Abstract
Skin disease is a prevalent condition all over the world. Computer vision-based technology for automatic skin lesion classification holds great promise as an effective screening tool for early diagnosis. In this paper, we propose an accurate and interpretable deep learning pipeline to achieve such a goal. Comparing with existing research, we would like to highlight the following aspects of our model. 1) Rather than a single model, our approach ensembles a set of deep learning architectures to achieve better classification accuracy; 2) Generative adversarial network (GAN) is involved in the model training to promote data scale and diversity; 3) Local interpretable model-agnostic explanation (LIME) strategy is applied to extract evidence from the skin images to support the classification results. Our experimental results on real-world skin image corpus demonstrate the effectiveness and robustness of our method. The explainability of our model further enhances its applicability in real clinical practice.
Introduction
Skin disease is one of the leading causes for global disease burden. It is reported that about 85 million Americans (27% of population; more than 1 in 4 individuals) were seen by dermatologists in 2013 and more than 9,500 people in the U.S. are diagnosed with skin cancer every day1,2. The diagnostic criteria of the skin diseases usually involve visual inspection of the skin lesions from their dermoscopic images as the first step. One such example is melanoma, which is one of the deadliest cancers. The diagnosis of melanoma includes two steps: visual inspection and biopsy. Because of the invasiveness of biopsy to patients, the accuracy of visual inspection is crucial. ABCDE (Asymmetric Shape, Border, Color, Diameter and Evolution) has been a popular rule for visual screening of melanoma based on the geometric characterization of the skin lesions3. However, without the equipment of these clinical knowledge, melanoma patients may not be aware of the severity of the disease they have and thus miss the best timing for treating their conditions.
In recent years, because of the rapid development of both computer hardware and software technologies, a large volume of skin images has been collected and sophisticated deep-learning based models have been trained to perform automatic analysis of these skin images. These models hold great promise as screening tools for skin diseases because of their superior capabilities of image analysis. For example, Esteva et al. proposed to adapt the Google Inception v3 model for melanoma detection from skin images4,5. The model, with parameters fine-tuned on 130K skin images, achieved classification performance comparable to human dermatologists. Kawahara et al. proposed a Fully Convolutional Neural Network (FCNN) model to extract multi-scale features skin images and perform lesion classification and achieved an accuracy of 81.8% on a 10-class skin disease classification problem, while the best reported classification accuracy on the same dataset is 67%6. Bi et al. proposed to leverage the deep Residual Network (ResNet) to perform melanoma detection and achieved state-of-the-art performance7,8.
Despite the good quantitative performance of these existing models, there are still some challenges to be addressed.
Model robustness. Existing research usually adapted well-established deep learning models (e.g., Inception or ResNet) trained on general computer vision tasks. It is difficult to guarantee any single model can work consistently well on different skin lesion images.
Sample limitation. In most of the existing studies, the amount of available skin images for training the complicated deep learning model is not enough9,10. Thus, these models typically will need to be pre-trained on other large-scale image data sets and the limited skin images will be used for fine-tuning to facilitate convergence.
Decision interpretation. Only quantitative classification results would not be enough for decision support in real clinical practice. As we introduced above, the dermatologists have specific diagnostic criteria to follow when they make the diagnosis. Therefore, the model we developed should also be able to generate the necessary evidence to support the classification results.
With the above considerations, we propose a novel deep learning pipeline for skin lesion classification in this paper.
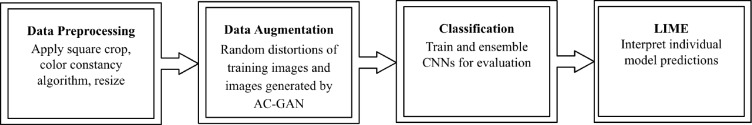
It is worthwhile to highlight the following aspects of our pipeline, as shown in Figure 1:
Figure 1.
Deep learning pipeline for skin lesion classification
Rather than using a single model, we ensembled a set of deep learning models (including VGG16, DenseNet, Xception, and Inception-ResNet v2) that have demonstrated to be effective on different computer vision tasks. In this way, we can sufficiently leverage the strength of different individual models and enhance the overall model robustness. We have also investigated different ensemble methods such as simple majority voting and model stacking.
In order to augment the training image set, we adopted the condition-based generative adversarial networks (GANs) on top of the traditional image augmentation approaches such as rotation and resizing11.
Local interpretable model-agnostic explanation (LIME) strategy is implemented to interpret final model predictions, so that image features that lead to the final classification are highlighted12.
We evaluate the effectiveness of the proposed pipeline using the Human Against Machine with 10000 training images (HAM10000) data set, which consists of 10015 dermoscopic images collected from different populations13.
Preliminaries
Convolutional Neural Network (CNN)14. CNN is a popular deep neural network model whose architecture design is inspired by the biological fact that the neuron connectivity patterns resemble the anima visual cortex, and for specific stimuli only the cortical neurons in a small region (a.k.a. receptive field) will respond. CNN has been shown to be very effective on computer vision tasks such as image and video analysis15,16. Two basic operations in CNN are convolution and pooling. Convolution convolves the whole image with a small filter mimicking the response of the receptive field (defined by the filter) centered around every pixel, and the resultant map after convolution will further go through a nonlinear activation function to obtain the final response. The pooling layer is for downsizing the image maps. A typical CNN architecture includes a series of convolution and pooling layers. The final resultant feature map will be stretched into a vector and fed to a fully connected layer for endpoint task (such as classification).
Generative Adversarial Network (GAN)17. GAN is a class of machine learning models that generates by mimicking real data distribution. There are two networks in the GAN model: a generative network that generates the data and a discriminative network to evaluate the generated data. The goal of GAN is to generate data objects as real as possible through zero-sum gaming. Many variants of GAN have been proposed in recent years. Particularly, in our case, we want to leverage the GAN technique for generating more skin image samples, and those samples should be not only real, but also can help improve the lesion classification performance. Mirza and Osindero proposed conditional GAN which makes both the data generation and discrimination probability conditioned on the data classes11. Odena et al. further proposed “Auxiliary Classifier GANs” (AC-GAN) by expanding the conditional GAN18. In the AC-GAN framework, the generator accepts a class label or condition along with the noise, while the discriminator estimates the probability that the image is real or fake along with the class probability. The authors note that although this modification is not tremendously different from previous works, it provides stability to the GAN’s notoriously unstable training process, in avoidance of problems like mode collapse, where the model collapses to a single mode; and training collapse, where the model stops improving.
Methods
As illustrated in Figure 1, there are 4 modules in our pipeline, and we will introduce them in detail in this section.
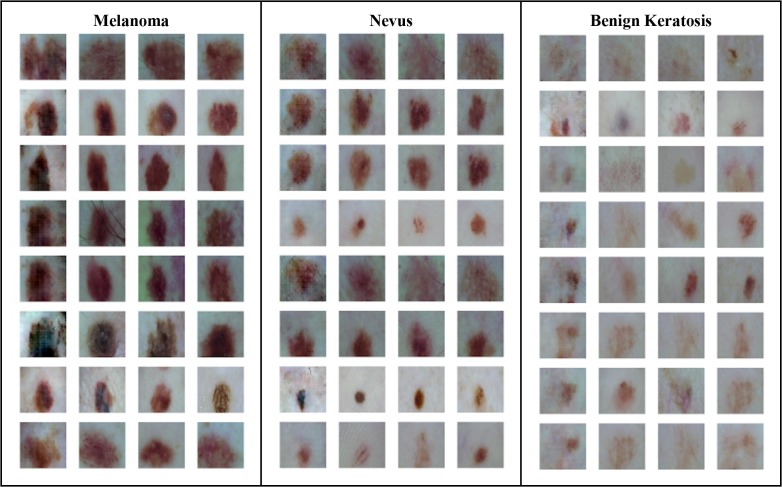
Data Preprocessing. The HAM10000 skin lesion dataset used for this work contains about 10,000 RGB images of skin lesions at a resolution of 450x600 pixels. For the purposes of this work we used 3 of the 7 skin lesion classes represented in the dataset that provided adequate samples for our GAN to learn conditional distributions, leaving a relatively small dataset of around 8900 images belonging to the disease classes of melanoma, nevus, and benign keratosis (the excluded images belonged to the 4 other classes).
Of the selected 8900 samples, 80% were used for purposes of training the models and 20% were set aside as for a final, testing set (Table 1), with both sets stratified. While training the CNNs we set aside 20% of the training dataset for internal validation.
Table 1.
Modified HAM10000 Dataset
| Melanoma | Nevus | Benign Keratosis | |
| Training | 890 | 5364 | 879 |
| Testing | 223 | 1341 | 220 |
The training set was preprocessed by application of the Gray World color constancy algorithm followed by a random 90% square crop (for more focus on the regions of interest), while for the testing dataset we took a 90% square center crop of the images. For training the CNNs, all images were then resized to 224x224 pixels, while for training the GAN, all images were resized to 64x64 pixels (later upsampled to match the CNN input size).
Data Augmentation. We first augment the data with traditional methods, which have been shown to reduce overfitting when the number of samples is lacking19. For each training sample we applied random distortions in contrast and brightness, clipped zoom, rotations, and flips, generating 4 samples per training sample. The validation and test sets were not augmented.
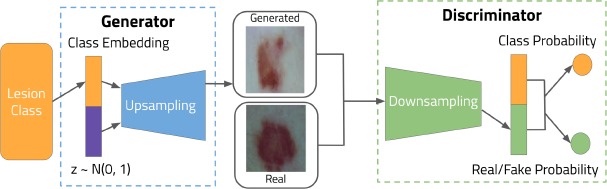
In addition, we also implemented the AC-GAN model for generating additional images. We followed the framework of a two-network minimax game. The generator accepted the product of random normal noise and a class embedding, outputting an image of dimensions 64x64x3, with a tanh function bringing the pixel values into the range [-1, 1]. The discriminator accepted both real and fake images and output both the probabilities of the images being real, with a sigmoid function; and the probability of the image belonging to a specific class, with a softmax function.
In our implementation, we used convolutional transpose layers in the generator network and convolutional layers in the discriminator network. For both of them we used Leaky Rectified Linear Units (Leaky ReLU) activation functions and batch normalization layers, both of which have been found to be able to help improve the training stability in GANs20,21,22. The generator and discriminator models are displayed in Figure 2.
Figure 2.
AC-GAN generator and discriminator framework. Batch diversity, Leaky ReLU, batch discrimination, and activations functions are not shown.
In order to address the problem of mode collapse, where the generator only learns one or a few modes of the training data’s distribution, we implemented mini-batch discrimination layers in the discriminator network23. During the training process, when the discriminator is fed both real and generated mini-batches, the mini-batch discrimination layers compute the entropy of the mini-batch. Since mini-batches with much lower entropy than real mini-batches are more likely to be generated, the discriminator can provide feedback to the generator in order to increase sample diversity.
Classification Model. In order to promote the robustness of the classification model, we adopted a model ensemble scheme, instead of just using a single model. Popular image classification architectures, including VGG-16, DenseNet, Xception, and Inception ResNet v2, are adopted as base classification models24, 25, 26, 27. As VGG simply stacks convolutional and pooling layers it is greatly inefficient, having an unfavorable proportion of parameters to accuracy. More recent architectures employ microarchitectures to improve performance such as the Inception module, residual connections, and dense connections between layers, aiming to allow networks to be deeper and more accurate. Xception modifies the Inception v3 architecture by stacking depthwise convolutions with residual connections and demonstrated a more efficient use of model parameters. Inception ResNet v2 adds residual connections to Inception v3, aiming to accelerate the training process and perhaps improve accuracy. DenseNet, by using dense connections between layers, encourages parameter sharing and feature reuse, thus achieving higher performance with fewer parameters. The different approaches taken by the different architectures allow them to have variance in their test-time errors, allowing an ensemble to of architectures to reduce variance and improve accuracy.
Because we are handling a multi-class classification problem, the categorical cross entropy (CCE) is adopted as the loss function for training the individual deep learning classifiers. Moreover, to further consider the size of different classes, we weight the cross entropy of different classes by their empirical prior probabilities calculated by Eq.(2). The final objective loss is shown in Eq. (1), where and are vectors representing the predicted softmax labels and true one-hot labels, respectively, with j as the index of the classes and J as the number of classes. is computed from softmax as shown in Eq.(3), where is the input to the softmax layer for class j.
| (1) |
| (2) |
| (3) |
In order to evaluate the performance of the models, we adopted balanced multiclass accuracy (BACC), defined in Eq.(4). The true positive predictions of each class are denoted by TP, the false positive predictions of each class are denoted by FP, c denotes a class, and N denotes the number of classes.
| (4) |
For the ensemble strategy, we implemented mean ensemble, where the softmax layers of the CNNs are averaged to get the image’s predicted class labels; and an RBF-kernel support vector machine (SVM)28. With the SVM ensemble method, we trained the ensemble on concatenations of softmax layers from the validation data, evaluating on the holdout test set.
For a baseline, we compared our model to scale-invariant feature transform (SIFT) with an SVM29. In this process, the RGB images are converted to grayscale. The images are then clustered with the k-means algorithm by their SIFT keypoints (128-length vectors), in order to perform vector quantization. An RBF-kernel SVM is then fit and evaluated with the dictionary of keypoints.
Model Interpretation. LIME is a package built to provide model-agnostic interpretations for classifiers. Being model-agnostic means that LIME acts without knowledge of the internal workings of a classifier, treating it as a black box and locally learning a mapping of input to output.
For interpretation of our models’ decisions, LIME also treats the CNNs as black boxes, perturbing an image it feeds to a given CNN and estimating the CNN’s decision function. The CNN’s decision function is estimated through a sparse linear model around a single image. By learning this decision function, LIME is able to highlight superpixels of images that lead to certain diagnoses by CNNs based on the importance of these superpixels.
Experiments
For all models we started their training with the ImageNet weights (available from Keras Applications)30. We used the Adam optimizer with a mini-batch size of 32 and learning rate of 0.0001, training for 20 epochs with early stopping. As the models displayed a tendency to overfit the training data, training was stopped when the validation accuracy and loss no longer improved. Moreover, DenseNet-169 was trained with GAN-generated samples (upscaled to model input size with Lanczos resampling) and traditional data augmentation.
Table 2 displays the balanced accuracies for each model and ensembles of models. In the ensembles we excluded SIFT with SVM, VGG-16, Xception, and DenseNet-121 as including these models decreased the ensemble accuracy.
Table 2.
Classification Results
| Model | Validation Accuracy | Test Accuracy |
| SIFT with SVM | .5444 | .5326 |
| VGG-16 | .7276 | .6838 |
| Xception | .7344 | .7275 |
| Inception v3 | .7537 | .7446 |
| Inception ResNet-v2 | .7819 | .7839 |
| DenseNet-121 | .7369 | .7367 |
| DenseNet-169 (no GAN Augmentation) | .7849 | .7821 |
| DenseNet-169 (with 40% GAN Augmentation) | .8089 | .8009 |
| DenseNet-201 (no GAN Augmentation) | .8236 | .8030 |
| DenseNet-201 (with 50% GAN Augmentation) | .8126 | .8156 |
| Mean Ensemble (without GAN-trained CNNs) |
.8352 | .8130 |
| Mean Ensemble | .8599 | .8478 |
| SVM Ensemble | .8577 | .8569 |
Our results show that, with 20% of the image set aside as independent validation, our pipeline can achieve a classification accuracy of 0.8569 over three skin lesion classes, while the traditional image classification method with Support Vector Machine (SVM) trained on Scale-Invariant Feature Transformation (SIFT) features can only get a classification accuracy of 0.532628,29.
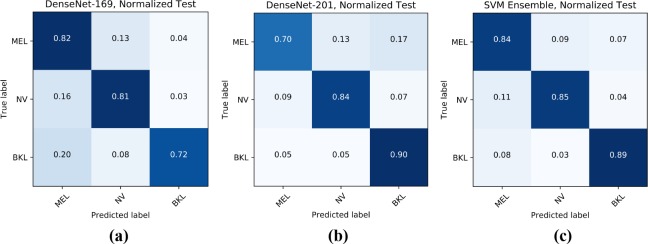
Figure 3 displays the normalized confusion matrices for various models on the testing set. The confusion matrices show the accuracy in predicting each disease across each row, e.g. in the first matrix, DenseNet-169 had 82% accuracy in predicting melanoma, 81% accuracy in predicting nevus, and 72% accuracy in predicting benign keratosis. Since the different CNNs learn different mappings of input to output, they have demonstrated different strengths in predicting the three diagnoses. While the final ensemble in the third confusion matrix is composed of more models than just DenseNet-169 and DenseNet-201 (50% GAN Augmentation Level), these confusion matrices demonstrate how an ensemble can balance out individual model predictions to improve classification accuracy. For instance, DenseNet-201’s poor melanoma classification accuracy being balanced out by DenseNet-169’s melanoma classification accuracy, and likewise, DenseNet-169’s poor benign keratosis classification accuracy is balanced out by DenseNet-201’s benign keratosis classification accuracy.
Figure 3.
(a) DenseNet-169, (b) DenseNet-201 (50%), (c) SVM Ensemble Confusion Matrices
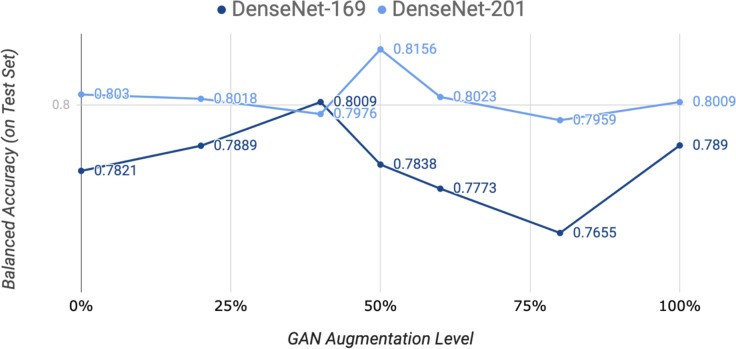
During the training of the CNNs, we evaluated different levels of augmentation with generated images over DenseNet-169 and DenseNet-201. The results are shown in Figure 4. Each level indicates the proportion of images that were generated and added to the training set, i.e. a level of 50% indicates the number of images equivalent to 50% of the original dataset (before traditional augmentation) were generated by the AC-GAN and added to the training set. For both DenseNet-169 and DenseNet-201, we employed augmentation levels of 0% (no additional augmentation), 20%, 40%, 50%, 60%, 80%, and 100%. DenseNet-169 achieved the highest balanced accuracy of 0.8009 at a level of 40%, and DenseNet-201 achieved the highest balanced accuracy of 0.8156 at a level of 50%. This indicates that a deeper model such as DenseNet-201 has more capacity for additional features provided by generated images compared to DenseNet-169. The trend for both models indicates an optimal level of augmentation at a moderate level with a performance drop with more generated data. As the level of augmentation increases, the classification accuracies of the models return to baseline levels, which may be attributed to the imprecision in generated data causing the models to lose generalizability to the testing set.
Figure 4.
DenseNet-169 and DenseNet-201 with different levels of GAN augmentation
During training of AC-GAN, we did not use an imbalanced training procedure (e.g., training the discriminator on 10 batches for every time the generator is trained). Instead, we used a balanced training procedure, training the discriminator and generator equally, with a batch size of 64, with the AC-GAN converging in about 20,000 updates.
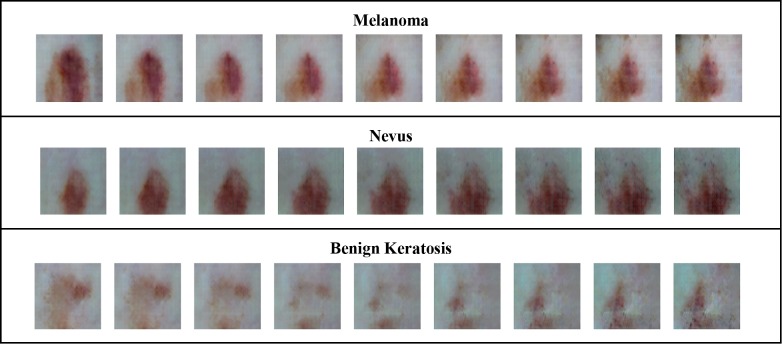
Figure 5 displays generated images side by side with their nearest neighbors (measured by Euclidean distance) in the training dataset. While we can see that skin lesions generated by the AC-GAN are similar in size, shape, and color to skin lesions from the actual training set, the AC-GAN seems to have produced original skin lesions rather than memorizing and reproducing skin lesions from the training set. Additionally, although some samples within classes are highly similar, there is still some diversity in the generated skin lesions both within each class and between the three classes, indicating the capability of AC-GAN for capturing of multiple modes of the data distribution and the absence of mode collapse.
Figure 5.
Images Generated by AC-GAN*
*Leftmost column of each class of images holds the generated images, while the 3 columns to the right of it hold the 3 nearest neighbors for each generated image.
In order to validate the acquired representation of the AC-GAN, we used latent space interpolation, which performs linear interpolation between two noise vectors (with the same class label). The results are shown in Figure 6. As the generated samples displayed smooth transitions with an absence of “holes” in the latent space of the AC-GAN, we can conclude the AC-GAN has learned meaningful features of the training distribution, rather than overfitting and producing discrete transitions.
Figure 6.
Latent Space Interpolation. The leftmost and rightmost images were generated from vectors sampled from Nevus Benign Keratosis Melanoma a normal distribution, and the other images were generated from interpolations between the two original vectors.
For diagnosis algorithms such as a CNN to be used in real clinical decision support, we cannot just provide the classification score, but also demonstrate the evidence on how the classification decision is made. We use a model-agnostic method, named LIME, to “open a black-box” of CNN and achieve such a goal.
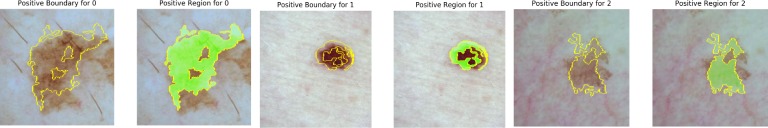
Figure 7 displays a few examples of positive diagnoses by Inception ResNet-v2 explained by LIME. The region bounded by a yellow line or highlighted for each lesion contain the top groups of pixels for each prediction. While LIME appears to highlight meaningful regions of the skin lesions, it may be lacking in specificity due to the difficulty of the classification task for both machines and humans. However, LIME does show that regions of the skin lesions are considered foremost before the backgrounds of the images, validating that the CNN is using relevant information for its diagnoses. There are also inherent limitations of the LIME’s model agnostic approach that come with its wide applicability, including its inability to use information from the weight activations of the CNN.
Figure 7.
True positive diagnoses for melanoma (0), nevus (1), and benign keratosis (2) from the test set with Inception ResNet-v2.
All CNNs and the AC-GAN were trained on an Amazon Web Service p3.2xlarge EC2 cloud instance, which has an NVIDIA Tesla V100 GPU, using the Keras deep learning framework with a TensorFlow backend. We release our source code and a demo web interface at https://github.com/alxiang/lesion-GAN.
Conclusions and Discussions
From Table 2, the results of the classification task, the top individual model was DenseNet-201 with GAN augmentation, with a test-time accuracy of .8156, while the less complex SIFT with SVM and VGG-16 models were unable to compete. For model ensemble strategies, averaging and SVM led to better classification accuracies, and we can also observe the expected decrease in variance obtained from combining multiple predictions.
We also found models tended to overfit without the regularization effect of data augmentation and converged faster by starting with the pre-trained ImageNet weights. We still observed overfitting with data augmentation and early stopping, which suggests that our exploitation of the data is still suboptimal, and more extensive data augmentation (e.g., with GAN-based methodologies) may improve results.
The results of the AC-GAN, displayed in Figures 5 and 6, showed that it can capture meaningful representations of the data, which correspond to the multiple modes of the data distribution in different classes. Moreover, the latent space interpolation experiments with the AC-GAN provide additional validation for the generator’s learned distribution. While generating 64x64 images with the AC-GAN was stabilized by inclusion of the class condition and batch diversity during training, we were unable to scale up the GAN to 256x256, closer to the desired input size of the model.
For the classification task, there is possible room for improvement through modifications to the CNN models including test-time augmentation, more advanced ensemble strategies, and more extensive data augmentation. Even deeper models are also promising, as they may have the useful capacity for deeper features in the training dataset.
Additionally, scaling up the resolution of images generated by the AC-GAN while preserving meaningful features of the training data could improve training of the CNNs, though in our experience, higher-resolution GANs were highly unstable to train. In future attempts of high-resolution image generation, generation prowess of different GANs can be evaluated through their aid in improving the accuracy on the classification task.
Acknowledgements
The work of Fei Wang is supported by NSF IIS-1750326
Figures & Table
References
- 1.Lim H.W., Collins S.A., Resneck J.S., Jr, Bolognia J.L., Hodge J.A., Rohrer T.A., Van Beek M.J., Margolis D.J., Sober A.J., Weinstock M.A., Nerenz D.R. The burden of skin disease in the United States. Journal of the American Academy of Dermatology. 2017 May 1;76(5):958–72. doi: 10.1016/j.jaad.2016.12.043. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019 doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Rigel D.S., Friedman R.J., Kopf A.W., Polsky D. ABCDE—an evolving concept in the early detection of melanoma. Archives of dermatology. 2005 Aug 1;141(8):1032–4. doi: 10.1001/archderm.141.8.1032. [DOI] [PubMed] [Google Scholar]
- 4.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017 Feb;542(7639):115. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition 2016 [Google Scholar]
- 6.Kawahara J., BenTaieb A., Hamarneh G. Deep features to classify skin lesions. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 2016 Apr 13;:pp. 1397–1400. IEEE. [Google Scholar]
- 7.Bi L., Kim J., Ahn E., Feng D. Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv preprint arXiv:1703.04197. 2017 Mar 12; [Google Scholar]
- 8.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition 2016. :pp. 770–778. [Google Scholar]
- 9.Romero Lopez A., Giro-i-Nieto X., Burdick J., Marques O. 2017 13th IASTED International Conference on Biomedical Engineering (BioMed) Innsbruck, Austria: 2017. “Skin lesion classification from dermoscopic images using deep learning techniques,”; pp. pp. 49–54. doi: 10.2316/P.2017.852-053. [Google Scholar]
- 10.Codella N., Nguyen Q.D., Pankanti S., Gutman D., Helba B., Halpern A., Smith J.S. Deep learning ensembles for melanoma recognition in dermoscopy images. ArXiv, abs/1610.04662. 2017 [Google Scholar]
- 11.Mirza M., Osindero S. Conditional generative adversarial nets. CoRR, abs/1411.1784. (2014) [Google Scholar]
- 12.Ribeiro M., Singh S., Guestrin C. “Why Should I Trust You?”: Explaining the predictions of any classifier. Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations. (2016) doi:10.18653/v1/n16-3020. [Google Scholar]
- 13.Tschandl P., Rosendahl C., Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data. 2018 Aug 14;5:180161. doi: 10.1038/sdata.2018.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015 May;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 15.Krizhevsky A., Sutskever I., Hinton G. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012 [Google Scholar]
- 16.Karpathy A., Toderici G., Shetty S., Leung T., Sukthankar R., Li F. Large-scale video classification with convolutional neural networks. IEEE Conference on Computer Vision and Pattern Recognition. 2014 [Google Scholar]
- 17.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. Generative adversarial nets. In Advances in neural information processing systems. 2014 [Google Scholar]
- 18.Odena Augustus, Olah Christopher, Shlens Jonathon. Conditional image synthesis with auxiliary classifier GANs. (2016) [Google Scholar]
- 19.Wong S. C., Gatt A., Stamatescu V., Mcdonnell M. D. Understanding data augmentation for classification: when to warp? 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA) (2016) doi:10.1109/dicta.2016.7797091. [Google Scholar]
- 20.Xu B., Wang N., Chen T., Li M. Empirical evaluation of rectified activations in convolutional network. arXiv preprint arXiv:1505.00853. 2015 May 5; [Google Scholar]
- 21.Ioffe S., Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning. 2015 Jun 1;:pp. 448–456. [Google Scholar]
- 22.Xiang S., Li H. On the effect of batch normalization and weight normalization in generative adversarial networks. CoRR, abs/1704.03971. (2017) [Google Scholar]
- 23.Salimans T., Goodfellow I.J., Zaremba W., Cheung V., Radford A., Chen X. Improved techniques for training GANs. NIPS. (2016) [Google Scholar]
- 24.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. CoRR, abs/1409.1556. (2014) [Google Scholar]
- 25.Huang G., Liu Z., Maaten L. V., Weinberger K. Q. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017) [Google Scholar]
- 26.Chollet F. Xception: Deep learning with depthwise separable convolutions. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017) doi:10.1109/cvpr.2017.195. [Google Scholar]
- 27.Szegedy Christian, Ioffe Sergey, Vanhoucke Vincent. Inception-v4, Inception-ResNet and the impact of residual connections on learning. AAAI Conference on Artificial Intelligence. (2016) [Google Scholar]
- 28.Cortes C., Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- 29.Lowe D. G. Method and apparatus for identifying scale invariant features in an image and use of same for locating an object in an image. U.S. Patent. 6(711):293. [Google Scholar]
- 30.Chollet Francois. Keras. 2015 https://github.com/ fchollet/keras. [Google Scholar]