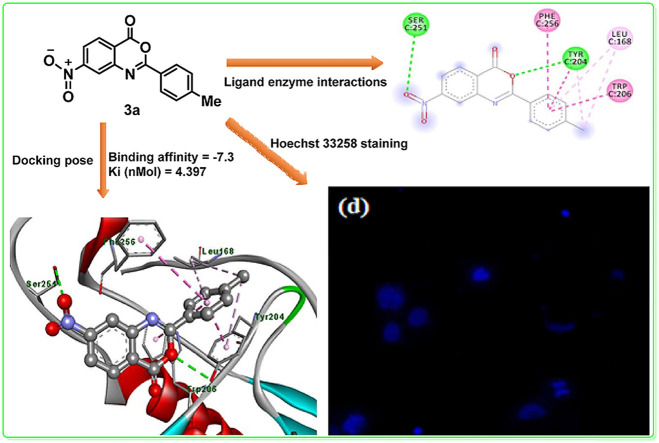
Abstract
A group of new nitro substituted benzoxazinones (3a-k) were synthesized from easily available 4-nitroanthranilic acid. All the synthesized compounds were characterized by FT-IR, 1H NMR, 13C NMR, mass spectrometry and elemental analysis. Anti-proliferative and pro-apoptotic potential of all the synthesized compounds (3a-k) was evaluated by MTT and Hoechst 33258 staining assay respectively whereas their antioxidant properties were determined via DPPH free radical scavenging assay. The most active compounds (3a, 3c and 3k) showed significant cytotoxic potential against HeLa cells with an inhibition of cell viability that ranged between 28.54 and 44.67% (P < 0.001). Albeit statistically different, the anti-proliferative effect of 3c was in close match with that of the reference drug doxorubicin. Likewise, the test compounds showed profound pro-apoptotic potential with an apoptotic index that ranged between 52.86 and 75.61%. Besides, the docking studies revealed a higher efficiency for compounds (3a and 3h) owing to their better affinity and inhibition constant (Ki = 4.397 and 3.713 nmol) respectively. The antioxidant potential of synthesized benzoxazinones (3a-k) was in close agreement with the experimental anticancer results with a percent inhibition from 34.45 to 85.93% as compared to standard (90.56%).
Keywords: Benzoxazin-4-ones, Human cervical carcinoma, DPPH free radical scavenging assay
Graphical abstract
A new series of benzoxazinones (3a-k) have been synthesized, characterized by FT-IR, 1H NMR, and 13C NMR and screened for their in vitro anticancer activity against HeLa cancer cell line. All synthesized compounds demonstrated good anticancer activity. Kinetic and docking study of potent compound 3a were also performed.
Highlights
-
•
Various benzoxazine-4-ones were synthesized by simple methodology.
-
•
Synthesized benzoxazinones were evaluated for their anticancer potential against HeLa cancer cells through cell proliferation assay.
-
•
Benzoxazinones presented good anticancer potential.
-
•
Computational studies were performed to understand their interactions.
1. Introduction
Cell being the structural and functional unit of human body have a genetic code that governs the proliferation, differentiation and apoptosis [1]. Before the cell proliferation, DNA duplication is a controlled process, where any defect in base pair sequencing is repaired. However, an impaired DNA replication may elicit a range of aberrant cell signaling with a potential to trigger the transformation of cells from normal to neoplastic lineage. These defiant cells tend to divide at an uncontrolled pace that precipitates into a tumor [2]. Tumor cells rewire their metabolism to promote growth, survival, proliferation and long-term maintenance through increased uptake of glucose and its fermentation to lactate (Warburg effect) [3]. These cells in spite of competing for resources and space also cooperate with each other by secreting diffusible factors that promote tumor growth and invasion through metastasis [[4], [5], [6]]. Metastasis is the primary cause of mortality in cancer comprising sequential steps for cancer cells dissemination from primary sites and secondary tumor growth at distant region. Disseminated cancer cells can grow out and form secondary tumors in distant area through invasion into local stroma and intravasation into vascular circulation [7]. The interaction between the cancer cells and microenvironment is essential for cancer progression. However, only a small fraction of the disseminated cells called metastatic stem cells are able to grow and overlap the important signaling pathways of normal cells such as Wnt, Notch and transforming growth factor-β (TGF-β) essential for metastatic progression [8]. The overexpression of c-erbB2 gene a member of erbB receptor tyrosine kinase family leads to cell proliferation, migration and survival [[9], [10], [11]]. For many decades, clinicians are trying to encounter cancerous cells within the biological system, but they are camouflaged in billions of normal cells making them difficult to wipe out completely.
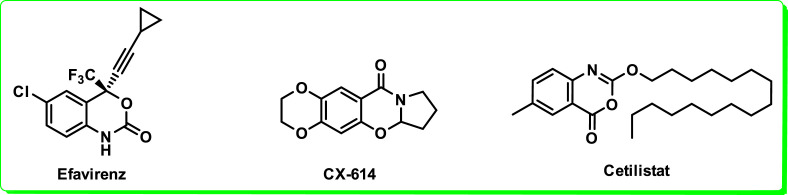
Benzoxazinones widely used in pharmaceutics are endowed with a wide range of biological activities such as antiphlogistic, antifungal, antibacterial [7], anti-human coronavirus [12], inhibitor of human leucocyte elastase [13], anti-cathepsin G [14], complement protein 1 receptor blocker [15] and α-chymotrypsin antagonist [4]. Fig. 1 represents some viable drugs containing benzoxazinone moiety such as CX-614 developed by Cortex pharmaceuticals has great potential in treating Parkinson’s and Alzheimer’s disease [16]. Efavirenz (Sustiva™) non-nucleotide HIV-1 reverse transcriptase inhibitor has been approved by FDA for the treatment of AIDS [17]. The 4H-benzo[d] [1,3]oxazin-4-one is a major bioactive core in Cetilistat, a phase III clinical trial drug to treat obesity [18].
Fig. 1.
Chemical structures of some pharmaceutical drugs containing benzoxazinone nucleus.
Considering the above observations, development of easy method for production of benzoxazinone skeleton from easily available reactants under mild reaction condition remains worthwhile preposition. In continuation of our efforts to develop anticancer agents, we have reported the synthesis of 7-nitro-2-aryl-4H-benzo[d] [1,3]oxazin-4-ones (3a-k) from easily available anthranilic acid along with their cytotoxic and pro-apoptotic evaluation in HeLa (human cervical cancer cell lines). The possible binding sites with the protease caspase, binding energy and inhibition constant (Ki) of the tested compounds were assessed by docking studies.
2. Experimental
2.1. Chemical reagents
All chemicals and reagents were procured from Sigma/Aldrich and Alfa Aesar and used without further purification. The solvents were dried by using standard procedures. Melting points were measured by Büchi 434 melting point apparatus. FT-IR spectra (KBr discs) were recorded on a Bruker FT-IR IFS48 spectrophotometer. 1H NMR and 13C NMR spectra were recorded in CDCl3 using Bruker AC400 (400 MHz) spectrometer. Mass spectrometry was carried out via negative electrospray ionization mode (mass spectrometer ESI Micromass ZMD-2000). The signals are given chemical shifts δ in ppm and splitting pattern is given in s = singlet, d = doublet, dd = doublet of doublet, t = triplet etc. The progression of the reactions was supervised by thin layer chromatography (TLC). TLC was performed on 2 × 5 cm aluminum sheets preloaded with silica gel 60F254 to a thickness of 0.25 mm (Merck). The chromatograms were visualized under UV light irradiation.
2.2. Chemistry
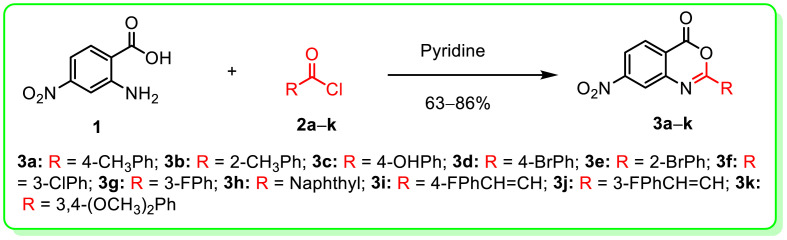
In current study, synthesis of 7-nitro-2-aryl-4H-benzo[d] [1,3]oxazin-4-ones (3a-k) is depicted in Scheme 1 . 4-Nitroanthranilic acid (1) (1.0 equiv.) was reacted with a variety of benzoyl chlorides (2a-k) (2.0 equiv.) in pyridine to give benzoxazinones (3a-k) in 63–86% yields [19b].
Scheme 1.
Schematic representation for the synthesis of benzoxazinones (3a-k).
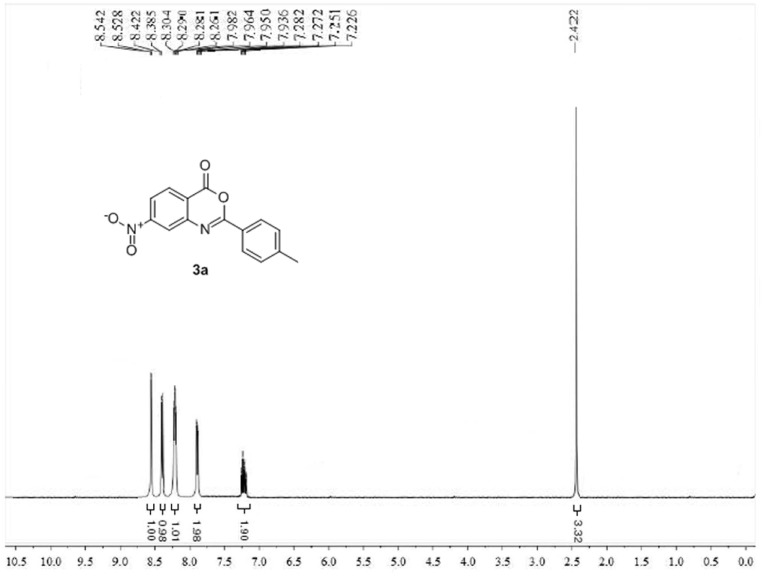
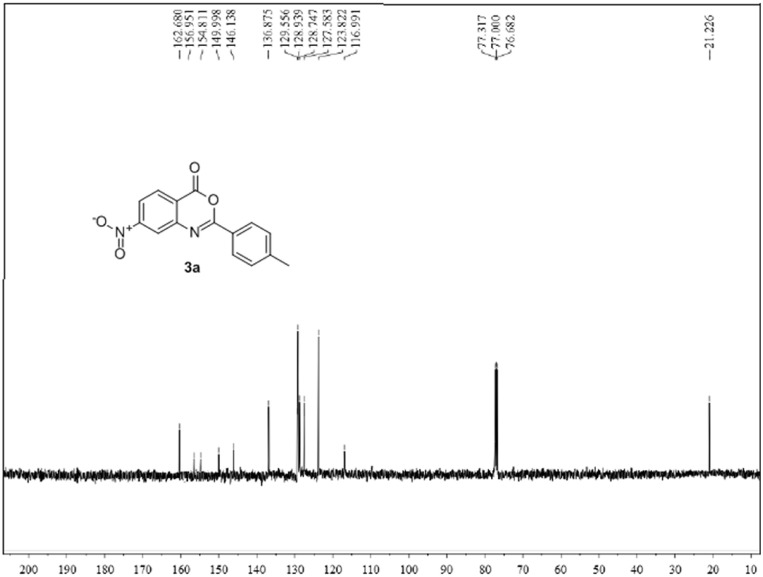
The structures of new benzoxazinones (3a-k) were determined by probing the FT–IR, 1H NMR, 13C NMR and mass spectral results. The FT-IR spectra showed two strong bands in the range of 1733–1772 cm−1 and 1624–1670 cm−1 which confirmed the structure according to literature. In case of compounds (3i-3k) a strong band in the range of 972–982 cm−1 attributed to out of plane bending vibration of C–H bond of E-ethylene. 1H NMR spectra also confirmed the required number of protons in the title compounds. The characteristic chemical shift (δ) values for olefinic moiety in compounds (3i-3k) were clearly observed and their trans-geometry was confirmed by the appearance of two doublets in spectra with large coupling constants (J = 16.0–16.2 Hz). 13C NMR also confirmed the structures of synthesized benzoxazinones (3a-k). 1H NMR and 13C NMR spectra of representative compound 3a are shown in Fig. 2, Fig. 3 .
Fig. 2.
1H NMR spectrum of compound 3a.
Fig. 3.
13C NMR spectrum (CDCl3) of compound 3a.
2.2.1. General procedure (GP-1) for the preparation of benzoxazinones (3a-k)
To a solution of 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) in pyridine (20 mL) was added respective acid chlorides (0.01 mol) under continuous stirring at 0 °C for 10 min. The resulting reaction mixture was further stirred at room temperature for half an hour. The reaction mixture was poured into ice cold water after completion of reaction as indicated by TLC analysis. The appeared precipitates were filtered and washed with cold water and dried. The crude product was crystallized with ethanol.
2.2.2. 7-Nitro-2-(p-tolyl)-4H-benzo[d] [1,3]oxazin-4-one (3a)
The compound (3a) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 4-methylbenzoyl chloride (1.54 g, 0.01 mol) by following general procedure GP-1. Yield: 74%; Grey solid; mp: 96 °C; FT-IR (υmax, KBr, cm−1):1750, 1629; 1H NMR (CDCl3, 400 MHz) δ: 8.53 (d, 1H, Ar–H, J = 2.2 Hz), 8.39 (d,1H, Ar–H, J = 8.4 Hz), 8.28 (dd,1H, Ar–H, J = 2.1, 8.7 Hz), 7.98–7.92 (m, 2H, Ar–H), 7.28–7.21 (m, 2H, Ar–H), 2.42 (s, 3H, CH3); 13C NMR (CDCl3, 75 MHz) δ: 160.2, 156.9, 154.7, 146.3, 143.3, 137.1, 129.5, 128.9, 128.4, 127.9, 127.4, 116.9, 20.9; ESI-MS: 281.04 [M−H]−; Anal. Calcd. For C15H10N2O4 (MW: 282.15 g/mol): C, 63.83%; H, 3.57%; N, 9.92%; Found: C, 63.85%; H, 3.55%; N, 9.91%.
2.2.3. 7-Nitro-2-(o-tolyl)-4H-benzo[d] [1,3]oxazin-4-one (3b)
The compound (3b) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 2-methylbenzoyl chloride (1.54 g, 0.01 mol) by following general procedure GP-1. Yield: 72%; Grey solid; mp: 79 °C; IR (υmax, KBr, cm−1):1751, 1624; 1H NMR (CDCl3, 400 MHz) δ: 8.49 (d, 1H, Ar–H, J = 2.1 Hz), 8.37 (d,1H, Ar–H, J = 8.2 Hz), 8.25 (dd, 1H, Ar–H, J = 2.1, 8.4 Hz), 7.65 (d, 1H, Ar–H, J = 8.1 Hz), 7.41–7.32 (m, 3H, Ar–H), 2.44 (s, 3H, CH3); 13C NMR (CDCl3, 75 MHz) δ: 159.9, 157.4, 154.6, 153.4, 141.2, 138.6, 131.5, 130.2, 129.2, 128.8, 128.6, 127.3, 124.7, 119.8, 20.1; ESI-MS: 281.08 [M−H]−; Anal. Calcd. For C15H10N2O4 (MW: 282.15 g/mol): C, 63.83%; H, 3.57%; N, 9.92%; Found: C, 63.84%; H, 3.56%; N, 9.90%.
2.2.4. 2-(4-Hydroxyphenyl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3c)
The compound (3c) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 4-hydroxybenzoyl chloride (1.56 g, 0.01 mol) by following general procedure GP-1. Yield: 83%; Yellow solid; mp: 90 °C; IR (υmax, KBr, cm−1): 3345,1733, 1626; 1H NMR (CDCl3, 400 MHz) δ: 8.52 (d,1H, Ar–H, J = 2.0 Hz), 8.39 (d,1H, Ar–H, J = 8.4 Hz), 8.29 (dd,1H, Ar–H, J = 2.0, 8.3 Hz), 7.90–7.81 (m, 2H, Ar–H), 7.07–6.99 (m, 2H, Ar–H), 4.22 (s, 1H, OH); 13C NMR (CDCl3, 75 MHz) δ:158.8, 156.9, 154.4, 154.2, 152.3, 136.6, 132.4, 130.8, 128.5, 122.6, 120.3, 118.6; ESI-MS: 283.05 [M−H]−; Anal. Calcd. For C14H8N2O5 (MW: 284.21 g/mol): C, 59.16%; H, 2.84%; N, 9.86%; Found: C, 59.15%; H, 2.83%; N, 9.88%.
2.2.5. 2-(4-Bromophenyl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3d)
The compound (3d) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 4-bromobenzoyl chloride (2.2 g, 0.01 mol) by following general procedure GP-1. Yield: 86%; Grey solid; mp: 118 °C; IR (υmax, KBr, cm−1): 1749, 1624; 1H NMR (CDCl3, 400 MHz) δ: 8.49 (d, 1H, Ar–H, J = 2.1 Hz), 8.37 (d, 1H, Ar–H, J = 8.2 Hz), 8.24 (dd, 1H, Ar–H, J = 2.2, 8.6 Hz), 7.84 (t, 1H, Ar–H, J = 7.2 Hz), 7.78–7.65 (m, 3H, Ar–H); 13C NMR (CDCl3, 75 MHz) δ: 157.9, 155.8, 154.6, 152.5, 143.8, 134.6, 133.4, 132.6, 131.4, 129.6, 128.9, 122.4; ESI-MS: 346.02 [M−H]−; Anal. Calcd. For C14H7BrN2O4 (MW: 347.10 g/mol): C, 48.44%; H, 2.03%; N, 8.07%; Found: C, 48.45%; H, 2.02%; N, 8.06%.
2.2.6. 2-(2-Bromophenyl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3e)
The compound (3e) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 2-bromobenzoyl chloride (2.2 g, 0.01 mol) by following general procedure GP-1. Yield: 81%; Yellow solid; mp: 89 °C; IR (υmax, KBr, cm−1): 1761, 1629; 1H NMR (CDCl3, 400 MHz) δ: 8.54 (d, 1H, Ar–H, J = 2.0 Hz), 8.40 (d,1H, Ar–H, J = 8.4 Hz), 8.21 (dd,1H, Ar–H, J = 2.4, 8.4 Hz), 7.70–7.62 (m,2H, Ar–H), 7.44–7.33 (m,2H, Ar–H); 13C NMR (CDCl3, 75 MHz) δ:159.2, 156.4, 154.6, 154.2, 142.6, 138.2, 134.1, 132.8, 132.2, 131.6, 129.9, 129.4, 128.4, 124.2; ESI-MS: 346.03 [M−H]−; Anal. Calcd. For C14H7BrN2O4 (MW: 347.10 g/mol): C, 48.44%; H, 2.03%; N, 8.07%; Found: C, 48.43%; H, 2.04%; N, 8.07%.
2.2.7. 2-(3-Chlorophenyl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3f)
The compound (3f) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 3-chorobenzoyl chloride (1.75 g, 0.01 mol) by following general procedure GP-1. Yield: 75%; Brown solid; mp: 98 °C; IR (υmax, KBr, cm−1): 1758, 1628; 1H NMR (CDCl3, 400 MHz) δ: 8.53 (d, 1H, Ar–H, J = 2.1 Hz), 8.29 (dd,1H, Ar–H, J = 2.2, 8.6 Hz), 8.22–8.12 (m, 3H, Ar–H) 7.49–7.44 (m,2H, Ar–H); 13C NMR (CDCl3, 75 MHz) δ: 159.6, 156.8, 154.6, 152.2, 141.5, 137.6, 133.6, 132.6, 131.3, 129.8, 129.5, 128.6; ESI-MS: 301.46 [M−H]−; Anal. Calcd. For C14H7ClN2O4 (MW: 302.64 g/mol): C, 55.56%; H, 2.33%; N, 9.26%; Found: C, 55.53%; H, 2.35%; N, 9.27%.
2.2.8. 2-(3-Fluorophenyl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3g)
The compound (3g) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) and 3-fluorobenzoyl chloride (1.58 g, 0.01 mol) by following general procedure GP-1. Yield: 79%; Yellow solid; mp: 104 °C; IR (υmax, KBr, cm−1): 1770, 1670; 1H NMR (CDCl3, 400 MHz) δ: 8.47 (d,1H, Ar–H, J = 2.2 Hz), 8.39 (d, 1H, Ar–H, J = 8.2), 8.24 (dd,1H, Ar–H, J = 2.4, 8.7 Hz), 7.76–7.68 (m, 3H, Ar–H) 7.28 (m,1H, Ar–H); 13C NMR (CDCl3, 75 MHz) δ: 164.3, 159.3, 154.4, 154.3, 152.6, 137.8, 132.2, 131.8, 130.2, 128.4, 124.2, 122.4, 119.6, 116.3; ESI-MS: 285.06 [M−H]−; Anal. Calcd. For C14H7FN2O4 (MW: 286.19 g/mol): C, 58.75%; H, 2.47%; N, 9.79%; Found: C, 58.71%; H, 2.50%; N, 9.80%.
2.2.9. 2-(Naphthalen-1-yl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3h)
The compound (3h) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) 2-naphthoyl chloride (1.9 g, 0.01 mol) by following general procedure GP-1. Yield: 73%; Grey solid; mp: 92 °C; IR (υmax, KBr, cm−1):1751, 1625; 1H NMR (CDCl3, 400 MHz) δ: 8.56 (d,1H, Ar–H, J = 2.3 Hz), 8.41 (d, 1H, Ar–H, J = 8.0), 8.28 (dd, 1H, Ar–H, J = 2.1, 8.2 Hz), 8.38–7.62 (m, 7H, Ar–H); 13C NMR (CDCl3, 75 MHz) δ: 159.6, 156.4, 154.8, 152.4, 138.8, 134.6, 133.2, 132.4, 131.7, 131.5, 130.2, 129.6, 129.2, 129.0, 127.9, 125.6, 122.2, 117.4; ESI-MS: 317.12 [M−H]−; Anal. Calcd. For C18H10N2O4 (MW: 318.25 g/mol): C, 67.92%; H, 3.17%; N, 8.80%; Found: C, 67.94%; H, 3.14%; N, 8.81%.
2.2.10. (E)-2-(4-Fluorostyryl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3i)
The compound (3i) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) (E)-3-(4-fluorophenyl)acryloyl chloride (1.84 g, 0.01 mol) by following general procedure GP-1. Yield: 68%; Grey solid; mp: 138 °C; IR (υmax, KBr, cm−1): 1772, 1660, 973; 1H NMR (CDCl3, 400 MHz) δ: 8.55 (d,1H, Ar–H, J = 2.2 Hz), 8.37 (d, 1H, Ar–H, J = 8.1), 8.20 (dd,1H, Ar–H, J = 2.2, 8.2 Hz), 7.79 (d, 1H, CH CH, J = 16.1 Hz), 7.63–7.32 (m, 4H, Ar–H), 6.76 (d, 1H, CH CH, J = 16.2 Hz); 13C NMR (CDCl3, 75 MHz) δ:164.7, 159.5, 157.2, 156.4, 154.6, 147.1, 141.8, 136.4, 130.2, 128.1, 126.8, 119.2, 116.9, 116.4; ESI-MS: 311.04 [M−H]−; Anal. Calcd. For C16H9FN2O4 (MW: 312.21 g/mol): C, 61.54%; H, 2.91%; N, 8.97%; Found: C, 61.55%; H, 2.89%; N, 8.98%.
2.2.11. (E)-2-(3-Fluorostyryl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3j)
The compound (3j) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) (E)-3-(3-fluorophenyl)acryloyl chloride (1.84 g, 0.01 mol) by following general procedure GP-1. Yield: 65%; Yellow solid; mp: 129 °C; IR (υmax, KBr, cm−1):1770, 1655, 982; 1H NMR (CDCl3, 400 MHz) δ: 8.51 (d, 1H, Ar–H, J = 2.1 Hz), 8.40 (d, 1H, Ar–H, J = 8.2 Hz) 8.18 (dd, 1H, Ar–H, J = 2.1, 8.0 Hz), 7.82 (d, 1H, CH CH, J = 16.2 Hz), 7.33–7.05 (m, 4H, Ar–H), 6.81 (d, 1H, CH CH, J = 16.0 Hz); 13C NMR (CDCl3, 75 MHz) δ: 165.8, 159.9, 157.8, 156.4, 155.6, 146.2, 138.6, 135.2, 133.4, 132.6, 130.2, 128.8, 124.2, 118.2, 116.4, 115.3; ESI-MS: 311.04 [M−H]−; Anal. Calcd. For C16H9FN2O4 (MW: 312.21 g/mol): C, 61.54%; H, 2.91%; N, 8.97%; Found: C, 61.56%; H, 2.89%; N, 8.97%.
2.2.12. (E)-2-(3,4-Dimethoxystyryl)-7-nitro-4H-benzo[d] [1,3]oxazin-4-one (3k)
The compound (3k) was synthesized from 4-nitroanthranilic acid (1) (0.9 g, 0.005 mol) (E)-3-(3,4-dimethoxyphenyl)acryloyl chloride (2.26 g, 0.01 mol) by following general procedure GP-1. Yield: 63%; Brown solid; mp: 152 °C; IR (υmax, KBr, cm−1):1764, 1651, 972; 1H NMR (CDCl3, 400 MHz) δ: 8.57 (d, 1H, Ar–H, J = 2.2 Hz), 8.36 (d, 1H, Ar–H, J = 8.0 Hz), 8.19 (dd, 1H, Ar–H, J = 2.2, 8.4 Hz), 7.74 (d, 1H, CH CH, J = 16.0 Hz), 7.23–7.09 (m, 3H, Ar–H), 6.79 (d, 1H, CH CH, J = 16.0 Hz), 3.90 (s, 6H, OCH3); 13C NMR (CDCl3, 75 MHz) δ: 158.6, 157.8, 156.4, 156.2, 152.4, 146.9, 136.0, 133.1, 132.5, 131.2, 128.3, 127.1, 120.4, 120.3, 116.6, 112.0, 56.1; ESI-MS: 353.18 [M−H]−; Anal. Calcd. For C18H14N2O6 (MW: 354.31 g/mol): C, 61.02%; H, 3.98%; N, 7.91%; Found: C, 61.01%; H, 3.98%; N, 7.92%.
2.3. Biological activities
2.3.1. Antioxidant activity
The newly synthesized benzoxazinones (3a-k) were evaluated for their antioxidant potential by DPPH free radical scavenging activity following the protocol described by Ref. [19].
2.3.2. Cell proliferation assay
HeLa cells were cultured in RPMI-1640 containing antibiotics (100 units/mL penicillin and 50 units/mL streptomycin) and fetal bovine serum (10%), in an atmosphere with a relative humidity of 95% and CO2 concentration of 5%. The cytotoxic effects of different formulations against HeLa cancer cells was measured by MTT assay [20]. The cells were seeded in 96-well plates (1 × 104 cells/well) in 200 μL medium and subsequently cultured overnight. The 50 μL of each synthesized compound at the concentration of 250 μg/mL and the standard drug (Doxorubicin) were added into the wells and incubated for 6 h. After that, the culture medium was replaced by complete culture media and cells were incubated for another 24 h or 48 h. At the end of the incubation time, 50 μL of prepared MTT solution in PBS buffer (2 mg/mL) was added to each well and incubated for another 4 h. Then, 200 μL of DMSO was added to dissolve the formazan crystals formed by the viable cells. In the end, the spectrophotometric plate reader (BioTek Instruments Inc, Vermont, USA) was used for UV absorbance measurements at 570 nm. All experiments were repeated thrice.
2.3.3. Apoptosis by Hoechst 33258 staining assay
As the newly synthesized benzoxazinones (3c, 3k and 3a) were demonstrated the significant cytotoxic potential on HeLa cancerous cells; therefore, apoptotic effect was further evaluated for understanding the cytological alterations following the protocol as previously described [1]. The apoptotic index was calculated by using the following formula:
2.4. Computational approach
2.4.1. Repossession of protein and ligands
The relevant x-ray structure was found out through literature review and caspase protease was chosen with PDBID: 3DEI which is pronounced to be caspase [21]. Protein data bank was searched for the 3D structure and the downloaded structure was eviscerated after the heteroatom removal in Biovia discovery studio 2016 [22]. The different compound structure as ligands were drawn using Pub Chemsketcher V2.4 [23] after observing their effects in laboratory work. The structures were then optimized in pdb form using “Biovia discovery studio visualizer 2016”. The standard used was downloaded from pubchem with CID: 31703 as doxorubicin [24].
2.4.2. Identification and conformation of binding sites
The binding site in a protease is vital part for its proper working. The activity of a protease or an enzyme is referred by its binding pocket and type of interaction it expresses. Thus, the binding site was analyzed by 3D ligand site and equated with the previous literature [25].
2.4.3. Docking
The molecular docking is assumed to be a powerful way to find out the binding of any compound computationally and then applied it in laboratory work. It clears out the ambiguity of binding a compound with any enzyme for practical application. The docking was applied on different compounds (3a-k) to find its placement in the desired protein. The docking was performed using autodock, vina autodock and later viewed in discovery studio 2016 resulting in 2D and 3D representation. The grid was designed after conversion the files into pdbqt format using autodock which was later used in script writing in vina autodock [26,27].
2.4.4. Statistical analysis
Analysis of data was carried out using IBM statistical package for social sciences (SPSS) software, version 23.0. All computations were executed in triplicate and the results were expressed as mean ± SEM (n = 3). Antioxidant and anticancer assays were computed with suitable dilutions for each sample. Statistical analysis was performed by one way analysis of variance (ANOVA) followed by Duncan multiple range Post-Hoc analysis. The statistical differences between the control and treatment groups were considered significant at p < 0.05 (∗), p < 0.01 (∗∗) and p < 0.001 (∗∗∗).
3. Results and discussion
3.1. Chemistry
Benzoxazinone analogues revealed significant biological potential for the development of new drugs such as CX-614, Efavirenz (Sustiva™) and Cetilistat [[16], [17], [18]]. Owing to promising biological potential of benzoxazinone scaffold, a small library of benzoxazinone with a range of substituents were designed and synthesized. Synthesis of 7-nitro-2-aryl-4H-benzo[d] [1,3]oxazin-4-ones (3a-k) was accomplished under convenient reaction conditions. In the present work target benzoxazinones (3a-k) were formed in good yields when 4-nitroanthranilic acid (1) was reacted with a range of benzoyl chlorides (2a-k) in the presence of pyridine as organic base. Various functional group were introduced onto benzoxazinone scaffold to access desired library of synthetic compounds (3a-k). These newly synthesized compounds were evaluated for their anti-proliferative and pro-apoptotic potential in HeLa (human cervical cancer cell lines). For this purpose, MTT assay was utilized for anti-proliferative potential of all synthesized in the current study. Pro-apoptotic potential was investigated by using Hoechst 33258 staining assay. Whereas antioxidant properties of synthesized benzoxazinones (3a-k) were determined via DPPH free radical scavenging assay to compare antioxidant activity with anticancer potential of synthesized compounds. Proliferation of cells in percentage was investigated by MTT assay. This assay clearly demonstrates significance of anti-proliferative and cytotoxic potential of synthesized compounds (3a-k) on HeLa cancerous cells. Therefore, apoptotic effect was further evaluated by using Hoechst 33258 staining assay and fluorescence microscope was employed to further understand the cytological alterations in cancerous cells. Finally, docking studies was also performed for compounds to evaluate binding affinity and inhibition constant (Ki).
3.2. Biological studies
3.2.1. Antioxidant activity
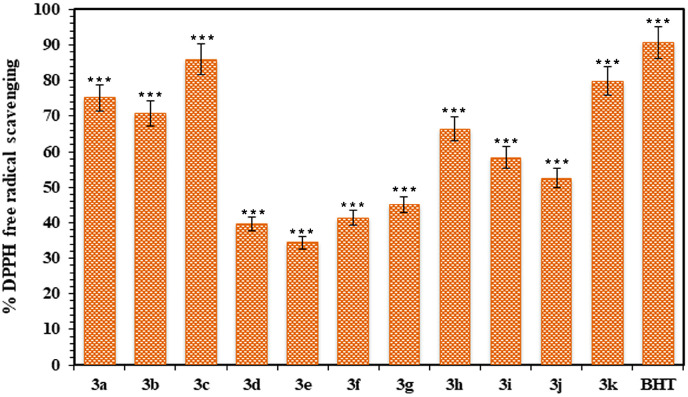
The antioxidant activity of the synthesized benzoxazinones (3a-k) was determined by scavenging the DPPH free radical in comparison to standard butylated hydroxy toluene (BHT). The antioxidant activity results (Fig. 4 ) depicted that BHT showed the highest percentage inhibition of DPPH (90.56%). Among all the tested compounds of synthesized benzoxazinones, the maximum percentage of DPPH free radicals scavenging (85.93%) was presented by compound (3c) comparable to standard while the compound (3e) demonstrated the least activity (34.45%) as shown in Fig. 4. Moreover, DPPH free radical scavenging results were also shown that the compounds (3c, 3k, 3a and 3b) with electron donating functionalities (-OH, –OCH3 and –CH3) at ortho and para positionsscavenged the DPPH free radicals more strongly. On the other hand, the compound (3h) with naphthalene ring in the molecule was found to display good antioxidant activity. Moderate to lower DPPH scavenging was observed with the compounds having halo group (-F, -Cl, and -Br) moieties at ortho, meta and para positions but among them fluorinated compounds (3g, 3i and 3j) revealed better antioxidant results because of weak destabilizing agent than others.
Fig. 4.
The antioxidant activity of synthesized benzoxazinones (3a-k) in terms of DPPH free radical scavenging in comparison to standard BHT.
3.2.2. In vitro anticancer activity
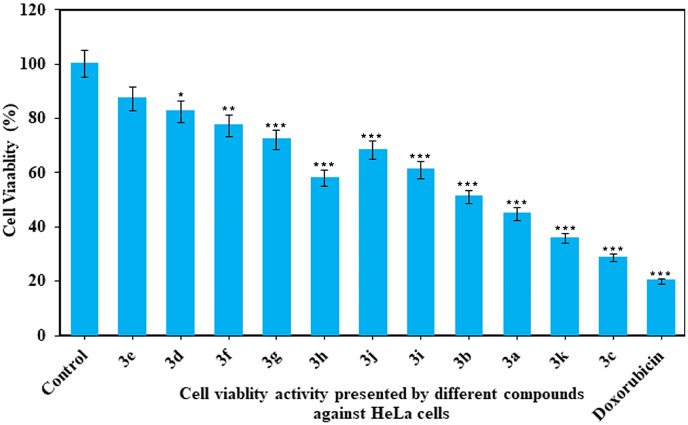
Chemotherapy has been considered as emerging and widely used approach for the treatment of both localized and metastasized cancerous cells. Due to antioxidant and cytotoxic potential of benzoxazinones analogues, they can prohibit the growth of several types of malignant cells [28,29]. Therefore, the synthesized benzoxazinones (3a-k) have been evaluated for their in vitro cytotoxic effect on the HeLa cells in comparison to standard anticancer drug (Doxorubicin). The cytotoxicity results demonstrated that most of the compounds were successfully inhibited the growth of HeLa cancer cells. The cell viability percentage of HeLa cancer cells upon interaction with the synthesized compounds were ranged from 28.53% to 87.22% (Fig. 5 ). The standard anticancer drug exhibited maximum cytotoxic effect (19.98%). Among all the synthesized benzoxazinones, compound (3c) exhibited the highest cytotoxic effect while least was shown by (3e). Moreover, it has been observed that the compounds with the substitution of strong to moderate electron donating groups (-OH, –OCH3, and –CH3) have demonstrated the superior to good cytotoxic activity. On the other hand, the compounds with substitution of halo group elements (-F, -Cl, and -Br) were found to present average to lower cytotoxicity propensity; however, among them fluoro substituted compounds manifested better results.
Fig. 5.
The cell viability percentage presented by synthesized benzoxazinones (3a-k) against HeLa cancerous cells in comparison to doxorubicin.
3.2.3. Apoptosis by Hoechst 33258 staining assay
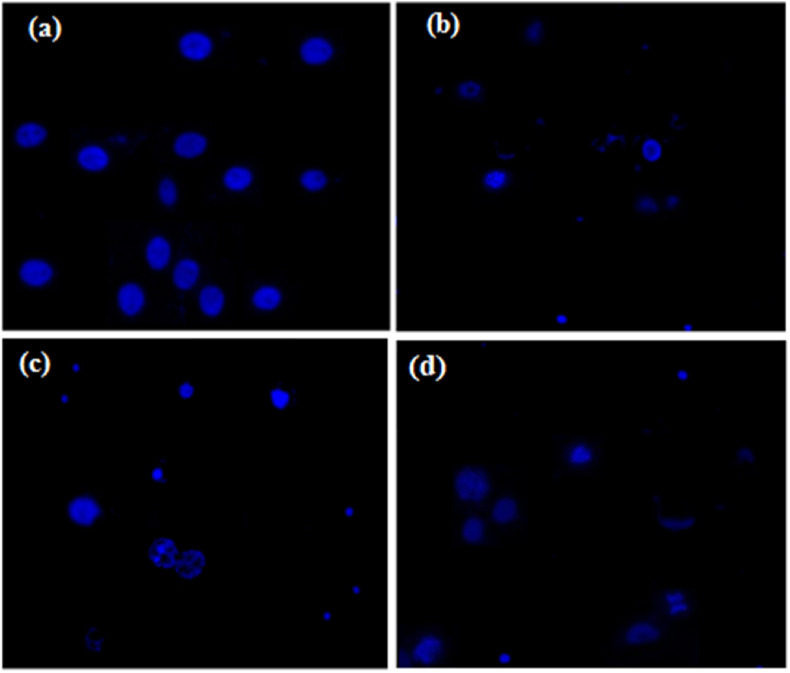
The apoptosis in HeLa cells triggered by the synthesized benzoxazinones (3c, 3k and 3a) was corroborated with the Hoechst 33258 staining assay and the cytological alterations in cancerous cells upon benzoxazinones treatment were analyzed by employing the fluorescence microscope. Substantial modifications including DNA fragmentation, chromosomal apoptotic bodies and chromatic condensation, nuclear fragmentation, shrinkage of cell, vacuolation of cytoplasm, binucleation, blebbing of plasma membrane, externalization, and activation of caspases were appeared in HeLa cancerous cells (Fig. 6 ).
Fig. 6.
Apoptotic cytological alterations in HeLa cells induced by the synthesized compound 3c (b), compound 3k (c) and compound 3a (d) in comparison to control (a).
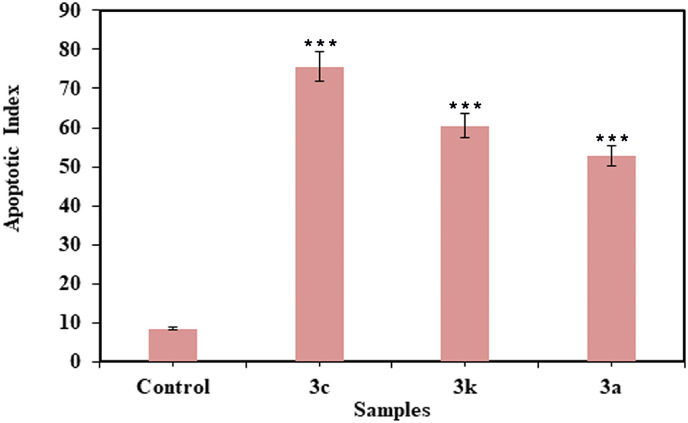
Apoptosis results further showed that upon increasing the incubation time, increase in plasma membrane blebbing with the rounded shaped cells was resulted. The apoptotic cell percentage was further calculated by counting the 500 cells randomly with light microscope and the results are presented in Fig. 7 .
Fig. 7.
The apoptotic cell percentage in HeLa cells treated with the synthesized benzoxazinones (3c), (3k) and (3a).
3.3. Computational approach
3.3.1. Binding sites
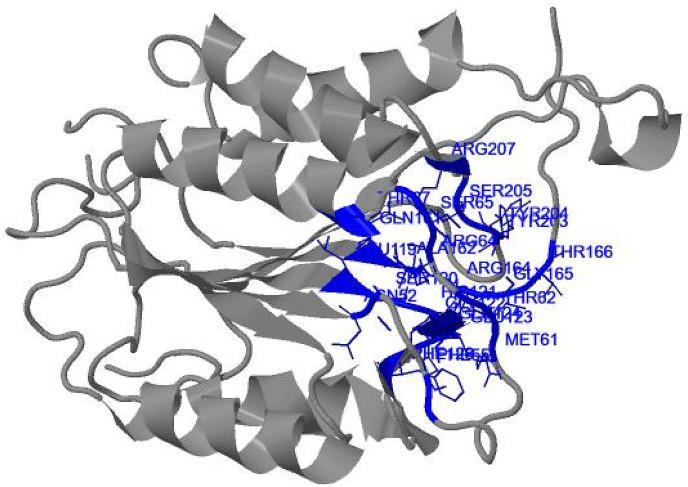
Three binding sites were analyzed by 3D ligand site. The predicted binding sites were having different residues including Met61, Thr62, Ser63, Arg64, Ser65, Gly66, Ser120, His121, Gly122, Glu123, Glu124, Phe128, Gln161, Ala162, Arg164, Gly165, Thr166, Glu167, Leu168, Thr199, Tyr204, Ser205, Trp206, Arg207, Asn208, Ser209, Trp214, Phe247, Glu248, Ser249, Phe250,Thr255 and Phe256 (Fig. 8 ). All residues from receptor caspase were according to the literature review.
Fig. 8.
Residues of receptor protein.
3.3.2. Docking
As the docking was performed by autodock and vina autodock, the compounds (3a-k) gave positive results as compared to standard doxorubicin against caspase protein. The interaction of receptor caspase and compounds (3h and 3a) revealed the binding of important residue leucine, which is considered important in treating cancer [30,31]. The caspase enzyme activity is typically controlled by the presence of leucine residue. Literature also reveals that the mutation of the leucine residues greatly decreases the enzyme activity. The presence of leucine residue on the L2 loop plays an exceptional role in maintaining the catalytic activity of caspases [30]. Moreover, leucine deficiency also triggers apoptotic death of cancerous cells [31]. Several interactions like hydrogen boding, π–π stacking and other hydrophobic interactions play a key role in the binding of compound with enzyme active sites (Table 1 ).
Table 1.
The results of compounds (3a-k) with docking score, inhibition constant value (Ki) and nature of binding interactions.
| Ligands/Compounds | Binding affinity | Ki (nMol) | Nature of binding interactions |
|---|---|---|---|
| 3a | −7.3 | 4.397 | Leu168-Ar(π-alkyl), Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π) |
| 3b | −6.7 | 12.118 | Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Tyr204-Ar(π-alkyl), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π) |
| 3c | −6.6 | 14.348 | Leu168-Ar(π-alkyl), Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π) |
| 3d | −6.9 | 8.643 | Leu168-Ar(π-alkyl), Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π) |
| 3e | −6.6 | 14.348 | Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Trp206-Ar(π-π), Ser251(H–B), Phe256-Ar(π-π) |
| 3f | −6.7 | 12.118 | Leu168-Ar(π-alkyl), Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Tyr204-CH3(π-alkyl), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π) |
| 3g | −6.7 | 12.118 | Leu168-Ar(π-alkyl), Tyr204-Oxa(H–B), Tyr204-Ar π-π), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π). |
| 3h | −7.4 | 3.713 | Leu168-Ar(π-alkyl), Tyr204-Oxa(H–B), Tyr204-Ar(π-π), Trp206-Ar(π-π), Phe256-Ar(π-π) |
| 3i | −6.7 | 12.118 | Leu168-Ar(π-alkyl), Trp204-Ar π-π) |
| 3j | −6.7 | 12.118 | Thr166-R(C–H), Trp204-Ar(π-π), Trp206-Ar(π-π), Ser252-ONO(H–B), Phe256-Ar(π-π) |
| 3k | −6.5 | 16.989 | Thr166-F(Halogen), Asn208-ONO(H–B), Trp206-Ar(π-π), Ser251-ONO(H–B), Phe256-Ar(π-π) |
| Standard (doxorubicin) | −7.4 | 3.713 | Tyr204-OH(H–B), Tyr204-hexanone(π-π), Ser209-ONO(H–B), His121-Ar(π-π), His121-hexanone(π-π) |
H–B = hydrogen bonding.
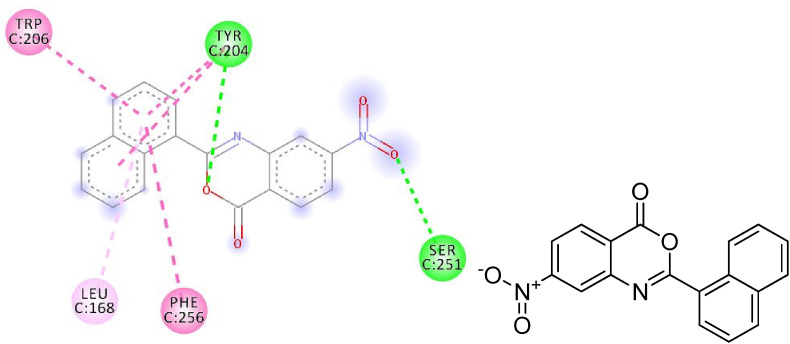
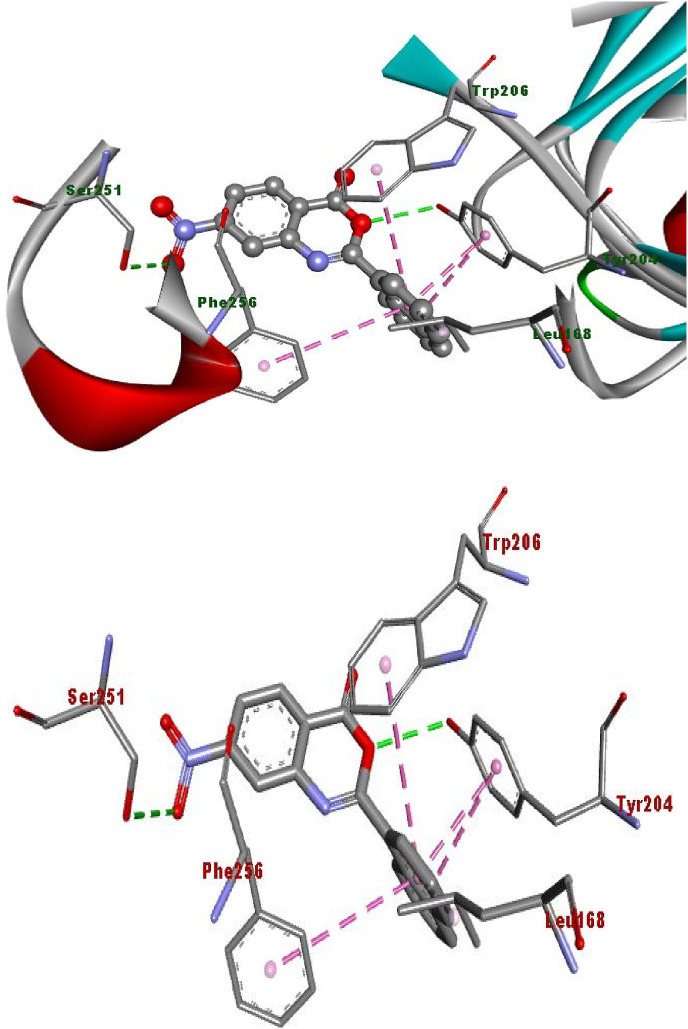
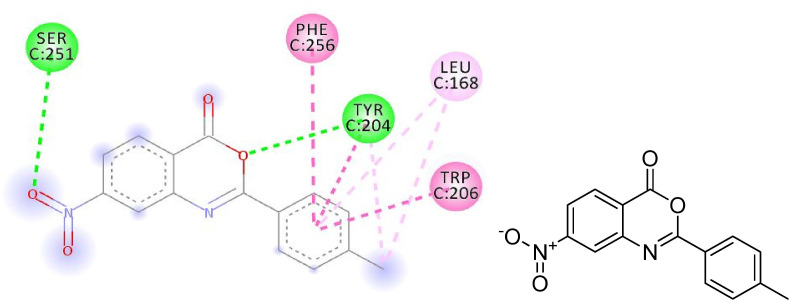
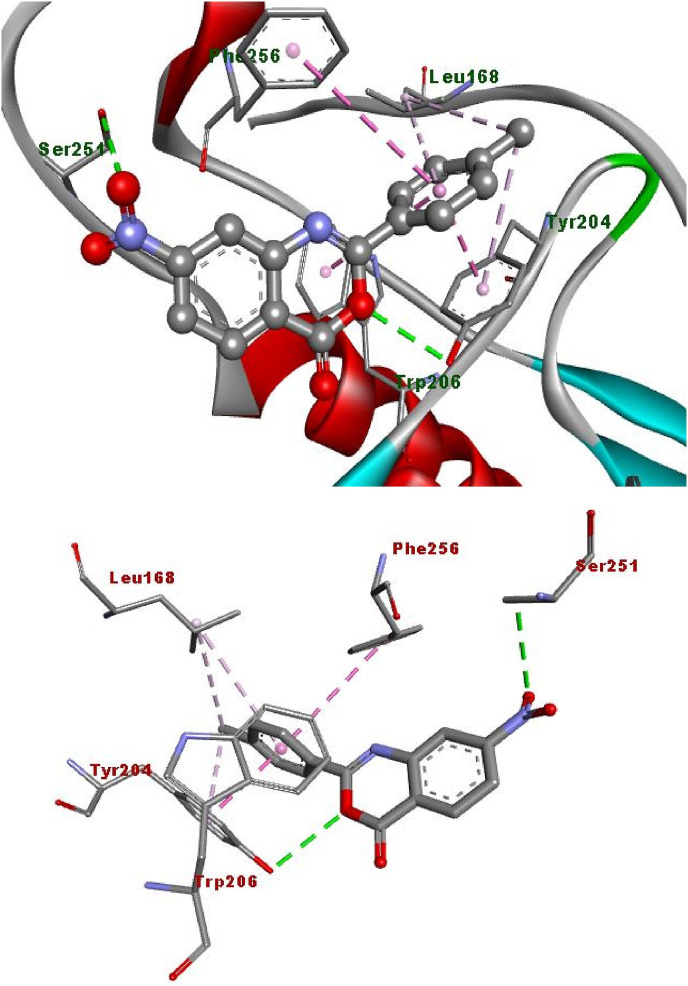
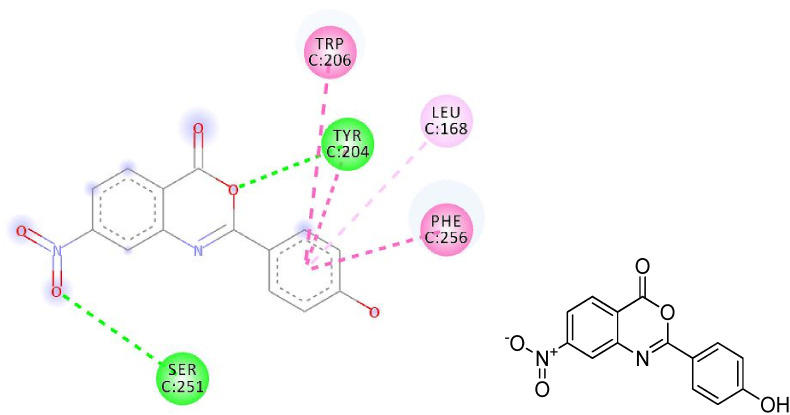
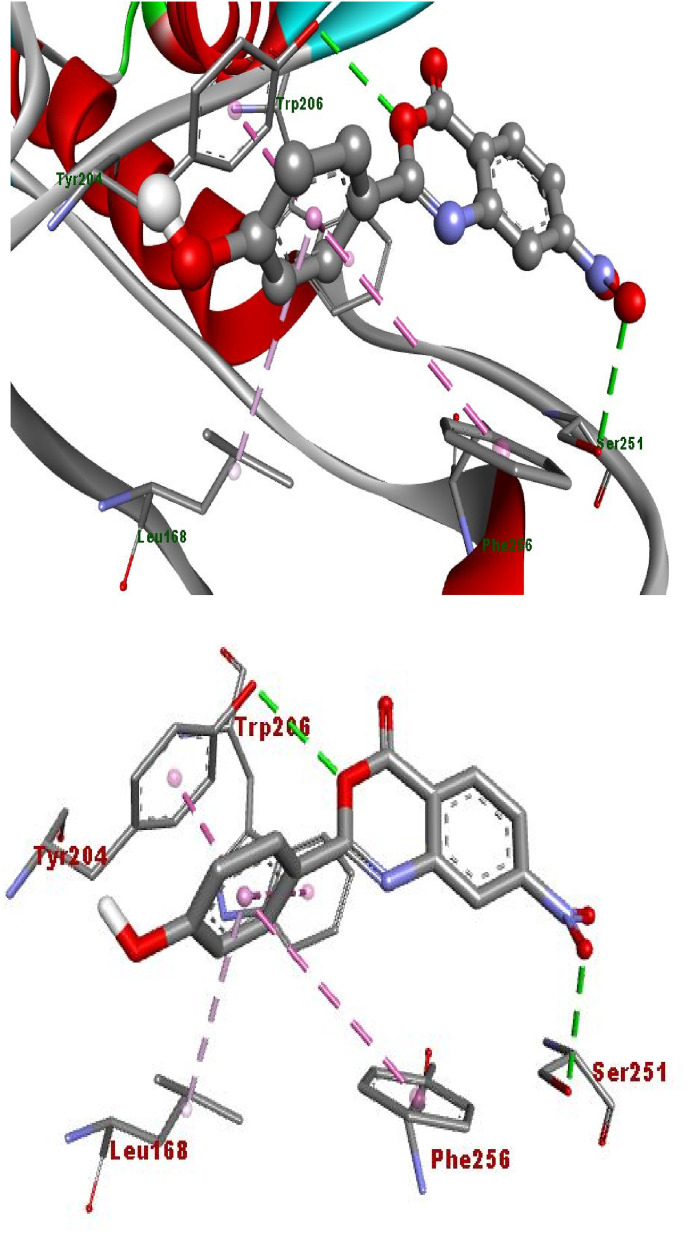
The hydrogen bonding occurs owing to the presence of electronegative atom (for example oxygen, nitrogen and fluorine) indicates strong binding while π–π and π–alkyl interactions are also occurred due to the presence of π electrons/bonds in the benzoxazinone group (Table 1). All the ligands with different groups bound with same binding pocket and almost with the same residues with slight difference in number of residues and bond type. The docking score indicated the strength of bond formed and helped us to find the inhibitor constant (Ki). The inhibitor constant has vital importance in drug designing because it specifies the inhibition potential to yield half concentration after inhibition. The Ki has inevitable effect on IC50 affecting the mode of action of drug. Smaller the Ki value better will be the IC50 [1]. Table 1 showed the results of docking score, Ki values and nature of binding interactions. The binding poses of compounds (3a, 3c and 3h) at their maximum bound cavity are shown in Fig. 9, Fig. 10, Fig. 11, Fig. 12, Fig. 13, Fig. 14 . The binding affinity and inhibitory constant (ki) of compound (3h) was same as that of standard doxorubicin which is best value i.e. −7.4 while the second-best value was given by compound (3a) i.e. −7.3. The residues in compound (3a, 3c and 3h) were Leu168, Tyr204, Trp206, Ser251 and Phe256 while the residues in standard were Tyr204, Ser251 and His121. Hence the main residues like Tyrosine and Serine were present in most of the compounds. While in experimental results compound (3c) with para hydroxyl and compound (3k) with 3,4-dimethoxy moieties showed highest activity. This difference in computational and experimental data might be due to different binding interaction between ligands and protein in docking and anticancer experiment.
Fig. 9.
2D plot, binding affinity of compound (3h) at highly bound cavity.
Fig. 10.
Binding of compound (3h) into protein active site, as assessed by computer modelling studies.
Fig. 11.
2D plot, binding affinity of compound (3a) at highly bound cavity.
Fig. 12.
Binding of compound (3a) into protein active site, as assessed by computer modelling studies.
Fig. 13.
2D plot, binding affinity of compound (3c) at highly bound cavity.
Fig. 14.
Binding of compound (3c) into protein active site, as assessed by computer modelling studies.
The docking score revealed that compounds (3d-3g and 3i-3j) with halogen (F, Cl and Br) moiety showed small values of binding affinities leading to more binding energies which was obviously due to their deactivating nature. The substitution of –CH3 group at para position (compound 3a) showed better activity than ortho substituted –CH3 (compound 3b). Hence, close agreement was observed between the experimental results of anticancer activity and the docking results of the synthesized compounds.
4. Conclusion
A small library of 7-nitro-2-aryl-4H-benzo[d] [1,3]oxazin-4-ones (3a-k) were synthesized by condensation of 7-nitroanthranilic acid with acid chlorides. The synthesized compounds were evaluated for antioxidant activity via DPPH free radical scavenging assay and anticancer activity by cell proliferation assay and apoptosis by Hoechst 33258 staining assay. The results of this study indicated that compounds 3a, 3c and 3k possess potent antioxidant and anticancer properties. Docking studies further revealed the active sites, binding, mode of action and bioactivity.
CRediT authorship contribution statement
Ayesha Bari: Conceptualization, Methodology, Investigation, Writing - original draft. Zulfiqar Ali Khan: Conceptualization, Methodology, Supervision, Validation, Project administration, Writing - review & editing, Resources. Sohail Anjum Shahzad: Methodology, Formal analysis, Supervision, Project administration, Funding acquisition, Writing - review & editing. Syed Ali Raza Naqvi: Validation, Data curation. Shakeel Ahmad Khan: Data curation, Investigation, Formal analysis. Hira Amjad: Software, Data curation, Formal analysis. Ahsan Iqbal: Visualization, Methodology. Muhammad Yar: Visualization, Writing - review & editing.
Declaration of competing interest
No conflict of interest is associated with this work.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors (Dr. Sohail A. Shahzad & Dr. Zulfiqar A. Khan) are very grateful to the Higher Education Commission (HEC) of Pakistan for providing financial support vide No. 5290/Federal/NRPU/R&D/HEC/2016.
Contributor Information
Zulfiqar Ali Khan, Email: zulfiqarchem@gamil.com.
Sohail Anjum Shahzad, Email: sashahzad@cuiatd.edu.pk.
References
- 1.Iqbal A., Khan Z.A., Shahzad S.A., Khan S.A., Naqvi S.A.R., Bari A., Amjad H., Umar M.I. Synthesis, modeling studies and evaluation of E-stilbene hydrazides as potent anticancer agents. J. Mol. Struct. 2019;1197:271–281. [Google Scholar]
- 2.Usman M., Volpi E.V. DNA damage in obesity: initiator, promoter and predictor of cancer. Mutat. Res. 2018;778:23–37. doi: 10.1016/j.mrrev.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Mohapatra D.K., Datta A. Efficient synthesis of biologically important chiral 2-alkylamino benzoxazinones. Bioorg. Med. Chem. Lett. 1997;7:2527–2530. [Google Scholar]
- 4.Marasini B.P., Rahim F., Perveen S., Karim A., Khan K.M. Atta-ur-Rahman, M.I. Choudhary, Synthesis, structure-activity relationships studies of benzoxazinone derivatives as α-chymotrypsin inhibitors. Bioorg. Chem. 2017;70:210–221. doi: 10.1016/j.bioorg.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox P.E. Chymotrypsinogens-chymotrypsins. Methods Enzymol. 1970;19:64–108. [Google Scholar]
- 6.Turk B. Targeting proteases: success, failures and future prospects. Nat. Rev. Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 7.Shariat M., Abdollahi S. Synthesis of benzoxazinone derivatives: a new route to 2-(N-phthaloylmethyl)-4H-3,1-benzoxazin-4-one. Molecules. 2004;9:705–712. doi: 10.3390/90800705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pein M., Oskarsson T. Microenvironment in metastasis: roadblocks and supportive niches. Am. J. Physiol. Cell Physiol. 2015;309:C627–C638. doi: 10.1152/ajpcell.00145.2015. [DOI] [PubMed] [Google Scholar]
- 9.Bonazzoli E., Cocco E., Lopez S., Bellone S., Zammataro L., Bianchi A., Manzano A., Yadav G., Manara P., Perrone E., Haines K., Espinal M., Dugan K., Menderes G., Altwerger G., Han C., Zeybek B., Schwartz P.E., Santin A.D. P13K oncogenic mutations mediate resistance to afatinib in HER2/neu overexpressing gynecological cancers. Gynecol. Oncol. 2019;153:158–164. doi: 10.1016/j.ygyno.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes N.E., Stern D.F. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim. Biophys. Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 11.Okines A., Cunningham D., Chau I. Targeting the human EGFR family inesophagogastric cancer. Nat. Rev. Clin. Oncol. 2011;8:492–503. doi: 10.1038/nrclinonc.2011.45. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh P.-W., Chang F.-R., Chang C.-H., Cheng P.-W., Chiang L.-C., Zeng F.-L., Lin K.-H., Wu Y.-C. 2-substituted benzoxazinone analogues as anti-human coronavirus (anti-HCoV) and ICAM-1 expression inhibition agents. Bioorg. Med. Chem. Lett. 2004;14:4751–4754. doi: 10.1016/j.bmcl.2004.06.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krantz A., Spencer R.W., Tam T.F., Liak T.J., Copp L.J., Thomas E.M., Rafferty S.P. Design and synthesis of 4H-3,1-benzoxazin-4-ones as potent alternate substrate inhibitors of human leukocyte elastase. J. Med. Chem. 1990;33:464–479. doi: 10.1021/jm00164a002. [DOI] [PubMed] [Google Scholar]
- 14.Gutschow M., Neumann U. Inhibition of cathepsin G by 4H-3,1-benzoxazin-4-ones. Bioorg. Med. Chem. 1997;5:1935–1942. doi: 10.1016/s0968-0896(97)00128-4. [DOI] [PubMed] [Google Scholar]
- 15.Hay S.J., Caprathe B.W., Gilmore J.L., Amin N., Emmerling M.R., Michael W., Nadimpalli R., Nath R., Raser K.J., Stafford D., Watson D., Wang K., Jaen J.C. 2-Amino-4H-3,1-benzoxazin-4-ones as inhibitors of C1r serine protease. J. Med. Chem. 1998;41:1060–1067. doi: 10.1021/jm970394d. [DOI] [PubMed] [Google Scholar]
- 16.Gao B., Chen K., Bi X., Wang J. Intramolecular functionalization of C(SP3)-H bonds adjacent to an amide nitrogen atom: metal-free synthesis of 2-hydroxy-benzoxazinone derivatives. Tetrahedron. 2017;73:7005–7010. [Google Scholar]
- 17.Patel M., McHugh R.J., Jr., Cordova B.C., Klabe R.M., Erickson-viitanen S., Trainor G.L., Ko S.S. Synthesis and evaluation of benzoxazinones as HIV-1 reverse transcriptase inhibitors. Analogs of Efavirenz (SustivaTM), Bioorg. Med. Chem. Lett. 1999;9:3221–3224. doi: 10.1016/s0960-894x(99)00565-x. [DOI] [PubMed] [Google Scholar]
- 18.(a) Yamada Y., Kato T., Ogino H., Ashina S., Kato K. Cetilistat (ATL-962), a novel pancreatic lipase inhibitor, ameliorates body weight gain and improves lipid profiles in rats. Horm. Metab. Res. 2008;40:539–543. doi: 10.1055/s-2008-1076699. [DOI] [PubMed] [Google Scholar]; (b) Padwal R. Cetilistat, a new lipase inhibitor for the treatment of obesity. Curr. Opin. Invest. Drugs. 2008;9:414–421. [PubMed] [Google Scholar]; (c) Kopelman P., de Groot H.G., Rissanen A., Rossner S., Toubro S., Palmer R., Hallam R., Bryson A., Hickling R.I. Weight loss, HbA1c reduction, and tolerability of cetilistat in a randomized, placebo-controlled phase 2 trial in obese diabetics: comparison with orlistat (Xenical) Obesity. 2010;18:108–115. doi: 10.1038/oby.2009.155. [DOI] [PubMed] [Google Scholar]
- 19.(a) Khan S.A., Rizwan K., Shahid S., Nomaan M.A., Rasheed T., Amjad H. Synthesis, DFT, computational exploration of chemical reactivity, molecular docking studies of novel formazan metal complexes and their biological applications. Appl. Organomet. Chem. 2020:e5444. [Google Scholar]; (b) Bain D.I., Smalley R.K. Synthesis of 2-substituted 4H-3,1-benzoxazin-4-ones. J. Chem. Soc. 1968:1593. [Google Scholar]
- 20.Ghaffari M., Dehghan G., Baradaran B., Zarebkohan A., Mansoori B., Soleymani J., Dolatabadi J.E., Hamblin M.R. Co-delivery of curcumin and Bcl-2 siRNA by PAMAM dendrimers for enhancement of the therapeutic efficacy in HeLa cancer cells. Colloids Surf. B Biointerfaces. 2020;188:110762. doi: 10.1016/j.colsurfb.2019.110762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.http://www.rcsb.org.
- 22.Dassault Systèmes Biovia . Dassault Systèmes; San Diego: 2016. Biovia Discovery Studio Visualizer, v16.1.0. [Google Scholar]
- 23.Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 24.Ihlenfeldt W.D., Bolton E.E., Bryant S.H. The PubChem chemical structure sketcher. J. Cheminf. 2009;1:20. doi: 10.1186/1758-2946-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wass M.N., Kelley L.A., Sternberg M.J. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris G.M., Ruth H., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function: efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gholami H.R., Asghari S., Mohseni M. Synthesis, characterization, and evaluation of antibacterial and antioxidant activities of novel benzoxazinones and benzoxathiinones. J. Heterocycl. Chem. 2019;56:1505–1513. [Google Scholar]
- 29.Hekal M.H., El-Azm F.S.A. New potential antitumor quinazolinones derived from dynamic 2-undecyl benzoxazinone: synthesis and cytotoxic evaluation. Synth. Commun. 2019;48:2391–2402. [Google Scholar]
- 30.Kang H.J., Lee Y.-m., Jeong M.S., Kim M., Bae K.-H., Kim S.J., Chung S.J. Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7. Biosci. Rep. 2012;32:305–313. doi: 10.1042/BSR20120009. [DOI] [PubMed] [Google Scholar]
- 31.Sheen J.-H., Zoncu R., Kim D., Sabatini D.M. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Canc. Cell. 2011;19:613–628. doi: 10.1016/j.ccr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]