Abstract
Accurate identification of temporal information such as date is crucial for advancing cancer research which often requires precise date information associated with related cancer events. However, there is a gap for existing natural language processing (NLP) systems to identify dates for specific cancer research studies. Illustrated with two case studies, we investigated the feasibility, evaluated the performances and discussed the challenges of date information extraction for cancer research.
Introduction
Cancer is the second leading cause of mortality in the US and despite extensive research and rapid advancements made in understanding this complex and heterogeneous disease, further innovative research leveraging real-world patient data could spearhead major therapeutic development. Observational studies, classically used to identify risk factors and prognostic indicators, were shown to be effective in estimating treatment effects1. Observational studies have the advantage of reduced cost, being timely and offering a broad range of observable patterns making them an appealing method for cancer research. However, in order to draw conclusive results, labor- and time-intensive conventional data collection methods are required for observational studies2.
Over the last decade, Electronic Health Records (EHR) systems have been increasingly implemented at US hospitals. Huge amounts of longitudinal and detailed patient information, including lab tests, medications, disease status, and treatment outcome, have been accumulated and are available electronically. One of the challenges faced in conducting EHR-based cancer research is to extract information from clinical narratives. Cancer-related events can vary among different cancers. For example, “transplant’ and “conditioning” before bone marrow transplant therapy are specific events for multiple myeloma. Traditional chart review has been labor intensive and expensive. With the advancement of cancer informatics research, natural language processing (NLP) techniques have been explored to extract cancer- related events with some success. In cancer research, accurate identification of temporal information associated with cancer-related events is crucial3. For example, diagnosis dates and treatment start dates of cancers are both important for survival analysis4,5, postsurgical death evaluation within 30 days6 or evaluation of post treatment side effect7. Pinpointing diagnosis dates of cancer at various stages are crucial for selecting therapy8,9, therefore, it’s very important for personalized cancer therapy10. Earlier diagnosis dates were also associated with more favorable outcomes among specific cancer types11. In addition, initial treatment date is an important factor for effective cancer management12 and better health outcomes13.
Existing clinical NLP efforts focus on profiling time line instead of pinpointing exact dates associated with specific events for unstructured data. For example, the Sixth Informatics for Integrating Biology and the Bedside (i2b2) Natural Language Processing Challenge for Clinical Records14 was to extract three types (before, after and overlap) of temporal relations between the given events and between events and temporal expressions with a focus on temporal reasoning in clinical narratives. The Clinical TempEval challenges in SemEval from 2015 to 2017 also addressed temporal information extraction and temporal relation tasks. The evaluation was conducted separately for time expression, event and temporal relations. Temporal relations (before, after, overlap, before_overlap), which is most related to the above issue of extracting dates associated with specific cancer events, were evaluated at two-level through 1) the most coarse level, i.e., relating events to the document creation time; 2) narrative container relations, i.e., the event occurred within a certain time frame, which is not specifically associated with a date.15-18
Systems produced19-22 based on the above tasks are capable to extract temporal relations only on document level between events and document creation time or between events and a certain time frame. Gaps exist in identifying date information for specific EHR-based cancer studies: 1) they mostly use documentation dates as anchor dates; 2) they are not capable to extract exact dates associated with specific events across clinical documents of the same patient; and 3) they have not been practically used and evaluated in real clinical context.
There are very few existing studies focusing on date extraction for observational studies. One study attempted to automatically generate an epidemiological line list for real-time monitoring and responses to emerging public health threats, disease onset date was identified with the accuracy of 0.01 to 0.37, outcome date with the accuracy between 0.36 to 0.66 using word embeddings23. However, this was a population-level study, without emphasis on exact date extraction for precise research purpose. Ruud et al. used the SAS text mining tool (SAS Text Miner) to extract date, time, physician, and location information of follow-up appointment arrangements from 6,481 free-text dismissal records at Mayo Clinic. The 6,481 free-text dismissal records were manually reviewed by a health services analyst to determine whether the instructions contained follow-up appointment arrangements. Sensitivity of date extraction achieved 0.996 (0.994–0.998) and specificity achieved 0.842 (0.828–0.856)24. Nevertheless, the study only focused on dismissal records of in-patient encounters.
In this study, we investigated the date extraction task using two case studies: one from malignant solid tumors (lung cancer) and one from malignant liquid tumors (multiple myeloma), this study investigated the feasibility, evaluated the performances and discussed the challenges of date information extraction for EHR-based cancer research, which to our knowledge has not previously been studied.
Methods
Figure 1 shows the overall study design for the two case studies, i.e., lung cancer and multiple myeloma. We developed a rule-based information extraction system using the open source clinical NLP pipeline MedTagger as the platform25. The system consists of two steps: (1) identifying events: we first identified sentences with events from electronic health records (EHR). Negated events were removed. (2) extracting dates associated with events: we then extracted dates within or around these sentences and linked the dates with extracted events. Evaluation was conducted against the reference standards after normalizing dates with MedTime26 and heuristic rules. The following sections will elaborate each part in detail.
Figure 1.
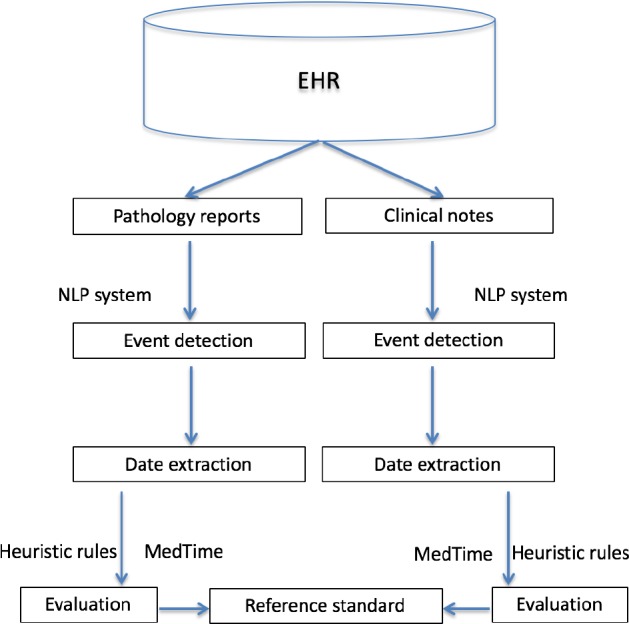
Overall study design.
Date sets
For lung cancer, an existing lung cancer cohort containing 4,110 patients definitively diagnosed with primary lung cancer from 1997 to 2016 was used for this study. Previously human abstractor manually reviewed charts to extract diagnosis dates for each patient. In this study, clinical narratives from various data sources including pathology reports and clinical notes were retrieved from Mayo Clinic clinical data warehouse (CDW). We randomly selected 499 lung cancer patients and associated clinical narratives as the training set for deriving language expression patterns. The remaining 3,611 patients were used as the test set to evaluate the NLP system.
For multiple myeloma, we randomly selected 318 patients with a treatment date after year 2000 from an existing multiple myeloma database consisting of 11,656 patients collected over the past 50 years. Human abstractors have manually reviewed charts to extract treatment start dates for each patient. Similar to lung cancer, we retrieved the associated clinical narratives from Mayo Clinic CDW. Among the 318 patients, 185 patients have information on chemotherapy and 133 patients have information on transplant. For chemotherapy, 93 patients were used for development and 92 were used for testing, for bone marrow transplant 70 were used for development and 63 were used for testing. Transplant data elements include transplant type, transplant mobilization and transplant conditioning. However, there were gaps in the data abstraction as for the patients who underwent stem cell transplant, not all patients had their mobilization and conditioning information extracted. In the training set, only 17 out of 70 patients had mobilization fields completed and 26 had conditioning fields completed. In the testing set, the respective numbers were 26 out of 63 and 41 of 63.
NLP method
For lung cancer, the event to be extracted was lung cancer diagnosis. We first identify the sentences with mentions of lung cancer diagnosis by developing a custom dictionary of terms that described the histological cell types of lung cancer, examinations of lung, symptoms of lung cancer and positive malignancy of lung tumor. This dictionary was developed using the training data set. For pathology reports, histological cell types of lung cancer and positive malignancy of lung tumor were used to identify lung cancer in the diagnosis section. For clinical notes, the whole dictionary was used associated with the mention of “diagnosis” to identify the sentence with lung cancer diagnosis. After extracting lung cancer diagnosis, we then extracted dates associated with this event from both pathology reports and clinical notes.
To extract dates associated with lung cancer diagnosis in pathology reports. A two-step strategy was developed:
If the lung cancer diagnosis was identified in pathology reports without the section of “Path Review of Outside Specimen”, then extract the dates of the pathology reports. Outside specimen refers to the specimen prepared by other institutions instead of Mayo Clinic.
If there is a section of “Path Review of Outside Specimen” in the pathology report, and the lung cancer diagnosis was identified from the outside specimen, then extract the date associated with the outside specimen.
To extract dates associated with lung cancer diagnosis in clinical notes. A three-step strategy was developed:
If a date was appearing in the same sentence with lung cancer diagnosis, then extract the date.
If there is no date appearing in the same sentence with lung cancer diagnosis, but there is a date appearing in the sentences surrounding that sentence with lung cancer diagnosis, then extract the date.
If there is no date appearing in the same sentence or surrounding sentences with lung cancer diagnosis, then use the clinical date as the date of lung cancer diagnosis.
For multiple myeloma, events to be extracted included chemotherapy and bone marrow transplant. Bone marrow transplant has three data elements, i.e., conditioning, mobilization and transplant type, each of them has multiple values (Table 1). To identify sentences with mentions of treatments, we also employed the dictionary lookup approach by constructing a dictionary consisting of chemotherapy drugs and transplant data element values. Additional keywords found during the training process were added into the dictionary.
Table 1.
Values of three transplant data elements.
| Values of Transplant type | Values of Conditioning | Values of Mobilization |
| Allogeniec | 1=Mel 200mg/m2 | 1=Cyclophosphamide |
| Autologous | 2=Mel 140mg/m2 | 2=G-CSF |
| 3=Mel othr/unk dose | 3=G-CSF+Plerixafor | |
| 4=Mel-TBI | ||
| 5=Cyclophosphamide/TBI | ||
| 6=BEAM | ||
| 7=Melphalan +other | ||
| 8=Other | ||
| 9=Mel 100mg/m2 | ||
| 10=Fludarabine +Mel |
After identifying the values of chemotherapy and transplant data elements using the dictionary, a four-step strategy was developed to extract dates:
If a date was appearing in the same sentence with values of chemotherapy or transplant data elements, then extract the date.
If there is no date appearing in the same sentence with values of chemotherapy or transplant data elements, but there is a date inference in the sentence such as “currently receiving bortezomib, day +7”, then we use the clinical note date as the anchor date to infer the actual date.
If there is no date or date inference appearing in the same sentence with chemotherapy or transplant data elements, but there is a date appearing in the sentences surrounding that sentence, then extract the date. In our study, we first label the anchor sentences with any mentions of values of chemotherapy or transplant data elements. Then one sentence before the anchor sentence and two following sentences after the anchor sentence were extracted.
If there is no date appearing in the same sentence or surrounding sentences with values of chemotherapy or transplant data elements, then use the clinical note date as the anchor date of values of chemotherapy or transplant data elements.
After the above steps, the extracted date information can be linked to chemotherapy as conditioning and mobilization are pre-transplant procedures. Therefore, after extracting all dates associated with the values of the three transplant data elements, dates of transplant type were used as the anchor dates to identify dates associated with mobilization and conditioning. Conditioning usually was done two days before transplant, we used a time window of 4 days to identify date of conditioning associated with transplant type. Mobilization may be done as early as more than one month before transplant, we used a time window of 80 days to identify date of mobilization with transplant type.
Date Normalization
Dates extracted from pathology reports are all in the formats like “4/10/12” or “2012-04-10”. In contrast, dates extracted from clinical notes could be expressed in different formats, for example, “6/12/14”, “05-02-2016”, “Jan 7th, 2013”, “June 2015”, “April 17th”, or even in unprecise way such as “the end of March”, “early July”, “late 2012”, etc. Therefore, it is necessary to normalize the extracted dates before evaluating against the reference standards which are in the format of “2013-10-28”. A strategy of two-level date normalization was utilized. For the extracted dates with “yy-mm-dd”, “mm-dd”, “mm” or “yy-mm”, MedTime was used to normalize the dates. For the unprecise time, a set of heuristic rules were developed to map them to exact dates.
MedTime is the open source NLP pipeline for clinical temporal information extraction26. It uses clinical note dates as anchor dates and can be used to normalize the extracted dates. Input dates with year, month and day in various formats as mentioned above will be normalized to the format of “YYYY-MM-DD”. If the input date has only information of year and month, it will be normalized to the day of “15”. For example, “June 2015” will be normalized to “2015-06-15”. If the input date has only information of month and day, the clinical note year will be used as the diagnosis year. For example, if “April 17th” appears in the clinical note recorded on “2014-02-14”, it will be normalized to “2014-04-17”.
The heuristic rules were primarily used to normalize obscure time of year, season and month. The following rules were used to normalize obscure time of year. Using such heuristics, a substitute date can be generated for all cancer events, from aggressive cancer types to multiple treatments that could occur over a short period of time, to prevent any missing data in data analysis.
Exact mention of year was normalized to the middle date of the year, July 1, e.g., 1998 was normalized to July 1 1998.
Early year: Feb 15 of the year, e.g., Early 2000 was normalized to Feb 15 2000.
Beginning of year: January 15 of the year, e.g., Beginning of 2002 was normalized to January 15 2002.
Middle of year: July 1 of the year, e.g., Middle of 2004 was normalized to July 1 of 2004.
Late year: November 15, e.g., late 2006 was normalized to November 15 2006.
End of year: December 15, e.g., end of 2008 was normalized to December 15 2008.
Four seasons were defined based on a three-month interval with spring starts from March and ends after May considering the weather in Rochester MN. The following rules were used to normalize obscure time of season.
Exact mention of seasons was normalized to the middle date of the season, e.g., spring 2012 was normalized to April 15 2012.
-
Taking “winter” as an example of obscure mention of seasons:
Early winter: Dec 31, e.g., early winter 2001 was normalized to Dec 31 2001.
End of winter: Feb 15, e.g., end of winter 2003 was normalized to Feb 15 2003.
Late winter: Feb 1, e.g., late winter 2005 was normalized to Feb 1 2005.
Middle of winter: Jan 15, e.g., middle of winter 2007 was normalized to Jan 15 2007.
The following rules were used to normalize obscure time of month.
Exact mention of months was normalized to the middle date of the month, e.g., January 2011 was normalized to January 15 2011.
-
Taking “June” as an example of obscure mention of months:
Early or beginning of June: June 5, e.g., Early June 2012 was normalized to June 5 2012.
Middle of June: June 15, e.g., Middle of June 2013 was normalized to June 15 2013.
Late or end of month: June 25, e.g., Late June 2014 was normalized to June 25 2014.
Evaluation
Evaluation was conducted based on event value and date match within a time window of 6 days or 30 days. For lung cancer, it was counted as a match if extracted lung cancer diagnosis and date match with the reference standard within a time window of 6 days or 30 days. We randomly selected 20 from non-matched patients within 30 days and analyzed the reasons. For multiple myeloma, it was counted as a match if extracted values of chemotherapy, transplant type, transplant mobilization and transplant conditioning match with the reference standard within a time window of 6 days or 30 days. All un-matched cases within 30 days were reviewed for error analysis.
Results
Among the whole lung cancer cohort of 4,110 patients, 4,034 have 14,890 pathology reports, with an average of 3.7 notes per patient. 4,051 patients have 173,070 clinical notes, with an average of 42.7 notes per patient. Table 2 shows the evaluation results of diagnosis date extraction from pathology reports and clinical notes for the 3,611 testing patients. From the results matched within 6 days, diagnosis dates of 1,228 (34.0%) patients came from pathology reports done at Mayo, additional 1,777 (49.2%) came from the pathology reports with outside specimen reviewed at Mayo Clinic, combining clinical notes, 3,206 could be found achieving 88.8%. From the results matched within 30 days, diagnosis dates of 1,627 (45.1%) patients came from pathology reports done at Mayo, additional 1,638 (45.3%) came from the pathology reports with outside specimen reviewed at Mayo Clinic, combining clinical notes, 3,414 could be found achieving 94.5%.
Table 2.
Evaluation results of diagnosis date extraction. Path (Mayo) refers to pathology report done at Mayo, Path (Outside) refers to pathology reports with outside specimen reviewed at Mayo.
| Match range | Data sources | ||
| Path (Mayo) | Path (Mayo)+Path (Outside) | Path (Mayo)+Path (Outside)+Clinical Notes | |
| 6 days | 1228, 34.0% | 3005, 83.2% | 3206, 88.8% |
| 30 days | 1627, 45.1% | 3265, 90.4% | 3414, 94.5% |
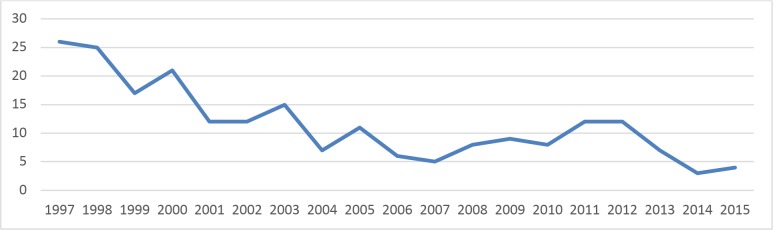
We analyzed the distribution of the missing diagnosis dates of 197 patients across different years. Figure 2 shows the time distribution of the missing diagnosis dates. Further analysis revealed that 13 patients have no clinical notes available. We randomly selected 20 patients in the remaining 184 patients and analyzed the reasons. Table 3 shows the error analysis results.
Figure 2.
Time distribution of diagnosis dates not found from either pathology reports or clinical notes.
Table 3.
Results of error analysis for lung cancer diagnosis date.
| Types of Error | Subtypes of Error | Number, % | Total |
| Outside material | None | 9, 45% | 20 |
| Human errors | None | 5, 25% | |
| Dictionary insufficiency | None | 1, 5% | |
| NLP rules | Missing inference for lung cancer diagnosis | 3, 15% | |
| Missing inference for dates | 1, 5% | ||
| Date ambiguity | 1, 5% |
The most common errors are due to outside materials (45%) and human errors (25%) (Table 3). When patients came from another hospital or country, the information of diagnosis date was often not complete in clinical notes and pathology reports. Human error may primarily result from typing process where a very near keyboard number was mistakenly typed in. For example, diagnosis date of “2013-01-22” maybe typed as “2011-01-22”. Dictionary insufficiency accounted for 5%. For those errors from NLP rules, the primary challenge came from the inference for lung cancer diagnosis, accounting for 15%. This mainly came from patients who were referred from other institutions and experience a long journey for lung cancer diagnosis. We only integrated date inference piece into the NLP tool without “year” inference, the case (5%) missing from NLP needed to use “year” inference, i.e., “Since her last evaluation one year ago, she was diagnosed as having a non-small lung cancer”. Date ambiguity happens when two dates appeared in one sentence, such as clinical note date and diagnosis date, it’s hard to be certain which date should be used.
For multiple myeloma, 312 out of 318 patients have 44,976 clinical notes, with an average of 144 notes per patient. Table 4 shows the evaluation results of treatment dates extracted from clinical notes for the testing patients. 60 (65.2%) out of 92 chemotherapy treatment start dates matched with the reference standard within 6 days, and additional 11 (12%) were matched within 30 days. For transplant type, only 1 more case was found when using 30 days match range compared with 6 days match range. For mobilization, 2 more cases were found when using 30 days match range compared with 6 days match range. For conditioning, no more cases were found when using 30 days match range compared with 6 days match range.
Table 4.
Evaluation results of treatment start date extraction.
| Match range | Chemotherapy (92) | Transplant | ||
| Type (63) | Mobilization (26) | Conditioning (41) | ||
| 6 days | 60, 65.2% | 49, 77.8% | 17, 65.4% | 24, 58.5% |
| 30 days | 71, 77.2% | 50, 79.4% | 19, 73.1% | 24, 58.5% |
Table 5 shows the error analysis for chemotherapy treatment date. Around half of 21 unmatched cases were due to human errors and outside materials (52.4%). In the remaining unmatched cases, the most challenge part came from lacking semantic inference for dates in NLP rules (33.3%). These cases were mostly referral patients who had a history of chemotherapy treatment in other institutions, and the clinical notes at Mayo Clinic only recorded the obscure time. For example, “She initially took thalidomide. She took treatment for about one to two years and then was observed until 2012”, or “He has been on ibrutinib for nearly three years”. When the chemotherapy in the reference standard was “Other” without clear definition of drug names, it is also challenging to extract corresponding chemotherapies. The last error came from the unprecise date mentioned in clinical note where only “year” was recorded. Through our heuristic rules, the year was normalized to “07-01” of the year, but the reference standard used “01-01”.
Table 5.
Results of error analysis-chemotherapy start date.
| Types of Error | Subtypes of Error | Number, % | Total No. |
| Human errors | None | 6, 28.6% | 21 |
| Outside material | None | 5, 23.8% | |
| Dictionary insufficiency | None | 2, 9.5% | |
| NLP rules | lacking semantic inference for dates | 7, 33.3% | |
| Heuristic rules for unprecise dates | 1, 4.8% |
Table 6 shows the error analysis for dates of transplant data elements. Outside material accounts for 75% for transplant type. Human error accounts for 71.4% for mobilization and 76.5% for conditioning. The “other” value was not clearly defined in dictionary. Thus, it was hard to extract. Since date mentions were very diverse, one was missed in the NLP rules.
Table 6.
Results of error analysis-transplant date.
| Data elements | Types of Error | Number, % | Total No. |
| Transplant type | Outside material | 9, 75% | 12 |
| No notes | 3, 25% | ||
| Mobilization for transplant | Human error | 5, 71.4% | 7 |
| No notes | 2, 28.6% | ||
| Conditioning for transplant | Human error | 13, 76.5% | 17 |
| Dictionary insufficiency | 2, 11.8% | ||
| Outside | 1, 5.9% | ||
| NLP rule | 1, 5.9% |
Discussion
Temporal extraction systems have not been transportable from one condition (e.g., colon cancer) to another (e.g., brain cancer) as shown in the SemEval challenge16-18. When training and testing on only notes from colon cancer patients, the top system achieved F1s of 0.76 for document time relations in 2016 with an increase of 0.058 compared with 2015, and 0.48 for narrative containers with an increase of 0.22 compared with 201516,17. When training on notes from colon cancer patients and testing on notes from brain cancer patients, the best F1 achieved 0.50 in linking events to document creation time and above 0.30 F1 for linking events to their narrative containers, with a 20+ point drops in performance18. Different patient cohorts and linguistic patterns behind different cancers are the underlying reasons for the results.
In the two case studies used in this research, lung cancer and multiple myeloma, all clinical notes were obtained from Mayo Clinic CDW which recorded not only events happened at recording time, but also recorded historical events with associated dates, if any. But different features of date expressions can be found due to distinct differences of various events in the two different cancers. Most multiple myeloma patients have clear records with treatment start dates. Since almost half of lung cancer patients in the cohort were referred from other institutions, learnt from the results (Table 2), in the clinical notes the history of lung cancer may be recorded very unclearly with more obscure time used than the multiple myeloma case.
Therefore, extracting dates associated with various cancer events required different event extraction dictionaries, different date extraction strategies as well as different approaches for linking dates with extracted events. Our study demonstrated that it is feasible to extract exact dates associated with specific cancer events for EHR-based cancer research. However, some challenges exist as illustrated in the following.
To extract precise dates associated with cancer events, it’s crucial to assert events first followed by temporal information extraction. However, it’s hard to accurately extract events as the events scatter in different data sources and there are no definite event mentions, as shown in the lung cancer case study. When using different data sources, different NLP strategies were necessary to extract events. Pathology reports record lung cancer diagnosis mainly through specific histologic cell types or assertion of positive malignancy in lung related biopsy sites. While in clinical notes lung cancer diagnosis is often represented by other miscellaneous terms, e.g., from lung examinations and symptoms of lung cancer in addition to specific histologic cell types or assertion of positive malignancy. This needs a large amount of time for developing dictionary during the training process if there was no one available. Second, the diagnosis of lung cancer may be a long journey and lacking definite diagnosis mentions was common. When the initial examinations provided uncertain results, the suspicious results would just remain undecided in one facility until it was ascertained in another facility. During the process, the mention of date and diagnosis are often existing in sentences far away from each other. To capture this complex situation, the date and diagnosis across multiple sentences were extracted using NLP. However, it’s hard to accurately extract diagnosis and date when too many sentences were involved, especially when there was no definite diagnosis mentions and there was a need to infer.
Since a lot of patients have been referred to Mayo Clinic for further treatment from other institutions, recording of exact diagnosis dates are not very clear in some cases. For example, lung cancer diagnosis date may be mentioned as “early winter 2000”. Given the long history of cancer registry databases, inconsistency of date normalization may exist among human abstractors to normalize the obscure time to specific dates. The gap may also exist between the heuristic rules we developed and the rules human abstractors used. In addition, different physicians have different styles for recording notes and there were no unified rules for recording dates. For example, it’s hard to normalize the chemotherapy start time recorded as “05/12”, because it’s hard to determine if “05” was month or year, and if “12” was month or “day”.
Linking dates to the associated events lies in the fact that events may be dependent to each other. As shown in the multiple myeloma case study, both transplant mobilization and conditioning affiliated to transplant type. Extracted mobilization and conditioning dates were linked to transplant type dates using inference rules, which may result in errors. Linking mobilization and conditioning values to associated dates was hard because 1) some patients may have several transplants within short time intervals, 2) different mobilization and conditioning values may appear repeatedly in clinical notes. This poses challenges not only for NLP but also human. In fact, as shown in evaluations we found human made many errors when linking mobilization and conditioning values to associated dates which NLP had successfully avoided.
In addition, events may have multiple related dates and it’s hard to choose the optimal date, as shown in both case studies. For lung cancer, multiple dates associated with definite lung cancer diagnosis existed in pathology reports. For example, one patient may have the same lung cancer diagnosis from different pathology reports on different dates using different specimens. For multiple myeloma, some patients may have as many as more than 20 times of chemotherapies, some of which were within very short time intervals. Thus, NLP techniques can accelerate the data abstraction process but not completely replace human efforts.
Conclusion
It is feasible to extract exact dates associated with specific cancer events for EHR-based cancer research using automatic NLP method to facilitate cancer registry curation. However, data extraction in various cancer registry databases has been ad hoc for specific research purpose. To gear with the demands for research-oriented information collection, developing NLP algorithms needs to consider the differences among various cancers in event extraction dictionaries, date extraction strategies as well as approaches for linking dates with extracted events. We plan to extend our investigation to include more cancer patients and more cancer types.
Acknowledgement
This work was supported by National Cancer Institute 1U24CA194215-01A and U01TR02062. We acknowledge Dr. Han Liu, Dr. Yanqi He, Dr. Lin Du, Dr. Yung-Hung Luo and Kathryn M Johanns for data annotation.
Figures & Table
References
- 1.Benson K, Hartz AJ. A comparison of observational studies and randomized, controlled trials. New England Journal of Medicine. 2000;342(25):1878–1886. doi: 10.1056/NEJM200006223422506. [DOI] [PubMed] [Google Scholar]
- 2.Silverman SL. From randomized controlled trials to observational studies. The American journal of medicine. 2009;122(2):114–120. doi: 10.1016/j.amjmed.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Koleck TA, Dreisbach C, Bourne PE, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. Journal of the American Medical Informatics Association. 2019 doi: 10.1093/jamia/ocy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroenke CH, Chen WY, Rosner B, Holmes MD. Weight, weight gain, and survival after breast cancer diagnosis. Journal of clinical oncology. 2005;23(7):1370–1378. doi: 10.1200/JCO.2005.01.079. [DOI] [PubMed] [Google Scholar]
- 5.Motzer RJ, Bacik J, Schwartz LH, et al. Prognostic factors for survival in previously treated patients with metastatic renal cell carcinoma. Journal of clinical oncology. 2004;22(3):454–463. doi: 10.1200/JCO.2004.06.132. [DOI] [PubMed] [Google Scholar]
- 6.Myrdal G, Gustafsson G, Lambe M, Hörte L, Ståhle E. Outcome after lung cancer surgery. Factors predicting early mortality and major morbidity. European Journal of cardio-thoracic surgery. 2001;20(4):694–699. doi: 10.1016/s1010-7940(01)00875-2. [DOI] [PubMed] [Google Scholar]
- 7.MacDonald V. Chemotherapy: managing side effects and safe handling. The Canadian Veterinary Journal. 2009;50(6):665. [PMC free article] [PubMed] [Google Scholar]
- 8.Timmerman R, Paulus R, Galvin J, et al. Stereotactic body radiation therapy for inoperable early stage lung cancer. Jama. 2010;303(11):1070–1076. doi: 10.1001/jama.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwan ML, Habel LA, Flick ED, Quesenberry CP, Caan B. Post-diagnosis statin use and breast cancer recurrence in a prospective cohort study of early stage breast cancer survivors. Breast cancer research and treatment. 2008;109(3):573–579. doi: 10.1007/s10549-007-9683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tate AR, Martin AG, Murray-Thomas T, Anderson SR, Cassell JA. Determining the date of diagnosis–is it a simple matter? The impact of different approaches to dating diagnosis on estimates of delayed care for ovarian cancer in UK primary care. BMC Medical Research Methodology. 2009;9(1) doi: 10.1186/1471-2288-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neal R, Tharmanathan P, France B, et al. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. British journal of cancer. 2015;112(s1) doi: 10.1038/bjc.2015.48. S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guadagnolo BA, Dohan D, Raich P. Metrics for evaluating patient navigation during cancer diagnosis and treatment: Crafting a policy-relevant research agenda for patient navigation in cancer care. Cancer. 2011;117(S15):3563–3572. doi: 10.1002/cncr.26269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed N, Kestin L, Ghilezan M, et al. Comparison of acute and late toxicities for three modern high- dose radiation treatment techniques for localized prostate cancer. International Journal of Radiation Oncology* Biology* Physics. 2012;82(1):204–212. doi: 10.1016/j.ijrobp.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. Journal of the American Medical Informatics Association. 2013;20(5):806–813. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pustejovsky J, Stubbs A. Increasing informativeness in temporal annotation. Paper presented at: Proceedings of the 5th Linguistic Annotation Workshop. 2011 [Google Scholar]
- 16.Bethard S, Derczynski L, Savova G, Pustejovsky J, Verhagen M. Semeval-2015 task 6: Clinical tempeval. Paper presented at: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) 2015 [Google Scholar]
- 17.Bethard S, Savova G, Chen W-T, Derczynski L, Pustejovsky J, Verhagen M. Semeval-2016 task 12: Clinical tempeval. Paper presented at: Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016) 2016 [Google Scholar]
- 18.Bethard S SG, Palmer M, et al. SemEval-2017 Task 12: Clinical TempEval. Proc 11th Int Work Semant Eval 2017, the 55th annual meeting of the Association for Computational Linguistics (ACL); 2017; Vancouver, Canada [Google Scholar]
- 19.Lin C, Dligach D, Miller TA, Bethard S, Savova GK. Multilayered temporal modeling for the clinical domain. Journal of the American Medical Informatics Association. 2015;23(2):387–395. doi: 10.1093/jamia/ocv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y-K, Chen H, Brown RA. MedTime: A temporal information extraction system for clinical narratives. Journal of biomedical informatics. 2013;46:S20–S28. doi: 10.1016/j.jbi.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Wang Y, Liu T, Tsujii J, Chang EI-C. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. Journal of the American Medical Informatics Association. 2013;20(5):849–858. doi: 10.1136/amiajnl-2012-001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherry C, Zhu X, Martin J, de Bruijn B. A la Recherche du Temps Perdu: extracting temporal relations from medical text in the 2012 i2b2 NLP challenge. Journal of the American Medical Informatics Association. 2013;20(5):843–848. doi: 10.1136/amiajnl-2013-001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh S, Chakraborty P, Lewis BL, et al. Gell: Automatic extraction of epidemiological line lists from open sources. Paper presented at: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2017 [Google Scholar]
- 24.Ruud KL, Johnson MG, Liesinger JT, Grafft CA, Naessens JM. Automated detection of follow-up appointments using text mining of discharge records. International Journal for Quality in Health Care. 2010;22(3):229–235. doi: 10.1093/intqhc/mzq012. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Bielinski SJ, Sohn S, et al. An information extraction framework for cohort identification using electronic health records. AMIA Summits on Translational Science Proceedings. 2013;2013:149. [PMC free article] [PubMed] [Google Scholar]
- 26.Sohn S, Wagholikar KB, Li D, et al. Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification. Journal of the American Medical Informatics Association. 2013;20(5):836–842. doi: 10.1136/amiajnl-2013-001622. [DOI] [PMC free article] [PubMed] [Google Scholar]