Abstract
It has been reported that SARS-CoV-2 may use ACE2 as a receptor to gain entry into human cells, in a way similar to that of SARS-CoV. Analyzing the distribution and expression level of ACE2 may therefore help reveal underlying mechanisms of viral susceptibility and post-infection modulation. In this study, we utilized previously uploaded information on ACE2 expression in various conditions including SARS-CoA to evaluate the role of ACE2 in SARS-CoV and extrapolate that to COVID-19. We found that the expression of ACE2 in healthy populations and patients with underlying diseases was not significantly different. However, based on the elevated expression of ACE2 in cigarette smokers, we speculate that long-term smoking may be a risk factor for COVID-19. Analysis of ACE2 in SARS-CoV infected cells suggests that ACE2 is not only a receptor but is also involved in post-infection regulation, including immune response, cytokine secretion, and viral genome replication. Moreover, we constructed Protein-protein interaction (PPI) networks and identified hub genes in viral activity and cytokine secretion. Our findings may help clinicians and researchers gain more insight into the pathogenesis of SARS-CoV-2 and design therapeutic strategies for COVID-19.
Keywords: 2019-nCoV, ACE2, Susceptibility, Immune response, Protein-protein interactions, COVID-19, SARS-CoV-2
Highlights
-
•
SARS-CoV-2 may use ACE2 as a receptor to gain entry into human cells, in a way similar to that of SARS-CoV.
-
•
PPI networks identified RPS3, RPS8 and RPS9 as hub genes in viral activity and SRC and CASP1 in cytokine secretion.
-
•
RPS3 plays a key role in viral replication.
-
•
SRC non-receptor protein kinase has a role in macrophage mediated innate immunity and cytokine release.
-
•
Blocking the ACE2 receptor may be protective or lead to worse lung injury due to the unmitigated action of angiotensin 2.
1. Introduction
In early December 2019, several cases of pneumonia of unknown etiology were reported in Wuhan, Hubei, China [1]. Later, a novel coronavirus, SARS-CoV-2 was identified based on sequencing of respiratory tract samples from these patients. As of April 12, 2020, more than 80,000 confirmed patients and more than 3000 deaths have been reported in China, with additional patients being identified in a rapidly growing number internationally. Analysis of the genome of SARS-CoV-2 showed that this new virus shares about 80–90% sequence identity to the original SARS-CoV. Both bioinformatics modeling and in vitro experiments indicate that 2019-nCoV likely utilizes angiotensin-converting enzyme 2 (ACE2) as a receptor to gain entry into human cells [2,3].
ACE2, discovered in 2000, shares 40% identity and 61% similarity with ACE [4]. The full-length protein structure of ACE2 consists of an N-terminal and a C-terminal domain with a single transmembrane helix and an intracellular segment [5]. Considering the difference in active sites between ACE2 and ACE, the functions of these two enzymes are distinct. Unlike ACE, which converts angiotensin I (Ang I) to angiotensin II (AngII), ACE2 converts Ang II to Ang-(1-(1 and Ang I to Ang-(1–9) [6,7]. ACE2 is expressed in different tissues, such as renal, cardiovascular and gastrointestinal [8,9]. ACE2 is also present in lung alveolar epithelial cells, enterocytes of the small intestine, arterial and venous endothelial cells and arterial smooth muscle cells [10]. ACE2 was previously identified as an entry receptor for SARS-CoV and HCoV-NL63 [11,12]. Considering that the sequence of SARS-CoV-2 is similar to the SARS-CoV and that SARS-CoV-2 appears to use the same ACE2 receptor to enter cells, analyses of ACE2 expression and distribution in lung and related biological processes may help us understand the pathogenesis and design therapeutic strategies for COVID-19.
Recent advances in bioinformatics methodology has enabled researchers to discern the underlying mechanisms of various diseases. In this study, we used new approaches in bioinformatics to identify ACE2 expression features in lung and the potential regulation networks.
2. Materials and methods
2.1. Data collection
Data from six independent studies, GSE37768 (n = 38), GSE73395 (n = 57), GSE89809 (n = 145), GSE63127 (n = 230), GSE97010 (n = 126), and GSE17400 (n = 27), covering healthy volunteers, COPD patients, asthma patients and smokers were accessed from the GEO data repository (https://www.ncbi.nlm.nih.gov/geo/). The samples consist of lung tissue, bronchoalveolar lavage samples, bronchial epithelial cells, small airway epithelial cells, and SARS infected cells.
2.2. Functional enrichment analysis
The gmt file containing Gene Ontology gene sets was downloaded from Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb/). Gene Ontology gene sets consists of genes annotated by the same Gene Ontology terms. Gene Set Enrichment Analysis (GSEA) was performed to analyze the possible biological processes related to ACE2 in healthy people using the clusterProfiler package, a statistical analysis and visualization of function profiles for genes and gene clusters [13]. The parameters were nPerm = 1000, minGSSize = 10, maxGSSize = 500. The biological processes with p-value < 0.05 were considered significant. Gene Set Variation Analysis (GSVA) was performed using GSVA R package. Immune cell infiltration was quantified using single sample (ss) GSEA methodology in the GSVA R package. The gene list for immune cells was derived from Bindea G et al. [14].
2.3. PPI
All proteins involved in the viral-related biological process and cytokine secretion-biological process were extracted from the gmt file. Cytoscape v3.7.2 was used to construct the PPI network using BisoGenet application. The PPI sources include DIP, BIOGRID, HPRD, INTACT, MINT and BIND. The nodes with topological importance in the interaction network were screened by calculating Degree Centrality (DC) with the Cytoscape plugin CytoNCA. Hub proteins were identified using Cytoscape plugin CytoHubba.
3. Results
3.1. ACE2 expression features in lung
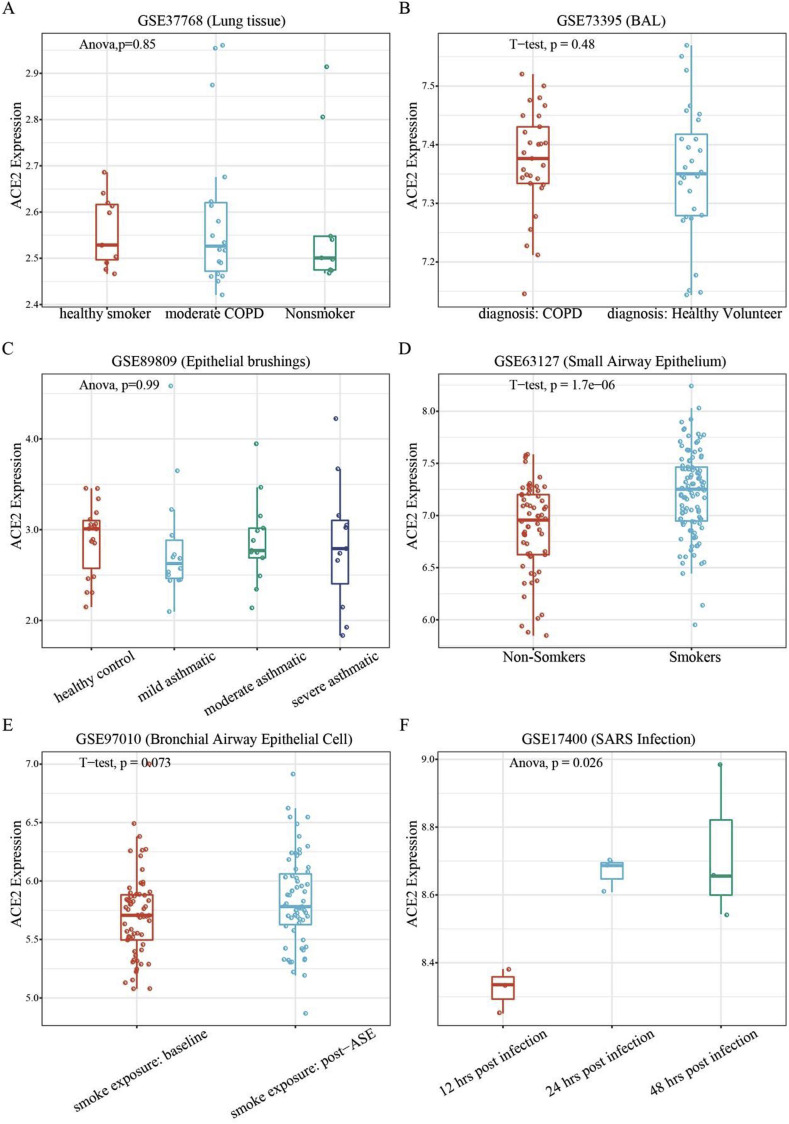
COVID-19 patients often have underlying diseases, including chronic respiratory disease and cardiovascular disease. It is important to evaluate whether patients with underlying diseases are more susceptible to coronavirus infection than the healthy population. To do this, we analyzed the expression of ACE2 in lung in different populations. The expression level of ACE2 was not significantly different between healthy populations and patients with chronic respiratory diseases including chronic obstructive pulmonary diseases (COPD) and asthma (Fig. 1 A, B, and 1C, p-value > 0.05).
Fig. 1.
Expression of ACE2 in lung tissues and epithelial cells. A. Expression of ACE2 in lung tissue from healthy smokers, moderate COPD patients and non-smokers. B. Expression of ACE2 in bronchoalveolar lavage samples from COPD patients and healthy volunteers. C. Expression of ACE2 in asthma patients. D. Expression of ACE2 in small airway epithelial cells in smokers (S) and non-smokers (NS). E. Expression of ACE2 after ASE. F. Expression of ACE2 in SARS-CoV infected bronchial cells. ASE = acute smoke exposure.
We also found that the level of ACE2 expression was markedly upregulated in long-term smokers (Fig. 1D, p-value < 0.05). However, the p-value between baseline and post-acute smoke exposure (ASE) (24 h after smoking 3 cigarettes) was 0.073 (Fig. 1E).
Finally, we analyzed the expression of ACE2 in airway epithelial cells after being infected with SARS-CoV. The results suggest that 24 h after SARS-CoV infection, the expression of ACE2 dramatically increases compared to the expression at 12 h. After 48 h, the expression of ACE2 remained at a high level. This indicates that ACE2 not only plays a critical role in viral susceptibility but may also be involved in post-infectious regulation.
3.2. Biological functions of ACE2 in coronavirus infection
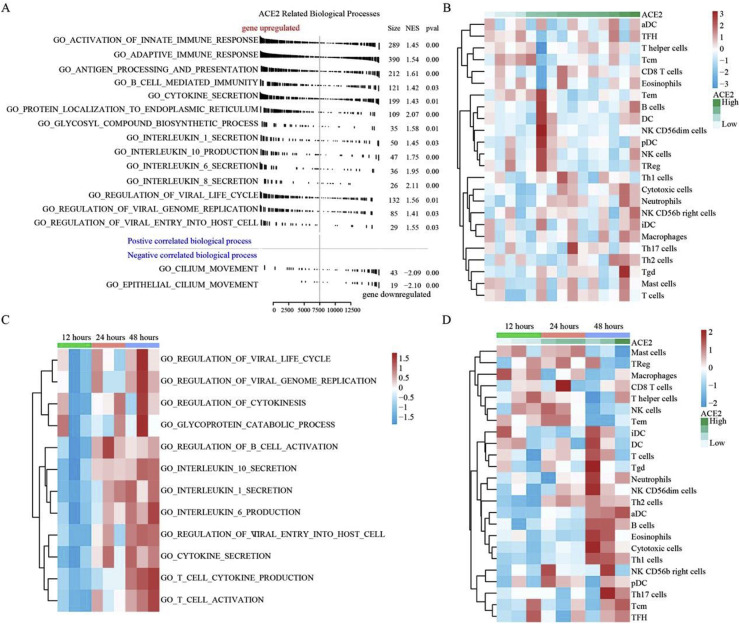
To investigate the virus-related potential biological processes associated with ACE2, we extracted the expression profiles of 15 healthy non-smokers from dataset GSE89809 and divided them into two groups: high and low-expression level of ACE2. We performed GSEA (Gene Set Enrichment Analysis) and found that the expression of ACE2 was mainly associated with innate and acquired immune responses, regulation of B cell mediated immunity, as well as cytokines IL-1, IL-10, IL-6, and IL-8 secretion (Fig. 2 A). Moreover, higher expression of ACE2 may prolong the virus life cycle, enhance virus replication and mediate penetration of the virus into the host cell.
Fig. 2.
Functional analysis of ACE2 revealing related biological processes. A. GSEA analysis showing the related biological processes of ACE2 in healthy non-smokers. B. Immune cell infiltration in healthy population. C. GSVA score of key processes showing time-dependent alterations. D. Immune cell infiltration in SARS-CoV infected cells.
To further explore whether the above activities could be triggered after coronavirus infection, we analyzed the expression profiles of epithelial cells which were infected by SARS-CoV. The biological process scores were obtained using GSVA and the Pearson correlation between process scores and ACE2 expression was calculated. Similarly, while ACE2 expression was increased after infection, the production of cytokines, IL-1, IL-10 and IL-6 was also increased, and regulation of B cell activated was seen. After 48 h, virus activities such as viral entry into the host cell, virus life cycle, and viral transcription were enhanced.
In addition, T-cell cytokine secretion was increased and T-cell activation was stimulated (Fig. 2C). We used ssGSEA to quantify the immune infiltrates in epithelial cells from healthy people and those following infection. We found that in normal samples, the immune cells were not activated and the correlation between infiltration and ACE2 expression was not significant (Fig. 2B). However, in the SARS-CoV infected cells, ACE2 was significantly correlated with neutrophils, Natural Killer (NK) cells, Th17 cells, Th2 cells, Th1 cells, and dendritic cells (Fig. 2D).
3.3. Regulation networks after infection
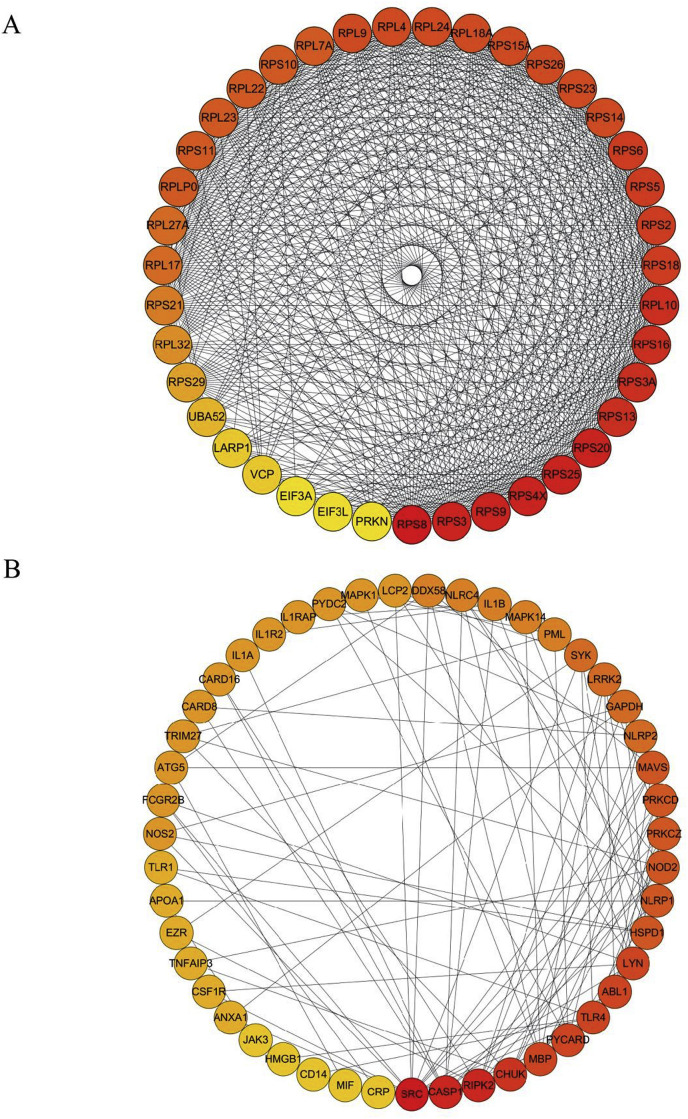
Considering that higher expression of ACE2 was involved in mediating inflammatory responses, immune activities, and viral replication in both healthy individuals and infected cells, we further explored the protein-protein regulation network in viral activity-related proteins and cytokine secretion, respectively. We extracted the viral activity-associated and cytokine secretion-associated proteins from the Gene ontology (GO) biological process gmt file. Construction of the Protein-protein interaction (PPI) showed that these proteins were tightly interactive among each other and here we discuss the most significant PPIs.
In terms of viral activity, we found that ribosomal proteins such as RPS3, RPS8 and PRS9 were the most important genes in the PPI network. In addition, VCP, LARP1, UBA52, PRKN, EIF3A and EIF3L were also important in the network (Fig. 3 A). In terms of the cytokine secretion associated proteins, we found that the proteins SRC, CASP1, and RIPK were Hub proteins in the regulating network of cytokine secretion (Fig. 3B), indicating that these molecules were critically important in ACE2-associated inflammatory responses.
Fig. 3.
PPI network. A: PPI network of viral replication-related proteins. B: PPI network of cytokine secretion-related proteins.
4. Discussion
The SARS-CoV-2 infection in Wuhan is a serious global health problem and has been declared a pandemic by the WHO. Based on data from the first 41 patients, a reduction of lymphocytes was a prominent feature of the disease. The main symptoms of patients with COVID-19 infection were interstitial changes in the lung as demonstrated by chest CT. Severe pulmonary interstitial disease leads to hypoxemia and type I respiratory distress, which is related to the prognosis of patients. SARS-CoV-2 invades the lungs through the respiratory tract and causes severe pneumonia, which is the most frequent presentation.
It has been found that SARS-CoV-2 is able to bind to the ACE2 receptor on the surface of epithelial cells. Zhou et al. further confirmed that ACE2 is necessary for infection of cells by SARS-CoV-2 [2]. We accessed and analyzed six independent studies (GSE37768, GSE73395, GSE89809, GSE63127, GSE97010, and GSE17400), covering a healthy population, chronic obstructive pulmonary disease patients, bronchial asthma patients and smokers. The samples included lung tissue, BALF, airway epithelial cells, small airway epithelial cells and coronavirus infected epithelial cells.
We showed that ACE2 was found in all of the samples including the healthy population and patients with chronic airway diseases. Thus, there may be no significant difference in the susceptibility to SARS-CoV-2 infection between a healthy population and patients with chronic respiratory diseases. In contrast, current limited data on COVID-19 suggests that patients with underlying lung conditions may be at greater risk for severe chronic airway disease or death.
Previous studies found that ACE2 is related to the severity of acute respiratory syndrome induced by the SARS-CoV infection, and mediates the production of cytokines associated with acute respiratory distress syndrome [15,16] (Fig. 4 ). Moreover, ACE2 is also related to adaptive immune responses [17]. In this current study, GSEA analysis showed that the high expression of ACE2 was related to innate immune responses, adaptive immune responses, B cell regulation and cytokine secretion, as well as an enhanced inflammatory response induced by IL-1, IL-10, IL-6, IL-8. We speculate that the immune system dysfunction involved in the high expression of ACE2 is related to the symptoms of a cytokine storm. A clinical study in Wuhan pointed out that the levels of IL-1β, IL-10 and IL-8 were significantly increased in critically ill patients with new coronavirus infection, indicating that the pathologic process may involve an exaggerated pro-inflammatory cytokine response [1]. This may be related to pyroptosis, which has been suggested as another pathogenic mechanism involved in COVID-19 infection. Pyroptosis is an inflammatory form of apoptosis. The fact that tissue injury and death may be related to a pro-inflammatory process resulting from the viral infection suggests that the use of IL-1β blockers or IL-18 blockers may have some benefit in COVID-19 patients.
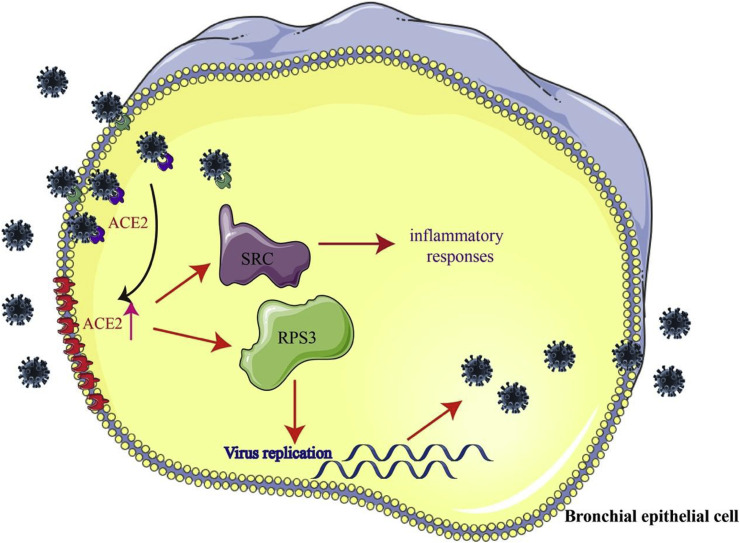
Fig. 4.
A schematic model of SARS-CoV-2 infection. SARS-CoV-2 uses ACE2 as a cellular entry receptor in airway epithelial cells. At the same time, the expression of ACE2 is increased by the infection. Furthermore, the increased expression of ACE2 affected RPS3 and SRC, the two hub genes involved in viral replication and inflammatory responses.
Our results also showed that the high expression of ACE2 increased the expression of genes involved in viral replication, which may enhance the ability of the virus to enter the host cells. Our calculations support the hypothesis that the transcriptome of epithelial cells is altered after infection by SARS-CoV-2 through increased ACE2 expression, which is beneficial to the replication and assembly of virus, and also to the entry of virus into host cells. T cell activation and inflammatory responses mediated by T cell factors are also significantly induced by the altered of transcriptome.
It has been speculated that the increased levels of IL1β, IFN-γ, IP10, and MCP1 in patients infected with SARS-CoV-2 are linked to the activation of T-helper-1 (Th1) cell responses [1]. However, there is a lack of pathological data of lung tissue infected with SARS-CoV-2. Through ssGSEA analysis, we found that high expression of ACE2 in lung tissue induced a cytotoxic reaction, neutrophil inflammation and a Th2-dominated immune response. Moreover, the expression of ACE2 varies in a time-dependent manner after SARS-CoV infection. ACE2 also mediates the activation of neutrophils, NK cells, Th17 cells, Th2 cells, Th1 cells, dendritic cells and TNFα secreting cells, leading to a severe inflammatory response.
Graph-based biological networks models capture the topology of the functional relationships between molecular entities such as gene, protein and small compounds, and achieve a finer grain representation of the bioentities and their relationships. In order to further analyze the core genes involved in viral replication and inflammatory responses mediated by ACE2, we constructed a protein-protein interaction network by using BisoGenet. It was found that RPS3 played a key role in viral replication, and SRC non-receptor protein kinase was the hub gene in the inflammatory response. It has been shown that RPS3 plays an important role in cell mitosis by interacting with α-tubulin. When cells are infected with HIV-1, HIV-1 Tat protein can interact with RPS3 and disturb its localization on the spindle, and ultimately inhibit cell proliferation [18]. Moreover, the function of RPS3 depends on its own nuclear translocation, which is regulated by protein kinases such as SRC [19]. We speculate that the increased expression of ACE2 affects RPS3 and SRC, which were the two hub genes involved in viral replication and the inflammatory response.
In view of the role of the ACE2 receptor in the ability of SARS-CoV-2 to enter host cells [20], the question arises as to the effect of ACE inhibitors and angiotensin-receptor blockers (ARBs) in the pathogenesis of COVID-19. This turns out to be a very complex relationship and is currently still very controversial. On the one hand, it has been found that in animal models, ACE inhibitors [21] and ARBs can increase the expression of ACE2 receptors [22], which theoretically can increase the ability of SARS-CoV-2 to enter cells. However, the data on this is inconsistent, and may in fact be tissue dependent, dosage dependent and may vary among different ARBs [22,23].
In addition, with SARS-CoV, it was discovered that infection with SARS-CoV led to downregulation of the ACE2 receptor [24]. From a physiologic standpoint, the ACE2 receptor serves as a negative regulator of severe lung edema and lung injury by decreasing Ang2 levels [25]. Ang2 induced vascular permeability and lung injury is facilitated by binding to the Angiotensin-2 receptor type 1 (AT1R) [26]. In a mouse model, SARS-CoV induced lung injury can be attenuated by blocking AT1R [27]. If SARS-CoV (and SARS-CoV-2) binding to ACE2 leads to a downregulation of ACE2, this could result in higher Ang2 levels and increased lung injury. Multiple ACE2 knockout mouse models have demonstrated the protective effects of ACE2 from lung injury and vascular inflammation [28,29].
Since ACE inhibitors do not bind the ACE2, but instead bind to ACE1, they theoretically should have no beneficial effect of blocking SARS-CoV-2 from entering cells. In fact, ACE inhibitors such as lisinopril and captopril have been shown to have no effect on activity of ACE2 [30,31]. At present there is no strong evidence to discontinue ACE inhibitors or ARBs in patients with COVID-19, nor conversely, to use these drugs as treatment for COVID-19.
It is perhaps significant to note that ACE2 is also a significant negative regulator of the renin-angiotensin system (RAS) in the cardiovascular system and plays a significant role in the control of blood pressure. It is postulated that this may be related to the higher mortality in Chinese patients with COVID-19 who have hypertension as a comorbidity. Additionally, ACE2 null mice display reduced cardiac contractility. If ACE2 is downregulated by SARS-CoV-2 S protein binding to ACE2, this may explain why cardiac impairment is observed to be a significant characteristic of patients who die from COVID-19 [32] Fig. 5 .
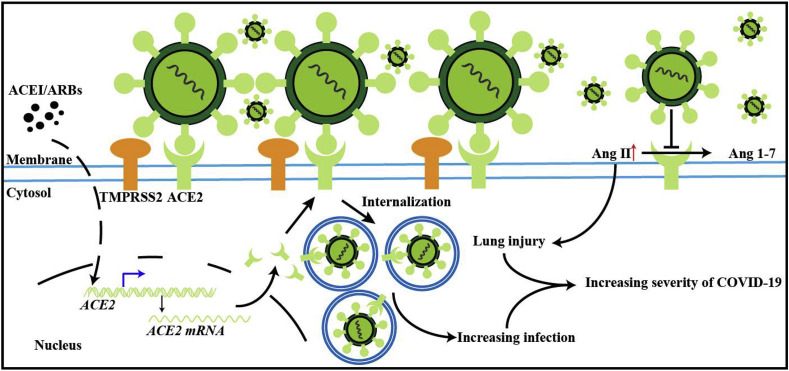
Fig. 5.
Effect of ACE2 and ARBs on SARS-CoV-1 or SARS-CoV-2 infection. This illustrates a proposed mechanism of the effects of ACE2 in COVID-19 infection. SARS-COV-2 virus uses the ACE2 receptor to gain entry into the cell, leading to the increase in proinflammatory cytokines and the development of cytokine storm, as well as increased viral replication (see Fig. 4). TMPRSS2 assists in S protein priming. ARBs may potentially increase the expression of ACE2, leading increased binding of SARS-CoV-2 and greater proinflammatory cytokine production. SARS-CoV-2 may at the same time downregulate ACE2, which leads to an increase in angiotensin 2 mediated lung injury. The negative regulatory activity of ACE2 is reduced by SARS-CoV-2 and leads to worsening severity of illness.
Incidentally, besides the ACE2 receptor, the TMPRSS2 serine protease has been implicated in S protein priming, and it has been proposed as a potential therapeutic target. Additionally, DPP-4 was found to be an entry receptor for MERS, but this has not yet been shown in SARS-CoV-2. A further consideration and potential target for treatment may relate to the role of ADAM-17. While ACE2 is a membrane bound protein, it is cleaved by ADAM-17 leading to shedding to form soluble ACE2. ADAM-17 therefore may lead to a decrease in membrane bound ACE2, reducing its regulatory activity. AT1R upregulates ADAM-17, so use of an ARB may potentially block shedding and reduction of membrane bound ACE2. However, it is believed that the majority of ACE2 is membrane bound and that the decrease resulting from shedding would not significantly affect ACE2 activity to begin with [33].
In conclusion, COVID-19 is a global pandemic that has already caused over 110,000 deaths worldwide so far. Due to human-human transmission of the virus, cases have easily spread across borders and has led to the need to implement disruptive quarantine procedures. Our current study found that there was no significant difference in ACE2 expression between a healthy population and patients with chronic airway disease. The activation of neutrophils, NK cells, Th17 cells, Th2 cells, Th1 cells, dendritic cells, TNFα secreting cells can be induced by overexpression of ACE2 leading to a severe inflammatory response. A schematic model is showed in Fig. 4. These findings may help clinicians and researchers gain more insight into the pathogenesis and use this to design therapeutic strategies for COVID-19. Note that all results in this study are based on data mining and basic science and translational studies are required to confirm these models.
Author statement
This work has not been submitted elsewhere for consideration. All figures are original. No permissions are required.
Ethical statement
The authors declare that they have no competing interests.
Declaration of competing interest
None.
Acknowledgements
The work was supported by 2019-nCoV tackling project of Chengdu Science and Technology Bureau, China (2020-YF05-00003-SN) and CAMS Innovation Fund for Medical Sciences, China (2016-I2M-1-003).
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong N., Yang X., Ye L., Chen K., Chan E.W.-C., Chen S. Genomic and protein structure modelling analysis depicts the origin and pathogenicity of 2019-ncov, a new coronavirus which caused a pneumonia outbreak in wuhan, China. F1000Res. 2020;9:121. [Google Scholar]
- 4.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 5.Ye Y.H., Chen K., Jin K.K., Zhang Y.F., Chen L. [progress on surgical treatment for femoral head-preservering in the precollapse stage of femoral head necrosis] Zhong Guo Gu Shang. 2017;30:287–292. doi: 10.3969/j.issn.1003-0034.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Batlle D., Wysocki J., Soler M.J., Ranganath K. Angiotensin-converting enzyme 2: enhancing the degradation of angiotensin ii as a potential therapy for diabetic nephropathy. Kidney Int. 2012;81:520–528. doi: 10.1038/ki.2011.381. [DOI] [PubMed] [Google Scholar]
- 7.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ace2) converts angiotensin i to angiotensin 1-9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 8.Dan, H.; Gilbert, M.; Borman, R.; Clark, K.L. Quantitative mrna expression profiling of ace 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 532, 0-110. [DOI] [PubMed]
- 9.Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275, 33238-33243. [DOI] [PubMed]
- 10.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Goor H.V. Tissue distribution of ace2 protein, the functional receptor for sars coronavirus. A first step in understanding sars pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Sui J., Huang I.-C., Kuhn J.H., Radoshitzky S.R., Marasco W.A., Choe H., Farzan M. The s proteins of human coronavirus nl63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ace2. Virology. 2007;367:367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L., Ding Y., Zhang Q., Che X., He Y., Shen H., Wang H., Li Z., Zhao L., Geng J. Expression of elevated levels of pro‐inflammatory cytokines in sars‐cov‐infected ace2+ cells in sars patients: relation to the acute lung injury and pathogenesis of sars. J. Pathol.: J. Pathol. Soc. Great Britain and Ireland. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G., Wang L.-G., Han Y., He Q.-Y. Clusterprofiler: an r package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Rockx B., Baas T., Zornetzer G.A., Haagmans B., Sheahan T., Frieman M., Dyer M.D., Teal T.H., Proll S., van den Brand J. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83:7062–7074. doi: 10.1128/JVI.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L., Ding Y., Zhang Q., Che X., He Y., Shen H., Wang H., Li Z., Zhao L., Geng J. Expression of elevated levels of pro-inflammatory cytokines in sars-cov-infected ace2+ cells in sars patients: relation to the acute lung injury and pathogenesis of sars. J. Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer D.D., Kandasamy S., Paim F.C., Langel S.N., Alhamo M.A., Shao L., Chepngeno J., Miyazaki A., Huang H.-C., Kumar A. Protein malnutrition alters tryptophan and angiotensin-converting enzyme 2 homeostasis and adaptive immune responses in human rotavirus-infected gnotobiotic pigs with human infant fecal microbiota transplant. Clin. Vaccine Immunol. 2017;24 doi: 10.1128/CVI.00172-17. e00172-00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J., Kim Y.-S. Effect of hiv-1 tat on the formation of the mitotic spindle by interaction with ribosomal protein s3. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-27008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang C.-Y., Kim H.D., Zhang X., Chang J.-S., Kim J. Ribosomal protein s3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 2012;429:57–62. doi: 10.1016/j.bbrc.2012.10.093. [DOI] [PubMed] [Google Scholar]
- 20.Qaradakhi T., Gadanec L.K., McSweeney K.R., Tacey A., Apostolopoulos V., Levinger I., Rimarova K., Egom E.E., Rodrigo L., Kruzliak P. The potential actions of angiotensin-converting enzyme ii (ace2) activator diminazene aceturate (dize) in various diseases. Clin. Exp. Pharmacol. Physiol. 2020;47:751–758. doi: 10.1111/1440-1681.13251. [DOI] [PubMed] [Google Scholar]
- 21.Ferrario C.M., Jessup J., Chappell M.C., Averill D.B., Brosnihan K.B., Tallant E.A., Diz D.I., Gallagher P.E. Effect of angiotensin-converting enzyme inhibition and angiotensin ii receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Ye Y., Gong H., Wu J., Yuan J., Wang S., Yin P., Ding Z., Kang L., Jiang Q. The effects of different angiotensin ii type 1 receptor blockers on the regulation of the ace-angii-at1 and ace2-ang(1-7)-mas axes in pressure overload-induced cardiac remodeling in male mice. J. Mol. Cell. Cardiol. 2016;97:180–190. doi: 10.1016/j.yjmcc.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Soler M.J., Ye M., Wysocki J., William J., Lloveras J., Batlle D. Localization of ace2 in the renal vasculature: amplification by angiotensin ii type 1 receptor blockade using telmisartan. Am. J. Physiol. Ren. Physiol. 2009;296:F398–F405. doi: 10.1152/ajprenal.90488.2008. [DOI] [PubMed] [Google Scholar]
- 24.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F. Differential downregulation of ace2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus nl63. J. Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia H. Pulmonary angiotensin-converting enzyme 2 (ace2) and inflammatory lung disease. Shock. 2016;46:239–248. doi: 10.1097/SHK.0000000000000633. [DOI] [PubMed] [Google Scholar]
- 27.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ace2) in sars coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu H., Xie Z., Li T., Zhang S., Lai C., Zhu P., Wang K., Han L., Duan Y., Zhao Z. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 2016;6:19840. doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas M.C., Pickering R.J., Tsorotes D., Koitka A., Sheehy K., Bernardi S., Toffoli B., Nguyen-Huu T.P., Head G.A., Fu Y. Genetic ace2 deficiency accentuates vascular inflammation and atherosclerosis in the apoe knockout mouse. Circ. Res. 2010;107:888–897. doi: 10.1161/CIRCRESAHA.110.219279. [DOI] [PubMed] [Google Scholar]
- 30.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R. A novel angiotensin-converting enzyme-related carboxypeptidase (ace2) converts angiotensin i to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 31.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 32.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ace2: a peptidase in the renin-angiotensin system, a sars receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bitker L., Burrell L.M. Classic and nonclassic renin-angiotensin systems in the critically ill. Crit. Care Clin. 2019;35:213–227. doi: 10.1016/j.ccc.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]