Highlights
-
•
A comprehensive genome and codon usage analysis of SARS-CoV-2 was performed.
-
•
SARS-CoV-2 has a distinct genome composition in relation to other βCoV strains.
-
•
The overall codon usage in SARS-CoV-2 is slightly biased.
-
•
Most of the highly frequent codons are A- and U-ending codons.
-
•
Different codon preferences in SARS-CoV-2 genes in relation to human ones were found.
Keywords: Coronavirus, Evolution, Codon usage, Wuhan, 2019-nCoV, SARS-CoV-2
Abstract
An outbreak of atypical pneumonia caused by a novel Betacoronavirus (βCoV), named SARS-CoV-2 has been declared a public health emergency of international concern by the World Health Organization. In order to gain insight into the emergence, evolution and adaptation of SARS-CoV-2 viruses, a comprehensive analysis of genome composition and codon usage of βCoV circulating in China was performed. A biased nucleotide composition was found for SARS-CoV-2 genome. This bias in genomic composition is reflected in its codon and amino acid usage patterns. The overall codon usage in SARS-CoV-2 is similar among themselves and slightly biased. Most of the highly frequent codons are A- and U-ending, which strongly suggests that mutational bias is the main force shaping codon usage in this virus. Significant differences in relative synonymous codon usage frequencies among SARS-CoV-2 and human cells were found. These differences are due to codon usage preferences.
1. Introduction
The family Coronaviridae consists of four genera, namely, Alphacoronavirus (αCoV), Betacoronavirus (βCoV), Gammacoronavirus (γCoV) and Deltacoronavirus (δCoV) (Chen et al., 2020). Viruses from this family possess a single stranded, positive-sense RNA genome ranging from 26 to 32 kilobases in length (Su et al., 2016). Coronaviruses (CoV) have been identified in several avian hosts, as well as in mammals, including humans, bats, civets, mice, dogs, cats, cows and camels (Clark, 1993; Cavanagh, 2007; Zhou et al., 2018). Although several CoV are pathogenic to humans, most of them are associated with mild clinical symptoms (Su et al., 2016). Nevertheless, two notable exceptions have been described: severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV), a novel βCoV that emerged in southern China in 2002 (Peiris et al., 2004) and Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV), which was first detected in Saudi Arabia in 2012 (Zaki et al., 2012). Before the SARS epidemic, bats were not known to be hosts for CoVs. In the last 15 years, bats have been found to be hosts of more than 30 CoVs. Interactions among various bat species themselves, bat-animal and bat-human interactions, such as the presence of live bats in wildlife wet markets in China, have been proved to be important for interspecies transmission of CoVs (Wong et al., 2019). In fact, both SARS-CoV and MERS-CoV likely originated in bats, and genetically diverse CoVs that are related to these viruses were discovered in bats worldwide (Cui et al., 2019). Previous studies in different bat species from China permitted the identification of at least 41 new βCoV, all of them were Rhinolophus spp. bat-CoV (Lin et al., 2017).
At this moment, in Wuhan, the capital city of Hubei province of the People’s Republic of China, an outbreak of atypical pneumonia caused by a novel coronavirus (SARS-CoV-2) is currently underway. The outbreak appears to have started from a zoonotic transmission at a market in Wuhan where animals and meat were sold (Chan et al., 2020a). As February 26th, 2020, there have been 77,780 cases of SARS-CoV-2 confirmed in China, including 2666 deaths (WHO, 2020a). In addition, SARS-CoV-2 has been reported in 33 countries outside China, with 2459 cases confirmed and 34 deaths (WHO, 2020a). Very recent studies revealed that SARS-CoV-2 can be considered a new human-infecting βCoV (Lu et al., 2020).
The World Health Organization declare this 2019-nCoV outbreak as a public health emergency of international concern on January 30th, 2020 (WHO, 2020b) and the disease caused by this specific virus species have recently been designated as COVID-19 (Coronavirus Infectious Disase-19) (WHO, 2020a). In this sense, the Coronavirus Study Group of the International Committee on Taxonomy of Viruses (ICTV), formally recognizes this virus as a relative to severe acute respiratory syndrome SARS-CoVs and designates it as severe acute respiratory syndrome coronavirus 2: SARS-CoV-2 (Gorbalenya et al., 2020).
In order to gain insight into the emergence, evolution, adaptation and spread of the SARS-CoV-2 viruses, a comprehensive analysis of genome composition and codon usage of βCoV circulating in China was performed.
2. Material and methods
2.1. Sequences
Available and comparable complete genome sequences of 81 βCoV strains isolated in China, including SARS-CoV-2 as well as βCoV isolated from different hosts, were obtained from GenBank database (available at: http://www.ncbi.nlm.nih.gov). For accession number, strain name, host and date of isolation, see Supplementary material Table 1. For each strain ORFs1a + ORF1b + S+E + N+M were concatenated. Sequences were aligned using the MACSE program (Ranwez et al., 2011). MACSE algorithm is a useful tool for accommodating sequencing errors and other biological deviations from the coding frame (Ranwez et al., 2011). The alignment is available upon request. The dataset comprised a total of 725,433 codons.
2.2. Data analysis
Codon usage, amino acid usage, dinucleotide frequencies, base composition, the relative synonymous codon usage (RSCU) (Sharp and Li, 1986), total GC content, GC content in the third position of the codon (GC3s) and effective numbers of codons (ENC) were calculated using the program CodonW (written by John Peden) as implemented in the Galaxy server version 1.4.4 (Afgan et al., 2018).
The relationship between compositional variables and samples was obtained using multivariate statistical analyses. Principal component analysis (PCA) is a type of multivariate analysis that allows a dimensionality reduction. Singular Value Decomposition (SVD) method was used to calculate the principal components (PC). Unit variance was used as scaling method. By the same approach, Heatmaps were also constructed, which is a data matrix for visualizing values in the dataset by the use of a color gradient. This gives a good overview of the largest and smallest values in the matrix. Rows and/or columns of the matrix are clustered so that sets of rows or columns rather than individual ones can be interpreted. PCA and Heatmaps analysis were done using the ClustVis program (Metsalu and Vilo, 2015).
The RSCU values of human cells were obtained from Kazusa database (available at: http://www.kazusa.or.jp/codon/).
To study codon usage preferences in SARS-CoV-2 in relation to the codon usage of human cells, we employed the codon adaptation index (CAI) (Sharp and Li, 1987). CAI was calculated using the approach of Puigbo et al. (2008a) (available at: http://genomes.urv.es/CAIcal) for human cells. This method allows to compare a given codon usage (in our case, SARS-CoV-2) to a predefined reference set (human). A statistically significant difference among CAI values was addressed applying a Wilcoxom & Mann-Whitney test (Wessa, 2012). In order to discern if statistically significant differences in the CAI values arise from codon preferences, we used e-CAI (Puigbo et al., 2008b) to calculate the expected value of CAI (eCAI) at the 95 % confident interval. A Kolmogorov-Smirnov test for the expected CAI was also performed (Puigbo et al., 2008b).
3. Results
3.1. Trends in compositional properties across βCoV strains circulating in China
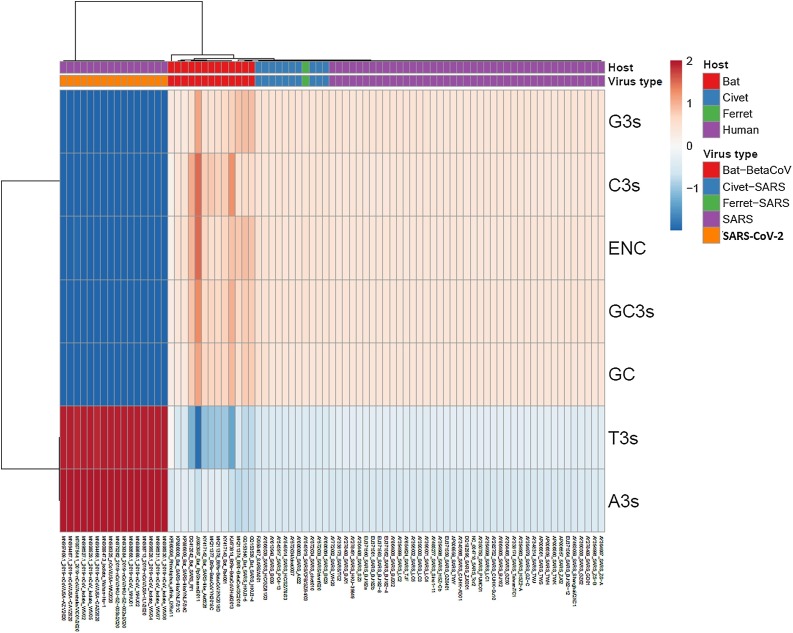
In order to study the genetic composition of SARS-CoV-2 emerging in China, 81 ORFs sequences from βCOV strains isolated in China (including SARS-CoV-2 and SARS-CoV from humans and βCoV isolated from bats, civets and ferrets) were aligned and the nucleotide frequencies at third codon position, total GC content, GC content at the third codon position, ENC and dinucleotide frequencies were established for all strains ORFs and PCA was performed. The results of these studies are shown in Fig. 1 .
Fig. 1.
Genome composition of βCoV strains. Heatmap of frequencies of G, A, T and C at the third codon position, GC total content, GC content at the third codon position and effective number of codons (ENC) in βCoV ORFs is shown. Unit variance scaling was applied. Each column corresponds to a different βCoV strain, who’s host and virus type are shown in the upper part of the figure. Both rows and columns are clustered using correlation distance and average linkage.
Heatmap analysis on genomic composition of all βCoV enrolled in these studies revealed a distinct genome composition of SARS-CoV-2 strains isolated in China, in relation to all other strains isolated from humans as well as bats, civets or ferrets (see Fig. 1).
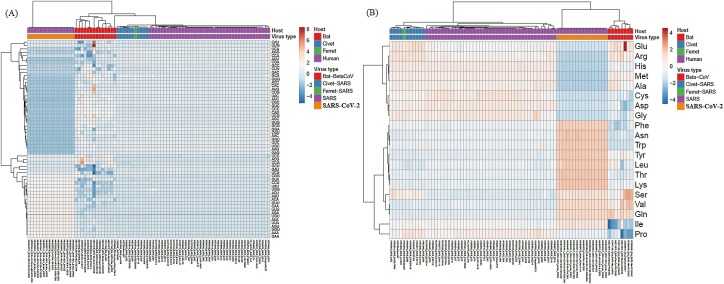
To study if these differences in genomic composition can be observed at codon and amino acid usage, these variables were established for all βCoV strains enrolled in these studies and their relations were observed by Heatmap analysis. The results of these studies are shown in Fig. 2 . Significant differences in codon and amino acid usage was observed among SARS-CoV-2 strains and all other βCoV strains included in these studies (see Fig. 2). Among β-CoV strains isolated from bats, a significant variation was also observed. Linkage analysis suggests a closer relation among SARS-CoV-2 and β-CoV isolated from bats, and a more distant relation with SARS-CoV and other β-CoV isolated from civet and ferret enrolled in these analyses (Fig. 2).
Fig. 2.
Heatmaps of codon and amino acid usage in βCoV ORFs. Unit variance scaling was applied. Each column corresponds to a different βCoV strain, who’s host and virus type are shown in the upper part of the figures. Both rows and columns are clustered using correlation distance and average linkage. In (a) and (b) codon and amino acids usage is shown, respectively.
3.2. The nucleotide frequencies in SARS-CoV-2 genome
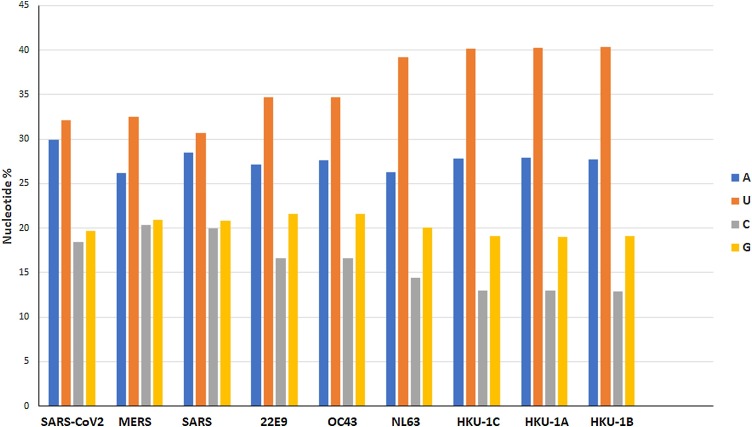
To gain insight into the genomic composition of SARS-CoV-2 strains, the nucleotide frequencies found for this virus where compared to the nucleotide frequencies found for other human CoVs. The results of these studies are shown in Fig. 3 .
Fig. 3.
Nucleotide composition of CoV genomes. Nucleotide composition for SARS-CoV-2 (accession number NC_045512), MERS (JX869059), SARS (NC004718), CoV 229E (KF514433), CoV OC43 (NC005147), CoV NL63 (JX504050), HKU-1A (DQ415914), HKU-1B (DQ415911) and HKU-1C (DQ415912) are shown.
Some general characteristics of CoVs were observed, since the U-frequencies is significantly above average, while the C-frequencies are below the expected frequencies. In the case of purines, A is preferred over G (Fig. 3). As it can be seen in Fig. 3, most variation occurs in the C/U and not the A/G section. Moreover, the results of these studies also revealed the presence of species-specific trends, since the C/U ratio differs profoundly per coronavirus type (see Fig. 3). Comparison of SARS-CoV-2 with the other two recent zoonotic transmission to the human population (SARS and MERS-CoVs) reveals that SARS-CoV-2 is quite extreme with a C-count of 18.4 % (Fig. 3). On the other hand, SARS-CoV-2 has the highest A-count among CoVs enrolled in these studies (29.9 %, mean A-count of 27.6 ± 1.1 for all human CoVs enrolled in these studies) (see Fig. 3).
3.3. General codon usage patterns in βCoV
To study the extent of codon usage bias in SARS-CoV-2, the ENC’s values were calculated for all SARS-CoV-2 strains enrolled in these studies. A mean value of 48.54 ± 2.34 was obtained. Due to the fact that all values obtained were >40, the results of these studies suggest that the overall codon usage among SARS-CoV-2 is similar among themselves and slightly biased. Mean ENC values of 49.11 ± 0.02,
49.66 ± 0.52 and 49.21 ± 0.05 were found for SARS and βCoV isolated from bats and civets, respectively. ENC quantifies how far a codon usage departs from equal usage of synonymous codons and is a measure of codon usage biases in genomes that ranges from 20 (maximal bias) to 61 (unbiased) (Wright, 1990). Although βCoV ENC values are roughly similar, in the case of SARS-CoV-2 strains we observed a range of ENC values from 45.18 to 50.09.
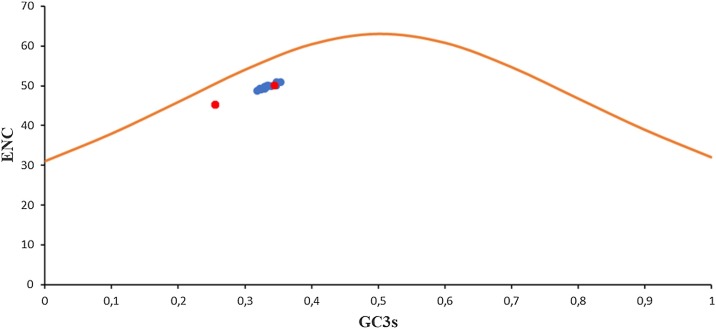
Since codon usage by its very nature is multivariate, it is necessary to analyze the data using different and complementary approaches. An ENC–GC3S plot (ENC plotted against GC content at the third codon position) can be used as a method that quantifies how far the codon usage of a gene departs from equal usage of synonymous codons (Wrigth, 1990). If GC3S is the only determinant factor shaping the codon usage pattern, the values of ENC would fall on a continuous curve, which represents random codon usage. If G + C compositional constraint influences the codon usage, then the GC3S and ENC correlated spots would lie on or below the expected curve (Tsai et al., 2007).
When the ENC–GC3S plot was constructed with values obtained for all 81 βCoV strains enrolled in this analysis (including SARS-CoV-2 strains), all spots lie below the expected curve, indicating that G + C compositional constraints may play a role in all βCoVs codon usage (see Fig. 4 ).
Fig. 4.
Effective number of codons (ENC) used in βCoV ORFs plotted against the GC3S. The orange curve plots the relationship between GC3S and ENC in absence of selection. Red dots show the results obtained for SARS-CoV-2 strains. Blue dots show the results obtained for the rest of βCoV strains enrolled in these studies. N = 81 datapoints.
3.4. Codon usage preferences in SARS-CoV-2
In order to compare the codon usage preferences of SARS-CoV-2 with those of human cells, the RSCU values of the codons in SARS-CoV-2 ORFs were calculated and compared with those of human. The results of these studies are shown in Table 1 .
Table 1.
Codon usage in SARS-CoV-2, displayed as RSCUa values.
| AA | Cod | HC | CoV-2 | SARS | MERS | AA | Cod | HC | CoV-2 | SARS | MERS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 0.92 | 1.42 | 1.23 | 1.28 | Ser | UCU | 1.14 | 2.00 | 1.96 | 2.11 |
| UUC | 1.08 | 0.58 | 0.77 | 0.71 | UCC | 1.32 | 0.44 | 0.42 | 0.71 | ||
| Leu | UUA | 0.48 | 1.66 | 1.04 | 1.21 | UCA | 0.90 | 1.63 | 1.70 | 1.21 | |
| UUG | 0.78 | 1.06 | 1.10 | 1.43 | UCG | 0.30 | 0.11 | 0.23 | 0.18 | ||
| CUU | 0.78 | 1.75 | 1.79 | 1.69 | Pro | CCU | 1.16 | 1.92 | 1.74 | 1.94 | |
| CUC | 1.20 | 0.57 | 0.83 | 0.70 | CCC | 1.28 | 0.31 | 0.40 | 0.65 | ||
| CUA | 0.42 | 0.68 | 0.64 | 0.70 | CCA | 1.12 | 1.63 | 1.70 | 1.21 | ||
| CUG | 2.40 | 0.28 | 0.60 | 0.48 | CCG | 0.44 | 0.14 | 0.16 | 0.18 | ||
| Ile | AUU | 1.08 | 1.54 | 1.72 | 1.71 | Thr | ACU | 1.00 | 1.77 | 1.66 | 1.95 |
| AUC | 1.40 | 0.54 | 0.67 | 0.57 | ACC | 1.44 | 0.39 | 0.59 | 0.68 | ||
| AUA | 0.51 | 0.92 | 0.62 | 0.71 | ACA | 1.12 | 1.65 | 1.57 | 1.18 | ||
| Met | AUG | 1.00 | 1.00 | 1.00 | 1.00 | ACG | 0.44 | 0.15 | 0.18 | 0.17 | |
| Val | GUU | 0.72 | 1.93 | 1.71 | 1.78 | Ala | GCU | 1.08 | 2.19 | 2.08 | 2.06 |
| GUC | 0.96 | 0.58 | 0.67 | 0.76 | GCC | 1.60 | 0.57 | 0.58 | 0.63 | ||
| GUA | 0.48 | 0.89 | 0.83 | 0.72 | GCA | 0.92 | 1.09 | 1.13 | 0.98 | ||
| GUG | 1.84 | 0.59 | 0.78 | 0.73 | GCG | 0.44 | 0.15 | 0.22 | 0.30 | ||
| Tyr | UAU | 0.88 | 1.23 | 1.12 | 1.26 | Cys | UGU | 0.92 | 1.59 | 1.27 | 1.19 |
| UAC | 1.12 | 0.77 | 0.88 | 0.72 | UGC | 1.08 | 0.41 | 0.73 | 0.80 | ||
| TER | UAA | ** | ** | ** | ** | TER | UGA | ** | ** | ** | ** |
| UAG | ** | ** | ** | ** | Trp | UGG | 1.00 | 1.00 | 1.00 | 1.00 | |
| His | CAU | 0.84 | 1.43 | 1.29 | 1.31 | Arg | CGU | 0.48 | 1.45 | 1.77 | 1.82 |
| CAC | 1.16 | 0.57 | 0.71 | 0.68 | CGC | 1.08 | 0.60 | 0.72 | 1.10 | ||
| Gln | CAA | 0.54 | 1.40 | 1.16 | 1.14 | CGA | 0.66 | 0.31 | 0.44 | 0.45 | |
| CAG | 1.46 | 0.60 | 0.84 | 0.86 | CGG | 1.20 | 0.20 | 0.09 | 0.43 | ||
| Asn | AAU | 0.94 | 1.36 | 1.24 | 1.39 | Ser | AGU | 0.90 | 1.46 | 1.17 | 1.33 |
| AAC | 1.06 | 0.64 | 0.76 | 0.60 | AGC | 1.44 | 0.36 | 0.52 | 0.43 | ||
| Lys | AAA | 0.86 | 1.29 | 1.04 | 1.00 | Arg | AGA | 1.26 | 2.64 | 2.08 | 1.34 |
| AAG | 1.14 | 0.71 | 0.96 | 0.99 | AGG | 1.26 | 0.82 | 0.90 | 0.84 | ||
| Asp | GAU | 0.92 | 1.29 | 1.24 | 1.28 | Gly | GGU | 0.64 | 2.36 | 2.02 | 2.05 |
| GAC | 1.08 | 0.71 | 0.76 | 0.72 | GGC | 1.36 | 0.70 | 0.95 | 1.00 | ||
| Glu | GAA | 0.84 | 1.45 | 1.04 | 1.05 | GGA | 1.00 | 0.81 | 0.85 | 0.64 | |
| GAG | 1.16 | 0.55 | 0.96 | 0.95 | GGG | 1.00 | 0.12 | 0.17 | 0.29 | ||
RSCU, relative synonymous codon usage; AA, amino acid; Cod, codons; HC, human cells; CoV-2, SARS-CoV-2; SARS, SARS-CoV; MERS, MERS-CoV. **, termination codons. Highly increased codons in SARS-CoV-2 with respect to human (Δ≥0.30) are shown underlined and in italics. Underrepresented codons are shown in bold.
The frequencies of RSCU in SARS-CoV-2 ORFs are significantly different in relation to human cells. Highly biased frequencies were found for UUU (Phe), UUA (Leu), CUU (Leu), AUU (Ile), AUA (Ile), GUU (Val), UAU (Tyr), CAU (His), CAA (Gln), AAU (Asp), GAA (Glu), UCU (Ser), CCU (Pro), CCA (Pro), GCU (Ala), UGU (Cys), CGU (Arg), AGA (Arg), and GGU (Gly). As it can be seen, most of the highly preferred codons are A and U-ending, while most of the highly underrepresented codons are C and G-endind codons, particularly CG containing codons. These results strongly suggest that mutational bias is a main force shaping codon usage in SARS-CoV-2 (see Table 1). In fact, when the occurrences of dinucleotides are established, they are not random and no dinucleotide is present at the expected frequencies (see Supplementary material Table 2). The relative abundances of CpG and CpC and GpG showed a strong deviation from the expected frequencies (i.e. 1.0) (mean ± S.D. = 0.22 ± 0.00, 0.48 ± 0.00 and 0.66 ± 0.00, respectively) and were markedly under-represented. Cytosine deamination and selection against CpG motifs have been proposed as two independent selection forces that shape codon usage bias in CoV (Woo et al., 2007), suggesting that immune selection may play a role in SARS-CoV-2 codon usage bias. On the other hand, UpU, ApA, UpG are markedly over-used (mean ± S.D. = 1.96 ± 0.00, 1.55 ± 0.00 and 1.40 ± 0.00, respectively) (Supplementary material Table 2). These results indicate that the composition of dinucleotides also determines the variation in synonymous codon usage among SARS-CoV-2 strains. Comparison of SARS-CoV-2 RSCU values with the ones of SARS-CoV (Gu et al., 2004) and MERS-CoV (Chen et al., 2017) revealed a similar pattern for most of the codons (Table 1). Highly underrepresented codons ACG (Thr), GCG (Ala) and GGG (Gly) were observed in all three viruses in relation to human cells (Table 1). On the other hand, significant differences in frequencies of two glutamine codons (GAG and GAA) were observed among SARS-CoV-2 and SARS and MERS-CoVs (Table 1). While GAG codon frequency is significantly different from the expected frequency (i.e. 1.0) in SARS-CoV-2 (0.55), it is not in SARS-CoV and MERS-CoV viruses (0.96 and 0.95, respectively). In the case of GAA, this codon is highly expressed in SARS-CoV-2 (1.45) and its frequency is very near the expected frequency in SARS-CoV and MERS-CoVs (1.04 and 1.05, respectively). These results suggest that although a general codon usage pattern among human βCoV can be found, each virus may evolve to a unique codon usage in its adaptation to the host’cells.
3.5. Codon usage adaptation in SARS-CoV-2
In order to compare the codon usage preferences of SARS-CoV-2 with those of humans, CAI values for all triplets were calculated, using human codon usage as reference set. The results of these studies are shown in Table 2 .
Table 2.
Codon adaptation of SARS-CoV-2 genes in relation to human codon usage, displayed as CAIa values.
| CAI-Hs | |
|---|---|
| SARS-CoV-2 genes | 0.710 ± 0.003 |
| Human genes | 0.809 ± 0.038 |
CAI, codon adaptation index. CAI-Hs; codon adaptation index in relation to H. sapiens reference codon usage set. In all cases, mean ± standard deviation values are shown.
CAI index ranges from 0 to 1, being 1 if the frequency of codon usage by SARS-CoV-2 equals the frequency of usage of the reference set. A mean value of 0.710 was obtained for SARS-CoV-2 genes in relation to human (see Table 2). To evaluate if the differences were statistically significant, we performed a Wilcoxon & Mann-Whitney test. The results of this tests revealed that the differences in CAI values were statistically significant (T = 256, p-value < 0.001).
To discern if the statistically significant differences in CAI values arise from codon preferences (Puigbo et al., 2008a), the expected CAI (e-CAI) values were calculated for SARS-CoV-2 sequences in relation to human codon usage reference set. The e-CAI algorithm (Puigbo et al., 2008b) generated 500 random sequences with the same nucleotide and amino acid composition as the sequences of interest (in this case SARS-CoV-2 sequences). Then, we calculated the CAI values for all of them, and applied a Kolmogorov-Smirnov test for the e-CAI of these random sequences in order to show whether the generated sequences follow a normal distribution. The results of these studies revealed an e-CAI value of 0.719 (p < 0.05). Kolmogorov-Smirnov test revealed a normal distribution of the generated sequences (Kolmogorov-Smirnov test of e-CAI value of 0.028, which is below critical value of 0.061). Taking all these results together, our studies revealed that the CAI values for SARS-CoV-2 genes are different from the CAI values obtained from human cells. Again, these results suggest that these differences are related to codon usage preferences.
4. Discussion
On January 30th 2020, the World Health Organization declared the current SARS-CoV-2 outbreak a public health emergency of international concern (WHO, 2020a). To gain insight into the biology and evolution of emerging SARS-CoV-2, a comprehensive analysis of genome composition, codon and amino acid usage of βCoV strains isolated in China from humans, bats, civets and ferret hosts was performed, including SARS-CoV-2 strains recently isolated from current outbreak.
The results of these studies revealed that SARS-CoV-2 strains enrolled in these analyses have a distinct genome composition in relation to other βCoV strains isolated from human (SARS-CoV), bats, civets and ferrets (see Fig. 1). This is in agreement with very recent studies revealing that SARS-CoV-2 is sufficiently divergent from SARS-CoV to be considered a new human-infecting βCoV (Lu et al., 2020; Zhu et al., 2020). This distinct genomic composition is also reflected in its codon and amino acid usage patterns (see Fig. 2). Moreover, correlation distances and average linkage suggests a closer relation among SARS-CoV-2 and bats βCoV isolated in China and a more distant relation to SARS-CoV or SARS-like βCoV isolated from civets or ferrets (see Fig. 2). This is in agreement with recent results suggesting that bats might be the original host of this virus, an animal sold at the seafood market in Wuhan (Lu et al., 2020; Chan et al., 2020b). This speaks of the importance of bats as a reservoir of potential emerging CoV. Moreover, significant numbers of new βCoV have been discovered in Chinese bats species, particularly Rhinolophus affinis (Lin et al., 2017). Interestingly, a clear degree of variation in codon and amino acid usage was observed among βCoV isolated from bats included in these studies (see Fig. 2). Nevertheless, bats might represent an intermediate host facilitating the emergence of the SARS-CoV-2 in humans (Wong et al., 2019). More studies will be needed to address this important issue.
In these studies, a biased nucleotide composition was found for SARS-CoV-2 genome (Fig. 3). This bias can also have a major influence on derived parameters, as previously demonstrated for other CoVs (Berkhout and Van Hemert, 2015). This is in agreement with the results of this work, since the nucleotide composition of SARS-CoV-2 has a strong influence in the codons that are used by this virus for the translation of its RNA genome. Previous studies on codon usage in RNA viruses have shown that mutational pressure is the major factor in shaping codon usage patterns in comparison with natural selection (Jenkins and Holmes, 2003; Wang et al., 2011). Although mutational pressure is still a major driving force, it is certainly not the only evolutionary force that might be considered in RNA viruses. In these studies, a significant A genomic content was found in SARS-CoV-2 in comparison with other human CoVs (Fig. 3). Previous studies done in Human Immunodeficiency virus (HIV) found that this virus has an A-rich genome and this property has been proposed to help the virus to avoid recognition by the innate immune system (Vabret et al., 2012). This could provide a strong selective pressure on retroviruses as well as many others RNA viruses, including CoVs (Kindler and Thiel, 2014; van Hemert et al., 2014). Moreover, previous studies done in HIV suggest that an RNA genome with A-rich domains may provide a molecular signature that is recognized during virus replication (van Hemert et al., 2013). In the context of CoV, A-rich regions, like the Transcription Regulation Sequence (TRS), which is involved in transcription of sub-genomics RNAs have been established (Pyrc et al., 2004). This suggest that nucleotide bias may serve distinct biological function in SARS-CoV-2 as well as in other CoVs and is in direct relation to the characteristic codon usage of these viruses (Berkhout and Van Hemert, 2015).
A mean ENC value of 48.54 ± 2.34 was obtained for SARS-CoV-2 strains enrolled in these studies, suggesting that the overall codon usage among SARS-CoV-2 is similar among themselves and slightly biased. This is in agreement with mean ENC values obtained for other CoV, like SARS-CoV (ENC = 48.99) (Gu et al., 2004); Bovine Coronavirus (BCoV) (ENC = 43.78) (Castells et al., 2017); MERS-CoV (ENC = 55.50) (Alnazawi et al., 2017) or Avian CoV (ENC = 51.33) (NSP2; Brandao, 2013). ENC-GC3s plot of the values obtained for SARS-CoV-2 revealed that all spots cluster below the expected curve, suggesting that G + C compositional constraints play a role in SARS-CoV-2 (see Fig. 3).
In these studies, significant differences in RSCU frequencies among SARS-CoV-2 and human cells were found (Table 1). A strong bias toward A and U ending codons was found. This in agreement with very recent studies revealing a significant predominance of A and U at third codon positions in CoV genomes (Sheikh et al., 2020; Kandeel et al., 2020). These results also suggest that these differences are related to codon usage preferences. Previous studies have shown that both cytosine deamination and selection of CpG-suppressed clones are the major factors that shape codon bias in CoV genomes (Woo et al., 2007).
MERS-CoV codon usage revealed a bias among hydrophobic amino acids, being CCG (Pro) and GUU (Val) the least and most frequently used codons (Chen et al., 2017). The same results were found in this work for SARS-CoV-2 ORFs, since CCG y GUU resulted to be the least and most frequently hydrophobic codons (11 and 352 times, respectively). Regarding hydrophilic amino acids, the least and most frequently used codons in MERS-CoV were CGG (Arg) and GAU (Asp), respectively (Chen et al., 2017). These codons were also the least and most frequently hydrophilic codons used in SARS-CoV-2 ORF’s (11 and 310 times, respectively). Similarly, CpG and UpU dinucleotide frequencies resulted to be the lowest and highest frequencies found in MERS-CoV (Chen et al., 2017). The same results were obtained in these studies on SARS-CoV-2, since CpG and UpU were the lowest and highest dinucleotide frequencies found (0.22 and 1.96, respectively) (see Supplementary material Table 2). Again, these analyses revealed that genomic composition affects the codon usage pattern in both, MERS-CoV and SARS-CoV-2 viruses.
5. Conclusions
The results of these studies revealed that SARS-CoV-2 strains enrolled in these analyses have a distinct genome composition in relation to other βCoV strains. This distinct genomic composition is also reflected in its codon and amino acid usage patterns. Most of the highly frequent codons are A- and U-ending, which strongly suggests that mutational bias is the main force shaping codon usage in this virus. Significant differences in RSCU frequencies among SARS-CoV-2 and human cells were found. These differences are due to codon usage preferences.
Funding
This research was funded by Agencia Nacional de Investigación e Innovación; Comisión Sectorial de Investigaciones Científica, Universidad de la República, Uruguay, through Grupos I + D grant and PEDECIBA, Uruguay.
CRediT authorship contribution statement
Fernando L. Tort: Visualization, Investigation, Data curation. Matías Castells: Visualization, Investigation, Data curation. Juan Cristina: Conceptualization, Methodology, Data curation, Writing - review & editing.
Declaration of Competing Interest
None.
Acknowledgements
We acknowledge Drs. Pilar Moreno, Gonzalo Moratorio and Rodney Colina for critical reading of this work.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.197976.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Afgan E., Baker D., Batut B. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnazawi M., Altaher A., Kandeel M. Comparative genomic analysis MERS CoV isolated from humans and camels with special reference to virus encoded helicase. Biol. Pharm. Bull. 2017;40:1289–1298. doi: 10.1248/bpb.b17-00241. [DOI] [PubMed] [Google Scholar]
- Berkhout B., van Hemert F. On the biased nucleotide composition of the human coronavirus RNA genome. Virus Res. 2015;202:41–47. doi: 10.1016/j.virusres.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao P.E. The evolution of codon usage in structural and non-structural viral genes: the case of avian coronavirus and its natural host Gallus Gallus. Virus Res. 2013;178:264–271. doi: 10.1016/j.virusres.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells M., Victoria M., Colina R., Musto H., Cristina J. Genome-wide analysis of codon usage bias in Bovine Coronavirus. Virol. J. 2017;14:115. doi: 10.1186/s12985-017-0780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xu Q., Yuan X. Analysis of the codon usage pattern in Middle East Respiratory Syndrome Coronavirus. Oncotarget. 2017;8:110337–110349. doi: 10.18632/oncotarget.22738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;2020(January (22)) doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.A. Bovine coronavirus. Br. Vet. J. 1993;149:51–70. doi: 10.1016/S0007-1935(05)80210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S. Severe acute respiratory syndrome-related Coronavirus: the species and its viruses – a statement of The Coronavirus Study Group. bioRxiv. 2020 doi: 10.1101/2020.02.07.937862. [DOI] [Google Scholar]
- Gu W., Zhou T., Ma J., Sun X., Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Holmes E.C. The extent of codon usage bias in human RNAviruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Kandeel M., Ibrahim A., Fayez M., Al‐Nazawi M. From SARS and MERS CoVs to SARS‐CoV‐2, moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V. To sense or not to sense viral RNA-essentials of coronavirusinnate immune evasion. Curr. Opin. Microbiol. 2014;20C:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.D., Wang W., Hao Z.Y. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T., Vilo J. Clustvis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P., Bravo I.G., Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct. 2008;3:e38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P., Bravo I.G., Garcia-Vallve S. E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI) BMC Bioinf. 2008;9:e65. doi: 10.1186/1471-2105-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V., Harispe S., Delsuc F., Douzery E.J.P. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0022594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/bf02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. The codon adaptation index – a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh A., Al‐Taher A., Al‐Nazawi M., Al‐Mubarak A.I., Kandeel M. Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J. Virol. Methods. 2020;277 doi: 10.1016/j.jviromet.2019.113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.T., Lin C.H., Chang C.Y. Analysis of codon usage bias and base compositional constraints in iridovirus genomes. Virus Res. 2007;126:196–206. doi: 10.1016/j.virusres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Vabret N., Bailly-Bechet M., Najburg V., Muller-Trutwin M., Verrier B., Tangy F. The biased nucleotide composition of HIV-1 triggers type I interferon response and correlates with subtype D increased pathogenicity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hemert F.J., van der Kuyl A.C., Berkhout B. The a-nucleotide preference of HIV-1 in the context of its structured RNA genome. RNA Biol. 2013;10:211–215. doi: 10.4161/rna.22896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hemert F., van der Kuyl A.C., Berkhout B. On the nucleotide compositionand structure of retroviral RNA genomes. Virus Res. 2014;193:16–23. doi: 10.1016/j.virusres.2014.03.019. [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang J., Zhou J.H., Chen H.T., Ma L.N., Ding Y.Z., Liu W.Q., Liu Y.S. Analysis of codon usage in bovine viral diarrhea virus. Arch. Virol. 2011;156:153–160. doi: 10.1007/s00705-010-0848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessa P. Office for Research Development and Education, version 1.1.23-r7; 2012. Free Statistics Software.http://www.wessa.net [Google Scholar]
- Wong A.C., Li X., Lau S.K., Woo P.C. Global Epidemiology of Bat Coronaviruses. Viruses. 2019;11 doi: 10.3390/v11020174. pii: E174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wong B.H., Huang Y., Lau S.K., Yuen K.Y. Cytosine deamination and selection of CpG suppressed clones are the two major independent biological forces that shape codon usage bias in coronaviruses. Virology. 2007;369:431–442. doi: 10.1016/j.virol.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease (COVID-19) Outbreak. Situation Report – 36, 25 February 2020. Available at: https://www.who.int (Accessed 26 February 2020) [Google Scholar]
- World Health Organization . 2020. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV) Available at: https://www.who.int (Accessed 26 February 2020) [Google Scholar]
- Wrigth F. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;8:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W. A novel Coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;2020(January (24)) doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.