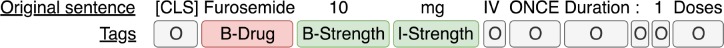Abstract
Natural language processing (NLP) is useful for extracting information from clinical narratives, and both traditional machine learning methods and more-recent deep learning methods have been successful in various clinical NLP tasks. These methods often depend on traditional word embeddings that are outputs of language models (LMs). Recently, methods that are directly based on pre-trained language models themselves, followed by fine-tuning on the LMs (e.g. the Bidirectional Encoder Representations from Transformers (BERT)), have achieved state-of-the-art performance on many NLP tasks. Despite their success in the open domain and biomedical literature, these pre-trained LMs have not yet been applied to the clinical relation extraction (RE) task. In this study, we developed two different implementations of the BERT model for clinical RE tasks. Our results show that our tuned LMs outperformed previous state-of-the-art RE systems in two shared tasks, which demonstrates the potential of LM-based methods on the RE task.
Introduction
Electronic Health Records (EHRs) contain massive free-text data documented by healthcare professionals during the course of patient care, such as clinical notes, discharge summaries, lab reports, and pathology reports1. Compared to structured data, free-text data often records detailed information of clinical events and communications between teams in hospital settings. Much important patient information often exists in unstructured format only, and manual extraction of such information is expensive and time consuming. Therefore, applying natural language processing (NLP) technologies to extract related patient information from clinical notes is highly recommended to support clinical research and applications2.
Relation extraction (RE), one of the essential tasks of information extraction, aims to identify semantic connections between mentions of concepts in a document3,4. For example, in the sentence “an MRI revealed a C5-6 disc herniation with cord compression,” we attempt to identify that the test “MRI” reveals two medical problems “a C5-6 disc herniation” and “cord compression”. Previous studies have investigated diverse types of relations, such as disease-attribute pair extraction5-6, temporal relation identification7, adverse drug event detection8-9, etc. Recently, the clinical NLP community started a series of shared tasks on relation extraction from clinical notes, including the Informatics for Integrating Biology and the Bedside (i2b2) challenges7,10, the Semantic Evaluation (SemEval) challenges11-12, the BioCreative V task13 and the most recent 2018 National NLP Clinical Challenge (n2c2)14. These open challenges greatly facilitate the development resources (e.g., corpora) and methods for RE in the medical domain.
Early RE systems can be classified into two major categories: (1) rule-based methods; and (2) machine learning-based methods. Rule-based methods such as dependency trees15 and coreference chains16 are used to extract relations. Machine learning-based methods such as Support Vector Machine (SVM)17 and Conditional Random Field (CRF)4are widely applied. However, current existing RE methods largely rely on heavy preprocessing steps that require extracting features from text, such as lexical, syntactic and semantics information18. Those extraction at early steps might introduce sources of errors that would accumulate to the following RE task19. These limitations severely restrict the portability and generalizability of RE to other novel resources.
In the past few years, deep learning methods have demonstrated their effectiveness and often achieved the state-of-the-art in diverse NLP tasks. Sequential-based deep learning methods including recurrent neural networks (RNN), convolutional neural networks (CNN) and their variants have been applied to clinical relation extraction20-22. More recently, Bidirectional Encoder Representations from Transformers (BERT)23 introduced masked language models and reported superior performance on multiple benchmark datasets of diverse NLP tasks. BERT is pre-trained on a large corpus in an unsupervised manner and can then be fine-tuned on a downstream task with a simple layer on top of its architecture. By fine-tuning as a whole, the network adjusts the entire language model and thus encodes more contextualized information. BERT has shown its benefits in downstream NLP tasks such as concept extraction24-25, text classification26, question answering27, and text generation28, especially pre-trained on a large clinical or biomedical corpus. Due to their ability to capture contextualized information, deep language representation approaches achieve a task-specific architecture by simply appending the deep learning model for the downstream task.
Despite BERT’s success in the open domain and biomedical literature, it has not yet been applied to clinical RE tasks. In this study, we aim to investigate how to apply BERT to clinical RE tasks by (1) comparing two different implementations of the BERT model for clinical RE; (2) evaluating BERT models across different clinical RE tasks; and (3) assessing four BERT models trained from the open domain, biomedical literature, and clinical text on the clinical RE task. To the best of our knowledge, this is the first study to apply pre-trained language models of BERT to clinical RE tasks and our evaluation results show that it outperformed the previous state-of-the-art systems in clinical RE.
Material and Method
Tasks and Datasets
In this study, we used two datasets: the 2018 National NLP Clinical Challenges (n2c2) corpus14 and the 2010 Informatics for Integrating Biology & the Bedside (i2b2) challenge corpus10. All named entities (clinical concepts) were given, and the task here was to identify relations between entities.
The n2c2 corpus included 505 discharge summaries, which came from the MIMIC-III (Medical Information Mart for Intensive Care III) clinical care database29. The corpus contained nine types of clinical concepts including drug and eight attributes (reason, frequency, ADE, strength, duration, route, form and dosage). The relations between drug and the eight attributes were also provided (statistics of relations are in table 1). The task is to recognize all relations between drugs and attributes. The training set included 303 discharge summaries and the test set included 202 discharge summaries.
Table 1.
Statistics of relations in the n2c2 and the i2b2 corpora.
| Dataset | Type | Example | Number |
| n2c2 | Strength → Drug | Patient has been switched to lisinopril iOmg 1 tablet PO QD. iOmg → Lisinopril | 10946 |
| Duration → Drug | Patient prescribed 1-2 325 mg / 10 mg Norco pills every 4-6 hours as needed for pain.4-6 hours → Norco | 1069 | |
| Route → Drug | Patient has been switched to lisinopril 10mg 1 tablet PO QD. PO → lisinopril | 9084 | |
| Form → Drug | Patient has been switched to lisinopril 10mg 1 tablet PO QD. tablet → lisinopril | 11028 | |
| ADE → Drug* | Patient is experiencing muscle pain, secondary to statin therapy for coronary artery disease. muscle pain → statin |
1840 | |
| Dosage → Drug | Patient has been switched to lisinopril 10mg i tablet PO QD. i → lisinopril | 6920 | |
| Reason → Drug | Patient prescribed 1-2 325 mg / 10 mg Norco pills every 4-6 hours as needed for pain.pain → Norco | 8578 | |
| Frequency → Drug | Patient has been switched to lisinopril 10mg 1 tablet PO QD. QD → lisinopril | 10344 | |
| Description | |||
| i2b2 | TrIP | Treatment improves medical problem | 203 |
| TrWP | Treatment worsens medical problem | 133 | |
| TrCP | Treatment causes medical problem | 526 | |
| TrAP | Treatment is administered for medical problem | 2616 | |
| TrNAP | Treatment is not administered because of medical problem | 174 | |
| TeRP | Test reveals medical problem | 3051 | |
| TeCP | Test conducted to investigate medical problem | 504 | |
| PIP | Medical problem indicates medical problem | 2203 |
*ADE: adverse drug event.
The other dataset used in this study came from the 2010 i2b2 challenge relation extraction task, including 426 discharge summaries collected from 2 hospitals. The dataset is a subset of the original dataset used in the challenge, since the University of Pittsburgh Medical Center’s data is not available to the public and was removed from the original dataset after the challenge. Only 170 of the original 394 training documents and 256 of 477 test documents were available for download, so we combined and re-split them in this study. The dataset includes 3 types of entities (medical problem, lab test and treatment) and 8 types of relations between them (Table 1).
BERT-based relation extraction
Given entities annotated in sentences, the relation extraction task can be transformed into a classification problem. A classifier can be built to determine categories of all possible candidate relation pairs (e1, e2), where entities e1 and e2 are from the same sentence. For the n2c2 dataset, we generated candidate pairs by pairing each of the drugs with each of the attributes in a sentence. For example, given sentence “Furosemide 10 mg IV ONCE Duration : 1 Doses”, there were five entities “Furosemide”, “10 mg”, “IV”, “ONCE” and “1”. All possible candidate pairs were (10 mg, Furosemide), (IV, Furosemide), (ONCE, Furosemide) and (1, Furosemide). For the i2b2 dataset, we generated candidate pairs by pairing each of the problems with the other entities: including problems, treatments and tests. Before we build the classifiers for relation extraction, all documents went through a pre-processing procedure that includes basic steps such as sentence boundary detection and tokenization, which were done using the CLAMP (Clinical Language Annotation, Modeling, and Processing) Toolkit30.
In this study, we developed two BERT-based methods: Fine-Tuned BERT and Feature Combined BERT to determine relation categories for these candidate pairs, described below.
Fine-Tuned BERT (FT-BERT)
Input representation
In order to represent a candidate relation pair in an input sentence, we used the semantic type of an entity to replace the entity itself. For example, as described above, there were four possible candidate pairs in the sentence “Furosemide 10 mg IV ONCE Duration: 1 Doses”, and it would be transformed into four samples (Figure 1). Each of them contained one candidate relation pair. In Sample 1, two entities “Furosemide” and “10 mg” were replaced by their semantic type labels “@Drug$” and “@Strength$”. Note that even if an entity contains multiple words, it is still replaced by one label.
Figure 1.
An example of transformed samples from an original sentence used in FT-BERT.
FT-BERT model
Devlin et al.’s BERT model23 was used, and a linear classification layer was added on top to predict the label of a candidate pair in sentential context (Figure 2). In detail, a classification token [CLS] was added at the beginning of a sentence, whose output vector was used for classification. As typical with BERT, we used a [CLS] vector as input to a classification layer. Then a softmax layer was added to output labels for the sentence.
Figure 2.
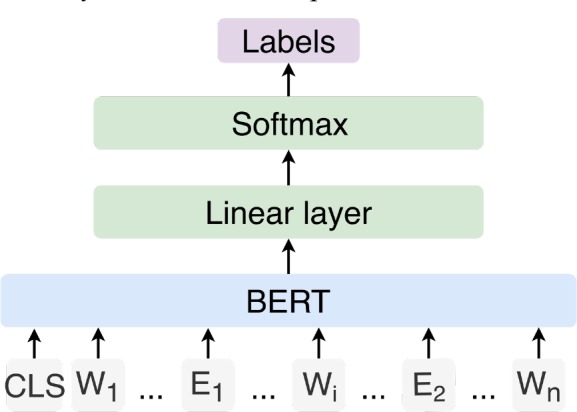
The architecture of FT-BERT. CLS represents the [CLS] token; Wi represents words in sentence; E1 and E2 represent replaced semantic type labels of entities.
Feature Combined BERT (FC-BERT)
Input representation
Instead of replacing entities with semantic type labels, we used additional BIO tags to represent entities in sentence, where “B” represents the beginning of an entity, “I” represents other words inside an entity, and “O” represents all other non-entity words. Compared with the input representation in FT-BERT, the BIO representation keeps entity word information so that the model has more information to classify. Figure 3 shows how to use BIO tags to represent sample (1) from Figure 1. Both sentence words and their tags are used together as input for FC-BERT.
Figure 3.
An example of the input representation in FC-BERT. The traditional BIO (Beginning, Inside, Outside) tags are augmented with the semantic types of the respective tokens.
FC-BERT Model
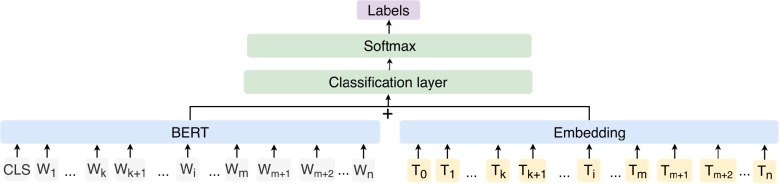
We utilized the BERT model to generate vectors for all words in sequence; in parallel to the BERT model, the BIO tag sequence of the sentence was represented in an embedding layer. The vectors for words and the vectors for tags were concatenated (according to the original index position) and then sent to a classification layer, which was a BI-LSTM neural network with attention31. Output labels per sentence were obtained via softmax.
Baseline methods
We also included a few baseline methods for comparison with the BERT models. For the n2c2 dataset, two strong baseline methods, the CNN-RNN method and the JOINT method were used, which were developed by our group and achieved the best performance on relation classification task in the n2c2 challenge32. For the i2b2 dataset, the baseline methods used were the Seg-CNN method from Luo et al.,33 and the SDP method from Li et al,34 which both outperformed the best systems that participated in the i2b2 challenge. We also compare with Li et al.’s recent Seg-GCRN method35, which improves upon the Seg-CNN results incrementally. The Seg-CNN and Seg-GCRN evaluations used the original 2010 i2b2 dataset (including data from the University of Pittsburgh Medical Center) to develop and evaluate their methods, while the SDP method (and our experiments) excluded the Pittsburgh data. All results of baseline methods came from their original paper without re-running.
Experiments
For each BERT-based method, we evaluated four pre-trained language models, namely, (1) uncased BERT-large23, (2) cased BERT-large23, (3) the BioBERT model that was pre-trained using PubMed Central full text articles and PubMed abstracts24 and (4) the cased MIMIC BERT model that was pre-trained using the MIMIC III dataset25. The difference between uncased BERT-large model and cased BERT-large model was that the former converted all words into lower case and the latter did not. For the n2c2 dataset, the original training set was randomly split into a new training set and a development set of 242 and 61 documents respectively (about 4:1), and the original test set (202 documents) was still used as test set for evaluation. For the i2b2 dataset, we mixed the original training set and test set together, then randomly split it into new training, development and test sets (with a ratio about 3:1:1). The development set was used for optimizing parameters, and the test set was used for evaluation. Note that this split of data implies that our i2b2 results are not directly comparable with the published literature using the original corpus. The evaluation metrics used in this study were as follows.
| (1) |
| (2) |
| (3) |
Model parameters. The parameters of BERT were not frozen during training. The parameter maximum sequence length for both of methods were 128. The hidden layer size for Bi-LSTM in FC-BERT was 100 and embedding size of tag in FC-BERT was 100. For other parameters, we used the default parameters in the BERT.
Results
Table 2 shows results of the FT-BERT/FC-BERT models and baseline methods using four different pre-trained models on the n2c2 dataset. On the n2c2 data all four FT-BERT models that used different pre-trained BERT models outperformed the baseline method CNN-RNN. Three of them (except the FT-BERT that used cased BERT) outperformed the JOINT method on F1 score (up to 0.0023). All FC-BERT models performed worse than baseline method JOINT, but the uncased BERT model was better than the CNN-RNN method. Among FT-BERTs that used different pre-trained models, the FT-BERT that used the MIMIC BERT model was better than the one that used other pre-trained BERT models on F1 score and precision. The pre-trained BERT model using biomedical literature (BioBERT) showed no improvement on the clinical datasets compared with the models trained on data from the open domain. This may be because the BioBERT model trained based on the original BERT-base model includes fewer parameters than the original BERT-large models used in the study.
Table 2.
The overall performances of the FT-BERT and the FC-BERT methods on the n2c2 dataset. The Cased and Uncased models are original Google models; BioBERT corresponds to a model trained from PMC and PubMed data; and MIMIC BERT is trained on MIMIC III. P, R, and F represent precision, recall and F1 score, respectively. The boldface represents the best performance for each metric.
| Dataset | Baseline | FT-BERT | FC-BERT | |||||||
| CNN-RNN | JOINT | Cased | Uncased | Bio | MIMIC | Cased | Uncased | MIMIC | ||
| n2c2 | P | 0.9673 | 0.9715 | 0.9811 | 0.9803 | 0.9798 | 0.9838 | 0.9632 | 0.9653 | 0.9662 |
| R | 0.8878 | 0.9079 | 0.899 | 0.9021 | 0.9018 | 0.9015 | 0.8524 | 0.8952 | 0.8769 | |
| F | 0.9258 | 0.9386 | 0.9383 | 0.9396 | 0.9392 | 0.9409 | 0.9044 | 0.9289 | 0.9194 | |
All FT-BERT models outperformed the baseline method SDP on the i2b2 dataset under all three metrics of precision, recall and F1 score (Table 3). Only the FT-BERT models that used uncased BERT and MIMIC BERT outperformed the other two baseline methods Seq-CNN and Seq-GCRN. All FC-BERT models were worse than baseline methods. Among the FT-BERT models, the one that used MIMIC BERT was still the best on the i2b2 dataset.
Table 3.
The overall performances of the FT-BERT and the FC-BERT methods on the i2b2 dataset. The Cased and Uncased models are original Google models; BioBERT corresponds to a model trained from PMC and PubMed data; and MIMIC BERT is trained on MIMIC III. P, R, and F represent precision, recall and F1 score, respectively. The boldface represents the best performance for each metric.
| Dataset | Baseline | FT-BERT | FC-BERT | ||||||||
| SDP | Seq-CNN* | Seq-GCRN* | Cased | Uncased | Bio | MIMIC | Cased | Uncased | MIMIC | ||
| i2b2 | P | 0.7569 | 0.748 | 0.772 | 0.7489 | 0.7673 | 0.7346 | 0.7624 | 0.6885 | 0.6843 | 0.6850 |
| R | 0.7303 | 0.736 | 0.743 | 0.7606 | 0.7681 | 0.7681 | 0.7734 | 0.6991 | 0.7056 | 0.7521 | |
| F | 0.7434 | 0.742 | 0.758 | 0.7547 | 0.7677 | 0.751 | 0.7679 | 0.6938 | 0.6948 | 0.7170 | |
*These two methods were developed and evaluated on the original dataset of the 2010 i2b2 challenge.
Table 4 shows the F1scores of our methods on each category of the n2c2 dataset. All four FT-BERT models performed better on all categories than CNN-RNN. The JOINT was slightly better than FT-BERT on four categories (Strength®Drug, Frequency ®Drug, Form®Drug and Route ®Drug). Compared with JOINT, three of four FT-BERT models improved the performance on the most difficult categories, reason and ADE; the FT-BERT models that used MIMIC BERT performed best (0.7674 vs. 0.7552, JOINT; 0.8124 vs. 0.7871, JOINT). The FC-BERT models performed better than the CNN-RNN but worse than the JOINT on these two categories.
Table 4.
Fl score of the FT-BERT and the FC-BERT methods on each category of the n2c2 dataset. The Cased and Uncased models are original Google models; BioBERT corresponds to a model trained from PMC and PubMed data; and MIMIC BERT is trained on MIMIC III. The boldface represents the best performance for each category.
| Relation type | Baseline | FT-BERT | FC-BERT | ||||||
| CNN-RNN | JOINT | Cased | Uncased | Bio | MIMIC | Cased | Uncased | MIMIC | |
| Strength→ Drug | 0.9743 | 0.9875 | 0.9832 | 0.9850 | 0.9842 | 0.9850 | 0.9348 | 0.9760 | 0.9598 |
| Dosage → Drug | 0.9572 | 0.9709 | 0.9685 | 0.9717 | 0.9693 | 0.9708 | 0.9320 | 0.9542 | 0.9402 |
| Duration → Drug | 0.8436 | 0.8798 | 0.8815 | 0.8939 | 0.8833 | 0.8920 | 0.8279 | 0.8674 | 0.8655 |
| Frequency → Drug | 0.9568 | 0.9674 | 0.9624 | 0.9631 | 0.9632 | 0.9637 | 0.9364 | 0.9611 | 0.9511 |
| Form → Drug | 0.9707 | 0.9761 | 0.9748 | 0.9754 | 0.975 | 0.9750 | 0.9656 | 0.9728 | 0.9702 |
| Route → Drug | 0.9653 | 0.9732 | 0.9706 | 0.9717 | 0.9704 | 0.9724 | 0.9324 | 0.9560 | 0.9468 |
| Reason → Drug | 0.7271 | 0.7552 | 0.7598 | 0.7603 | 0.7642 | 0.7674 | 0.7106 | 0.7409 | 0.7338 |
| ADE → Drug | 0.7329 | 0.7871 | 0.8027 | 0.8000 | 0.8084 | 0.8124 | 0.7483 | 0.7744 | 0.7644 |
Table 5 shows the F1scores of our methods on each category of the i2b2 dataset. The FT-BERT method performed better on 6 of all categories than SDP. On the one hand, PIP was a larger category (of 2203 relations), so there were enough positive examples for the model to learn. But, on the other hand, there were too many possible candidate relation pairs that made it difficult for the model to classify. FT-BERT improved the PIP performance from 0.6333 to 0.7260 (BioBERT). For the categories with less relations (TrWP, TrNAP and TrIP), FT-BERT improved the performance on TrNAP, but failed to improve the other two. FC-BERT showed an improvement on TrWP (0.5217 vs. 0.4457).
Table 5.
F1 score of the FT-BERT and the FC-BERT methods on each category of the i2b2 dataset. The Cased BERT, Uncased BERT, BioBERT, and MIMIC BERT represent the methods that uses pre-trained models of cased BERT-large, uncased BERT-large, BERT model trained from the PMC and PubMed and BERT model trained from the MIMIC III respectively. The boldface represents the best performance.
|
Relation type |
Baseline | FT-BERT | FC-BERT | |||||
| SDP | Cased | Uncased | Bio | MIMIC | Cased | Uncased | MIMIC | |
| TeCP | 0.6117 | 0.5833 | 0.6537 | 0.6146 | 0.6462 | 0.5486 | 0.6238 | 0.6114 |
| TeRP | 0.8444 | 0.8547 | 0.8681 | 0.8535 | 0.8763 | 0.8122 | 0.8577 | 0.8303 |
| PIP | 0.6333 | 0.7204 | 0.7188 | 0.7260 | 0.721 | 0.6277 | 0.5963 | 0.6591 |
| TrCP | 0.6213 | 0.6369 | 0.6087 | 0.5650 | 0.6162 | 0.5904 | 0.6145 | 0.5146 |
| TrAP | 0.7974 | 0.7918 | 0.7989 | 0.7762 | 0.7965 | 0.7121 | 0.7222 | 0.7437 |
| TrWP | 0.4457 | 0.3051 | 0.3729 | 0.4231 | 0.3673 | 0.4138 | 0.5217 | 0.4687 |
| TrNAP | 0.4227 | 0.4333 | 0.6286 | 0.3662 | 0.5079 | 0.3175 | 0.3235 | 0.4416 |
| TrIP | 0.6159 | 0.4167 | 0.5152 | 0.5128 | 0.5275 | 0.5250 | 0.4865 | 0.5783 |
In order to compare our methods with previous studies, we also merged all eight types of relations into three. Relations between tests and problems (TeCP and TeRP) were merged as Test-Problem, relations between treatments and problems (TrCP, TrAP, TrWP, TrNAP and TrIP) were merged as Treatment-Problem, and relations between problems and problems were kept unchanged. The performances of our methods were calculated on the merged categories. Table 6 shows the results. On overall performance, FT-BERT was 0.01 higher than the best baseline method Seg-GCRN. FT-BERT was better than baseline methods on two of three merged categories (Test-Problem and Treatment-Problem).
Table 6.
F1 score of the FT-BERT on merged categories of the i2b2 dataset. The Uncased BERT and MIMIC BERT represent the methods that uses uncased BERT-large and BERT model trained from the MIMIC III respectively. The boldface represents the best performance.
| Baseline | FT-BERT | |||
| Seg-CNN | Seg-GCRN | Uncased | MIMIC | |
| Test-Problem | 0.820 | 0.827 | 0.836 | 0.843 |
| Problem-Problem | 0.702 | 0.741 | 0.719 | 0.721 |
| Treatment-Problem | 0.686 | 0.692 | 0.736 | 0.728 |
| Overall | 0.742 | 0.758 | 0.768 | 0.768 |
Discussion
In this study, we investigated deep learning methods that made use of pre-trained language models on relation extraction from clinical narratives. It fills the gap of applying novel deep learning methods with pre-trained language models to the clinical RE task. Our results showed that the FT-BERT pre-trained language model achieved an F1 score of 0.9409 and 0.7679 on the n2c2 and the i2b2 datasets respectively, which outperformed previous state-of-the-art systems in the both challenges, demonstrating the advantage of using BERT models. A pre-trained BERT model that used MIMIC III corpus improved the overall performance, including performance in some difficult categories (e.g. Reason→Drug, ADE→Drug, etc.).
Research about deep language representations for clinical RE is still in its early stages, and pre-trained models like BERT show promise as a successful paradigm for future work. Previous deep learning approaches require further work on issues like parameter optimization during deployment. BERT, on the other hand, is a pre-trained language model on a large corpus, which can be used for multiple downstream tasks. Each task simply adds a layer on top of the basic BERT model, and then fine-tunes the parameters for that task-specific layer. It is thus easier to deploy than feature-based methods that build complex models on top of features (e.g. word vectors from word2vec). In this study, we also show that the fine-tune method (FT-BERT) performed better than the feature-based method (FC-BERT), indicating the simple architecture built on BERT is promising.
Although some improvements were observed, the FC-BERT method performed worse than the FT-BERT method. There could be several reasons. First, the FC-BERT method used all entity words as inputs; but low frequency entity words may not be easily learned by the model. In contrast, FT-BERT made the patterns clear by replacing all entities with their semantic types. For example, in sentence “… given nebs , IV solumedrol and dose of levoquin IV and admitted for copd exacerbation .”, FT-BERT recognizes all relations correctly and FC-BERT fails to recognize all of them. The fact that FC-BERT uses all information including words may also cause the failure of recognizing some parallel structure. For example, in sentence “… Vancomycin / Cefepime for Staph …”, FC-BERT can only recognize relation between Cefepime and Steph, but fails to recognize relation between Vancomycin and Staph.”. Second, in the FC-BERT method, a random initialized embedding layer was used to represent BIO tags, which may be too simple to capture the difference and connections between BIO tags. A pre-trained vector may improve the performance of FC-BERT.
The MIMIC FT-BERT model performed the best on both of these two datasets, but on the i2b2 dataset it was only slightly better than the uncased BERT model. It may be because the n2c2 dataset was a subset of the MIMIC III corpus, so that MIMIC BERT was trained on in-domain data. Both cased BERT and uncased BERT were pre-trained on corpora from the open domain, but the performance of the latter was better. It may be because the word shape feature doesn’t help for RE. The BERT model that pre-trained on biomedical literature only slightly improved the overall performance on two datasets. One possible reason for this was that it is pre-trained on BERT-base instead of BERT-large, and the language used in biomedical literature is different from the language used in clinical narratives.
This is just the first attempt to apply BERT to clinical RE and there are different aspects that can be further improved. Our study followed the traditional framework of classifying candidate relation pairs, using the FT-BERT and FC-BERT models. Because the architecture of the JOINT method showed good performance on the n2c2 challenge, in the future, it’s possible to improve performance by combining BERT with JOINT. Moreover, we plan to further evaluate pre-trained language models from clinical corpora, e.g., supplement additional medical vocabulary and train uncased MIMIC III models.
Conclusion
In this study, we developed and evaluated BERT-based methods for clinical relation extraction. Our results show our RE methods based on pre-trained language models outperformed previous state-of-the-art RE systems in two shared tasks. In addition, our evaluation shows using clinical data to pre-train BERT models can benefit clinical RE.
Conflicts of Interest
Dr. Xu and The University of Texas Health Science Center at Houston have research-related financial interests in Melax Technologies, Inc.
Acknowledgement
This work is supported by NLM 5R01LM010681, NLM R00LM012104 , NCI U24 CA194215, NIGMS 5U01TR002062 and the Cancer Prevention Research Institute of Texas (CPRIT) Training Grant #RP 160015.
Figures & Table
Figure 4.
The architecture of FC-BERT. “Wi” represents word, and “Ti” represents corresponding BIO tag. “Wk Wk+1” and “Wm Wm+1 Wm+2” represents two entities of the sentence.
References
- 1.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nature Reviews Genetics. [Internet] 2012 Jun; doi: 10.1038/nrg3208. [cited 2019 Mar 11];13(6):395-405. Available from: http://www.nature.com/articles/nrg3208. [DOI] [PubMed] [Google Scholar]
- 2.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2009.08.007. [Internet]. 2009 Oct [cited 2019 Mar 11];42(5):760-72. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1532046409001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culotta A, McCallum A, Betz J. In: Proceedings of the main conference on Human Language Technology Conference of the North American Chapter of the Association of Computational Linguistics -. [Internet] New York, New York: Association for Computational Linguistics; 2006. Integrating probabilistic extraction models and data mining to discover relations and patterns in text. [cited 2019 Mar 11]. p. 296-303. Available from: http://portal.acm.org/citation.cfm?doid=1220835.1220873. [Google Scholar]
- 4.Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. Journal of the American Medical Informatics Association. 2011;18(5):594–600. doi: 10.1136/amiajnl-2011-000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Q, Ji Z, Li Z, Du J, Wang J, Tiryaki F, Wu S, Zhang Y, Xu H. UTH: Identifying Medications and Corresponding Attributes in Electronic Health Record. In AMIA n2c2 Shared-Task and Workshop. 2018 [Google Scholar]
- 6.Si Y, Roberts K. A Frame-Based NLP System for Cancer-Related Information Extraction. AMIA Annu Symp Proc. 2018;2018:1524–33. [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. Journal of the American Medical Informatics Association. 2013;20(5):806–813. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Lee H-J, Ji Z, Wang J, Wei Q, Xu H. UTHCCB System for Adverse Drug Reaction Extraction from Drug Labels at TAC-ADR 2017. In: TAC. 2017 [Google Scholar]
- 9.Aramaki E, Miura Y, Tonoike M, Ohkuma T, Masuichi H, Waki K, et al. Extraction of adverse drug effects from clinical records. Stud Health Technol Inform. 2010;160(Pt 1):739–43. [PubMed] [Google Scholar]
- 10.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bethard S, Savova G, Chen WT, Derczynski L, Pustejovsky J, Verhagen M. Semeval-2016 task 12: Clinical tempeval. In Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016) 2016:pp. 1052, 1062. [Google Scholar]
- 12.Bethard S, Derczynski L, Savova G, Pustejovsky J, Verhagen M. Semeval-2015 task 6: Clinical tempeval. In Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) 2015:pp. 806–814. [Google Scholar]
- 13.Wei CH, Peng Y, Leaman R, Davis AP, Mattingly CJ, Li J, Wiegers TC, Lu Z. Overview of the BioCreative V chemical disease relation (CDR) task. In Proceedings of the fifth BioCreative challenge evaluation workshop. 2015:pp. 154–166. [Google Scholar]
- 14.Harvard Medical School. 2018 n2c2 shared-task and workshop. track 2: Adverse drug events and medication extraction in ehrs. doi: 10.1093/jamia/ocz166. URL https://n2c2.dbmi. hms.harvard.edu/track2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fundel K, Küffner R, Zimmer R. RelEx—Relation extraction using dependency parse trees. Bioinformatics. 2006 Dec 1;23(3):365–71. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 16.Lavergne T, Grouin C, Zweigenbaum P. The contribution of co-reference resolution to supervised relation detection between bacteria and biotopes entities. BMC bioinformatics. 2015;16(10):S6. doi: 10.1186/1471-2105-16-S10-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HJ, Xu H, Wang J, Zhang Y, Moon S, Xu J, Wu Y. UTHealth at SemEval-2016 task 12: an end-to-end system for temporal information extraction from clinical notes. In Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016) 2016:pp. 1292–1297. [Google Scholar]
- 18.Konstantinova N. In International Conference on Analysis of Images, Social Networks and Texts. Springer, Cham: 2014. Apr 10, Review of relation extraction methods: What is new out there? pp. pp. 15–28. [Google Scholar]
- 19.De Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. Journal of the American Medical Informatics Association. 2011 May 12;18(5):557–62. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y. Recurrent neural networks for classifying relations in clinical notes. Journal of biomedical informatics. 2017 Aug 1;72:85–95. doi: 10.1016/j.jbi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahu SK, Anand A, Oruganty K, Gattu M. Relation extraction from clinical texts using domain invariant convolutional neural network. arXiv preprint arXiv:1606.09370. 2016 Jun 30; [Google Scholar]
- 22.Raj D, SAHU S, Anand A. Learning local and global contexts using a convolutional recurrent network model for relation classification in biomedical text. In Proceedings of the 21st conference on computational natural language learning (CoNLL 2017) 2017:pp. 311–321. [Google Scholar]
- 23.Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805. 2018 Oct 11; [Google Scholar]
- 24.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. BioBERT: pre-trained biomedical language representation model for biomedical text mining. arXiv preprint arXiv:1901.08746. 2019 Jan 25; doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si Y, Wang J, Xu H, Roberts K. Enhancing Clinical Concept Extraction with Contextual Embedding. Journal of the American Medical Informatics Association. 2019 Jul 2; doi: 10.1093/jamia/ocz096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kant N, Puri R, Yakovenko N, Catanzaro B. Practical Text Classification With Large Pre-Trained Language Models. arXiv preprint arXiv:1812.01207. 2018 Dec 4; [Google Scholar]
- 27.Alberti C, Lee K, Collins M. A BERT Baseline for the Natural Questions. arXiv preprint arXiv: 1901.08634. 2019 Jan 24; [Google Scholar]
- 28.Gero KI, Karamanolakis G, Chilton L. Transfer Learning for Style-Specific Text Generation [Google Scholar]
- 29.Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Scientific data. 2016 May 24;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, Xu H. CLAMP-a toolkit for efficiently building customized clinical natural language processing pipelines. Journal of the American Medical Informatics Association. 2017 Nov 24;25(3):331–6. doi: 10.1093/jamia/ocx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P, Shi W, Tian J, Qi Z, Li B, Hao H, Xu B. Attention-based bidirectional long short-term memory networks for relation classification. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers) 2016;Vol. 2:pp. 207–212. [Google Scholar]
- 32.Wei Q, Ji Z, Li Z, Du J, Wang J, Xu J, et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. Journal of the American Medical Informatics Association. 2019 May 28; doi: 10.1093/jamia/ocz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y, Cheng Y, Uzuner O, Szolovits P, Starren J. Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. Journal of the American Medical Informatics Association. 2017 Aug 31;25(1):93–8. doi: 10.1093/jamia/ocx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Yang Z, Shen C, Xu J, Zhang Y, Xu H. Integrating shortest dependency path and sentence sequence into a deep learning framework for relation extraction in clinical text. BMC medical informatics and decision making. 2019 Jan;19(1):22. doi: 10.1186/s12911-019-0736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Jin R, Luo Y. Classifying relations in clinical narratives using segment graph convolutional and recurrent neural networks (Seg-GCRNs) Journal of the American Medical Informatics Association. 2018 Dec 27;26(3):262–8. doi: 10.1093/jamia/ocy157. [DOI] [PMC free article] [PubMed] [Google Scholar]