Abstract
Measuring and quantifying the binding of a drug to a protein target inside living cells and thereby correlating biochemical or biophysical activity with target engagement in cells or tissue represents a key step in target validation and drug development. A prototypic target engagement assay should allow for unbiased determination of small molecule–protein interactions in order to confirm cellular mechanism-of-action (MoA) while avoiding major artificial perturbations of cellular homeostasis and integrity. Recently, several new additions to the chemical biology toolbox have expanded our ability to study drug action in intact cells and enabled surveying of intracellular residence time and binding kinetics, which are particularly important for potent receptor ligands and therapeutic moieties with limited therapeutic index.
Introduction
In 1937, Alfred Joseph Clark noted the following in his seminal textbook General Pharmacology: “It is important in the first place to remember that our direct knowledge is limited to the fact that when tissues are exposed to these drugs certain responses are produced. We have no direct knowledge as to the manner in which the drugs act on the cells.” Fast forward more than 80 years from the time of the publishing of the book, this observation is still true for many approved drugs and uncovering the many ways by which small molecules perturb cellular systems represents a formidable challenge for chemical biology and drug discovery. However, recent advances in genomic and proteomic approaches have greatly broadened our ability to survey the complex actions of small molecules and drugs in living systems.
Cellular Target Engagement in Drug Discovery
Drugs achieve their effects through interaction with specific proteins in accordance with physicochemical laws (in agreement with the receptor theory which nota bene Clark was instrumental in formulating). The demonstration of, in Clark’s words, “certain responses” in cells upon exposure to a drug is insufficient without clear mechanistic evidence of drug target engagement. The immediate relevance of such studies is highlighted by case studies such as the bona fide PARP inhibitor iniparib; structurally unrelated to other established PARP-targeting small molecules, iniparib exhibited cytotoxic effects toward a number of breast cancer cell lines in preclinical studies. This prompted the progression of the compound into clinical trials that revealed an unexpected lack of efficacy. Following termination of the program, several studies confirmed that iniparib does in fact not engage PARP1 in cells and instead generates reactive oxygen species likely contributing to the initial antiproliferative effects observed with the compound.1 Failure of a compound to reach its designated target, i.e. inadequate exposure, can therefore result in a perceived lack of efficacy even for established targets.
Likewise, off-target activity can lead to similar or even far more reaching consequences. A clinical trial investigating the fatty acid amide hydrolase (FAAH) inhibitor BIA 10-2474 was abruptly halted in January 2016 when six individuals developed serious neurological lesions that led to one patient fatality. A subsequent comprehensive proteomic and lipidomic analysis of the compound by the Cravatt group revealed that the compound indeed binds FAAH within cells but also a whole host of other enzymes, resulting in a heavy disruption of lipid metabolism, potentially contributing to the clinical outcomes.2
These examples highlight the fact that demonstration of compound binding to a given target in a biophysical or biochemical assay using recombinantly expressed protein (or domain thereof) is in no way a guarantee of the same outcome being achieved when the drug is administered to a cell, much less a patient. Biological systems, and tumor cells in particular, have evolved a myriad of ways to regulate the trafficking of xenobiotics, among them being modulation of membrane permeability, compound sequestration, export, or protein compartmentalization,3 thus necessitating target engagement assays in a cellular- or tissue-relevant context. Retrospective analyses conducted by pharma companies such as Pfizer and AstraZeneca uncovered that close to a fifth of phase II failures due to efficacy lacked conclusive demonstration of adequate target exposure.4,5
Methods to Study Drug–Target Engagement
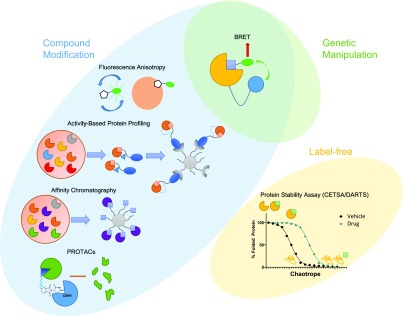
The physical phenomenon of bioluminescence resonance energy transfer, or BRET, whereby a bioluminescent donor can transfer energy to a proximal fluorophore, can be exploited to study cellular target engagement. By using a fluorescently tagged protein (acceptor) or small molecule (often referred to as a tracer or energy transfer probe) and transfecting cells with the target fused with a luciferase, BRET can be achieved in living cells. Disruption of this interaction by e.g. outcompeting the tracer with increasing doses of a putative inhibitor of the target enables quantitative monitoring of drug–protein interactions, including kinetic studies (Figure 1). The assay can be adapted to a microplate format6 and recent studies have successfully harnessed CRISPR/Cas9 to tag endogenous proteins, thereby circumventing the need for transfection with an exogenous copy of the gene.7
Figure 1.
Overview of cell target engagement approaches.
Another biophysical technique to examine cellular target engagement relies on fluorescence anisotropy, which occurs when a fluorescently tagged molecule engages a protein and thus, following excitation, begins to emit light in a directional fashion, compared to free-flowing fluorophores. In a typical experiment, cells are dosed with the compound and after incubation, alterations in the isotropy of emission are measured by fluorescence polarization. The advantage of this technique is that it can be used to study target engagement in biopsies taken from live organisms8 or, when using window chambers, even directly in live animals.9
Protein mass spectrometry (MS)-based approaches offer a potential route to both investigating target engagement as well as selectivity profiling on a global level. Classic methods involve affinity chromatography, where cellular lysates are treated with resins functionalized with immobilized compounds and, following affinity capture and digestion, the bound proteins are analyzed by MS. Activity-based protein profiling (ABPP) and photoaffinity labeling (PAL) rely on the synthesis of functionalized compounds bearing a reactive capturing group, as well as an affinity tag for subsequent enrichment.10 While the principle is identical to classical drug affinity chromatography, ABPP and PAL allow for the use of live cells instead of cell lysates, and the use of covalent warheads increases retention. The separation of true hits from false positives, which can occur through proteins binding nonspecifically to the scaffold or the resin, can be achieved by repeating the experiment with an analogue of the drug that has been inactivated by minimal modification (oftentimes in the case of chiral compounds, the distomer can elegantly fulfill this role).
In recent years, the cellular thermal shift assay (CETSA) has emerged as a versatile target engagement and identification assay. CETSA is based on the principle of protein denaturation upon a heat pulse and alterations in protein thermal stability upon ligand binding. This often leads to an increase of the temperature needed for unfolding, thus causing the observed “thermal shift”. CETSA can be performed with intact cells, where, following the heat pulse and subsequent lysis, the aggregated and denatured proteins are removed by centrifugation and target engagement is confirmed by the presence of an increased amount of protein for the compound-treated sample. The coupling of the technique to quantitative proteomics enables the profiling of thousands of proteins for their capacity to engage various types of small molecules.11−13 A key advantage is that CETSA does not require chemical modification of the compound of interest. It is readily adaptable to a microplate screening format for high-throughput target engagement studies, either through fusion to a reporter including enzyme complementation strategies,14 the use of AlphaLISA double antibody proximity assays,15,16 or an imaging readout.17 Alternatively, the denaturing thermal heat pulse may be replaced by a chaotrope followed by pulse proteolysis as exemplified by the DARTS method (drug affinity responsive target stability).18 Notably, a recent study took advantage of the emerging field of targeted protein degradation to demonstrate a tool compound could bind its target by designing a bifunctional proteolysis-targeting chimera (PROTAC) which induced degradation of the putative target.19
Conclusion
The development of novel methods to quantify drug–target engagement in living systems in recent years has helped to close in on the gap between in vitro assay results and in vivo observations. The complementarity of these approaches to classical screening campaigns using purified proteins lies in the ability to study natural full-length proteins with posttranslational modifications in the presence of endogenous competitors and cofactors. Target engagement assays are becoming more miniaturized and adaptable to screening formats, allowing them to augment or even replace conventional high-throughput screening campaigns, and MS-based techniques can provide a proteome-wide view on drug action in cells and tissue samples. Current and future challenges include improving our ability to interrogate membrane proteins, which comprise many important drug targets such as transporters, ion channels, GPCRs, and finally yet importantly compounds that do not act via direct interactions with proteins. Within the pages of Applied Pharmacology, Clark writes: “The great complexity of living cells is one of the chief difficulties in the analysis of the action of drugs” and while we may not be at the stage of being able to completely elucidate the MoA of every drug from chemotype to phenotype, we are closer than ever before.
This work was supported by funding from the Engineering and Physical Sciences Research Council (EPSRC) and the Medical Research Council (MRC) [grant number EP/L016044/1]. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z].
Views expressed in this editorial are those of the authors and not necessarily the views of the ACS.
The authors declare no competing financial interest.
References
- Mateo J.; Ong M.; Tan D. S. P.; Gonzalez M. A.; de Bono J. S. Appraising Iniparib, the PARP Inhibitor That Never Was—What Must We Learn?. Nat. Rev. Clin. Oncol. 2013, 10 (12), 688–696. 10.1038/nrclinonc.2013.177. [DOI] [PubMed] [Google Scholar]
- van Esbroeck A. C. M.; Janssen A. P. A.; Cognetta A. B.; Ogasawara D.; Shpak G.; van der Kroeg M.; Kantae V.; Baggelaar M. P.; de Vrij F. M. S.; Deng H.; et al. Activity-Based Protein Profiling Reveals off-Target Proteins of the FAAH Inhibitor BIA 10–2474. Science 2017, 356 (6342), 1084–1087. 10.1126/science.aaf7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- César-Razquin A.; Snijder B.; Frappier-Brinton T.; Isserlin R.; Gyimesi G.; Bai X.; Reithmeier R. A.; Hepworth D.; Hediger M. A.; Edwards A. M.; et al. A Call for Systematic Research on Solute Carriers. Cell 2015, 162 (3), 478–487. 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Cook D.; Brown D.; Alexander R.; March R.; Morgan P.; Satterthwaite G.; Pangalos M. N. Lessons Learned from the Fate of AstraZeneca’s Drug Pipeline: A Five-Dimensional Framework. Nat. Rev. Drug Discovery 2014, 13 (6), 419–431. 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- Bunnage M. E.; Chekler E. L. P.; Jones L. H. Target Validation Using Chemical Probes. Nat. Chem. Biol. 2013, 9 (4), 195–199. 10.1038/nchembio.1197. [DOI] [PubMed] [Google Scholar]
- Vasta J. D.; Corona C. R.; Wilkinson J.; Zimprich C. A.; Hartnett J. R.; Ingold M. R.; Zimmerman K.; Machleidt T.; Kirkland T. A.; Huwiler K. G.; et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem. Biol. 2018, 25 (2), 206–214. 10.1016/j.chembiol.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; e11.
- White C. W.; Vanyai H. K.; See H. B.; Johnstone E. K. M.; Pfleger K. D. G. Using NanoBRET and CRISPR/Cas9 to Monitor Proximity to a Genome-Edited Protein in Real-Time. Sci. Rep. 2017, 7 (1), 3187. 10.1038/s41598-017-03486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubach J. M.; Vinegoni C.; Mazitschek R.; Fumene Feruglio P.; Cameron L. A.; Weissleder R. In Vivo Imaging of Specific Drug–Target Binding at Subcellular Resolution. Nat. Commun. 2014, 5 (1), 3946. 10.1038/ncomms4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubach J. M.; Kim E.; Yang K.; Cuccarese M.; Giedt R. J.; Meimetis L. G.; Vinegoni C.; Weissleder R. Quantitating Drug-Target Engagement in Single Cells in Vitro and in Vivo. Nat. Chem. Biol. 2017, 13 (2), 168–173. 10.1038/nchembio.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara D.; Ichu T.-A.; Jing H.; Hulce J. J.; Reed A.; Ulanovskaya O. A.; Cravatt B. F. Discovery and Optimization of Selective and in Vivo Active Inhibitors of the Lysophosphatidylserine Lipase α/β-Hydrolase Domain-Containing 12 (ABHD12). J. Med. Chem. 2019, 62 (3), 1643–1656. 10.1021/acs.jmedchem.8b01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K. V. M.; Olek K. M.; Müller A. C.; Tan C. S. H.; Bennett K. L.; Colinge J.; Superti-Furga G. Proteome-Wide Drug and Metabolite Interaction Mapping by Thermal-Stability Profiling. Nat. Methods 2015, 12 (11), 1055–1057. 10.1038/nmeth.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski M. M.; Reinhard F. B. M.; Franken H.; Werner T.; Savitski M. F.; Eberhard D.; Molina D. M.; Jafari R.; Dovega R. B.; Klaeger S.; et al. Tracking Cancer Drugs in Living Cells by Thermal Profiling of the Proteome. Science (Washington, DC, U. S.) 2014, 346 (6205), 1255784. 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- Becher I.; Werner T.; Doce C.; Zaal E. A.; Tögel I.; Khan C. A.; Rueger A.; Muelbaier M.; Salzer E.; Berkers C. R.; et al. Thermal Profiling Reveals Phenylalanine Hydroxylase as an Off-Target of Panobinostat. Nat. Chem. Biol. 2016, 12 (11), 908–910. 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- McNulty D. E.; Bonnette W. G.; Qi H.; Wang L.; Ho T. F.; Waszkiewicz A.; Kallal L. A.; Nagarajan R. P.; Stern M.; Quinn A. M.; et al. A High-Throughput Dose-Response Cellular Thermal Shift Assay for Rapid Screening of Drug Target Engagement in Living Cells, Exemplified Using SMYD3 and IDO1. SLAS Discovery Adv. Life Sci. R&D 2018, 23 (1), 34–46. 10.1177/2472555217732014. [DOI] [PubMed] [Google Scholar]
- Martinez N. J.; Asawa R. R.; Cyr M. G.; Zakharov A.; Urban D. J.; Roth J. S.; Wallgren E.; Klumpp-Thomas C.; Coussens N. P.; Rai G.; et al. A Widely-Applicable High-Throughput Cellular Thermal Shift Assay (CETSA) Using Split Nano Luciferase. Sci. Rep. 2018, 8 (1), 9472. 10.1038/s41598-018-27834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J.; Leveridge M.; Norling C.; Karén J.; Molina D. M.; O’Neill D.; Dowling J. E.; Davey P.; Cowan S.; Dabrowski M.; et al. Determining Direct Binders of the Androgen Receptor Using a High-Throughput Cellular Thermal Shift Assay. Sci. Rep. 2018, 8 (1), 163. 10.1038/s41598-017-18650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson H.; Almqvist H.; Otrocka M.; Vallin M.; Lundqvist S.; Hansson P.; Karlsson U.; Lundbäck T.; Seashore-Ludlow B. In Situ Target Engagement Studies in Adherent Cells. ACS Chem. Biol. 2018, 13 (4), 942–950. 10.1021/acschembio.7b01079. [DOI] [PubMed] [Google Scholar]
- Lomenick B.; Hao R.; Jonai N.; Chin R. M.; Aghajan M.; Warburton S.; Wang J.; Wu R. P.; Gomez F.; Loo J. A.; et al. Target Identification Using Drug Affinity Responsive Target Stability (DARTS). Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (51), 21984–21989. 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessum N. E. A.; Sharp S. Y.; Caldwell J. J.; Pasqua A. E.; Wilding B.; Colombano G.; Collins I.; Ozer B.; Richards M.; Rowlands M.; et al. Demonstrating In-Cell Target Engagement Using a Pirin Protein Degradation Probe (CCT367766). J. Med. Chem. 2018, 61 (3), 918–933. 10.1021/acs.jmedchem.7b01406. [DOI] [PMC free article] [PubMed] [Google Scholar]