Abstract
Mitomycin C (MC), an antitumor drug, and decarbamoylmitomycin C (DMC), a derivative of MC, alkylate DNA and form deoxyguanosine monoadducts and interstrand crosslinks (ICLs). Interestingly, in mammalian culture cells, MC forms primarily deoxyguanosine adducts with a 1”-R stereochemistry at the guanine-mitosene bond (1”-α) whereas DMC forms mainly adducts with a 1”-S stereochemistry (1”-β) The molecular basis for the stereochemical configuration exhibited by DMC has been investigated using biomimetic synthesis. Here, we present the results of our studies on the monoalkylation of DNA by DMC. We show that the formation of 1”-(β-deoxyguanosine adducts requires bifunctional reductive activation of DMC, and that monofunctional activation only produces 1”-α-adducts. The stereochemistry of the deoxyguanosine adducts formed is also dependent on the regioselectivity of DNA alkylation and on the overall DNA CG content. Additionally, we found that temperature plays a determinant role in the regioselectivity of duplex DNA alkylation by mitomycins: At 0°C, both deoxyadenosine (dA) and deoxyguanosine (dG) alkylation occur whereas at 37 °C, mitomycins alkylate dG preferentially. The new reaction protocols developed in our laboratory to investigate DMC-DNA alkylation raise the possibility that oligonucleotides containing DMC 1”-β-deoxyguanosine adducts at a specific site may be synthesized by a biomimetic approach.
Keywords: Mitomycins, Biomimetic synthesis, Antitumor agent, Oligonucleotides, Diastereoselectivity
Graphical abstract
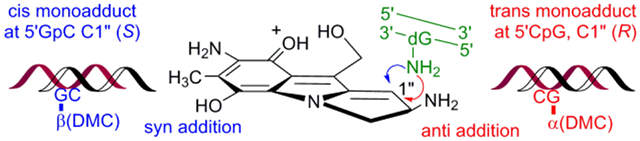
Decarbamoylmitomycin C (DMC) forms DNA deoxyguanosine monoadducts both at GpC and CpG sequences. The major deoxyguanosine adducts produced have the opposite stereochemical configuration at each sequence in mammalian DNA. The GpC sequence is the major target for mammalian DNA (42% CG) in culture cells by this drug and the major adduct has 1”-β (cis) stereochemistry at C1”. In CG rich DNA, formation of 1”-α (trans) adduct is favored.
Introduction
Mitomycin C (MC) is a DNA alkylating agent currently used as a cancer chemotherapeutic drug.[1,2] Its cytotoxicity is attributed to the formation of DNA-adducts, in particular the formation of interstrand crosslinks (ICLs). MC monoadducts and ICLs are formed at the exocyclic amino group of 2′-deoxyguanosine (dG) in DNA.[3,4] 10-decarbamoylmitomycin C (DMC) is a derivative of MC obtained from chemical removal of the 10-carbamoyl group. Despite lacking a carbamate group, DMC is able to form ICLs and generates 10 to 20 times more dG adducts than MC in mammalian culture cells.[5–7] Furthermore, the major dG adducts generated by MC and DMC have opposite stereochemistry at C1 of the guanine-mitosene bond: C1”-R (trans or 1”-α) for MC and C1”-S (cis or 1”-β) for DMC (Figure 1: MC major adducts are 1a, 2a and the α-ICL 3a,[8,9] whereas DMC forms primarily the epimers 2b and the β-ICL 3b).[6,7] Another common DNA adduct formed by MC and DMC at the 2-amino group of dG results from the alkylation of DNA by 2,7-diaminomitosenes 7a and 7b (Figure 1), the major metabolites of MC and DMC.[10]
Figure 1.
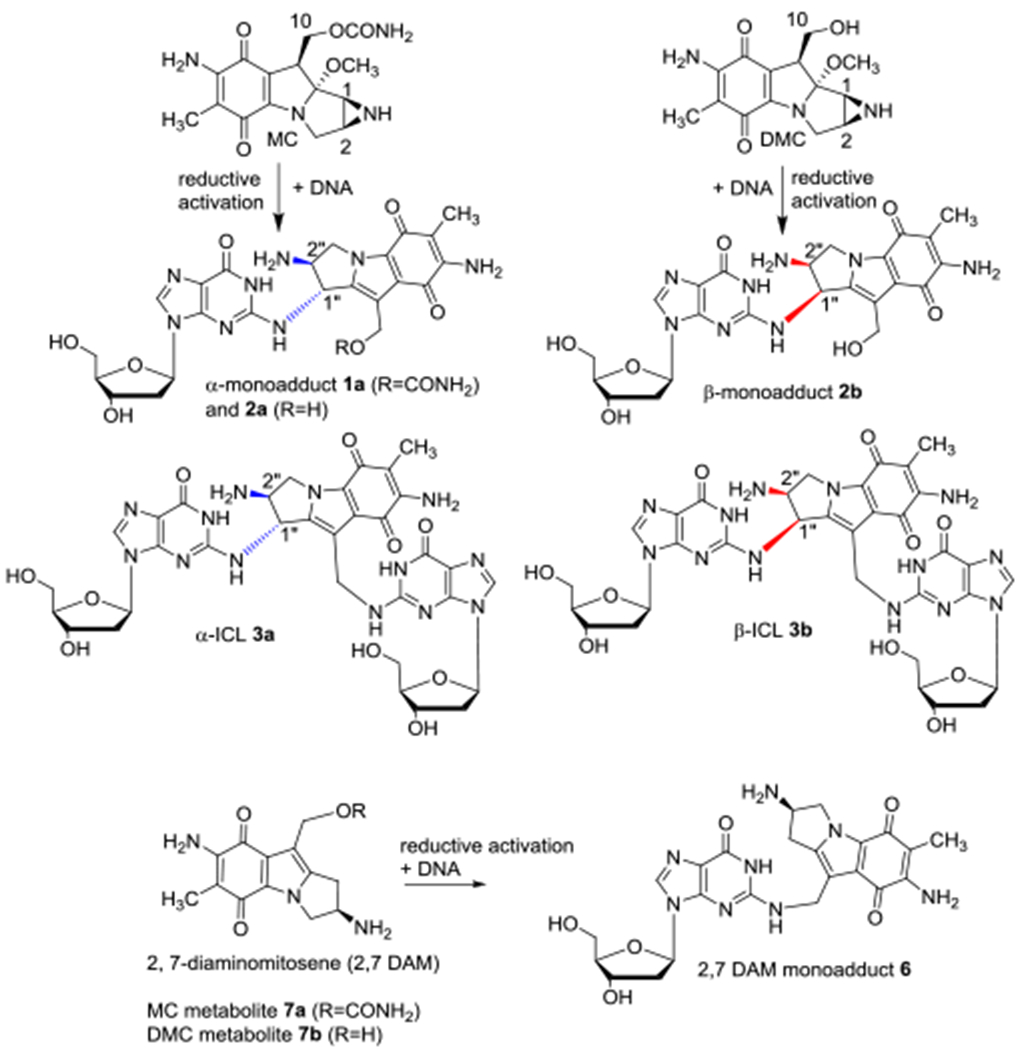
Structure of the major mitomycin C (MC), decarbamoylmitomycin C (DMC) and 2, 7-diaminomitosenes 7a and 7b deoxyguanosine adducts detected in cells.
Both MC and DMC alkylation processes are dependent on the reductive activation of their quinone systems. These complex processes consist of 2 competing activation pathways (monofunctional and/or bifunctional) yielding 2 different DNA-reactive electrophiles (illustrated in Scheme 1A i.e., species 11 and 13).[11,12] The DNA adducts formed in cells by MC indicate that the reductive activation occurs through the bifunctional mechanism. In this case, both mitomycins’ alkylating centers, the 1,2-aziridine and 10-carbamate for MC or 10-hydroxyl for DMC, can react with the exocyclic amino groups of deoxyguanosine residues located on opposing DNA strands to generate ICLs.[12]
Scheme 1.
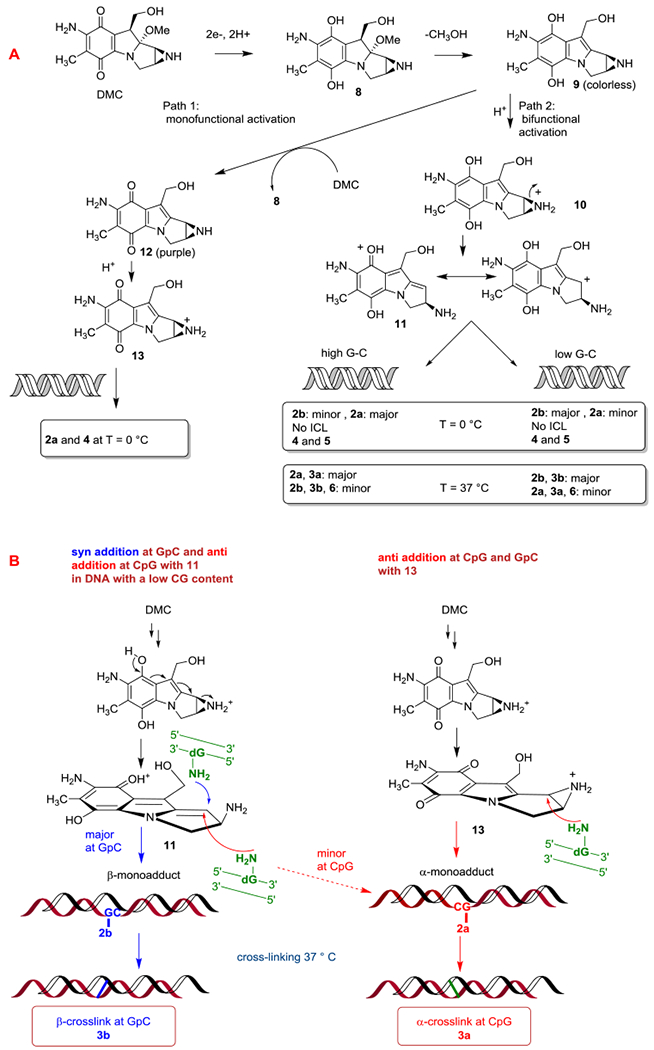
(A) DMC-DNA adducts produced from the monofunctional and bifunctional activation pathways of DMC with different types of DNA, (B) syn and anti addition of dG with 11 and anti addition only with 13.
The role of the opposite stereochemistry of MC and DMC DNA adducts in the cytotoxicity of the two drugs is not fully understood. Strong evidence supports the hypothesis that the structure of ICLs generated by chemotherapeutics plays an important role in the mode of action of these drugs. The importance of the structure of ICLs in repair and recognition processes during replicative and non-replicative events has been previously evidenced.[13,14] In line with this hypothesis, the structure of the two diastereomeric ICLs generated by MC and DMC (3a and 3b, Figure 1) could be at the origin of the contrasting biochemical responses exhibited by the two drugs,[15] exemplified as follows: (i) DMC induces cell cycle arrest at lower concentrations when compared with MC[16] (ii) Removal of DMC 1”β adducts 2b and 3b is faster than removal of MC 1” α adducts 2a and 3a in EMT6 cells;[7] (iii) The DNA-adducts generated by DMC treatment (1”-β-ICL) rapidly activate a p53-independent signal transduction pathway.17
The comparison of the local DNA structure of MC and DMC adducts with the biological activity of both drugs should provide invaluable insight into the structure-activity relationship of the stereoisomeric DNA lesions. Such studies require the use of modified oligonucleotides containing a specific adduct at a single position of their base sequences.[18–22]
However, examination of the biochemical effects of DMC adducts has been severely limited by the inability to prepare sufficient quantities of oligonucleotides containing 2b and 3b. Our long-term objective is to synthesize duplex oligonucleotides bearing DMC 1”- β-monoadduct 2b and ICL 3b to elucidate the role of the adducts’ β stereochemistry at C1” in the drug biological activity. Consistent with this objective, the present work focuses on the molecular mechanisms responsible for the stereochemical outcome of DMC-DNA adducts formation. Previous DNA alkylation reactions between DMC and oligonucleotides or plasmids yielded only trans (1”-α) adducts (2a and 3a, Figure 1) as major adducts.[23–25] However, we have recently developed experimental conditions capable of generating DMC cis (1”-β) adducts 2b and 3b (Figure 1). We have established that DMC is able to generate crosslinks 3a and 3b both at CpG and GpC sequences and that the crosslinking reaction is diastereospecific and diastereodivergent: Only the 1”S- diastereomer of the initially formed monoadduct can form crosslinks at GpC sequences, and only the 1”R- diastereomer of the monoadduct can form crosslinks at CpG sequences.[26] We report here, that the diastereoselective formation of 1”-β adducts by DMC requires bifunctional activation of DMC. Furthermore, by using oligonucleotides with C·G base pairs flanked by different base sequences, we establish that: (i) DMC guanine monoadducts 2b (1”-β) and 2a (1”-α) are produced simultaneously in most sequence contexts; (ii) the regioselectivity and diastereoselectivity of DNA monoalkylation by DMC are interdependent; (iii) single-stranded DNA favors the α-monoadduct 2a. We also report that, in DNA with a low CG content, formation of 2b (1”-β adduct) is favored overall whereas formation of 2a (1”-α adduct) is favored in DNA with a high CG content. Finally, we found that temperature plays a determinant role in the regioselectivity of duplex DNA alkylation by mitomycins: At 0 °C, both deoxyadenosine (dA) and deoxyguanosine (dG) alkylation occur whereas at 37 °C, DMC alkylates dG preferentially.
Results and Discussion
Results
DMC 1”-β Monoadduct (2b) is Produced only under Bifunctional Activation Conditions (Figure 2 and Table S1, supporting information).
Figure 2.
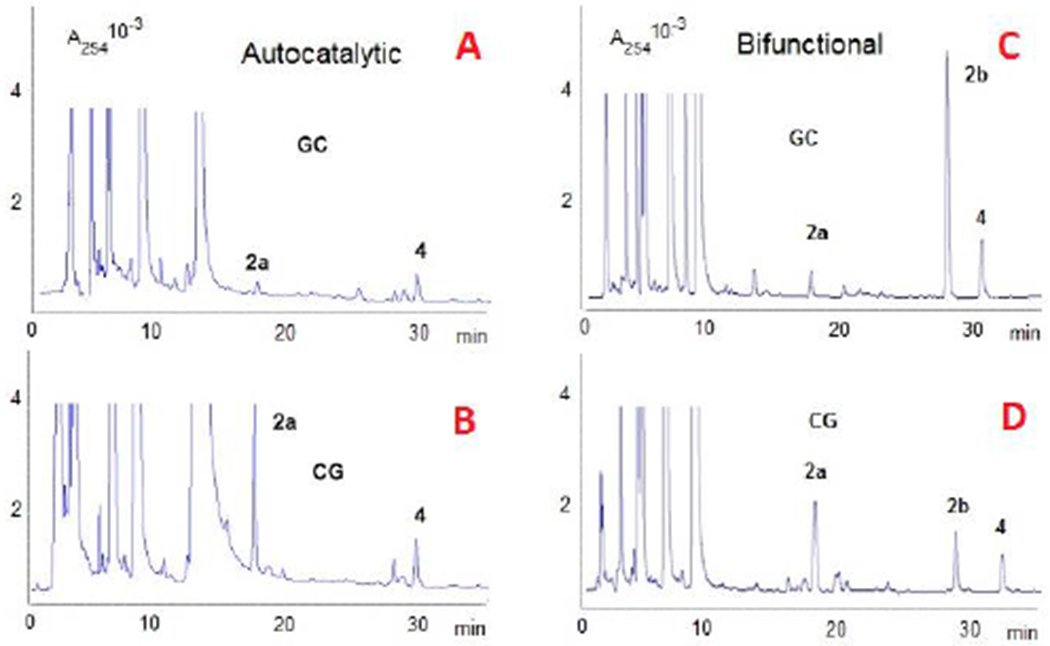
HPLC analysis of digests of DMC-oligonucleotide complexes formed under DMC autocatalytic activation (A and B) and bifunctional activation (C and D). Oligonucleotides used: duplex 5’-ATATAGCTATAT-3’ (A and C) and duplex 5’-ATATACGTATAT-3’ (B and D). 4: adenine adduct.25
The α-adducts 1a (from MC) and 2a (from DMC) were formed in the reaction of duplex oligonucleotides a: 5’-ATATACGTATAT-3’ and b: 5’-ATATAGCTATAT-3’ with MC or DMC under autocatalytic conditions at 0 °C (Figures 2A, 2B and Table S1, supporting information). In both cases, the frequency of adduct formation was higher at 5’-CpG steps. The 1”-β adducts 1b (1b (C1” S) is the stereoisomer of 1a (C1” R)) and 2b (Figure 1) were not detected, and the frequency of adduct formation was higher in the case of MC compared to DMC. No interstrand crosslink (3a or 3b, Figure 1) was detected at that temperature. These results are in agreement with previous observations.[23]
We then used a newly developed bio-mimetic protocol to reduce DMC fully and quickly so that DNA alkylation is achieved by 11 (i.e., under bifunctional conditions, scheme 1) rather than 13 (i.e., under auto-catalytic conditions, scheme 1). This protocol is described in the experimental section (Oligonucleotide Alkylation by DMC under Bifunctional Conditions, protocol 1). Bifunctional conditions require a complete and fast reduction of DMC to minimize the monofunctional pathway; path 1 (Scheme 1A). Reduction under bifunctional conditions was achieved by adding an excess of sodium dithionite and a low concentration of DMC under anaerobic conditions. This can be monitored visually: The reaction mixture becomes colorless as soon as sodium dithionite is added and remains so during the course of the reaction, indicating that compound 9, which is colorless, is not reoxidized to aziridomitosene 12, which is purple (Scheme 1A).
Under such bifunctional conditions, DMC monoadduct 2b was formed both at 5’-CpG and at 5’-GpC steps (Figure 2C and 2D). Oligonucleotide b (GpC central sequence) yielded adduct 2b as the major adduct, in contrast to oligonucleotide a (CpG central sequence). The formation of DMC 1”-β-monoadduct 2b was favored at low concentrations of DMC (Figure S1, Supporting information). As the concentration of DMC decreases, the autocatalytic pathway is necessarily disfavored and this correlates with an increase in the ratio 2b/2a. Taken together, these results indicate that the formation of 2b (1”-β stereochemistry) is only observed when the dG alkylation step at C1” is achieved by intermediate 11 (generated by bifunctional activation) and that the monoalkylation of duplex DNA by aziridinomitosene 13 (generated under autocatalytic conditions) produces mainly 2a (1”-α stereochemistry) at 0 °C (Figure 2 and Scheme 1B). Additionally, deoxyadenosine (dA) monoadducts 4 and 5 (Chart 1) were also detected at 0 °C. A full description of the formation of these dA adducts is the subject of a separate manuscript.[27]
Chart 1.
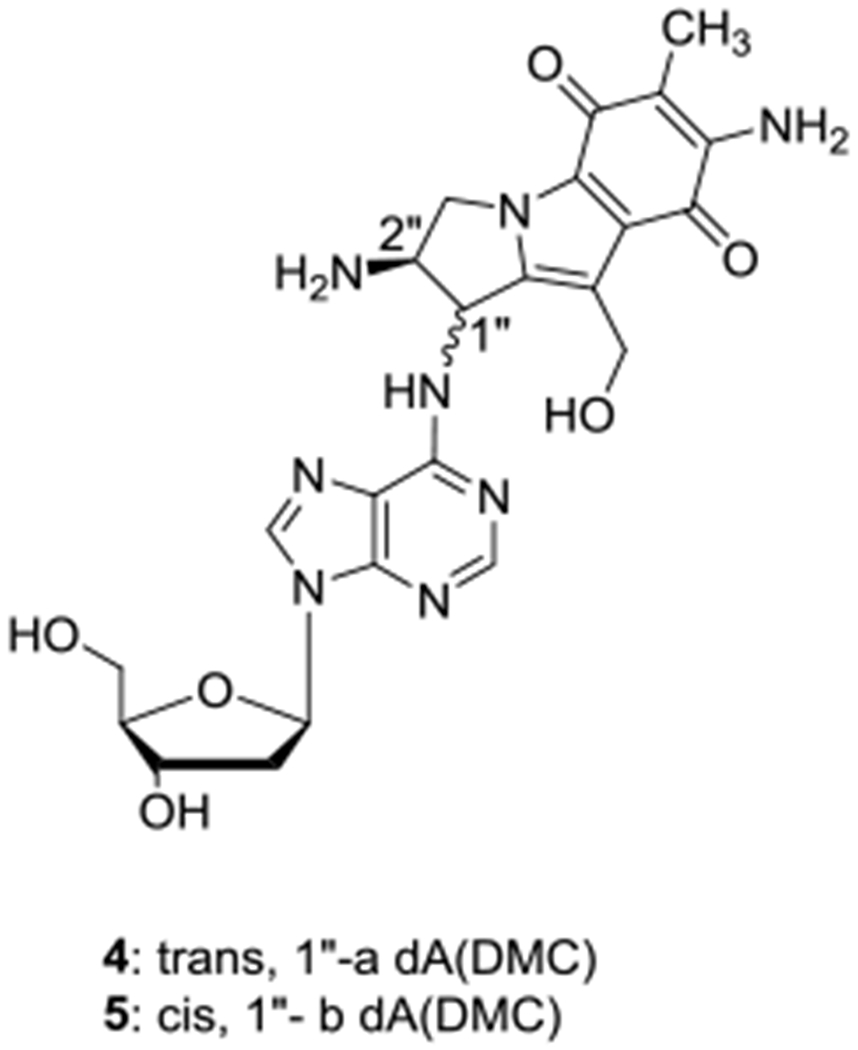
Structure of deoxyadenosine-DMC adducts detected in reactions between DMC and oligonucleotides at 0 °C.
Enzymatic activation (NADH-cytochrome c reductase) at pH 5.8 yielded adduct 2b as a minor adduct and only at 5’-GpC steps (Supporting information, Table S2). This implies that adduct 2b is produced less efficiently under enzymatic activation in our model system. This is probably due to the slower kinetic of enzymatic reduction and the presence of unreduced DMC in the reaction mixture. Consequently, the autocatalytic pathway is probably also activated (path 1, Scheme 1A). This supports the rationale that both 11 (from bifunctional activation, path 2, Scheme 1A) and 13 (from auto-catalytic activation, path 1, Scheme 1A) contribute to dG monoalkylation at pH 5.8 in this case.
Finally, alkylation of single stranded oligonucleotides under bifunctional conditions yielded both 2a and 2b. However, in single stranded oligonucleotides, the frequency of adducts 2a and 2b was similar both in a 5’-CpG and 5’-GpC sequence context and, in both cases, the major adduct detected was 2a (Table 1).
Table 1.
Frequencies of Guanine Adducts in Single Strand DNA and Duplex DNA under Bifunctional Conditions (0.32 μM DMC). Calculation of Percent Yields of Adducts 2a and 2b is Described in the Experimental Section and in the Supporting Information Section (page S8).
| Single Strand DNA | 2a | 2b | 2a+2b | 2b/2a |
|---|---|---|---|---|
| 5’-ATTATCGTTATT-3’ | 0.36 (± 0.04) | 0.076 (± 0.007) | 0.44 | 0.21 |
| 5’-ATTATTGCTATT-3’ | 0.31 (± 0.02) | 0.12 (± 0.02) | 0.43 | 0.36 |
| Duplex DNA | ||||
| 5’-ATATAGCTATAT-3’ | 0.12 (± 0.03) | 1.25 (± 0.13) | 1.38 | 10 |
| 5’-ATATACGTATAT-3’ | 0.79 (± 0.06) | 0.37 (± 0.04) | 1.16 | 0.47 |
In summary, when duplex DNA is alkylated at 0 °C by monofunctionally activated DMC, the 1”α adduct 2a is the only dG adduct observed, and it is formed more efficiently at 5’-CpG steps than at 5’-GpC steps. When the same reactions are performed under bifunctional activation conditions both the 1”-β dG adduct 2b and the 1”-α adduct 2a are formed. The diastereoselectivity of the alkylation reaction is sequence-dependent: Isomer 2b is strongly favored at 5’-GpC steps (10-fold higher frequency), while isomer 2a is favored at CpG steps, but with lower diastereoselectivity (2-fold higher frequency). These results prove that the structure of the alkylating agent generated by reduction of DMC determines the stereochemical outcome of DNA alkylation by DMC. Aziridinomitosene 13 only produces α adducts whereas reduced DMC species 11 produces both α and β adducts (Scheme 1B). No crosslink was detected at 0 °C.
The Adduct Profile of DNA Alkylation is Temperature and pH Dependent. Development of Reaction Conditions for the Formation of DNA Crosslinks by DMC (Table S3, supporting information and Figures 3 and 4).
Figure 3.
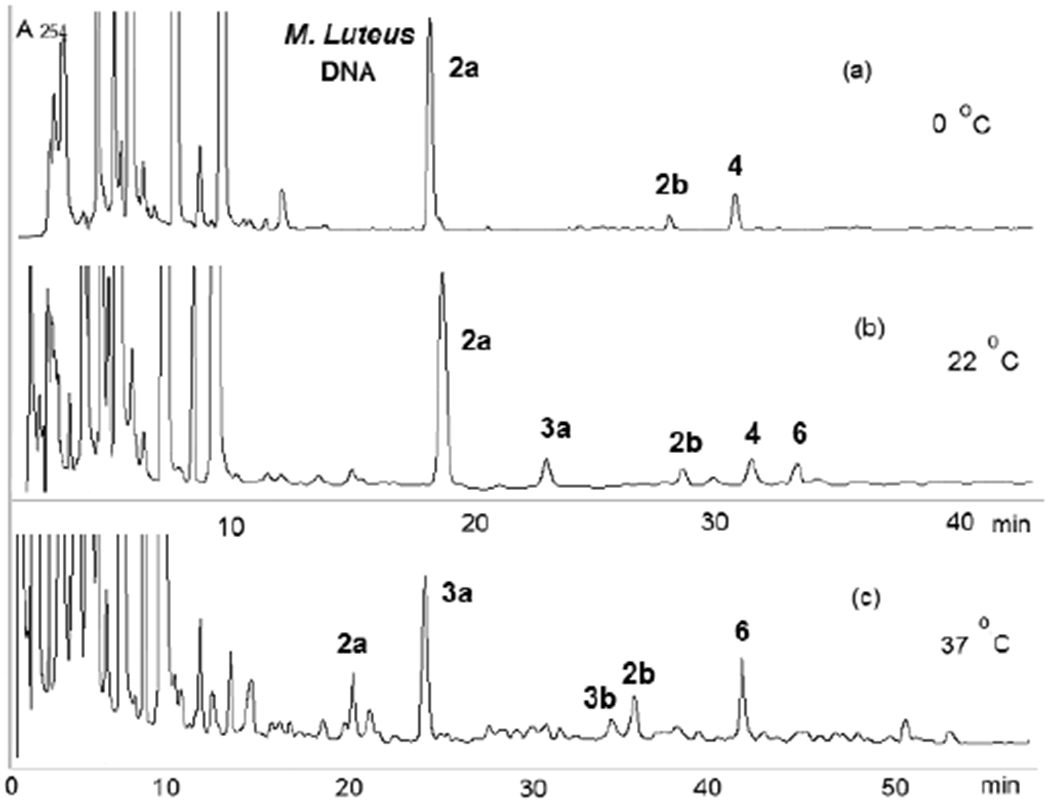
HPLC analysis of digests of DMC-oligonucleotide complexes formed under bifunctional conditions with M. Luteus DNA at various temperatures. (c) The elution gradient was modified at 37 °C (6 to 18% acetonitrile in 105 minutes) for a better separation of 3b and 2b.
Figure 4.
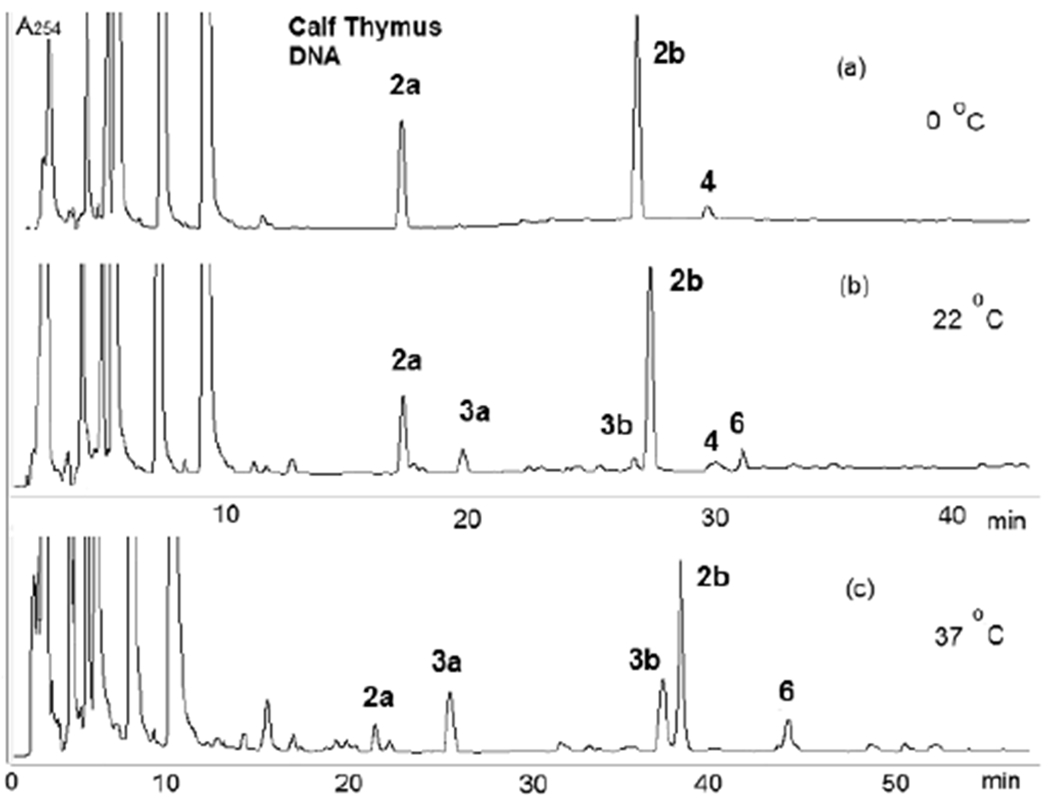
HPLC analysis of digests of DMC-oligonucleotide complexes formed under bifunctional conditions with calf thymus DNA at various temperatures. (c) The elution gradient was modified at 37 °C (6 to 18% acetonitrile in 105 minutes) for a better separation of 3b and 2b. Reprinted from reference 26.
Treatment of Calf Thymus DNA and M. Luteus DNA with DMC at Various Temperatures (Figures 3 and 4).
Treatment of calf thymus DNA and M. Luteus DNA with DMC under bifunctional conditions resulted in the formation of dG adducts 2a, 2b, 3a, 3b, 6 (Figure 1) and dA adducts 4 and 5 (Chart 1). The adduct profile of the alkylation reaction depends on the DNA CG content, the reaction temperature, and the pH. The transmonoadduct 2a was the major adduct formed in M. luteus DNA (72% GC) but the cis-monoadduct 2b was formed preferentially in calf thymus DNA (42% GC). This is a strong indication that the stereoselectivity exhibited by DMC is dependent and the CG content of the DNA substrate. The formation of the diastereomeric ICLs 3a and 3b with both types of DNA is temperature dependent. At 0 °C, ICLs 3a and 3b (Figures 3 and 4) did not form. In contrast, MC crosslinking is possible at 0-5 °C.[24] Interestingly, adducts 4 and 5 (deoxyadenosine (dA) adducts, Chart 1) were observed at 0 °C in the reaction mixtures.[27] At room temperature, crosslinking was observed and the trans crosslink 3a was formed with both M. luteus DNA and calf thymus DNA (Figures 3 and 4). The cis crosslink 3b was detected with calf thymus DNA, but not with M. luteus DNA. In addition, dG-N2-2,7 DAM adduct 6 (Figure 1) was detected with both types of DNA. The 2,7-DAM adduct 6 was also observed in cells treated with DMC as a minor adduct.[10] Higher temperature (37 °C) promoted the formation of ICLs and adducts 3a and 3b were formed in higher proportion (Figures 3 and 4). The trans adduct 3a was the major ICL adduct formed with M. Luteus DNA, while the cis-adduct 3b was the major ICL adduct formed with calf thymus DNA. No deoxyadenosine adducts were detected at higher temperature in duplex DNA. The adduct pattern resulting from the alkylation of calf thymus DNA with DMC (Figure 4) shows a remarkable resemblance to the adduct pattern from treatment of EMT6, FAA-A fibroblasts, and MCF-7 cells with DMC.[7] The major difference is that the ratio 2a/3a is higher in cells than in our model systems.
Influence of pH on DMC Alkylation of Calf Thymus DNA (Table S3, supporting information).
A higher ratio crosslink/monoadduct (3b/2b) was observed when reactions were performed at a more acidic pH (pH 4.5). However, under those conditions, the adduct profile presented a high proportion of products derived from N7 guanine monoalkylation, in addition to the expected N2 adducts (data not shown). This is in agreement with previous observations on the reaction of acid activated mitomycins with DNA.[28] At higher pH (pH 7.5), as anticipated, the ratio crosslinks/monoadducts was slightly lower than at pH 5.8 and the 2,7 DAM adduct 6 could not be detected.[12] At pH 5.8, the adduct profile from DMC treated calf thymus DNA was very similar to the adduct profile observed in cells (Figure 4). In addition, this pH also maximized the amount of crosslinks and it was chosen for further reactions (Table S3, supporting information). The study of the influence of temperature and pH on the alkylation of DNA by bifunctionally activated DMC discussed above resulted in the development of reaction conditions that produce significant yields of crosslink adducts.
These reaction conditions permitted us to perform a rigorous study on the regioselectivity and diastereospecificity of the formation of DNA interstrand crosslinks by DMC. The results of this work have been presented recently.[26]
Sequence-Selectivity of Guanine Monoalkylation by Bifunctionally Activated DMC.
The results discussed above indicate that formation of 2b is favored in DNA with a low CG content (as in calf thymus DNA) but that formation of 2a is favored in DNA with a high proportion of CG base pairs (as in M. Luteus DNA). In order to gain insight into the regioselectivity of deoxyguanosine monoalkylation in mammalian DNA (42% CG), oligonucleotides in 17 different sequence contexts (Table 2) were hybridized with their complementary strand and reacted with reduced DMC.
Table 2.
Oligonucleotides used for the Regioselectivity and Stereoselectivity of DMC-DNA monoalkylation
| Sequence | Sequence | ||
|---|---|---|---|
| 14 | 5’-ATTATGGCTATT 3’-TAATACCGATAA |
23 | 5’-ATTATCGATATT 3’-TAATAGCTATAA |
| 15 | 5’-ATTATCGCTATT 3’-TAATAGCGATAA |
24 | 5’-ATTATAGATATT 3’-TAATATCTATAA |
| 16 | 5’-ATTATAGCTATT 3’-TAATATCGATAA |
25 | 5’-ATTATTGATATT 3’-TAATAACTATAA |
| 17 | 5’-ATTATTGCTATT 3’-TAATAACGATAA |
26 | 5’-ATTATGGGTATT 3’-TAATACCCATAA |
| 18 | 5’-ATTATGGTTATT 3’-TAATACCAATAA |
27 | 5’-ATTATCGGTATT 3’-TAATAGCCATAA |
| 19 | 5’-ATTATCGTTATT 3’-TAATAGCAATAA |
28[a] | 5’-ATTATAGGTATT 3’-TAATATCCATAA |
| 20 | 5’-ATTATAGTTATT 3’-TAATATCAATAA |
29 | 5’-ATTATGGTATT 3’-TAATACCATAA |
| 21 | 5’-ATTATTGTTATT 3’-TAATAACAATAA |
30 | 5’-TTATTGCAATTA 3’-AATAACGTTAAT |
| 22 | 5’-ATTATGGATATT 3’-TAATACCTATAA |
31 | 5’CGCGCGCGCGCG 3’-CGCGCGCGCGCG |
29 = 18.
Further Optimization of Reaction Conditions for the Alkylation of Oligonucleotides by DMC.
We performed an optimization of the bifunctional activation of DMC with the aim of increasing the frequency of adducts, thus facilitating their detection and improving the precision in their quantification. These reaction conditions ensured that DMC was kept fully reduced in solution to minimize the autocatalytic route. We hypothesized that repeated additions at low concentrations of fully reduced DMC would result in a significant increase of the yield of adducts, and we tested our hypothesis using the conditions described as protocol 2 in the experimental part. This new protocol gave consistently higher yields and was used in our subsequent investigations of DMC regioselectivity. Reactions were performed at least twice and the adduct pattern observed with oligonucleotides 14-31 was highly reproducible. Reactions were performed at 0 °C because carrying the reaction at this temperature prevented the formation of crosslinks, thus facilitating the study of the monoalkylation reaction. The 1”-β dG adduct 2b was always produced alongside 1”-α dG adduct 2a and the alkylation frequency of 2a and 2b was determined in each case (Figures 5 and 7).
Figure 5.
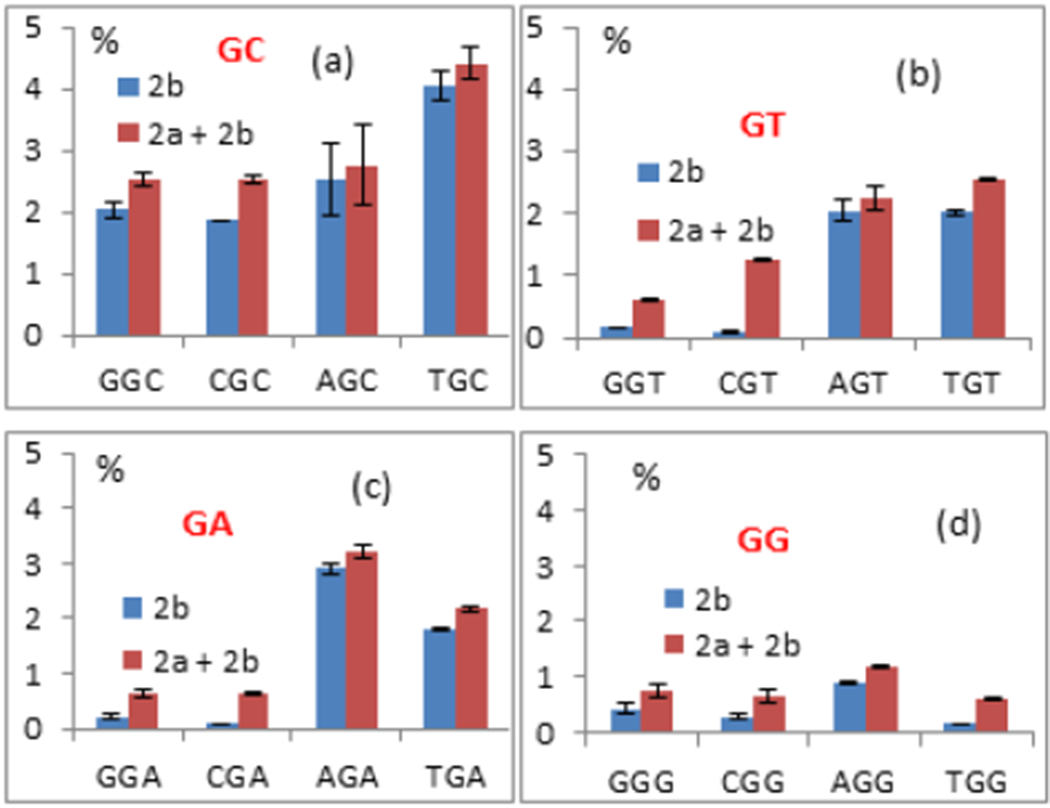
Figures a, b, c and d: Frequency of DMC monoadduct 2b and of the sum of adducts (2b + 2a) formed under bifunctional conditions with various oligonucleotides. The central sequence of each oligonucleotide is indicated on each horizontal axis: (a) oligonucleotides 14, 15, 16, 17; GpC step (b) oligonucleotides 18, 19, 20, 21; GpT step (c) oligonucleotides 22, 23, 24, 25; GpA step (d) oligononucleotides 26, 27, 28, 29; GpG step
Figure 7.
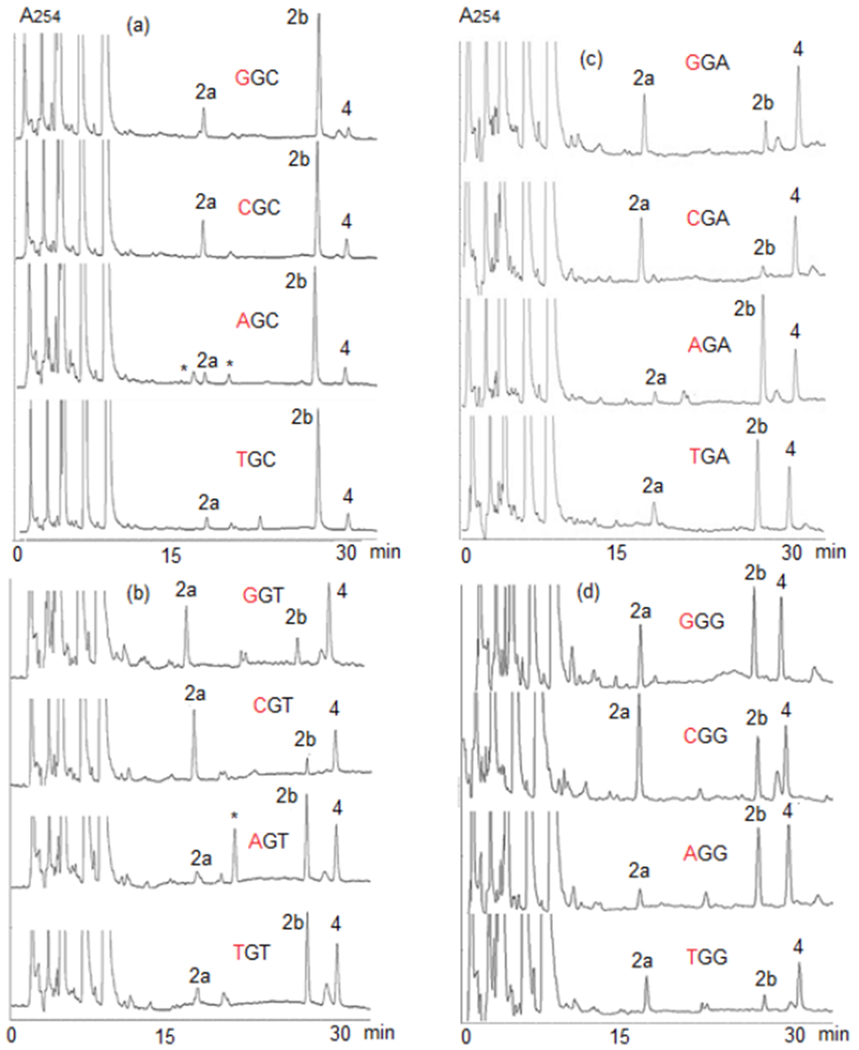
HPLC of digests of DMC-oligonucleotide complexes formed under bifunctional conditions (a) GpC step; from top to bottom: oligonucleotides 14, 15, 16, 17 (b) GpT step; from top to bottom: oligonucleotides 18, 19, 20, 21 (c) GpA step; from top to bottom: oligonucleotides 22, 23, 24, 25 (d) GpG step; from top to bottom: oligonucleotides 26, 27, 28, 29=18.
Regioselectivity and Diastereoselectivity in the Alkylation of Oligonucleotides by DMC.
DMC generated adducts 2b and 2a in all oligonucleotides studied. Calculation of percent yields for the formation of 2a and 2b is described in the experimental section as well as in the supporting Information section (page S8) and in our previous publication.[26] The frequency of guanine alkylation was highest at XpGpC steps (X = C, G, A, T) and XpGpY (X, Y = A, T) (Figures 5 and 7). TpGpC steps produced the highest yield of dG alkylation and also the highest diastereoselectivity in the formation of 2b: Oligonucleotide 17 (TpGpCpT) yielded 4.0% of 2b (for a total of 4.4% of guanine adducts, Figure 5a and 6) and oligonucleotide 30 (TpGpCpA) 4.3% of 2b (for a total of 4.8% of guanine adducts, data not shown). Both 5’-GpG and 5’-CpG sequences gave the lowest yield of dG adducts and the highest diastereoselectivity for 2a (Figures 5, 6 and 7). This indicates that there is a competition between guanine-N2 nucleophilic attack on the a and β face of 11 at 0 °C in all sequence contexts studied. In oligonucleotides 14-30, DMC regioselectivity and stereoselectivity are interdependent, i.e., oligonucleotides with a high frequency of dG alkylation are also oligonucleotides yielding a high ratio 2b/2a (Figures 5–7 and S2, supporting information). Formation of 2b and therefore, guanine-N2 nucleophilic attack on the β face of reduced DMC is favored at XpGpC steps (X=C, G, A, T; oligonucleotides 14, 15, 16, 17, 30, Figures 6, 7 and Figure S2, supporting information) and XpGpY (X,Y=A,T; oligonucleotides 20, 21, 24, 25, Figures 6, 7 and S2, supporting information).
Figure 6.
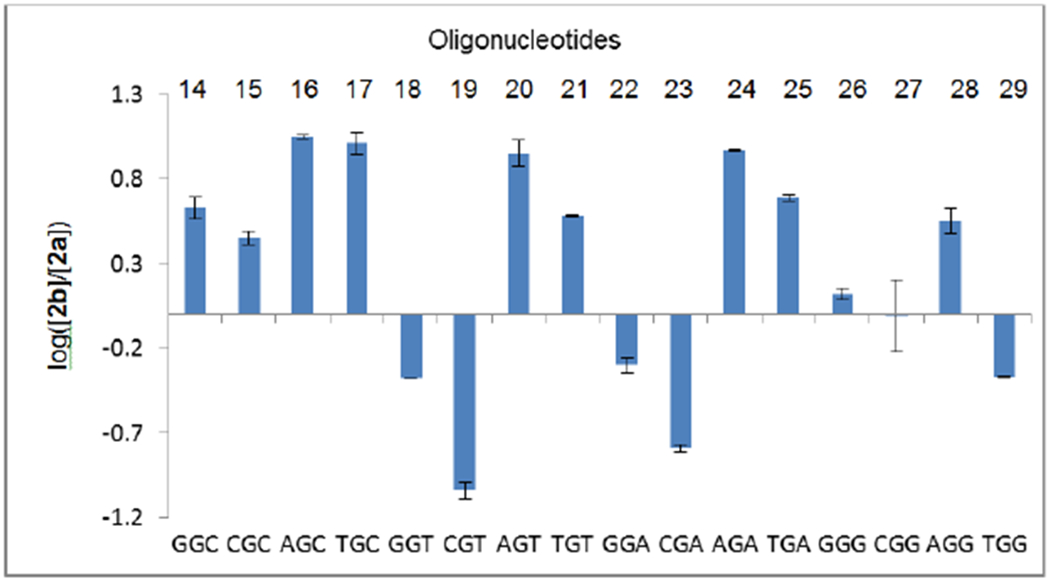
Log([2b/2a]) in oligonucleotides 14-29 (18=29).
Formation of 1”-α adduct 2a and, therefore, guanine-N2 nucleophilic attack on the β face of reduced DMC is favored at CpGpX (with X = A, T, G; oligonucleotides 19, 23 and 27, Figures 6, 7 and S2, supporting information) and TpGpGpY (with Y = A, T; oligonucleotides 18 and 22, Figures 6, 7 and S2, supporting information). In oligonucleotides 14-30 (low CG content), formation of 1”-β adduct 2b is favored overall. Taken together, these results indicate that there is interdependence between regioselectivity and diastereoselectivity in the alkylation of guanine residues by DMC. The alkylation of dG is favored at NpGpN (where N = A or T) and at GpC steps, and this effect correlates with the stereoselective formation of the 1”-β monoadduct. Conversely, the stereoselective formation of the 1”-α monoadduct is favored at CpG and GpG steps.
The Ratio α/β Adducts is a Function of DNA G-C Content.
Results from CT DNA and M. Luteus DNA alkylation with DMC above, indicate that the overall DNA CG content has an influence on the stereoselectivity of DMC guanine alkylation. This phenomenon was further investigated by examining the ratio α/β adducts versus the CG content of all DNA substrates studied (Figure 8).
Figure 8.
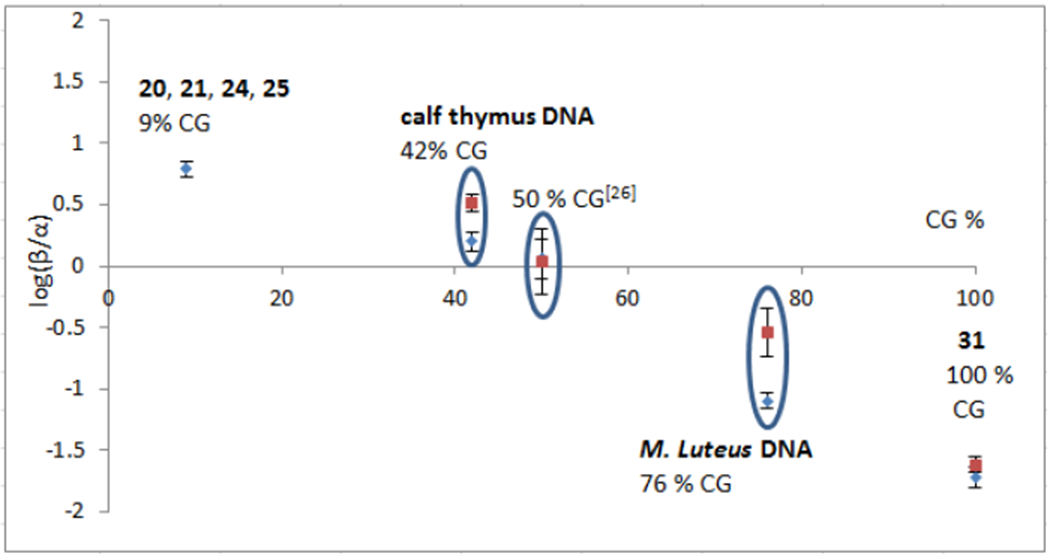
Variation of the ratio between DMC alpha adducts and beta adducts (log (([2a]+[3a])/([2b]+[3b]))) formed under bifunctional conditions according to DNA C-G content. Data are taken from experiments with oligonucleotides 20, 21, 24, 25 (9% C-G), with calf thymus DNA (42% C-G), oligonucleotides with a 50% CG content,26 M. Luteus DNA (76% C-G content) and oligonucleotide 31 (100% C-G content). Blue diamonds represent the ratio 2b/3b for 0 °C reactions (only monoadducts formed) and red squares represent the ratio (2b+3b)/(2a+3a) for 37 °C reactions (monoadducts and crosslinks formed).
The data shown in Figure 8 highlights the interdependence between the CG content of DNA and the diastereoselectivity of dG alkylation by DMC: An increase of C-G content correlates with a decrease with the ratio β/α. In DNA with a low CG content, such as calf-thymus DNA, the formation of 1”-β adducts 2b and 3b is favored. However, in DNA with a high C-G content, such as M. Luteus DNA, the formation of 1”-α adducts 2a and 3a is favored. The only crosslink detected with oligonucleotide 31 (100% CG) at 37 °C was 1”-α crosslink 3a. Although the regioselectivity of dG monoalkylation with DMC in DNA with a high C-G content has not been systematically determined in this work, the fact that α-crosslink 3a is the major adduct formed with M. Luteus DNA suggests that, for DNA with a high CG content, the alkylation of DNA by DMC is selective for CpG steps since the crosslink adduct 3a can only be formed at CpG sequences.[26]
Influence of the Exocyclic Amino Group on the Opposite or Adjacent Guanine on DMC Stereoselectivity.
The results discussed above indicate that the 1”-α adduct is favored at most CpG sequences and the 1”-β adduct is favored at most GpC steps. These observations prompted an investigation into the role played by the 2-NH2 group of guanine in the opposite strand (at CpG and GpC steps) in the regio- and stereospecificity of the alkylation of dG by DMC. To this end, one deoxyguanosine in duplexes 17 and 19 was replaced by deoxyinosine either at CpG (duplex 17) or GpC steps (duplex 19). The replacement of deoxyguanosine by deoxyinosine in the opposite strand at GpC in duplex 17 resulted in a significant decrease in the frequency of dG alkylation (−2%), and a significant decrease in the amount of 2b versus 2a (Table 3 and Figure S3, supporting information). In contrast, the replacement of deoxyguanosine by deoxyinosine in the opposite strand at the CpG step in duplex 19 resulted in a small increase in the global yield of dG adducts (+0.5%), and a significant increase in the amount of 2b versus 2a (Table 3 and Figure S4, supporting information). These results show that the 2-amino group on deoxyguanosine of the nonbonding strand contributes to the regio- and stereoselectivity exhibited by DMC at GpC and CpG steps and also suggest the presence of other contributing factors.
Table 3.
Frequencies of Guanine Adducts upon Replacement of the Non-Alkylated Guanine by Inosine at CpG, GpC and GpG steps in oligonucleotides 17, 18, and 19.
| Frequency (%) of the major adducts detected | |||||
|---|---|---|---|---|---|
| Oligonucleotides | 2a | 2b | 2a + 2b | log(2b/2a) | |
| 17 | 5’-ATTATTGCTATT 3’-TAATAACGATAA |
0.36 (±0.04) | 4.04 (±0.3) | 4.41 | 1.05 |
| 5’-ATTATTGCTATT 3’-TAATAACIATAA |
0.52 (±0.05) | 2.13 (±0.19) | 2.64 | 0.61 | |
| 19 | 5’ATTATTCGTATT 3’-TAATAAGCATAA |
1.17 (±0.03) | 0.1 (±0.01) | 1.27 | −1.05 |
| 5’-ATTATTCGTATT 3’-TAATAAICATAA |
0.94 (±0.04) | 0.64 (±0.05) | 1.58 | −0.18 | |
| 18 | 5’-ATTATTGGTATT 3’-TAATAACCATAA |
0.43 (±0.03) | 0.18 (±0.005) | 0.6 | −0.38 |
| 5’-ATTATTIGTATT 3’-TAATAACCATAA |
0.40 (±0.04) | 1.27 (±0.2) | 1.68 | 0.50 | |
| 5’-ATTATTGITATT 3’-TAATAACCATAA |
0.53 (±0.04) | 0.40 (±0.012) | 0.93 | −0.11 | |
Similarly, the role played by the 2-NH2 group on the adjacent deoxyguanosine at GpG steps was investigated (table 3 and Figure S5, supporting information). One deoxyguanosine in duplex 18 was replaced by deoxyinosine either at the 5’ or at the 3’ position. Replacement of deoxyguanosine by deoxyinosine at the 5’ position at GpG resulted in an increase in the frequency of dG alkylation (+1%) by DMC, and in a reversal of stereoselectivity, as 2b became the major adduct (Table 3 and Figure S5, supporting information). This suggests that the presence of the exocyclic 2-NH2 group of the 5’ guanine at GpG steps must be essential for the 1”-α stereoselectivity exhibited by DMC at GpG steps. On the other hand, replacement of deoxyguanosine by deoxyinosine at the 3’ position at GpG steps resulted in smaller variations both in the frequency of guanine alkylation (+0.3%) and in the ratio 2b/2a (2a was still the major adduct) (Table 3 and Figure S5, supporting information). Considered together, these results demonstrate that the exocyclic amino group on deoxyguanosine located at the 5’ position at GpG steps and on the opposite strand at GpC and CpG steps plays an important but not exclusive role in the determination of DMC alkylation frequency and stereoselectivity.
Discussion
The identification of DNA adducts generated in mammalian culture cells upon treatment with MC and DMC showed that the two drugs generate stereoisomeric DNA adducts. The DMC major dG monoadduct and ICL, 2b and 3b, have a 1”-β (cis) stereochemistry at C1” whereas the MC major DNA adducts, 1a, 2a, and 3a have a 1”-α (trans) stereochemistry. DMC also generates 20 to 30 times more monoadducts than MC when administered at the same doses.[7] The aim of this work was to provide a rationale for these observations in order to understand the molecular basis of DMC cytotoxicity. Previous reactions between short oligonucleotides, poly (dG-dC), or plasmids in vitro with reduced DMC produced the 1”-α (trans) adduct 2a as the major monoadduct, while the diastereomeric adduct 2b was not formed or formed in minimal proportions.[25] This is in contrast to what happens in mammalian cultured cells where 2b is the major DMC-DNA adduct formed. We hypothesized that the discrepancy between the adduct patterns observed with cultured cells and the ones obtained using in vitro chemical reactions was a direct consequence of the reductive conditions under which previous DNA alkylation reactions had been performed. Until now, research on the alkylation of DNA by MC or DMC has mostly used sequential additions of substoichiometric amounts of dithionite or slow enzymatic reductions of MC or DMC.[25] Under such conditions, the auto-catalytic monofunctional pathway is promoted (path 1, Scheme 1A),[30] and DNA alkylation is effected by aziridinomitosene 13.[12] To test our hypothesis, we reacted short oligonucleotides containing a CpG or GpC central sequence with DMC using autocatalytic or bifunctional reductive conditions and analyzed the adducts formed. The results showed that when autocatalytic conditions were used, the alkylation of short oligonucleotides by 13 was selective for CpG sequences and more importantly, produced mostly α-adducts (Figures 2A and 2B, Table S1). However, when DMC was reduced under conditions that minimize the formation of 13 and favor the formation of 11 (bifunctional pathway, Scheme 1A), the 1”-β dG adduct 2b was observed, and its formation appeared to be selective for 5’-GpC sequences. These results clearly show that, in the case of DMC, the structure of the electrophilic species influences the stereochemical outcome of DMC DNA alkylation, meaning that 13 (generated under auto-catalytic conditions) produces only 1”-α dG adducts (Figure 2, left) but 11 (generated under bifunctional conditions) is able to generate 1”-β dG adducts (Figure 2, right). In contrast, in the case of MC, the major dG adducts produced under auto-catalytic,[23–25] or bifunctional conditions,[9] have the same α stereochemistry at C1”.
Although reduction of DMC under bifunctional conditions (to generate 11) is a necessary condition for the formation of 1”-β dG adducts, it is not sufficient. The amount of CG base pairs in the DNA substrate also determines the stereochemical outcome of DNA alkylation by DMC (Figure 8). Substrates with a high proportion of CG base pairs favor the formation of 1”-α dG adducts and 1”-α ICLs, whereas substrates with a low proportion of CG base pairs favor the formation of 1 ”-β dG adducts and 1” β -ICLs. The reason for this phenomenon is not clear. AT rich segments tend to adopt very narrow minor grooves and this has implications for drug-DNA interactions.[31] Consequently, the difference in minor groove structures between AT-rich and CG-rich segments may influence DMC stereoselectivity. In general, GC-rich DNA in the highest multicellular organisms constitutes gene-rich actively-transcribed genomic regions. It was recently established that cis-platin, another DNA alkylating agent, targets promoters and regions harboring transcription termination sites in genomic DNA.[32] Our results suggest that, upon treatment with DMC, regions of mammalian DNA with a high CG content are likely to be rich in 1”-α dG adducts whereas regions with a low CG content are likely to be rich in 1”-β dG adducts. Further work is necessary to verify if this uneven ratio of 1”-α and 1”-β adducts is indeed present in DMC treated mammalian DNA and, if so, to thereby understand its biological relevance.
We have previously established that the formation of DNA-DNA crosslinks by DMC is diastereospecific and diastereodivergent.[26] In this manuscript we further investigated the regioselectivity and stereoselectivity of dG monoalkylation by DMC in oligonucleotides with a low CG content. To do so, we treated short oligonucleotides 14–30 with reduced DMC under optimized bifunctional conditions. The results show that the formation of either 1”-α-guanine 2a or 1”-β-guanine 2b is dependent on the local sequence context (Figures 5 and 6) and follows the general rule in our model systems (oligonucleotides 14–30, Figures 5–7): Formation of 2b and dG alkylation are favored at 5’-NpGpN (N = A or T) steps and at 5’-GpC steps (Figures 5–7, oligonucleotides 16, 17, 21, 24, 25). In contrast, at 5’-CpG and 5’-GpG steps, formation of 2a is favored (Figures 5–7, oligonucleotides 18, 19, 22, 23). This demonstrates an interdependence between regioselectivity and stereoselectivity and implies that both the nature of the 3’ and 5’ side of the alkylated dG play a role in determining the regio- and diastereoselectivity for DMC alkylation in DNA with a low CG content (such as mammalian DNA; 42% CG). In contrast, MC alkylates mammalian DNA selectively at 5’-CpG steps and generates the 1”-α-guanine adduct 1a as the major adduct.[7]Our results also show that the frequency of alkylation is higher at GpC versus CpG in oligonucleotides 14–30 (Figure 5) and support the hypothesis that DMC targets GpC sequences over CpG sequences for dG monoalkylation in DNA. However, DMC is overall less regioselective than MC because it also targets NpGpN (N = A or T) sites with high frequency. Therefore, if DMC targets more sequence contexts than MC, the frequency of DMC DNA adducts should be higher than MC when equivalent drug treatment is performed, specifically since the rate of bioactivation of the 2 drugs is similar (assuming cellular uptake is similar for both drugs).[6] This is in agreement with the fact that DMC forms 20–30-fold more DNA monoadducts than MC in mammalian cells.[7] Results from our work also provide a rationale for the observed higher ratio monoadducts/crosslinks displayed by DMC versus MC in culture cells:[6,7] On one hand, DMC targets dG in non-crosslinkable sequences (i.e., NpGpN; N = A, T) with high frequency; on the other hand, the formation of crosslinks is less efficient at GpC steps (the major target of DMC) than at GpC steps (the major target of MC), as we reported earlier.[26] On average, the conversion of monoadduct to crosslink occurs with 31% efficiency at GpC steps and with 80% efficiency at CpG steps. As a result (and because DMC targets GpC sequences over CpG sequences in mammalian DNA), only a low proportion of monoadducts will be able to form interstrand crosslinks.
Finally, we experimentally verified the role of the 2-NH2 group of the non-alkylated guanine in the formation of 2b and 2a at CpG, GpC and GpG steps when DNA is treated by 11. It has been postulated that pre-covalent interactions between the hydroxyl oxygen of 11 (Figure 2) and the amino group of the non-alkylated guanine were responsible for the orientation of DMC in duplex DNA leading to the formation of 1”-β monoadduct 2b.[33] Our results show that the 2-amino group on the guanine of the non-reacting strand contributes to the stereoselectivity exhibited by DMC at GpC and CpG steps, but that it is not the only contributing factor (Tables 3 and 4 and Figures S3–S4, supporting information). We propose that pre-covalent interactions between the exocyclic 2-NH2 group of guanine in the opposite strand and activated DMC largely contribute to placing DMC in an orientation which favors nucleophilic attack on the β face of the drug’s tetrahydropyrrole ring at GpC steps and on the α face of the drug’s tetrahydropyrrole ring at CpG steps. However, the diastereoselectivity is not fully abolished upon replacement of guanosine by inosine in either case, suggesting that other factors may contribute to DMC diastereoselectivity. In contrast, the exocyclic 2-NH2 group on the 5’ guanine is essential for the 1”-α stereoselectivity exhibited by DMC at GpG steps (Table 4 and Figure S5, supporting information). We propose therefore, that pre-covalent interactions involving the 2-NH2 group on the 5’guanine at GpG steps and activated DMC are the main contributors to the orientation of DMC in the duplex so that nucleophilic attack on the α face of the drug occurs preferentially (Table 3 and Figure S5, Supporting information). Therefore, the exocyclic amino groups described above play an important, but not exclusive, role in the diastereoselectivity of the alkylation of DNA by DMC. Two observations indicate that 11 may also form competing pre-covalent interactions with bases other than guanine, in particular, interactions involving deoxyadenosine are likely to occur because: i) DMC 1”-β adducts are always favored when at least one dA is in the 5’ position of dG (as seen with oligonucleotides 14, 15, 16, 17, 20, 24, 28, Figures 5–7) and ii) adenine can react with DMC under certain conditions in duplex DNA to generate stereoisomeric adducts.[27]
What is the biological significance of the fact that MC and DMC target different sequence contexts in mammalian DNA and of the stereochemistry of the DNA adducts formed? We, and others have observed that the biochemical responses triggered by MC and DMC are widely different in cultured cells.[15–17] Our results suggest that the recognition of damaged DNA is likely to occur differently when DNA is alkylated by MC or DMC. Proteins recognize damaged DNA by two mechanisms: The base read out and the shape read out.[34] Since both drugs target different sequences, the recognition process of DNA adducts by base read out is likely to differ in the case of MC and DMC. The shape read out mechanism is also likely to vary if MC and DMC adducts induce different structural DNA distortion.[13] In addition, trans-adducts 2a and 3a formed at CpG by MC and cis-adducts 2b and 3b formed at GpC by DMC may also confer different conformational flexibility near damaged DNA sites. The efficiency of DNA repair has been linked to DNA flexibility near a damaged site.[35] If ICL 3a (CpG) and ICL 3b (GpC) do indeed confer different conformational flexibility to DNA, this may partly explain why the rate of removal of 3b is enhanced compared to the rate of removal of 3a in EMT6 cells treated with MC.[7]
Previous research indicates that the MC and DMC major dG adducts are also likely to be processed differently because of the different stereochemistry of the DNA adducts formed in each case (1”-α versus 1”-β). Various steps in the processing of ICLs in cells and cell extracts are dependent on the structure of the ICL itself during replicative events.[14,36] Furthermore, in the case of DNA monoadducts, recognition and incision by base excision repair are different for stereoisomeric lesions.[37] The structure of oligonucleotides modified with adducts 2a and 3a are known (both monoadduct 2a and crosslink 3a lie in the slightly widened minor groove of a minimally distorted DNA helix)[38,39] but there is no structural information on DMC adducts 2b and 3b at the oligonucleotide level. Therefore, further efforts to produce DMC 1”-β dG adducts 2b and 3b on a larger scale and examination of their local DNA structures are warranted, with the aim of linking the local DNA structure of 1”-α and 1”-β adducts with the DNA damage response triggered by the two drugs.
Lastly, the 2,7 DAM adduct formed by DMC (adduct 6, figure 1) was only detected as a minor adduct and at a low frequency in this work. Previous work has also established that adduct 6 is formed at much lower frequency with DMC compared to MC in culture cells;[7] a likely consequence of the better leaving group ability of the carbamoyl at C10 in 7a compared to the hydroxyl in 7b (Figure 1). The relative low abundance of 6 in culture cells treated with DMC compared to MC correlates with DMC higher toxicity. This is in agreement with the hypothesis stating that formation of adduct 6, a non-cytotoxic DNA adduct, is part of the reductive detoxification pathway of mitomycins.[22,40]
Conclusion
The aim of this work was to determine the factors governing the formation of the 1”-β dG adducts produced by DMC. We have successfully developed conditions which reproduce the pattern of DNA alkylation by DMC observed in cells: In order to produce 1”-β (cis) DNA adducts as major adducts in vitro upon chemical reductive activation, DMC must be reduced in a bifunctional way, DNA must be in the duplex form and the C-G DNA content must be low so that CG base pairs are flanked by A, T residues. Our results also revealed the importance of temperature in the reaction outcome. At 0 °C, only monoadducts are formed and N2-adenine adducts are observed. However, high temperatures promote the formation of crosslinks and of 2,7-DAM adduct. The influence of the structure of DNA adducts generated by MC and DMC with respect to DNA repair and cytotoxicity is still not completely understood. The different sequence selectivity of DMC versus MC and the opposite stereochemistry of the major adducts produced by MC and DMC may play a role in the different biological activity of DMC and MC since it is likely to affect the processing of DNA adducts by cells. While the structure of 1”-β adducts has been elucidated at the nucleoside level, there are no available routes to prepare oligonucleotides bearing a single DMC monoadduct or crosslink with a 1”-β (cis) stereochemistry. The synthesis of such site-specifically modified oligonucleotides is necessary to understand the relationship between the structure of DMC and MC adducts and the activity of the drugs. Our results raise the possibility that such oligonucleotides containing DMC-dG 1”-β adducts may be synthesized by a biomimetic approach.
Experimental Section
Materials.
MC was generously gifted by Professor Maria Tomasz. DMC was synthesized from MC by a published procedure[41]. Phosphodiesterase I (snake venom diesterase (SVD), Crotalus adamanteus venom, E.C. 3.1.4.1.) and alkaline phosphatase (Escherichia coli, EC 3.1.3.1) were obtained from Worthington Biochemical Corp (Freehold, NJ). Nuclease P1 (penicillium citrinum, EC 3.1.30.1), NADH-cytochrome c reductase, NADH, 2’-deoxyadenosine, M. lysodeicticus DNA (M. luteus DNA, type XI; 72% GC) and calf thymus DNA (type XV; 42% GC) were from Sigma Life Sciences (St. Louis, MO). Sep-Pak C-18 cartridges were purchased from Waters Corp (Milford, MA). Oligonucleotides were purchased from Midland Certified Reagent (Midland, TX).
Methods. Quantitative Analysis.
Quantitation of DNA, DMC or MC-DNA adducts was based on UV spectrophotometry using the following molar extinction coefficients: MC and DMC, 21800 (367 nm); M. luteus-DNA, 6000 (260 nm, mononucleotide unit); calf thymus DNA, 6500 (260 nm, mononucleotide); dT, 6600 (254 nm); dA, 14200 (254 nm); MC and DMC monoadducts (1a, 2a, 2b, 4, 5, 6), 24000 (254 nm); MC and DMC ICLs (3a, 3b), 30000 (254 nm).The percent yields of DMC and MC DNA-adducts (1a, 2a, 2b, 3a, 3b, 4, 5 and 6) designate the mole percent adducted guanine (for 1a, 2a, 2b, 3a, 3b and 6) or adenine (for 4 and 5) of total guanine or adenine nucleotide residues present in the oligonucleotides or DNA substrates. A full description of the method is available in previous publications and in the supporting information.[12],[13]
Reproducibility.
Reactions under bifunctional conditions were repeated at least twice independently and average deviation from the mean value is represented with error bars on graphs or in brackets in tables. Autocatalytic activation reactions were only performed once as they were repeats of previous experiments.[23]
Identification of Adducts 1a, 2a, 2b, 3a, 3b, 4, 5 and 6 in Enzymatic Digests.
Adducts 1a, 2a, 2b, 3a, 3b, 4, 5 and 6 were identified by their UV spectra, retention times and co-elution with authentic standards synthesized previously in our laboratory.[10b, 7, 26, 27, 42, 43]
Oligonucleotide Alkylation by DMC and MC under Autocatalytic Conditions.
Oligonucleotide alkylation by DMC and MC under autocatalytic conditions has been previously described and is also available in the supporting information section.[23]
Oligonucleotide Alkylation by DMC under Bifunctional Conditions, (protocol 1)
Self-complimentary duplex oligonucleotides (5’-ATATACGTATAT-3’or 5’-ATATAGCTATAT-3’: 10 A260 units; 330 μg; corresponding to 0.083 μmol) were annealed (55 °C heating for 10 min followed by slow cooling to RT) and then mixed with 0.648 μmol of DMC in 10 mM potassium phosphate buffer, pH 5.8 (355 μL). The mixture was cooled down to 0°C in an ice bath while deaerating via argon bubbling (30min). Freshly prepared Na2S2O4 solution (1.62 μmol in 10 μL of 10 mM potassium phosphate buffer, pH 5.8) was added at once. After 1 hr at 0 °C, the reaction was then removed from ice and opened to air, followed by gentle stirring until a consistent purple color was obtained. The mixture was stirred for 20 min and chromatographed on a 2.5*56 cm Sephadex G-25 column using 20 mM NH4HCO3 as eluent. Oligonucleotide containing fractions were lyophilized. In the case of single stranded oligonucleotides, 5’-ATTATCGTTATT-3’ and 5’-ATTATTGCTATT-3’. the reaction was performed without the annealing step.
Oligonucleotide Alkylation by DMC under Bifunctional Conditions, Repeated Addition of Fully Reduced DMC (Protocol 2)
Oligonucleotides 13-30 (5’-ATTATXGYTATT-3’, 10 A260 unit scale; corresponding to about 0.072 μmol) were mixed with an equal amount of their complementary strand (5’-AATAXGYATAAT-3’, 10 A260 unit scale, about 0.072 μmol). The mixture was lyophilized. Oligonucleotides were then annealed by heating (55°C, 10 min) after the addition of 10 mM potassium phosphate buffer, pH 5.8 (355 μL) followed by slow cooling. The reaction mixture was then put under ice and deaerated via argon bubbling (30 min, 0°C). Excess Na2S2O4 (5 eq; 6.48 μmol in 20 μL of potassium phosphate buffer, pH 5.8) from a freshly prepared anaerobic solution in water was added to the mixture quickly followed by a dropwise addition of DMC (1.3 μmol in 200 μL of potassium phosphate buffer, pH 5.8). The reaction was allowed to stir for 10 min before another addition of Na2S2O4 (2.5 eq, 3.24 μmol) was immediately followed by a dropwise addition of an anaerobic DMC solution (1.3 μmol in 200 μL). This process was repeated 3 more times until the reaction was treated with a total of 6.48 μmol of DMC. The reaction mixture was stirred in ice with argon bubbling for 1 hr and then was removed from ice and opened to air, followed by gentle stirring until a consistent purple color was obtained. The mixture was then stirred for 20 min under air and then chromatographed on a 2.5 x 56 cm Sephadex G-25 column using 20 mM NH4HCO3 as eluent. Oligonucleotide containing fractions were lyophilized. In the case of duplex oligonucleotide 31, the reaction was performed both at 0 and 37 °C.
Oligonucleotide Alkylation by Enzymatic Activation.
Self-complimentary duplex oligonucleotides (5’-ATATACGTATAT-3’ or 5’-ATATAGCTATAT-3’: 10 A260 units; 330 μg; corresponding to 0.083 μmol) were mixed with 4.0 μmol of DMC in 100 mM potassium phosphate buffer, pH 5.8 (500 μL) at 0 °C in an ice bath while deaerating (30min). NADH (8 μmol in 515 μL of 10 mM potassium phosphate, pH 5.8) was added to the reaction followed by cytochrome c reductase (6 units in 12μL of deaerated 100 mM potassium phosphate, pH 7.4). The reaction was left in an ice bath with argon bubbling for 1hr and then stirred open to air for 20min. The mixture was chromatographed on a 2.5 x 56 cm Sephadex G-25 column using 20 mM NH4HCO3 as eluent.
M. luteus and calf thymus DNA Alkylation by DMC under Na2S2O4 Activation.
A solution of sonicated DNA (either M. luteus or calf thymus; 12 mM) and DMC (1mM) was deaerated in 10 mM potassium phosphate-0.001 M EDTA, pH 5.8, 4.5 or 7.5 (1 mL). A deaerated solution of Na2S2O4 (2mM, from a freshly prepared anaerobic 120 mM solution in water) was added incrementally following protocol 2 until a total of 5 mM of DMC was added. The drug-DNA complexes were extracted by adding 10 mM potassium phosphate, pH 9.0 to adjust the pH to 8.0 and then adding 1 mL of phenol:CHCl3:isoamyl alcohol extraction solution (25:24:1 v/v). The solution was then vortexed vigorously and allowed to settle before transferring into vials and centrifuged (13,000 rpm, 10 min). DNA was isolated by applying only the top aqueous layer on a Sephadex G-25 column as described above.
Enzymatic Digestion of the DMC-DNA Complexes to Nucleosides and DMC-Nucleoside Adducts.
The lyophilized DMC-DNA complex (M. luteus or calf thymus) was dissolved in 20 mM ammonium acetate, pH 5.5 (2.5 A260 units/mL). Nuclease P1 (1.0 unit/A260 unit of complex) was added to the mixture followed by incubation for 4 hr at 37 °C. The pH was adjusted to 8.2 by addition of 200 mM NaOH (25 μL/mL of ammonium acetate), and MgCl2 was added to a concentration of 0.9 mM. Addition of snake venom diesterase (2.25 units/A260 unit of complex) and 2 hr incubation at 37 °C were followed by the addition of alkaline phosphatase (1.6 units/A260 unit of complex) and incubation overnight at 37 °C. Samples were lyophilized and redissolved for HPLC analysis.
Enzymatic Digestion of Alkylated Oligonucleotides.
Nuclease P1/SVD/AP protocol: 1 A260 unit of oligonucleotide and 1 unit of nuclease P1 were incubated at 37 °C for 2 hr in 0.8 mL of 20 mM ammonium acetate, pH 5.5; 100 mM MgCl2 (20μL) was added, and the pH was adjusted to 8.2 by addition of 20 μL of 200 mM NaOH. SVD (2 units) and AP (2 units) were added and incubation was continued at 37 °C for 2.5 hr.
Analysis of DNA Adducts after Enzymatic Digestion of Alkylated Oligonucleotides and Calf Thymus or M. Luteus DNA.
Digestion mixtures were directly analyzed by HPLC using an Agilent 1200 HPLC system and a Kromasil C-18 reverse phase column (0.46 x 25 cm). The elution system was 6-18% acetonitrile in 30 mM potassium phosphate, pH 5.4, in 60 min, 1 mL/min flow rate.
Supplementary Material
Acknowledgements
This research was supported by a grant from the National Institute of Health (5SC3GM105460-04) to E.C. The authors would like to thank the PRISM (Program for Research Initiatives in Science and Math) program for support to A. Vargas and M. Zheng.
References
- [1].a) Bass PD, Gubler DA, Judd TC, Williams RM, Chem Rev. 2013, 113, 6816–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hata T, Hoshi T, Kanamori K, Matsumae A, Sano Y, Shima T, Sugawara R, J. Antibiot 1956, 9, 141–146 [PubMed] [Google Scholar]; c) Wakaki S, Marumo H, Tomioka K, Shimizu G, Kato E, Kamada H, Kudo S, Fujimoto Y, Antibiot. Chemother 1958, 8, 228–240. [PubMed] [Google Scholar]
- [2].Bradner WT, Cancer Treat. Rev 2001, 27, 35–50. [DOI] [PubMed] [Google Scholar]
- [3].Iyer VN, Szybalski W, Proc. Natl. Acad. Sci. U. S. A 1963, 50, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Iyer VN, Szybalski W, Science 1964, 145, 55–58 [DOI] [PubMed] [Google Scholar]
- [5].Rockwell S, Kim SY, Oncol. Res 1995, 7, 39–47. [PubMed] [Google Scholar]
- [6].Palom Y, Suresh Kumar G, Tang LQ, Paz MM, Musser SM, Rockwell S, Tomasz M, Chem. Res. Toxicol 2002, 15, 1398–1406. [DOI] [PubMed] [Google Scholar]
- [7].Paz MM, Ladwa S, Champeil E, Liu Y, Rockwell S, Boamah EK, Bargonetti J, Callahan J, Roach J, Tomasz M, Chem. Res. Toxicol 2008, 21, 2370–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tomasz M, Chowdary D, Lipman R, Shimotakahara S, Veiro D, Walker V, Verdine GL, Proc. Natl. Acad. Sci. U. S. A 1986, 83, 6702–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tomasz M, Lipman R, Chowdary D, Pawlak J, Verdine GL, Nakanishi K, Science 1987, 235, 1204–1208. [DOI] [PubMed] [Google Scholar]
- [10].a) Paz MM, Pritsos CA, “The Molecular Toxicology of Mitomycin C” in Adv. Mol. Toxicol Vol. 6 (Ed.: Fishbein JC) Elsevier, 2012. pp. 244–286 [Google Scholar]; b) Champeil E, Paz MM, Ladwa S, Clement CC, Zatorski A, Tomasz M, J. Am. Chem. Soc 2012,130, 9556–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tomasz M, Chawla AK, Lipman R, Biochemistry 1988, 27, 3182–3187. [DOI] [PubMed] [Google Scholar]
- [12].Suresh Kumar G, Lipman R, Cummings J, Tomasz M, Biochemistry 1997, 36, 14128–14136. [DOI] [PubMed] [Google Scholar]
- [13].Kato N, Kawasoe Y, Williams H, Coates E, Roy U, Shi Y, Beese LS, Schärer OD, Yan H, Gottesman ME, Takahashi TS, Gautier J, Cell Reports 2017, 21, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC, Cell 2016, 167, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bargonetti J, Champeil E, Tomasz M, J. Nucleic Acids 2010, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheng S-Y, Seo J, Huang BT, Napolitano T, Champeil E, Int. J. Oncol 2016, 49, 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Boamah EK, White DE, Talbott KE, Arva NC, Berman D, Tomasz M, Bargonetti J, ACS Chem. Biol 2007, 2, 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boamah EK, Breckman A, Tomasz M, Myeku N, Figueiredo-Pereira M, Hunter S, Meyer J, Bhosle RC, Bargonetti J, Chem. Res. Toxicol 2010, 23, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xiao G, Kue P, Bhosle R, Bargonetti J, Cell Cycle 2015, 14, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weng M-W, Zheng Y, Jasti VP, Champeil E, Tomasz M, Wang Y, Basu AK, Tang M-S, Nucleic Acids Res. 2010, 38, 6976–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bose A, Surugihalli C, Pande P, Champeil E, Basu AK, Chem. Res. Toxicol 2016, 29, 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ramos LA, Lipman R, Tomasz M, Basu AK, Chem. Res. Toxicol 1998, 11, 64–69. [DOI] [PubMed] [Google Scholar]
- [21].Basu AK, Hanrahan CJ, Malia SA, Kumar S, Bizanek R, Tomasz M, Biochemistry 1993, 32, 4708–4718. [DOI] [PubMed] [Google Scholar]
- [22].Utzat CD, Clement CC, Ramos LA, Das A, Tomasz M, Basu AK, Chem. Res. Toxicol 2005, 18, 213–223. [DOI] [PubMed] [Google Scholar]
- [23].Kumar S, Lipman R, Tomasz M, Biochemistry 1992, 31, 1399–1407. [DOI] [PubMed] [Google Scholar]
- [24].Borowy-Borowski H, Lipman R, Tomasz M, Biochemistry 1990, 29, 2999–3006. [DOI] [PubMed] [Google Scholar]
- [25].a) Li V-S, Kohn H, J. Am. Chem. Soc 1991, 113, 275–183 [Google Scholar]; b) Warren AJ, Hamilton JW, Chem. Res. Toxicol 1996, 9, 1063–1071 [DOI] [PubMed] [Google Scholar]; c) Teng SP, Woodson SA, Crothers DM, Biochemistry 1989, 28, 3901–3907. [DOI] [PubMed] [Google Scholar]
- [26].Aguilar W, Paz MM, Vargas A, Cheng S-Y, Clement CC, Champeil E, Chem. Eur. J 2018, 24, 6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zacharias O, Aguilar W, Paz MM, Vargas A, Zheng M, Cheng S-Y, Pradhan P, Champeil E, Manuscript in preparation. [Google Scholar]
- [28]. https://pubchem.ncbi.nlm.nih.gov/compound/mitomycin_C#.
- [29].Tomasz M, Lipman R, Lee MS, Verdine GL, Nakanishi K, Biochemistry 1987, 26, 2010–2017. [DOI] [PubMed] [Google Scholar]
- [30].Schiltz P, Kohn H, J. Am. Chem. Soc 1993, 115, 10497–10509. [Google Scholar]
- [31].Oguey C, Foloppe N, Hartmann B, Plos One 5, 2010, e15931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shu X, Xiong X, Song J, He C, Yi C, Angew. Chem 2016, 128, 14458–14461 [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. Engl 2016, 55, 14246–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bueren-Calabuig JA, Negri A, Morreale A, Gago F, Org. Biomol. Chem 2012, 10, 1543–1552. [DOI] [PubMed] [Google Scholar]
- [34].a) Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS, Annu. Rev. Biochem 2010, 79, 233–269 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ramachandran S, Temple B, Alexandrova AN, Chaney SG, Dokholyan NV, Biochemistry 2012, 51, 7608–7617. [DOI] [PubMed] [Google Scholar]
- [35].Noll DM, Webba da Silva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS, Biochemistry 2005, 44, 6764–6775. [DOI] [PubMed] [Google Scholar]
- [36].a) Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, D. O, Walter JC, Cell 2008, 134, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Minko IJ, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS, J Biol. Chem 2008, 283, 17075–17082 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Scharer OD, Nucl. Acids Res 2011, 39, 7455–7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].a) Khutsishvili I, Zhang N, Marky LA, Crean C, Patel DJ, Geacintov NE, Shafirovich V, Biochemistry 2013, 52, 1354–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Krishnamurthy N, Zhao X, Burrows CJ, David SS, Biochemistry 2008, 47, 7137–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jia L, Shafirovich V, Geacintov NE, Broyde S, Biochemistry 2007, 46, 5305–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sastry M, Fiala R, Lipman R, Tomasz M, Patel DJ, J. Mol. Biol 1995, 247, 338–359. [DOI] [PubMed] [Google Scholar]
- [39].Norman D, Live D, Sastry M, Lipman R, Hingerty BE, Tomasz M, Broyde S, Patel DJ, Biochemistry 1990, 29, 2861–2875. [DOI] [PubMed] [Google Scholar]
- [40].Kumar G. Suresh, Musser SM, Cummings J, Tomasz M, J. Am. Chem. Soc 1996, 118, 9209–9217. [Google Scholar]
- [41].Kinoshita S, Uzu K, Nakano K, Takahashi KT, J. Med. Chem 1971, 14, 109–112. [DOI] [PubMed] [Google Scholar]
- [42].Champeil E, Paz MM, Lukasiewicz E, Kong W, Watson S, Sapse AM, Bioorg. Med. Chem. Lett 2012, 22, 7198–7200. [DOI] [PubMed] [Google Scholar]
- [43].Champeil E, Cheng S-Y, Huang BT, Conchero-Guisan M, Martinez T, Paz AM, Sapse AM, Bioorg. Chem 2016, 65, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


