Abstract
With the increased adoption of electronic health records, data collected for routine clinical care is used for health outcomes and population sciences research, including the identification of phenotypes. In recent years, research networks, such as eMERGE, OHDSI and PCORnet, have been able to increase statistical power and population diversity by combining patient cohorts. These networks share phenotype algorithms that are executed at each participating site. Here we observe experiences with phenotype algorithm portability across seven research networks and propose a generalizable framework for phenotype algorithm portability. Several strategies exist to increase the portability of phenotype algorithms, reducing the implementation effort needed by each site. These include using a common data model, standardized representation of the phenotype algorithm logic, and technical solutions to facilitate federated execution of queries. Portability is achieved by tradeoffs across three domains: Data, Authoring and Implementation, and multiple approaches were observed in representing portable phenotype algorithms. Our proposed framework will help guide future research in operationalizing phenotype algorithm portability at scale.
Introduction and Background
As the proliferation of electronic health records (EHRs) has increased the secondary use of healthcare data, it has also catalyzed the establishment of collaborative research networks where multiple institutions are working on large studies to increase their sample size and diversity of the study population. This has been demonstrated via multi-site prospective clinical trials and large-scale observational studies, where participant eligibility criteria queries - which, typically take the form of phenotype algorithms (i.e., structured selection criteria designed to produce research-quality phenotypes)1,2 - are run at each participating institution to identify the study cohort. More recently, large collaborative networks such as the Health Care Systems Research Network (HCSRN, formerly the HMORN)3, Mini-Sentinel4, the electronic Medical Records and Genomics (eMERGE) Network5, the National Patient-Centered Clinical Research Network (PCORnet)6, the Accrual to Clinical Trials (ACT) Network7, the Observational Health Data Sciences and Informatics (OHDSI) program8, and the Electronic Health Records for Clinical Research (EHR4CR) European project9 have shown different models for how a single phenotype algorithm may be run across heterogeneous EHRs and database systems. A desired state from many of these networks is to have “computable phenotype algorithms” that are portable, and can be executed in a high-throughput manner10.
In our observations, across both clinical trials and research networks, two primary models have emerged for the implementation of phenotype algorithms at multiple sites: 1) sharing a human-readable narrative of the algorithm that is re-implemented at each site (e.g., clinical trials, eMERGE), and 2) using a pre-coordinated common data model (CDM) which allows sharing of machine interpretable executable code (e.g., OHDSI, PCORnet, HCSRN). The first approach often reduces the burden on the algorithm authoring institution, as they may describe the phenotype algorithm logic and required medical vocabulary codes in a more generic manner or in a manner that meets their local institution-specific settings (e.g., local, non-standardized laboratory measurement codes). This algorithm logic can then be disseminated widely via public repositories - the eMERGE Network uses the Phenotype KnowledgeBase (PheKB)11, and clinical trial criteria are made available on ClinicalTrials.gov12. Sites implementing the phenotype algorithm then interpret and convert the written description to an executable format that can run against their local data, which often requires iterative communication with the original author(s) to ensure accuracy. Local validation is an important component to ensure the implementation is correct, and that the phenotype algorithm implementation is sufficiently sensitive and/or specific (depending on the specific study needs) when implemented across multiple sites13. To this end, phenotype algorithms may be easily shared as a narrative document, but this limits scalability as each algorithm must be transformed from a narrative to local queries. This has been shown to be a cumbersome, resource-intensive and error-prone process14.
The second approach, the use of a CDM, uses a pre-coordinated structure for the data such that the phenotype algorithm – represented as a database query – may be directly shared and executed at different sites using the same CDM. Several CDMs have been defined, including Informatics for Integrating Biology and the Bedside (i2b2)15, PCORnet6, Mini-Sentinel CDM16, OHDSI’s Observational Medical Outcomes Partnership (OMOP)17, the Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM)18, and the HCSRN’s Virtual Data Warehouse (VDW)19,20. CDMs by their nature are optimized for a specific set of uses and data sources (e.g., clinical vs. research, claims data vs. EHR data)21. Furthermore, no CDM is “universally common,” meaning that translation and reimplementation are required when crossing CDM boundaries. A more recent project led by the Food and Drug Administration, “Harmonization of Various Common Data Models and Open Standards For Evidence Generation”22, is developing mappings from several of these CDMs to the BRIDG23 domain analysis model to address this need. In addition, local adjustment of the phenotype algorithm is still sometimes needed to account for differences that may not be addressed by a CDM, such as different numeric ranges for abnormal lab results across sites. Similar to the first approach, sensitivity and specificity still need to be assessed to ensure the phenotype algorithm is complete (e.g., includes all relevant vocabulary codes). Because of this, portability of phenotype algorithms is not solved solely by a CDM, although a CDM provides many advantages10.
The Phenotype Execution and Modeling Architecture (PhEMA) project has proposed an architectural model24 that supports the authoring of standards-based EHR-based phenotype algorithms, and their subsequent execution across multiple CDMs. This is done by the use of a canonical, standard representation of the phenotype algorithm logic and value sets (collections of standard medical vocabulary codes). PhEMA then automates the translation steps to convert these pieces to computable phenotype algorithms for individual CDMs. Most recently, PhEMA was used to execute a benign prostatic hyperplasia phenotype algorithm across multiple institutions’ data warehouses and i2b2 instances25, demonstrating a basis for a single representation of a phenotype algorithm that can span multiple data models. In addition to crossing data model boundaries, we recognize that many other facets of portability exist. In this work, we build on the reported experiences of collaborative research networks, as well as our experiences in implementing PhEMA across three academic medical centers, by further exploring the considerations needed for cross-site portability of phenotype algorithms.
Methods
We identified large research networks published in the health and biomedical informatics literature that have focused on building or leveraging infrastructure to support cross-site execution of EHR-based phenotype algorithms. Identification of the networks was done from on a cursory search of PubMed for consortia describing EHR-based phenotyping, supplemented by the authors’ collective experience. Within this paper, we are broadly considering an “EHR-based phenotype” as a definition of a disease or trait that is ascertained primarily from data stored in an EHR. We limited consideration to only those networks describing a distributed model, in which participating institutions retain access to their respective data, and those that have an established infrastructure to demonstrate portable phenotype algorithm execution. In total, we selected a purposive sample of seven collaborative networks: ACT Network, EHR4CR, eMERGE, HSCRN, Mini-Sentinel, OHDSI, and PCORnet. We reviewed the literature published by the respective networks, as well as documentation available online regarding CDMs and technical architecture. This was supplemented by the authors’ experience from participation in some (but not all) of the networks, and personal communication with members in other networks. From this, we created maps showing the elements used to compose a phenotype algorithm, the methods by which phenotype algorithms are shared across sites, and how they are executed at each site. Multiple approaches may be used within a single network; however, as our objective was to evaluate the distinct approaches, we did not exhaustively document all possible permutations across all networks. These models were consolidated into a final framework to capture and model the considerations needed for portable EHR-based phenotype algorithms.
Results
From review and consolidation of the respective research network models, we developed a framework for phenotype algorithm portability that has three main components: 1) a phenotype algorithm workflow model, 2) a portability tradeoff model, and 3) a portability representation model.
Phenotype Algorithm Workflow Model
The phenotype algorithm workflow model is comprised of 8 steps across 3 broad domains that we identified as being involved within the phenotype modeling and execution process, and that have potential impact on portability (Table 1). As described, these steps identify the core work needed to establish a portable phenotype algorithm, and are part of an iterative process.
Table 1.
Phenotype algorithm workflow model
| Domain | Step | Description | Potential Challenges to Portability |
| Data | Data Collection | The processes by which data is collected within the source EHR, and its intended purpose. | Only data that is collected can be used for electronic phenotyping. How data is collected at a local institution (vocabulary used, frequency of collection, etc.) determines how that institution authors a phenotype algorithm. Modality of data collection (e.g., structured, narrative text, images) can affect how and if the data used in executing a phenotype algorithm. |
| Data Preparation | Extract-Transform-Load (ETL) processes through which data is consolidated into an integrated data repository (IDR). | The need to transform the shape of the data from an IDR data into a common data model (CDM). Effort to convert data from one modality to another (e.g., natural language processing to obtain structured results). Mapping of local terms to a standard vocabulary term (national standard or prescribed by CDM), and potential lossy mappings or semantic drift. | |
| Authoring | Define Value Sets | Identifying the medical terms that are used to represent data elements within the phenotype algorithm logic. | Not all terminologies/vocabularies are fully implemented at all institutions. Value sets may list all codes, or may list codes at the top level of a hierarchy that need to be expanded. |
| Define Logic | Create a representation of the required data elements, and how the elements are related by different operators (e.g., Boolean, temporal) to create a phenotype algorithm. | The modality of the logic representation (narrative, intermediate representation, programming language), and what system(s) may understand it. Strictness of the logic, considering local instead of broader data availability. |
|
| Implementation | Distribution | The mechanism by which a phenotype algorithm is transmitted from the author to an implementing site. | Automated vs. manual approach. Policies that require human review and approval before execution. |
| Translation | How the phenotype algorithm is converted into an executable representation that may be directly applied to the institutional data model. | Automated vs. manual approach. Technology-specific customizations (e.g., database schema names, table names). Information loss when elements of a data model do not have a direct translation or differ in granularity. | |
| Execution | The computation process by which the executable representation is applied to an institutional data warehouse, and results are retrieved. | Syntax errors that require human intervention and correction. | |
| Validation | A formal or informalcomparison of the execution results against a reference standard. | Lack of detailed information concerning the inclusion and exclusion implications across multiple phenotype algorithm steps. Lack of access to source data to evaluate results. |
Data Collection - A reality for any research network is that if a particular data element is not collected as part of an institution’s care process, it will never be available to a phenotype algorithm. Similarly, if a data element is available within a source system but is not made available to a site’s integrated data repository (IDR) in a way that it can be queried, a phenotype algorithm will not be able to include it. This may be observed with data collected as clinical narratives or images, as opposed to structured data. We note that even with a common data model that is capable of representing all medical concepts, these potential barriers would keep the data not represented in the CDM from being provided. This remains a consistent barrier across all networks.
Data Preparation - We observed that many networks adopted a CDM to aid with portability. This requires those involved with the Data Preparation step to spend additional time “data wrangling” (i.e., cleaning, mapping and translating their source EHR data into the CDM). This reduces the downstream effort on both authors and implementers. There are variations in the pre-coordination of vocabularies within the CDMs, however. For example, within i2b2 (used in the ACT network), each site may have a local ontology specified for their data elements. This allows for custom data elements (e.g., status within a biobank or clinical trial), as well as individual site selection of the vocabulary to use (e.g., NDC codes for medications instead of RxNorm codes). Other networks, such as OHDSI, provide stronger guidance on the vocabularies to use, potentially requiring additional time and effort in the data mapping process. They still allow flexibility by preserving the source system’s native vocabulary code, and allow phenotype algorithms to be defined using those codes. Some CDMs account for processing steps to convert data between modalities, such as the NOTE_NLP table within OMOP to store structured results from natural language processing (NLP). This requires additional effort to perform NLP on clinical text, but produces structured data that is more amenable to processing. Depending on the amount of effort needed, not all sites adopting a CDM may perform this pre-processing, or may do it on an ad hoc basis as new categories of data are needed for a study (e.g., processing just echocardiogram reports). On the other hand, it was pointed out that existing NOTE_NLP table lacks flexibility to store relations between medical concepts26.
Defining Value Sets - In networks where a pre-coordinated set of vocabularies have been implemented (as described in Data Preparation), time and effort are reduced within the authoring process as the phenotype algorithm author can write the phenotype algorithm for just those vocabularies. For example, within OMOP, diagnoses may be represented entirely using SNOMED CT. Within networks such as eMERGE where sites re-implement the phenotype algorithm locally, additional effort is needed by the author or implementation site to expand a value set. For example, an existing phenotype algorithm developed using just ICD-9-CM codes only would need to be updated for an implementer wishing to include ICD-10-CM codes (ICD-10 codes were used in the US only after October 2015). We note that the reduced authoring time in networks having a pre-coordinated vocabulary does not necessarily guarantee portability across research network boundaries. For example, an HSCRN query representing a medication value set with NDC codes would require mapping to RxNorm to support execution in ACT.
Defining Logic - Research networks use three general approaches to make their phenotype algorithms portable. The first is the use of a specific programming language to represent the query (e.g., SAS within HSCRN, R and SQL within OHDSI). A second approach is the use of a system-specific intermediary representation for the logic, agnostic to underlying implementation. For example, the ATLAS JSON format within OHDSI, the XML query definition in i2b2, and the Eligibility Criteria Language for Clinical Trial Investigation and Construction (ECLECTIC) within EHR4CR27. The third approach is the development of a written narrative and/or visual flowchart to describe the phenotype logic (e.g., phenotype logic posted to PheKB for eMERGE). This last format is based on a computable implementation at the authoring site, which may be done against any data model and using any technology the authoring site chooses. The description of the phenotype algorithm may be easier for the author to provide as it can convey the intent of the logic without prescribing specific tables or fields to be used, but requires each implementing site to read, evaluate and map the intent of the logic to their local institutional data model.
Distribution – Phenotype algorithms were distributed manually or automatically, and the choice of distribution method often depends on how the logic was represented as well as constraints within the research network. Within the eMERGE network, the author will manually publish a phenotype algorithm to PheKB, and inform implementing sites when it is ready. Similarly within OHDSI, phenotype algorithms may be tracked within a central GitHub repository28, where they can be reviewed and manually imported into a local OHDSI instance. The HSCRN and PCORnet have infrastructure to electronically share the phenotype algorithm to implementing sites, where it may be reviewed from a worklist before execution. Federated architectures, such as EHR4CR’s Orchestrator, ACT’s use of the Shared Health Research Information Network (SHRINE)29 on top of i2b2, or the ARACHNE system within OHDSI, allow phenotype algorithms to be automatically distributed. Within this step, automation can reduce burden on implementers, and also reduce burden on authors if a technical solution exists to link authoring to distribution.
Translation - Varying levels of transformation are needed across the networks, driven by technical capabilities. Within HSCRN (where SAS files were directly shared) and within OHDSI (where SQL and R code are shared), some manual translation may be needed to configure the code to run locally. As an example, SQL code for OHDSI may require specifying a local database schema name before it can be run. Within OHDSI where the JSON representation of a phenotype algorithm is shared from the ATLAS system, the translation is done automatically. Similarly, within ACT, SHRINE performs real-time mapping of ontology terms in the XML message to the local i2b2 instance, and i2b2 itself manages translation of XML to local SQL queries.
Execution - This step is consistent across all research networks, as all translations have been completed and the phenotype algorithm is now in an executable format for the local data repository. During the execution phase, considerations for portability are driven largely by the results of the execution. A phenotype algorithm may be appropriately translated to SQL code, but requires a significant amount of time or resource to run. This may be to the point where the SQL code is unable to complete before a system-level timeout causes it to halt; thus, it is not truly executable and is no longer viewed as a portable implementation. Likewise, a phenotype algorithm may have execution paths that are entirely dependent on a category of data that is not present due to considerations from steps in the Data domain. Although a syntactically valid definition of the phenotype algorithm exists, it is also not viewed as portable given that it returns no records when some records are expected to be found.
Validation – This step is carried out in multiple ways across the different research networks, often depending on the technical approach used to perform actual execution. Usually the first pass of the execution is a simple cohort count (“how many cases and controls with X?”), which may be supplemented with a demographic breakdown. An initial face-validity check of the result can be used to ascertain if the results seem feasible, given expected prevalence of a condition. To perform more in-depth validation, or to troubleshoot why a phenotype algorithm is returning an empty cohort, access to the underlying data model is typically needed. As this is a manual process, it requires additional coordination and time across implementation sites by the author site to conduct chart reviews. Automation of this type of analysis can be done in the Execution step, although this increases overall execution time30.
Portability Tradeoff Model
The second component of our proposed framework is a portability tradeoff model that represents the interlinked nature of the three domains in the workflow model. Across the seven reviewed research networks, we observed that different weights are given to Data, Authoring and Implementation. Within the portability tradeoff model, we consider a higher weight in a domain as having an increased overall cost (although we note that a high cost is not inherently “good” or “bad”). These costs are in turn linked to the “iron triangle” of project management: time, features and resource31. This results in what we have termed the “nested iron triangle of phenotype algorithm portability” (Figure 1). For example, OHDSI and PCORnet put additional weight on the Data domain, such that more up-front work is needed at each institution to adhere to the CDM. This reduces the time spent on Authoring, as the author only needs to consider a single data model and a limited set of vocabularies. Similarly, less effort is then needed at each site for Implementation, as less localization is needed during the Translation step. As a second example, sharing phenotype algorithms as narratives (employed within the eMERGE network) allows flexibility in the data model, often leveraging existing data warehousing infrastructure (decreased weight on Data). However, additional time may be required during Authoring, if the author is considering portability and thoroughly investigating comprehensive value sets. Furthermore, more weight is given to steps in the Implementation domain, as each site must put more effort into the Translation step to arrive at an executable definition.
Figure 1.
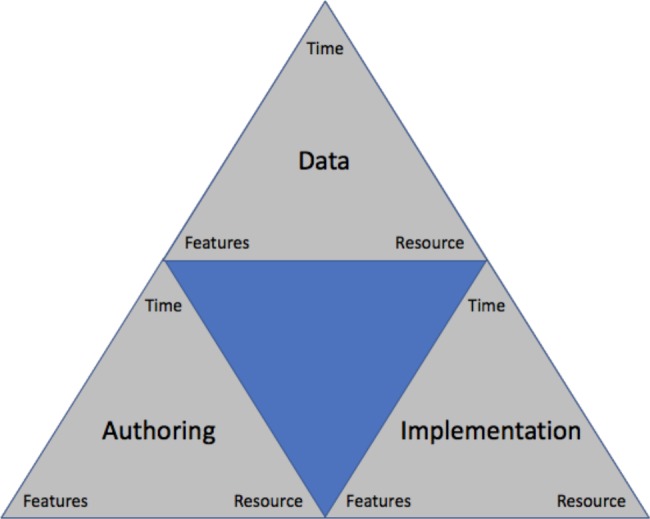
Portability tradeoff model: the “nested iron triangle of phenotype algorithm portability”.
Portability Representation Model
During our review, we observed how the seven research networks approached the representation of the phenotype algorithms, and the impact this representation had on portability across steps in the Authoring and Implementation domains. This is defined in its own model given the additional considerations needed. Four general approaches were identified (Figure 2), with their own benefits and limitations.
Figure 2.
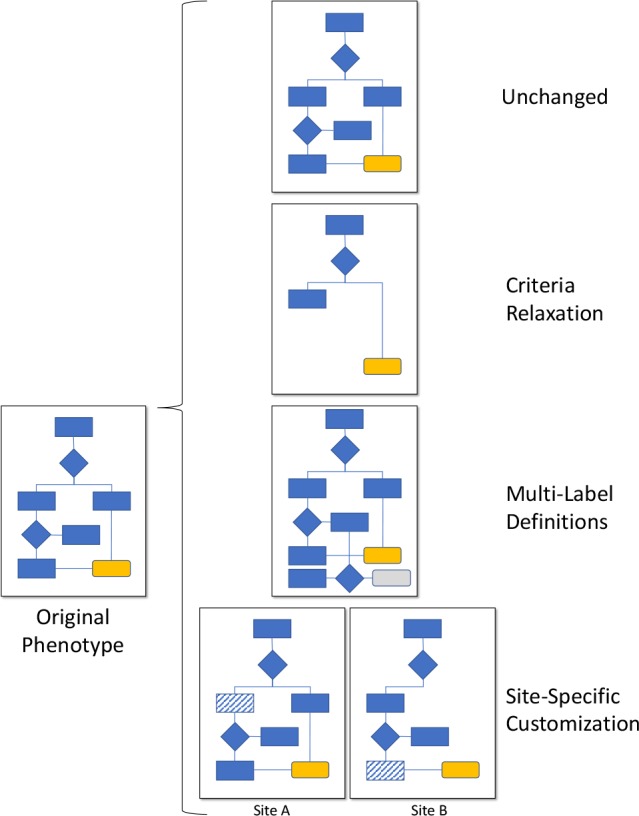
Portability representation model: Four approaches to represent computable phenotype algorithms that have been executed across multiple sites.
Unchanged – This approach is the ideal state of portable representation, as a single definition (the originally developed phenotype algorithm) may be distributed for use at the implementing sites. Technically this approach is currently available in our reviewed networks, with the exception of the non-computable narrative phenotype algorithms in the eMERGE network.
Criteria Relaxation - This approach allows making a phenotype algorithm more general by reducing or removing constraints, while monitoring and balancing sensitivity and specificity for the intended use case25. This results in a single phenotype algorithm that is distributed, and only includes the logic known to be runnable at each implementing site. We observed this as being universally available across all of the research networks from a technical standpoint. All reviewed systems across the research networks allow criteria to be removed entirely (if overly-exclusive), or constraints broadened (if insufficiently inclusive). This requires creating a revision of the phenotype algorithm with the modified logic and/or value sets. This may result in a phenotype algorithm with overall lower sensitivity and higher specificity across multiple sites, but allows the phenotype algorithm to remain a singular entity that eases distribution, as the same phenotype algorithm is shared universally.
Multi-Label Specification - This approach involves implementing a single version of the phenotype algorithm, but allowing different labels to be applied to the cohort, depending on quality of evidence (e.g., “gold standard” and “silver standard”)32. Unlike criteria relaxation, this allows the single phenotype algorithm to be run at each site and produce results, but may increase overall complexity of the phenotype algorithm depending on the number of logic paths. This is not directly supported outside of narrative descriptions (eMERGE phenotype algorithms) and executable implementations (e.g., HSCRN SAS, OHDSI SQL and R). In networks that provide an intermediary platform to author phenotype algorithms (ACT’s i2b2 and OHDSI’s ATLAS), no solution was observed. Instead, authors must create multiple versions of a phenotype algorithm - one for each label. This causes duplication of logic, which must now be maintained across multiple phenotype algorithms, and these platforms do not provide a way to group the disparate phenotype algorithms together to show they are related.
Site-Specific Customizations - In this approach, the implementing site changes the logic or translates executable code of the phenotype algorithm so that it will run and produce acceptable results25. This may be done at the discretion of the implementing site, or done in conjunction with the original author to assess tradeoffs. The result is multiple versions of the phenotype algorithm, which are (at best) loosely linked together. This approach is also universally available across all of the research networks. We note that while it provides flexibility to implementers (at the tradeoff of additional work), it severs provenance to the original phenotype algorithm. This makes subsequent changes by the original author difficult or impossible to incorporate automatically, as well as changes from the implementers that may be useful to the author and other implementers.
Discussion
Here we have presented a framework of phenotype algorithm portability based on the reported experiences of seven research networks. It is comprised of three interlinked models, which together provide researchers and system evaluators with a comprehensive framework against which to consider barriers and solutions to the portability of EHR-based phenotype algorithms. The framework demonstrates that the concept of phenotype algorithm portability is multi-faceted, as shown by the number of steps and potential impact points, and by the observation that the approaches within each model are not necessarily mutually exclusive within a single network. For example, within the portability tradeoff model, an institution adopting a CDM such as PCORnet may put less up-front investment in the Data domain (reducing it compared to other sites), which then would increase Implementation resource needs for that site. Similarly, within the portability representation model, multiple approaches may be observed within the same network. Within OHDSI, an ATLAS-authored phenotype algorithm may be shared across sites as unchanged, but a SQL script written for a specific database vendor may require site-specific customization to port the syntax to another vendor system. While in our framework and respective models we consider automation and computability, we note that the concept of portability is not dependent on them. For example, several of the research networks that we evaluated require the manual review of a phenotype algorithm before it could be executed (driven by that network’s operating policies). We recognize that this mandated manual review step impacts how scalable that portability solution is to a large number of phenotype algorithms.
Our proposed framework is supported by previous studies reported in the literature, but to our knowledge, no previous study has considered all aspects of portability. Our framework supports the ongoing need for localization of phenotype algorithms, which was also identified within a previously reported desiderata33. We believe the need for localization will remain for the foreseeable future, given the number of disparate CDMs that exist, and the complexities of data collection and representation within the EHR34. As CDMs continue to evolve, and as more work is done to harmonize CDMs35, that need will lessen. However, previous studies have demonstrated the impact of data heterogeneity on phenotype algorithms36,37, which we believe will be an ongoing issue. Recognizing this, we propose adjusting the view of a phenotype algorithm as a single, immutable object. This view is currently reflected in practice, as demonstrated by three of the four approaches in our portability representation model (criteria relaxation, multi-label specification, and site-specific customization). No system to date has fully addressed how to represent this in a computable way, although we note systems like PheKB provide a communication hub to centralize storing multiple site implementations for a phenotype algorithm.
Instead, as more phenotype algorithms are made computable, they should incorporate localization considerations centrally, regardless of the approach needed. For example, criteria relaxation allows for a central computable definition, as it involves changing a single definition. The inclusion of version tracking (an approach used by PCORnet’s ADAPTABLE trial38 and OHDSI28) would also allow tracking changes to the phenotype algorithm as criteria are relaxed, and allow documentation of the tradeoff decisions considered over time. The resulting phenotype algorithm would then be a single repository with the full history available. For multi-label specification, a computable phenotype algorithm should provide a means by which the label may be assigned during authoring, and automatically tracked during implementation. This will require adjusting tools and specifications used in the authoring and execution processes. We note that within the PhEMA project, work is ongoing to develop a representation that meets these requirements. The final approach, site-specific customization, may benefit from lessons learned in version control systems for source code management. Based on a single, original phenotype algorithm, implementers may make a copy (“branch”) which retains a link back to the original. The implementer may propose changes (via “pull request”) or the author may review localized copies of the phenotype algorithm to determine if changes could be incorporated into the original definition.
With particular respect to controlled medical vocabularies, a previous review noted the overlap in available terms across vocabularies, although no clear rationale was ascertained as to why different vocabularies (with incomplete mappings observed in some instances) were used in a phenotype algorithm14. We posit, based on our experiences, that this is in part due to the availability of data at a local institution (the Data phase of our proposed model). That is, the rules developed or features learned in the creation of a phenotype algorithm are driven by the data collected and mapped to an IDR or CDM. While some phenotype algorithm authors do consider the most comprehensive value set definition up-front, and CDMs such as OMOP use a prescribed set of vocabularies to obviate the need for heterogeneous vocabulary mapping, they are not singular solutions. This further highlights the importance of implementing and evaluating phenotype algorithms at multiple institutions, which will naturally uncover gaps in the original definition. Portability, in turn, becomes a critical consideration.
Although our review primarily considered rules-based phenotype algorithms, similar parallels can be drawn with respect to machine learning (ML)-based data-driven algorithms14,39. ML-based approaches have demonstrated potential to achieve high-throughput phenotyping by learning relevant features of the EHR data (including features derived from narrative text), thereby reducing the time to establish a phenotype algorithm as opposed to manually curated, expert-driven rules-based phenotype algorithms at each institution40,41. Additional considerations for portability within the phenotype algorithm workflow model would include an expansion of our proposed Translation and Execution steps. This is because collection of a relevant corpus and re-training of the algorithm is needed locally before the phenotype algorithm may perform optimally42,43. For the portability tradeoff model, we believe our proposed criteria relaxation and site-specific customization approaches would apply to ML-based phenotypes, as features used within the model may be removed (relaxed) from the model for all implementers, or require other local changes depending on factors such as data availability. More recent advances in ML have included the concept of “federated learning”44, whereby predictive models may be trained across disparate data sources that are not centrally available to the model developer (e.g., to preserve privacy). This has been demonstrated in the healthcare space45, and future work in this area will be an important consideration for portable ML-based phenotype algorithms that can automate the evolution of phenotype algorithm by iterating through all sites’ data.
We do acknowledge some limitations of this work. Although we are proposing our framework as a global one, we recognize that this is not based on a systematic literature review, and is focused on the health and biomedical domain. As our reconciliation of the network-specific models was based on multiple permutations, and we reached a saturation of new permutations during the review, we do believe the model represents the current state of the field. Similarly, as new knowledge is gained within the health and biomedical domain and beyond, the model can (and should) be adjusted accordingly. We also recognize that no evaluation of the reviewed research networks is presented using this framework. In development of the framework, we observed that heterogeneity within each network can and does exist (as previously mentioned). This precluded us from classifying each network as a whole, and instead requires formal study within and across the respective networks. We believe this is important future work to be undertaken, but was outside of the scope of this paper. Given current knowledge, this model provides health and biomedical informatics researchers with a framework by which to formally evaluate and further study aspects of portability of EHR-based phenotype algorithms.
Conclusion
Based on a review of seven multi-site research networks, we have identified multiple steps and considerations that impact how an EHR-based phenotype algorithm may be made portable. We have provided a more comprehensive definition of phenotype algorithm portability by proposing a framework with eight steps across three domains (Data, Authoring, Implementation). Within these domains, we have also identified and illustrated how optimizing one can result in more or less effort required in another, and how multiple representations for portable phenotype algorithms have been used. We believe this framework will help guide future research in the area of phenotype algorithm portability.
Acknowledgements
This work was supported in part by NIGMS grant R01GM105688, NHGRI grants U01HG008673 and U01HG6379, and NCATS grant UL1TR001422. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures & Table
References
- 1.Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e2):e206–11. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH. Advances in Electronic Phenotyping: From Rule-Based Definitions to Machine Learning Models. Annual Review of Biomedical Data Science. 2018;1(1):53–68. doi: 10.1146/annurev-biodatasci-080917-013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogt TM, Elston-Lafata J, Tolsma D, Greene SM. The role of research in integrated healthcare systems: the HMO Research Network. Am J Manag Care. 2004;10(9):643–8. [PubMed] [Google Scholar]
- 4.McGraw D, Rosati K, Evans B. A policy framework for public health uses of electronic health data. Pharmacoepidemiology and Drug Safety. 2012;21:18–22. doi: 10.1002/pds.2319. Suppl 1. [DOI] [PubMed] [Google Scholar]
- 5.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Califf RM. The Patient-Centered Outcomes Research Network: a national infrastructure for comparative effectiveness research. North Carolina Medical Journal. 2014;75(3):204–10. doi: 10.18043/ncm.75.3.204. [DOI] [PubMed] [Google Scholar]
- 7.NCATS. Welcome to the ACT Network! 2019 [March 13, 2019]. Available from: http://www.actnetwork.us. [Google Scholar]
- 8.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Studies in health technology and informatics. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 9.De Moor G, Sundgren M, Kalra D, Schmidt A, Dugas M, Claerhout B, et al. Using electronic health records for clinical research: the case of the EHR4CR project. Journal of Biomedical Informatics. 2015;53:162–73. doi: 10.1016/j.jbi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Richesson RL, Sun J, Pathak J, Kho AN, Denny JC. Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artificial Intelligence in Medicine. 2016;71:57–61. doi: 10.1016/j.artmed.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. Journal of the American Medical Informatics Association : JAMIA. 2016;23(6):1046–52. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.U.S. National Library of Medicine. Home -ClinicalTrials.gov 2019 [March 13, 2019]. Available from: https://clinicaltrials.gov/ [Google Scholar]
- 13.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2014;21(2):221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. Journal of the American Medical Informatics Association: JAMIA. 2012;19(2):181–5. doi: 10.1136/amiajnl-2011-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sentinel Coordinating Center. Sentinel Common Data Model 2019 [March 13, 2019]. Available from: https://www.sentinelinitiative.org/sentinel/data/distributed-database-common-data-model. [Google Scholar]
- 17.FitzHenry F, Resnic FS, Robbins SL, Denton J, Nookala L, Meeker D, et al. Creating a Common Data Model for Comparative Effectiveness with the Observational Medical Outcomes Partnership. Applied Clinical Informatics. 2015;6(3):536–47. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical Data Interchange Standards Consortium. Study Data Tabulation Model 2018 [March 13, 2019]. Available from: https://www.cdisc.org/standards/foundational/sdtm. [Google Scholar]
- 19.Platt R, Davis R, Finkelstein J, Go AS, Gurwitz JH, Roblin D, et al. Multicenter epidemiologic and health services research on therapeutics in the HMO Research Network Center for Education and Research on Therapeutics. Pharmacoepidemiology and Drug Safety. 2001;10(5):373–7. doi: 10.1002/pds.607. [DOI] [PubMed] [Google Scholar]
- 20.Ross TR, Ng D, Brown JS, Pardee R, Hornbrook MC, Hart G, et al. EGEMS. 1. Vol. 2. Washington,DC: 2014. The HMO Research Network Virtual Data Warehouse: A Public Data Model to Support Collaboration. p. 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garza M, Del Fiol G, Tenenbaum J, Walden A, Zozus MN. Evaluating common data models for use with a longitudinal community registry. Journal of Biomedical Informatics. 2016;64:333–41. doi: 10.1016/j.jbi.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.U.S.Department of Health and Human Services. Harmonization of Various Common Data Models and Open Standards for Evidence Generation 2017 [July 10, 2019] Available from: https://aspe.hhs.gov/harmonization-various-common-data-models-and-open-standards-evidence-generation. [Google Scholar]
- 23.Clinical Data Interchange Standards Consortium. BRIDG 2019. 2019 Jul 10; Available from: https://www.cdisc.org/standards/domain-information-module/bridg. [Google Scholar]
- 24.Rasmussen LV, Kiefer RC, Mo H, Speltz P, Thompson WK, Jiang G, et al. A Modular Architecture for Electronic Health Record-Driven Phenotyping. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science. 2015;2015:147–51. [PMC free article] [PubMed] [Google Scholar]
- 25.Pacheco JA, Rasmussen LV, Kiefer RC, Campion TR, Speltz P, Carroll RJ, et al. A case study evaluating the portability of an executable computable phenotype algorithm across multiple institutions and electronic health record environments. Journal of the American Medical Informatics Association : JAMIA. 2018;25(11):1540–6. doi: 10.1093/jamia/ocy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Szolovits P, editors. Implementing a Portable Clinical NLP System with a Common Data Model -- a Lisp Perspective. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2018 Dec 3-6; doi: 10.1109/bibm.2018.8621521. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bache R, Miles S, Taweel A. An adaptable architecture for patient cohort identification from diverse data sources. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e2):e327–33. doi: 10.1136/amiajnl-2013-001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.OHDSI. OHDSI/PhenotypeLibrary: A repository to store validated phenotype cohort definitions, with all associated source code, documentation, and validation results. 2019. 2019 Mar 13; Available from: https://github.com/OHDSI/PhenotypeLibrary. [Google Scholar]
- 29.McMurry AJ, Murphy SN, MacFadden D, Weber G, Simons WW, Orechia J, et al. SHRINE: Enabling Nationally Scalable Multi-Site Disease Studies. PLOS ONE. 2013;8(3):e55811. doi: 10.1371/journal.pone.0055811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo H, Jiang G, Pacheco JA, Kiefer R, Rasmussen LV, Pathak J, et al. A Decompositional Approach to Executing Quality Data Model Algorithms on the i2b2 Platform. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science. 2016;2016:167–75. [PMC free article] [PubMed] [Google Scholar]
- 31.Atkinson R. Project management: cost, time and quality, two best guesses and a phenomenon, its time to accept other success criteria. International Journal of Project Management. 1999;17(6):337–42. [Google Scholar]
- 32.Jackson KL, Mbagwu M, Pacheco JA, Baldridge AS, Viox DJ, Linneman JG, et al. Performance of an electronic health record-based phenotype algorithm to identify community associated methicillin-resistant Staphylococcus aureus cases and controls for genetic association studies. BMC Infectious Diseases. 2016 doi: 10.1186/s12879-016-2020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo H, Thompson WK, Rasmussen LV, Pacheco JA, Jiang G, Kiefer R, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. Journal of the American Medical Informatics Association : JAMIA. 2015;22(6):1220–30. doi: 10.1093/jamia/ocv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girardeau Y, Doods J, Zapletal E, Chatellier G, Daniel C, Burgun A, et al. Leveraging the EHR4CR platform to support patient inclusion in academic studies: challenges and lessons learned. BMC medical research methodology. 2017;17(1):36. doi: 10.1186/s12874-017-0299-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang G, Kiefer RC, Sharma DK, Prud’hommeaux E, Solbrig HR. A Consensus-based Approach for Harmonizing the OHDSI Common Data Model with HL7 FHIR. Studies in health technology and informatics. 2017;245:887–91. [PMC free article] [PubMed] [Google Scholar]
- 36.Wei WQ, Leibson CL, Ransom JE, Kho AN, Caraballo PJ, Chai HS, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association : JAMIA. 2012;19(2):219–24. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim Choi Keung SN, Zhao L, Tyler E, Taweel A, Delaney B, Peterson KA, et al. Cohort identification for clinical research: querying federated electronic healthcare records using controlled vocabularies and semantic types. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science. 2012;2012:9. [PMC free article] [PubMed] [Google Scholar]
- 38.National Patient-Centered Clinical Research Network (PCORnet). ADAPTABLETRIAL/PHENOTYPE: Eligibility phenotype for potential participant identification 2017. 2019 Mar 13; Available from: https://github.com/ADAPTABLETRIAL/PHENOTYPE. [Google Scholar]
- 39.Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ : British Medical Journal. 2015;350:h1885. doi: 10.1136/bmj.h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho JC, Ghosh J, Steinhubl SR, Stewart WF, Denny JC, Malin BA, et al. Limestone: High-throughput candidate phenotype generation via tensor factorization. Journal of Biomedical Informatics. 2014;52:199–211. doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farjah F, Halgrim S, Buist DS, Gould MK, Zeliadt SB, Loggers ET, et al. EGEMS 2016. 1. Washington,DC; 2016. An Automated Method for Identifying Individuals with a Lung Nodule Can Be Feasibly Implemented Across Health Systems. p. 1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association: JAMIA. 2012;19(e1):e162–9. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrell DS, Schoen RE, Leffler DA, Morris M, Rose S, Baer A, et al. Challenges in adapting existing clinical natural language processing systems to multiple, diverse health care settings. Journal of the American Medical Informatics Association : JAMIA. 2017;24(5):986–91. doi: 10.1093/jamia/ocx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahan B, Ramage D. Google AI Blog. [Internet] 2017 Available from: https://ai.googleblog.com/2017/04/federated-learning-collaborative.html. [Google Scholar]
- 45.Brisimi TS, Chen R, Mela T, Olshevsky A, Paschalidis IC, Shi W. Federated learning of predictive models from federated Electronic Health Records. International Journal of Medical Informatics. 2018;112:59–67. doi: 10.1016/j.ijmedinf.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


