Abstract
Circular RNA (circRNA) plays an important role in the development of diseases, and it provides a novel idea for drug development. Accurate identification of circRNAs is important for a deeper understanding of their functions. In this study, we developed a new classifier, CirRNAPL, which extracts the features of nucleic acid composition and structure of the circRNA sequence and optimizes the extreme learning machine based on the particle swarm optimization algorithm. We compared CirRNAPL with existing methods, including blast, on three datasets and found CirRNAPL significantly improved the identification accuracy for the three datasets, with accuracies of 0.815, 0.802, and 0.782, respectively. Additionally, we performed sequence alignment on 564 sequences of the independent detection set of the third data set and analyzed the expression level of circRNAs. Results showed the expression level of the sequence is positively correlated with the abundance. A user-friendly CirRNAPL web server is freely available at http://server.malab.cn/CirRNAPL/.
Abbreviations: circRNA, circular RNA; RF, random forest; PSO, particle swarm optimization algorithm; lncRNAs, long non-coding RNAs; PCGs, protein coding genes; DAC, Dinucleotide-based auto-covariance; DCC, Dinucleotide-based cross-covariance; DACC, Dinucleotide-based auto-cross-covariance; MAC, Moran autocorrelation; GAC, Geary autocorrelation; NMBAC, Normalized Moreau–Broto autocorrelation; PC-PseDNC-General, General parallel correlation pseudo-dinucleotide composition; SC-PseDNC-General, General series correlation pseudo-dinucleotide composition; Triplet, Local structure-sequence triplet element; PseSSC, Pseudo-structure status composition; PseDPC, Pseudo-distance structure status pair composition; ELM, extreme learning machine; RBF, radial basis function; CNN, Convolutional Neural Networks; SVM, support vector machine; SE, Sensitivity; SP, Specifity; ACC, Accuracy; MCC, Matthews Correlation Coefficient; MRMD, Maximum-Relevance-Maximum-Distance
Keywords: Circular RNA, Extreme learning machine, Particle swarm optimization algorithm, Identification, Expression level
1. Introduction
Circular RNA (circRNA) is a newly identified RNA type that differs from conventional linear RNA in humans. It is a noncoding RNA molecule without having a 5-end cap or a 3-end tail, instead, forming a circular structure [1], [2] (Fig. 1). CircRNA was first discovered in 1969 by Diener, while researching potato spindle tuber disease [3]. Electron microscopy revealed the formation of such closed-loop RNA, also known as a viroid. The subsequent emergence of high-throughput sequencing techniques (RNA-seq) enabled improved sequencing of circRNAs of various species, and many circRNAs have now been identified [4], [5]. To date, more than 10,000 different circRNAs have been successfully identified from fruit flies and worms to mice and humans [6], [7].
Fig. 1.
CircRNA splicing structure. CircRNA is a new class of RNA that differs from traditional linear RNA. It does not have a 5′ cap or a 3′ tail and is not easily degraded by exonuclease. In humans, it is more stable than linear RNA. Most circRNAs are formed by exons, while a few are derived from intron fragments.
Against background researches and applications of circRNA, a series of databases have been built, such as circBase [8], circNet [9], circ2Traits [10], TSCD [11], circRNADb [12], and CircInteractome [13]. Studying the structure and function of circRNAs, researchers have revealed their importance in the pathogenesis of arteriosclerosis, nervous system disorders, diabetes, and tumors [14], [15], [16]. The unique endogenous characteristics of circRNAs enable their value as biomarkers, providing a new approach for drug development [6], [17], [18] and a new direction for the evolution of life. Xu used Qualcomm sequencing to detect the expression profiles of three wheat samples (LH9, XN979, and YN29) and identified 33 differentially expressed circRNA [19]. Ye used published RNA-Seq data to perform genome-wide identification of circRNA in rice and Arabidopsis. Based on this, they compared the characteristics of plant and animal circRNA [20]. Moreover, Memczak analyzed circRNA from humans, mice, and nematodes, and detected thousands of well-represented, stable circRNAs, demonstrating their regulatory capabilities [21]. Given this landscape, accurate detection and quantification of circRNAs are essential for a deeper understanding of their function.
In addition to the abundance of RNA-seq data, a variety of algorithms for calculating and visualizing circRNA have recently been developed [2], [17], [22]. Li Chen developed the CIRCexplorer software based on RNA-seq data of Arabidopsis and rice, providing a more comprehensive and sensitive circRNA prediction method for plants [23]. Song et al. developed a computational pipeline, called UROBORUS, to detect circRNA in RNA-seq data. They successfully verified 24 circRNA from 27 randomly selected circRNA [6]. Additionally, based on RNA-seq, Danan developed a circRNA-seq method for reading circRNA in an unbiased, genome-wide manner. Moreover, they mapped the transcriptome of solfataricus [24]. In another study, Jeck et al. used high-throughput sequencing of ribosomal-deficient RNA and identified more than 25,000 loops in human fibroblasts containing non-collinear exons (“reverse splicing”) [25]. Gao et al. proposed a new algorithm, CIRI, based on cross-shear signal, which uses a variety of filtering strategies to accurately detect circRNA from transcriptome data [26]. Zhang et al. used non-polyadenylation and rnaser-treated RNA-seq data from H9 human embryonic stem cells to predict reverse splicing junctions, and they systematically characterized circRNA using the newly developed pipeline CIRCexplorer [27]. Furthermore, Vo et al. used the exome capture RNA sequencing protocol to detect and characterize circRNA across >2000 cancer samples, establishing the most comprehensive catalog of circRNA species to date (MiOncoCirc) [28]. You et al. proposed the method, Acfs, allowing for the redefinition and accurate and rapid identification of circRNA from single-ended and double-ended RNA-seq data, as well as their abundance [29]. Furthermore, Zhang et al. sequenced Branchiostoma belcheri circRNA and identified 1859 circRNAs using the find_circ and CIRI algorithms [30]. At present, many available recognition methods are based on RNA-Seq data, and it is very important to use bioinformatics with direct circRNA sequence training to achieve more accurate recognition. In contrast to the tools that use RNA-seq data as input, Pan et al. extracted the characteristics of the sequence and used the random forest (RF) algorithm to identify circRNAs, subsequently building the online identification server WebCircRNA [31]. PredcircRNA classified circRNA and lncRNA based on a computational method of a multi-core learning framework with multiple functional training [32]. H-ELM used a hierarchical extreme learning machine algorithm with feature selection to extract the same features from other lncrna and classify circular RNAs [33]. Mohamed Chaabane proposed an end-to-end deep learning framework circDeep, which integrates shared representations across different modes and improves circularRNA for classification [34].
Many studies for protein recognition [35] and site detection [36] have been performed based on machine learning, such as RF [37] and Artificial Neural Network [38]. By contrast, few studies focus on the identification of circRNAs. Therefore, there is a need for studies on how to use sequence information to achieve more accurate identification by using the characteristics of RNA sequences. We propose a new method that mainly uses the structural features of RNA and nucleotide composition to optimize the extreme learning machine (ELM) based on the particle swarm optimization algorithm (PSO). Based on these, a classification system was built to optimize the effect of circRNA identification.
2. Material and methods
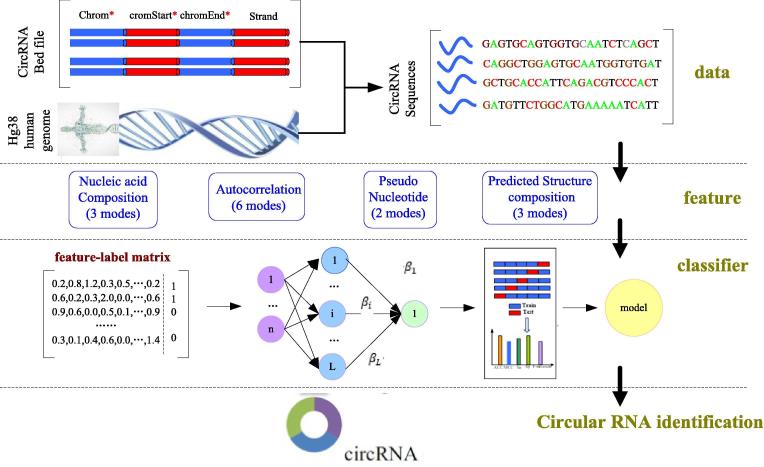
In this study, we used the ELM method to identify circRNA. The flowchart of the identification framework is shown in Fig. 2.
Fig. 2.
Flowchart of CirRNAPL. CirRNAPL identifies circRNA in four main steps: data, feature, classifier, and circRNA identification Data: This involves dataset construction. According to the bed data file and the hg38 human genome, we write Python script files to extract the corresponding sequence data. Features: This involves the extraction of features. In this work, information such as the structural characteristics of the RNA sequence is used as the feature to be extracted, including four parts, with a total of 14 calculation models. Classification: This involves the construction of the classifier. Here, the extreme learning machine based on particle swarm optimization is used as the classification algorithm. The classifier CirRNAPL is constructed by a tenfold cross-validation method, and the final classification model is output. CircRNA identification: The RNA sequence to be labeled is identified using the classifier CirRNAPL.
2.1. Datasets
circRNAs are evolutionarily conserved among different species, and the detection of circular RNAs is important for further understanding the biological origin and purpose of circular RNAs [39], [40]. The datasets that we used were presented in the literature of Pan et al. [31] (Table 1). The literature downloaded 92,375 circRNA transcripts from circBase. After deleting transcripts of less than 200nt in length and the overlapping circular RNA transcripts, 14,084 circRNA data were obtained. The database source of the 9533 PCGs and 19,723 lncRNAs dataset is GENCODv19, in which overlapping circRNA sequences were removed from the PCG data. The third dataset includes 2082 circRNAs from stem cells expressed in H1hsec and the same number of identically derived circRNAs from other cells that were not expressed in H1hsec. And the circRNA vs lncRNA dataset also applied to the literature of Mohamed et al. [34], Pan et al. [32] and Chen et al. [33].
Table 1.
The details of the datasets.
| Datasets | Positive Data | Negative Data |
|---|---|---|
| circRNA vs PCG | 14,084 circRNAs | 9533 PCGs |
| circRNA vs lncRNA | 14,084 circRNAs | 19,722 lncRNAs |
| Stem cell vs not | 2082 circRNAs | 2082 circRNAs |
The construction of the classifier used in this study involved ten-fold cross-validation. To verify the classifier, independent training and test set verification was also performed. For the dataset of 14,084 circRNAs, the number used for the training set was 10,000, and that for the test set was 4084. For the lncRNA dataset of 19,722, the number for the training set was 10,000, and that for the test set was 9722. For the PCG training set of 9533, the number for the training set was 8000, and that for the test set was 1533. Finally, for the two datasets of 2082 circRNAs of each of stem cells and other cells, the training set consisted of 1800 circRNAs, and the test set consisted of the remaining 282 circRNAs.
2.2. Feature representation methods
To identify circRNAs, four features of the sequence data were extracted, including Ribonucleic acid composition, Autocorrelation, Pseudo-ribonucleic acid composition, and Predicted structure composition [41]. These four features comprised a total of 15 modes, wherein Ribonucleic acid composition included k-mer (the parameter k is 2 and 3), Mismatch, and Subsequence; Autocorrelation included DAC, DCC, DACC, MAC, GAC, and NMBAC; Pseudo-nucleotide composition included General parallel correlation pseudo-dinucleotide composition (PC-PseDNC-General) and General series correlation pseudo-dinucleotide composition (SC-PseDNC-General); and Predicted structure composition included Local structure-sequence triplet element (Triplet), PseSSC Pseudo-structure status, and PseDPC. A total of 520-dimensional features were obtained through these 15 modes(Fig. 3).
Fig. 3.

Feature expression method and feature dimension histogram.
2.2.1. Ribonucleic acid composition
For the nucleic acid composition feature of RNA sequences, three features, including Basic k-mer, Mismatch, and Subsequence, were used, of which k-mer [42] is the simplest method for expressing RNA. Basic k-mer can then be used to calculate the frequency of occurrence of k adjacent nucleic acids. Mismatch [43], [44] can calculate the frequency of k-length adjacent nucleic acids, which differ by up to m mismatches (m < k); Subsequence is a method that allows discontinuous matching [45].
Suppose an RNA sequence R is as follows:
| (1) |
wherein R1 represents the first nucleic acid in R, R2 represents the second nucleic acid in R, and so on.
Then, the sequence feature vector obtained by using Mismatch is:
| (2) |
where represents the number of occurrences of the j-th k-mer type in RNA sequence R,.
Lastly, the sequence feature vector obtained by using Subsequence is:
| (3) |
When aj is exact matching, ; when aj is non-contiguous matching, . Here, represents the attenuation coefficient, ,.
2.2.2. Autocorrelation
For the autocorrelation feature of RNA sequences, Dinucleotide-based auto-covariance (DAC) [46], Dinucleotide-based cross-covariance (DCC), Dinucleotide-based auto-cross-covariance (DACC) [47], Moran autocorrelation (MAC), Geary autocorrelation (GAC), and Normalized Moreau–Broto autocorrelation (NMBAC) are used [48]. DAC measures the correlation of the same physicochemical index between two dinucleotides along the sequence interval l. DCC measures the correlation of two different physicochemical indicators between two dinucleotides separated by l nucleic acids. MAC measures the correlation of the same property between two residues along with sequence interval l. GAC measures the correlation of the same properties between two residues of distance l. NMBAC measures the correlation of the same properties between two residues at a distance l.
2.2.3. Pseudo-ribonucleic acid composition
For the Pseudo-ribonucleic acid composition feature of RNA sequences, PC-PseDNC-General and SC-PseDNC-General are used. In the PC-PNC-General method, the user can select not only 22 built-in physical and chemical indicators, but also upload their own indicators to generate PC-PNC-General feature vectors. SC-PseDNC-General is a variant of PC-PseDNC-General [49].
2.2.4. Predicted structure composition
For the Predicted structure composition feature of RNA sequences, Triplet, Pseudo-structure status composition (PseSSC), and Pseudo-distance structure status pair composition (PseDPC) are used. Triplet (24) is an early method based on RNA sequence-structure information, and it shows better circRNA identification performance than other sequence-based methods [50].
2.3. Extreme machine learning
To identify circRNAs, the ELM is used here as the basic classification algorithm. It has also been applied to biometric identification [40], [51], [52], [53], [54], [55].
ELM is a generalized single hidden layer feedforward network. This algorithm randomly assigns input weights and hidden layer thresholds and directly calculates output layer weights by least squares. The entire learning process is completed once, and no iteration is required. Therefore, the algorithm learns rapidly.
2.4. Particle swarm optimization algorithm
Particle swarm optimization (PSO) is often used to neural network optimization due to its simple rules, rapid convergence, less parameter adjustability, and strong ability for search [56], [57]. PSO is based on the behavior of group foraging, where the particles are the solution to be optimized [58]. PSO is used to optimize the input weight and hidden layer deviation of ELMs, which can improve the generalizability of the methods. The particle swarm ELM algorithm relies on fewer hidden layer nodes to achieve higher precision.
2.5. ELM of PSO optimization
The kernel function of the extreme learning machine has a significant influence on the performance of the algorithm. The kernel parameter σ and penalty coefficient C in the kernel function have an important impact on the performance of the ELM. σ affects the scope of the kernel function, and C affects the stability of the model. The paper used PSO to optimize the parameters σ and C. The search space of the PSO corresponds to the parameters of the ELM. The position of the particles represents the parameter value, and the accuracy is used as the fitness function of PSO. The steps of the PSO optimization ELM are as follows.
-
(1)
Initialization. The number of iterations and the overall size were set to 50 and 50, respectively. A particle population was randomly generated. Each particle in the population consists of a set of σ and C.
-
(2)
Calculation of fitness function value. The fitness function value of the PSO algorithm is the accuracy of the ELM.
-
(3)
Updating the best position and particle position of the population, as well as the speed and position of the particles according to the formula (4), (5) [58].
| (4) |
| (5) |
where and are the position and velocity of the t-th iteration of particle, respectively; is the optimal solution for particle I; is the optimal solution for the population; is the weight; c1 and c2 are the acceleration factors; R1 and R2 are random numbers between 0 and 1; t is the number of iterations.
-
(4)
Checking termination conditions. If the maximum fitness value or the maximum number of iterations is reached, go to step (5). Otherwise, return to step (2).
-
(5)
Obtaining the optimal ELM parameter with the largest fitness value. In this way, the parameters of the ELM model can be obtained, and the circRNA is identified using the optimized ELM.
2.6. Performance measurement
To evaluate the identification effect of the constructed classifier, four commonly used indicators were selected here: SE (Sensitivity), SP (Specifity), ACC (Accuracy), and MCC (Matthews Correlation Coefficient). SE indicates the rate of correct prediction of positive sequences. SP indicates the rate of correct prediction of the counterexample. ACC indicates the correct rate of classification. MCC indicates the reliability of the classifier, which can reflect the prediction ability more fairly. A larger MCC reflects better reliability.
| (6) |
| (7) |
| (8) |
| (9) |
Here, TP indicates the number of circRNAs predicted correctly; FP indicates the number of non-circRNAs predicted correctly; TN indicates the number of incorrectly predicted circRNAs; FN indicates the number of incorrectly predicted non-circRNAs.
3. Results
3.1. Feature importance analysis
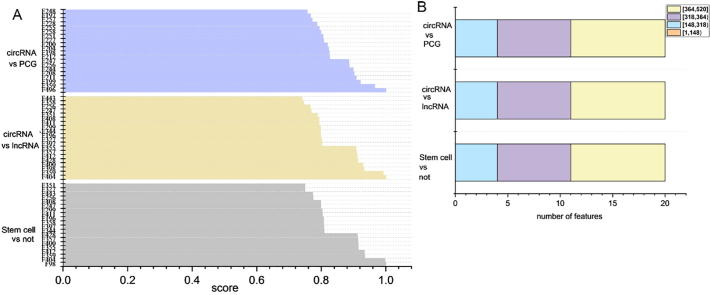
Taking into account the efficiency of the classifier operation, MRMD (Maximum-Relevance-Maximum-Distance) was used for feature selection and the output of feature score sorting results [59]. We analyzed the top 20 features based on the output feature scores (Fig. 4(A)), and get the 20-dimensional feature set obtained by feature extraction. On the circRNA vs PCG dataset, the first dimension is F496, and the 17-dimensional distribution is around F200. On the CircRNA vs IncRNA dataset, the first dimension feature is F404, and most are located between F200 and F300-F450. For the last dataset, the first dimension is characterized by F98 and F404 is the second dimension. Most of the feature distributions are the same as the second one. Moreover, we can get the common points of the 20-dimensional feature distribution on the three data sets. That is, most of the features are around F200 and F356. Finally, we analyzed which features are important for the identification of circRNA based on feature extraction.
Fig. 4.
Feature importance analysis. A) Top 20-dimensional feature distribution on three data sets. B) Feature Importance Analysis: On the three data sets, 520-dimensional features were obtained by feature selection. The 520-dimensional feature distribution was organized.
To analyse the distribution of local features, we divided the 520 features into four intervals ([1,148), [148,318), [318,364), and [364,520]) according to the order and results of using the feature expression method. The distribution of the top 20 features in each interval is given in Fig. 4(B). According to the top 20 features of the score and the interval map of the distribution, four falls into the interval [148,318), seven into [318,364), and nine into [364,520], in which the scores in the intervals [318,364) and [364,520] were also ranked higher. This shows that the structural features of RNA and Pseudo-ribonucleic acid composition are most important, and the proportions of Autocorrelation and Ribonucleic acid composition are relatively less important. Therefore, we concluded that the structural features and Pseudo-ribonucleic acid composition contain more feature information than the other two feature regions.
3.2. Optimization of kernel function of ELM
For ELM with excellent classification performance, it is important to choose an appropriate and stable hidden layer activation function. This section presents a discussion of the different classification performances of different activation functions and how to find the optimal activation function. For optimization of the kernel function, five common activation functions are selected here: sigmoid, sine, hardlim, tribas, and radial basis function (RBF) [60].
Five activation functions were used to the three datasets, respectively. The classification results are evaluated based on four indicators: ACC, SE, SP, and MCC. The recognition results were obtained under tenfold cross-validation (Fig. 5(A)). We found that the values of MCC, SP, and SE of RBF are not the best for the dataset of circRNAs expressed and not expressed in stem cells. However, the effect is better than the other four functions for the other two datasets. The results for the three datasets are as follows: accuracy of 0.784, 0.794, and 0.749; SE of 0.762, 0.679, and 0.749; and SP of 0.75, 0.839, and 0.739, respectively. In general, the RBF function achieves the best recognition of the overall effect and proves the validity of RBF for recognizing circRNA. Therefore, RBF was selected as the activation function of ELM.
Fig. 5.
Classifier validity verification. A) Identification results of five activation functions under tenfold cross-validation. B) Validation of the kernel function on the independent test set. C) Optimization of the experimental results of the extreme learning machine. D) Results of comparison with other classifiers under ten-fold cross-validation. E) Validation of classifiers on independent test sets. F) Comparison of identification results compared with traditional blast sequences.
To further prove the stability of the classification effect, the activation function was verified on an independent test set. The results are shown in Fig. 5(B). According to the trend of the fold line, the RBF function is at a higher position for the three datasets in terms of accuracy, and the accuracy of the independent test set is higher than the ten-fold cross-validation effect. This also proves that the recognition effect of RBF has certain stability and effectiveness. Therefore, in the following experiments, RBF is used as an activation function of the hidden layer.
3.3. Optimization of ELM using PSO algorithm
The traditional ELM lacks effective training methods, and it has the disadvantage of poor prediction accuracy [61]. In contrast, PSO, as an intelligent optimization algorithm, can improve the performance of ELM. Here, the PSO algorithm was used to optimize ELM and it was compared with basic ELM results and the optimization effect of genetic algorithm [62]. The results of experiments of tenfold cross-validation show that, compared with the recognition results of the optimized ELM and the basic ELM algorithm, GA-ELM and CirRNAPL improved to some extent (Fig. 5(C)). In terms of the classification effect, ELM and CirRNAPL achieved better results than GA-ELM. On the three datasets, CirRNAPL achieved accurate ACC values of 0.815, 0.822, and 0.782. Experiments showed that the PSO effectively improved the prediction accuracy and generalizability of the ELM network. Therefore, the improved ELM was used as a classification algorithm to identify circRNA.
3.4. Comparison with other classifiers
To verify the validity of the CirRNAPL, the classifier was compared to other classification algorithms. Based on Section 3.2 and Section 3.3, RBF is the selected activation function and the ELM was improved using the PSO algorithm. Next, the classification effect of the improved ELM algorithm was compared with those commonly used algorithms, such as CNN (Convolutional Neural Networks) [63], RF, support vector machine (SVM), J48 [64], and ZeroR [65]. The comparison results are shown in Fig. 5(D).
As shown in Fig. 5(D), from the comparison of the results of ACC, SE, SP, and MCC, compared with the CNN, RF, SVM, J48, and ZeroR algorithms, the classifier CirRNAPL constructed here achieved good results. For the three datasets, CirRNAPL achieves recognition accuracy of 0.815, 0.822, and 0.782, and also demonstrates the effectiveness of PSO-ELM for identifying circRNA.
After verifying the classifier CirRNAPL under ten fold cross-validation, to further explain the effect, CirRNAPL was further validated here on an independent test set. The results of the independent test set verification are shown in Fig. 5(E). According to the accuracy values of the five algorithms on the three datasets, the classifier CirRNAPL constructed in this study achieved a high accuracy rate. The accuracy rates for the three datasets are 0.887, 0.906, and 0.841, and the accuracy of the independent test set is higher than the accuracy of the tenfold cross-validation. The findings also prove that the classification accuracy of the tenfold cross-validated classifier is reliable and stable.
3.5. Comparison of the results with the traditional blast method
This section mainly describes a comparison of the recognition effects of the constructed classifier CirRNAPL and the traditional blast sequence alignment. In biological research, if a previously unknown gene is found, then a standard method, such as the base alignment search tool blast, is applied to obtain useful information through comparative analysis. With the development of machine learning algorithms and information technology, an increasing abundance of methods is becoming available for applying machine learning algorithms to identify unknown genes. To combine the effect of the traditional blast and the identification effect of the machine learning method, we compared the identification effect between CirRNAPL and blast. When performing blast, all sequence files are database files queried by blast, and each sequence is used as a query file in turn. Each sequence can get a blast result file, and the results are arranged in descending order of identity/length. Then, the prediction category of the query sequence is the category corresponding to the sequence with the largest identity/length value. The final result is shown in Fig. 5(F). We found that the classification accuracy of blast is clearly lower than that of CirRNAPL. The recognition accuracy of blast is 0.439, 0.605, and 0.611, while the classification accuracy of CirRNAPL is 0.815, 0.802, and 0.782, respectively. Given that BLAST only compares certain keywords that are more or less important in the sequence, it is not surprising that the accuracy is slightly lower. Thus, there is no doubt that the sequence data-based CirRNAPL classification methods will have increasingly broad validity and usability in research.
3.6. Comparison with state-of-the-art methods
To test the effectiveness of the identification of the proposed classifier CirRNAPL, we compared its identification performance with that of existing classifiers: WebCircRNA, PredcircRNA, H-ELM, circDeep (Fig. 6(A)). Among these models, the literature [31] only used SE and SP indicators among the four commonly used evaluation indicators. PredcircRNA, H-ELM, and circDeep only used the circRNAA vs lncRNA dataset.
Fig. 6.
A) Comparison with state-of-the-art methods. B) Analysis of the importance of violin diagram features.
Firstly, we compared the results of CirRNAPL with WebCircRNA. From Fig. 6(A), we found that cirRNAPL achieved better performance on the “stem cell vs not” and “circRNA vs PCG” datasets than WebCircRNA, and in contrast, the performance on “circRNA vs lncRNA” was slightly worse. We analyzed the causes of the performance on the “circRNA vs lncRNA” data set from the perspective of features. According to the score generated after feature selection, the paper selected the two-dimensional features that effect is better in the results of each feature expression method and used violin plots to show the data distribution on the positive and negative example data sets (Fig. 6(B)). From the Fig. 6(B), we can see that the difference of feature distribution is relatively small between positive and negative examples, showing that the feature representation method we used cannot represent the characteristics of circRNA and lncRNA well. This result indicates that the feature representation method, which can make a significant difference in the distribution of positive and negative samples, would greatly contribute to the final classification result. It brings new challenges on how to use more efficient features and improve the performance of the classifier.
Secondly, we compared the results of CirRNAPL with PredcircRNA, H-ELM, and circDeep, respectively. From Fig. 6(A), we can know that the performance of cirRNAPL is better than PredcircRNA and H-ELM in the three indicators of ACC, SE, and MCC, while lower than the latest circDeep. The ACC value of the circDeep method is higher than cirRNAPL. By comparison, we can know that although cirRNAPL has achieved some progress, more advanced techniques are required for better identification results in future research.
3.7. Blast sequence alignment expression analysis based on RNA-seq data
After the prediction of circular RNA, we further consider whether it is possible to analyze the expression of circular RNA based on the available dataset. We explore the relationship between the level of sequence expression and the abundance of the sequence (the number of reads on the alignment). So, we downloaded the RNA-seq data from the GEO database (https://www.ncbi.nlm.nih.gov/geo), which is used for similarity detection. The independent test set of the third dataset in the paper was used as a query sequence and included 282 positive samples that were expressed as H1hsec in the stem cell and 282 negative samples that were not in the stem cell and were not expressed as H1hesc.
Similarity sequence alignment was then performed by local blast, with different E values, including 1.0, 0.1, 0.01, 1e−3, and 1e−4, respectively. For the two groups of 282 query sequences, we recorded the values of different sequence alignments under different E-values (Fig. 7(A)). When the E-value was at least 1e−3, the numbers of sequence alignments on the two datasets were 64 and 69. In the case of the stem and no stem, the changing trend of e value and expression level is the same. It can be seen that the influence. Then we calculated the abundance of each circRNA and then obtained a scatter plot of the relationship between expression and abundance (Fig. 7(B)). The scatter plot implicates that the abundance of the sequence is high when the expression level of the sequence is high. Then, the polynomial fitting of the obtained data points was carried out, and two fitting curves were obtained. According to the trend analysis of the fitted curve of Fig. 7(B), the relationship between the expression level of circRNA and the sequence abundance is the same in both cases: the expression level increases with the increase of abundance. The correlation coefficients in the two cases were 0.840 and 0.899, respectively, and the correlation of not stem was higher. We can also see that there are more sequence abundances on the not stem data. When the expression reaches a certain level, it does not change even though the sequence abundance increases. It shows that the expression level and abundance of the sequence are positively correlated.
Fig. 7.
A) Relationship between sequence alignment and E value under stem and non-stem. B) Analysis of the relationship between the expression level and abundance of the sequence. C) Sequence alignment partial conservative region display and Consensus log.
After analyzing the expression level, we randomly selected 8 sequences for multiple sequence alignment. The multi-sequence alignment of the circRNA was obtained by the Clustal Omega online alignment tool, and some of the conserved regions were displayed (Fig. 7(C)). The results showed there are different colorations of nucleic acids in conserved regions while the consensus logo in the conservative region [66]. The nucleic acid distribution characteristics of the conserved regions of the circRNA were revealed by the displayed logo map.
4. Conclusion
We constructed a new classifier for the accuracy of identification of circRNA, and the effectiveness of the identification of circRNA has been demonstrated based on three publicly available datasets. Through the analysis of feature importance, we found that the structural features and Pseudo-ribonucleic acid composition feature showed better performance. However, the performance to distinguish circRNA and lncRNA is not very good, and it is still a widespread problem to distinguish them. More effective features and data are urgently required. Additionally, sequence alignments of circRNAs were preferably analyzed based on RNA-seq data. The comparison with the circDeep method also prompted us to use more efficient and updated techniques to improve the performance of our model.
Planned future work includes not only finding more useful features but also using advanced parallel technology to identify the growing number of circRNA sequences. Both of them will improve the efficiency of identification. Meanwhile, given that the functions of most circRNAs are still unknown, and little work has been performed on the large-scale discovery of disease-related circRNAs, future studies should focus on the biological significance of circRNAs.
5. Data accession numbers
The RNA-seq data is from the GEO database (https://www.ncbi.nlm.nih.gov/geo). The GEO number is GSE63823.
CRediT authorship contribution statement
Mengting Niu: Writing - original draft, Methodology. Jun Zhang: Conceptualization, Software. Yanjuan Li: Project administration. Cankun Wang: Formal analysis, Validation. Zhaoqian Liu: Writing - review & editing. Hui Ding: Data curation. Quan Zou: Funding acquisition. Qin Ma: Supervision.
Acknowledgments
Acknowledgements
We thank all members in the Zou lab for helping in paper writing and server support. We also thank Marlena Merling for language help.
Funding
The work was supported by the National Key R&D Program of China, China (2018YFC0910405), and the Natural Science Foundation of China, China (No. 61771331, 61300098). The work was supported by the Fundamental Research Funds for the Central Universities, China (No. 2572017CB33).
Contributor Information
Quan Zou, Email: zouquan@nclab.net.
Qin Ma, Email: qin.ma@osumc.edu.
References
- 1.Hao S., Lv J., Yang Q., Wang A., Li Z., Guo Y. Identification of key genes and circular RNAs in human gastric cancer. J Med Sci Monitor. 2019;25:2488–2504. doi: 10.12659/MSM.915382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogard B., Francastel C., Hubé F. A new method for the identification of thousands of circular RNAs. J Non-coding RNA Investigation. 2018:2. [Google Scholar]
- 3.Diener T. Potato spindle tuber “virus”: IV. A replicating, low molecular weight RNA. J Virol. 1971;45:411–428. doi: 10.1016/0042-6822(71)90342-4. [DOI] [PubMed] [Google Scholar]
- 4.Cheng J., Metge F., Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. J Bioinformatics. 2015;32:1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- 5.Dori M., Alieh L.H.A., Cavalli D., Massalini S., Lesche M., Dahl A. Sequence and expression levels of circular RNAs in progenitor cell types during mouse corticogenesis. J Life Sci Alliance. 2019;2 doi: 10.26508/lsa.201900354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song X., Zhang N., Han P., Moon B.-S., Lai R.K., Wang K. Circular RNA profile in gliomas revealed by identification tool UROBORUS. J Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S., Han L. Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. J Genome Med. 2019;11:15. doi: 10.1186/s13073-019-0629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. J Rna. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y.-C., Li J.-R., Sun C.-H., Andrews E., Chao R.-F., Lin F.-M. CircNet: a database of circular RNAs derived from transcriptome sequencing data. J Nucleic Acids Res. 2015;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosal S., Das S., Sen R., Basak P., Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. J Front Gen. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia S., Feng J., Lei L., Hu J., Xia L., Wang J. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. J Briefings Bioinformatics. 2016;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 12.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. J Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. J RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong X., Wu G., Zeng C. Role of circular RNAs in cardiovascular diseases. J Experimental Biol Med. 2019;244:73–82. doi: 10.1177/1535370218822988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian M., Chen R., Li T., Xiao B. Reduced expression of circ RNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32 doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao T., Chen Q., Shao Z., Song Z., Fu L., Xiao B. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. 2018;32 doi: 10.1002/jcla.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J.-T., Chen J.-N., Gong L.-P., Bi Y.-H., Liang J., Zhou L. Identification of virus-encoded circular RNA. J Virol. 2019;529:144–151. doi: 10.1016/j.virol.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B, Yang W, Tang J, RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. J Lupus, 2019. 0961203319830493. [DOI] [PubMed]
- 19.Xu Y., Ren Y., Lin T., Cui D. Identification and characterization of CircRNAs involved in the regulation of wheat root length. J Biol Res. 2019;52:19. doi: 10.1186/s40659-019-0228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye C.Y., Chen L., Liu C., Zhu Q.H., Fan L. Widespread noncoding circular RNA s in plants. J New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 21.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. J Nature. 2013;495:333. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 22.Hansen T.B. Improved circRNA identification by combining prediction algorithms. J Front Cell Devel Biol. 2018;6:20. doi: 10.3389/fcell.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Yu Y., Zhang X., Liu C., Ye C., Fan L. PcircRNA_finder: a software for circRNA prediction in plants. J Bioinformatics. 2016;32:3528–3529. doi: 10.1093/bioinformatics/btw496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danan M., Schwartz S., Edelheit S., Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. J Nucleic Acids Res. 2011;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. J Nature biotechnology. 2014;32:453. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. J Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X.-O., Wang H.-B., Zhang Y., Lu X., Chen L.-L., Yang L. Complementary sequence-mediated exon circularization. J Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y. The landscape of circular RNA in cancer. J Cell. 2019;176(869–881) doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You X., Conrad T.O. Acfs: accurate circRNA identification and quantification from RNA-Seq data. J Sci Rep. 2016;6:38820. doi: 10.1038/srep38820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q.-L., Ji X.-Y., Li H.-W., Guo J., Wang F., Deng X.-Y. Identification of circular RNAs and their altered expression under poly (I: C) challenge in key antiviral immune pathways in amphioxus. J Fish Shellfish Immunol. 2019;86:1053–1057. doi: 10.1016/j.fsi.2018.12.061. [DOI] [PubMed] [Google Scholar]
- 31.Pan X., Xiong K., Anthon C., Hyttel P., Freude K., Jensen L. WebCircRNA: Classifying the circular RNA potential of coding and noncoding RNA. J Genes. 2018;9:536. doi: 10.3390/genes9110536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan X., Xiong K. PredcircRNA: computational classification of circular RNA from other long non-coding RNA using hybrid features. J Molecular Biosyst. 2015;11:2219–2226. doi: 10.1039/c5mb00214a. [DOI] [PubMed] [Google Scholar]
- 33.Chen L., Zhang Y.-H., Huang G., Pan X., Wang S., Huang T. Discriminating cirRNAs from other lncRNAs using a hierarchical extreme learning machine (H-ELM) algorithm with feature selection. J Molecular Gen Genomics. 2018;293:137–149. doi: 10.1007/s00438-017-1372-7. [DOI] [PubMed] [Google Scholar]
- 34.Chaabane M., Williams R.M., Stephens A.T., Park J.W. circDeep: deep learning approach for circular RNA classification from other long non-coding RNA. J Bioinformatics. 2020;36:73–80. doi: 10.1093/bioinformatics/btz537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei L., Liao M., Gao X., Zou Q. An improved protein structural classes prediction method by incorporating both sequence and structure information. J IEEE Trans Banobiosci. 2015;14:339–349. doi: 10.1109/TNB.2014.2352454. [DOI] [PubMed] [Google Scholar]
- 36.Wei L., Xing P., Tang J., Zou Q. PhosPred-RF: a novel sequence-based predictor for phosphorylation sites using sequential information only. J IEEE Trans Nanobiosci. 2017;16:240–247. doi: 10.1109/TNB.2017.2661756. [DOI] [PubMed] [Google Scholar]
- 37.Niu M., Li Y., Wang C., Han K. RFAmyloid: a web server for predicting amyloid proteins. J Int J Molecular Sci. 2018;19:2071. doi: 10.3390/ijms19072071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L., Zhang J., Xuan P., Zou Q. BP neural network could help improve pre-miRNA identification in various species. J BioMed Res Int. 2016;2016 doi: 10.1155/2016/9565689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong R., Ma X.-K., Li G.-W., Yang L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. J Genomics, Proteomics Bioinform. 2018;16:226–233. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji P., Wu W., Chen S., Zheng Y., Zhou L., Zhang J. Expanded expression landscape and prioritization of circular RNAs in mammals. J Cell Rep. 2019;26(3444–3460) doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 41.Liu B., Liu F., Wang X., Chen J., Fang L., Chou K.-C. Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. J Nucleic Acids Res. 2015;43:W65–W71. doi: 10.1093/nar/gkv458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei L., Liao M., Gao Y., Ji R., He Z., Zou Q. Improved and promising identification of human microRNAs by incorporating a high-quality negative set. J IEEE/ACM Trans Comput Biol Bioinform (TCBB) 2014;11:192–201. doi: 10.1109/TCBB.2013.146. [DOI] [PubMed] [Google Scholar]
- 43.Luo L., Li D., Zhang W., Tu S., Zhu X., Tian G. Accurate prediction of transposon-derived piRNAs by integrating various sequential and physicochemical features. J PloS One. 2016;11 doi: 10.1371/journal.pone.0153268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Niu Y. Predicting flexible length linear b-cell epitopes using pairwise sequence similarity. In: 2010 3rd International Conference on Biomedical Engineering and Informatics, Volume 6. IEEE; 2010. p. 2338–42.
- 45.Lodhi H., Saunders C., Shawe-Taylor J., Cristianini N., Watkins C. Text classification using string kernels. J Machine Learning Res. 2002;2:419–444. [Google Scholar]
- 46.Dong Q., Zhou S., Guan J. A new taxonomy-based protein fold recognition approach based on autocross-covariance transformation. J Bioinformatics. 2009;25:2655–2662. doi: 10.1093/bioinformatics/btp500. [DOI] [PubMed] [Google Scholar]
- 47.Friedel M., Nikolajewa S., Sühnel J., Wilhelm T. DiProDB: a database for dinucleotide properties. J Nucleic Acids Res. 2008;37:D37–D40. doi: 10.1093/nar/gkn597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W., Zhang X., Brooker J., Lin H., Zhang L., Chou K.-C. PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. J Bioinformatics. 2014;31:119–120. doi: 10.1093/bioinformatics/btu602. [DOI] [PubMed] [Google Scholar]
- 49.Chen W., Feng P.-M., Lin H., Chou K.-C. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. J Nucleic Acids Res. 2013;41:e68. doi: 10.1093/nar/gks1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B., Fang L., Liu F., Wang X., Chen J., Chou K.-C. Identification of real microRNA precursors with a pseudo structure status composition approach. J PloS One. 2015;10 doi: 10.1371/journal.pone.0121501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You Z.H., Lei Y.K., Zhu L., Xia J., Wang B. Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. J Bmc Bioinformatics. 2013;14:S10. doi: 10.1186/1471-2105-14-S8-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao J., Lin Z., Huang G.B., Liu N. Voting based extreme learning machine. J Inform Sci. 2012;185:66–77. [Google Scholar]
- 53.Cao J., Xiong L. Protein sequence classification with improved extreme learning machine algorithms. J Biomed Res Int. 2014;2014 doi: 10.1155/2014/103054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D, Huang GB. Protein sequence classification using extreme learning machine. In: IEEE International Joint Conference on Neural Networks, 2005. IJCNN '05. Proceedings. vol. 1403; 2005. p. 1406–11.
- 55.Huang G.B., Zhu Q.Y., Siew C.K. Extreme learning machine: theory and applications. J Neurocomputing. 2006;70:489–501. [Google Scholar]
- 56.Pham D., Karaboga D. Springer Science & Business Media; 2012. Intelligent optimisation techniques: genetic algorithms, tabu search, simulated annealing and neural networks. [Google Scholar]
- 57.Li Y., Niu M., Guo J. An inductive logic programming algorithm based on artificial bee colony. J Inform Technol Res (JITR) 2019;12:89–104. [Google Scholar]
- 58.Bai Q. Analysis of particle swarm optimization algorithm. J Comp Inform Sci. 2010;3:180. [Google Scholar]
- 59.Zou Q., Zeng J., Cao L., Ji R. A novel features ranking metric with application to scalable visual and bioinformatics data classification. J Neurocomputing. 2016;173:346–354. [Google Scholar]
- 60.Javed K., Gouriveau R., Zerhouni N. SW-ELM: a summation wavelet extreme learning machine algorithm with a priori parameter initialization. J Neurocomputing. 2014;123:299–307. [Google Scholar]
- 61.Wang Y., Cao F., Yuan Y. A study on effectiveness of extreme learning machine. J Neurocomputing. 2011;74:2483–2490. [Google Scholar]
- 62.Deb K., Pratap A., Agarwal S., Meyarivan T. A fast and elitist multiobjective genetic algorithm: NSGA-II. J IEEE Trans Evolution Comput. 2002;6:182–197. [Google Scholar]
- 63.Fiannaca A., La Rosa M., La Paglia L., Rizzo R., Urso A. nRC: non-coding RNA Classifier based on structural features. J BioData Mining. 2017;10:27. doi: 10.1186/s13040-017-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patil T.R., Sherekar S. Performance analysis of Naive Bayes and J48 classification algorithm for data classification. J Int J Comp Sci Appl. 2013;6:256–261. [Google Scholar]
- 65.Aher S.B., Lobo L. Comparative study of classification algorithms. J Int J Inform Technol. 2012;5:239–243. [Google Scholar]
- 66.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. J Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]