Summary
The ongoing pandemic of COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), underscores the urgency to develop experimental systems for studying this virus and identifying countermeasures. We report a reverse genetic system for SARS-CoV-2. Seven complimentary DNA (cDNA) fragments spanning the SARS-CoV-2 genome were assembled into a full-genome cDNA. RNA transcribed from the full-genome cDNA was highly infectious after electroporation into cells, producing 2.9 × 106 plaque-forming unit (PFU)/mL of virus. Compared with a clinical isolate, the infectious-clone-derived SARS-CoV-2 (icSARS-CoV-2) exhibited similar plaque morphology, viral RNA profile, and replication kinetics. Additionally, icSARS-CoV-2 retained engineered molecular markers and did not acquire other mutations. We generated a stable mNeonGreen SARS-CoV-2 (icSARS-CoV-2-mNG) by introducing this reporter gene into ORF7 of the viral genome. icSARS-CoV-2-mNG was successfully used to evaluate the antiviral activities of interferon (IFN). Collectively, the reverse genetic system and reporter virus provide key reagents to study SARS-CoV-2 and develop countermeasures.
Keywords: coronavirus, SARS-CoV-2, COVID-19, SARS-CoV, vaccine, antiviral
Graphical Abstract

Highlights
-
•
A reverse genetic system has been established for SARS-CoV-2
-
•
Recombinant SARS-CoV-2 replicates as efficiently as the original clinical isolate
-
•
A stable mNeonGreen reporter SARS-CoV-2 has been developed
-
•
The mNeonGreen SARS-CoV-2 can be used to screen antiviral inhibitors
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a devastating global pandemic. Xie et al. generated an infectious cDNA clone of SARS-CoV-2 and a mNeonGreen reporter virus. Recombinant SARS-CoV-2 and reporter virus replicate as efficiently as the original clinical isolate.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in early 2020 with human cases in Wuhan, China (Zhou et al., 2020, Zhu et al., 2020). It has rapidly rampaged worldwide, causing a pandemic of coronavirus disease (COVID-19) that ranges from fever and breathing difficulty to acute respiratory distress and death (Huang et al., 2020, Zhu et al., 2020). With over 300,000 people infected in less than 3 months, SARS-CoV-2 causes the most severe disease in older patients and people with co-morbidities, including heart disease, diabetes, and other health conditions (Wu and McGoogan, 2020). Before 2019, six α- and β-coronaviruses were known to cause respiratory diseases of different severity, including four common cold coronaviruses (229E, NL63, OC43, and HKU1) and two highly pathogenic coronaviruses (severe acute respiratory syndrome [SARS-CoV] and Middle East respiratory syndrome [MERS-CoV], which emerged in 2003 and since 2012, respectively) (Assiri et al., 2013, Huang et al., 2020). Importantly, with massive hospitalization rates and high mortality, SARS-CoV-2 remains a major threat to humankind and intervention strategies are being rapidly pursued.
A key tool in responding to emergent viruses is the generation of reverse genetic systems to explore and characterize new pathogens. Classically, reverse genetic systems for coronaviruses have been complicated by their large genome size (~30,000 nucleotides) and the existence of bacteriotoxic elements in their genome that make them difficult to propagate (Almazán et al., 2014). Several approaches have been devised to overcome this barrier, such as multiple plasmid systems to disrupt toxic elements and to reduce deletions and mutations (Yount et al., 2002). Using this approach, researchers have developed infectious clones for several coronaviruses, including SARS-CoV, MERS-CoV, and others (Menachery et al., 2015, Menachery et al., 2016, Scobey et al., 2013, Yount et al., 2003). Thao et al., (2020) recently reported a yeast-based synthetic genomics platform for rapid construction of infectious clones for murine hepatitis coronavirus (MHV-CoV), MERS-CoV, and SARS-CoV-2. However, the yeast-platform-produced SARS-CoV-2 has not been fully characterized for its biological properties (e.g., replication kinetics) in comparison with its original clinical isolate. Such characterization is essential for ensuring the quality of the genetic system to rescue recombinant viruses that recapitulate the biological features of their corresponding clinical isolates. Once validated, the reverse genetic systems allow rapid characterization of novel viruses, development of reporter viruses, and generation of live-attenuated vaccine candidates to respond to emerging infections. Together with animal pathogenesis models, reverse genetic systems offer powerful tools needed to characterize, understand, and respond to emerging virus outbreaks.
In response to the ongoing pandemic of SARS-CoV-2, we have developed a robust reverse genetic system for SARS-CoV-2 and a mNeonGreen reporter virus. Recombinant virus derived from the system recapitulates the replication kinetics of the original clinical isolates. In addition, the mNeonGreen reporter remains stable for at least five passages, allowing its use in long-term studies. Using type I interferon (IFN), we demonstrated that the mNeonGreen virus could be reliably used to study viral replication and pathogenesis as well as to develop vaccines and antiviral drugs.
Results
Design of a SARS-CoV-2 Full-Length cDNA
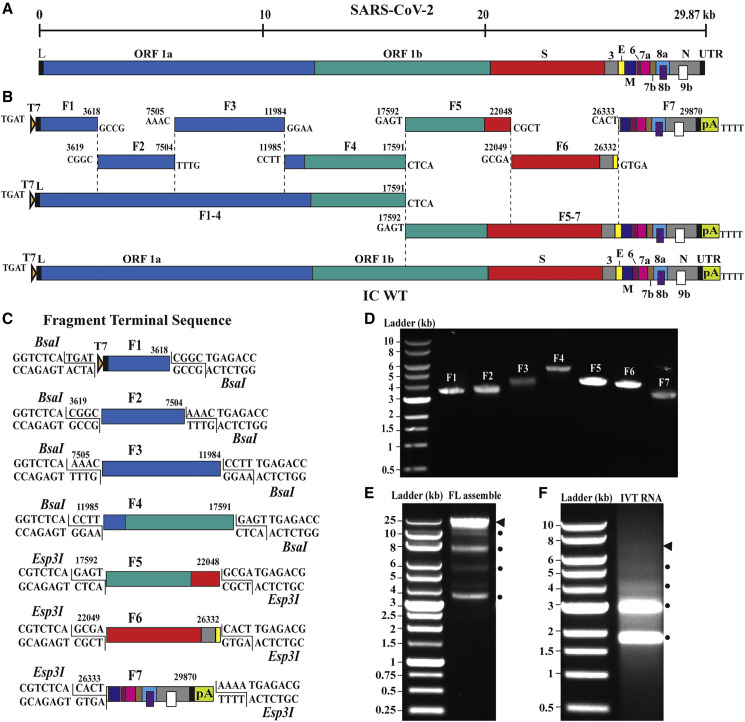
We designed an in vitro ligation approach, similar to that used for constructing the infectious clones of SARS-CoV and MERS-CoV (Scobey et al., 2013, Yount et al., 2003), to directionally assemble the full-length complimentary DNA (cDNA) of the SARS-CoV-2 genome (Figure 1 A). Our reverse genetic system was based on the virus strain (2019-nCoV/USA_WA1/2020) isolated from the first reported SARS-CoV-2 case in the US (Harcourt et al., 2020, Holshue et al., 2020). Viral RNA, extracted from the passage 4 virus from Vero E6 cells, was used as a template for RT-PCR to produce cDNA fragments. Seven contiguous cDNA fragments were constructed to cover the entire viral genome (Figure 1B). Some of the seven cDNA fragments were prepared through RT-PCR, whereas others were generated by chemical synthesis (see Method Details). All cDNA fragments were individually cloned into plasmid vectors. For facilitating directional assembly of genome-length cDNA, each cDNA fragment was flanked by a class IIS restriction endonuclease site (BsaI or Esp3I). The class IIS endonucleases recognize asymmetric DNA sequences, cleave outside their recognition sequences, and generate unique cohesive overhangs (Figure 1C). After digestion with BsaI or Esp3I, the seven fragments were directionally ligated to assemble the genome-length cDNA. The unique cohesive ends of each fragment ensured one directional, seamless assembly of the seven fragments with the concomitant loss of the restriction enzyme sites. Figure 1C depicts the details of the seven fragments: F1 (T7 promoter sequence plus nucleotides 1–3,618), F2 (nucleotides 3,619–7,504), F3 (nucleotides 7,505–11,984), F4 (nucleotides 11,985–17,591), F5 (nucleotides 17,592–22,048), F6 (nucleotides 22,049–26,332), and F7 (nucleotides 26,333–29,870 plus a poly(A)29 sequence). We engineered a T7 promoter and a poly(A)29 tail at the upstream end of F1 and the downstream end of F7, respectively. In vitro transcription of the ligated F1–F7 DNA was expected to produce a 5′ capped (because cap analog was included in the in vitro transcription reaction) and 3′ polyadenylated genome-length RNA. To differentiate the infectious clone-derived virus from the parental clinical isolate, we engineered three synonymous nucleotide mutations as markers.
Figure 1.
Assembly of a Full-Length SARS-CoV-2 Infection cDNA Clone
(A) Genome structure SARS-CoV-2. The open reading frames (ORFs) from the full genome are indicated.
(B) Strategy for in vitro assembly of an infectious cDNA clone of SARS-CoV-2. The nucleotide sequences and genome locations of the cohesive overhangs are indicated. The WT full-length (FL) cDNA of SARS-CoV-2 (IC WT) was directionally assembled using in vitro ligation.
(C) Diagram of the terminal sequences of each cDNA fragment recognized by BsaI and Esp3I.
(D) Gel analysis of the seven purified cDNA fragments. Individual fragments (F1–F7) were digested from corresponding plasmid clones and gel purified. Seven purified cDNA fragments (50–100 ng) were analyzed on a 0.6% native agarose gel. The 1-kb DNA ladders are indicated.
(E) Gel analysis of cDNA ligation products. About 400 ng of purified ligation product was analyzed on a 0.6% native agarose gel. Triangle indicates the FL cDNA product. Circles indicate the intermediate cDNA products.
(F) Gel analysis of RNA transcripts. About 1 μg of in-vitro-transcribed (IVT) RNAs were analyzed on a 0.6% native agarose gel. DNA ladders are indicated. Because this is a native agarose gel, the DNA size is not directly corelated to the RNA size. Triangle indicates the genome-length RNA transcript. Circles show the shorter RNA transcripts.
Assembly of a SARS-CoV-2 Full-Length cDNA
We cloned each of the seven cDNA fragments into a plasmid and sequenced them to ensure no undesired mutations. For assembly of full-length cDNA, the seven cDNA plasmids were digested with BsaI or Esp3I. The resulting cDNA fragments were gel-purified (Figure 1D) then in vitro ligated to assemble the genome-length cDNA in three steps: (1) ligation of F1, F2, F3, and F4 to produce F1–4 cDNA; (2) ligation of F5, F6, and F7 to produce F5–7 cDNA; and (3) ligation of F1–4 and F5–7 to produce the full-length F1–7 cDNA. Agarose gel analysis of the ligation (3) reaction showed a major DNA product representing the size of genome-length cDNA (~29.87 kb, indicated by an arrow in Figure 1E) in addition to several smaller intermediate cDNA products (indicated by circles). In vitro transcription using the cDNA template (directly from ligation (3) without gel purification) generated multiple RNA bands, among which a faint high molecular band might represent the genome-length RNA (indicated by an arrow in Figure 1F) together with several smaller RNA transcripts (indicated by circles).
Recovery of Recombinant SARS-CoV-2
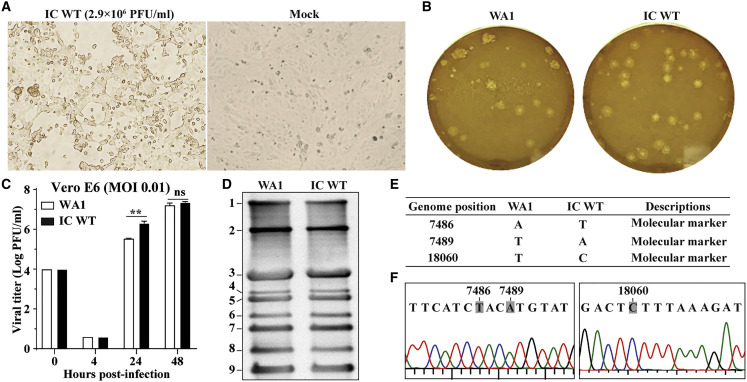
To recover recombinant SARS-CoV-2 from the infectious cDNA clone (icSARS-CoV-2), we electroporated in-vitro-transcribed genome-length RNA into Vero E6 cells. We directly electroporated the RNA transcription mixture from Figure 1F into cells without purification. Given that N protein was reported to enhance the infectivity of coronavirus RNA transcripts (Curtis et al., 2002, Yount et al., 2003, Yount et al., 2002), we co-electroporated an mRNA encoding the SARS-CoV-2 N protein with the full-length RNA. The transfected cells developed cytopathic effects (CPEs) on day 4 after transfection and produced infectious virus (denoted as passage 0 (P0) virus) with a titer of 2.9 × 106 plaque-forming units (PFU)/mL (Figure 2 A). It is worth emphasizing that such a high titer of recombinant virus was produced directly from the electroporated cells without additional rounds of cell culture passaging, indicating the robustness of the system and also suggesting a lack of any errors. Next, we compared the replication properties between the recombinant virus and the original clinical isolate. The wild-type icSARS-CoV-2 (icSARS-CoV-2-WT) developed plaques similar to the original clinical isolate (Figure 2B) and exhibited equivalent replication kinetics on Vero E6 cells (Figure 2C). We did not extend the time points of replication beyond 48 h because CPEs were observed at 40–48 h after infection. Northern blot analysis showed that viral messenger RNA (mRNA) species from the clinical-isolate-infected cells and the icSARS-CoV-2-infected cells were identical to the predicted set of genome-length RNA and eight subgenomic RNAs (Figure 2D). Full-genome sequencing showed that the recombinant virus retained the three engineered synonymous mutations with no other sequence changes, demonstrating the rescued virus did not result from contamination by the parental virus isolate (Figure 2E). Furthermore, the DNA sequencing chromatogram did not show any partial reversion of the three engineered molecular markers (Figure 2F). Collectively, the results demonstrate that (1) the in-vitro-transcribed full-length RNA is highly infectious upon electroporation into cells, and (2) the recombinant virus recapitulates the replication properties of the original clinical isolate on Vero E6 cells.
Figure 2.
Characterization of the IC WT
(A) Bright-field images of the Vero E6 cells electroporated with RNA transcripts. CPEs appeared in the IC-WT-RNA-transfected cells on day 4 after transfection. The titer of the P0 virus (directly from the transfected cells) is shown in PFUs per ml.
(B) Plaque morphology of the original clinical isolate (WA1 = 2019-nCoV/USA_WA1/2020) and the recombinant P1 IC WT virus. Plaques were developed in Vero E6 cells on day 2 after infection.
(C) Replication kinetics. Vero E6 cells were infected with the clinical isolate or recombinant P1 IC WT virus at MOI 0.01. Viruses in culture fluids were quantified by plaque assay. Results from triplicate experiments were presented with error bars indicating standard deviations.
(D) Northern blot analysis of FL and subgenomic RNAs. Numbers indicated the FL (band 1) and eight subgenomic RNAs (bands 2–9).
(E) Sequence differences between the original clinical isolate WA1 and the recombinant P1 IC WT. The three silent nucleotide changes were engineered as molecular markers.
(F) Chromatograms of Sanger sequencing results. The engineered molecular maker mutations are indicated.
Development and Characterization of mNeonGreen SARS-CoV-2
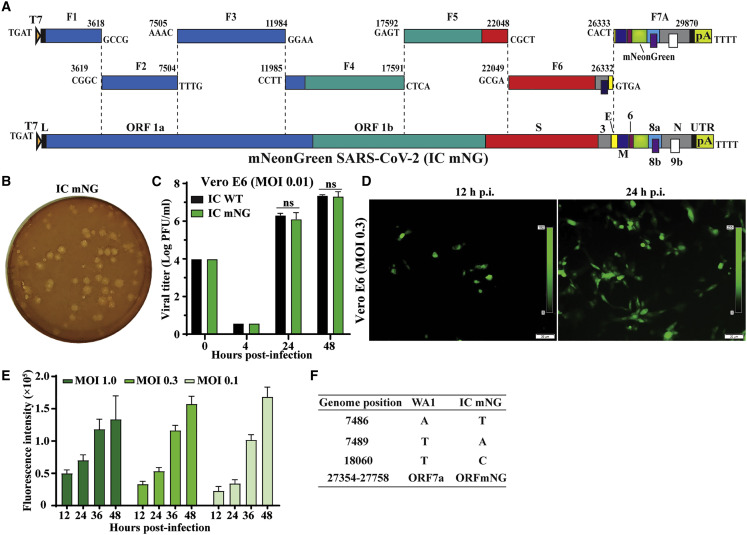
Reporter viruses are useful tools to study viral replication and pathogenesis and to develop countermeasures. To establish a reporter SARS-CoV-2 infectious clone, we engineered an mNeonGreen (mNG) gene into the ORF7 of viral genome (Figure 3 A), similar to the SARS-CoV reporter (Sims et al., 2005). The same in vitro ligation and transcription protocols (described above) were used to prepare the mNG full-length RNA. After electroporation, we recovered icSARS-CoV-2-mNG (6.9 × 106 PFU/mL). To examine whether the reporter gene attenuates viral replication, we compared the replication properties between the WT and reporter viruses on Vero E6 cells. The icSARS-CoV-2-mNG produced plaques similar to those of the icSARS-CoV-WT (compare Figure 3B with 2B). Indistinguishable replication kinetics were observed for the icSARS-CoV-2-mNG and icSARS-CoV-WT (Figure 3C). Infection with icSARS-CoV-2-mNG developed increasing numbers of mNG-positive cells over time (Figure 3D). Concurrently, the fluorescent signals increased from 12 to 48 h after infection (Figure 3E). At 12–36 h after infection, the level of fluorescent signal correlated with the initial multiplicities of infection (MOIs), whereas a reverse trend was observed at 48 h after infection, most likely due to earlier CPEs caused by the higher MOI. Full-genome sequencing showed that icSARS-CoV-2-mNG retained the three engineered markers with no additional mutations (Figure 3F). These results indicate that icSARS-CoV-2-mNG is initially stable, maintains the WT replication, and expresses robust mNG in Vero E6 cells.
Figure 3.
Generation of a mNeonGreen SARS-CoV-2
(A) Assembly of the FL mNG SARS-CoV-2 cDNA. The mNG gene was placed downstream of the regulatory sequence of ORF7 to replace the ORF7 sequence (Sims et al., 2005) in the subclone F7.
(B) Plaque morphology of the P1 IC mNG virus. Plaques were developed in Vero E6 cells on day 2 after infection.
(C) Replication kinetics. Vero E6 cells were infected with the IC WT or reporter icSARS-CoV-2-mNG (IC mNG) at MOI of 0.01. Viruses in culture medium were quantified by plaque assay.
(D) Fluorescence microscopy analysis of P1-mNG-virus-infected cells. Vero E6 cells were infected with P1 mNG viruses at MOI of 0.3. Representative mNG-positive (green) images are shown.
(E) Kinetics of fluorescence intensity. Vero E6 cells were infected with MOIs of 1.0, 0.3, or 0.1. After background signal subtraction, the fluorescence intensities from 12 to 48 h after infection are shown. Results from triplicate experiments were presented with bars representing standard deviations.
(F) Summary of full-genome sequence of mNG virus (P1 IC mNG). Nucleotides different from the original clinical isolate (WA1) are indicated.
Stability of icSARS-CoV-2-mNG
To examine the longer-term stability of icSARS-CoV-2-mNG, we serially passaged the reporter virus on Vero cells for 5 rounds (1–2 days per round). Cells infected with equal PFU of passage 1 (P1) or passage 5 (P5) viruses produced comparable numbers of mNG-positive cells (Figure 4 A). Next, RT-PCR was performed to verify the retention of mNG in the P1 and P5 viral genomes by using two primers targeting the insertion junctions (corresponding to nucleotides 25,068–28,099 of the viral genome). As expected, the RT-PCR products derived from both P1 and P5 mNG viruses were larger than those from the WT icSARS-CoV-2 (Figure 4B, lanes 1–3). Digestion of the RT-PCR products with BsrGI (located upstream of the mNG insertion site) and StuI (in the mNG gene) developed distinct cleavage patterns between the reporter and WT viruses, whereas P1 and P5 viruses produced an identical digestion pattern (Figure 4B, lanes 4–6). Finally, sequencing the P1 and P5 RT-PCR products confirmed the retention of the mNG gene (data not shown). Altogether, the results demonstrate the stability of icSARS-CoV-2-mNG after five rounds of passaging on Vero E6 cells. Given that Vero E6 cells are defective in type 1 IFN production, it remains to be tested whether the reporter virus is stable when passaged on interferon-competent cell lines.
Figure 4.
Stability and Application of mNeonGreen Virus
The stability of mNG virus was analyzed by comparing the fluorescent signals between the cells infected with P1 and P5 reporter viruses. The presence of mNG gene in the P1 and P5 reporter viruses was also verified using RT-PCR. The application of mNG virus was examined by testing the antiviral activity of IFN-α treatment.
(A) Fluorescence microscopy analysis of the P1- and P5-mNG-virus-infected cells. Vero E6 cells were infected with P1 or P5 virus at an MOI of 0.3. The cells were monitored for mNG-positive signals at 24 h after infection. Color representations are as follows: green, mNG; blue, nucleus.
(B) Gel analysis of mNG virus stability. The top graphic depicts the theoretical results of RT-PCR followed by restriction enzyme digestion. The bottom graphic shows the gel analysis of the RT-PCR products before (lanes 1–3) and after BsrGI/StuI digestion (lanes 4–6). About 100 ng DNA samples were analyzed on a 0.6% agarose gel. The DNA sizes are indicated.
(C) Schematic diagram of IFN-α treatment.
(D) Representative fluorescence images of reporter-virus-infected cells after IFN-α treatment. The doses of IFN-α treatment are indicated.
(E) Dose response curve of mNG signal inhibited by IFN-α. The Hillslope and EC50 values are indicated. Results from triplicate experiments were presented with bars representing standard deviations.
Application of icSARS-CoV-2-mNG
To explore the utility of icSARS-CoV-2-mNG, we used the reporter virus to rapidly screen the antiviral activity of a known inhibitor of coronaviruses. We previously showed that pre-treatment of Vero cells with type 1 IFN inhibits SARS-CoV-2 replication (Lokugamage et al., 2020). Here, we explored the dose-responsive effect of IFN-α pre-treatment on icSARS-CoV-mNG replication (Figure 4C). No mNG expression was visually observed when the infected cells were pre-treated with the highest dose of IFN-α (1,000 u/mL), whereas a dose-dependent reduction of mNG signal was detected at an intermediate dose (111 u/mL) (Figure 4D). Quantification of the fluorescent readouts estimated a concentration inhibiting 50% of viral replication (EC50) of 101 u/mL, confirming the inhibition of SARS-CoV-2 by IFN-α (Figure 4E). This result is consistent with previous findings that SARS-CoV-2 is sensitive to type 1 IFN inhibition. The reporter virus assay required fewer days and less labor than with the conventional plaque-reduction assay. Collectively, the results indicate that icSARS-CoV-2-mNG could be reliably used to study SARS-CoV-2 replication and to screen antiviral inhibitors.
Discussion
We report the development of a full-length infectious clone and a reporter virus for SARS-CoV-2. One of the key utilities for the reverse genetic system is to facilitate antiviral testing and therapeutic development. The icSARS-CoV-2-mNG reporter virus allows the use of fluorescence as a surrogate readout for viral replication. Compared with a standard plaque assay or median tissue culture infectious dose (TCID50) quantification, the fluorescent readout shortens the assay turnaround time by several days. In addition, the fluorescent readout offers a quantitative measure that is less labor-intensive than the traditional means of viral titer reduction. Furthermore, the mNG-virus-based assay could be automated in a high-throughput format to screen compounds against viral replication. As a proof-of-concept, we demonstrated that, after treatments with type 1 IFN, the reporter virus reliably revealed efficacy in a rapid and efficient manner. In addition, the stability of the mNG reporter virus allows it to be used for longer-term studies and in vivo without fear of losing its fluorescent marker. Thus, this reporter virus offers a huge advantage for the research community and pharmaceutical companies to develop therapeutics for COVID-19.
Our reverse genetic system represents a major reagent in the pursuit of understanding SARS-CoV-2 and COVID-19. Compared with the clinical isolate, the recombinant WT SARS-CoV-2 has no deficit in terms of viral RNA species produced, plaque morphology, or replication kinetics. Therefore, it might be used as an equivalent to the clinical strain, and mutant viruses can be generated to characterize mutational effect on viral infection. This approach has allowed researchers to identify key viral antagonists of innate immunity for SARS-CoV and MERS-CoV (Menachery et al., 2015, Totura and Baric, 2012). Several of these mutant viruses have subsequently been employed as live-attenuated vaccine candidates for SARS-CoV and MERS-CoV (de Wit et al., 2016, Schindewolf and Menachery, 2019). Using our system, this knowledge might now be applied to the current SARS-CoV-2. Characterizing these mutations might provide insight into SARS-CoV-2 pathogenesis.
Our reverse genetic system also allows exploration of research questions fundamental to understanding the SARS-CoV-2 pandemic. As additional genomic sequences become available, evolutionary mutations can be interrogated for their effect on viral transmission and disease outcome. For example, a 382-nucleotide deletion covering almost the entire ORF8 of SARS-CoV-2 was observed in eight hospitalized patients in Singapore; virus isolation of the deletion strains has not been reported in the study (Su et al., 2020). A four-amino-acid insertion (conferring a possible furin cleavage site) was reported in the spike (S) protein of SARS-CoV-2 but is absent in the S protein of SARS-CoV and other group-2B CoVs (Coutard et al., 2020). Using the infectious clone, we can now evaluate the effect of these genetic changes by removing the reported sequences from SARS-CoV-2 and examining their effect on virus replication and S protein processing. In addition, mouse models for SARS-CoV-2 have been limited by the absence of viruses capable of binding to mouse angiotensin-converting enzyme 2 (ACE2) (Zhu et al., 2020). Point mutations in the receptor binding domain of the SARS-CoV-2 S protein might facilitate mouse adaptation and development of a model that recapitulates human diseases in a standard mouse strain. Altogether, the above questions are a few examples of how our infectious clone can be used to advance SARS-CoV-2 research.
In summary, we have developed a robust reverse genetic system for SARS-CoV-2 that can be used to study viral replication and pathogenesis. We have also established an mNG reporter SARS-CoV-2 that is a reliable surrogate for high-throughput drug discovery. The reverse genetic system represents a major tool for the research community and significantly advances opportunities for countermeasure development for COVID-19.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli strain Top10 | ThermoFisher Scientific | Cat#C404006 |
| TransforMax™ EPI300™ Chemically Competent E. coli | Lucigen Corporation, Middleton, WI 53562 | Cat#C300C105 |
| SARS-CoV strain 2019-nCoV/USA_WA1/2020 (WA1) | World Reference Center of Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| IFN-α A Protein, Recombinant human | Millipore Sigma | Cat#IF007 |
| Critical Commercial Assays | ||
| T7 mMessage mMachine kit | Thermo Fisher Scientific | Cat#AM1344 |
| Ingenio® Electroporation solution | Mirus Bio LLC | Cat#MIR 50117 |
| SuperScript™ IV First-Strand Synthesis System | Thermo Fisher Scientific | Cat#18091300 |
| Platinum™ SuperFi II DNA Polymerase | Thermo Fisher Scientific | Cat#12361010 |
| Experimental Models: Cell Lines | ||
| Vero E6 cells | ATCC | Cat# CRL-1586; RRID: CVCL_0574 |
| Oligonucleotides | ||
| primer Cov-T7-N-F (TACTGTAATACGA CTCAC TATAGGATGTCTGATAATGGACCCCAAAATC) |
Integrated DNA Technologies (Skokie, Illinois) | N/A |
| primer polyT-N-R (TTTTTTTTTTTTTTTT TTT TTTT TTTTTTTTTTTTTTTAGGCCT GAGTTGAGTCAGCAC) |
Integrated DNA Technologies (Skokie, Illinois) | N/A |
| Recombinant DNA | ||
| pUC57-CoV2-F1 | This paper | N/A |
| pCC1-CoV2-F2 | This paper | N/A |
| pCC1-CoV2-F3 | This paper | N/A |
| pUC57-CoV2-F4 | This paper | N/A |
| pUC57-CoV2-F5 | This paper | N/A |
| pUC57-CoV2-F6 | This paper | N/A |
| pCC1-CoV2-F7 | This paper | N/A |
| pCC1-CoV2-F7-mNG | This paper | N/A |
| Synthesized mNeonGreen gene (sequence-optimized) | This paper and Shaner et al., 2013 | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | N/A |
| Prism 8.0 software | GraphPad | N/A |
| Illustrator CC | Adobe | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pei-Yong Shi (peshi@utmb.edu). Plasmids and virus generated in this study will be made available on request, but we might require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Experimental Model and Subject Details
Virus and Cell Lines
The stock of SARS-CoV-2 strain 2019-nCoV/USA_WA1/2020 was derived from the first patient diagnosed in the US. The virus isolate was originally provided by Dr. Natalie Thornburg from the Centers for Disease Control and Prevention in Atlanta, GA as described previously (Holshue et al., 2020), and amplified on Vero E6 cells at the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch at Galveston (UTMB). The P5 passage was used in this study.
African green monkey kidney epithelial cells (Vero E6; CRL-1586) were purchased from the American Type Culture Collection (ATCC, Bethesda, MD) and maintained in a high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT) and 1% penicillin/streptomycin (P/S). Cells were grown at 37°C with 5% CO2. All culture media and antibiotics were purchased from ThermoFisher Scientific (Waltham, MA). All cell lines were tested negative for mycoplasma.
Method Details
Cloning the SARS-CoV-2 cDNAs
Two approaches were taken to rapidly obtain stable cDNAs of SARS-CoV-2. First, the cDNAs of fragments F1, F4, F5, and F6 were successfully synthesized from the GenScript company (Piscataway, NJ) and cloned into a high-copy plasmid pUC57. The F1 contains a T7 promoter sequence at the upstream of the 5′ end of the SARS-CoV-2 sequence. Other cDNA fragments were also synthesized but found unstable after cloning into plasmid pUC57. For overcoming this hurdle, the cDNAs of fragments F2, F3, and F7 were obtained by reverse transcription and PCR (RT-PCR). RT was performed by using the SuperScript IV First-Strand Synthesis System (ThermoFisher Scientific) with random hexamer primers and extracellular viral RNA (extracted from the supernatants of SARS-CoV-2-infected Vero E6 cells). The cDNA was used as a template to amplify the fragments F2, F3, and F7 by high fidelity PCR with the Platinum SuperFi II DNA Polymerase (ThermoFisher Scientific) according to the manufacturer’s instructions. A poly(T)29 sequence was introduced by PCR to the 3′ end of the untranslated region of viral genome. The amplicons were cloned into a single-copy vector pCC1BAC (Epicenter) to increase the stability of the cDNA plasmids when propagated in E. coli. To ensure a seamless assembly of the full-length cDNA, we introduced two cleavage sites of class IIS restriction enzymes (BsaI and Esp3I) at both ends of each sibling cDNA during PCR or gene synthesis. To differentiate the infectious clone-derived virus from the parental clinical isolate 2019-nCoV/USA_WA1/2020, we engineered three silent mutations at nucleotide positions 7,486 (A-to-T change), 7,489 (T-to-A change), and 18,058 (T-to-C change). For constructing the pCC1-F7-mNG, the gene of mNeonGreen (sequence-optimized) was synthesized and inserted at the downstream of the regulatory sequence of ORF7a to replace the entire ORF7a, according to the study as described previously (Sims et al., 2005). All subclones were finally validated by Sanger sequencing.
Assembly of a Full-Length SARS-CoV-2 cDNA
To assemble the full-length cDNA, we digested individual cDNA plasmids and purified each cDNA fragment. Specifically, F1, F2, F3 and F4 cDNA fragments were obtained by digesting the corresponding plasmids with enzyme BsaI. F5 and F6 fragments were obtained by digesting the plasmids with enzymes Esp3I and PvuI. F7 and F7-mNG cDNA fragments were obtained by digesting the corresponding plasmids by Esp3I and SnaBI. PvuI and SnaBI were included in the digestion to eliminate undesired DNA bands that co-migrated with the targeted fragments on agarose gels. All fragments after restriction enzyme digestion were separated on 0.6% agarose gels, visualized under a darkreader lightbox (Clare Chemical Research, Dolores, CO), excised, and purified using the QIAquick Gel Extraction Kit (QIAGEN, Germantown, MD). To assemble the full-length cDNA, we ligated the seven cDNA fragments in a three-step manner. First, equimolar amounts of F1 (0.61 μg), F2 (0.65 μg), F3 (0.75 μg), and F4 (0.94 μg) were ligated in a PCR tube using T4 DNA ligase in a 40 μl-reaction at 4°C for 18 h, resulting in F1-4 DNA. Second, equimolar amounts of fragments F5 (0.75 μg), F6 (0.72 μg), and F7 (0.60 μg) were ligated in a separate PCR tube to produce F5-7 DNA using the same ligation conditions. Third, without any DNA purification, the two reactions (containing F1-4 and F5-7) were combined (total 80 μl) and topped with additional T4 ligase (2 μl), buffer (2 μl) and nuclease-free water (16 μl) to a 100 μL reaction. The final reaction was incubated at 4°C for 18 h to produce the full-length F1-7 DNA. Afterward, the full-length cDNA was phenol/chloroform extracted, isopropanol precipitated, and resuspended in 10 μL nuclease-free water.
RNA Transcription, Electroporation, Virus Production and Quantification
RNA transcript was in vitro synthesized by the mMESSAGE mMACHINE T7 Transcription Kit (ThermoFisher Scientific) according to the manufacturer’s instruction with some modifications. A 50 μL reaction was set up by adding 1 μg DNA template and 7.5 μL GTP (cap analog-to-GTP ratio of 1:1). The reaction was incubated at 32°C for 5 h. After removing the template DNA by nuclease per manufacturer’s protocol, the RNA was phenol/chloroform extracted and isopropanol precipitated. A SARS-CoV-2 N gene transcript was in vitro transcribed from a DNA template using the mMESSAGE mMACHINE T7 Transcription Kit with a 2:1 ratio of cap analog to GTP. The N gene DNA template was prepared by PCR using primer Cov-T7-N-F (tactgTAATACGACTCACTATAGGatgtctgataatggaccccaaaatc; the uppercase sequence represents T7 promoter; the underlined sequence represents the 5′ end of N gene) and primer polyT-N-R [(t)37aggcctgagttgagtcagcac].
RNA transcripts were electroporated into Vero E6 cells using a protocol as previously described (Shan et al., 2016) with some modifications. Twenty micrograms of total RNA transcripts (containing both full-length RNA and short RNAs) and 20 μg N gene transcript were mixed and added to a 4-mm cuvette containing 0.8 mL of Vero E6 cells (8 × 106) in Ingenio® Electroporation Solution (Mirus). Single electrical pulse was given with a GenePulser apparatus (Bio-Rad) with setting of 270V at 950 μF. After 5 min recovery at room temperature, the electroporated cells were seeded into a T-75 flask and incubated at 37°C with 5% CO2. On the next day, the culture fluid was replaced with 2% FBS DMEM medium. The cells were monitored daily for virus-mediated cytopathic effect (CPE). One milliliter of the P0 virus was inoculated to a T-175 flask containing 80% confluence Vero E6 cells. The infected cells were incubated at 37°C with 5% CO2 for 2-3 days. Culture supernatants (P1) were harvested when CPE occurred. The amount of infectious virus was determined by a standard plaque assay on Vero E6 cells. All virus cultures were performed in a biosafety level 3 (BSL-3) laboratory with redundant fans in the biosafety cabinets. All personnel wore powered air purifying respirators (Breathe Easy, 3M) with Tyvek suits, aprons, booties, and double gloves.
Interferon Treatment
Vero E6 cells were plated as 1.5 × 104 cells/well in a black 96-well plate (Greiner). For interferon treatment, at 6 h post-seeding, cells were treated with various doses of IFN-α (Millipore Sigma). After 14 h of treatment, the culture fluids were replaced with 2% FBS medium, and P1 IC mNG viruses were added to the cells at MOI 0.3 with additional IFN-α at corresponding concentrations. At 24 h post-infection, Hoechst 33342 (ThermoFisher Scientific) was added to a final concentration of 0.1% to counterstain the nucleus. The green fluorescence signals were detected by Cytation 5 (BioTek) and the infection rate was calculated according to the manufacturer’s instructions.
RNA Extraction, RT-PCR and Sanger Sequencing
250 μL of culture fluids were mixed with three volumes of TRIzol LS Reagent (Thermo Fisher Scientific). Viral RNAs were extracted per manufacturer’s instructions. The final RNAs were dissolved in 30 μL nuclease-free water. 11 μL of RNA sample was used for reverse transcription by using the SuperScript IV First-Strand Synthesis System (ThermoFisher Scientific) with random hexamer primers. Nine DNA fragments covering the entire viral genome were amplified by PCR with specific primers. The resulting DNAs were cleaned up by the QIAquick PCR Purification Kit and Sanger sequencing was performed by GENEWIZ (South Plainfield, NJ).
Northern Blot
Vero E6 cells were infected with clinical isolate WA1 or the infectious clone-derived SARS-CoV-2 (IC WT) at MOI 0.01. At 48 h post-infection, total intracellular RNAs were isolated using TRIzol reagent (Invitrogen). Northern blot analysis was performed using total intracellular RNAs as described previously (Narayanan et al., 2008). A digoxigenin (DIG)-labeled random-primed probe, corresponding to nucleotides 28,999 to 29,573 of the SARS-CoV-2 genome, was used to detect SARS-CoV-2 mRNAs and visualized by DIG luminescent detection kit (Roche, Indianapolis, IN) according to the manufacturer’s protocol.
Quantification and Statistical Analysis
All numerical data are presented as the mean ± SD (standard deviations). Group comparisons of viral growth kinetics in Figures 2 and 3 were performed using multiple t test with Bonferroni-Dunn correction in software Prism 8.0 (GraphPad). ∗p < 0.05, significant; ∗∗p < 0.01, significant; p > 0.05, ns (not significant). The 50% effective concentration (EC50) in Figure 4 was estimated by using a four-parameter logistic regression model from the GraphPad Prism 8 software (GraphPad Software Inc., San Diego CA). Minimal adjustment was made in the software ImageJ to enhance the contrast for bright-field images in Figures 1, 2, and 3. Blue- and green-fluorescence images were merged in ImageJ. Figures were finally assembled using the software Adobe illustrator CC.
Data and Code Availability
All data are present in this study.
Acknowledgments
We thank Natalie Thornburg and other colleagues from the Centers for Disease Control and Prevention for providing the clinical virus isolate. We also thank colleagues at University of Texas Medical Branch (UTMB) for support and discussions. Research was supported by grants from NIA and NIAID of the NIH (U19AI100625 and R00AG049092 to V.D.M.; R24AI120942 (WRCEVA) to S.C.W.; AI114657 and AI146081 to S.M.; and 5UC7AI094660 to J.W.L.). Research was also supported by a STARs Award provided by the University of Texas System to V.D.M., trainee funding provided by the McLaughlin Fellowship Fund at UTMB, and an IHII Pilot grant to S.M. P.-Y.S. was supported by NIH grants AI142759, AI145617, AI127744, AI136126, AI134907 and UL1TR001439 and by awards from the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation. A.M. is supported by a Clinical and Translational Science Award NRSA (TL1) Training Core (TL1TR001440) from the NIH.
Author Contributions
Conceptualization, X.X., V.D.M., and P.-Y.S.; Methodology, X.X., A.M., K.G.L., K.N., X.Z., J.Z., J.L., C.S., N.B., P.A., K.S.P., S.W, S.M., J.W.L., V.D.M, and P.-Y.S.; Investigation, X.X., A.M., K.G.L., K.N., X.Z., J.Z., J.L., C.S., N.B., and P.A.; Resources, K.S.P., S.W., and C.-T.K.T.; Data Curation, X.X., A.M., K.G.L., K.N., J.L., and N.B.; Writing-Original Draft, X.X., K.N., V.D.M., and P.-Y.S.; Writing-Review & Editing, X.X., V.D.M., and P.-Y.S.; Visualization, X.X., A.M., K.G.L., N.B., and P.-Y.S.; Supervision, X.X., V.D.M., and P.-Y.S.; Funding Acquisition, P.A., S.W., S.J., J.W.L., V.D.M., and P.-Y.S.
Declaration of Interests
X.X., V.D.M, and P.-Y.S. have filed a provisional patent on the reverse genetic system of SARS-CoV-2. Other authors have no conflicts of interest to declare.
Published: April 13, 2020
Contributor Information
Xuping Xie, Email: xuxie@UTMB.edu.
Vineet D. Menachery, Email: vimenach@UTMB.edu.
Pei-Yong Shi, Email: peshi@UTMB.edu.
References
- Almazán F., Sola I., Zuñiga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K.M., Yount B., Baric R.S. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002;76:1422–1434. doi: 10.1128/JVI.76.3.1422-1434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease, United States. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Washington State 2019-nCoV Case Investigation Team First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. SARS-CoV-2 sensitive to type I interferon pretreatment. bioRxiv. 2020 doi: 10.1101/2020.03.07.982264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindewolf C., Menachery V.D. Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism. Viruses. 2019;11:E74. doi: 10.3390/v11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Xie X., Muruato A.E., Rossi S.L., Roundy C.M., Azar S.R., Yang Y., Tesh R.B., Bourne N., Barrett A.D. An Infectious cDNA Clone of Zika Virus to Study Viral Virulence, Mosquito Transmission, and Antiviral Inhibitors. Cell Host Microbe. 2016;19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Lambert G.G., Chammas A., Ni Y., Cranfill P.J., Baird M.A., Sell B.R., Allen J.R., Day R.N., Israelsson M. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.C., Baric R.S., Yount B., Burkett S.E., Collins P.L., Pickles R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005;79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Zhu F., Linster M., Kalimuddin S., Low J.G.H., Yan Z., Jayakumar J., Sun L. Discovery of a 382-nt deletion during the early evolution of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.11.987222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao T.T.N., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. bioRxiv. 2020 doi: 10.1101/2020.02.21.959817. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are present in this study.