Abstract
With advances in Machine Learning (ML), neural network-based methods, such as Convolutional/Recurrent Neural Networks, have been proposed to assist terminology curators in the development and maintenance of terminologies. Bidirectional Encoder Representations from Transformers (BERT), a new language representation model, obtains state-of-the-art results on a wide array of general English NLP tasks. We explore BERT’s applicability to medical terminology-related tasks. Utilizing the “next sentence prediction” capability of BERT, we show that the Fine-tuning strategy of Transfer Learning (TL) from the BERTBASE model can address a challenging problem in automatic terminology enrichment – insertion of new concepts. Adding a pre-training strategy enhances the results. We apply our strategies to the two largest hierarchies of SNOMED CT, with one release as training data and the following release as test data. The performance of the combined two proposed TL models achieves an average F1 score of 0.85 and 0.86 for the two hierarchies, respectively.
Introduction
Insertion of new concepts into their proper positions in a terminology is a challenging problem in automatic enrichment of terminologies. Traditionally, for terminologies based on a Description Logic, curators tend to use a reasoner such as Snorocket [1] or HermiT [2] to insert a new concept into a terminology’s hierarchy. Recently, researchers have proposed to use Deep Learning models such as Convolutional/Recurrent Neural Network models [3] to verify an IS A relationship between a new concept and an existing concept, which will recommend the location of the new concept in the hierarchy. In such an approach, concepts are represented by various language embeddings from Natural Language Processing (NLP) [4-6].
However, training such an ML model from scratch is expensive and time-consuming, as it requires large data sets and extensive computing resources. Instead, it is common and efficient to conduct Transfer Learning from pre-trained language representation models to the task of interest. In this paper, we utilize a new general language representation model called Bidirectional Encoder Representations from Transformers (BERT) [7]. BERT is a pre-trained model which obtains state-of-the-art results on a wide range of NLP tasks. It has been integrated into applications [8,9] for clinical tasks in biomedical domain. We experiment with two Transfer Learning (TL) strategies, Fine-tuning and Pre- training, from the pre-trained BERTBASE model to address the terminology enrichment problem. In Fine-tuning, we train a classifier on top of a BERTBASE network with IS-A relationships as training data. This training fine-tunes the weights of the pre-trained BERTBASE network to enable it to classify relationships between new concepts and existing concepts. The fine-tuning strategy innovatively utilizes the “next sentence prediction” of BERT, to train BERT to distinguish which pairs of concepts should be connected by IS-A relationships. In Pre-training, we accommodate BERT to medical data, by training it from scratch using terminology data from SNOMED CT.
To measure the performance of the two proposed strategies, we use the two largest hierarchies of the SNOMED CT [10] terminology, the Clinical Finding and the Procedure hierarchies, as our testbed. The SNOMED CT release of July 2017 is used as training data. For testing, we use 911 and 2005 new concepts from the Procedure and Clinical Finding hierarchies of the January 2018 release, respectively. The results of this experiment for the Fine-tuning strategy and for the combined strategy starting with Pre-training and continuing with Fine-tuning are reported.
Background
SNOMED CT is an internationally leading clinical terminology, managed by SNOMED International. The Clinical Finding and the Procedure hierarchy of the January 2018 release of SNOMED CT consist of 111,081 and 57,806 active concepts, respectively. SNOMED CT is released twice every year on January and July. All the content for a given release of SNOMED CT terminology is defined in a “snapshot” file. In addition, a “delta” file identifies the individual changes that occurred between the previous release and the current release. A full history of concepts and relationships that are added, changed, or removed is also provided. By comparing the Procedure and Clinical Finding hierarchies of January 2018 with the previous July 2017 release, we found that 911 new concepts were added into the Procedure hierarchy and 2005 new concepts were added into the Clinical Finding hierarchy and placed in the proper positions in the hierarchy by SNOMED CT’s curators, based on their structural and semantic definitions stated in Description Logic. Liu et al. [11] proposed a methodology to automatically determine the placement of a new concept in the ontology’s hierarchy if a new concept’s name and one of the concept’s parents is given. The solution is based on training a Convolutional Neural Network (CNN) model to distinguish between those pairs of concepts that are connected by IS-A links and those pairs that are not.
For the language representation model, there are two main research streams: Context-free and Contextual representations. Traditional word embeddings such as word2vec [12], GloVe [13], or fastText [14], are Context-free embeddings, which generate a single “word embedding” representation for each token in the vocabulary. Therefore, they are not likely to capture any word meaning changes caused by surrounding context changes. Contextual models, instead, generate a representation of each word that is based on the other words in the context. Contextual representations can further be categorized into unidirectional or bidirectional.
BERT is the first unsupervised, deeply bidirectional system that outperforms previous methods. BERT is a general- purpose “language understanding” model trained on a large text corpus (like Wikipedia), which can be used for various downstream NLP tasks without heavy task-specific engineering. BERT’s model architecture is a multi-layer bidirectional transformer encoder, based on the original implementation proposed by Vaswani et al. [15]. BERT has advanced the state-of-the-art for several major NLP benchmarks, including named entity recognition on CoNLL-2003 [16], question answering on SQuAD [17], and sentiment analysis on SST-2 [18].
Methods
The main contribution of this paper is suggesting a way to harness the high performance of BERT for a critical task in medical terminology enrichment, in spite of the fact that BERT was not trained with medical literature. One approach is to add medical knowledge to the general knowledge learning of BERT. For this we use the SNOMED CT knowledge, providing a “document” for each concept of SNOMED CT. The second approach is to train BERT to be able to distinguish between concept pairs which should be connected by IS-A relationships and pairs that shouldn’t. This kind of learning utilizes the “next sentence prediction” feature of BERT. In the following we describe the technical issues and the details involved in implementing these ideas.
We experimented with two strategies of using BERT. 1) Fine-tuning BERTBASE by supervised training a relationship classifier on top of BERT with concept pairs connected by IS-A relationships and pairs not connected (non-IS-A pairs) taken from SNOMED CT. 2) Pre-training BERTBASE with unsupervised concept-based “documents” from SNOMED CT, and then fine-tuning it as an IS-A relationship classifier. We implemented the experiments with Tensorflow [19] and ran the testcases on a computer with two Intel Xeon E5-2630-v4 CPUs with processor speed 2.2 GHz; 128 GB memory per CPU and two Nvidia Tesla P100 “Pascal” video cards with 16 GB RAM per GPU.
BERTBASE (12 Transformer layers) and BERTLARGE (24 Transformer layers) are two models trained on English Wikipedia (2,500M words) and BooksCorpus [20] (800M words) for one million update steps. Due to limited GPU resources, we only used BERTBASE in this experiment, since BERTLARGE requires resources currently beyond our high- performance hardware. The configuration parameters of the pre-trained BERTBASE model are L=12, H=768, A=12, total Parameters=110M, where L is the number of layers (i.e., Transformer blocks), H is the hidden size, and A is the number of self-attention heads. The feed-forward/filter size is set to 4H, i.e., 3072 for H = 768.
The two strategies of our research are as follows:
Strategy 1
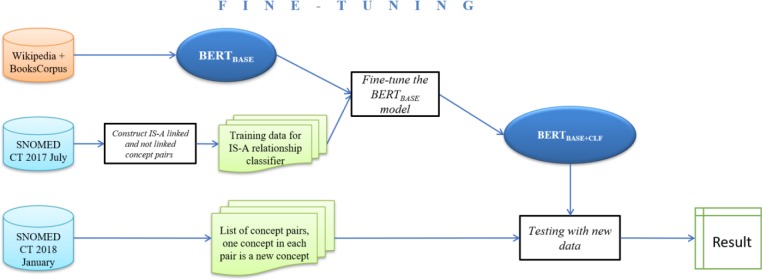
Step 1. Fine-tuning the BERTBASE model (Figure 1): We extracted IS-A linked and not linked concept pairs as supervised fine-tuning data from the SNOMED CT July 2017 release. We will refer to these pairs as IS-A and non- IS-A pairs. Then we trained a relationship classifier on top of BERTBASE with the IS-A and non-IS-A pairs to obtain the BERTBASE+CLF model.
Figure 1:
The pipeline for Strategy 1 Fine-tuning. CLF is short for Classifier.
Step 2. Prediction on new release data (illustrated in the rightmost process of Figure 1): We tested the trained BERTBASE+CLF model to verify IS-A links and the absence of IS-A links for newly added concepts in the SNOMED CT January 2018 release.
Strategy 2
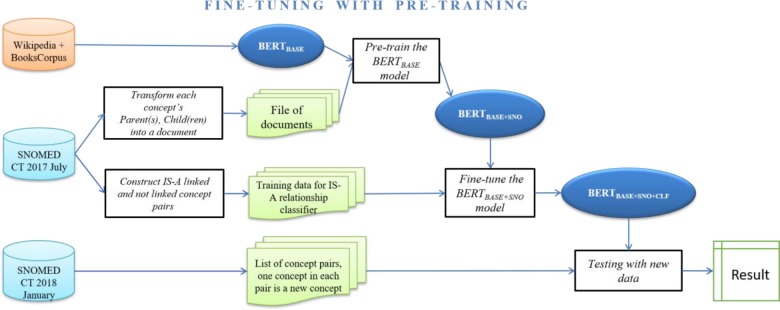
Step 1. Pre-training the BERTBASE model (Figure 2): We preprocessed concept-related information from the July 2017 release to generate documents that were used as unsupervised pre-training data. Then we trained BERTBASE with unsupervised concept level data so that the trained BERTBASE+SNO model integrated terminology information. Then we applied the fine-tuning process (of Strategy 1) to train a classifier on top of BERTBASE+SNO to derive the BERTBASE+SNO+CLF model.
Figure 2:
The pipeline for Strategy 2 combining Fine-tuning with Pre-training. SNO is short for SNOMED CT.
Step 2. Prediction on new release data (illustrated in the rightmost process of Figure 2): We tested the trained BERTBASE+SNO+CLF model to verify IS-A links and non-IS-A pairs for newly added concepts in the SNOMED CT 2018 January release.
Details of the specific stages of the two strategies are discussed in the following:
Fine-tuning strategy
Data preparation: The training samples passed to the Fine-tuning process are a set of IS-A and non-IS-A concept pairs. In SNOMED CT, the IS-A concept pairs are given. Thus, we can use them as a positive sample. On the other hand, the negative sample could be all the non-IS-A pairs of concepts. This creates an imbalance between the positive and negative samples, because there are many more pairs not connected by IS-A links. Thus, we pick non-IS-A pairs for the negative sample as follows. For each IS-A pair (A, B) we look for the siblings C1, C2, …Ck of B. Then we choose non-IS-A pairs (A, Ci) with i=1, 2, … k. The advantage of such pairs is that they are closely related to the corresponding IS-A pair. This will sharpen the distinction between IS-A and non-IS-A pairs in training. For example, (Crushing injury of back, Crushing Injury) defines an IS-A link, while (Crushing injury of back, Shear injury) is a similar pair that should not be connected by an IS-A link. The reason is that Shear injury is a sibling of Crushing Injury, with the same parent Injury by mechanism.
In the data preparation for Fine-tuning, we first extracted the positive and negative samples, and randomly downsampled the negative sample to the size of the positive sample, at the beginning of each training round. We shuffled our dataset and then used 90% of it for training and kept 10% as the test set. The samples went through three preprocessing steps: Text normalization (E.g., Excision of Reinke’s edema, → excision of reinke’s edema), Punctuation splitting (E.g., excision of reinke’s edema, → excision of reinke ‘ s edema), and WordPiece tokenization (excision of reinke ‘ s edema, → ex ##cision of rein ##ke ‘ s ed ##ema). Then the samples were processed by BERTBASE, which performed its own preprocessing, including input embeddings, segment masking, labeling, etc. [7]. The input embeddings are the sum of the token embeddings, the segmentation embeddings and the position embeddings. Due to space limitations, we concentrate on the operations that are essential for Fine-tuning.
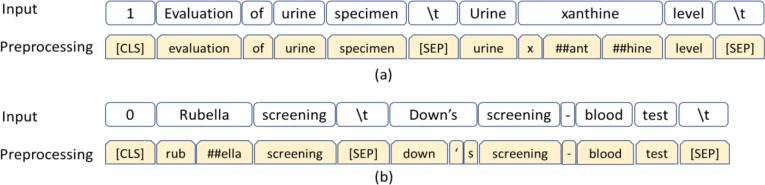
For example, Urine xanthine level is the child of Evaluation of urine specimen in the SNOMED CT Procedure hierarchy. The input sequence will be “1 Evaluation of urine specimen (\t) Urine xanthine level (\t)”. This input will be converted as one training instance to “[CLS] evaluation of urine specimen [SEP] urine x ##ant ##hine level [SEP]” as shown in Figure 3(a). The first token of the sequence is the classification embedding ([CLS]), representing a classification label. A special token ([SEP]) is used to separate sentences. Out-of-vocabulary words are split into word pieces and denoted with ##. For example, “xanthine” is denoted as three items “x”, “##ant”, and “##hine.” Similarly, Rubella screening is not a child of Down’s screening – blood test. The input sequence “0 Rubella screening (\t) Down’s screening - blood test (\t)” will be converted to “[CLS] rub ##ella screening [SEP] down ‘ s screening - blood test [SEP]” in Figure 3(b).
Figure 3:
Fine-tuning data: Preprocessing (a) IS-A and (b) non-IS-A concept pairs
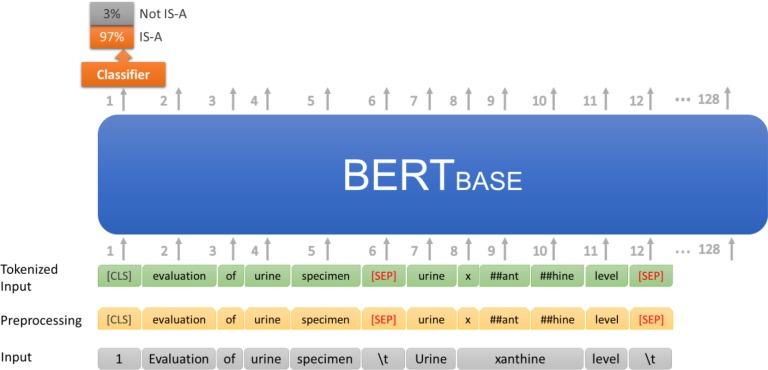
Fine-tuning the BERTBASE model: We fine-tuned the BERTBASE model to predict the IS-A and non-IS-A linking for the concept pairs in the test data. This is similar to a binary sentence-pair classification task. We used the sentence prediction capability of BERTBASE, trained as BERTBASE+CLF, to predict IS-A links between concept pairs of a terminology. We employed a classifier using softmax with categorical cross-entropy on top of BERTBASE. The parameters of BERTBASE were fine-tuned to maximize the log-probability of the correct label (IS-A or non-IS-A). Given two concepts A and B, described as two “sentences,” the classifier learned to determine whether B is the next “sentence” following A. This was modeled as equivalent to classifying whether the concept B should be a child of A, i.e., B is-a A (Figure 4). The input “1 Evaluation of urine specimen (\t) Urine xanthine level (\t)” was converted as one training instance to “[CLS] evaluation of urine specimen [SEP] urine x ##ant ##hine level [SEP]” with Class label = 1. Class 1 means that there should be an IS-A link between the two concepts, and Class 0 means that there shouldn’t be such a link. The BERTBASE model computed the probabilities for Class 0 and Class 1, and recorded the result as a 2 × 1 vector. The classifier reported the class label with the higher probability. The error between the true label and the label predicted by the model was back-propagated through the model to improve the network’s parameters. The obtained model is denoted as BERTBASE+CLF (CLF = classifier), the model after Fine-tuning. For this we used the default model hyperparameters in pre-trained BERTBASE, with the exception of the sequence length (=128), batch size (=64), learning rate (=2e-5), and number of training epochs (=3).
Figure 4:
Fine-tuning the BERTBASE model with concept pairs to obtain BERTBASE+CLF model.
Pre-training and fine-tuning strategy
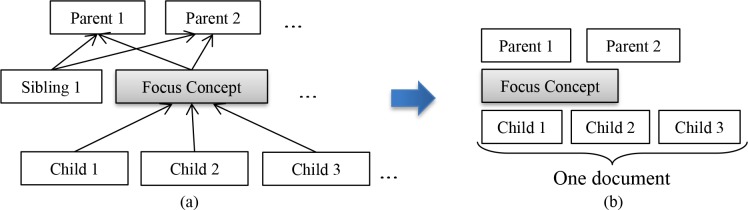
Data Preparation: In our setup of unsupervised Pre-training, BERTBASE is not trained for a specific task, but the purpose is integrating medical knowledge into its representation. This is done by training with non-task related samples taken from SNOMED CT. BERT was originally trained with millions of documents that are composed of sentences. Similarly, we generated a list of terminology-oriented documents by creating one “document” per focus concept F (Figure 5), with related concept(s) as “sentences” of such documents. The ID for a document is the corresponding SNOMED CT concept ID. The content of this document consists of the concepts that are hierarchically related to F (Figure 5(a)). Specifically, we chose F’s parents (targets of IS-A links from F), F itself, and its children (sources of IS-A links to F). Thus, the whole document text is: Parent(s) – Focus concept – Child(ren) (Figure 5(b)). The concept groups are separated into lines, e.g., “(Parents) finding of abnormal level of heavy metals in blood, finding of trace element level –NEW LINE– (Focus concept) blood copper abnormal –NEW LINE– (Children) raised blood copper level, serum copper level abnormal” is the document for the focus concept blood copper abnormal. This construction is based on the idea that a concept is the topic of a document and that the closely related concepts are descriptions of the meaning of this concept in the terminology hierarchy. To feed sentences into the BERTBASE model, all the documents are concatenated in one text file, separated by empty lines.
Figure 5:
Pre-training data: Serializing (a) the hierarchical structure of one concept into (b) one document
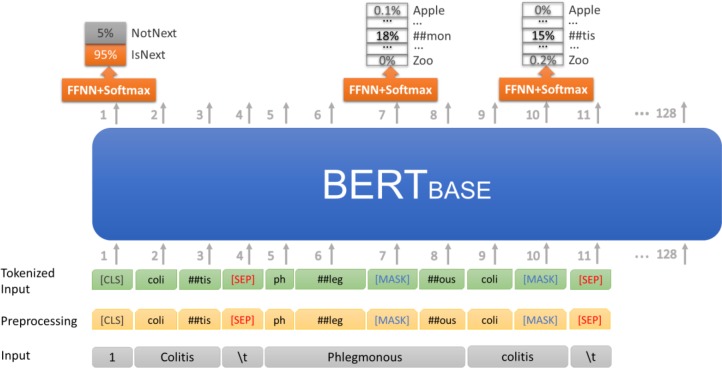
Pre-training the BERTBASE model: To utilize BERT’s powerful language representation, we started with BERTBASE and embedded terminology knowledge with new training data. BERT was originally trained for two unsupervised prediction tasks: Masked Language Modeling (MLM) and Next Sentence Prediction (NSP) on an arbitrary text corpus. We adopted the same two training tasks and objective with concept-based documents from SNOMED CT. In the MLM phase, the training objective is to predict only the masked words. We randomly masked out 15% of the words across all the concept-based documents, and then trained the complete BERTBASE model to output the masked words. In the NSP phase, the objective is to learn relationships between concepts: Given two concepts A and B, is B a child of A, or not (Figure 6). We extracted two “sentences” Colitis and Phlegmonous colitis from the document for the focus concept Colitis. After preprocessing these two concepts (treated as two “sentences”) as shown in the middle level, we masked out two token – “##mon” and “##tis”. The BERTBASE model was trained to raise the probabilities of two correct tokens “##mon” and “##tis” over other tokens in the vocabulary. In addition, as Phlegmonous colitis IS-A Colitis, the BERTBASE model was also trained to output the correct classification label “IsNext.” The obtained model is denoted as BERTBASE+SNO (SNO=SNOMED CT). Then we applied the Fine-tuning process (see above) to the BERTBASE+SNO model to get the BERTBASE+SNO+CLF model.
Figure 6:
Pre-training the BERTBASE model with concept-based documents to obtain BERTBASE+SNO model. FFNN is short for Feedforward neural network.
The training parameters used for Pre-training are as follows: batch size = 64, sequence length =128, training steps = 15,000 for Procedure and 200,000 for Clinical Finding. learning rate =2e-5, dropout rate = 0.1, and activation function = gelu (Gaussian error linear unit).
Testing the prediction on new release data
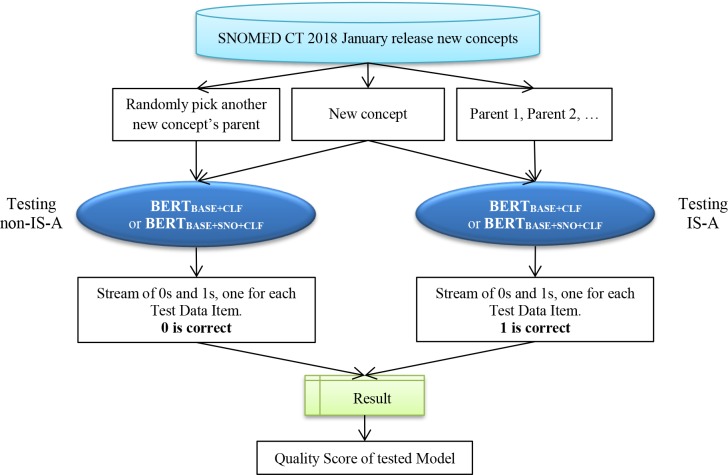
To evaluate the BERTBASE+CLF and the BERTBASE+SNO+CLF models on previously unseen data, we created separate test tasks, using new concepts from the Procedure and the Clinical finding hierarchy of the January 2018 release (Figure 7). This description will focus on the Procedure hierarchy. For each new concept that was added to the Procedure hierarchy in this release, we extracted it and its parents as positive sample pairs. For example, Local excision of lesion of kidney has two parents Local excision and Excision of lesion of kidney. The corresponding positive testing samples are “Local excision (\t) Local excision of lesion of kidney” and “Excision of lesion of kidney (\t) Local excision of lesion of kidney” with the true Class label = 1. For the negative sample, we paired each new concept with a randomly chosen concept from the other new concepts’ parents. For example, we randomly select Ultrasonography of left lower limb, which is the parent of Ultrasonography of left knee region, and paired it with Local excision of lesion of kidney to form the instance “Ultrasonography of left lower limb (\t) Local excision of lesion of kidney” with the label = 0.
Figure 7:
Data flow for testing the trained BERTBASE+CLF or BERTBASE+SNO+CLF models with unseen data
The concept pairs were randomly arranged into a sequence and sent to the trained BERTBASE+CLF and BERTBASE+SNO+CLF models. The tested model processed each input pair, using the weights that it had learned before, returning a class label (0 or 1) as prediction result. For negative samples, label 0 is correct, indicating that there is no IS-A link between these two concepts in the new SNOMED CT release. Label 1 is correct for positive samples, indicating the existence of an IS-A link. The predicted result labels were compared with the true labels to calculate prediction accuracy in terms of Precision, Recall, and F1 score.
Results
We first report the prediction results of the Fine-tuning model and Pre-training & Fine-tuning model with samples extracted from the Procedure hierarchy of the SNOMED CT 2018 January release, with 15,000 training steps. The Precision, Recall, and F1 scores for ten tests are presented in Table 1. For the Procedure hierarchy, the model was tested against 3,908 pairs (1,954 positives and 1,954 negatives). For example, in Test 7 for IS-A classification, the Precision is 0.69, Recall is 0.98, and F1 score is 0.81 for Fine-tuning. When adding Pre-training, Precision is 0.73, Recall is 0.98, and F1 score is 0.84. The F1 score improved by about 3.7%. Similarly, for Non-IS-A tests, the F1 score increased from 0.71 to 0.77, an 8.5% improvement. On average, by adding Pre-training, there are 6.3% (from 0.80 to 0.85) and 14.5% (from 0.69 to 0.79) improvements of F1 for IS-A and Non-IS-A classifications, respectively.
Table 1.
Precision, Recall, and F1 score for ten tests of Procedure hierarchy (training steps = 15,000).
| Procedure | IS-A Classification | Non-IS-A Classification | ||||||||||
| Fine-tuning | Pre-training & Fine-tuning | Fine-tuning | Pre-training & Fine-tuning | |||||||||
| No. | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| 1 | 0.67 | 0.98 | 0.79 | 0.75 | 0.98 | 0.85 | 0.96 | 0.51 | 0.67 | 0.97 | 0.68 | 0.80 |
| 2 | 0.66 | 0.98 | 0.79 | 0.77 | 0.98 | 0.86 | 0.97 | 0.48 | 0.64 | 0.98 | 0.70 | 0.82 |
| 3 | 0.70 | 0.98 | 0.81 | 0.75 | 0.98 | 0.85 | 0.96 | 0.58 | 0.72 | 0.97 | 0.68 | 0.80 |
| 4 | 0.66 | 0.98 | 0.79 | 0.74 | 0.98 | 0.84 | 0.97 | 0.50 | 0.66 | 0.97 | 0.66 | 0.79 |
| 5 | 0.69 | 0.98 | 0.81 | 0.74 | 0.98 | 0.84 | 0.96 | 0.56 | 0.70 | 0.97 | 0.66 | 0.79 |
| 6 | 0.71 | 0.97 | 0.82 | 0.74 | 0.98 | 0.85 | 0.96 | 0.59 | 0.73 | 0.97 | 0.66 | 0.79 |
| 7 | 0.69 | 0.98 | 0.81 | 0.73 | 0.98 | 0.84 | 0.97 | 0.56 | 0.71 | 0.97 | 0.64 | 0.77 |
| 8 | 0.67 | 0.98 | 0.80 | 0.71 | 0.98 | 0.82 | 0.96 | 0.52 | 0.68 | 0.97 | 0.59 | 0.73 |
| 9 | 0.69 | 0.98 | 0.81 | 0.73 | 0.98 | 0.84 | 0.96 | 0.56 | 0.71 | 0.97 | 0.63 | 0.77 |
| 10 | 0.66 | 0.98 | 0.79 | 0.76 | 0.98 | 0.86 | 0.96 | 0.49 | 0.65 | 0.97 | 0.69 | 0.81 |
| Average | 0.68 | 0.98 | 0.80 | 0.74 | 0.98 | 0.85 | 0.96 | 0.54 | 0.69 | 0.97 | 0.66 | 0.79 |
| Standard Deviation | 0.02 | 0.00 | 0.01 | 0.02 | 0.00 | 0.01 | 0.00 | 0.04 | 0.03 | 0.00 | 0.03 | 0.03 |
Due to space limitations, we only report the summary of the Precision, Recall, and F1 scores from ten tests for the Procedure hierarchy with 10,000 training steps (Table 2). In each test, the model was tested against 3,908 pairs (1,954 positives and 1,954 negatives). On average, by adding Pre-training to Fine-tuning, the improvements are 3.75% (from 0.80 to 0.83) and 10.1% (from 0.69 to 0.76) for IS-A and Non-IS-A classifications, respectively.
Table 2.
Precision, Recall, and F1 score for ten tests of Procedure hierarchy (training steps = 10,000).
| Procedure | IS-A Classification | Non-IS-A Classification | ||||||||||
| Fine-tuning | Pre-training & Fine-tuning | Fine-tuning | Pre-training & Fine-tuning | |||||||||
| Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | |
| Average | 0.68 | 0.98 | 0.80 | 0.72 | 0.98 | 0.83 | 0.96 | 0.54 | 0.69 | 0.97 | 0.62 | 0.76 |
| Max | 0.71 | 0.98 | 0.82 | 0.75 | 0.99 | 0.85 | 0.97 | 0.59 | 0.73 | 0.98 | 0.67 | 0.79 |
| Min | 0.66 | 0.97 | 0.79 | 0.71 | 0.98 | 0.82 | 0.96 | 0.48 | 0.64 | 0.96 | 0.59 | 0.73 |
| Standard Deviation | 0.02 | 0.00 | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.04 | 0.03 | 0.01 | 0.02 | 0.02 |
For the Clinical finding hierarchy, the summary of ten tests results with training steps = 200,000 is reported in Table 3. In each test, the model was tested against 8,574 pairs (4,287 positives and 4,287 negatives). On average, by adding Pre-training, the improvements are 7.5% (from 0.80 to 0.86) and 15.3% (from 0.72 to 0.83) for IS-A and Non-IS-A classifications, respectively.
Table 3.
Precision, Recall, and F1 score for ten tests of Clinical Finding hierarchy (training steps = 200,000).
| Clinical Finding | IS-A Classification | Non-IS-A Classification | ||||||||||
| Fine-tuning | Pre-training& Fine-tuning | Fine-tuning | Pre-training& Fine-tuning | |||||||||
| Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | |
| Average | 0.70 | 0.94 | 0.80 | 0.79 | 0.94 | 0.86 | 0.90 | 0.60 | 0.72 | 0.93 | 0.76 | 0.83 |
| Max | 0.72 | 0.94 | 0.82 | 0.82 | 0.95 | 0.88 | 0.91 | 0.64 | 0.75 | 0.94 | 0.80 | 0.86 |
| Min | 0.69 | 0.93 | 0.79 | 0.77 | 0.94 | 0.85 | 0.89 | 0.58 | 0.70 | 0.92 | 0.73 | 0.81 |
| Standard Deviation | 0.01 | 0.00 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 | 0.02 | 0.02 | 0.01 | 0.02 | 0.01 |
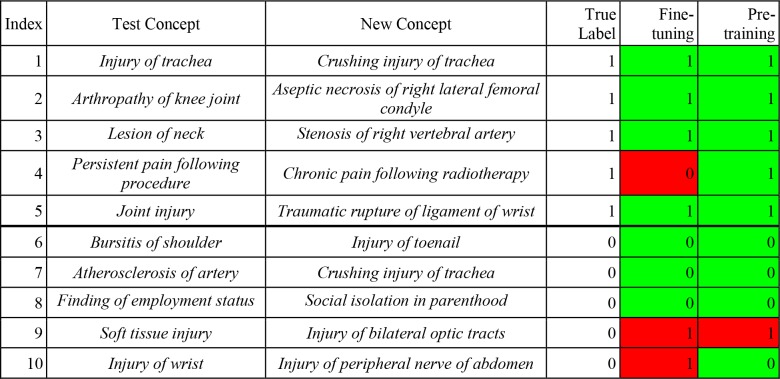
Regarding the prediction of IS-A links for new concepts, we show ten examples of our two models’ prediction results (Table 4) for ten pairs for which the second concept was newly added to SNOMED CT’s Clinical finding hierarchy in the 2018 January release. For each test, we paired one Test Concept with one New Concept as one test instance, then we let the model predict IS-A links between them. For instance, for Example 2, we chose Arthropathy of knee joint as the first concept and paired it with Aseptic necrosis of right lateral femoral condyle. Then the task became to predict whether there is an IS-A link between the two concepts. Both the Fine-tuning and Pre-training & Fine-tuning models returned the correct label (=1). For Example 4, the Fine-tuning model is wrong, and the combined model is correct that Chronic pain following radiotherapy IS-A Persistent pain following procedure. Both models are wrong about Injury of bilateral optic tracts, because it is not a Soft tissue injury (Example 9).
Table 4.
Prediction results of two models on five IS-A & five non-IS-A examples from Clinical Finding hierarchy.
 |
Discussion
In this paper we set out to investigate whether the “next sentence prediction” capability of BERT can be fine-tuned to verify the parent(s) of new concepts added to a terminology. Such a capability can be utilized in automatic enrichment of terminologies. The results for the Procedure and Clinical finding hierarchy confirm that our technique, which utilizes this capability of BERT, is indeed able to verify the IS-A relationships from new concepts to their parents with a 0.80 F1 average value. When enhancing Fine-tuning of BERT with Pre-training the average F1 score grows to 0.85 (0.86).
However, looking at the details we see that the recall to identify IS-A relationships is very high (0.98) while the precision is only 0.74. The outcome for identifying the Non-IS-A pairs of concepts is the opposite, with high (0.97) precision and low (0.66) recall, yielding an F1 value of 0.79. These results indicate that our technique verifies almost all the IS-A relationships, but wrongly identifies some Non-IS-A pairs as having IS-A relationships.
Thus, the challenge for future research is to improve the precision. In previous research [11, 21], we have shown that using summarization techniques of terminologies [22] can improve the training of ML techniques to better distinguish between IS-A relationships and pairs that are not connected by IS-A relationships. In future research, we will investigate whether utilizing summarization techniques can improve the precision and thus the F1 value.
Another issue is the number of training steps required for Pre-training. For the Procedure hierarchy 15,000 training steps provided the best results for enhancing the process by Pre-training. The enhancement was about double than for 10,000 training steps. Experiments with 20,000 and 25,000 training steps showed a leveling off of F1 at 20,000 and a lower F1 value at 25,000. However more research is needed to confirm this behavior beyond 15,000 training steps. For the Clinical finding hierarchy, which is about twice the size of the Procedure hierarchy, 200,000 training steps were required to obtain the same enhancement as for the Procedure hierarchy, which had required just 15,000 training steps. Hence, much more pre-training steps are required when the hierarchy is larger. The two transfer learning models using BERT are superior to our previously proposed CNN model [11], which was trained with the whole SNOMED CT consisting of 473,756 concepts. That CNN model achieved an average F1 score of 0.70, and could only verify IS A links for new concepts with multiple parents. As a consequence of this observation, it is preferable to perform automatic enrichment for each hierarchy of SNOMED CT separately, rather than for the whole SNOMED CT. Another reason for enriching each hierarchy separately is that the content of each hierarchy is different, and ML is likely more effective by modularizing the learning into uniform hierarchies than learning a large, non-uniform body of knowledge in one process.
In the BERTBASE model, features are more generic/linguistic in the early network layers and more dataset-specific in the later layers. Thus, fine-tuning is normally inexpensive, because one only needs to modify the later layers or train one or two task-specific layers on top. All of the results in the paper can be replicated in at most 3 to 5 hours on a single GPU, starting from the same pre-trained BERTBASE model. Pre-training is more expensive than Fine-tuning. For example, it took about four days to run 200,000 steps to pre-train the model with the Clinical finding hierarchy data on a single GPU. However, this is a one-time procedure for each hierarchy. We plan to release the two pre-trained models of this paper for future research work, to save other researchers the effort and time to pre-train their own models from scratch.
Limitations: The BERTBASE model was trained with the concatenation of the BooksCorpus (800M words) [20] and English Wikipedia (2,500M words). It employs the WordPiece embeddings [23] with a 30,522 tokens vocabulary, which does not include most medical terms. Thus, medical terms that are not in the WordPiece vocabulary are split into word pieces denoted by ##. For example, “adenoid” is split into “aden” and “##oid.” The lack of medical terms in BERT’s vocabulary limited its applicability to support insertion of new concepts into a medical terminology such as SNOMED CT, and would probably impair other NLP tasks within the medical domain. In future work, we will expand the vocabulary to include common medical terms selected from terminologies such as SNOMED CT. We will investigate whether pre-training BERT with medical terms can help improve its performance in some common NLP tasks in the medical domain, such as tagging and named entity recognition in EHRs.
Conclusion
We have shown that one can fine-tune the BERT model and obtain an effective technique for correctly placing new concepts in the right positions of a terminology. Furthermore, by pre-training BERT with SNOMED CT content, we improved the precision while preserving the high recall and thus we improved the F1 value.
Acknowledgments:
Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA190779. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures & Table
References
- 1.Jimenez AM, Lawley MJ. Snorocket 2.0: Concrete Domains and Concurrent Classification. OWL Reasoner Evaluation Workshop (ORE 2103) 2013 [Google Scholar]
- 2.Shearer R, Motik B, Horrocks I. HermiT: a highly-efficient OWL reasoner. Proceedings of the Fifth OWLED Workshop on OWL: Experiences and Directions. 2008 [Google Scholar]
- 3.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 4.McInnes BT, Pedersen T, Liu Y, Pakhomov SV, Melton GB. Using second-order vectors in a knowledge-based method for acronym disambiguation. Proceedings of the 15th Conf on Computational Natural Language Learning; 2011: Association for Computational Linguistics [Google Scholar]
- 5.Pakhomov SV, Finley G, McEwan R, Wang Y, Melton GB. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics. 2016;32(23):3635–44. doi: 10.1093/bioinformatics/btw529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajani NF, Bornea M, Barker K. Stacking with Auxiliary Features for Entity Linking in the Medical Domain. BioNLP. 2017;2017:39–47. [Google Scholar]
- 7.Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805. 2018 [Google Scholar]
- 8.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. Biobert: pre-trained biomedical language representation model for biomedical text mining. arXiv preprint arXiv:190108746. 2019 doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Si Y, Wang J, Xu H, Roberts K. Enhancing Clinical Concept Extraction with Contextual Embedding. arXiv preprint arXiv:190208691. 2019 doi: 10.1093/jamia/ocz096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SNOMED CT [1/17/2019]. Available from: https://www.snomed.org/ [Google Scholar]
- 11.Liu H, Geller J, Halper M, Perl Y. Using Convolutional Neural Networks to Support Insertion of New Concepts into SNOMED CT. AMIA Annual Symposium Proceedings; 2018: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 12.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems. 2013 [Google Scholar]
- 13.Pennington J, Socher R, Manning C. Glove: Global vectors for word representation. Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP) 2014 [Google Scholar]
- 14.Joulin A, Grave E, Bojanowski P, Douze M, Jegou H, Mikolov T. Fasttext. zip: Compressing text classification models. 2016 arXiv preprint arXiv:161203651. [Google Scholar]
- 15.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Advances in Neural Information Processing Systems. 2017 [Google Scholar]
- 16.Sang EF, De Meulder F. Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. 2003 arXiv preprint cs/0306050. [Google Scholar]
- 17.Rajpurkar P, Zhang J, Lopyrev K, Liang P. Squad: 100,000+ questions for machine comprehension of text. 2016 arXiv preprint arXiv:160605250. [Google Scholar]
- 18.Socher R, Perelygin A, Wu J, Chuang J, Manning CD, Ng A, et al. Recursive deep models for semantic compositionality over a sentiment treebank. Proceedings of the conference on empirical methods in NLP. 2013 [Google Scholar]
- 19.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. TensorFlow: A System for Large-Scale Machine Learning. OSDI. 2016 [Google Scholar]
- 20.Zhu Y, Kiros R, Zemel R, Salakhutdinov R, Urtasun R, Torralba A, et al. Aligning books and movies: Towards story-like visual explanations by watching movies and reading books. Proceedings of the IEEE international conference on computer vision. 2015 [Google Scholar]
- 21.Liu H, Zheng L, Perl Y, Geller J, Elhanan G. Can a Convolutional Neural Network Support Auditing of NCI Thesaurus Neoplasm Concepts? International Conference on Biomedical Ontology (ICBO-2018) 2018 [Google Scholar]
- 22.Halper M, Gu H, Perl Y, Ochs C. Abstraction networks for terminologies: Supporting management of “big knowledge”. Artif Intell Med. 2015;64(1):1–16. doi: 10.1016/j.artmed.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Schuster M, Chen Z, Le QV, Norouzi M, Macherey W, et al. Google’s neural machine translation system: Bridging the gap between human and machine translation. 2016 arXiv preprint arXiv:160908144. [Google Scholar]