Abstract
Surgical Site Infection surveillance in healthcare systems is labor intensive and plagued by underreporting as current methodology relies heavily on manual chart review. The rapid adoption of electronic health records (EHRs) has the potential to allow the secondary use of EHR data for quality surveillance programs. This study aims to investigate the effectiveness of integrating natural language processing (NLP) outputs with structured EHR data to build machine learning models for SSI identification using real-world clinical data. We examined a set of models using structured data with and without NLP document-level, mention-level, and keyword features. The top-performing model was based on a Random Forest classifier enhanced with NLP document-level features achieving a 0.58 sensitivity, 0.97 specificity, 0.54 PPV, 0.98 NPV, and 0.52 F0.5 score. We further interrogated the feature contributions, analyzed the errors, and discussed future directions.
Introduction
Healthcare-associated infections (HAIs) affect 10% of patients undergoing surgical procedures in the United States and contribute a significant amount of morbidity to patients1. For patients, the development of an HAI can be associated with increased risk of mortality, longer length of stay, and increased risk of readmission2. Given the significance of HAIs to healthcare utilization, several surgical quality improvement programs have been designed to aid in the surveillance of HAIs. These programs include the American College of Surgeons National Surgical Quality Improvement Program (NSQIP) and the Centers for Disease Control (CDC) National Healthcare Safety Network3. The backbone of these programs is the ascertainment of clinical data through a manual chart review processes4. These processes, while providing high-quality information, are inherently limited by the cost and effort of chart review5. Additionally, these processes are under high risk of underreporting HAIs6,7, caused by biased interpretation of guideline or data, use of different criteria, insufficient training, etc. To account for these limitations, the programs rely on a sampling methodology either by selecting specific procedures or by a random sampling of procedures, thus limiting the scalability and generalizability of the manual chart review process.
Surgical site infections (SSI) are the most common type of HAI occurring after surgical procedures2. According to the CDC, an SSI is an infection in the surgical wound occurring within 30 days of the operative procedure and can be classified as superficial, deep, or organ space SSI depending on the location of the infection with the surgical wound8. One important distinction for classification of SSIs is to identify the cases with an infection present at the time of surgery (PATOS) as PATOS infections significantly increase the risk of a recurrence of an SSI after the procedure. The diagnosis of SSI can occur through multiple pathways, including provider diagnosis, provider-initiated treatment, or isolation of organisms from a microbiology culture. Given the heterogeneous nature of diagnosing SSIs, manual chart review is currently required to verify or exclude diagnoses4.
Over the past ten years, in response to the Health Information Technology for Economic and Clinical Health Act, there has been rapid adoption of Electronic Health Records (EHR) in US hospitals9. As of 2017, 96% of federal acute care hospitals have adopted a Certified EHR capable of computerized decision support, computerized provider order entry and electronic physician documentation. The almost universal adoption of certified EHRs has to potential to leverage the secondary use of EHR data for quality assurance programs and reduce or depreciate the effort of the manual chart review for surveillance of SSIs.
In previous work, several groups have leveraged electronic health care data to create automated surveillance systems for SSIs. Hu et al. used EHR data from a single institution along with machine learning models to detect the occurrence of HAIs after surgical procedures10. They used International Classification of Diseases (ICD) Codes, laboratory results, microbiology orders and results, medication administration, radiology orders, and vital signs to develop logistic regression models, using manually chart reviewed data as the gold standard. Overall, the area under the curve (AUC) for the classification of SSI was 0.91. The most predictive features included microbiology results and imaging orders. The limitation of this study is the lack of clinical text information in the development of the classification models. Given that the diagnosis of SSI is mainly dependent on physician diagnosis that is usually described in clinical notes, the inclusion of text information may improve the performance of the classification model.
Grundmeier et al.11 expanded upon this work to include limited text information to build classification models for SSI. Pre-specified keywords related to SSI that had been identified from clinical narratives were added using regular expression matching. The overall performance of the classification model significantly improved with the addition of keywords (AUC 0.97 vs. 0.94). Their best model performed with a 0.9 sensitivity and 0.28 positive predictive value (PPV). The most informative keyword features they found included: “pain,” “infection,” “warm,” “drainage.” While the inclusion of keywords improves the model performance, ignoring the context of keywords is a significant limitation of this approach, which we elaborated further in the discussion.
Other groups have focused exclusively on natural language processing methodology. FitzHenry et al. developed a rules-based NLP system using SNOMED-CT concepts from free text notes in the Veterans Affairs Healthcare sys-tem12. They developed rules for classifying ten postoperative complications after surgical procedures, including SSI. The performance of the system for detection of SSI was modest, with a sensitivity of 77% and specificity of 63%. The advantage of this approach is the inclusion of text-based features for SSI classification. However, they did not include other structured data into their algorithms, such as medications and laboratory values.
In the present study, we aim to build upon previous work on automated detection of SSIs using EHR data. We combine machine learning techniques with a rules-based natural language processing system. We hypothesize the inclusion of NLP-derived features in structured data-based machine learning models will improve the classification performance for detection of SSIs after surgical procedures. To aim at real-world use, we target our cohort to all surgical patients without any complication code filtering.
Methods
In this study, we applied a rule-based natural language processing (NLP) to extract the information related to surgical site infections (SSI) from the clinical notes, and infer the infection status for each document. Then we combined the keywords or the NLP derived features with structured EHR data to build machine learning models to classify the patients’ SSI status.
Machine learning data set development
The patient data was collected through the University of Utah Health Enterprise Data Warehouse (EDW), including patients from the University Hospital and Clinics, the University of Utah from July 1st, 2013 to August 31st, 2017. The inclusion criteria were patients whose surgical course underwent review by a trained surgical clinical reviewer (SCR) as part of the ACS-NSQIP program. Patients were excluded if they expired within 30 days of the operation. For patients included in the present study, we obtained the following structured EHR data from their records between 2-30 days after their operative procedure: hospital encounters, ICD 9th/10th Edition Diagnosis Codes, medication administration, radiology orders, laboratory and microbiology results, and vital signs. A total of 5,795 patients were included, among which 291 patients were labeled as SSI positive by manual chart review performed by an SCR. For the text data, we created a full corpus by obtaining all clinical notes from 0-30 days after the operative procedure of the following note types: admission notes, history and physical notes, discharge notes and progress notes. We excluded notes that were incomplete or not signed by a credentialed provider (Physician or Advanced Practice Provider).
Structured EHR Data Feature Engineering
For each category of features from the structured EHR data, we performed feature engineering for the development of machine learning models. ICD codes were mapped to the Agency for Healthcare Research and Quality Clinical Classification software and treated as a binary variable. Medications were mapped to RxNorm, and the number of days of each medication class was used as features. For radiology orders, we grouped common orders and used the number of orders for that particular group as a feature. Laboratory results and vital signs were grouped based on clinical meanings. For example, Potassium levels from both serum or plasma were grouped. Due to the temporal nature of laboratory and vital sign measurement, we used only the minimum, median, and maximum values of each group as features. Groups with greater than 25% missing elements were excluded. For groups with <25% missing values, we imputed using the median of the group. For the microbiology results, we created binary features based on the culture type and organism isolated.
Development of NLP Annotation Guidelines
Based on the ACS NSQIP definitions and clinical expertise, we developed an annotation guideline to perform iterative manual annotation of the NLP development corpus. We defined a mention-level annotation type Evidence of SSI and a document-level annotation type SSI_Status. The Evidence_of_SSI was used to annotate the sentences that explicitly or implicitly mentioned any evidence of infection of a surgical wound. Infection mentions without explicitly referring to an incision were not annotated. For instance, we ignored “he doesn’t have signs of infection.” The Evidence_of_SSI had two attributes: Infection_Status and Temporality. The Infection_Status was used to clarify if an SSI statement was affirmative, probable, or negated. Temporality was used to distinguish current and past SSIs. The document-level annotation type SSI_Status has one attribute Infection_Status with the same three eligible values above: affirmative, probable, or negated. At this step, we did not distinguish between SSI and SSI PATOS.
Creation of Gold-Standard NLP-Development Corpus
We created an NLP-development corpus for manual annotation from a random sample of 78 patients from the ACS NSQIP flagged SSI positive cases. The notes were selected using the same criteria mentioned above and cut off at total 2000 notes. We elected to only include SSI positive cases in our NLP development corpus due to the low prevalence of SSIs. Sampling data from positive patients improved the efficiency of both annotation and NLP rule development. In addition, because SSIs develop over the postoperative course, clinical notes from patients with SSIs contain a combination of SSI positive and negative documents.
These clinical notes were pre-annotated using Knowledge Author13 with broad vocabularies consolidated based on CDC and NSQIP definitions and using vocabulary expansion from the Unified Medical Language System (UMLS). The corpus was then split into 50 document batches and imported into the annotation tool: eHOST (The Extensible Human Oracle Suite of Tools)14. Two surgeons with expertise in SSI (LCCP, CLL) were trained using the annotation guidelines and evaluated using inter-annotator agreement. After training on five batches, the inter-annotator agreement reached over 80% for mention level and over 90% for document-level annotation. A third reviewer (BTB) checked the disagreements and performed adjudication. We subsequently performed a single-person annotation on the remaining 1750 documents with 10% overlap between the annotators to ensure consistent agreement. On the overlapped notes the annotators had an inter-annotator agreement of 70% at the mention level and 85% at the document level. The annotated gold-standard NLP-development corpus was split 50% for NLP pipeline training (1000 documents) and testing (1000 documents) corpus.
NLP Pipeline Development and Evaluation
The NLP pipeline was developed using a rule-based tool, Easy Clinical Information Extractor (EasyCIE)15. The main components of EasyCIE are listed in Table 1. These rule-based components were built on an optimized rule processing engine16 to allow high-speed processing of large rule sets and data sets. The rules for section detector, sentence segmenter, and negation context detector were imported from previous projects with minor tuning. The rest of the rules were developed based on the 1,000 NLP pipeline training documents. The developed pipeline was assessed on the training set for its rule coverage and then evaluated against the NLP pipeline test corpus (1000 notes). Because the rule-based NLP system used more granular and additional mention types to infer document-level conclusions, its mention-level annotation types did not match with the reference standard. Thus, we did not evaluate the mention-level performance, and only document-level F1 scores are reported.
Table 1:
NLP components and corresponding functionality of EasyCIE
| NLP Components | Functionality Description |
| Section detector | Identify the sections, e.g., History of Present Illness, Family History |
| Sentence Segmenter | Detect sentence boundaries |
| Named entity recognizer | Identify target concepts using dictionaries |
| Context detector | Attach the context information as feature values to the corresponding target concepts |
| Attribute inferencer | Make mention-level conclusions based on target concepts and corresponding attributes |
| Document inferencer | Make a document-level conclusion from the corresponding mention-level conclusions |
Machine Learning Model Development and Evaluation
After the NLP system was developed from the NLP corpus, we processed all the text data in the machine learning corpus to develop NLP features. These features included two binary feature sets: an NLP document feature set(ND) and an NLP mention feature set (NM). The document feature set included two features: ND_SSI_POS to cover whether a patient’s documents contain any positive SSI note; and ND_SSI_PROB to represent whether a probable SSI note was included. The mention feature set includes 28 features corresponding to the 28 mention-level annotation types generated by NLP. For instance, NM_ABDOMEN represents the feature describing if there is any mention that can indicate the abdomen body site; NM_INCISION represents the feature describing if any incision mention exists.
In addition, two numeric feature sets were also created from NLP document-level results. Frequency (FRE): the total number of the positive SSI notes or the possible SSI notes of a patient during the observation time window. Normalized frequency (NOR): the FRE divided by the total notes of that patient during the observation time window. In total, we obtained six document-level features. We performed the same conversion to mention-level annotations, resulting in 84 mention-level features. As another baseline comparison, we simply used the named entity recognizer without any context detection and inferences to extract the keywords in the notes. The extracted mentions were used to generated keyword features (KW). For readers’ convenience, we listed the abbreviations of different feature sets in Table 2 below.
Table 2:
Abbreviations of the feature sets used in this study
| Abbreviations* | Feature level | Feature type |
| NO NLP | - | - |
| ND BIN | Document | Binary |
| ND FRE | Document | Frequency |
| ND NOR | Document | Normalized frequency |
| ND ALL | Document | All three types above |
| NM BIN | Mention | Binary |
| NM FRE | Mention | Frequency |
| NM NOR | Mention | Normalized frequency |
| NM ALL | Mention | All three types above |
| KW BIN | Keywords | Binary |
| KW FRE | Keywords | Frequency |
| KW NOR | Keywords | Normalized frequency |
| KW ALL | Keywords | All three types above |
Note: Each of these abbreviations represents a set offeatures instead of a single feature.
* All the above feature sets include structured data features
We performed a Chi-Square feature selection to select the top performing variables. Logistic regression, support vector machines (SVM), and random forest (RF) models were developed. We applied nested cross-validation to the data set to avoid overfitting17. Because the data set was highly imbalanced with relatively sufficient positive cases, we performed down-sampling rather than up-sampling to avoid overfitting. We used three folds in the outer loop and five folds in the inner loop. We used a grid search for parameters in the inner loop. The best estimators from the inner loop were selected based on F1 score because the data used for inner loop cross-validation were down sampled. Then these best estimators were evaluated on test sets of the outer loop (without down-sampling). We used the following performance metrics for evaluation: sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), plus F0.5. We choose the F measure with β =0.5, because we prefer to weight precision higher than recall for practical surveillance consideration. Due to the imbalanced data set, we elected not to use the area under the receiver operating curve (AUC) in the assessment. We compared the sensitivity and specificity of each model to the NO NLP baseline machine learning model using McNemar’s test. In addition, we used bootstrapping with 1000 replications to obtain the distribution of F0.5 score and compared the distribution of each model to the NO NLP baseline machine learning model.
Results
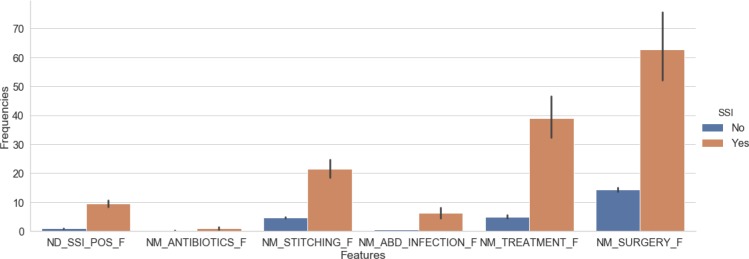
Once the rule-based NLP solution was developed, we assessed the rule coverage by calculating the performance on the training set. Our final micro-average of precision, recall, and F1 scores were 0.83, 0.83, 0.83 on the training corpus, respectively. Then we evaluated the NLP system on the 1000 blind test notes. The micro-average of the precision, recall, and F1 scores were 0.75, 0.75, 0.75 in the testing corpus, respectively. Next, we use the developed NLP solution to process all the text data of our full corpus to generate NLP features for machine learning. To demonstrate the value of the NLP-derived features, Figure 1 plots the frequency of selected NLP-derived features in patients with and without SSI. There are significantly more SSI-related NLP features in SSI positive compared to SSI negative patients (p<0.01).
Figure 1:
The means and standard deviations of example frequency features within SSI positive and negative cohorts
We subsequently used the NLP-derived features to develop the machine learning models for patient-level classification of SSI. We evaluated models using the features described in Table 2 and compared the performance of each model to the baseline structured data model without NLP-derived features. The results are shown in Table 3. The RF models with document features have the highest average Specificity (0.97) and significantly improved compared to the baseline RF NO_NLP model (p<0.001). The best PPV (0.54) and best F0.5 (0.52) comes from the RF models enhanced with binary document features (ND_BIN). Compared with the RF models with structured data features alone (NO_NLP), the PPV significantly improved by 8% and F0.5 improved by 7% (p<0.001). The addition of mention-level features or keyword features did not significantly improve the F0.5 compared the baseline RF model with structured data alone. Although the logistic regression models and the SVM models have higher NPV values (0.99), their F0.5 scores are lower than the corresponding RF models, because they are lower in PPV. The SVM models with binary mention features (NM_BIN), all mention features (NM_All) and all NLP features (ND_NM) also have the highest sensitivity, but much worse F0.5. The highest F0.5 of all the models enhanced with keywords features is 0.480, which is not significantly different from the models with structured data alone.
Table 3:
Machine learning models’ performances (average scores) on the test sets without balancing
| Models | Feature sets* | Sensitivity (p-value)# | Specificity (p-value)# | PPV | NPV | F0.5 (p-value)# |
| Random | NO NLP | 0.70 | 0.95 | 0.46 | 0.98 | 0.47 |
| Forest | ND BIN | 0.58 (<0.001) | 0.97 (<0.001) | 0.54 | 0.98 | 0.54 (<0.001) |
| ND FRE | 0.59 (<0.001) | 0.97 (<0.001) | 0.53 | 0.98 | 0.53 (<0.001) | |
| ND NOR | 0.59 (<0.001) | 0.97 (<0.001) | 0.53 | 0.98 | 0.52 (<0.001) | |
| ND ALL | 0.57 (<0.001) | 0.97 (<0.001) | 0.53 | 0.98 | 0.52 (<0.001) | |
| NM BIN | 0.59 (<0.001) | 0.96 (<0.001) | 0.51 | 0.98 | 0.50 (0.99) | |
| NM FRE | 0.70 (0.56) | 0.94 (0.02) | 0.45 | 0.98 | 0.46 (0.08) | |
| NM NOR | 0.69 (0.65) | 0.95 (0.81) | 0.46 | 0.98 | 0.47 (0.93) | |
| NM ALL | 0.58 (<0.001) | 0.96 (<0.001) | 0.51 | 0.98 | 0.50 (0.73) | |
| ND NM | 0.59 (<0.001) | 0.96 (<0.001) | 0.52 | 0.98 | 0.51 (0.14) | |
| KW BIN | 0.58 (<0.001) | 0.96 (<0.001) | 0.51 | 0.98 | 0.50 (0.63) | |
| KW FRE | 0.70 (0.32) | 0.95 (0.29) | 0.45 | 0.98 | 0.47 (0.57) | |
| KW NOR | 0.60 (<0.001) | 0.96 (<0.001) | 0.52 | 0.98 | 0.51 (0.06) | |
| KW ALL | 0.59 (<0.001) | 0.96 (<0.001) | 0.51 | 0.98 | 0.50 (0.84) | |
| SVM | NO NLP | 0.80 | 0.91 | 0.32 | 0.99 | 0.37 |
| ND BIN | 0.80 (0.48) | 0.91 (0.61) | 0.33 | 0.99 | 0.37 (0.63) | |
| ND FRE | 0.79 (0.65) | 0.91 (0.04) | 0.33 | 0.99 | 0.38 (0.11) | |
| ND NOR | 0.80 (1.00) | 0.91 (0.09) | 0.32 | 0.99 | 0.36 (1.00) | |
| ND ALL | 0.80 (1.00) | 0.91 (0.68) | 0.32 | 0.99 | 0.37 (0.01) | |
| NM BIN | 0.81 (0.53) | 0.91 (0.60) | 0.33 | 0.99 | 0.37 (0.01) | |
| NM FRE | 0.78 (0.06) | 0.91 (0.10) | 0.33 | 0.99 | 0.37 (0.15) | |
| NM NOR | 0.80 (1.00) | 0.92 (<0.001) | 0.34 | 0.99 | 0.38 (1.00) | |
| NM ALL | 0.81 (0.39) | 0.92 (0.16) | 0.34 | 0.99 | 0.38 (0.01) | |
| ND NM | 0.81 (0.49) | 0.91 (0.21) | 0.34 | 0.99 | 0.38 (0.01) | |
| KW BIN | 0.80 (0.81) | 0.91 (0.48) | 0.32 | 0.99 | 0.36 (0.62) | |
| KW FRE | 0.79 (0.26) | 0.92 (<0.001) | 0.35 | 0.99 | 0.39 (0.01) | |
| KW NOR | 0.79 (0.26) | 0.91 (0.05) | 0.33 | 0.99 | 0.38 (0.34) | |
| KW ALL | 0.80 (1.00) | 0.91 (0.86) | 0.32 | 0.99 | 0.36 (0.86) | |
| Logistic | NO NLP | 0.77 | 0.95 | 0.43 | 0.99 | 0.48 |
| Regression | ND BIN | 0.78 (0.18) | 0.95 (1.00) | 0.44 | 0.99 | 0.48 (0.92) |
| ND FRE | 0.77 (0.32) | 0.95 (0.04) | 0.45 | 0.99 | 0.49 (0.15) | |
| ND NOR | 0.77 (1.00) | 0.95 (1.00) | 0.43 | 0.99 | 0.48 (0.15) | |
| ND ALL | 0.79 (0.10) | 0.94 (<0.001) | 0.40 | 0.99 | 0.45 (0.70) | |
| NM BIN | 0.79 (0.18) | 0.94 (<0.001) | 0.40 | 0.99 | 0.44 (0.47) | |
| NM FRE | 0.78 (0.65) | 0.95 (<0.001) | 0.45 | 0.99 | 0.49 (0.60) | |
| NM NOR | 0.77 (1.00) | 0.95 (1.00) | 0.43 | 0.99 | 0.48 (0.01) | |
| NM ALL | 0.79 (0.18) | 0.94 (<0.001) | 0.40 | 0.99 | 0.44 (0.11) | |
| ND NM | 0.79 (0.11) | 0.94 (<0.001) | 0.40 | 0.99 | 0.44 (0.17) | |
| KW BIN | 0.79 (0.17) | 0.93 (<0.001) | 0.39 | 0.99 | 0.43 (<0.001) | |
| KW FRE | 0.77 (1.00) | 0.95 (1.00) | 0.43 | 0.99 | 0.48 (1.00) | |
| KW NOR | 0.77 (1.00) | 0.95 (1.00) | 0.43 | 0.99 | 0.48 (1.00) | |
| KW ALL | 0.79 (0.17) | 0.93 (<0.001) | 0.39 | 0.99 | 0.43 (<0.001) |
* The abbreviations of feature sets are explained in Table 2. NO NLP, structured data features alone.
ND, NLP document-level features; NM, NLP mention-level features; BIN, binary features;
FRE, frequency features; NOR, normalized frequency features; ALL, use all document or mention features.
# Computed against the models using structured data alone.
Discussion
The value of NLP in SSI detection
In previously reported studies of surgical site infection surveillance, structured variables (e.g., ICD Codes, laboratory results, microbiology orders and results, medication administration, etc.) are commonly used to generate machine learning features for disease classification. These variables can reflect a significant amount of information related to SSIs, including diagnostic information as well as treatment modalities for SSI. However, given the complex nature of SSI diagnosis, without the inclusion of textual information, these models are inherently limited in performance. For example, the documentation of SSI may only be located in the clinical notes if the infection did not result in any change of laboratory orders, vital sign or medication administration, e.g., a mild surgical wound infection. Also, laboratory results or medication administration may not exist in the structured database if a patient has received emergent care outside the index hospital. This type of missing information varies a lot across different health organizations; it is especially prevalent in tertiary medical centers where patients commonly travel a long distance to receive care.
Previous studies have investigated incorporating free text into machine learning models using keywords; however, without contextual information the use of keywords does not maximize the potential of NLP. In this study, we observed that it is common that clinical notes mentioned the keywords related to SSI but not in the way of demonstrating evidence of an SSI. For example, surgical procedures in the past medical history or any medical conditions mentioned as patient education for future precautions. Of all the mentions (980,045 mentions) that we extracted using our NLP systems, 7.3% are negated concepts (e.g., “Midline incision well approximated with staples, no erythema, no drainage”), 1.5% are within irrelevant context (e.g., “Stay away from people with infections”), 3.5% are about other infections other than abdominal SSIs (e.g., “septic arthritis of the bilateral wrists”), and 0.5% are historical concepts (e.g., “Sepsis 05/2011”). Ignoring the context of these mentions adds in a significant amount of noise (12.8% in total) to machine learning models. Additionally, a document-level conclusion of an SSI may not stem from the explicit mention of SSI, but instead from inferencing multiple mentions of other different concepts. For instance, a note with mentions of “post-op s/p” and “intraabdominal infection” in separated sentences. Neither of these two concepts alone can conclude SSI, while together, they suggest a positive SSI. These cases require additional inference from mention-level information to draw document-level conclusions, whereas the simple keywords approach is incapable of.
The results of the comparative experiments above (Table 3 suggest that handling these subtle contexts and logic correctly does provide additional information gains in the development of machine learning models. The best F0.5 score using structured data alone is 0.48 (Logistic regression model). Adding keywords features improves the performance on the RF models compared with RF models using structured data alone, but the best F0.5 of all keywords enhanced models is still 0.48. Using NLP features, the best F0.5 score reaches 0.52 – an 8.3% improvement (p<0.05).
NLP error analyses and indications for future directions
The NLP errors occur at two levels: mention level, document level. The most common mention-level errors are from the context not seen in the training set. For instance, “site color red” is mentioned in a list of description of a surgical incision. Our NLP solution did not capture the earlier context about the incision, thus failed to detect the “red” as a sign of infection. In the document-level errors, the most common cause is that our developed rules do not capture all the logic at the document level. For instance, we did not observe the rare co-occurrence of other healthcare-associated infection, such as pneumonia or urinary tract infection (UTI) in the NLP training set. In these cases, it is common to refer to pneumonia or UTI as “infection” in general. Within a context of abdominal condition description, NLP tends to interpret these general infection mentions as possible abdomen infections. However, in the NLP test set, these cases were more prevalent. Among all the 180 false negative SSI notes, 35 of them have a diagnosis of pneumonia, and 64 notes have other infections without an explicit mention of an anatomical site. These are the errors that we will address in future studies.
Additionally, we also found some annotation errors in the reference standard. Often these errors happened within the notes where doctors use templates or copy a section of text from other notes, while changing a few words that support different document conclusions. The annotators tended to skip the snippets that they have seen before, and these slightly modified text pieces become difficult to identify. On the other hands, it suggests that pre-annotating these notes using developed NLP solutions might improve the human review accuracy and reduce the workload. Implementing these into real practice workflow could speed up the human chart review process, reduce errors and human labor, even if a completely automated surveillance system is not available.
Machine learning models analyses and indications for future directions
As the results show, NLP features do provide additional value to the machine learning models. Notably, our champion RF models using NLP document features not only scored better than the others but also have statistically significant differences compared with the models using structured data alone.
The logistic regression models generated sub-optimal results among the three classifiers. We examined the best performing models built from NLP document frequency features (ND_FRE). Because we used nested cross-validation, we trained 15 logistic regression models enhanced with the ND_FRE feature set (5 models in each outer loop). Then we got three best models (one per each outer loop). We identified the common features among these three models, by using the average of the corresponding coefficients and ranked them in descending order (see Table 4). The results rank the NLP derived document-level feature — the frequency of SSI positive documents, as the No. 1 weighted feature.
Table 4:
The average coefficients of the common features used in the three best Logistical Regression models
| Feature names | Average coefficients |
| ND SSI POS F | 5.21 |
| Anaerobic Culture | 3.74 |
| Abscess | 2.97 |
| Guided | 2.68 |
| CCS Dx Postoperative Infection | 2.56 |
| Wound Culture | 1.83 |
| CCS Dx Peritonitis & intestinal abscess | 0.87 |
| CCS Dx Complications of surgical procedures or medical care | 0.62 |
We also examined the Random Forest classifier by estimating the importance of features (See Table 5). It shows that the NLP document feature is ranked as the most important feature. We chose one of the decision trees within the RF classifier to visualize its tree structure. Figure 2 visualizes a decision tree’s root from one of the top RF models, which identified the NLP binary document feature stands as the tree root. 56.7% of the test cases are classified as positive, and 43.3% are classified as negative. Moreover, 80% of the cases on both sides are correctly classified. Other trees show similar functions to these features. These visualizations indicate that NLP does play an essential role in our champion RF models. Because of the overall performance of SVM models (they are the lowest among the three classifiers in this study), and the difficult nature of feature interpretation for SVM, we do not present the SVM models analyses here.
Table 5:
The feature permutation importance from the best RF models using NLP document binary features
| Feature names | Feature importance |
| ND_SSI_POS | 0.20 |
| Med_DIAGNOSTIC_PRODUCTS | 0.19 |
| CCS_Dx_Postoperative_Infection | 0.16 |
| CT_abd_pelvis | 0.13 |
| Anaerobic_Culture | 0.11 |
| Wound_Culture | 0.09 |
| CCS_Dx_Peritonitis_and_intestinal_abscess | 0.04 |
| CCS_Dx_Peritonitis | 0.03 |
| CCS_Dx_Complications_of_surgical_procedures_or_medical_care | 0.02 |
| Guided | 0.02 |
| Abscess | 0.01 |
Figure 2:
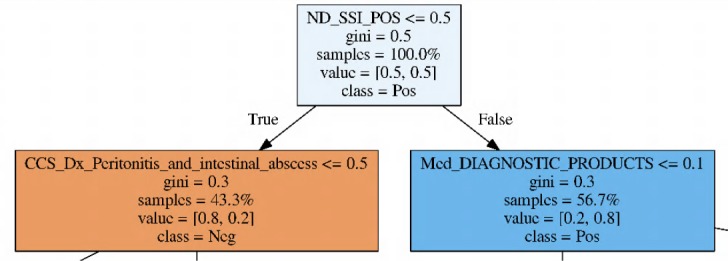
Visualizing the root of a decision tree from one of the best Random Forest models enhanced with document binary features. “samples” means the percentage of cases reaching the node. “value”: left is the percentage of true negative cases, right is the percentage of true positive cases. “class” is the conclusion from the parent node.
Additionally, we investigated the errors predicted by machine learning models and NLP. We found a significant number of errors are caused by misclassifying the patients who admitted with infections present at the time of surgery (PATOS), e.g., perforated appendix or cholecystitis. Among these patients, some patient went on to develop infectious complications after surgery and are labeled as SSI PATOS. Others do not develop infectious complications and are not labeled as SSI or SSI PATOS. Our structured data features neither include any temporal information such as when a lab was ordered, lab values were collected, nor anti-infection treatments were administrated. Also, our current NLP system does not discriminate the infection status at the time of surgery or later in the postoperative course. Thus our machine learning models and NLP system have difficulty correctly classifying these patients. Since these patients are not rare in general surgery, we will address the issue in future studies. This barrier might be overcome either by integrating temporal features into machine learning models or adding higher level inferencer of NLP from document-level conclusions to patient level conclusions.
Compared with previously published studies10,12, our methods are inherently different. The reported scores are not directly comparable. FitzHenry et al. used manipulated input data (patients with complications mixed with 10% of the ones without complications) to develop and evaluate their system. Thus the reported performance does not reflect the system’s real performance on real-world data. Hu et al. compared several different models using data without any manipulation, but only reported AUC. Because both of our data are highly skewed with negative cases, only a small portion of ROC AUC can effectively reflect true performance. For this reason, we did not report ROC AUC. Hu et al. did not report the exact AUC scores for SSI detection other than a plot. Our several models (e.g., SVM enhanced with document binary features, Logistic regression with all document features or mention features) reach 0.92 in AUC, which seems comparable to their best model. However, they are not the best model that we choose using F0.5 score. Because we built and evaluated our models on a real-world data set, these models can be more practically used in the real environment.
Conclusion
We assessed the value of applying NLP to enhance machine learning models for SSI detection. The results show improved performance of the NLP enhanced models compared with the models using structured data alone or models enhanced with keywords features. A close investigation of these enhanced models confirms that NLP features play an important role in classification. Our error analysis suggests that future studies should include temporal status into consideration to further improve the performance.
Acknowledgments
We acknowledge funding and resources: the Agency for Healthcare Research and Quality, 1K08HS025776 (BTB) and the VA Salt Lake City Center of Innovation Award (COIN) #I50HX001240 from the US Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Health Services Research and Development (AVG). The views expressed in this article are those of the authors and do not necessarily represent the position or policy of the US Department of Veterans Affairs or the US government.
Figures & Table
References
- 1.Battles J B, Farr S L, Weinberg D A. From research to nationwide implementation: the impact of AHRQ’s HAI prevention program. Med Care. 2014;52((2 Suppl 1)):91–6. doi: 10.1097/MLR.0000000000000037. [DOI] [PubMed] [Google Scholar]
- 2.Merkow R P, Ju M H, Chung J W, Hall B L, Cohen M E, Williams M V, Tsai T C, Ko C Y, Bil-imoria K Y. Underlying reasons associated with hospital readmission following surgery in the United States. JAMA. 2015;313(5):483–495. doi: 10.1001/jama.2014.18614. [DOI] [PubMed] [Google Scholar]
- 3.Mila H. Ju, Clifford Y. Ko, Bruce L. Hall, Charles L. Bosk, Karl Y. Bilimoria, and Elizabeth C. Wick. A comparison of 2 surgical site infection monitoring systems. JAMA Surgery. 2015;150(1):51–57. doi: 10.1001/jamasurg.2014.2891. [DOI] [PubMed] [Google Scholar]
- 4.Hall B L, Hamilton B H, Richards K, Bilimoria K Y, Cohen M E, Ko C Y. Does surgical quality improve in the American College of Surgeons National Surgical Quality Improvement Program: an evaluation of all participating hospitals. Ann Surg. 2009;250(3):363–376. doi: 10.1097/SLA.0b013e3181b4148f. [DOI] [PubMed] [Google Scholar]
- 5.Hollenbeak C S, Boltz M M, Wang L, Schubart J, Ortenzi G, Zhu J, Dillon P W. Cost-effectiveness of the National Surgical Quality Improvement Program. Ann Surg. 2011;254(4):619–624. doi: 10.1097/sla.0b013e318230010a. [DOI] [PubMed] [Google Scholar]
- 6.Marti R., Rosenthal W.P., Weber W.R., Reck H., Misteli S., Dangel M., Oertli D., Widmer A.F. Surveillance of surgical site infections by surgeons: biased underreporting or useful epidemiological data? Journal of Hospital Infection. 2010;75(3):178–182. doi: 10.1016/j.jhin.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Aplin J., Tanner D., Khan C., Ball J., Thomas M., Bankart J. Post-discharge surveillance to identify colorectal surgical site infection rates and related costs. Journal of Hospital Infection. 2009;72(3):243–250. doi: 10.1016/j.jhin.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 8.Ban Kristen A, Minei Joseph P, Laronga Christine, Harbrecht Brian G, Jensen Eric H, Fry Donald E, Itani Ka-mal M.F., Dellinger E. Patchen, Ko Clifford Y, Duane Therese M. American College of Surgeons and Surgical Infection Society: Surgical Site Infection Guidelines, 2016 Update. Journal of the American College of Surgeons. 2017;224(1):59–74. doi: 10.1016/j.jamcollsurg.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Joseph S, Sow M, Furukawa M F, Posnack S, Chaffee M A. HITECH spurs EHR vendor competition and innovation, resulting in increased adoption. Am J Manag Care. 2014;20(9):734–740. [PubMed] [Google Scholar]
- 10.Hu Z, Melton G B, Moeller N D, Arsoniadis E G, Wang Y, Kwaan M R, Jensen E H, Simon G J. Accelerating Chart Review Using Automated Methods on Electronic Health Record Data for Postoperative Complications. AMIA Annu Symp Proc. 2016;2016:1822–1831. [PMC free article] [PubMed] [Google Scholar]
- 11.Grundmeier Robert W, Xiao Rui, Ross Rachael K, Ramos Mark J, Karavite Dean J, Michel Jeremy J, Gerber Jeffrey S, Coffin Susan E. Identifying surgical site infections in electronic health data using predictive models. Journal of the American Medical Informatics Association. 2018;25(9):1160–1166. doi: 10.1093/jamia/ocy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry Fern Fitz, Murff Harvey J, Matheny Michael E, Gentry Nancy, Fielstein Elliot M, Brown Steven H, Reeves Ruth M, Aronsky Dominik, Elkin Peter L, Messina Vincent P, Speroff Theodore, Henry F F, Murff Harvey J, Matheny Michael E, Gentry Nancy, Fielstein Elliot M, Brown Steven H, Reeves Ruth M, Aronsky Dominik, Elkin Peter L, Messina Vincent P, Speroff Theodore. Exploring the frontier of electronic health record surveillance: the case of postoperative complications. Medical care. 2013;51(6):509–16. doi: 10.1097/MLR.0b013e31828d1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scuba William, Tharp Melissa, Mowery Danielle, Tseytlin Eugene, Liu Yang, Frank A. Drews, and Wendy W. Chapman. Knowledge Author: facilitating user-driven, domain content development to support clinical information extraction. Journal of Biomedical Semantics. 2016;7((1)):42. doi: 10.1186/s13326-016-0086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.South Brett R, Shen Shuying, Leng Jianwei, Forbush Tyler B, Duvall Scott L, Chapman Wendy W. A Prototype Tool Set to Support Machine-Assisted Annotation. Proceedings of the 2012 Workshop on Biomedical Natural Language Processing (BioNLP 2012) 2012:130–139. (BioNLP) [Google Scholar]
- 15.Shi Jianlin, Mowery Danielle, Zhang Mingyuan, Sanders Jessica, Chapman Wendy, Gawron Lori. Extracting Intrauterine Device Usage from Clinical Texts Using Natural Language Processing. Proceedings - 2017 IEEE International Conference on Healthcare Informatics, ICHI 2017. 2017 [Google Scholar]
- 16.Shi Jianlin. and John F. Hurdle. Trie-based rule processing for clinical NLP: A use-case study of n-trie, making the ConText algorithm more efficient and scalable. Journal of Biomedical Informatics. 2018;85:106–113. doi: 10.1016/j.jbi.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cawley Gavin C., Talbot Nicola L. On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. Journal of Machine Learning Research. 2010;11:2079–2107. [Google Scholar]