Abstract
MEDLINE is the National Library of Medicine’s premier bibliographic database for biomedical literature. A highly valuable feature of the database is that each record is manually indexed with a controlled vocabulary called MeSH. Most MEDLINE journals are indexed cover-to-cover, but there are about 200 selectively indexed journals for which only articles related to biomedicine and life sciences are indexed. In recent years, the selection process has become an increasing burden for indexing staff, and this paper presents a machine learning based system that offers very significant time savings by semi-automating the task. At the core of the system is a high recall classifier for the identification ofjournal articles that are in-scope for MEDLINE. The system is shown to reduce the number of articles requiring manual review by 54%, equivalent to approximately 40,000 articles per year.
Introduction
MEDLINE® is the U.S. National Library of Medicine’s (NLM) journal citation database. It contains over 25 million citations and is growing at rate of over 900,000 citations per year. The subject scope of MEDLINE is biomedicine and health, broadly defined to encompass the information needs of those working in healthcare and life sciences. A distinctive feature of MEDLINE is that records are manually indexed with NLM Medical Subject Headings (MeSH ® ). MEDLINE currently covers over 5,200 international journals, and the majority of these journals are indexed cover-to-cover; that is, all articles, substantive editorials, and letters are indexed. About 200 journals are selectively indexed, meaning that only articles related to biomedicine and life sciences are indexed. These selectively indexed journals are typically multidisciplinary journals, such as general science or general chemistry titles.
Between 2000 and 2017, the number of articles from selectively indexed journals has increased rapidly from approximately 13,000 to 78,000, while at the same time the fraction of articles selected for indexing has decreased from about 90% to 25%†. As a consequence, selecting articles for MEDLINE indexing is an increasing burden for NLM indexers, distracting them from their core task of indexing the biomedical literature.
This paper presents a machine learning based system that has been developed to assist indexers with the selection process. The core component of the system is a high recall classifier for the identification of journal articles that are in-scope for MEDLINE. The main inputs to the classifier are the article title and abstract, and the output is a prediction of whether the article is in-scope for MEDLINE. The classifier is used to pre-filter articles that are likely to require MEDLINE indexing and offers significant time-savings by reducing the number of articles requiring manual review. Indexers require the classifier to have close to 100% recall as they do not want to miss in-scope articles.
Related Work
We are not aware of any prior work on selecting biomedical journal articles for indexing, however triage of relevant PubMed articles is often the first step in database curation. An example of such an approach is the TREC 2004 genomics track categorization task2. This task simulated the curation of the Mouse Genome Informatics system and required the triage of articles that contain evidence supporting the assignment of a GO code to a specific gene. The goal was to limit the number of articles sent to human curators for more exhaustive analysis. A similar example is the selection and ranking of articles for the Comparative Toxicogenomics Database (CTD) in the BioCreative evaluations3. The CTD database captures chemical-gene-disease relationships and triage effectiveness was measured by gene, disease, and chemical named entity recognition performance. A different application, that also requires the selection of a subset of PubMed articles, is the identification of scientifically rigorous articles for clinicians practicing evidence-based medicine. Several machine learning based approaches have been developed to solve this important problem4–6. More generally, topic-based text classification has been extensively studied in many domains. A relevant and well-studied topic-based text classification problem is the automatic indexing of biomedical articles using the MeSH vocabulary7–9.
Methods
Dataset
The dataset is comprised of citation data for MEDLINE articles published in selectively indexed journals before September 2018. It was constructed using a list of selectively indexed journals (and associated start/end years of selective indexing) automatically extracted from the 2018 NLM List of Serials Indexed for Online Users file10. Citation data was downloaded from the 2018 MEDLINE/PubMed annual baseline1 and daily update files, and articles from selectively indexed journals were filtered based on their journal and publication date. Articles published in the same year that their journal was selected or deselected for selective indexing were excluded as selective indexing may have started or ended part way through the year.
Articles from selectively indexed journals have typically been reviewed by a single NLM indexer, and whether or not an article has been selected for indexing can be determined from the MEDLINE citation status. Indexed articles are assigned “MEDLINE” status and out-of-scope articles are assigned “PubMed-not-MEDLINE” status. There are 33 journals that are known to have problematic determinations before 2015, and the affected articles were removed from the dataset. The validation and test sets only contain articles published in 2018, as we want to evaluate how the system performs on recent articles. The test set only contains articles from a subset of 132 selectively indexed journals that the indexing team are particularly interested in‡. Some special citation types (Comment, Erratum, Expression of Concern, Republished, Retraction, Update, Reprint, and Patent Summary) are also excluded from the validation and test sets because they follow special indexing rules. The final dataset contains 1.5 million training set articles, 14,346 validation set articles, and 29,833 test set articles. As discussed previously, the fraction of articles selected for indexing has decreased over time: the training set contains articles published before 2018 and has an overall indexing rate of 64%, while the validation and test sets contain articles published in 2018 and have indexing rates of 18%.
Classifier for In-Scope Biomedical Journal Articles
The developed classifier combines the predictions of an ensemble of traditional machine learning algorithms and a Convolutional Neural Network (CNN). These two component classifiers are described in detail below.
Ensemble of Traditional Machine Learning Algorithms
The ensemble is implemented using scikit-learn11 (v0.20.2) and uses averaging to combine the predictions of Bernoulli Naive Bayes, Logistic Regression, Stochastic Gradient Descent, and Random Forest classifiers. The model was trained on 2017 data and the input features are concatenated term frequency inverse document frequency (TF-IDF) representations of the title, abstract, and author affiliations. Model hyperparameters were optimized using a grid search for F2-score and are listed in Table 1.
Table 1:
Hyperparameters selected using a grid search. Any parameters not listed in the table were set to the scikit-learn default values.
| Classifier | Hyperparameters |
| Bernoulli Naive Bayes | alpha=.01 |
| Logistic Regression | C=2 |
| Stochastic Gradient Descent | loss=modified huber, alpha=.0001, max_iter=1000 |
| Random Forest Classifier | n estimators=100, criterion=‘gini’ |
The individual algorithms for the ensemble were chosen by first evaluating their standalone performance on the validation set. Only algorithms available in scikit-learn, and known to perform well on natural language processing (NLP) tasks, were considered. Individual algorithms that performed well were chosen as candidates for the final ensemble model, and the best performing combination of algorithms was determined using a grid search.
Convolutional Neural Network
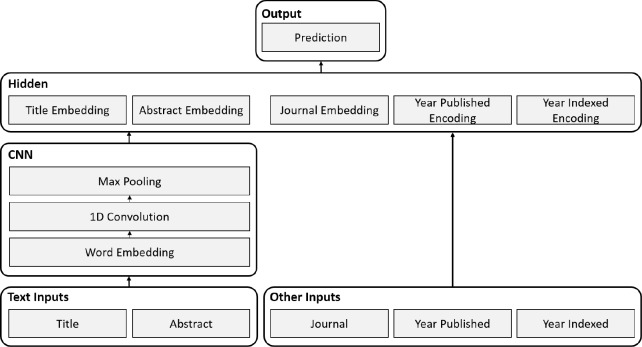
A Convolutional Neural Network is a type of deep neural network that is commonly applied to image processing tasks. Recently, however, CNNs have also been shown to be effective for various NLP problems, including text classification. The neural network architecture used in this paper (Figure 1) is based on the CNN architecture presented by Kim et al.12. In their paper, Kim et al. demonstrate that this architecture is effective for sentence classification. The architecture represents words as vectors and input text as the concatenation of word vectors. The network learns a set of convolutional filters that are convolved along the length of the input text to produce an activation map; filters learn to activate when they detect a specific type of feature (e.g. discriminative words or phrases) at some position in the text. The convolution operation is followed by a max pooling operation that keeps only the maximum activation of each filter. The result is a fixed length representation of the input text that is invariant to the position of the detected features. The final layer of the network is a task specific classification layer.
Figure 1:
CNN architecture.
This paper presents a custom neural network architecture that uses a CNN to process text inputs. There are five inputs to the network: the article title, abstract, journal, publication year, and indexing year. The network generates a fixed length representation of each input and then concatenates them to construct the input to the hidden layer. The final classification layer uses a sigmoid activation function to generate a single output value between zero and one, which can be interpreted as the probability of an article being in-scope for MEDLINE. The model uses randomly initialized word vectors, dropout regularization, and batch normalization for the hidden and convolution layers.
The title and abstract inputs are processed separately using the same word embeddings and convolutional filter weights. Standard max pooling is used for the title, whereas dynamic max pooling13 is used for the abstract. Dynamic max pooling is implemented by first dividing the abstract into five equal length sections, and then standard max pooling is applied to each of these sections. The intention is to create a richer representation of the abstract by retaining some position information.
The journal is treated as a categorical input, and each journal is represented by a fixed length vector. Like the word embeddings, the journal embeddings are learned during training. The two year inputs are represented using the special encoding scheme shown in (Figure 2). The encoding is similar to one-hot encoding; however, positions for the year and preceding years are activated. The encoding is intended to capture the sequential nature of time and allow for better generalization between years.
Figure 2:
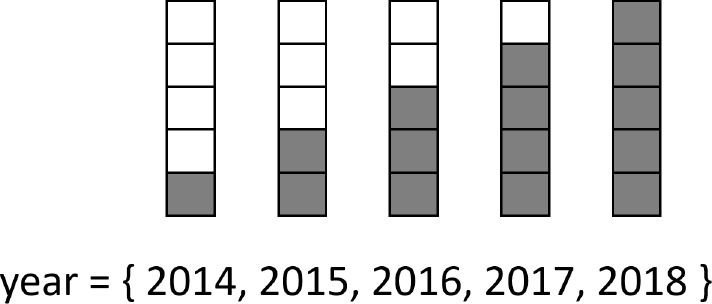
Illustration of the special encoding used for year inputs. The example shows how years between 2014 and 2018 would be encoded.
The model was trained on the full training set using binary cross entropy loss, the Adam optimizer, and mini-batch gradient descent. The F1-score metric was monitored on the validation set, and training was stopped early when it stopped increasing. Model hyperparameters (Table 2) were chosen based on the literature 12–14 and manual optimization for F1-score.
Table 2:
CNN hyperparameters.
| Hyperparameter | Value |
| vocabulary size | 400,000 |
| word embedding size | 300 |
| title max words | 64 |
| abstract max words | 448 |
| number of convolution filters | 350 |
| convolution filter sizes | 2, 5, 8 |
| dynamic max pooling number of regions | 5 |
| activation function for classification layer | sigmoid |
| activation function for all other layers | relu |
| hidden layer size | 3365 |
| journal embedding size | 50 |
| dropout rate | 0.5 |
| vocabulary dropout rate | 0.25 |
| batch size | 128 |
| learning rate | 0.001 |
Final Combined Model
The final model combines the predictions of the ensemble of traditional machine learning algorithms and the CNN. To compute the output probability of the combined model, we assume that the component model predictions are independent and take the product of their output probabilities.
The final prediction of whether an article should be selected for indexing can only be made after the selection of a decision threshold. There is a trade-off between precision and recall: a high threshold will result in high precision but low recall, whereas a low threshold will result in low precision but high recall. The classifier developed in this paper is required to have close to 100% recall, and therefore a relatively low threshold was selected on the validation set. All the code, datasets and trained models required to reproduce the results of this paper are publically available on GitHub at http://github.com/indexing-initiative/selective_indexing.
Results
This section presents a performance evaluation of the developed classifier on the 2018 test set. We first compare the performance of the combined model to the standalone performance of its component models. We then breakdown the performance of the combined model by journal topic.
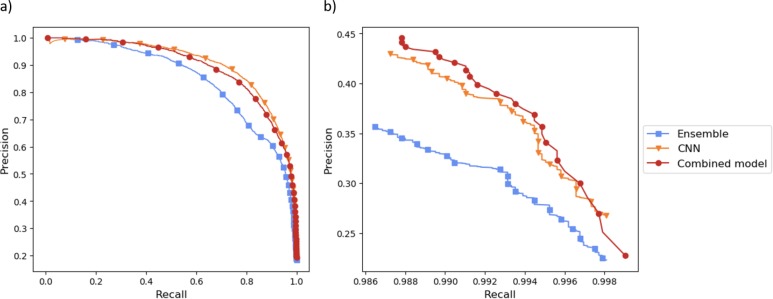
Figure 3 shows precision-recall curves for the ensemble of traditional machine learning algorithms, CNN, and combined models. Figure 3a is the full plot and Figure 3b is a zoomed in plot showing recall values close to 100%. Figure 3a shows that, for intermediate values of recall, the CNN is the best performing model. In this recall range, the relatively low performance of the ensemble degrades the performance of the combined model. We are, however, interested in performance at high recall values. For the high recall values shown in Figure 3b, it can be seen that the predictions of the ensemble and the CNN model are complementary and that the combined model has the highest precision for most values of recall.
Figure 3:
Precision-recall curves for the ensemble of traditional machine learning algorithms, CNN, and combined model a) full plot b) zoomed in plot showing precision at high recall.
After discussion with NLM indexers, it was decided that recall of about 99.5% resulted in a good balance between classifier precision and false negative error rate. After selecting a decision threshold on the validation set, the final combined model precision and recall was measured to be 38.0% and 99.4% respectively, on the test set.
In order to breakdown model performance by journal topic, each selectively indexed journal was assigned to one of four journal groups: Chemistry, Science, Jurisprudence, or Biotech. Both the compilation of the list of journal groups and the assignment of journals to groups was done based on human judgement; the MeSH terms that are assigned to MEDLINE journals15 were used as guidance. Figure 4 plots precision-recall curves for the combined model by journal group and shows that the model performance varies significantly between journal groups. Specifically, the model performs better on Science and Jurisprudence articles and worse on Biotech and Chemistry articles.
Figure 4:
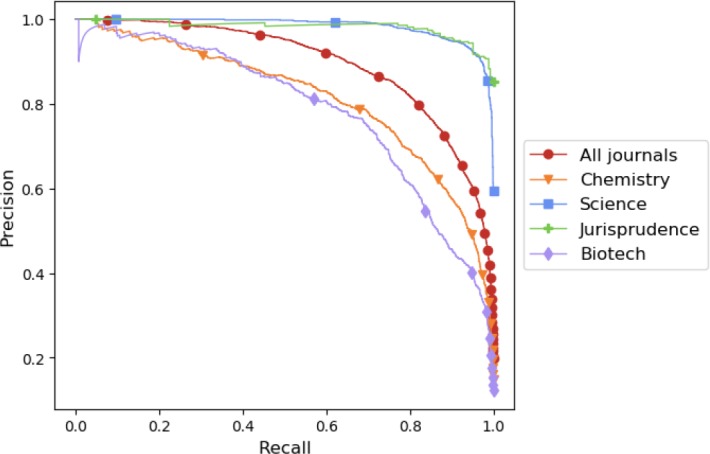
Precision-recall curves for combined model by journal group.
Discussion
At the measured precision of 38.0%, the implemented system offers NLM indexers considerable time and cost savings by allowing them to automatically discard the 54% of articles that are very unlikely to require indexing. In 2017 there were approximately 80,000 articles processed from selectively indexed journals, and we would therefore expect the system to exclude approximately 40,000 articles from manual review each year. The measured recall value of 99.4% indicates that approximately 0.6% of in-scope articles will be missed by the system, but this is considered acceptable given the expected time-savings. It is important to realize that even human indexers may miss articles or disagree on whether an article is in-scope.
In the results section it was shown that the model performance varies considerably with article topic. The reason for this variation in performance is not explored in this paper, but the analysis highlights that the model could potentially be deployed for a subset of journals for which it is particularly effective.
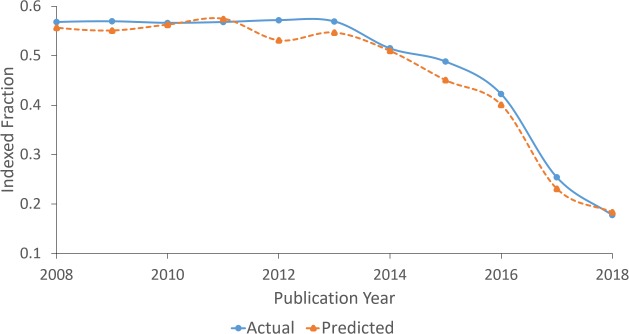
One of the key challenges that was faced when developing the presented classifier was the time-variance of the dataset. There are many factors that cause time-variance and these include changes to selective indexing policy, changes to the list of selectively indexed journals, and concept drift due to scientific progress and trends. Figure 5 offers one view of the time-variance of the dataset: it shows variation in the fraction of indexed articles from selectively indexed journals over time. The recent drop in indexing rate may be attributed to many factors, but a recent tightening of selection criteria is likely to be the most significant.
Figure 5:
Fraction of indexed articles from selectively indexed journals against publication year. Shows the actual fraction and the fraction predicted by the CNN model.
The observed time-variance is especially problematic for the training of the CNN because it requires a large amount of training data to achieve high performance. There is insufficient data from recent years, and consequently the CNN is forced to model the time-variance of the dataset and to generalize across years if possible. The publication year and indexing year inputs were added for this purpose and were found to improve performance on the 2018 validation set. Figure 5 shows that the indexing rate predicted by the CNN closely follows the true indexing rate, and this provides further evidence that the CNN is effectively modeling the time-variance of the dataset.
Conclusion
This paper presents a machine learning based system that has been developed to assist indexers with the selection of articles for MEDLINE indexing. At the core of the system is a high recall classifier for the identification of journal articles that are in-scope for MEDLINE that combines the predictions of traditional machine learning algorithms and a Convolutional Neural Network. The system is shown to offer very significant time and cost savings by allowing indexers to discard 54% of articles that are very unlikely to require indexing.
For future work, we plan to further explore the effect of the dataset time-variance on model performance. It is claimed that new language model based text representations16 require less task specific training data. It may therefore be possible to achieve better performance by training models using these new representations only on recent data. We would also like to understand why the model performance varies so much with article topic. Our motivation is that the two worst performing groups (Biotech and Chemistry) make up over two thirds of 2018 articles. Any performance improvements for these two groups will result in a significant increase in overall performance.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC).
We would like to thank Susan Schmidt, Melanie Huston, and Olga Printseva for their invaluable advice on selecting articles for indexing.
Footnotes
Statistics computed using the 2018 MEDLINE/PubMed baseline1
List of selectively indexed journals of interest to NLM indexers can be downloaded from the paper GitHub repository (http://github.com/ indexing-initiative/selective_indexing)
Figures & Table
References
- 1.MEDLINE/PubMed baseline; 2018. Available from: https://mbr.nlm.nih.gov/Download/ Baselines/2018/ [Google Scholar]
- 2.Cohen AM, Hersh WR. The TREC 2004 genomics track categorization task: classifying full text biomedical documents. Journal of Biomedical Discovery and Collaboration. doi: 10.1186/1747-5333-1-4. 2006 Mar;1(1):4 Available from: https: //doi.org/10.1186/1747-5333-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiegers TC, Davis AP, Mattingly CJ. Collaborative biocuration - text-mining development task for document prioritization for curation. Database. doi: 10.1093/database/bas037. 2012 Nov;2012. Available from: https://dx.doi.org/10.1093/ database/bas037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilicoglu H, Demner-Fushman D, Rindflesch TC, Haynes RB, Wilczynski NL. Towards automatic recognition of scientifically rigorous clinical research evidence. Journal of the American Medical Informatics Association. 2009 Jan;16(1):25–31. doi: 10.1197/jamia.M2996. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Fiol G, Michelson M, Iorio A, Cotoi C, Haynes RB. A deep learning method to automatically identify reports of scientifically rigorous clinical research from the biomedical literature comparative analytic study. J Med Internet Res. 2018 Jun;20(6):e10281. doi: 10.2196/10281. Available from: http://www.jmir.org/2018/6/e10281/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bian J, Abdelrahman S, Shi J, Fiol GD. Automatic identification of recent high impact clinical articles in PubMed to support clinical decision making using time-agnostic features. Journal of Biomedical Informatics. 2019;89:1–10. doi: 10.1016/j.jbi.2018.11.010. Available from: http://www.sciencedirect.com/science/article/pii/ S1532046418302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mork J, Aronson A, Demner-Fushman D. 12 years on – Is the NLM medical text indexer still useful and relevant? Journal of Biomedical Semantics. 2017 Feb;8(1):8. doi: 10.1186/s13326-017-0113-5. Available from: https://doi.org/10.1186/ s13326-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You R, Peng S, Zhu S, Wang H, Zhai C, Mamitsuka H. DeepMeSH: deep semantic representation for improving large-scale MeSH indexing. Bioinformatics. 2016 Jun;32(12):i70–i79. doi: 10.1093/bioinformatics/btw294. Available from: https://dx.doi. org/10.1093/bioinformatics/btw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Q, Dhingra B, Cohen W, Lu X. AttentionMeSH: simple, effective and interpretable automatic MeSH indexer. BioASQ 2018: Proceedings of the 6th BioASQ Workshop - a challenge on large-scale biomedical semantic indexing and question answering; 2018 Nov 1; Brussels, Belgium ACL. :p. 47–56. Available from: http: //aclweb.org/anthology/W18-5306. [Google Scholar]
- 10.List of serials indexed for online users; 2018. Available from: ftp://ftp.nlm.nih.gov/online/ journals/archive/lsi2018.xml. [Google Scholar]
- 11.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 12.Kim Y. Convolutional neural networks for sentence classification. EMNLP 2014: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing 2014 Oct 25-29; Doha, Qatar. ACL. 2014:p. 1746–1751. Available from: http://aclweb.org/anthology/D/D14/D14-1181.pdf. [Google Scholar]
- 13.Liu J, Chang WC, Wu Y, Yang Y. Deep learning for extreme multi-label text classification. SIGIR ’17: Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval; 2017 Aug 7-11; Tokyo, Japan. ACM. 2017:p. 115–124. Available from: http://doi.acm.org/ 10.1145/3077136.3080834. [Google Scholar]
- 14.Zhang Y, Wallace B. A sensitivity analysis of (and practitioners’ guide to) convolutional neural networks for sentence classification. Proceedings of the Eighth International Joint Conference on Natural Language Processing; 2017 Nov 27 - Dec 1; Taipei, Taiwan. Asian Federation of Natural Language Processing. 2017:p. 253–263. Available from: http://aclweb.org/anthology/I17-1026. [Google Scholar]
- 15.Humphrey SM, Lu CJ, Rogers WJ, Browne AC. Journal descriptor indexing tool for categorizing text according to discipline or semantic type. Proceedings of the 2006 AMIA Annual Symposium; 2006 Nov 11-15; Washington, DC, USA. AMIA. 2006:p. 960. [PMC free article] [PubMed] [Google Scholar]
- 16.Devlin J, Chang M, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding; 2018. arXiv:1810.04805v1 [Preprint] Available from: https://arxiv.org/abs/1810. 04805v1. [Google Scholar]