | 0.010 | −0.003 | 0.013 | −0.006 | 0.014 | −0.014 | 0.012 | −0.020 | 0.015 | −0.003 | 0.015 | 0.008 | 0.015 |
| rs34040697 | 15 | 97,125,666 | − | A | G | 0.016 | 0.003 | −0.009 | 0.009 | 0.005 | 0.013 | −0.022 | 0.013 | −0.014 | 0.011 | −0.012 | 0.014 | −0.026 | 0.014 | −0.009 | 0.014 |
| rs142354201 | 15 | 99,524,022 | PGPEP1L | G | A | 0.034 | 0.006 | −0.020 | 0.022 | −0.037 | 0.031 | −0.006 | 0.032 | −0.025 | 0.026 | −0.059 | 0.033 | −0.001 | 0.034 | 0.005 | 0.034 |
| rs4988483 | 16 | 1,129,010 | SSTR5 | A | C | −0.172 | 0.006 | −0.048 | 0.026 | −0.039 | 0.036 | −0.054 | 0.037 | −0.015 | 0.031 | 0.016 | 0.040 | −0.041 | 0.041 | −0.119 | 0.041 |
| rs72761177 | 16 | 1,833,508 | NUBP2 | A | G | 0.077 | 0.004 | −0.020 | 0.016 | −0.018 | 0.022 | −0.024 | 0.023 | −0.027 | 0.019 | −0.026 | 0.025 | −0.030 | 0.026 | −0.019 | 0.025 |
| rs11077337 | 16 | 3,492,048 | AC025283.2/ZNF597 | T | G | 0.015 | 0.003 | 0.006 | 0.009 | 0.014 | 0.012 | 0.000 | 0.013 | 0.007 | 0.011 | 0.014 | 0.014 | 0.002 | 0.014 | 0.012 | 0.014 |
| rs8182173 | 16 | 4,420,787 | CORO7−PAM16 | C | T | −0.018 | 0.003 | −0.009 | 0.010 | −0.017 | 0.015 | 0.001 | 0.015 | −0.016 | 0.013 | −0.046 | 0.016 | 0.009 | 0.017 | 0.000 | 0.016 |
| rs74774288 | 16 | 5,922,263 | RBFOX1 | G | T | 0.027 | 0.003 | 0.020 | 0.012 | 0.003 | 0.017 | 0.036 | 0.017 | 0.011 | 0.014 | 0.005 | 0.018 | 0.015 | 0.019 | 0.001 | 0.018 |
| rs4985062 | 16 | 8,996,636 | USP7 | T | C | 0.015 | 0.003 | 0.002 | 0.009 | 0.002 | 0.013 | 0.001 | 0.013 | 0.010 | 0.011 | 0.011 | 0.014 | 0.007 | 0.014 | −0.009 | 0.014 |
| rs1532824 | 16 | 10,532,211 | ATF7IP2 | C | A | −0.017 | 0.003 | 0.005 | 0.010 | 0.009 | 0.014 | 0.002 | 0.014 | 0.001 | 0.012 | −0.012 | 0.015 | 0.015 | 0.016 | 0.011 | 0.015 |
| rs12935465 | 16 | 17,476,853 | XYLT1 | T | C | 0.016 | 0.003 | −0.002 | 0.009 | −0.005 | 0.012 | 0.003 | 0.013 | −0.001 | 0.011 | 0.007 | 0.014 | 0.001 | 0.014 | 0.008 | 0.014 |
| rs2023762 | 16 | 19,276,597 | SYT17 | T | C | 0.015 | 0.003 | −0.001 | 0.009 | −0.020 | 0.013 | 0.017 | 0.013 | 0.000 | 0.011 | 0.002 | 0.015 | −0.002 | 0.015 | −0.005 | 0.015 |
| rs12927172 | 16 | 27,325,021 | IL4R | G | A | −0.015 | 0.003 | 0.006 | 0.009 | 0.021 | 0.013 | −0.013 | 0.013 | 0.015 | 0.011 | 0.002 | 0.014 | 0.033 | 0.015 | 0.012 | 0.014 |
| rs7498665 | 16 | 28,883,241 | SH2B1 | G | A | 0.019 | 0.003 | 0.006 | 0.009 | 0.009 | 0.013 | 0.003 | 0.013 | 0.008 | 0.011 | 0.015 | 0.014 | 0.013 | 0.014 | 0.008 | 0.014 |
| rs4788220 | 16 | 30,063,780 | FAM57B | A | G | −0.017 | 0.003 | −0.003 | 0.009 | −0.002 | 0.012 | −0.003 | 0.013 | −0.006 | 0.011 | 0.000 | 0.014 | −0.008 | 0.014 | 0.004 | 0.014 |
| rs750952 | 16 | 31,093,954 | ZNF646 | C | T | 0.032 | 0.003 | −0.014 | 0.009 | −0.012 | 0.013 | −0.015 | 0.013 | −0.015 | 0.011 | −0.034 | 0.014 | 0.000 | 0.014 | −0.008 | 0.014 |
| rs116971887 | 16 | 51,170,026 | SALL1 | G | T | 0.036 | 0.006 | −0.018 | 0.022 | 0.023 | 0.030 | −0.047 | 0.031 | −0.025 | 0.026 | −0.034 | 0.033 | −0.007 | 0.034 | −0.009 | 0.034 |
| rs12597502 | 16 | 53,170,069 | CHD9 | A | G | −0.015 | 0.003 | −0.001 | 0.010 | −0.002 | 0.014 | −0.001 | 0.014 | 0.005 | 0.012 | 0.004 | 0.015 | 0.006 | 0.015 | −0.018 | 0.015 |
| rs1548917 | 16 | 56,109,333 | CES5A | C | T | −0.015 | 0.003 | −0.010 | 0.009 | −0.010 | 0.012 | −0.010 | 0.013 | −0.009 | 0.011 | −0.021 | 0.014 | 0.003 | 0.014 | −0.002 | 0.014 |
| rs111792934 | 16 | 69,131,293 | HAS3 | C | T | 0.022 | 0.003 | 0.018 | 0.012 | 0.001 | 0.017 | 0.039 | 0.017 | 0.023 | 0.015 | 0.042 | 0.019 | 0.018 | 0.019 | 0.005 | 0.019 |
| rs17299478 | 16 | 69,775,500 | − | C | T | 0.032 | 0.003 | −0.008 | 0.012 | −0.009 | 0.017 | −0.009 | 0.018 | −0.021 | 0.015 | −0.018 | 0.019 | −0.014 | 0.020 | 0.003 | 0.019 |
| rs12935091 | 16 | 71,525,208 | ZNF19 | A | G | −0.035 | 0.006 | −0.022 | 0.023 | −0.080 | 0.032 | 0.032 | 0.032 | −0.022 | 0.027 | 0.001 | 0.036 | −0.053 | 0.036 | −0.061 | 0.035 |
| rs147491123 | 16 | 72,567,795 | LINC01572 | C | T | 0.036 | 0.007 | 0.026 | 0.024 | 0.025 | 0.034 | 0.030 | 0.035 | 0.025 | 0.029 | 0.004 | 0.037 | 0.060 | 0.039 | 0.062 | 0.038 |
| rs8059803 | 16 | 81,603,001 | CMIP | A | G | 0.031 | 0.003 | 0.005 | 0.010 | −0.010 | 0.014 | 0.019 | 0.014 | 0.001 | 0.012 | −0.005 | 0.015 | 0.003 | 0.015 | −0.008 | 0.015 |
| rs11149612 | 16 | 83,980,965 | AC009119.2 | C | T | 0.027 | 0.003 | 0.010 | 0.009 | −0.001 | 0.013 | 0.022 | 0.014 | 0.002 | 0.011 | 0.003 | 0.015 | 0.005 | 0.015 | 0.015 | 0.015 |
| rs8054322 | 16 | 85,201,405 | GSE1 | G | A | −0.015 | 0.003 | −0.009 | 0.009 | −0.009 | 0.013 | −0.009 | 0.013 | 0.000 | 0.011 | 0.004 | 0.014 | −0.002 | 0.014 | −0.014 | 0.014 |
| rs7502910 | 17 | 1,638,718 | WDR81 | A | G | 0.016 | 0.003 | 0.009 | 0.009 | 0.009 | 0.012 | 0.011 | 0.013 | 0.016 | 0.011 | 0.006 | 0.014 | 0.031 | 0.014 | 0.008 | 0.014 |
| rs2309401 | 17 | 5,471,902 | NLRP1 | T | G | 0.015 | 0.003 | 0.006 | 0.009 | 0.019 | 0.013 | −0.009 | 0.013 | −0.001 | 0.011 | −0.006 | 0.014 | 0.007 | 0.014 | 0.030 | 0.014 |
| rs9892862 | 17 | 7,439,014 | POLR2A | G | A | 0.022 | 0.003 | −0.002 | 0.010 | −0.002 | 0.015 | −0.006 | 0.015 | 0.008 | 0.013 | 0.007 | 0.017 | 0.014 | 0.017 | −0.017 | 0.016 |
| rs6416868 | 17 | 15,924,370 | TTC19 | G | A | −0.019 | 0.003 | −0.010 | 0.009 | −0.025 | 0.012 | 0.006 | 0.013 | −0.007 | 0.011 | −0.002 | 0.014 | −0.013 | 0.014 | −0.010 | 0.014 |
| rs8075153 | 17 | 17,622,666 | RAI1 | C | T | 0.021 | 0.003 | −0.020 | 0.009 | −0.021 | 0.012 | −0.017 | 0.013 | −0.022 | 0.011 | −0.034 | 0.014 | −0.006 | 0.014 | −0.004 | 0.014 |
| rs8079923 | 17 | 19,869,544 | AKAP10 | C | T | 0.016 | 0.003 | 0.004 | 0.010 | 0.000 | 0.014 | 0.010 | 0.015 | −0.007 | 0.012 | −0.028 | 0.016 | 0.004 | 0.016 | 0.021 | 0.016 |
| rs56030650 | 17 | 38,131,187 | GSDMA | A | C | −0.022 | 0.003 | 0.005 | 0.009 | −0.007 | 0.012 | 0.022 | 0.013 | 0.004 | 0.011 | −0.010 | 0.014 | 0.013 | 0.014 | 0.023 | 0.014 |
| rs668799 | 17 | 40,716,235 | COASY | C | T | 0.018 | 0.003 | −0.022 | 0.010 | −0.038 | 0.014 | −0.004 | 0.014 | −0.032 | 0.012 | −0.022 | 0.016 | −0.036 | 0.016 | −0.027 | 0.016 |
| rs199525 | 17 | 44,847,834 | WNT3 | T | G | −0.020 | 0.003 | 0.014 | 0.011 | −0.006 | 0.015 | 0.034 | 0.016 | 0.011 | 0.013 | 0.006 | 0.017 | 0.016 | 0.017 | −0.002 | 0.017 |
| rs11079157 | 17 | 53,360,799 | HLF | G | T | −0.020 | 0.003 | 0.003 | 0.011 | −0.011 | 0.015 | 0.021 | 0.016 | 0.019 | 0.013 | −0.001 | 0.017 | 0.039 | 0.018 | −0.025 | 0.017 |
| rs2250014 | 17 | 57,836,134 | VMP1 | T | C | −0.021 | 0.003 | −0.029 | 0.012 | −0.030 | 0.017 | −0.022 | 0.017 | −0.038 | 0.014 | −0.056 | 0.018 | −0.011 | 0.019 | 0.001 | 0.019 |
| rs142377191 | 17 | 61,649,170 | DCAF7 | G | A | −0.125 | 0.009 | 0.028 | 0.036 | 0.012 | 0.050 | 0.049 | 0.052 | 0.059 | 0.043 | 0.110 | 0.057 | 0.004 | 0.056 | −0.025 | 0.055 |
| rs76708468 | 17 | 62,206,299 | ERN1 | T | C | −0.087 | 0.006 | 0.009 | 0.030 | −0.013 | 0.042 | 0.028 | 0.043 | 0.023 | 0.036 | 0.002 | 0.047 | 0.013 | 0.048 | −0.033 | 0.046 |
| rs78357146 | 17 | 64,305,051 | PRKCA | A | G | −0.090 | 0.007 | 0.048 | 0.028 | 0.018 | 0.039 | 0.083 | 0.041 | 0.027 | 0.033 | −0.002 | 0.043 | 0.072 | 0.044 | 0.100 | 0.044 |
| rs77542162 | 17 | 67,081,278 | ABCA6 | G | A | 0.054 | 0.009 | 0.031 | 0.037 | 0.055 | 0.052 | −0.008 | 0.054 | 0.025 | 0.044 | 0.024 | 0.057 | 0.055 | 0.058 | 0.049 | 0.057 |
| rs6501601 | 17 | 71,124,903 | SLC39A11 | G | A | 0.015 | 0.003 | −0.009 | 0.010 | −0.025 | 0.013 | 0.006 | 0.014 | −0.020 | 0.011 | −0.029 | 0.015 | −0.007 | 0.015 | −0.007 | 0.015 |
| rs4789227 | 17 | 73,794,354 | UNK | T | C | 0.015 | 0.003 | 0.004 | 0.009 | 0.018 | 0.013 | −0.012 | 0.013 | 0.002 | 0.011 | 0.001 | 0.014 | 0.011 | 0.015 | 0.012 | 0.014 |
| rs4075483 | 17 | 79,074,817 | BAIAP2 | C | T | 0.017 | 0.003 | 0.000 | 0.009 | −0.015 | 0.013 | 0.015 | 0.013 | 0.008 | 0.011 | 0.001 | 0.014 | 0.018 | 0.015 | −0.033 | 0.014 |
| rs8095538 | 18 | 1,616,505 | − | T | G | −0.020 | 0.003 | 0.003 | 0.010 | 0.011 | 0.013 | −0.005 | 0.014 | −0.004 | 0.011 | −0.012 | 0.015 | −0.005 | 0.015 | 0.009 | 0.015 |
| rs8084351 | 18 | 50,726,559 | DCC | G | A | 0.015 | 0.003 | −0.011 | 0.009 | −0.008 | 0.012 | −0.013 | 0.013 | −0.014 | 0.011 | −0.014 | 0.014 | −0.022 | 0.014 | −0.001 | 0.014 |
| rs11152071 | 18 | 56,087,417 | AC105105.3 | C | T | 0.020 | 0.003 | 0.007 | 0.010 | −0.003 | 0.014 | 0.016 | 0.015 | 0.007 | 0.012 | −0.007 | 0.016 | 0.020 | 0.016 | 0.012 | 0.016 |
| rs190102446 | 18 | 57,048,571 | − | C | T | 0.041 | 0.007 | −0.007 | 0.023 | −0.018 | 0.032 | 0.011 | 0.034 | −0.013 | 0.028 | 0.010 | 0.036 | −0.044 | 0.036 | 0.002 | 0.036 |
| rs585187 | 18 | 58,177,124 | MRPS5P4 | T | G | 0.015 | 0.003 | −0.008 | 0.009 | −0.010 | 0.012 | −0.005 | 0.013 | −0.013 | 0.011 | −0.026 | 0.014 | 0.001 | 0.014 | −0.006 | 0.014 |
| rs12454712 | 18 | 60,845,884 | BCL2 | T | C | 0.018 | 0.003 | 0.003 | 0.009 | −0.005 | 0.013 | 0.012 | 0.013 | 0.007 | 0.011 | 0.003 | 0.015 | 0.012 | 0.015 | −0.003 | 0.015 |
| rs8097893 | 18 | 74,983,055 | GALR1 | A | G | 0.058 | 0.006 | −0.009 | 0.021 | 0.018 | 0.030 | −0.034 | 0.031 | −0.007 | 0.026 | 0.023 | 0.034 | −0.020 | 0.034 | 0.041 | 0.034 |
| rs67868323 | 19 | 4,048,561 | ZBTB7A | T | G | 0.016 | 0.003 | −0.006 | 0.010 | 0.016 | 0.014 | −0.028 | 0.014 | 0.007 | 0.012 | 0.032 | 0.016 | −0.031 | 0.016 | −0.033 | 0.015 |
| rs2602717 | 19 | 4,902,950 | UHRF1/ARRDC5 | C | T | 0.019 | 0.003 | 0.024 | 0.011 | 0.034 | 0.016 | 0.017 | 0.017 | 0.030 | 0.014 | 0.019 | 0.018 | 0.033 | 0.018 | 0.033 | 0.018 |
| rs8112883 | 19 | 7,179,320 | INSR | G | T | 0.017 | 0.003 | 0.007 | 0.010 | 0.011 | 0.014 | 0.005 | 0.014 | 0.006 | 0.012 | 0.004 | 0.015 | 0.014 | 0.015 | 0.028 | 0.015 |
| rs8105174 | 19 | 10,347,032 | DNMT1 | C | T | 0.050 | 0.003 | 0.010 | 0.012 | 0.018 | 0.016 | 0.004 | 0.017 | 0.021 | 0.014 | 0.014 | 0.018 | 0.030 | 0.018 | −0.003 | 0.018 |
| rs6510033 | 19 | 30,710,785 | AC005597.1 | A | G | 0.020 | 0.003 | 0.007 | 0.010 | 0.001 | 0.014 | 0.015 | 0.014 | 0.012 | 0.012 | 0.000 | 0.015 | 0.022 | 0.016 | 0.002 | 0.015 |
| rs6510177 | 19 | 31,211,647 | ZNF536 | C | T | −0.023 | 0.003 | 0.032 | 0.013 | 0.024 | 0.017 | 0.042 | 0.018 | 0.035 | 0.015 | 0.021 | 0.019 | 0.063 | 0.020 | 0.040 | 0.020 |
| rs62102136 | 19 | 34,700,561 | LSM14A | C | T | 0.016 | 0.003 | −0.007 | 0.010 | 0.008 | 0.014 | −0.022 | 0.014 | −0.014 | 0.012 | −0.010 | 0.015 | −0.011 | 0.015 | −0.002 | 0.015 |
| rs7254601 | 19 | 36,147,315 | COX6B1 | A | G | −0.016 | 0.003 | 0.017 | 0.010 | 0.027 | 0.014 | 0.005 | 0.015 | 0.014 | 0.012 | 0.005 | 0.016 | 0.022 | 0.016 | −0.013 | 0.016 |
| rs58560372 | 19 | 38,758,752 | SPINT2 | C | T | 0.020 | 0.003 | 0.011 | 0.013 | 0.023 | 0.018 | −0.004 | 0.018 | −0.012 | 0.015 | −0.016 | 0.020 | −0.023 | 0.020 | 0.016 | 0.020 |
| rs15052 | 19 | 41,813,375 | HNRNPUL1/TGFB1 | T | C | −0.018 | 0.003 | −0.014 | 0.013 | −0.037 | 0.018 | 0.010 | 0.018 | −0.001 | 0.015 | −0.011 | 0.020 | 0.015 | 0.020 | −0.039 | 0.020 |
| rs11671304 | 19 | 47,564,643 | ZC3H4 | T | C | −0.018 | 0.003 | 0.007 | 0.010 | −0.006 | 0.014 | 0.022 | 0.014 | 0.003 | 0.012 | 0.013 | 0.015 | −0.012 | 0.015 | 0.009 | 0.015 |
| rs296361 | 19 | 48,389,363 | SULT2A1 | G | A | −0.025 | 0.003 | 0.008 | 0.012 | 0.018 | 0.017 | −0.005 | 0.017 | 0.016 | 0.014 | −0.001 | 0.019 | 0.021 | 0.019 | 0.004 | 0.019 |
| rs2287922 | 19 | 49,232,226 | RASIP1 | A | G | −0.030 | 0.003 | 0.022 | 0.009 | 0.020 | 0.013 | 0.025 | 0.013 | 0.034 | 0.011 | 0.044 | 0.014 | 0.023 | 0.014 | −0.006 | 0.014 |
| rs7256521 | 19 | 53,837,110 | ZNF845 | A | G | −0.015 | 0.003 | −0.002 | 0.009 | −0.005 | 0.013 | 0.001 | 0.013 | −0.006 | 0.011 | 0.008 | 0.014 | −0.013 | 0.015 | −0.003 | 0.014 |
| rs12975366 | 19 | 54,759,361 | LILRB5 | C | T | −0.020 | 0.003 | −0.002 | 0.009 | 0.014 | 0.013 | −0.021 | 0.013 | −0.005 | 0.011 | −0.002 | 0.014 | −0.018 | 0.015 | 0.006 | 0.014 |
| rs6037508 | 20 | 3,217,989 | SLC4A11 | T | G | −0.017 | 0.003 | −0.020 | 0.011 | −0.019 | 0.015 | −0.022 | 0.016 | −0.029 | 0.013 | −0.045 | 0.017 | −0.020 | 0.017 | 0.001 | 0.017 |
| rs7267595 | 20 | 10,643,850 | JAG1 | A | C | 0.015 | 0.003 | 0.005 | 0.009 | 0.023 | 0.012 | −0.012 | 0.013 | −0.004 | 0.011 | −0.015 | 0.014 | 0.006 | 0.014 | 0.012 | 0.014 |
| rs2273058 | 20 | 20,033,319 | CRNKL1 | G | A | −0.022 | 0.003 | −0.005 | 0.009 | −0.002 | 0.012 | −0.008 | 0.013 | −0.011 | 0.011 | −0.013 | 0.014 | −0.009 | 0.014 | −0.002 | 0.014 |
| rs6106324 | 20 | 20,964,988 | AL133465.1 | T | C | 0.019 | 0.003 | −0.011 | 0.009 | −0.009 | 0.013 | −0.012 | 0.013 | −0.008 | 0.011 | −0.011 | 0.014 | −0.007 | 0.014 | −0.010 | 0.014 |
| rs2424396 | 20 | 21,630,280 | LINC01726 | A | G | −0.033 | 0.004 | 0.016 | 0.016 | 0.003 | 0.022 | 0.026 | 0.022 | 0.025 | 0.019 | 0.034 | 0.024 | 0.022 | 0.025 | 0.020 | 0.024 |
| rs6088579 | 20 | 33,284,624 | PIGU/NCOA6 | G | A | 0.027 | 0.003 | −0.016 | 0.012 | −0.021 | 0.017 | −0.011 | 0.017 | −0.010 | 0.014 | −0.016 | 0.019 | −0.004 | 0.019 | −0.045 | 0.019 |
| rs2207132 | 20 | 39,142,516 | MAFB | G | A | 0.048 | 0.007 | 0.059 | 0.028 | 0.033 | 0.039 | 0.090 | 0.042 | 0.033 | 0.034 | 0.046 | 0.045 | 0.000 | 0.045 | 0.059 | 0.045 |
| rs17265513 | 20 | 39,832,628 | ZHX3 | C | T | −0.022 | 0.003 | −0.016 | 0.011 | −0.029 | 0.016 | −0.001 | 0.016 | −0.018 | 0.014 | −0.014 | 0.017 | −0.017 | 0.018 | −0.005 | 0.017 |
| rs16995311 | 20 | 49,201,102 | PTPN1 | A | C | 0.040 | 0.005 | 0.024 | 0.018 | 0.036 | 0.024 | 0.010 | 0.025 | 0.015 | 0.021 | 0.006 | 0.027 | 0.027 | 0.028 | 0.032 | 0.027 |
| rs2104476 | 20 | 54,852,856 | − | A | G | −0.020 | 0.003 | 0.006 | 0.010 | 0.012 | 0.014 | −0.001 | 0.014 | 0.003 | 0.012 | 0.006 | 0.015 | 0.003 | 0.016 | −0.004 | 0.015 |
| rs2738787 | 20 | 62,328,375 | RTEL1−TNFRSF6B/RTEL1 | G | A | −0.037 | 0.005 | −0.028 | 0.017 | −0.060 | 0.023 | 0.007 | 0.024 | −0.005 | 0.020 | 0.001 | 0.026 | 0.000 | 0.026 | −0.064 | 0.025 |
| rs9978775 | 21 | 40,694,526 | BRWD1−AS1 | G | A | 0.019 | 0.003 | 0.009 | 0.009 | 0.021 | 0.012 | −0.001 | 0.013 | 0.008 | 0.011 | 0.008 | 0.014 | 0.001 | 0.014 | 0.011 | 0.014 |
| rs8138950 | 22 | 29,448,643 | ZNRF3 | C | T | 0.015 | 0.003 | 0.016 | 0.009 | 0.013 | 0.012 | 0.020 | 0.013 | 0.005 | 0.011 | −0.004 | 0.014 | 0.010 | 0.014 | 0.032 | 0.014 |
| rs2412973 | 22 | 30,529,631 | HORMAD2 | C | A | −0.014 | 0.003 | 0.024 | 0.009 | 0.011 | 0.012 | 0.035 | 0.013 | 0.028 | 0.011 | 0.037 | 0.014 | 0.022 | 0.014 | 0.016 | 0.014 |
| rs12106594 | 22 | 31,885,316 | DRG1/EIF4ENIF1/SFI1 | C | T | −0.036 | 0.006 | 0.007 | 0.020 | −0.017 | 0.028 | 0.030 | 0.029 | 0.008 | 0.024 | 0.010 | 0.031 | −0.004 | 0.031 | 0.003 | 0.031 |
| rs5755948 | 22 | 36,179,095 | RBFOX2 | G | A | −0.028 | 0.004 | 0.003 | 0.013 | 0.008 | 0.018 | −0.005 | 0.019 | 0.011 | 0.016 | 0.028 | 0.020 | −0.005 | 0.021 | −0.001 | 0.020 |
| rs6519133 | 22 | 39,096,602 | JOSD1 | T | C | 0.029 | 0.003 | 0.011 | 0.009 | 0.012 | 0.013 | 0.011 | 0.013 | 0.005 | 0.011 | 0.013 | 0.014 | 0.002 | 0.015 | 0.024 | 0.014 |
| rs9611565 | 22 | 41,767,486 | TEF | T | C | 0.029 | 0.003 | 0.001 | 0.010 | 0.010 | 0.014 | −0.010 | 0.015 | 0.006 | 0.012 | −0.001 | 0.016 | 0.006 | 0.016 | −0.015 | 0.016 |
| rs4823324 | 22 | 46,238,123 | ATXN10 | T | C | 0.016 | 0.003 | 0.002 | 0.009 | 0.001 | 0.013 | 0.003 | 0.013 | 0.006 | 0.011 | 0.023 | 0.014 | −0.013 | 0.014 | 0.000 | 0.014 |
| IGFBP3 - n SNPs = 4 | |||||||||||||||||||||
| rs4234798 | 4 | 7,219,933 | SORCS2 | G | T | 0.095 | 0.011 | 0.012 | 0.009 | 0.020 | 0.013 | 0.003 | 0.013 | 0.005 | 0.011 | −0.003 | 0.014 | 0.015 | 0.014 | 0.015 | 0.014 |
| rs11977526 | 7 | 46,008,110 | IGFBP3 | A | G | 0.287 | 0.011 | 0.037 | 0.009 | 0.034 | 0.013 | 0.042 | 0.013 | 0.042 | 0.011 | 0.042 | 0.014 | 0.042 | 0.014 | 0.037 | 0.014 |
| rs700753 | 7 | 46,753,684 | TNS3 | G | C | 0.158 | 0.011 | −0.006 | 0.009 | −0.006 | 0.013 | −0.007 | 0.013 | −0.009 | 0.011 | 0.001 | 0.014 | −0.025 | 0.015 | −0.010 | 0.014 |
| rs1065656 | 16 | 1,838,836 | NUBP2 | G | C | 0.111 | 0.011 | 0.010 | 0.010 | 0.015 | 0.013 | 0.006 | 0.014 | −0.003 | 0.012 | 0.004 | 0.015 | −0.010 | 0.015 | 0.017 | 0.015 |
Chr, chromosome; SE, standard error.
Overlapped or nearest gene (sourced from: https://snp-nexus.org/index.html). Where blank, gene unknown.
Supplementary Table 3.
Risk (HRs) of Colorectal Cancer Associated With Circulating IGF1 Levels With Those Participants With Circulating HbA1c Levels ≥48 mmol/mol (or 6.5%, the Cutoff for Type 2 Diabetes) Excluded
| Q1 | Q2 | Q3 | Q4 | Q5 | P-trend | HR per 1-SD increment | HR per 1-SD increment (adjusted)a | |
|---|---|---|---|---|---|---|---|---|
| Colorectal cancer | ||||||||
| Both sexes | 1 | 1.13 (1.00–1.27) | 1.14 (1.01–1.29) | 1.12 (0.98–1.27) | 1.34 (1.18–1.53) | <0.0001 | 1.09 (1.04–1.14) | 1.12 (1.06–1.18) |
| Colon cancer | ||||||||
| Both sexes | 1 | 1.06 (0.91–1.22) | 1.09 (0.94–1.26) | 1.06 (0.91–1.24) | 1.34 (1.15–1.57) | 0.002 | 1.08 (1.03–1.14) | 1.11 (1.04–1.19) |
| Proximal colon cancer | ||||||||
| Both sexes | 1 | 1.15 (0.94–1.40) | 1.22 (1.00–1.50) | 0.97 (0.77–1.21) | 1.35 (1.08–1.68) | 0.09 | 1.08 (1.00–1.16) | 1.10 (1.00–1.21) |
| Distal colon cancer | ||||||||
| Both sexes | 1 | 0.96 (0.77–1.20) | 0.97 (0.77–1.23) | 1.18 (0.94–1.48) | 1.31 (1.03–1.65) | 0.01 | 1.09 (1.00–1.18) | 1.12 (1.01–1.24) |
| Rectal cancer | ||||||||
| Both sexes | 1 | 1.29 (1.05–1.59) | 1.27 (1.02–1.57) | 1.25 (1.00–1.56) | 1.36 (1.08–1.72) | 0.02 | 1.10 (1.02–1.19) | 1.14 (1.03–1.25) |
NOTE. Multivariable Cox regression model using age as the underlying time variable and stratified by sex, Townsend deprivation index (quintiles), region of the recruitment assessment center, and age at recruitment. Models adjusted for waist circumference (per 5 cm), total physical activity (<10, 10 to <20, 20 to <40, 40 to <60, ≥60 metabolic equivalent hours per week, unknown), height (per 10 cm), alcohol consumption frequency (never, special occasions only, 1–3 times per month, 1–2 times per week, 3–4 times per week, daily/almost daily, unknown), smoking status and intensity (never, former, current- <15 per day, current- ≥15 per day, current- intensity unknown, unknown), frequency of red and processed meat consumption (<2, 2 to <3, 3 to <4, ≥4 occasions per week, unknown), family history of colorectal cancer (no, yes, unknown), educational level (Certificates of secondary education [CSEs]/Ordinary [O]-levels/General Certificates of Secondary Education [GCSEs] or equivalent, National Vocational Qualification [NVQ]/Higher National Diploma [HND]/Higher National Certificate [HNC]/Advanced [A]-levels/Advanced Subsidiary [AS]-levels or equivalent, other professional qualifications, college/university degree, none of the above, unknown), regular aspirin/ibuprofen use (no, yes, unknown), ever use of hormone replacement therapy (no, yes, unknown), and circulating levels (sex-specific quintiles, missing/unknown) of CRP (mg/L), HbA1c (mmol/mol), testosterone (nmol/L), and SHBG (nmol/L).
HRs per SD increment were additionally corrected for regression dilution using a regression dilution ratio (0.76) obtained from the subsample of participants with repeat IGF1 measurements.
Supplementary Table 4.
MR Pleiotropy Sensitivity Tests for the Circulating IGF1 and IGFBP3 Levels and Risk of Colorectal Cancer
| n SNPs | Group | Weighted median approach |
MR-Egger test |
MR-PRESSO |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | LCI | UCI | P | Intercept | LCI | UCI | P | OR | LCI | UCI | P | P Horizontal pleiotropy | P distortion | Outlier SNPs | |||
| IGF1 | 413 | Colorectal cancer, both sexes | 1.12 | 1.04 | 1.20 | 2.9E−03 | −7.1E−04 | −3.0E−03 | 1.6E−03 | 0.54 | 1.11 | 0.98 | 1.27 | 0.11 | <1e−04 | .91 | rs1150752 rs174554 |
| Colorectal cancer, men | 1.07 | 0.97 | 1.18 | 0.15 | −1.4E−03 | −4.5E−03 | 1.8E−03 | 0.4 | 1.18 | 1.00 | 1.38 | 0.05 | <1e−04 | − | − | ||
| Colorectal cancer, women | 1.07 | 0.97 | 1.18 | 0.2 | −1.9E−05 | −3.3E−03 | 3.3E−03 | 0.99 | 1.06 | 0.89 | 1.25 | 0.53 | <1e−04 | .82 | rs174554 | ||
| Colon cancer, both sexes | 1.12 | 1.03 | 1.23 | 0.01 | −1.7E−03 | −4.5E−03 | 9.9E−04 | 0.21 | 1.14 | 0.98 | 1.32 | 0.09 | <1e−04 | .83 | rs174554 | ||
| Proximal colon cancer, both sexes | 1.08 | 0.97 | 1.21 | 0.16 | 5.8E−04 | −3.0E−03 | 4.1E−03 | 0.75 | 1.02 | 0.84 | 1.23 | 0.84 | <1e−04 | .90 | rs33932084 | ||
| Distal colon cancer, both sexes | 1.10 | 0.98 | 1.23 | 0.11 | −4.2E−03 | −7.8E−03 | −5.5E−04 | 0.02 | 1.26 | 1.05 | 1.50 | 0.01 | <1e−04 | .82 | rs174554 | ||
| Rectal cancer, both sexes | 1.01 | 0.90 | 1.13 | 0.88 | −4.2E−04 | −4.0E−03 | 3.1E−03 | 0.82 | 1.09 | 0.91 | 1.30 | 0.36 | <1e−04 | .85 | rs1150752 rs61867536 rs9267488 |
||
| IGFBP3 | 4 | Colorectal cancer, both sexes | 1.14 | 1.07 | 1.20 | 1.1E−05 | −0.01 | −0.05 | 0.03 | 0.35 | 1.18 | 1.07 | 1.31 | 0.08 | .29 | − | − |
| Colorectal cancer, men | 1.13 | 1.04 | 1.22 | 2.8E−03 | 0.00 | −0.06 | 0.05 | 0.82 | 1.14 | 0.98 | 1.34 | 0.23 | .72 | − | − | ||
| Colorectal cancer, women | 1.13 | 1.04 | 1.22 | 3.9E−03 | −0.02 | −0.08 | 0.04 | 0.26 | 1.25 | 1.18 | 1.32 | 0.02 | .18 | − | − | ||
| Colon cancer, both sexes | 1.13 | 1.05 | 1.21 | 7.9E−04 | −0.03 | −0.07 | 0.02 | 0.16 | 1.27 | 1.16 | 1.38 | 0.03 | .04 | − | − | ||
| Proximal colon cancer, both sexes | 1.12 | 1.02 | 1.23 | 0.01 | −0.02 | −0.08 | 0.04 | 0.28 | 1.25 | 1.21 | 1.29 | 0.01 | .21 | − | − | ||
| Distal colon cancer, both sexes | 1.13 | 1.03 | 1.24 | 0.01 | −0.03 | −0.10 | 0.03 | 0.17 | 1.29 | 1.01 | 1.65 | 0.18 | .03 | − | − | ||
| Rectal cancer, both sexes | 1.14 | 1.05 | 1.25 | 2.6E−03 | −0.01 | −0.07 | 0.05 | 0.57 | 1.18 | 1.01 | 1.39 | 0.18 | .62 | − | − | ||
LCI, lower confidence interval; UCI, upper confidence interval.
Supplementary Table 5.
MR leave-one-out analysis for IGFBP3 Levels and Risk of Colorectal Cancer
| MR | ||
|---|---|---|
| SNP excluded | Likelihood-based approach OR (95% CI) per 1-SD increment | P value |
| rs11977526 | 1.02 (0.92–1.13) | 0.7 |
| rs4234798 | 1.11 (1.04–1.18) | 1.E−03 |
| rs700750 | 1.15 (1.08–1.23) | 2.E−05 |
| rs9923699 | 1.12 (1.05–1.19) | 4.E−04 |
References
- 1.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 2.Samani A.A., Yakar S., LeRoith D. The role of the IGF System in Cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 3.Kelley K.M., Oh Y., Gargosky S.E. Insulin-like growth factor-binding proteins (IGFBPs) and their regulatory dynamics. Int J Biochem Cell Biol. 1996;28:619–637. doi: 10.1016/1357-2725(96)00005-2. [DOI] [PubMed] [Google Scholar]
- 4.Oh Y., Muller H.L., Ng L. Transforming growth factor- beta-induced cell growth inhibition in human breast cancer cells is mediated through insulin-like growth factor-binding protein-3 action. J Biol Chem. 1995;270:13589–13592. doi: 10.1074/jbc.270.23.13589. [DOI] [PubMed] [Google Scholar]
- 5.Baxter R.C. Signalling pathways involved in antiproliferative effects of IGFBP-3: a review. Mol Pathol. 2001;54:145–148. doi: 10.1136/mp.54.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma J., Pollak M.N., Giovannucci E. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J Natl Cancer Inst. 1999;91:620–625. doi: 10.1093/jnci/91.7.620. [DOI] [PubMed] [Google Scholar]
- 7.Kaaks R., Toniolo P., Akhmedkhanov A. Serum C-Peptide, Insulin-Like Growth Factor (IGF)-I, IGF-Binding Proteins, and Colorectal Cancer Risk in Women. J Natl Cancer Inst. 2000;92:1592–1600. doi: 10.1093/jnci/92.19.1592. [DOI] [PubMed] [Google Scholar]
- 8.Gunter M.J., Hoover D.R., Yu H. Insulin, insulin-like growth factor-i, endogenous estradiol, and risk of colorectal cancer in postmenopausal women. Cancer Res. 2008;68:329–337. doi: 10.1158/0008-5472.CAN-07-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otani T., Iwasaki M., Sasazuki S. Plasma C-peptide, insulin-like growth factor-I, insulin-like growth factor binding proteins and risk of colorectal cancer in a nested case-control study: the Japan public health center-based prospective study. Int J Cancer. 2007;120:2007–2012. doi: 10.1002/ijc.22556. [DOI] [PubMed] [Google Scholar]
- 10.Palmqvist R., Stattin P., Rinaldi S. Plasma insulin, IGF-binding proteins-1 and -2 and risk of colorectal cancer: a prospective study in northern Sweden. Int J Cancer. 2003;107:89–93. doi: 10.1002/ijc.11362. [DOI] [PubMed] [Google Scholar]
- 11.Rinaldi S., Cleveland R., Norat T. Serum levels of IGF-I, IGFBP-3 and colorectal cancer risk: results from the EPIC cohort, plus a meta-analysis of prospective studies. Int J Cancer. 2010;126:1702–1715. doi: 10.1002/ijc.24927. [DOI] [PubMed] [Google Scholar]
- 12.Probst-Hensch N.M., Yuan J.M., Stanczyk F.Z. IGF-1, IGF-2 and IGFBP-3 in prediagnostic serum: association with colorectal cancer in a cohort of Chinese men in Shanghai. Br J Cancer. 2001;85:1695–1699. doi: 10.1054/bjoc.2001.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovannucci E., Pollak M.N., Platz E.A. A prospective study of plasma insulin-like growth factor-1 and binding protein-3 and risk of colorectal neoplasia in women. Cancer Epidemiol Biomarkers Prev. 2000;9:345–349. [PubMed] [Google Scholar]
- 14.Wei E.K., Ma J., Pollak M.N. A prospective study of C-peptide, insulin-like growth factor-I, insulin-like growth factor binding protein-1, and the risk of colorectal cancer in women. Cancer Epidemiol Biomarkers Prev. 2005;14:850–855. doi: 10.1158/1055-9965.EPI-04-0661. [DOI] [PubMed] [Google Scholar]
- 15.Fred Hutchinson Cancer Research Center Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) https://www.fredhutch.org/en/research/divisions/public-health-sciences-division/research/cancer-prevention/genetics-epidemiology-colorectal-cancer-consortium-gecco.html Available at:
- 16.Allen N., Sudlow C., Downey P. UK Biobank: current status and what it means for epidemiology. Health Policy and Technology. 2012;1:123–126. [Google Scholar]
- 17.UK-Biobank UK Biobank. Protocol for a large-scale prospective epidemiological resources. 2010. http://www.ukbiobank.ac.uk/wp-content/uploads/2011/11/UK-Biobank-Protocol.pdf Available at: Accessed April 1, 2018.
- 18.Hosgood H.D., Gunter M.J., Murphy N. The relation of obesity-related hormonal and cytokine levels with multiple myeloma and non-Hodgkin lymphoma. Front Oncol. 2018;8:103. doi: 10.3389/fonc.2018.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UK-Biobank UK Biobank Biomarker Project - Companion Document to Accompany Serum Biomarker Data. Prepared for: UK Biobank Showcase. 2019;Volume 1 Available at: http://biobank.ctsu.ox.ac.uk/showcase/showcase/docs/serum_biochemistry.pdf. Accessed June 1, 2019. [Google Scholar]
- 20.Schoenfeld D. Partial residuals for the proportional hazards regression model. Biometrika. 1982;69:239–241. [Google Scholar]
- 21.Clarke R., Shipley M., Lewington S. Underestimation of risk associations due to regression dilution in long-term follow-up of prospective studies. Am J Epidemiol. 1999;150:341–353. doi: 10.1093/oxfordjournals.aje.a010013. [DOI] [PubMed] [Google Scholar]
- 22.MacMahon S., Peto R., Collins R. Blood pressure, stroke, and coronary heart disease: Part 1, prolonged differences in blood pressure: prospective observational studies corrected for the regression dilution bias. Lancet. 1990;335:765–774. doi: 10.1016/0140-6736(90)90878-9. [DOI] [PubMed] [Google Scholar]
- 23.Clarke R., Emberson J.R., Breeze E. Biomarkers of inflammation predict both vascular and non-vascular mortality in older men. Eur Heart J. 2008;29:800–809. doi: 10.1093/eurheartj/ehn049. [DOI] [PubMed] [Google Scholar]
- 24.Murphy N., Strickler H.D., Stanczyk F.Z. A prospective evaluation of endogenous sex hormone levels and colorectal cancer risk in postmenopausal women. J Natl Cancer Inst. 2015;107(10) doi: 10.1093/jnci/djv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aleksandrova K., Jenab M., Boeing H. Circulating C-reactive protein concentrations and risks of colon and rectal cancer: a nested case-control study within the European prospective investigation into cancer and nutrition. Am J Epidemiol. 2010;172:407–418. doi: 10.1093/aje/kwq135. [DOI] [PubMed] [Google Scholar]
- 26.Mori N., Sawada N., Iwasaki M. Circulating sex hormone levels and colorectal cancer risk in Japanese postmenopausal women: the JPHC nested case–control study. Int J Cancer. 2019;145:1238–1244. doi: 10.1002/ijc.32431. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldi S., Rohrmann S., Jenab M. Glycosylated hemoglobin and risk of colorectal cancer in men and women, the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol Biomarkers Prev. 2008;17:3108–3115. doi: 10.1158/1055-9965.EPI-08-0495. [DOI] [PubMed] [Google Scholar]
- 28.Harrell F. Springer; New York: 2001. Regression Modeling Strategies: With applications to linear models, logistic regression, and survival analysis. [Google Scholar]
- 29.Sinnott-Armstrong N., Tanigawa Y., Amar D. Genetics of 38 blood and urine biomarkers in the UK Biobank. bioRxiv. 2019:660506. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teumer A., Qi Q., Nethander M. Genomewide meta-analysis identifies loci associated with IGF-I and IGFBP-3 levels with impact on age-related traits. Aging Cell. 2016;15:811–824. doi: 10.1111/acel.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huyghe J.R., Bien S.A., Harrison T.A. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brion M.-J.A., Shakhbazov K., Visscher P.M. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genetic Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess S., Scott R.A., Timpson N.J. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J., Davey Smith G., Haycock P.C. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbanck M., Chen C.-Y., Neale B. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentinis B., Baserga R. IGF-I receptor signalling in transformation and differentiation. Mol Pathol. 2001;54:133–137. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekharam M., Zhao H., Sun M. Insulin-like growth factor 1 receptor enhances invasion and induces resistance to apoptosis of colon cancer cells through the Akt/Bcl-xL Pathway. Cancer Res. 2003;63:7708–7716. [PubMed] [Google Scholar]
- 41.Lahm H., Amstad P., Wyniger J. Blockade of the insulin-like growth-factor-I receptor inhibits growth of human colorectal cancer cells: evidence of a functional IGF-II-mediated autocrine loop. Int J Cancer. 1994;58:452–459. doi: 10.1002/ijc.2910580325. [DOI] [PubMed] [Google Scholar]
- 42.Levine Morgan E., Suarez Jorge A., Brandhorst S. Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab. 2014;19:407–417. doi: 10.1016/j.cmet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradbury K.E., Balkwill A., Tipper S.J. The association of plasma IGF-I with dietary, lifestyle, anthropometric, and early life factors in postmenopausal women. Growth Horm IGF Res. 2015;25:90–95. doi: 10.1016/j.ghir.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Zhu K., Meng X., Kerr D.A. The effects of a two-year randomized, controlled trial of whey protein supplementation on bone structure, IGF-1, and urinary calcium excretion in older postmenopausal women. J Bone Miner Res. 2011;26:2298–2306. doi: 10.1002/jbmr.429. [DOI] [PubMed] [Google Scholar]
- 45.Nishida Y., Matsubara T., Tobina T. Effect of low-intensity aerobic exercise on insulin-like growth factor-I and insulin-like growth factor-binding proteins in healthy men. Int J Endocrinol. 2010;2010:452820. doi: 10.1155/2010/452820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigneri P.G., Tirrò E., Pennisi M.S. The insulin/IGF system in colorectal cancer development and resistance to therapy. Front Oncol. 2015;5:230. doi: 10.3389/fonc.2015.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deal C., Ma J., Wilkin Fo. Novel promoter polymorphism in insulin-like growth factor-binding protein-3: correlation with serum levels and interaction with known regulators1. J Clin Endocrinol Metab. 2001;86:1274–1280. doi: 10.1210/jcem.86.3.7280. [DOI] [PubMed] [Google Scholar]
- 48.Kamat M.A., Blackshaw J.A., Young R. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui H., Cruz-Correa M., Giardiello F.M. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 50.Cruz-Correa M., Cui H., Giardiello F.M. Loss of imprinting of insulin growth factor II gene: a potential heritable biomarker for colon neoplasia predisposition. Gastroenterology. 2004;126:964–970. doi: 10.1053/j.gastro.2003.12.051. [DOI] [PubMed] [Google Scholar]
- 51.Unger C., Kramer N., Unterleuthner D. Stromal-derived IGF2 promotes colon cancer progression via paracrine and autocrine mechanisms. Oncogene. 2017;36:5341. doi: 10.1038/onc.2017.116. [DOI] [PubMed] [Google Scholar]
- 52.Morris J.K., George L.M., Wu T. Insulin-like growth factors and cancer: no role in screening. Evidence from the BUPA study and meta-analysis of prospective epidemiological studies. Br J Cancer. 2006;95:112–117. doi: 10.1038/sj.bjc.6603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young N.J., Metcalfe C., Gunnell D. A cross-sectional analysis of the association between diet and insulin-like growth factor (IGF)-I, IGF-II, IGF-binding protein (IGFBP)-2, and IGFBP-3 in men in the United Kingdom. Cancer Causes Control. 2012;23:907–917. doi: 10.1007/s10552-012-9961-6. [DOI] [PubMed] [Google Scholar]
References
- 1.Sinnott−Armstrong N., Tanigawa Y., Amar D. Genetics of 38 blood and urine biomarkers in the UK Biobank. bioRxiv. 2019:660506. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teumer A., Qi Q., Nethander M. Genomewide meta−analysis identifies loci associated with IGF−I and IGFBP3 levels with impact on age−related traits. Aging Cell. 2016;15:811–824. doi: 10.1111/acel.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]