Abstract
This paper describes a novel technique for annotating logical forms and answers for clinical questions by utilizing Fast Healthcare Interoperability Resources (FHIR). Such annotations are widely used in building the semantic parsing models (which aim at understanding the precise meaning of natural language questions by converting them to machine-understandable logical forms). These systems focus on reducing the time it takes for a user to get to information present in electronic health records (EHRs). Directly annotating questions with logical forms is a challenging task and involves a time-consuming step of concept normalization annotation. We aim to automate this step using the normalized codes present in a FHIR resource. Using the proposed approach, two annotators curated an annotated dataset of 1000 questions in less than 1 week. To assess the quality of these annotations, we trained a semantic parsing model which achieved an accuracy of 94.2% on this corpus.
Introduction
Fast Healthcare Interoperability Resources (FHIR)1 is an emerging standard developed by Health Level Seven International for storing and sharing information between different electronic health systems. Because of its RESTful architecture (adherence to REpresentational State Transfer style), it can be effectively used to view the data in different settings such as mobile and web applications. However, accessing some required information in electronic health records (EHRs) is still a cumbersome task2. Efficient query mechanisms such as question answering (QA) from medical records have the potential to reduce the delay between stored information and its prospective users.
EHRs contain information in both structured and unstructured format. Information extraction from unstructured data present in EHR (such as clinical notes and diagnostic reports) is well-researched and holds a promise to convert the free text data into structured format3,4. Efficient techniques to surface information from such structured EHR data have the potential to make the overall information access faster and reduce burdens related to system complexity.
Systems for QA from structured databases rely on unambiguous interpretation of natural language questions, often represented as machine-understandable logical forms. This process of converting natural language questions to their logical representations is called semantic parsing. Training semantic parsers usually requires a set of questions along with their annotated logical forms and/or answers5,6. Such corpora are widely available for the general domain but there is a scarcity of such datasets for EHR QA, mainly because of privacy issues and the complexity of EHR data.
Though annotating the questions with answers seems to be an easier task, one of the main difficulties involved in training a semantic parser using such dataset is ambiguity related to the realization of logical forms. Multiple logical forms can produce the correct answer but it becomes challenging for the semantic parser to separate the correct logical forms from the incorrect ones in such a scenario. Moreover, constructing a dataset of question-logical form pairs require annotating concept normalizations where medical concepts are recognized and mapped to a standard ontology. This is one of the hardest and time-consuming parts of creating a dataset for semantic parsing7.
Concept normalization alone is a challenging task as the same basic concept may legitimately map to multiple clinical codes and/or be stored in separate FHIR resources, even with the interoperability standards in place. For instance, the information that a patient has abdominal tenderness can be stored using FHIR Observation resource in at least the following ways8:
code: C4321457 (Examination), value: Abdomen tender
code: C0562238 (Examination of abdomen), value: Tenderness
code: C0232498 (Abdominal tenderness), value: found or true
code: C0232498 (Abdominal tenderness), value: no value
Moreover, a Condition resource can also be used to store this information if the symptoms are lasting9.
One of the main reasons behind such disparities is the use of different practices to store information at different organizations, which makes the process of normalizing concepts harder in a real setting. While there exist automated methods for concept normalization10, they are not tailored to the practices of a specific organization. Hence, it is important to learn these conventions at the organizational level where the QA system will be deployed.
In this paper, we propose a novel approach to construct a sizeable corpus of question-logical form-answer triplets using a FHIR server in a relatively short amount of time. Precisely, the list of available FHIR resources for a patient is shown to the annotators who, then, create a question, select the correct answer, and construct the corresponding logical form. We make use of the FHIR resource answers to automatically annotate concept normalizations for highlighted question phrases, which reduces the overall annotation time. Further, we train a semantic parser over the constructed dataset to learn the underlying logical forms. We also implement a concept normalizer tailored to the annotated concepts present in our corpus. Because of the shorter time and lesser prerequisite knowledge requirements for such corpus construction, the approach can be scaled across the organizations.
Background
We divide the related work into the following two sections based on the type of data used by medical QA systems.
1. Unstructured Data
Numerous studies in medical QA focused on the unstructured data sources such as biomedical literature, health-related social media data, and free text EHR data11,12. Several studies aimed at extracting relevant documents from the biomedical literature13–16. The BioASQ challenge17 introduced a decent sized dataset for machine comprehension (MC), i.e., answering a question on the basis of a given free text, from biomedical scientific articles. Similarly, a small MC dataset was released as part of the QA4MRE task18. Further, a recent study created large scale MC dataset using PubMed19. Another work constructed a small dataset using the resolved question-answer pairs from community-based health forums20. The MedQA task21 involved a large scale reading comprehension dataset which was built using data from certification exam for medical practitioners. Using the same certification exam dataset, a study proposed a framework for automatically generating question-answer pairs22. Further, with the MedicalQA task23, a dataset was developed utilizing various components of EHRs such as history of present illness. Raghavan et al.24 defined a process of annotating question-answer pairs using the EHR clinical notes.
Moreover, several efforts aimed at structuring the data present in unstructured form. Goodwin and Harabagiu25 proposed an approach for converting MIMIC-III medical records to knowledge graph for QA. Another study by Ayalew et al.26 created an ontology for the dataset of online frequently asked questions to assist in QA.
The majority of the datasets proposed for medical QA from unstructured data are based on biomedical literature whereas, only recently, the unstructured EHR data is gaining attention. Nevertheless, many studies are directed toward information extraction from free text present in EHRs3. Our aim, however, is to construct a corpus using the structured EHR data.
2. Structured Data
Very few works concentrated on QA from the structured biomedical data. Asiaee et al.27 and Amith28 worked on QA over a medical ontology. Pampari et al.29 constructed question-logical form and question-answer pairs using a template-based approach. QA dataset generation using templates limits the variety of questions to a certain extent and are not representative of the real world queries. Our previous work included annotating a set of EHR questions with their corresponding logical forms7. As stated earlier, logical form annotations are time-consuming to annotate, especially because of difficulties involved in annotating the concept normalizations. In this paper, we aim to quicken the process of concept annotations utilizing FHIR. To our knowledge, this is the first work to construct a dataset for EHR QA using FHIR resources.
Materials and Methods
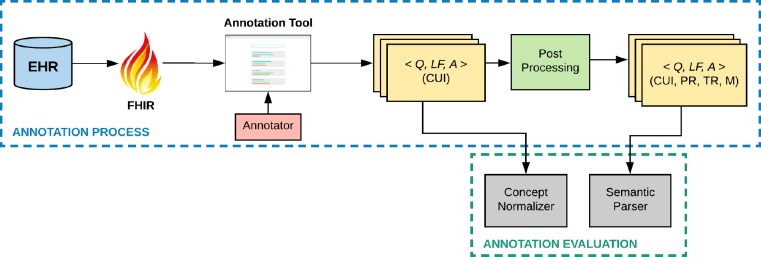
The following sections describe the methods used for constructing and evaluating the dataset. A graphical representation of our methods is shown in Figure 1.
Figure 1.
Overview of the methods. <Q, LF, A> - Triplet containing Question, Logical Form, and Answer resource. (●) – annotations over the triplets. CUI - Concept Unique Identifier, PR – Person Reference. TR – Temporal Reference. M - Measurement.
1. Dataset Construction
We set up a local FHIR server which serves as an underlying data source for constructing the dataset. An annotation tool is implemented to present the information contained in the FHIR server to the annotator. The annotators reviewed the available patient resources and constructed questions along with their corresponding simple logical forms. An institution-specific concept normalizer is trained based on these annotations. We further processed the constructed questions to be used in training a semantic parsing system. Each of these tasks is explained further in the following sub-sections:
1.1. FHIR Server
We deployed a FHIR server locally using an existing open source implementation by the MITRE Corporation for DSTU2 version (v1.0.2)*. To generate the data for this local server, we used Synthea, a tool for producing realistic health care records30. The FHIR resources generated by Synthea are added to the local FHIR server using the REST APIs provided by the server. We chose to use the local FHIR server instead of any online available sandbox servers to maintain stability and consistency.
1.2. Annotation Tool
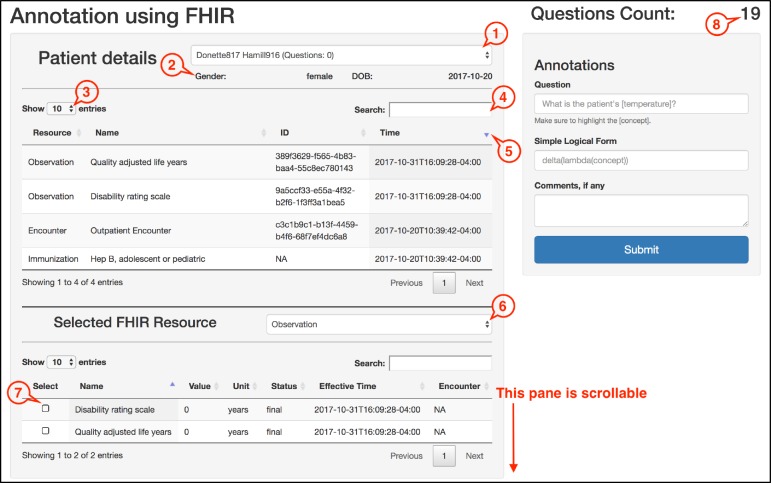
We implemented a web application to facilitate the browsing of FHIR resources present in the local server for an efficient annotation process. The tool displays all the available patients along with their resources from the server. The three main components of the tool interface allow, for a selected patient, viewing all the resources, viewing all the resources of a selected resource type, and entering the constructed question with its corresponding logical form. The user interface for the annotation tool is shown in Figure 2. The main components of the tool are explained below. Indices in the parentheses, (#), point to the corresponding elements in Figure 2.
Figure 2.
Components of the annotation tool. (1) Dropdown menu for viewing and changing patients. (2) Sub-section for patient information. (3) Dropdown for changing the number of visible entries. (4) Search box to filter any specific resources. (5) Sorting toggle buttons. (6) Dropdown for viewing and selecting from the available FHIR resource types. (7) Checkbox for selecting a set of resources as the answer. (8) Total count of the annotations by the current user.
I. Patient Details
This view gives an overview of all the FHIR resources for a selected patient. A list of available patients along with a count of questions constructed for them can be seen using the dropdown (1). The patient can be changed using this dropdown or by refreshing the browser. The gender and date of birth information are shown below the dropdown at (2) for a selected patient.
The table in this component shows the information common to all the FHIR resources, namely, resource type, name, ID, and time. All the entries in this table can be capped, filtered, and/or sorted using the corresponding functionalities at (3), (4), and (5). Such flexibility in viewing the resources facilitates the overview of patient history.
II. Selected FHIR Resource
This part enables the annotator to view more granular details for the resources of a selected type. All the available resource types for the selected patient can be seen in the dropdown (6). The table under this section has similar functionalities in terms of filtering and viewing the results. In addition to these, the table allows selecting one or more resources from the list using checkboxes (7). This selection serves as the answer to a constructed question.
III. Annotations
Above this section at (8) is an ongoing count of the total number of questions the annotator has constructed so far. This count takes into account the questions constructed for all the patients to show the overall annotation progress. The question text box in this component allows the annotator to input their constructed question. The concept boundaries can be highlighted using a set of opening and closed square brackets. The marked text is automatically normalized to the medical concept present in the selected answer. The text box for simple logical form is used for entering the logical form for the constructed question. There is another text box for entering the comments in regard to the annotation, if there are any. The submit button is used to save the annotation to the corpus.
The tool is equipped with certain validation checks which ensure that a correct annotation is saved. Specifically, the tool checks for the following:
At least one FHIR resource is selected as an answer
The question contains a highlighted concept
The simple logical form is valid (check for balanced parentheses and valid logical predicates)
1.3. Annotation Process
The description of the annotation tool provides some intuition to the annotation process. In this section, we explain the overall annotation process in detail. The annotator starts with a selected patient and views all the FHIR resources for them in the Patient Details component. The annotator decides to ask questions based on the available resources of a patient. After getting a quick overview of the patient, the annotator moves to the Selected FHIR Resource component and selects a resource type to construct some question. The annotator enters a question in the Question input and selects the corresponding FHIR resource(s) containing the answer. Finally, the user enters a corresponding simple logical form and hits submit. We follow the logical form structure as described in our previous work on annotating the EHR questions7. After the submission of an annotation, the highlighted concept in the question text is automatically normalized to the medical concept of the selected FHIR resource(s). The annotation is saved to the corpus with this normalized concept. For instance:
Answer Resource: Condition resource Rupture of appendix with concept code C0267628
Question: When was the [appendix ruptured]?
Normalized Question: When was the nn:concept(C0267628)?
Simple Logical Form: time(latest(lambda(concept)))
Logical Form: time(latest(λx.has_concept(x, C0267628)))
Again, only the Question and Simple Logical Form are typed by the annotator. The Answer Resource comes from the checkbox (7) and the Normalized Question is computed automatically.
1.4. Post Processing
After the annotation process is completed, the constructed corpus is further processed to add more annotations. Particularly, we employ rule-based approaches to identify the person references, temporal references, and measurements. These additional annotations are important to separate the task of semantic parsing from concept normalization6. The following question exemplifies such normalization categories:
Question: Did her hemoglobin A1c exceed 6% in the past 3 years?
Normalized: Did pos:person nn:concept exceed measurement(‘6%’) in the temporal_ref(‘ past 3 years’)?
These additional references are easier to annotate automatically because a small number of patterns can cover the majority of cases. For instance, the person references can be found in the question text using a set of patterns such as the patient, she, he, her, and him. The temporal references are identified using the regular expression patterns, e.g. yesterday, past n years, and last n days. Similarly, the measurements are extracted by employing patterns like n units where units can be C, F, lb and so on. These rules for identifying the additional concepts are motivated by our previous work7.
2. Evaluation
For assessing the quality of annotations using our tool, we employ a concept normalization and a semantic parsing model. Each of these models is separately explained in this section.
2.1. Concept Normalization
For assessing the effectiveness of learning an institution-specific concept normalizer, we use simple deep learning models based on convolutional neural network (CNN) and recurrent neural network (RNN) architectures. Specifically, we follow the technique inspired by Limsopatham and Collier31, which make use of the publicly available pre-trained embeddings to normalize medical concepts from health-related social media messages. We randomly initialized the embeddings for the words present in questions and concepts.
To train these models, we input the highlighted concept phrase in the question along with the automatically annotated concept term from FHIR. Using the above example of Rupture of Appendix,
Highlighted phrase in question: appendix ruptured
Annotated concept: Rupture of Appendix (C0267628)
The concept normalizer learns these mappings from the dataset and aims to translate the medical terms present in natural language text to the standard medical concepts. Note that we do not train a concept boundary classifier, as this is an extremely common clinical natural language processing (NLP) task32–36.
2.2. Semantic Parsing
We train a semantic parser over the question and logical form annotations following a hybrid approach using both rule-based and machine learning-based techniques. Full details of the employed method can be found in our previous work on semantic parsing6. We update the lexicon, which maps lexical phrases present in questions to the corresponding logical operations, to extend its coverage. Moreover, we use similar rules for candidate generation and same features for the machine learning model as used in our previous approach.
Results
Two annotators (a physician and a biomedical informatics doctoral student) were involved in constructing the corpus of 1000 questions. Each annotator independently created 500 questions following the annotation guidelines presented in the methods section. The individually constructed annotations were cross reviewed by both the annotators after the completion of the first 50 and all 500 annotations to ensure the completeness. We stored the timestamps for each annotation which gave us the ability to analyze our annotation times in detail. We kept track of the review times manually. The first checkpoint of 50 annotations was completed in 2.5 hours by each annotator and was reviewed by both the annotators together in 1 hour. Each annotator took about 24 hours to complete the remaining 450 annotations, which were then separately reviewed by both the annotators in 8 hours. The whole annotation process was completed in less than one calendar week. By contrast, Roberts and Demner-Fushman7 required multiple weeks to annotate less than half as much data, while their concept normalization choices were likely incompatible with many hospitals as it was not built from a reference FHIR instance.
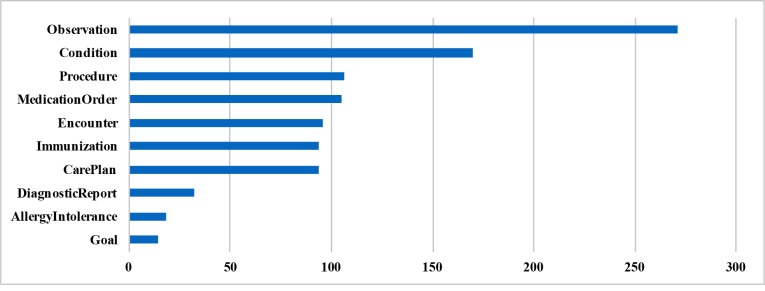
We observed a total of 10 unique FHIR resource types which were used as answers in our corpus. The distribution of these resource types is presented in Figure 3. We note that the most number of constructed questions were about Observation resource followed by Condition and Procedure resources.
Figure 3.
Frequency distribution of FHIR resource types selected as answer in the constructed corpus.
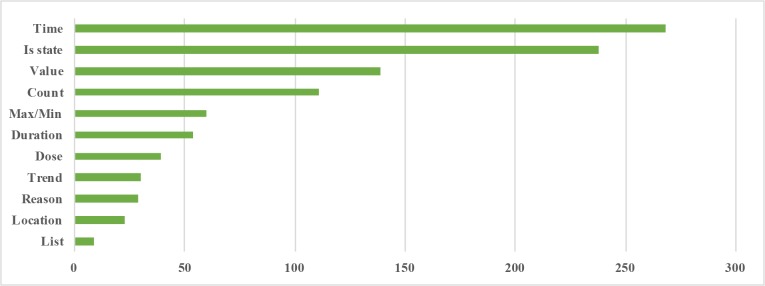
Based on the annotated logical forms, we enumerated 11 answer types. Figure 4 shows the distribution of these answer type frequencies in our annotated corpus. Each type can be perceived as a logical predicate applied to the FHIR resource(s). For e.g., Time answer type is assigned to questions which return the time of a specific resource. Similarly, Is state type aims to determine the state of some specific resource(s). It should be noted that these answer types are based upon the outermost logical predicates in logical forms.
Figure 4.
Frequency distribution of answer types in the annotated corpus. Types (y-axis) denote the final function applied over the answer resource(s).
We analyzed the combinations of FHIR resource and answer types in our dataset to get the impression of most frequent question varieties. We present the top 5 of these combinations with example question and logical forms in Table 1. The most frequent number of questions, specifically 92, were about querying the value of Observation resource, which succeeded by querying the Max/Min and Is state.
Table 1.
Most frequently co-occurring FHIR resource and answer types along with their corresponding counts. Q – Question, LF – Logical Form. Concepts in the questions are underlined.
| Resource type | Answer type | Frequency | Example |
| Observation | Value | 92 |
Q: What is his t-score? LF: latest(λx.has_concept(x, C1526354)) |
| Observation | Max/Min | 60 |
Q: What was the highest hemoglobin A1c value in the past 2 years? LF: max(λx.has_concept(x, C0366781) ^ time_within(x, ‘past 2 years’)) |
| Observation | Is state | 59 |
Q: Has her calcium level ever been less than 9 mg/dl? LF: delta(λx.has_concept(x, C1977516) ^ less_than(x, ‘9 mg/dl’)) |
| Condition | Is state | 54 |
Q: Was her ankle sprain healed? LF: is_healed(latest(λx.has_concept(x, C0160087))) |
| Condition | Time | 53 |
Q: When did he develop microalbuminuria? LF: time(latest(λx.has_concept(x, C3875084))) |
The results of training semantic parser and concept normalizer are shown in Table 2. Using rules and features of the best performing model from our previous approach6, we achieved an accuracy of 94.2% with leave-one-out validation. We trained the concept normalizer models based on RNN and CNN for 50 epochs each, as per the evaluation results presented in the original paper31, and achieved an accuracy of 76.0% and 70% respectively.
Table 2.
Results of evaluation. RNN – Recurrent Neural Network, CNN – Convolutional Neural Network.
| Model | Accuracy | |
| Semantic Parser | 94.2% | |
| Concept Normalizer | RNN | 76.0% |
| CNN | 70.0% | |
Discussion
Annotating concept normalizations during the logical form annotation is a challenging and time-consuming task. We aimed at automating this step using the highlighted concept text in question. This intervention reduced a significant amount of time for annotating the logical forms in comparison to our previous approach7 . Another advantage of this approach is that the annotated concepts in our corpus are well-aligned with concepts used in the referenced FHIR server. This enabled us to train a concept normalizer tailored to the FHIR server used for annotations.
The result of the semantic parsing evaluation highlights the quality of generated corpus. The accuracy of the semantic parser on our corpus is slightly less than that on the dataset it was originally built on. This difference might have resulted from increased vocabulary and variety of questions in our corpus.
The decent performance of the concept normalizer shows the potential of this annotation approach for concept normalization. In comparison to other methods32–36, we trained on less data. Also, our corpus contains a wider variety of concepts than just disorders or diseases. Other concept normalizers such as DNORM37 could be trained but that was not the focus of this work.
Using the proposed annotation approach, a large annotated dataset of questions can be created comparatively faster. This dataset can then be used for building a semantic parser to support natural language queries.
We used a rule-based approach to identify the temporal expression spans, as this approach was able to capture the variety of expressions in our corpus. More sophisticated temporal information extraction systems can be incorporated in the future38,39.
One of the limitations of our study is that we limit the answer resources selection to a single concept type. In other words, the constructed questions could be about a single type of concept. In the future, we aim to include multiple concept types during answer selection which can result in the creation of more complex questions.
Conclusion
We have described a novel approach for constructing an annotated corpus of EHR questions using FHIR. The process of annotating concept normalizations is automated which significantly reduced the overall annotation time. Using the proposed approach, two annotators created a corpus of 1000 questions in less than one week. We evaluated our constructed dataset by training a semantic parser and a concept normalizer, both of which showed promising results with accuracies of 94.2% and 76% respectively.
Acknowledgments
This work was supported by the U.S. National Library of Medicine, National Institutes of Health (NIH), under award R00LM012104, as well as the Bridges Family Doctoral Fellowship Award.
Footnotes
Figures & Table
References
- 1.Health Level Seven International. Welcome to FHIR. [Internet] [cited 2019 Jul 8]. Available from: https://www.hl7.org/fhir/ [Google Scholar]
- 2.Zhang J, Walji M, editors. National Center for Cognitive Informatics & Decision Making in Healthcare; 2014. Better EHR: Usability, workflow and cognitive support in electronic health records. [Google Scholar]
- 3.Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, et al. Clinical information extraction applications: A literature review. J Biomed Inform. 2018 Jan 1;77:34–49. doi: 10.1016/j.jbi.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 5.Kamath A, Das R. A Survey on Semantic Parsing. In: Automated Knowledge Base Construction. 2019 [Google Scholar]
- 6.Roberts K, Patra BG. A Semantic Parsing Method for Mapping Clinical Questions to Logical Forms. In: AMIA Annual Symposium Proceedings. American Medical Informatics Association. 2017 [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts K, Demner-Fushman D. In: Proceedings of the 10th International Conference on Language Resources and Evaluation, LREC 2016. NIH Public Access; 2016. Annotating logical forms for EHR questions; pp. p. 3772–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Health Level Seven International. Resource Observation. [Internet] 2018 [cited 2019 Mar 8]. Available from: https://www.hl7.org/fhir/observation.html. [Google Scholar]
- 9.Health Level Seven International. Observation vs Condition. [Internet] 2015 [cited 2019 Mar 8]. Available from: http://wiki.hl7.org/index.php?title=Observation_vs_Condition. [Google Scholar]
- 10.Pradhan S, Elhadad N, South BR, Martinez D, Christensen L, Vogel A, et al. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J Am Med Inform Assoc. 2015;22(1):143. doi: 10.1136/amiajnl-2013-002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athenikos SJ, Han H. Biomedical question answering: A survey. Comput Methods Programs Biomed. 2010 Jul 1;99(1):1–24. doi: 10.1016/j.cmpb.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Neves M, Leser U. Question answering for Biology. Methods. 2015 Mar 1;74:36–46. doi: 10.1016/j.ymeth.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 13.Hersh W, Cohen AM, Roberts P, Rekapalli HK. TREC 2006 genomics track overview. In: 15th Text REtrieval Conference, TREC. 2006 [Google Scholar]
- 14.Hristovski D, Dinevski D, Kastrin A, Rindflesch TC. Biomedical question answering using semantic relations. BMC Bioinformatics. 2015 Dec 16;16(1):6. doi: 10.1186/s12859-014-0365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts K, Simpson M, Demner-Fushman D, Voorhees E, Hersh W. State-of-the-art in biomedical literature retrieval for clinical cases: a survey of the TREC 2014 CDS track. Inf Retr J. 2016 Apr 18;19(1–2):113–48. [Google Scholar]
- 16.Noh J, Kavuluru R. Document Retrieval for Biomedical Question Answering with Neural Sentence Matching. Proc Int Conf Mach Learn Appl. 2018 Dec;2018:194–201. doi: 10.1109/ICMLA.2018.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsatsaronis G, Balikas G, Malakasiotis P, Partalas I, Zschunke M, Alvers MR, et al. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinformatics. 2015 Dec 30;16(1):138. doi: 10.1186/s12859-015-0564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morante R, Krallinger M, Valencia A, Daelemans W. In: CLEF 2012 Conference and Labs of the Evaluation Forum-Question Answering For Machine Reading Evaluation (QA4MRE) Rome/Forner; 2012. Machine reading of biomedical texts about Alzheimer’s disease; pp. p. 1–14. [Google Scholar]
- 19.Kim S, Park D, Choi Y, Lee K, Kim B, Jeon M, et al. A Pilot Study of Biomedical Text Comprehension using an Attention-Based Deep Neural Reader: Design and Experimental Analysis. JMIR Med informatics. 2018 Jan 5;6(1):e2. doi: 10.2196/medinform.8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wongchaisuwat P, Klabjan D, Jonnalagadda SR. A Semi-Supervised Learning Approach to Enhance Health Care Community-Based Question Answering: A Case Study in Alcoholism. JMIR Med informatics. 2016 Aug 2;4(3):e24. doi: 10.2196/medinform.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Wu J, He Z, Liu X, Su Y. Medical exam question answering with large-scale reading comprehension. In: Thirty-Second AAAI Conference on Artificial Intelligence. 2018 [Google Scholar]
- 22.Shen S, Li Y, Du N, Wu X, Xie Y, Ge S, et al. On the Generation of Medical Question-Answer Pairs. 2018 Nov 1; [Google Scholar]
- 23.Hao Y, Liu X, Wu J, Lv P. Exploiting Sentence Embedding for Medical Question Answering. 2018 Nov 14; [Google Scholar]
- 24.Raghavan P, Patwardhan S, Liang JJ, Devarakonda M V. Annotating Electronic Medical Records for Question Answering. 2018 May 17; [Google Scholar]
- 25.Goodwin TR, Harabagiu SM. Medical Question Answering for Clinical Decision Support. Proc ACM Int Conf Inf Knowl Manag. 2016 Oct;2016:297. doi: 10.1145/2983323.2983819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayalew Y, Moeng B, Mosweunyane G. Experimental evaluation of ontology-based HIV/AIDS frequently asked question retrieval system. Health Informatics J. 2018 May 23; doi: 10.1177/1460458218775147. 146045821877514. [DOI] [PubMed] [Google Scholar]
- 27.Asiaee AH, Minning T, Doshi P, Tarleton RL. A framework for ontology-based question answering with application to parasite immunology. J Biomed Semantics. 2015 Dec 17;6(1):31. doi: 10.1186/s13326-015-0029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amith M. Ontology-Based Dialogue Systems for Improved Patient HPV Vaccine Knowledge and Perception. In: Proceedings of the Doctoral Consortium at the 15th International Semantic Web Conference (ISWC) 2016:p. 9–16. [Google Scholar]
- 29.Pampari A, Raghavan P, Liang J, Peng J. Brussels, Belgium: Association for Computational Linguistics; 2018. emrQA: A Large Corpus for Question Answering on Electronic Medical Records; In: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing; pp. p. 2357–68. [Google Scholar]
- 30.Walonoski J, Kramer M, Nichols J, Quina A, Moesel C, Hall D, et al. Synthea: An approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J Am Med Informatics Assoc. 2018 Mar 1;25(3):230–8. doi: 10.1093/jamia/ocx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limsopatham N, Collier N. Stroudsburg, PA, USA: Association for Computational Linguistics; 2016. Normalising Medical Concepts in Social Media Texts by Learning Semantic Representation; In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) pp. p. 1014–23. [Google Scholar]
- 32.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Informatics Assoc. 2011 Sep 1;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suominen H, Salanterä S, Velupillai S, Chapman WW, Savova G, Elhadad N, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013. In: International Conference of the Cross-Language Evaluation Forum for European Languages. 2013:p. 212–31. [Google Scholar]
- 34.Kelly L, Goeuriot L, Suominen H, Schreck T, Leroy G, Mowery DL, et al. Overview of the share/clef ehealth evaluation lab 2014. In: International Conference of the Cross-Language Evaluation Forum for European Languages. 2014:p. 172–91. [Google Scholar]
- 35.Pradhan S, Elhadad N, Chapman W, Manandhar S, Savova G. In: Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014) Stroudsburg, PA, USA: Association for Computational Linguistics; 2014. SemEval-2014 Task 7: Analysis of Clinical Text; pp. p. 54–62. [Google Scholar]
- 36.Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, Savova G. In: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) Stroudsburg, PA, USA: Association for Computational Linguistics; 2015. SemEval-2015 Task 14: Analysis of Clinical Text; pp. p. 303–10. [Google Scholar]
- 37.Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013 Nov 15;29(22):2909–17. doi: 10.1093/bioinformatics/btt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Informatics Assoc. 2013 Sep 1;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bethard S, Savova G, Chen W-T, Derczynski L, Pustejovsky J, Verhagen M. Stroudsburg, PA, USA: Association for Computational Linguistics; 2016. SemEval-2016 Task 12: Clinical TempEval. In: Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016) pp. p. 1052–62. [Google Scholar]